Abstract
Human milk contains a number of nutritional and bioactive molecules including microorganisms that constitute the so-called “Human Milk Microbiota (HMM)”. Recent studies have shown that not only bacterial but also viral, fungal, and archaeal components are present in the HMM. Previous research has established, a “core” microbiome, consisting of Firmicutes (i.e., Streptococcus, Staphylococcus), Proteobacteria (i.e., Serratia, Pseudomonas, Ralstonia, Sphingomonas, Bradyrhizobium), and Actinobacteria (i.e., Propionibacterium, Corynebacterium). This review aims to summarize the main characteristics of HMM and the role it plays in shaping a child’s health. We reviewed the most recent literature on the topic (2019-2021), using the PubMed database. The main sources of HMM origin were identified as the retrograde flow and the entero-mammary pathway. Several factors can influence its composition, such as maternal body mass index and diet, use of antibiotics, time and type of delivery, and mode of breastfeeding. The COVID-19 pandemic, by altering the mother-infant dyad and modifying many of our previous habits, has emerged as a new risk factor for the modification of HMM.
HMM is an important contributor to gastrointestinal colonization in children and therefore, it is fundamental to avoid any form of perturbation in the HMM that can alter the microbial equilibrium, especially in the first 100 days of life. Microbial dysbiosis can be a trigger point for the development of necrotizing enterocolitis, especially in preterm infants, and for onset of chronic diseases, such as asthma and obesity, later in life.
Keywords: Milk human, microbiota; Dysbiosis; Microbiota; Necrotizing enterocolitis; Asthma; Obesity
INTRODUCTION
For a long time, human milk has been thought to be sterile, but recent studies have demonstrated that it is a rich source of microbes, that have the potential to influence a child’s health. Currently, the retrograde flow and the entero-mammary pathway are accepted as the two primary sources of origin of milk microbiota [1]. The former is the consequence of transmission of microbes from the infant’s oral cavity into the mammary duct during suckling [1], while the latter results from the translocation of maternal gut bacteria through the intestinal epithelial barrier to reach the mammary gland via the lymphatic circulation [1].
Several factors can influence the composition of the human milk microbiota (HMM), including the stage of lactation, maternal body mass index (BMI), age and diet [1], parity, geographical location, socioeconomic status, use of antibiotics or probiotics during pregnancy, and type of delivery [1,2].
Breast milk provides the infant with its own microbiota, as well as prebiotic, immunological, and other microbiota-shaping compounds that can indirectly alter colonization patterns in the baby [3]. Therefore, early microbial colonization of mucosal tissues is fundamental for the development, maintenance, and control of the immune system, with the bacterial diversity playing a pivotal role in the maintenance of a healthy immune balance [4]. HMM is responsible for shaping the composition of gut and respiratory microbiota in children [3,5,6] by supplying nutrients for bacterial growth as well as dictating the production of metabolites [3]. Microbiota of the respiratory and gastrointestinal (GI) tracts maintains human health through the so-called “gut-lung axis” [4]. It is quite clear that there is a cross-dialogue between the intestine and the lungs, which is vital for educating the host’s immune system, even if the mechanisms through which the gut impacts pulmonary health or disease and vice versa are only partially discovered [7].
Early intestinal microbial dysbiosis, as that occurring during pregnancy, can affect the composition of the newborn’s pioneer bacterial communities [8], and this can act as a risk factor for development of diseases such as obesity and asthma in the future [9], as well as for pathologies such as necrotizing enterocolitis (NEC) in the first few months of life, especially in preterm infants [10].
In recent years, bacterial culture techniques have been used to analyze HMM composition using standard morphological and biochemical characteristics of species identification [1]. These methods have limited accuracy as they are able to reveal only those bacteria that can survive the sampling procedures and grow under laboratory conditions [1]. For this reason, culture-independent methods, such as sequencing of 16S ribosomal RNA (16S rRNA), clone libraries and metataxonomics have been developed [1]. These analytical methods allow a more accurate identification of microbial communities that may dictate health or disease in humans [7].
The aim of this review is to summarize the latest knowledge regarding HMM with particular focus on its composition, changes, and impact on children’s health.
HUMAN MILK MICROBIOTA COMPOSITION
The term “microbiota” refers to a population of microorganisms that exists within a niche in the human body, with a mutualistic relationship with the host [11] and includes bacteria, viruses, fungi, parasites and archaea. The term “microbiome” refers to the collection of genomes from all these microorganisms [11]. In a recent review [12], it was found that the human breast milk microbiota is quite diverse, with more than 800 bacterial species, the most prevalent being facultative anaerobic or strictly aerobic groups. In general, the presence of anaerobic microbiota in breast milk is known to influence the infant’s health [13]. Taxonomic classification [14] of the major bacterial phyla and their predominant genera in the HMM is shown in Fig. 1.
Fig. 1. Human milk microbiota: Taxonomic classification of the major bacterial phyla and their predominant genera.
Staphylococcus, Streptococcus, Serratia, Pseudomonas, Corynebacterium, Ralstonia, Propionibacterium, Sphingomonas and Bradyrhizobium are the nine genera that constitute the “core” bacteriome of the HMM. They represent approximately half of the microbial milk community, although their abundance can vary across milk samples [14,15]. HMM is the second integral source of microbes for infants after the birth canal (during vaginal delivery) [13]; the mammary glands, periareolar skin, and infant’s mouth may influence its composition [11]. Every day, a breastfed infant ingests between 1×105 and 1×107 bacteria, which means that almost 30% of the infant’s bacteria are acquired through breast milk [16]. Although the actual mechanism of HMM formation is not yet known, two main patterns of origin have been hypothesized: the entero-mammary pattern and retrograde flow. In the entero-mammary mechanism [11], a specific homing system composed of dendritic cells and macrophages is responsible for transporting bacteria from the mother’s mucosal tissues to the lactating breast, along the mucosal-associated lymphoid tissues. On the other hand, the retrograde flow hypothesis states that there is an exchange of microorganisms between the breast and the mouth of the infant, with mutual sharing of microbes [1,11]. This could explain how bacteria commonly found in the infant’s oral cavity (i.e., Veillonella, Prevotella) or human vagina (i.e., Lactobacillus) can also be found in human milk [1,9]. In particular, vaginal bacteria acquired by the infant during natural delivery can be transferred to the human milk through retrograde flow [1]. At the same time, the presence of bacterial communities in colostrum collected before the first infant suckling demonstrates the existence of the entero-mammary pathway [9,13] and that human milk is not a sterile fluid [9]. Although several studies have demonstrated a difference in the bacterial compositions of colostrum and transitional milk as a direct consequence of the interaction with the infant’s oral microbiota during the first 1-6 months after birth, other studies have found no such difference [1]. In addition, bacteria present in the pre-colostrum might have an important role in the initial establishment of the infant oral microbiota [17].
