Abstract
Cancer cells utilize epithelial to mesenchymal transition (EMT) during invasion and metastasis. This program has intermediate cell states with retained epithelial and gained mesenchymal features together, referred to as partial EMT. Histone demethylase LSD1 forms a complex with the EMT master transcription factor Snail to modify histone marks and regulate target gene expression. However, little is known about the formation of this complex during the Snail-dependent transition between partial EMT and EMT. Here we visualized the nuclear complex of Snail and LSD1 as foci signals using proximity ligation assay. We demonstrated that the nuclear foci numbers varied with the transition of exogenous Snail-dependent partial EMT to EMT. Furthermore, we found that long exposure to dexamethasone could revert exogenous Snail-dependent EMT to partial EMT. In this reversion, the nuclear foci numbers also returned to previous levels. Therefore, we concluded that Snail might select partial EMT or EMT by altering its association with LSD1.
Keywords: Epithelial to mesenchymal transition (EMT), Partial EMT, Snail, LSD1
Highlights
-
•
Nuclear complexes of Snail was visualized by PLA.
-
•
Exogenous Snai1 differently induced pEMT and EMT in OM-1.
-
•
Dexamethasone reverted Snail-induced EMT to pEMT.
-
•
Nuclei showed distinct foci numbers of Snail/LSD1 and Snail/methylated H3 in EMT and pEMT.
Abbreviations
- LSD1
Lysine specific demethylase 1
- EMT
Epithelial to mesenchymal transition
- pEMT
Partial epithelial to mesenchymal transition
- SCC
Squamous cell carcinoma
- RNA
Ribonucleic acid
- cDNA
Complementary deoxyribonucleic acid
- RT-PCR
Reverse transcription polymerase chain reaction
- COC
Cyclic olefin copolymer
- PBS
Phosphate-buffered saline
- BSA
Bovine serum albumin
- DAPI
4′,6-diamidino-2-phenylindole
- PLA
Proximity ligation assay
- GR
Glucocorticoid receptor
- SNAG
Snail/Gfi-1
- ChIP-seq
Chromatin immunoprecipitaton sequencing
1. Introduction
The progression of oral squamous cell carcinoma is initiated by local invasion of cancer cells from the epithelium into the interstitium, followed by intravascular migration and metastatic lesion formation. Epithelial-to-mesenchymal transition (EMT) is a physiological program in which epithelial cells are reversibly converted to mesenchymal cells in epithelial organ formation and wound healing, and cancer cells infiltrate and metastasize distantly [1]. Recent studies demonstrated that the EMT process is not binary but exhibits an intermediate state, in which cells express both epithelial and mesenchymal markers [2]. This intermediate state has been referred to as partial, incomplete, or hybrid EMT, in contrast with EMT, which is a pure mesenchymal state [2]. In this study, we call the intermediate state partial EMT (pEMT) and the pure mesenchymal state, EMT. In addition, mesenchymal-to-epithelial transition, in which cells in EMT, mesenchymal cells, or stem cells acquire epithelial traits, has also been considered in physiological or metastatic cancer-developing processes [3]. Snail is a master transcription factor for EMT, and we previously reported several characterizations by exogenous Snail-dependent EMT using oral SCC cell lines, especially the established OM-1 [4]. Our previous studies focused on binary EMT using fully mesenchymal clones and Snail-expressing vector transduction [5,6], however, recent results indicated exogenous Snail-produced pEMT features in the majority, represented by retained CDH1 expression [7]. In gene repression at Snail-loaded chromatin, complex formation on the chromatin with histone demethylase LSD1 plays a crucial role [8]. Although inducible Snail nuclear translocation resulted in a transient chromatin loading and target gene expression for a short period (6–48 h) despite its continued existence, long-lasting histone modification alteration by its cofactors, including LSD1, may elicit a stable EMT phenotype [9]. Because little is known about how exogenous Snai1s differentially induced EMT or pEMT, we aimed to achieve a piece of evidence for the functional difference of Snail in our established cells showing stable pEMT and EMT. Furthermore, since our stable pEMT and EMT cells that invariably expressed exogenous Snail nuclei were predicted to have different chromatin changes with long-lasting Snail loading on chromatin, we investigated possible alterations in chromatin by monitoring whether Snail recruited its functional cofactor LSD1 as well as histone modifications at Snail-loaded chromatin regions in pEMT and EMT cells.
