Abstract
Coronavirus disease 2019 (COVID‐19) is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2), which has a high mortality rate and transmissibility. In this context, medicinal plants have attracted attention due to the wide availability and variety of therapeutic compounds, such as alkaloids, a vast class with several proven pharmacological effects, like the antiviral and anti‐inflammatory activities. Therefore, this scoping review aimed to summarize the current knowledge of the potential applicability of alkaloids for treating COVID‐19. A systematic search was performed on PubMed and Scopus, from database inception to August 2021. Among the 63 eligible studies, 65.07% were in silico model, 20.63% in vitro and 14.28% clinical trials and observational studies. According to the in silico assessments, the alkaloids 10‐hydroxyusambarensine, cryptospirolepine, crambescidin 826, deoxynortryptoquivaline, ergotamine, michellamine B, nigellidine, norboldine and quinadoline B showed higher binding energy with more than two target proteins. The remaining studies showed potential use of berberine, cephaeline, emetine, homoharringtonine, lycorine, narciclasine, quinine, papaverine and colchicine. The possible ability of alkaloids to inhibit protein targets and to reduce inflammatory markers show the potential for development of new treatment strategies against COVID‐19. However, more high quality analyses/reviews in this field are necessary to firmly establish the effectiveness/safety of the alkaloids here described.
Keywords: alkaloids, coronavirus, medicinal plant, natural product, SARS‐CoV‐2
Abbreviations
- 2‐O‐MTase
2′‐O‐ribose methyltransferase
- ACE1
angiotensin‐converting enzyme I
- ACE2
angiotensin‐converting enzyme II
- ACP
acid phosphatase assay
- ADMET
absorption, distribution, metabolism, excretion and toxicity.
- Anti‐CoV
anti‐coronaviruses
- AT1
angiotensin I receptor
- AT2
angiotensin II receptor
- Caco‐2
human colorectal adenocarcinoma cells
- Calu‐3
human lung adenocarcinoma cells
- CC50
half‐maximal cytotoxic concentration
- CCK8
Cell Counting kit‐8
- CLpro
chymotrypsin‐like protease
- COLCORONA
Colchicine Coronavirus SARS‐CoV‐2 Trial
- COVID‐19
Coronavirus disease 2019
- CQ
chloroquine
- CQN
chloroquine
- CRP
C‐reactive protein
- CTB‐ACE2
C‐terminal domain of S1 protein in complex with ACE2
- CTD‐ACE2
C‐terminal domain of S1 protein bound angiotensin‐converting enzyme II receptor
- CTG
CellTiter‐Glo® Luminescent Cell Viability Assay (Promega)
- DAMPs
damage‐associated molecular Patterns
- EC50
half‐maximal effective concentration
- FDA
Food and Drug Administration
- HCQ
hydroxychloroquine.
- HHT
homoharringtonine
- HIV
human immunodeficiency virus
- HSV1
hepatitis B virus and herpes simplex virus 1
- IC50
half‐maximal inhibitory concentration
- IFN‐α
interferon‐ α
- IFN‐β
interferon‐β
- IFN‐γ
interferon‐γ
- IL‐18
interleukin‐18
- IL‐1β
interleukin‐1β
- IL‐6
interleukin‐6
- LCR
lymphocyte‐to‐CRP ratio
- MERS‐CoV
Middle East Respiratory Syndrome Coronavirus
- MOI
multiplicity of infection
- MP
methylprednisolone
- Mpro
main protease
- MTS
3‐(4,5‐dimethylthiazol‐2‐yl)‐5‐(3‐carboxymethoxyphenyl)‐2‐(4‐sulfophenyl)‐2H‐tetrazolium
- MTT
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide
- ND
not determined
- NI
not informed
- NLRP3
NLR family pyrin domain containing 3
- NSP10
nonstructural protein 10
- NSP12
nonstructural protein 12
- NSP15
nonstructural protein 15
- NSP16
nonstructural protein 16
- NSP2
nonstructural protein 2
- OSF
Open Science Framework
- PAMPs
pathogen‐associated molecular patterns
- PCC
Problem, Concept and Context
- PDB
Protein Data Bank
- PLpro
Papain‐like protease
- PRISMA‐ScR
Systematic Reviews and Meta‐Analyses Extension for Scoping Reviews
- Q‐L
quinine‐sulfate extracted from a tablet
- qRT‐PCR
quantitative real‐time polymerase chain reaction
- Q‐S
quinine‐sulfate as solid material
- RBD‐ACE2
Complex between the SARS‐CoV‐2S protein receptor binding domain and ACE2 receptor
- RdRp
RNA‐dependent RNA polymerase
- RNA
Ribonucleic acid
- ROX score
respiratory oxygenation index
- RTC
replicase‐transcriptase complex
- sACE2‐INS1
soluble angiotensin‐converting enzyme II receptor in complex with spike (S) protein of Indian SARS‐CoV‐2