Bacterial, viral, fungal and archaeal components
Several bacterial components are present in HMM, and among these, Bifidobacterium and Lactobacillus species (spp.) are known to have important potential probiotic roles [12]. Researchers have isolated three potentially probiotic Lactobacillus strains from milk: Lactobacillus gasseri, Lactobacillus salivarius and Lactobacillus fermentum. These microbes induce interleukin (IL)-10 production by increasing the IgA concentration in feces. In a recent study, Rajoka et al. [18] isolated seven Lactobacillus rhamnosus strains from breast milk that had excellent antioxidant and anticancer activities [13]. Another strain isolated from human milk with a potential probiotic role was L. gasseri MA-4 [13]. Bifidobacterium spp. represent a large proportion of the species specifically found in the breast milk microbiota and not in other human secretions [12]. Arboleya et al. [19] and Solís et al. [20] isolated three Bifidobacterium breve and longum strains from human milk that demonstrated adherence to the intestinal mucosa. In a recent cohort study [17], a distinct strain of B. breve was identified in the maternal rectum, human milk, and in the stool of a cesarean section (C-section) delivered child, thus supporting the entero-mammary pathway of HMM formation. In particular, it constituted 28% of the maternal milk sample microbiome and 68% of the infant gut microbiome [17]. On the other hand, Staphylococcus aureus and Streptococcus agalactiae are commonly isolated from infected breast milk, but they are also the most frequent species found in healthy controls, thereby challenging the Koch’s postulates. This may be attributed to the differences in the virulence of distinct strains and the modifications of these microbes during immunological changes [12]. Recent studies [21,22] have also demonstrated the importance of the viral microbiota (virome) for a child’s health, where members of the Myoviridae, Siphoviridae and Podoviridae families were revealed to be dominant members of normal human microbiome [23]. It is believed that the newborn is first colonized by bacteriophages, induced by the bacteria constituting the primitive microbiota [21], followed by replication of these viruses in the infant’s intestine, which modulate and regulate bacterial growth and, consequently, the infant GI microbiota [21,23]. It has been demonstrated that, in breastfed infants, there is a reduction in the quantity and pathogenicity of these viruses. Recently, it has been shown [21] that human milk viruses are also transmitted from the mother to the child via breastfeeding, with a vertical transmission of bacteriophages. Accordingly, we can hypothesize that the mammary gland provides both the bacterial and viral parts of the microbiome [21], and that human breast milk favors the transmission of “good” viral particles, hindering the transmission and replication of pathogenic viral strains [21]. Non-phage viral sequences have also been found in human milk samples (Papillomaviridae, Retroviridae and Herpesviridae families) [23]. HMM of a healthy mother also contains a fungal fraction, known as “mycobiome”, with a conserved vertical transmission from the mother to the child [23,24]; its load has been estimated to be 105 cells/mL [16], with the major part being Saccharomyces, Malassezia, Alternaria, Rhodotorula and Candida spp. [14,16]. Malassezia and Davidiella spp. are known to constitute the “core” mycobiome; in particular, Malassezia globosa is detected in all healthy human milk samples, but it is absent from all mastitis-associated samples, demonstrating that mastitis can create a fungal dysbiosis [23]. Among yeasts, Debaryomyces hansenii is the dominant species during breastfeeding [16], whereas Saccharomyces cerevisiae becomes the dominant species during weaning [16]. In addition, some protozoal elements have also been isolated (Giardia intestinalis and Toxoplasma gondii) in the HMM [23]. Archaea are considered to one of the potential sources of microbes in the human milk [16]. Two metagenomic studies in the past have revealed the presence of archaeal DNA in human milk, with the presence of halophilic archaea (i.e., Haloarcula marismortui) [23]. Togo et al. [25] have shown that Methanobrevibacter smithii is the dominant human gut-associated archaeon, and is a critical commensal for human health, by playing a vital role in weight regulation. In particular, they demonstrated the presence of this microorganism in human colostrum and milk, with an essential role in seeding the infant’s future gut microbiota [25].
Factors influencing human milk microbiota composition
In recent years, researchers have focused their attention on the different factors that could influence HMM composition and, consequently, a child’s health [13]. Microbes present in human milk have an important role in the regulation of cytokine production in the enteric nervous system and maintenance of mucosal immune functions [15]. One of the variable factors that can influence microbiota composition of milk is maternal BMI. In obese mothers, there is a predominance of Lactobacillus spp. in the colostrum and Staphylococcus and Akkermansia spp. in the mature milk [1], with a concordant reduction of genera belonging to Bacteroidetes, Proteobacteria and Firmicutes phyla. Moreover, maternal diet, by altering the maternal gut microbiota could influence the microbes in human milk, and consequently their vertical transfer to the child [1]. In particular, vitamin C is associated with an increase in the bacteria of Staphylococcus genus, whereas the consumption of polyunsaturated fatty acids and linoleic acid leads to a rise in the number of bacteria of the Bifidobacterium genus. In addition, the consumption of B1, B2, and B9 vitamins can also influence breast milk microbiota composition [16]. High consumption of a calorie-rich diet in pregnant women has been shown to cause an increase in the number of organisms of Firmicutes phylum [14]. Few confirmatory studies have been conducted on the influence of consumption of antibiotics and/or probiotics during pregnancy on HMM composition [26]. Although studies have shown that antibiotic therapy can alter the balance between microorganisms, with a reduction in Bifidobacterium and Lactobacillus spp., none of these provide conclusive results [2]. In a recent analysis [27], it was found that bacteria of Bifidobacterium spp. were present only in the breast milk samples of mothers who did not receive intrapartum antibiotic administration [14]. This is an important observation since a reduction in Bifidobacteria in early infancy may be associated with a higher risk of atopy and obesity [14]. The time and type of delivery are also important in determining the composition of milk microbiota [2]. Studies have demonstrated a lower level of Bifidobacterium spp. in the preterm pregnancy group than in the term pregnancy group at all stages of lactation [13]. It has been suggested that labor influences the HMM composition by increasing intestinal permeability and facilitating the entero-mammary pathway. In particular, it has been observed in C-section deliveries that there is a higher relative abundance of Proteobacteria, with a reciprocal reduction of Firmicutes (i.e., Bifidobacterium and Lactobacillus) in the HMM [2]. It has been proven that emergency C-section milk samples have a microbial profile similar to that of vaginal deliveries, while samples from non-emergency C-sections have a microbial profile similar to that of the skin and oral microbial communities [16]. The infant’s mode of breastfeeding also plays an important role in shaping the HMM. Many studies have shown the relative abundance of Bifidobacteria in the feces of human milk-fed infants, whereas Enterococci and Clostridia are most prevalent in formula-fed infants [15]. In particular, numerous studies affirm that direct breastfeeding favors the acquisition of bacterial microbiota from the child’s mouth, whereas indirect breastfeeding (pumped-milk) increases acquisition of environmental bacteria [28]. In directly breast-fed infants, there is a relatively high prevalence of Bifidobacterium spp. On the contrary in infants fed with pumped-milk, there is an abundance of potential pathogens. Furthermore, the sex of the baby is also believed to play a pivotal role in determining HMM composition; the milk microbiota is partially derived from the infant’s oral cavity and while the host is always female (mother), there are differences between male and female infants in terms of contribution to the HMM (retrograde inoculation hypothesis) [9]. Additionally, the potential role of parity in influencing the HMM composition cannot be overlooked. Staphylococcus and Haemophilus are probably more abundant in the bacterial samples of multiparous mothers than in those of primiparous mothers [2]. Maternal psychosocial distress is also related to lower bacterial diversity in human milk three months postpartum, underlining the relationship between maternal psychosocial distress and milk microbiota [14]. It has been proven that there exists a difference in the HMM composition between populations of rural and urban countries, with a higher bacterial diversity being displayed in the rural milk samples [23]. In addition, a different microbial composition has been found in the milk samples from women with lactational acute mastitis [16]. All the factors that may influence the HMM have been systematically summarized by Zimmermann and Curtis [29].