2. Material and methods
2.1. Cell lines and cell culture
The human oral SCC cell line OM-1 [9,10,11] and its subclones [7] (pEMT OM-1snai1 and its derivate EMT OM-1snai1) were cultured at 37 °C in a humidified atmosphere of 5% CO2 in air and maintained in Dulbecco's modified Eagle's medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich). For exogenous Snai1 transduction, as described previously [7], parental OM-1 cells were infected with a retrovirus produced in host cell line HEK293gp packaging cells (RCB, Tsukuba, Japan), which was co-transfected with Snai1-encoding pBabe-puro [7] and pVSV-G (Invitrogen, Carlsbad, CA, USA). The infected OM-1 cells were selected using 3 μg/mL Puromycin dihydrochloride (InvivoGen, USA) to establish pEMT OM-1snai1, from which EMT OM-1snai1 cells were established by limited dilution.
2.2. RNA extraction, first-strand cDNA synthesis, semi-quantitative RT-PCR, and real-time RT-PCR
Total RNA was isolated using an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. First-strand cDNA libraries were synthesized using ReverTra Ace qPCR RT Kit (TOYOBO, Osaka, Japan) according to the manufacturer's protocol. RT-PCR was performed using a Go Taq Green Master Mix (Promega, Madison, WI, USA) for 30 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 40 s. The PCR products were analyzed using 2% agarose gel electrophoresis. Real-time PCR was performed using a Thunderbird SYBR qPCR Mix (Toyobo, Osaka, Japan) and signals were detected using a Thermal Cycler Dice Real-Time System III (TAKARA BIO, Shiga, Japan), according to the manufacturer's protocol. The collected data were analyzed by the delta Ct (2–ΔΔCt) method using GAPDH as an internal control [12]. The PCR primers and amplicon sizes are shown below.
CDH1 (131 bp): 5′-GCCTCCTGAAAAGAGAGTGGAAG-3′ and 5′-TGGCAGTGTCTCTCCAAATCCG-3′
Snai1 (321 bp): 5′-AATCGGAAGCCTAACTACAG-3′ and 5′-GGAAGAGACTGAAGTAGAG-3′
Vimentin (750 bp): 5′-TGGCACGTCTTGACCTTGAA-3′ and 5′-GGTCATCGTGATGCTGAGAA-3′
Ovol1 (138 bp): 5′-ACGATGCCCATCCACTACCTG-3′ and 5′-TTTCTGAGGTGCTGGTCATCATTC-3′
Ovol2 (141 bp): 5′-CCACAACCAGGTGAAAAGACACC-3′ and 5′-CGCTGGGTGAAGGCTTTATTGC-3′
LSD1 (114 bp): 5′-TCAGGAGTTGGAAGCGAATCCC-3′ and 5′-GTTGAGAGAGGTGTGGCATTAGC-3′
GAPDH (373 bp): 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-CAGCCCCAGCGTCAAAGGTG-3′
2.3. Reagents and antibodies
Dexamethasone (Tokyo Chemical Industry Co., Ltd, Tokyo, Japan) was applied in culture at 200 nM. RU-486 (Tokyo Chemical Industry Co., Ltd) was applied at 10 μM. The antibodies and dilution ratios used in immunocytochemistry were as follows: anti-E-cadherin rabbit monoclonal antibody (clone24E10, 1:200, #3195, Cell Signaling Technology, Danvers, MA, USA), anti-Snail mouse monoclonal antibody (clone L70G2, 1:200, #3895, Cell Signaling Technology), and anti-Vimentin mouse monoclonal antibody (clone RV202, 1:200, sc-32322, Santa Cruz Technology, Dallas, TX, USA), Anti-KDM1/LSD1 antibody (clone EPR6825, 1:200, ab129195, abcam, Cambridge, UK), anti-Histone H3K4 me2 mouse monoclonal antibody (clone MABI0303, 1:200, No: 39679, Active Motif, Carlsbad, CA, USA), anti-monomethyl Histone H3 (Lys9) mouse monoclonal antibody (clone MA306A, 1:200, No: 19010, MAB Institute, Inc., Iida, Japan), and anti-dimethyl Histone H3 (Lys9) mouse monoclonal antibody (clone MA307A, 1:200, No: 20006, MAB Institute, Inc.).