- SARS‐CoV‐2
Severe Acute Respiratory Syndrome Coronavirus 2
- SoC
standard‐of‐care
- TCID50
median tissue culture infectious dose
- TMPRSS2
human transmembrane protease
- TNF‐α
tumor necrosis factor‐α
- USA
United States of America
- WHO
World health Organization
- WST
water‐soluble tetrazolium salt
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), an enveloped‐type virus with positive‐sense single‐stranded ribonucleic acid (RNA), is the cause of coronavirus disease 2019 (COVID‐19), first reported in the city of Wuhan, Hubei province (China). This virus species belongs to Betacoronavirus genus and is capable of causing an intense inflammatory response especially in the respiratory system, which can lead to acute lung injury and respiratory distress, being associated with high rates of mortality and morbidity (Chaomin Wu et al., 2020).
In this context, the high transmissibility and impact of this disease on worldwide health and economy led the World health Organization (WHO) to declare a Public Health Emergency of International Concern (Li et al., 2020; Sohrabi et al., 2020). Scientific studies for the development of drugs that inhibit viral replication and disease progression have been carried out. Nonetheless, to date, there are no approved therapies with established efficacy and safety against SARS‐CoV‐2 (Dai et al., 2020).
In this field, in silico studies can speed up the traditional process of drug discovery by identifying a novel clinical use for compounds that have already proven to be safe and effective in humans, are approved for other indications, or traditionally used by some populations. This strategy can also reduce the costs required for the development of new drugs, with notable savings in preclinical studies, and has been previously used for the development of additional therapeutic options against Ebola, hepatitis C, and zika virus infection (Sultana et al., 2020; Venkatesan, 2021).
The natural products, including primary and secondary metabolites, work as a model for the synthesis of new antiviral drugs given their availability and variety of compounds with therapeutic potential. About 50% of all approved drugs for sale between 1981 and 2014 were derived from natural products (Newman & Cragg, 2020). In view of that, alkaloids, an extensive group of secondary metabolites (more than 12,000 compounds) mostly characterized by the presence of at least one nitrogen atom in a negative oxidation state (at any position in the structure, however it does not include nitrogen in an amide or peptide bond) may be useful in seeking drug treatment for COVID‐19 (Bribi, 2018).
These compounds are found mainly in flowering plants, bacteria, fungi, some animal species, among other sources, being classified according to the biosynthetic route as quinolines, isoquinolines, tropanes, purines, imidazoles, indoles, pyrrolidines, pyrrolizidines, pyridines and other types. The most known pharmacological activities of these compounds include antioxidant, anticarcinogenic, antimalarial, antibacterial, antifungal and antiviral effects (Simões, Schenkel, de Mello, Mentz, & Petrovick, 2017).
Previous narrative reviews focused on a broader description of the effects of some of the natural products or herbal medicines with antiviral effects. Among the alkaloids, homoharringtonine (HHT), lycorine and emetine were previously mentioned as promising therapies against COVID‐19 (Boozari & Hosseinzadeh, 2021; Verma et al., 2020).
On the basis of the prior discuss, the aim of this study was to summarize the current state of knowledge on the potential applicability of alkaloids for treating COVID‐19 by means of a scoping review.
2. METHODS
2.1. Research question
This scoping review was performed according to the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta‐Analyses Extension for Scoping Reviews (PRISMA‐ScR) Checklist (Tricco et al., 2018), the Cochrane Handbook for Systematic Reviews of Interventions version 6.2 (Higgins & Thomas, 2021) and the Joanna Briggs Institute methodology for scoping reviews (Joanna Briggs Institute, 2015), with Open Science Framework (OSF) register doi: 10.17605/OSF.IO/7QXV8.