Bacterial extracellular vesicles: what is known
Human milk contains abundant quantities of extracellular vesicles (EVs) that may originate from multiple cellular sources and contain different bioactive molecules [2]. These are generally classified into three subcategories based mainly on their size as follows: exosomes, microvesicles (also called microparticles), and apoptotic bodies [29]. It is well known that all bacteria can release EVs, and these have been designated as bacterial membrane vesicles produced by gram positive bacteria and outer membrane vesicles produced by gram negative bacteria [2]. In a study conducted by Kim and Yi [2], the dominant bacterial EVs in human breast milk belonged to the Bacteroides, Acinetobacter and Lactobacillus genera; in contrast, Streptococcus and Staphylococcus were predominant in bacterial samples. These results indicate that the bacteria releasing EVs do not necessarily match with the bacteria present in human breast milk and are not evenly distributed [2]. Moreover, this study suggests that EVs may contribute to the vertical transfer of commensal microbiota from mothers to infants and, since these vesicles are rich in microRNAs, they can influence mucosal immunity and increase the intestinal microbiome diversity of children [30]. Moreover, in a recent study by Wang et al. [31], a peptidomic analysis of human milk exosomes was performed by comparing the milk of term pregnancies to that of preterm pregnancies. The authors discovered that the preterm and term milk exosomes had comparatively different peptide compositions [32]. Based on these findings, we hypothesized that there could also be a difference in the composition of bacterial EVs in the human milk that might influence the future health related outcomes of a child.
COVID-19 and milk microbiota: what has changed
The recent coronavirus disease (COVID-19) pandemic has affected people’s health, economy, and society as a whole by changing a large part of our habits. A survey of mothers in European countries highlighted the significant stress and anxiety experienced by mothers during the pandemic [33]. In fact, at the beginning of the pandemic, there were several uncertainties regarding medical protocols and, in the presence of a COVID-19 positive mother, early direct breastfeeding and skin-to-skin contact were not followed in clinical settings, despite the recommendations of the World Health Organization [33,34]. It is well known that, for SARS-CoV-2, the detection of viral RNA in human milk is uncommon, and transmission along human milk has not been recorded [33]. Even if transmission of COVID-19 through breastfeeding appears unlikely, the infection could affect HMM, with breast milk being an expression of connection between mother and child [35]. During pregnancy, intestinal dysbiosis associated with COVID-19 inflammation could affect the composition of a newborn’s pioneer bacterial communities [8]. It was observed that greater the severity of the disease, greater was the intestinal dysbiosis. In affected patients, symbiotic bacteria were significantly reduced, with a corresponding enrichment of opportunistic flora [35]. Zuo et al. [36] have shown that in the context of Firmicutes phylum, there was a reduction in some bacterial genera and an increase in other genera (i.e., Coprobacillus spp.) [36]. Yeoh et al. [37], in their research, revealed a reduction in bacteria belonging to the Actinobacteria and an increase in those belonging to the Bacteroidetes and Verrucomicrobia phyla [37,38]. Interestingly, dysbiosis continues even after recovery from SARS-CoV-2 infection [35]. Moreover, the reduction of Ruminococcus and Lachnospira spp. (butyrate-producing bacteria) with a dramatic increase in Streptococcus spp. in COVID-19 affected patients is positively correlated with the levels of C-reactive protein and D-dimer [35]. Zhao et al. [39] demonstrated a change in proteomics and metabolomics in human milk samples of COVID-19 positive mothers; three primary pathways were thought to be involved: complement activation, platelet degranulation, and macrophage function [40]. To date, HMM has not been investigated in pregnant COVID-19 affected women, and therefore, we can only examine the existing studies that have been carried out in adults [35]. We can hypothesize that the above findings might be partially applicable to COVID-19 affected pregnant women, and since it is supposed that composition of the breast milk impacts the infant’s intestine, further studies on the changes in the microbiota of pregnant women affected by COVID-19 are necessary [35]. What is certain is that the pandemic has influenced HMM composition in several phases, both directly and indirectly, which can be substantiated by the fact that the placentas of women affected by severe acute respiratory syndrome have increased rates of vascular malperfusion features that can alter normal bacterial exchange between the mother and child [8]. Moreover, the presence of respiratory infection, enhances the probability of a C-section delivery, with a consequential reduction in maternal transfer of vaginal microbes to newborns [8]. Finally, increased hygiene and sanitization are the other two factors contributing to the minimization of transmission of maternal skin flora to the infant [8].