2.4. Immunocytochemistry
Cells were cultured in 96-well Half Area COC Film bottom plates (Corning Incorporated, Corning, NY, USA). The cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min. Following blocking and permeabilization with 0.3% Triton X-100, 5% BSA in PBS (all from Sigma-Aldrich) for 1 h, the cells were incubated with antibodies in 0.3% Triton X-100, 5% BSA in PBS at 4 °C for 12 h. Antibody binding was visualized with Alexa Fluor 568 goat anti-rabbit IgG (H + L) (A-11036, Invitrogen) or Alexa Fluor 488 goat anti-mouse IgG (A-11001, Invitrogen), diluted 1:10,000 in PBS for 1 h. After mounting with 4′,6-diamidino-2-phenylindole (DAPI) containing VECTASHIELD (Vector Laboratories Inc., Burlingame, CA, USA), images were analyzed with a BZ-9000 fluorescence microscope (KEYENCE, Osaka, Japan).
2.5. Proximity ligation assay (PLA)
For cell culture, fixation, and primary antibody reaction, the same procedures listed in section 2.4 were performed. PLA was performed to quantify protein–protein interactions in situ [13]. In this assay, two molecules located within 30 nm of each other develop a fluorescent signal spot [14]. The Duolink In Situ Detection Reagents Kit Red (Olink Bioscience, Uppsala, Sweden) was used to amplify specific signals. When either primary antibody for PLA was omitted, no signal was detected (data not shown). Images were acquired with a BZ-9000 microscope (KEYENCE).
2.6. Quantification and statistical analyses
Image data were quantified using Image J (https://imagej.nih.gov/ij/) [15]. Histogram plots were expressed using R Studio (https://www.rstudio.com/products/rstudio/download/) [16], where Welch's t- and f-tests were used for statistical comparison.
3. Results
3.1. Exogenous Snai1 led both stable pEMT and EMT in OM-1
As previously reported, exogenous Snai1 in OM-1 produces only a partial EMT phenotype, which was characterized by transcriptional activation of Vimentin but a lack of repression of CDH1 [7]. During passage of the culture cells, the cells stably exhibited the partial EMT phenotype (pEMT OM-1snai1), whereas we were able to isolate clones showing a stable EMT phenotype (Fig. 1A, EMT OM-1snai1). The newly established EMT OM-1snai1 also produced transcriptional repression of CDH1 and Ovol2 (Fig. 1B and C), the latter of which was recently reported to be a master transcription factor for mesenchymal-to-epithelial transition [17], suggesting that pEMT OM-1snai1 maintained the epithelial traits.
Fig. 1.
OM-1 with exogenous snai1. (A) Phase-contrasted microscope images of OM-1, pEMT OM-1snai1, and EMT OM-1snai1. (B) mRNA expression profiles of OM-1, pEMT OM-1snai1, and EMT OM-1snai1. (C) Quantified mRNA expression profiles of independently established pEMT OM-1snai1 and EMT OM-1snai1 with parental OM-1.