2.2. Search strategy
A systematic search was conducted in the electronic databases Medline (via PubMed) and Scopus with no restriction for publication date or language (the last recovery date of studies was August 18, 2021). The main descriptors used referred to “coronavirus,” “COVID‐19,” “SARS‐CoV‐2” and “alkaloids” (see full strategy search in Appendix A provided in the supplementary material). Manual search in the reference list of the included studies was also performed.
2.3. Inclusion and exclusion criteria
We included: studies designed as in silico, in vitro, in vivo, clinical trials and observational; Studies that evaluated the use of alkaloids (any subclasses) isolated or associated with other compounds for the treatment of infection caused by the SARS‐CoV‐2 virus (COVID‐19).
Studies that did not evaluate SARS‐CoV‐2; reviews of any type; letter to editor; comments; editorials or expert opinions were excluded. Synthetic and semi‐synthetic alkaloids were not included. Studies that analyzed crude extract or fractions of plants; articles written in non‐Roman characters were excluded from this scoping review (see complete inclusion and exclusion criteria in Appendix B provided in the supplementary material).
2.4. Eligibility and data extraction
The selection process of studies involved two stages, the first being the reading of titles and abstracts (screening phase) for exclusion of irrelevant registers, and the second the reading of full text (eligibility phase) aiming at selecting eligible studies for data extraction. Data was extracted using structured tables containing general characteristics of the trials, evaluated alkaloid, methodological aspects, and main findings in accordance with the study design. Given the nature of the data, evidence was qualitatively synthetized. All steps of this study were conducted by two reviewers independently and disagreements were solved by third referee whenever necessary during consensus meetings.
3. RESULTS
3.1. Study selection
A total of 1,012 registers were selected from the database after duplicates removal, of which 839 were excluded during screening (title and abstract reading). From the remaining 173 studies, 110 were excluded after full‐text appraisal (see the complete list in Appendix C provided in supplementary material). Finally, 63 studies meeting the eligibility criteria had their data extracted and analyzed. No articles were added by manual search (see Figure 1). Overall, 41 studies (65.07%) were in silico model, 13 (20.63%) in vitro models, 9 clinical trials and observational (14.28%) studies.
FIGURE 1.
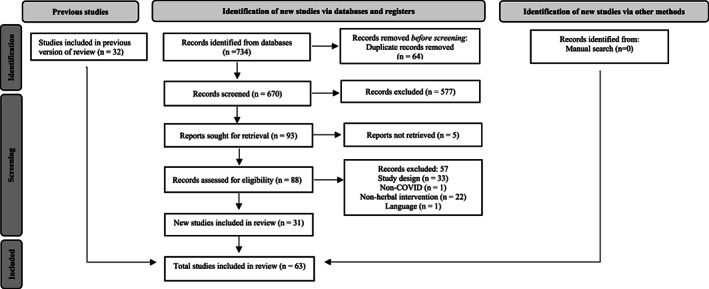
Flowchart of the scoping review
3.2. In silico studies
Table 1 presents the main findings from the 41 in silico studies on the alkaloids with higher binding energy with target proteins. Most trials (51.11%) were performed in India, whilst other studies were performed in other countries such as Brazil, Chile, Indonesia, Iran, Saudi Arabia, Turkey and United States of America (USA).
TABLE 1.