MICROBIAL DYSBIOSIS: A “TRIGGER POINT” FOR CHILDREN’S HEALTH
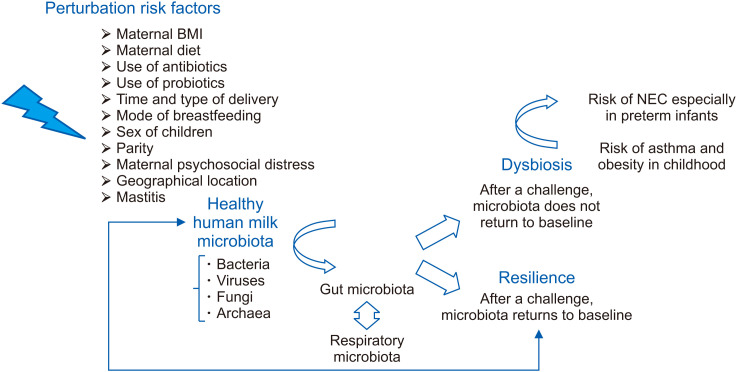
HMM composition can influence a child’s health in two main ways: by promoting intestinal immune homeostasis and facilitating digestive processes [16]. Bacterial diversity plays a pivotal role in the maintenance of immune balance in humans [4]. In particular, human milk bacteria provide early antigenic stimuli that promote intestinal immune system maturation; they also encourage a Th1/Th2 balanced response, promoting immune homeostasis [16]. Microbes produce short-chain fatty acids (SCFAs) through the fermentation of human milk oligosaccharides (HMOs) [16], which are the most extensively studied metabolites because of their immunomodulatory effects on several aspects of host physiology [7]. In vitro studies have demonstrated that some bacterial components of HMM (i.e., Lactobacillus spp.) can maintain an active immunological balance by inhibiting the proliferation of pathogenic microorganisms [4,16]. Several factors such as diet, ethnicity, medication use or geographic location can impart variability to the milk microbiota and can be a source of “stress” for it. Following a challenge, the microbiota of a healthy individual is able to return to baseline (resilience); however; if it does not happen, a new equilibrium is established with a drift towards dysbiosis [41]. Resilience can be used as a surrogate marker of healthy ecosystem as the microbiota of a healthy person resists changes under stress and fully recovers from perturbations [41]. As HMM is a fundamental contributor to GI colonization in children [15], establishing a healthy profile of intestinal bacteria is important for preventing or correcting dysbiosis and minimizing its impact on health [41]. In fact, the presence of dysbiosis can alter the balance between pathogens and commensal organisms, with a consequential life-threatening effect on the host [11]. In preterm infants, NEC results from an amplified and destructive inflammatory response to intestinal dysbiosis, with consequent tissue damage and loss of intestinal barrier integrity [42]. In fact, when the immature intestine of these neonates, which is accustomed to residing in the intrauterine environment where few microorganisms are available for interaction, encounters trillions of colonizing bacteria, excessive inflammation ensues, leading to NEC [10]. Dysbiosis in the microbiota can be a starter point for the development of some diseases not only in the first few months of life but also later during childhood; in particular, both GI and respiratory tract dysbiosis may have a causative role in the development of respiratory diseases such as asthma [4]. According to the latest studies a “gut-lung axis” exists in human beings [7], which is responsible for the systemic dissemination of bacterial-derived components and metabolic degradation products [43]. During intestinal dysbiosis, these bioactive compounds secreted by microbes and absorbed into the circulation can directly alter lung function [7], and each molecular pattern that is created (endotype) corresponds to a phenotype of asthma [44]. Dysbiosis of the intestinal microbiota is also associated with the onset of obesity in childhood and is characterized by alterations in gut microbiota diversity, with a relative abundance of certain microbial genera. The metabolites generated by microbes translocated from a disrupted intestinal barrier can influence several organs, contributing to systemic metabolic inflammation [45]. Therefore, both neonatal pathologies, such as NEC, and chronic diseases with a later onset in childhood, such as asthma and obesity, are clinical manifestations of an early dysbiosis of microbiota [9] ( Fig. 2).
Fig. 2. Pictorial representation of the response of human milk microbiota to stress factors.
BMI: body mass index, NEC: necrotizing enterocolitis.
Role in NEC onset
NEC is an intestinal disease with an unknown etiology that primarily affects preterm infants (approximately 10%) and is associated with an increased risk of morbidity (i.e., short-bowel syndrome, liver failure) and mortality (up to 40%) [42,46]. However, the precise pathogenesis of NEC remains unclear. It might be caused by a specific dysbiosis of the infant gut microbiota leading to an inflammatory state that alters the equilibrium of the intestinal immune system [10], with the consequent loss of bacterial diversity and increased abundance of pathogens [46]. Several factors that are able to induce enterocyte injury can contribute to the onset of NEC [47]. Among these, proton pump inhibitors and prenatal and postnatal antibiotic use can be risk factors for changes in the mother’s microbiota and, subsequently, in HMM composition [42,48]. It has been demonstrated that human milk from mothers of preterm infants is different from that of full-term infants, with higher levels of protein, fat, free amino acids, and sodium, which tend to decrease over time [49,50]. Moreover, bioactive components such as HMOs and lactoferrin are higher in preterm milk and colostrum [47]. Staphylococcus spp. are predominant in the milk samples of preterm mothers, which may be due to the higher abundance of this bacterial genus in the hospital environment and in the skin of hospitalized preterm infants [48]. Preterm neonates are indirectly breastfed using pumped-milk, often stored in a refrigerator or freezer in plastic containers and administered to the child via a nasogastric tube that ‘passes’ through the oral cavity. This further influences the HMM composition, with a reduction in Streptococcus spp., which are common bacteria in the oral cavity [47]. Stenotrophomonas and Acinetobacter spp. are known to be prevalent later in lactation [47,48]. It has been demonstrated that the gut microbiota of infants who develop NEC has reduced diversity and stability, with an overabundance of Proteobacteria (i.e., Pseudomonas) and a paucity of obligate anaerobic species, which are markers of healthy microbiota [42,47]. The same changes are present during the days immediately preceding NEC diagnosis [47,48]. In a recent study, Gopalakrishna et al. [51] discovered that preterm babies, who were exclusively formula-fed had a very low level of IgA-associated intestinal bacteria, and children with NEC had higher levels of IgA-unbound Enterobacter spp. compared to the healthy age-matched controls. It is probable that human milk, by developing a commensal infant gut microbiome, can decrease the risk of NEC [46]. The influence of mother’s own breastmilk on gut microbiome composition and the decreased risk of NEC is dose-dependent [46]. In particular, according to recent literature [10,47], the combination of expressed breast milk and a probiotic is the optimum approach to prevent NEC in premature infants. Milk from mothers who deliver prematurely has increased amounts of immune mediated and anti-inflammatory factors [10]. Moreover, it also contains bacteria translocated from the mother’s intestine that stimulate infant’s “pioneer” bacteria (i.e., Bifidobacterium infantis and Bacteroides fragilis) to produce metabolites with an anti-inflammatory role. Probiotics that interact with human milk can amplify this process [10,52].