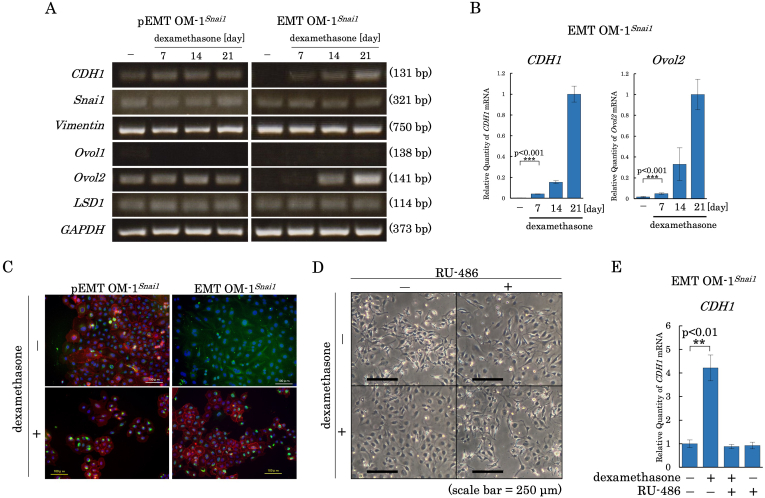
3.2. Dexamethasone reverted stable EMT to partial EMT
Since the EMT program is known to be reversible [18], we tried to revert the EMT profile of EMT OM-1snai1 to pEMT using extracellular stimuli. Since dexamethasone is known to induce mesenchymal-to-epithelial transition [19], we tested whether it could revert EMT to pEMT. As demonstrated in Fig. 2A, we found that long exposure to dexamethasone reverted the EMT profile of EMT OM-1snai1 to pEMT, with re-activation of CDH1 and Ovol2. An independent EMT OM-1snai1 clone also reproduced the result (Fig. 2B). While pEMT OM-1snai1 cells exhibited cytoplasmic unassembled vimentin and plasma membranous E-cadherin together (Fig. 2C, left top panel), EMT OM-1snai1 cells displayed assemblage of fibrous vimentin as a typical characteristic of mesenchymal cells, as well as the disappearance of E-cadherin (Fig. 2C, right top panel). We confirmed that this change was due to dexamethasone via glucocorticoid receptor (GR) signaling by using a GR antagonist, RU-486. RU-486 inhibited dexamethasone-induced changes at 7 days of EMT OM-1Snai1 cells in terms of its morphology (Fig. 2D) and CDH1 expression (Fig. 2E). Since pEMT OM-1snai1 did not diminish cytoplasmic unassembled vimentin after extended exposure to dexamethasone (Fig. 2A and right bottom panel of 2C), reversible transition by dexamethasone may be limited to pEMT, but not the epithelial state, in the presence of exogenous Snai1.
Fig. 2.
Dexamethasone reprogrammed stable EMT to partial EMT. (A) mRNA expression profiles for pEMT OM-1snai1 and EMT OM-1snai1 cultured for indicated periods with 200 nM dexamethasone. (B) Quantified mRNA expression profiles of independently established EMT OM-1snai1. (C) Immunocytochemical analysis of pEMT OM-1snai1 and EMT OM-1snai1. Controls (upper panels) were compared with 21 days cultures with dexamethasone (lower panels). All panels were double stained with rabbit anti-E-cadherin and mouse anti-vimentin antibodies (green: Vimentin, red: E-cadherin). Nuclei were visualized with DAPI (blue). (D) Phase-contrast microscope image of EMT OM-1Snai1. EMT OM-1Snai1 has a mesenchymal morphology (top left), but changes to an epithelial-like morphology with the addition of dexamethasone (bottom left). However, RU-486 inhibited dexamethasone-induced changes in cell morphology (bottom right). There was no change with the addition of RU-486 alone (top right). (E) Quantified mRNA expression profiles of independently established EMT OM-1snai1.
3.3. Colocalized nuclear foci of Snail and LSD1 reflected pEMT and EMT states
Next, we focused on histone demethylase LSD1, which is known to associate with nuclear Snail to regulate epigenetic histone marks [8]. The nuclear expressions of Snail and LSD1 were unaltered under dexamethasone exposure both in pEMT OM-1snai1 and EMT OM-1snai1 cells (Supplemental Figure). As demonstrated in Fig. 3A, we evaluated the in situ nuclear interaction foci between Snail and LSD1 in OM-1, pEMT OM-1snai1, and EMT OM-1snai1 by PLA visualization [14]. The Snail-LSD1 foci markedly increased in EMT OM-1snai1 (Fig. 3B and C), indicating that Snail-LSD1 foci numbers might represent the pEMT–EMT state. This was confirmed with long exposure to dexamethasone, which resulted in a specific decrease in Snail-LSD1 foci in EMT OM-1snai1 (Fig. 3D and E) concomitant with a transition from EMT to pEMT (Fig. 2).
Fig. 3.