Main characteristics of the 41 in silico studies
| Reference | Country | Compounds | Protein targets with PDB identification | Result of alkaloid with higher binding energy | Main programs |
|---|---|---|---|---|---|
| Maurya et al., 2020 | India |
Piperine Nafamostat a |
ACE2 (1R42); spike glycoprotein (6VXX) | Piperine alkaloid obtained a higher binding affinity with spike glycoprotein (−104.56 kcal/mol) and also with ACE2 (−112.83 kcal/mol). The antiviral Nafamostat a showed a binding affinity of ‐ 114.63 kcal/mol for spike glycoprotein. | Molegro virtual Docker 3.0.0 |
| Kumar & Wei, 2020 | India | Nicotine | sACE2(Q9BYF1); spike 1 (S1) protein (6VW1) | Nicotine showed a firmer bound to the active site of the sACE2‐INS1 complex than to the ACE2 protein alone. Affinity score of −5.24 kcal/mol for ACE2 alone and − 6.33 kcal/mol for the sACE2‐INS1 complex. | AutoDock 4.0 and CHIMERA |
| Lestari et al., 2020 | Indonesia | Quinine | ACE2 receptor (6VW1) | Quinine interacts with the Lys353 residue in the peptidase domain of the ACE2 receptor with a binding affinity of −4.89 kcal/mol. | AutoDock Vina 1.1 |
| Gyebi, Adegunloye, et al., 2020 | Nigeria |
10‐Hydroxyusambarensine Cryptospirolepine Camostat a Nafamostat a |
ACE2 a (1R42); ACE2‐RBDb (6VW1); TMPRSS2 (1OQ5); spike gylcoprotein (6VSB) | 10‐Hydroxyusambarensine obtained the highest binding energy with TMPRSS2 (−10.40 kcal/mol in Vina/ ‐10.70 in Bindsurf). Cryptospirolepine obtained the highest binding energy with ACE2‐RBD (−10.70 kcal/mol in Vina; −10.90 kcal/mol in Bindsurf) and with ACE2 (−10.70 kcal/mol in Vina; −10.80 kcal/mol in Bindsurf) and with spike glycoprotein (−10.60 kcal/mol in Vina; −10.90 kcal/mol in Bindsurf). Camostat a showed −7.6 kcal/mol in Vina/−7.8 kcal/mol in Bindsurf for TMPRSS2 target protein. Nafamostat a showed −7.0 kcal/mol in vina/−7.2 kcal/mol in Binsurf for spike glycoprotein target. | AutoDock Vina, AutoDock 4.2 BINDSURF; CHARMM‐GUI web server. |
| Mohammadi et al., 2020 | Iran |
Caffeine Nicotine |
RBD‐ACE2b (6VW1); CTD‐ACE2c (6LZG) | Nicotine + favipiravir + CTD‐ACE2 and caffeine + ribarivin + RBD‐ACE2 obtained binding energy equal to −7.13 kcal/mol and − 6.76 kcal/mol, respectively. | AutoDock 4.2 |
| Maiti et al., 2021 | India | Nigellidine | ACE1 (6EN5); ACE2 (4APH); ATI (6OS1); AT2 (5XJM); spike glycoprotein (6VSB); | Nigellidine showed the highest binding energy with ACE2 (−7.54 kcal/mol) and had strong‐binding energy with ACE1 (−5.48 kcal/mol). It also binds with angiotensin‐receptors AT1 (−5.96 kcal/mol) and AT2 (−6.61 kcal/mol), as well to spike glycoprotein (−5.31 kcal/mol). | PatchDock; AutoDock |
| Mao et al., 2021 | China |
Ergotamine Simeprevir a |
Spike glycoprotein (6VXX). | Ergotamine and simeprevir a obtained binding energy of −9.2 kcal/mol with spike glycoprotein. | AutoDock Vina |
| Skariyachan et al., 2021 | India | Hyoscyamine | Spike glycoprotein (1WYY) | Hyoscyamine showed the highest binding potential for spike glycoprotein with a binding energy of −8.14 kcal/mol, also a several stabilizing interactions were observed in comparison with other selected molecular targets. | AutoDock Vina |
| Sharma et al., 2020 | India |
Ergotamine Saquinavir a Indinavir a |