Role in asthma development
Asthma is one of the most widespread chronic respiratory diseases in childhood, especially under the age of five years. Its development is correlated with many factors, including genetic predisposition, environmental exposures, infections, nutritional factors, and the gut microbiome [53]. Gut microbiota are transferred from the mother to the child during breastfeeding [53] and an early-life microbial dysbiosis has been linked to an increased risk of asthma and allergic diseases in early childhood [4,53,54]. In particular, during the first few years of life, the microbiota composition changes dynamically, and a “window of opportunity” can be created to determine future health or disease [55]. A recent longitudinal study revealed that children at risk of asthma exhibited transient intestinal microbial dysbiosis during the first 100 days of life [56] and that the development of a healthy microbiome depends significantly on the first bacterial colonization. For a long time, the lung was believed to be sterile; however, several studies have demonstrated that it harbors its own microbiota [4]. Although the intestine and lungs are anatomically distinct, they can potentially communicate through the pharynx, and the complex pathways involving their microbiota have reinforced the existence of a “gut-lung” axis [4]. It can shape the immune response and interfere with the course of respiratory diseases, but the underlying mechanisms are not fully understood [3,4]. Maternal BMI, atopy and stress, delivery mode, and antibiotic exposure are important modifiers of the infant gut microbiota that contribute to the onset of asthma. An important risk factor from this point of view is the mother’s diet; for example, a high-fat maternal diet during pregnancy has been associated with lower levels of Bacteroidetes phyla in the offspring gut microbiota, which can be a predisposing factor for the development of asthma [53]. In the initial years of a child’s life, recurrent viral infections and asthma onset would be correlated to a dysbiotic nasopharyngeal microbiome; meanwhile, early colonization with a non-pathogenic and varied bacterial community may be protective for the lung microbiota [4]. With regard to the respiratory microorganisms that enter the oral cavity and arrive at the lung, the fluctuation of the mucous film that covers the upper respiratory tract (URT) and the airflow determine an equilibrium between microbial immigration and elimination. The main source of lower airway colonization is the microbiome residing in the upper airways, along with microaspiration and/or direct inhalation of oropharyngeal secretions [4]. A recent culture-independent method of analysis has revealed differences in the microbial communities of the healthy respiratory tract compared to those of patients with asthma. Firmicutes (i.e., Staphylococcus, Streptococcus, Veillonella), Proteobacteria (i.e., Moraxella, Haemophilus), Actinobacteria (i.e., Propionibacterium), and Bacteroidetes (i.e., Prevotella) are some of the principal phyla present in healthy URT. In the low respiratory tract there are six dominant bacterial phyla: Firmicutes, Bacteroidetes (i.e., Prevotella), Proteobacteria, Fusobacteria, Acidobacteria and Actinobacteria [55]. Along the entire respiratory tract, viral and fungal components are also present, but these form only a minor portion of the respiratory microbiome [55]. Although Firmicutes, Proteobacteria and Actinobacteria are abundant in asthmatic individuals too, there might be a shift towards certain genera within these phyla that could generate dysbiosis [4]. Therefore, regardless of the composition of the microbiota, dysbiosis can trigger asthma in genetically predisposed subjects. In the first few years of life, gut dysbiosis is characterized by a reduction in individual diversity in the microbial ecosystem, with colonization by opportunistic pathogens (i.e., Enterococcus spp.) [57]. Early colonization of the URT in children by Moraxella genus is associated with respiratory tract infections; nasal secretion samples from asthmatic children of ages 6 to 17 years showed a significant activation of eosinophils, especially by Moraxella catarrhalis [4,58]. Moreover, colonization by Clostridium difficile at 1 month of age correlates with the occurrence of wheezing and asthma in the first 6 years of life [4,57,59]. In infants at risk of developing asthma, there is usually a relative decline in the genera Lachnospira, Veillonella, Faecalibacterium and Rothia in the first 100 days of life, as well as a relative reduction in Clostridium neonatale in the first 3 months of life [4]. Finally, a role of the intestinal archaeal component in the dysbiosis-asthma paradigm has also been suggested [55,60], although further studies are necessary in this area.
Role in obesity development
Obesity is one of the most prevalent non-communicable diseases worldwide as well as in pediatric patients. Therefore, it is important to understand the potential causes to reduce the prevalence and associated morbidities [61]. In this regard, the mode of delivery, maternal health, antibiotic exposure, and breastfeeding are considered as factors that play a pivotal role in altering the infant gut microbiota and, subsequently, the probability of obesity onset in children [61]. It has been demonstrated that breastfed infants have a lower incidence and risk of obesity than formula-fed infants. This may be due to maturation and immune tolerance conferred by commensal bacteria present in the HMM [23]. In particular, a recent cohort study demonstrated that obese mothers provide their infants with less breast milk at 6 and 12 months postpartum than normal-weight mothers, perhaps due to physical, physiological, and psychological obstacles. A shorter breastfeeding period is associated with minor diversity in the gut microbiota of children, which could be a risk factor for obesity onset [61]. It is well known the human gut microbiota is characterized by the presence of Firmicutes and Bacteroidetes phyla, as well as Proteobacteria, Fusobacteria, Verrucomicrobia and Actinobacteria. At the compositional level, obesity seems to be the consequence of a change in the Firmicutes/Bacteroidetes ratio (F/B), especially the reduction in the relative abundance of the latter [62], with a higher proportion of Actinobacteria [63]. Nevertheless, there are still several controversies, especially concerning childhood [62] and it is not clear which bacteria are necessary and/or sufficient to assemble a healthy gut microbiota [61,64]. For example, a recent study by Bai et al. [65] highlighted a positive correlation between high Bifidobacteria levels and elevated BMI [62], detaching itself from the most commonly cited information in the literature, stating that Bifidobacteria spp. are more abundant in normal weight children [62,65]. A specific marker of the human milk lipid component, called alkylglycerol-type (AKG-type), is able to reduce weight by maintaining beige adipose tissue (BeAT) and avoiding its transformation into lipid-storing white adipose tissue, using the IL-6/STAT (Signal Transducer and Activator of Transcription) pathway [66]. There exists a negative correlation between BeAT content and obesity and a lack of AKG intake may lead to a premature loss of BeAT, leading to risk of developing obesity [66]. Another mechanism by which AKG aids in controlling obesity is the increase in Lactobacillus proliferation with the establishment of healthy intestinal microbiota [66]. It is likely that metabolic activity, and not gut microbiota composition, is more relevant in obesity onset [61]. A recent systematic review conducted by Kim et al. [67] showed that obese patients had higher levels of acetate, propionate, and butyrate SCFAs than lean controls [63,67]. The increased F/B ratio could be a result of dysbiosis in obese children, arising from adaptation to this long-term metabolic dysfunction [62]. For pediatric patients, no definitive data are available to date.
METHODS OF STUDYING MICROBIOTA COMPOSITION
HMM exploration relies on both culture-dependent and culture-independent approaches, including sequencing of 16S rRNA clone libraries and metataxonomics. The latter is based on 16S rRNA gene amplicon sequencing and has emerged as a useful tool for bacterial identification in the last 10 years [6]. Metataxonomics is one of the applications of next generation sequencing, which involves the shotgun sequencing of the entire metagenome, typing of the microbial genome, and sequencing of the entire microbial genome.
Culture-dependent methods
Traditional culture methods report on the presence of lactic acid bacteria such as Lactobacillus, Lactococcus, Bifidobacterium [16], Streptococcus, Propionibacterium and Staphylococcus spp. These methods have the advantage of allowing the detection of viable bacteria, providing data on cell viability, and paving the way for strain-level genomic analyses [23,68,69]; however, they can fail to detect fastidious bacteria, leading to an underestimation of the diversity within a sample [23,69]. These methods can only detect cultivable bacteria, and the results are strictly dependent on the media used, sample storage conditions, and growth conditions [6]. It is important to emphasize that a culture-dependent method can reveal a bacterial count between 102 and 104 bacterial cells/mL, whereas using a culture-independent method, can reveal a bacterial load of between 104 and 105 bacterial cells/mL in human milk, suggesting that a part of the milk microbiota corresponds to non-viable and non-cultivable bacteria [6].