Nuclear foci number of LSD1-carrying Snail reflected partial EMT and EMT states. (A) PLA results were analyzed using Image J. Image of nucleus stained with DAPI taken with a fluorescence microscope (top left). The number of nuclei was counted and set to display the outline (top right). The PLA spot is highlighted in red (bottom left). After merging the images, the red highlighted spots in the outline were calculated as pixel values for each nucleus (bottom right). The calculated pixel values in the nucleus are listed on the right side of the image. (B) A rabbit antibody (anti-LSD1) and a mouse antibody (anti-Snail) were used in this PLA assay for OM-1 (left), pEMT OM-1snai1 (middle), and EMT OM-1snai1 (right). (C) The images obtained in (B) were analyzed using Image J. The signal spots were quantified by pixel values, expressed using violin plots with included boxplot. Welch's t-test was used for analysis using R studio (***p < 0.001). EMT OM-1snai1 significantly increased the PLA signal compared with pEMT OM-1snai1. (D) PLA with a rabbit antibody (anti-LSD1) and a mouse antibody (anti-Snail) for pEMT OM-1snai1 and EMT OM-1snai1 with or without dexamethasone exposure for 21 days. (E) The images obtained in (D) were analyzed using Image J. The spots were quantified by pixel values, expressed as box plots. Welch's t-test was used for analysis using R studio (***p < 0.001). Addition of dexamethasone significantly reduced the PLA signal in EMT OM-1snai1.
3.4. Histone marks around Snail-loaded chromatin represented pEMT
As the Snail-LSD1 complex was expected to alter histone marks at the loaded chromatin, we analyzed histone marks at Snail-loaded chromatin regions by applying PLA between Snail and differentially methylated H3K4 or H3K9, which are substrates of LSD1 [20]. The nuclei of OM-1 cells displayed fewer foci for any histones because of the reduced expression of Snai1, confirming that the signal spots developed by PLA in pEMT OM-1snai1 and EMT OM-1snai1 were specific Snail-histone complexes (Fig. 4A). Dexamethasone-induced reversion from EMT to pEMT in EMT OM-1snai1 diminished H3K4 me2 and H3K9 me2 but increased H3K9 me1 around the Snail-loaded chromatins (Fig. 4B and C), suggesting dynamic changes in Snail-target gene expression via chromatin modification.
Fig. 4.
Histone marks alterations at Snail-loaded chromatin during dexamethasone-induced reversion of EMT OM-1snai1. (A) PLA was performed with anti-H3K4 me2 (top), anti-H3K9 me1 (middle), anti-H3K9 me2 (bottom) antibodies, and anti-Snail antibody (left: DAPI, right: PLA signals). PLA signal was not detected in OM-1 in any combination but was detected in pEMT OM-1Snai1 with exogenous Snail. (B) PLA was performed in the same combination as (A) in EMT OM-1Snai1 with or without 21 days dexamethasone exposure. Nuclei were visualized with DAPI (left), PLA signals were highlighted with red (right). (C) PLA was performed under the same conditions as (B) in EMT OM-1Snai1. The images obtained were analyzed with Image J. The spots were quantified by pixel values, expressed as box plots. Statistical analysis (f-test) was performed using R studio (***p < 0.001). Addition of dexamethasone changed the dispersion in EMT OM-1Snai1 cells, which was smaller in H3K4 me2 and H3K9 me2, and larger in H3K9 me1.
4. Discussion
Our previous studies indicated that exogenous Snai1 in OM-1 is able to induce mostly pEMT [7], of which subclones elicit EMT. The former (pEMT OM-1snai1) expressed E-cadherin at cell-to-cell contact and mRNA of Vimentin, but expressed little cytoplasmic Vimentin [7]. The latter (EMT OM-1snai1) was stably established by single cell cloning after Snai1-expressing vector transduction. The pEMT OM-1snai1 also underwent EMT conditionally (e.g., cytokine stimuli and low density culture) [7], which was characterized by a complete loss of E-cadherin, suggesting plasticity in the transition of pEMT to EMT. In the present study, we succeeded in reverting EMT to pEMT using EMT OM-1snai1, indicating bidirectional plasticity between both Snail-dependent EMT and pEMT. The reverted features of EMT OM-1snai1 to pEMT after three weeks’ exposure to dexamethasone were maintained after withdrawal of dexamethasone (data not shown), indicating some establishment of epigenetic modifications. We found that histone demethylase LSD1 [20] increased the foci complex with Snail along with progression to EMT. In reverting EMT to pEMT by dexamethasone, the complex foci of Snail and LSD1 decreased, indicating that LSD1 was the partner to regulate epigenetic modification by Snail between pEMT and EMT. Recently, it was reported that Ovol1 and Ovol2 regulate the mesenchymal-to-epithelial transition. Our data demonstrated that pEMT OM-1snai1 diminished Ovol1 but maintained Ovol2. In the reversion process, EMT OM-1Snai1 regained Ovol2 as well as E-cadherin. Interestingly, both Ovols and Snail possess a SNAG motif to interact with LSD1 [18,19,21,22], therefore, it may be possible that these SNAG proteins compete for LSD1 to select pEMT or EMT features. Since Ovol2 is also known to be a direct GR-target gene [23], it would be interesting to test its contribution in EMT to pEMT reversal.