2′‐O‐MTase (YP_009725311.1) | Ergotamine showed better binding energy of −10.00 kcal/mol for the evaluated target. Saquinavir a and indinavir a obtained a binding energy of −8.7 kcal/mol with 2′‐O‐MTase. | AutoDock Vina 4.2.6 and MGL tools 1.5.6 |
| Pandeya et al., 2020 | India | Protopine | RdRp (6 M71) | Protopine showed the highest binding energy (−6.07 kcal/mol) of all evaluated target. | AutoDock 4.2 |
| Ogunyemi et al., 2020 | Nigeria |
Cryptospirolepine Remdesivir a Sofosbuvir a |
RdRp (7BTF) | Cryptospirolepine obtained a higher binding energy with RdRp (−10.6 kcal/mol). Remdesivir a and sofosbuvir a showed a binding energies of −7.9 and −7.2 kcal/mol for RdRp target, respectively. | AutoDock Vina |
| Abd El‐Aziz et al., 2021 | Egypt |
Caffeine Remdesivir a Ribavirin a |
RdRp (6 M71) | Caffeine had a binding energy of −6.10 kcal/mol (estimated ΔG). Remdesivir a and ribavirin a showed a binding energies of −8.51 and −6.01 kcal/mol for RdRp target, respectively. | AutoDock 4.0; CHIMERA 1.8.1 |
| Gurung et al., 2021 | India |
Emetine Paritaprevir a Rilpivirine a Simeprevir a |
RdRp (7BV2) | The binding energy found is −8.81 kcal/mol to evaluated target. Paritaprevir a , rilpivirine a and simeprevir a obtained binding energies of −10.46, −8.25 and −8.08 kcal/mol with RdRp, respectively. | AutoDock 4.2 |
| Gul et al., 2020 | Turkey |
Dihydroergotamine Ergotamine Nelfinavir a Tipranavir a |
RdRp (6NUR); Mpro (3CLpro 6Y84) | Dihydroergotamine showed a binding energy of −16.22 kcal/mol (MM/GBSA BFE) with Mpro, and ergotamine −24.65 kcal/mol (MM/GBSA BFE) with RdRp. Nelfinavir a obtained a binding energy of −26.28 kcal/mol with Mpro and tipranavir a a value of −26.08 kcal/mol with RdRp. | AutoDock Vina 1.1.2; CHARMM36 force field |
| Borquaye et al., 2020 | Ghana |
Cryptomisrine Nelfinavir a Adenosine triphosphate (ATP) a Remdesivir a Lopinavir a |
RdRp (6NUR); Mpro (3CLpro 6Y84) | Cryptomisrine showed the best results for Mpro with binding energy of −10.60 kcal/mol and for RdRp with energy of −9.80 kcal/mol. Nelfinavir obtained a binding energy of −8.30 kcal/mol with Mpro. Remdesivir obtained a binding energy of −6.90 kcal/mol with RdRp target. Lopinavir showed a binding energy of −8.70 kcal/mol with Mpro and −7.80 kcal/mol with RdRp. A binding energy value of −7.40 kcal/mol was found for ATP with RdRp target. | AutoDock Vina |
| Devasia et al., 2021 | India |
Camptothecin Lopinavir a |
RdRp (7BTF); Mpro (6LU7) | Campotecin showed a binding energy of −7.50 kcal/mol for Mpro, and −7.30 for RdRp. Lopinavir a showed a binding energy of −8.00 and −8.40 kcal/mol for Mpro and RdRp, respectively. | AutoDock Vina; CHIMERA 1.13.1 |
| Alfaro et al., 2020 | Chile |
Schizanthin Z Schizantin Y Lopinavir a |
PLpro (6WX4) | Schizanthin Z showed the highest binding affinity with PLpro (−7.50 kcal/mol). Lopinavir a showed binding energy of 7.00 kcal/mol for PLpro target. | AutoDock Vina; CHARMM36 force field |
| Jade et al., 2021 | India |
Ergotamine Leuconicine F |
PLpro (6W9C); Mpro (6LU7) | Leuconicine F presented better binding free energy with Mpro (−224.322 kJ/mol or −53.61 kcal/mol) and ergotamine with PLpro (−200.06 kJ/mol or −47.82 kcal/mol) | AutoDock 4.0 |
| Sisakht et al., 2021 | Iran ‐ 215 |
Tubocurarine Nelfinavir a Lopinavir a |
Mpro (6LU7) | Tubocurarine showed better binding energy of −10.90 kcal/mol for the evaluated target. Nelfinavir a and lopinavir a obtained binding energies of −9.10 and −8.40 kcal/mol with Mpro, respectively. | AutoDock Vina |