Culture-independent methods
A recent culture-independent method has been able to demonstrate a complex and variable microbial composition in human milk [16,69]. This method can detect a broad range of microbes, including anaerobes (i.e., Clostridium spp.), which are not detected by traditional culture methods [23]. The extracted DNA from collected samples is amplified and then sequenced through a genome sequencer instrument (Illumina MiSeq) or through a technique known as pyrosequencing [11]. The results are then analyzed with a computer program that provides data on distinct operational taxonomic units (OTUs) [11]. Every OTU represents the number of distinct clusters of genetic sequences that meet 97% similarity in pairs of hypervariable regions [1,11]. Amplicon sequencing is a simple and inexpensive method; however, it has an important limitation of being unable to differentiate living microbes from dead ones or from cell-free DNA. For this reason, viability dyes are used, such as propidium monoazide [11,23]. The milk fraction used for the analysis, lysis method [6], purification kit, target gene used for amplicons, number of polymerase chain reaction cycles, and sequencing platform could be the factors that might introduce variability in the results of amplicon sequencing [6]. Another limitation is the impossibility of producing quantitative results with this method and only semi-quantitative data can be obtained at best. Recently, the whole-genome shotgun sequencing approach has been used, with superior resolution and the possibility of detecting all portions of HMM (bacterial, viral, fungal, and archaeal components) [11,23], and it is still considered the gold standard of microbiome research [1]. With this latest sequencing mode, 10 most frequent bacterial genera found in HMM include Staphylococcus, Streptococcus, Lactobacillus, Pseudomonas, Bifidobacterium, Corynebacterium, Enterococcus, Acinetobacter, Rothia and Cutibacterium spp. [1,69]. Nevertheless, this is a DNA-based method; therefore, it cannot demonstrate transcriptional activity or cell viability. Moreover, because of the high level of human DNA in the HMM, this method may limit the sequencing depth of microbial DNA. A human DNA depletion step is required in this type of analysis [23]. To date, only a few studies have been conducted on milk microbiota using shotgun metagenomic approaches [6].
FUTURE PERSPECTIVES AND CONCLUSIONS
Some human milk-derived bacterial strains can be considered as potential probiotics [14]; for example, Lactobacillus reuteri, which is found in gut and breast milk, has an anti-inflammatory role that is exerted by reduction of proinflammatory cytokines and microbial translocation across the intestinal epithelium [14]. Therefore, its supplementation could be an attractive preventive and/or therapeutic strategy against inflammatory diseases [14]. The same organism can attenuate overweight, hypertriglyceridemia, and hypercholesterolemia. On the other hand, L. fermentum can be used for prophylaxis against community-acquired infections. Another microbe that could be used as a potentially safe probiotic is Enterococcus faecium, which is always isolated from breast milk [14]. It has been shown that the use of a combination of Lactobacillus and Bifidobacterium spp. in preterm infants, reduces the relative risk for NEC, even if the time and dose of administration are not well defined at present. More studies are necessary to evaluate the beneficial effects of this strategy [42]. Formula-fed infants have markedly different GI microbiomes and health outcomes than their breastfed counterparts; therefore, it may be possible to identify keystone microbial species present in human milk that are fundamental for children’s health (i.e., Bifidobacterium spp.) and to selectively fortify infant formula with them, making the formula more similar to human milk [23]. The European Commission [70] recently evaluated the possibility of using probiotics in infant formulas. Bifidobacterium animalis subsp., Lactis INL-1 and Lactobacillus plantarum 73a have been isolated from human milk and tested as complementary strategies to prevent non-communicable diseases such as obesity [5,71,72]. To modulate HMM and early life GI colonization of the infant, some studies have been conducted in lactating mothers who were administered oral probiotics to prevent or treat mastitis [23]. A future perspective is to use topical probiotics to shape the maternal skin microbiota, and consequently, HMM [23]. At the same time, phage therapy may be utilized, and in bovines, a range of phages against S. aureus and Escherichia coli-related mastitis have been demonstrated [23]. The composition of HMM is a topic that has garnered a lot of interest from many researchers in recent years. It has been demonstrated that not only a bacterial component but also viral, fungal, and archaeal components are present in HMM. It is fundamental to define a healthy microbiota and to prevent microbial dysbiosis that could be correlated to NEC onset in the initial months of a child’s life as well as to chronic diseases, such as asthma and obesity, later in the childhood. However, the real relationship between HMM composition and these pathologies is largely unknown at present. Therefore, it is necessary to conduct future studies in human lactation settings with emphasis on transcriptional activity of HMM.
Footnotes
Conflict of Interest: The authors have no financial conflicts of interest.
References
- 1.Lopez Leyva L, Brereton NJB, Koski KG. Emerging frontiers in human milk microbiome research and suggested primers for 16S rRNA gene analysis. Comput Struct Biotechnol J. 2020;19:121–133. doi: 10.1016/j.csbj.2020.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SY, Yi DY. Analysis of the human breast milk microbiome and bacterial extracellular vesicles in healthy mothers. Exp Mol Med. 2020;52:1288–1297. doi: 10.1038/s12276-020-0470-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, et al. The gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol. 2020;10:9. doi: 10.3389/fcimb.2020.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hufnagl K, Pali-Schöll I, Roth-Walter F, Jensen-Jarolim E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol. 2020;42:75–93. doi: 10.1007/s00281-019-00775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez C, Franco L, Regal P, Lamas A, Cepeda A, Fente C. Breast milk: a source of functional compounds with potential application in nutrition and therapy. Nutrients. 2021;13:1026. doi: 10.3390/nu13031026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oikonomou G, Addis MF, Chassard C, Nader-Macias MEF, Grant I, Delbès C, et al. Milk microbiota: what are we exactly talking about? Front Microbiol. 2020;11:60. doi: 10.3389/fmicb.2020.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang AT, Marsland BJ. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019;12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 8.Romano-Keeler J, Zhang J, Sun J. COVID-19 and the neonatal microbiome: will the pandemic cost infants their microbes? Gut Microbes. 2021;13:1–7. doi: 10.1080/19490976.2021.1912562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25:324–35.e4. doi: 10.1016/j.chom.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Walker WA, Meng D. Breast milk and microbiota in the premature gut: a method of preventing necrotizing enterocolitis. Nestle Nutr Inst Workshop Ser. 2020;94:103–112. doi: 10.1159/000505337. [DOI] [PubMed] [Google Scholar]