In the present study, we identified nuclear foci numbers of Snail-LSD1 complexes that represented Snail-dependent pEMT or EMT states. Although nuclear Snail actually loaded on chromatin and altered its histone marks during dexamethasone-dependent transition from EMT to pEMT, the exact contribution of nuclear Snail-LSD1 complexes to the histone modification were not fully characterized here. Especially, reduced Snail–LSD1 numbers by dexamethasone might cause accumulation of methylated histones at the Snail-loaded chromatin. Indeed, H3K9me1 displayed this anticipated accumulation and its foci number also had decreased EMT tendency compared with pEMT, suggesting that H3K9me1-accumulated Snail might load on genes with LSD1 for pEMT or EMT determination. Moreover, the Snail-loaded chromatin unexpectedly lost H3K4me2 and H3K9me2 upon dexamethasone treatment, suggesting that other histone modifiers for this histone substrate might be recruited to the Snail-loaded chromatin regions via dexamethasone-dependent upregulations.
In this study, antibody-dependent PLA served as a powerful tool for in situ visualization but had obvious limitations in addressing specific genes on the Snail-loaded chromatin regions. Possibly, interference occurred between the GR directly and the Snail–LSD1 association on the chromatin. Therefore, it is necessary to perform triple-color PLA in the future. To elucidate the Snail-target genes that determine the pEMT or EMT state and Snail-loaded chromatin modification, especially colocalized LSD1 and its substrate histones, further genome-wide experiments, such as detailed transcriptome analysis with ChIP-seq for these nuclear factors, need to be conducted.
Funding
This work was supported by KAKENHI [JP20K10139, JP19K10309, JP19K10358, JP19K19161, and JP18K17198] by a Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, and Technology.
Declaration of competing interest
The authors have no potential conflicts of interest to declare.
Acknowledgements
We are grateful to Dr. Nobuyuki Kamata and Dr. Koichiro Higashikawa, who passed away after we had established a research group in Hiroshima.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101277.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
figs1.
References
- 1.Thiery J.P., Acloque H., Huang R.Y.J., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Pastushenko I., Blanpain C. EMT Transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Baum B., Settleman J., Quinlan M.P. Transitions between epithelial and mesenchymal states in development and disease. Semin. Cell Dev. Biol. 2008;19:294–308. doi: 10.1016/J.SEMCDB.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Uetsuki R., Higashikawa K., Okuda S., Yamakado N., Ishida F., Rizqiawan A., Ono S., Takechi M., Mizuta K., Shigeishi H., Kamata N., Tobiume K. The squamous cell carcinoma cell line OM-1 retains both p75-dependent stratified epithelial progenitor potential and cancer stem cell properties. Biochem. Biophys. Rep. 2021;26 doi: 10.1016/J.BBREP.2021.101003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higashikawa K., Yoneda S., Tobiume K., Saitoh M., Taki M., Mitani Y., Shigeishi H., Ono S., Kamata N. ΔNp63α-dependent expression of Id-3 distinctively suppresses the invasiveness of human squamous cell carcinoma. Int. J. Cancer. 2009;124:2837–2844. doi: 10.1002/ijc.24280. [DOI] [PubMed] [Google Scholar]
- 6.Higashikawa K., Yoneda S., Taki M., Shigeishi H., Ono S., Tobiume K., Kamata N. Gene expression profiling to identify genes associated with high-invasiveness in human squamous cell carcinoma with epithelial-to-mesenchymal transition. Cancer Lett. 2008;264:256–264. doi: 10.1016/j.canlet.2008.01.045. [DOI] [PubMed] [Google Scholar]