| Garg & Roy, 2020 | India | Emetine | Mpro (6LU7) | Emetine showed better binding energy of −10.17 kcal/mol for the evaluated target. | AutoDock 4.0 |
| Florez, 2020 | India | Escholtzine; (S)‐Stylopine | Mpro (6M03) | Both compounds showed good binding affinity (−8.80 kcal/mol) with Mpro. | PyRx 0.8; AutoDock Vina. |
| Gurung et al., 2020 | India |
18‐Hydroxy‐11‐methoxytabersonine Daclatasvir a Glecaprevir a Ledipasvir a Paritaprevir a Simeprevir a |
Mpro (3CLpro 6Y2F) | Among 46 compounds, it was 18‐Hidroxy‐3‐epi‐alpha‐yohimbine that obtained the highest binding energy (−8.10 kcal/Mol). Simeprevir a , ledipasvir a , paritaprevir a , glecaprevir a and daclatasvir a showed binding energies of −9.70, −9.30, −9.30, −9.30 and −9.20 kcal/mol with Mpro, respectively. | AutoDock Vina |
| Das et al., 2020 | India |
Emetine Lopinavir a Penciclovir a Ritonavir a |
Mpro (6Y84) | Emetine can bind to the active site of Mpro with a binding affinity of −9.07 kcal/mol (estimated ΔG). Penciclovir a , ritonavir a and lopinavir a showed binding energies of −6.57, −9.52 and −9.00 kcal/mol with Mpro, respectively. | SwissDock based on EADock DSS software |
| Elzupir, 2020 | Kindom of Saudi Arabia | Caffeine | Mpro (6Y2E) | Caffeine presents a good binding affinity (−8.91 kcal/mol) to the catalytic residues His41 and Cys145 of Mpro (ΔG). Remdesivir a shower a binding affinity of −32.57 kcal/mol (ΔG) with Mpro. | AutoDockVina |
| Ghosh et al., 2020 | India |
Anisotine Darunavir a Lopinavir a |
Mpro (GLU7) | Anisotine interacted with both the catalytic residues (His41 and Cys145) of Mpro, with binding affinity of −7.90 kcal/mol. Darunavir a and lopinavir a showed binding affinities of −7.40 and −7.30 kcal/mol with Mpro target, respectively. | AutoDock Vina |
| N. Kumar, Awasthi, et al., 2020 | India |
Noscapine Favipiravir a Hydroxychloroquine a |
Mpro (6LU7) | The conjugate composed by noscapine with hydroxychloroquine showed the most definite binding affinity towards the Mpro (−10.01 kcal/mol). Only noscapine attached to Mpro a value of −8.42 kcal/mol was found. Noscapine conjugated with favipiravir a showed binding affinity of −9.17 kcal/mol. Favipiravir a and hydroxychloroquine a had binding affinities of −6.16 and −8.56 kcal/mol with Mpro, respectively. | HEX 8.0 software |
| N. Kumar, Sood, et al., 2020 | India |
Noscapine Favipiravir a Ribavirin a |
Mpro (6LU7) | Noscapine binds closely to pocket‐3 of the Mpro enzyme with binding energy of −292.42 kJ/mol (−69.84 kcal/mol). Favipiravir a and ribavirin a obtained binding energies of −153.91 kJ/mol (36.78 kcal/mol) and −214.17 kJ/mol (51.18 kcal/mol) with Mpro, respectively. | HEX 8.0 software |
| D. Kumar, Kumari, et al., 2020 | India | Noscapine | Mpro (6LU7) | Noscapine showed binding energy of −118.65 kcal/mol to Mpro | ParDOCK web server; iGEMDOCK |
| Gyebi, Ogunro, et al., 2020 | Nigeria |
10‐Hydroxyusambarensine |
Mpro (6LU7). | 10‐Hydroxyusambarensine interacts with the binding site and catalytic dyad of 3CLpro (Mpro) with the best binding affinity (−10.00 kcal/mol). Ritonavir a showed biding affinity of −6.8 kcal/mol for 3CLpro (Mpro) and lopinavir a −8.3 kcal/mol. | AutoDock Vina 4.2;BINDSURF |
| Qiao et al., 2020 | USA | ErgotamineLopinavir a Nelfinavir a Indinavir a | Mpro (6LU7). |