- 11.Groer MW, Morgan KH, Louis-Jacques A, Miller EM. A scoping review of research on the human milk microbiome. J Hum Lact. 2020;36:628–643. doi: 10.1177/0890334420942768. [DOI] [PubMed] [Google Scholar]
- 12.Togo A, Dufour JC, Lagier JC, Dubourg G, Raoult D, Million M. Repertoire of human breast and milk microbiota: a systematic review. Future Microbiol. 2019;14:623–641. doi: 10.2217/fmb-2018-0317. [DOI] [PubMed] [Google Scholar]
- 13.Lyons KE, Ryan CA, Dempsey EM, Ross RP, Stanton C. Breast milk, a source of beneficial microbes and associated benefits for infant health. Nutrients. 2020;12:1039. doi: 10.3390/nu12041039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moubareck CA. Human milk microbiota and oligosaccharides: a glimpse into benefits, diversity, and correlations. Nutrients. 2021;13:1123. doi: 10.3390/nu13041123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demmelmair H, Jiménez E, Collado MC, Salminen S, McGuire MK. Maternal and perinatal factors associated with the human milk microbiome. Curr Dev Nutr. 2020;4:nzaa027. doi: 10.1093/cdn/nzaa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selma-Royo M, Calvo Lerma J, Cortés-Macías E, Collado MC. Human milk microbiome: from actual knowledge to future perspective. Semin Perinatol. 2021;45:151450. doi: 10.1016/j.semperi.2021.151450. [DOI] [PubMed] [Google Scholar]
- 17.Kordy K, Gaufin T, Mwangi M, Li F, Cerini C, Lee DJ, et al. Contributions to human breast milk microbiome and enteromammary transfer of Bifidobacterium breve. PLoS One. 2020;15:e0219633. doi: 10.1371/journal.pone.0219633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajoka MSR, Mehwish HM, Siddiq M, Haobin Z, Zhu J, Yan L, et al. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. Lebensm Wiss Technol. 2017;84:271–280. [Google Scholar]
- 19.Arboleya S, Ruas-Madiedo P, Margolles A, Solís G, Salminen S, de Los Reyes-Gavilán CG, et al. Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int J Food Microbiol. 2011;149:28–36. doi: 10.1016/j.ijfoodmicro.2010.10.036. [DOI] [PubMed] [Google Scholar]
- 20.Solís G, de Los Reyes-Gavilan CG, Fernández N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010;16:307–310. doi: 10.1016/j.anaerobe.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Morniroli D, Consales A, Crippa BL, Vizzari G, Ceroni F, Cerasani J, et al. The antiviral properties of human milk: a multitude of defence tools from mother nature. Nutrients. 2021;13:694. doi: 10.3390/nu13020694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohandas S, Pannaraj PS. Beyond the bacterial microbiome: virome of human milk and effects on the developing infant. Nestle Nutr Inst Workshop Ser. 2020;94:86–93. doi: 10.1159/000504997. [DOI] [PubMed] [Google Scholar]
- 23.Stinson LF, Sindi AS, Cheema ASM, Lai CT, Mühlhäusler BS, Wlodek ME, et al. The human milk microbiome: who, what, when, where, why, and how? Nutr Rev. 2021;79:529–543. doi: 10.1093/nutrit/nuaa029. [DOI] [PubMed] [Google Scholar]
- 24.Boix-Amorós A, Puente-Sánchez F, du Toit E, Linderborg KM, Zhang Y, Yang B, et al. Mycobiome profiles in breast milk from healthy women depend on mode of delivery, geographic location, and interaction with bacteria. Appl Environ Microbiol. 2019;85:e02994–e02918. doi: 10.1128/AEM.02994-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Togo AH, Grine G, Khelaifia S, des Robert C, Brevaut V, Caputo A, et al. Culture of methanogenic archaea from human colostrum and milk. Sci Rep. 2019;9:18653. doi: 10.1038/s41598-019-54759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinleyici M, Pérez-Brocal V, Arslanoglu S, Aydemir O, Sevuk Ozumut S, Tekin N, et al. Human milk virome analysis: changing pattern regarding mode of delivery, birth weight, and lactational stage. Nutrients. 2021;13:1779. doi: 10.3390/nu13061779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermansson H, Kumar H, Collado MC, Salminen S, Isolauri E, Rautava S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front Nutr. 2019;6:4. doi: 10.3389/fnut.2019.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notarbartolo V, Carta M, Insinga V, Giuffrè M. Human milk is not “merely nutritious”: how its bioactive role can influence child health. EMBJ. 2021;16:21–26. [Google Scholar]
- 29.Zimmermann P, Curtis N. Breast milk microbiota: a review of the factors that influence composition. J Infect. 2020;81:17–47. doi: 10.1016/j.jinf.2020.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Macia L, Nanan R, Hosseini-Beheshti E, Grau GE. Host- and microbiota-derived extracellular vesicles, immune function, and disease development. Int J Mol Sci. 2019;21:107. doi: 10.3390/ijms21010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Yan X, Zhang L, Cai J, Zhou Y, Liu H, et al. Identification and peptidomic profiling of exosomes in preterm human milk: insights into necrotizing enterocolitis prevention. Mol Nutr Food Res. 2019 doi: 10.1002/mnfr.201801247. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.Galley JD, Besner GE. The therapeutic potential of breast milk-derived extracellular vesicles. Nutrients. 2020;12:745. doi: 10.3390/nu12030745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spatz DL, Davanzo R, Müller JA, Powell R, Rigourd V, Yates A, et al. Promoting and protecting human milk and breastfeeding in a COVID-19 world. Front Pediatr. 2021;8:633700. doi: 10.3389/fped.2020.633700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.WHO. Breastfeeding and COVID-19: scientific brief. Geneva: WHO; 2020. [Google Scholar]
- 35.Bardanzellu F, Puddu M, Fanos V. Breast milk and COVID-19: from conventional data to “omics” technologies to investigate changes occurring in SARS-CoV-2 positive mothers. Int J Environ Res Public Health. 2021;18:5668. doi: 10.3390/ijerph18115668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–55.e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gulati M, Singh SK, Corrie L, Kaur IP, Chandwani L. Delivery routes for faecal microbiota transplants: available, anticipated and aspired. Pharmacol Res. 2020;159:104954. doi: 10.1016/j.phrs.2020.104954. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y, Shang Y, Ren Y, Bie Y, Qiu Y, Yuan Y, et al. Omics study reveals abnormal alterations of breastmilk proteins and metabolites in puerperant women with COVID-19. Signal Transduct Target Ther. 2020;5:247. doi: 10.1038/s41392-020-00362-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dogra SK, Doré J, Damak S. Gut microbiota resilience: definition, link to health and strategies for intervention. Front Microbiol. 2020;11:572921. doi: 10.3389/fmicb.2020.572921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nolan LS, Rimer JM, Good M. The role of human milk oligosaccharides and probiotics on the neonatal microbiome and risk of necrotizing enterocolitis: a narrative review. Nutrients. 2020;12:3052. doi: 10.3390/nu12103052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gautier T, David-Le Gall S, Sweidan A, Tamanai-Shacoori Z, Jolivet-Gougeon A, Loréal O, et al. Next-generation probiotics and their metabolites in COVID-19. Microorganisms. 2021;9:941. doi: 10.3390/microorganisms9050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma A, Laxman B, Naureckas ET, Hogarth DK, Sperling AI, Solway J, et al. Associations between fungal and bacterial microbiota of airways and asthma endotypes. J Allergy Clin Immunol. 2019;144:1214–27.e7. doi: 10.1016/j.jaci.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan X, Chen R, McCormick KL, Zhang Y, Lin X, Yang X. The role of the gut microbiota on the metabolic status of obese children. Microb Cell Fact. 2021;20:53. doi: 10.1186/s12934-021-01548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis JA, Baumgartel K, Morowitz MJ, Giangrasso V, Demirci JR. The role of human milk in decreasing necrotizing enterocolitis through modulation of the infant gut microbiome: a scoping review. J Hum Lact. 2020;36:647–656. doi: 10.1177/0890334420950260. [DOI] [PubMed] [Google Scholar]