- 7.Okui G., Tobiume K., Rizqiawan A., Yamamoto K., Shigeishi H., Ono S., Higashikawa K., Kamata N. AKT primes snail-induced EMT concomitantly with the collective migration of squamous cell carcinoma cells. J. Cell. Biochem. 2013;114:2039–2049. doi: 10.1002/jcb.24545. [DOI] [PubMed] [Google Scholar]
- 8.Skrypek N., Goossens S., De Smedt E., Vandamme N., Berx G. Epithelial-to-mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet. 2017;33:943–959. doi: 10.1016/J.TIG.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Javaid S., Zhang J., Anderssen E., Black J.C., Wittner B.S., Tajima K., Ting D.T., Smolen G.A., Zubrowski M., Desai R., Maheswaran S., Ramaswamy S., Whetstine J.R., Haber D.A. Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell Rep. 2013;5:1679–1689. doi: 10.1016/J.CELREP.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka F., Rizqiawan A., Higashikawa K., Tobiume K., Okui G., Shigeishi H., Ono S., Shimasue H., Kamata N. Snail promotes Cyr61 secretion to prime collective cell migration and form invasive tumor nests in squamous cell carcinoma. Cancer Lett. 2013;329:243–252. doi: 10.1016/j.canlet.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Rizqiawan A., Tobiume K., Okui G., Yamamoto K., Shigeishi H., Ono S., Shimasue H., Takechi M., Higashikawa K., Kamata N. Autocrine galectin-1 promotes collective cell migration of squamous cell carcinoma cells through up-regulation of distinct integrins. Biochem. Biophys. Res. Commun. 2013;441:904–910. doi: 10.1016/j.bbrc.2013.10.152. [DOI] [PubMed] [Google Scholar]
- 12.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Kubozono K., Mizuta K., Fujimoto S., Tran T.T., Kamata N., Tobiume K. Dysferlin-deficient myotubes show tethering of different membrane compartments characterized by TMEM16E and DHPRα. Biochem. Biophys. Res. Commun. 2020;529:720–725. doi: 10.1016/J.BBRC.2020.06.079. [DOI] [PubMed] [Google Scholar]
- 14.Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 15.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;97:671–675. doi: 10.1038/nmeth.2089. 2012, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: a Language and Environment for Statistical Computing.http://www.R-project.org [Google Scholar]
- 17.Watanabe K., Liu Y., Noguchi S., Murray M., Chang J.C., Kishima M., Nishimura H., Hashimoto K., Minoda A., Suzuki H. OVOL2 induces mesenchymal-to-epithelial transition in fibroblasts and enhances cell-state reprogramming towards epithelial lineages. Sci. Rep. 2019;9 doi: 10.1038/S41598-019-43021-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu W., Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell. 2019;49:361–374. doi: 10.1016/J.DEVCEL.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Lei W., Wang X., Tang Y., Song J. Glucocorticoid induces mesenchymal-to-epithelial transition and inhibits TGF-β1-induced epithelial-to-mesenchymal transition and cell migration. FEBS Lett. 2010;584:4646–4654. doi: 10.1016/J.FEBSLET.2010.10.038. [DOI] [PubMed] [Google Scholar]
- 20.Lin T., Ponn A., Hu X., Law B.K., Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29:4896–4904. doi: 10.1038/ONC.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y., Wu Y., Li J., Dong C., Ye X., Chi Y.I., Evers B.M., Zhou B.P. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/EMBOJ.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang C., Ayyanathan K. Snail/Gfi-1 (SNAG) family zinc finger proteins in transcription regulation, chromatin dynamics, cell signaling, development, and disease. Cytokine Growth Factor Rev. 2013;24:123–131. doi: 10.1016/J.CYTOGFR.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadmiel M., Janoshazi A., Xu X., Cidlowski J.A. Glucocorticoid action in human corneal epithelial cells establishes roles for corticosteroids in wound healing and barrier function of the eye. Exp. Eye Res. 2016;152:10–33. doi: 10.1016/J.EXER.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]