- 47.Granger CL, Embleton ND, Palmer JM, Lamb CA, Berrington JE, Stewart CJ. Maternal breastmilk, infant gut microbiome and the impact on preterm infant health. Acta Paediatr. 2021;110:450–457. doi: 10.1111/apa.15534. [DOI] [PubMed] [Google Scholar]
- 48.Asbury MR, Butcher J, Copeland JK, Unger S, Bando N, Comelli EM, et al. Mothers of preterm infants have individualized breast milk microbiota that changes temporally based on maternal characteristics. Cell Host Microbe. 2020;28:669–82.e4. doi: 10.1016/j.chom.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Carr LE, Virmani MD, Rosa F, Munblit D, Matazel KS, Elolimy AA, et al. Role of human milk bioactives on infants’ gut and immune health. Front Immunol. 2021;12:604080. doi: 10.3389/fimmu.2021.604080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buffet-Bataillon S, Bellanger A, Boudry G, Gangneux JP, Yverneau M, Beuchée A, et al. New insights into microbiota modulation-based nutritional interventions for neurodevelopmental outcomes in preterm infants. Front Microbiol. 2021;12:676622. doi: 10.3389/fmicb.2021.676622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gopalakrishna KP, Macadangdang BR, Rogers MB, Tometich JT, Firek BA, Baker R, et al. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat Med. 2019;25:1110–1115. doi: 10.1038/s41591-019-0480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker WA. Summary on microbiota of milk and lactation: influence on gut colonization. Nestle Nutr Inst Workshop Ser. 2020;94:113–114. doi: 10.1159/000505552. [DOI] [PubMed] [Google Scholar]
- 53.Alsharairi NA. The infant gut microbiota and risk of asthma: the effect of maternal nutrition during pregnancy and lactation. Microorganisms. 2020;8:1119. doi: 10.3390/microorganisms8081119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrante G, Carta M, Montante C, Notarbartolo V, Corsello G, Giuffrè M. Current insights on early life nutrition and prevention of allergy. Front Pediatr. 2020;8:448. doi: 10.3389/fped.2020.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The role of lung and gut microbiota in the pathology of asthma. Immunity. 2020;52:241–255. doi: 10.1016/j.immuni.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Espírito Santo C, Caseiro C, Martins MJ, Monteiro R, Brandão I. Gut microbiota, in the halfway between nutrition and lung function. Nutrients. 2021;13:1716. doi: 10.3390/nu13051716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ballarini S, Rossi GA, Principi N, Esposito S. Dysbiosis in pediatrics is associated with respiratory infections: is there a place for bacterial-derived products? Microorganisms. 2021;9:448. doi: 10.3390/microorganisms9020448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toivonen L, Hasegawa K, Waris M, Ajami NJ, Petrosino JF, Camargo CA, Jr, et al. Early nasal microbiota and acute respiratory infections during the first years of life. Thorax. 2019;74:592–599. doi: 10.1136/thoraxjnl-2018-212629. [DOI] [PubMed] [Google Scholar]
- 59.Stinson LF. Establishment of the early-life microbiome: a DOHaD perspective. J Dev Orig Health Dis. 2020;11:201–210. doi: 10.1017/S2040174419000588. [DOI] [PubMed] [Google Scholar]
- 60.Barnett DJM, Mommers M, Penders J, Arts ICW, Thijs C. Intestinal archaea inversely associated with childhood asthma. J Allergy Clin Immunol. 2019;143:2305–2307. doi: 10.1016/j.jaci.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 61.Haddad EN, Sugino KY, Kerver JM, Paneth N, Comstock SS. The infant gut microbiota at 12 months of age is associated with human milk exposure but not with maternal pre-pregnancy body mass index or infant BMI-for-age z-scores. Curr Res Physiol. 2021;4:94–102. doi: 10.1016/j.crphys.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petraroli M, Castellone E, Patianna V, Esposito S. Gut microbiota and obesity in adults and children: the state of the art. Front Pediatr. 2021;9:657020. doi: 10.3389/fped.2021.657020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moszak M, Szulińska M, Bogdański P. You are what you eat-the relationship between diet, microbiota, and metabolic disorders-a review. Nutrients. 2020;12:1096. doi: 10.3390/nu12041096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McBurney MI, Davis C, Fraser CM, Schneeman BO, Huttenhower C, Verbeke K, et al. Establishing what constitutes a healthy human gut microbiome: state of the science, regulatory considerations, and future directions. J Nutr. 2019;149:1882–1895. doi: 10.1093/jn/nxz154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bai J, Hu Y, Bruner DW. Composition of gut microbiota and its association with body mass index and lifestyle factors in a cohort of 7-18 years old children from the American Gut Project. Pediatr Obes. 2019;14:e12480. doi: 10.1111/ijpo.12480. [DOI] [PubMed] [Google Scholar]
- 66.Yu H, Dilbaz S, Coßmann J, Hoang AC, Diedrich V, Herwig A, et al. Breast milk alkylglycerols sustain beige adipocytes through adipose tissue macrophages. J Clin Invest. 2019;129:2485–2499. doi: 10.1172/JCI125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim KN, Yao Y, Ju SY. Short chain fatty acids and fecal microbiota abundance in humans with obesity: a systematic review and meta-analysis. Nutrients. 2019;11:2512. doi: 10.3390/nu11102512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernández L, Pannaraj PS, Rautava S, Rodríguez JM. The microbiota of the human mammary ecosystem. Front Cell Infect Microbiol. 2020;10:586667. doi: 10.3389/fcimb.2020.586667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beghetti I, Biagi E, Martini S, Brigidi P, Corvaglia L, Aceti A. Human milk’s hidden gift: implications of the milk microbiome for preterm infants’ health. Nutrients. 2019;11:2944. doi: 10.3390/nu11122944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van den Akker CHP, van Goudoever JB, Shamir R, Domellöf M, Embleton ND, Hojsak I, Lapillonne A, et al. Probiotics and preterm infants: a position paper by the European Society for Paediatric Gastroenterology Hepatology and Nutrition Committee on Nutrition and the European Society for Paediatric Gastroenterology Hepatology and Nutrition Working Group for Probiotics and Prebiotics. J Pediatr Gastroenterol Nutr. 2020;70:664–680. doi: 10.1097/MPG.0000000000002655. [DOI] [PubMed] [Google Scholar]
- 71.Oddi S, Binetti A, Burns P, Cuatrin A, Reinheimer J, Salminen S, et al. Occurrence of bacteria with technological and probiotic potential in Argentinian human breast-milk. Benef Microbes. 2020;11:685–702. doi: 10.3920/BM2020.0054. [DOI] [PubMed] [Google Scholar]
- 72.Oddi S, Huber P, Rocha Faria Duque AL, Vinderola G, Sivieri K. Breast-milk derived potential probiotics as strategy for the management of childhood obesity. Food Res Int. 2020;137:109673. doi: 10.1016/j.foodres.2020.109673. [DOI] [PubMed] [Google Scholar]