Ergotamine binds efficiently with Mpro (−8.80 kcal/mol). Lopinavir a , nelfinavir a and indinavir a showed binding energies of 7.70, −7.30 and 8.10 kcal/mol with Mpro. |
AutoDock 4.2.6, AutoDock Vina |
| Rahman et al., 2020 | Bangladesh | ErgotamineSimeprevir a Tipranavir a | Mpro (6LU7) | Ergotamine showed the highest binding affinity with Mpro (−9.80 kcal/mol). Simeprevir a and tipranavir a showed binding affinities with Mpro of −10.3 and −8.4 kcal/mol, respectively. | Autodock Vina; autodock 4.2.6 |
| de Sá et al., 2021 | Brazil | Epiisopiloturine | Mpro (6LU7) | Epiisopiloturine showed better binding energy with Mpro (−7.0 kcal/mol). | UCSF CHIMERA; AutoDock Vina |
| Mostafa et al., 2021 | Kingdom of Saudi Arabia |
Papaverine Ergotamine Theobromine |
Mpro complexs: Z45617795 (5R7Y); Z1220452176 (5R7Z); Z18197050 (5R80); Z1367324110 (5R81); Z219104216 (5R82). | Papaverine showed better binding energy with the Mpro complex 5R7Y (−19.2769 kcal/mol) and with 5R7Z (−23.1619 kcal/mol). Ergotamine showed better binding energy with the Mpro complex 5R80 (−24.7449 kcal/mol) and 5R81 (−20.4813 kcal/mol). Theobromine showed better binding energy with the Mpro complex 5R82 (−14.2779 kcal/mol). | Molecular operating environment (MOE) |
| Quimque et al., 2020 | Philippines | Norquinadoline A Quinadoline B Scedapin C Scequinadoline A | Mpro (3CLpro 6LU7); PLpro (6W9C);RdRp (6M71); NSP15 (6VWW); spike’s protein binding domain to GRP78 (6VXX). | Norquinadoline A showed better binding energy with PLpro (−10.90 kcal/mol), as well as Scedapin C (−10.90 kcal/mol). Quinadoline B showed the highest binding energy with RdRp (−9.80 kcal/mol), NSP15 (−9.10 kcal/mol) and protein S (−10.50 kcal/mol). Scequinadoline A showed the higher binding energy to Mpro (−8.70 kcal/mol). | AutoDock Vina version 1.1.2 |
| Maiti et al., 2020 | India | Nigellidine |
Mpro (6LU7) Spike glycoprotein (6VSB) NSP2 (QHD43415_2) nucleocapsid (QHD43423) |
Nigellidine showed the highest binding energy with NSP2 (−6.60 kcal/mol), Nucleocapsid (−6.24 kcal/mol) and spike glycoprotein (−6.11 kcal/mol). Nigellidine also showed a strong interaction with Mpro (−6.38 kcal/mol) and affinity to IL1R (−6.23 kcal/mol). |
AutoDock 4.2 |
| Firdiana et al., 2021 | Indonesia |
Norboldine Pallidine |
Mpro (1Z1J_B); PLpro (4MM3_B); spike (APF29063.1); ACE 2 (BAB40370.1) | Norboldine showed the highest binding affinity with 3CLpro (−7.9 kcal/mol), PLpro (−8.3 kcal/mol) and ACE2 (−8.2 kcal/mol).Pallidine showed the highest binding affinity with spike glycoprotein (−6.5 kcal/mol). | PyRx 0.8; PyMOL software 1.3.0.0 |
| Ismail et al., 2021 | Sudan |
Deoxynortryptoquivaline Trytoquivaline Quinadoline A |
Mpro (6LU7); spike glycoprotein (6LZG); ACE2 (IR42). | Deoxynortryptoquivaline had the highest binding energy with Mpro (−9.64 kcal/mol) and spike glycoprotein (−9.53 kcal/mol). Trytoquivaline and norquinadoline A showed the same and the highest binding energy with ACE2 (−11.01 kcal/mol). | AutoDock 4.0 |
| de Leon et al., 2021 | Philippines | Michellamine B | RdRp (6M71); helicase (6JYT); NSP16‐NSP10 complex (6W4H); | Michellamine B showed the highest binding affinity with NSP16 (−10.6 kcal/mol), also had good binding affinity with RdRp (−8.8 kcal/mol), helicase (−8.8 kcal/mol) and NSP10 (−7.2 kcal/mol). | CHIMERA 1.14 with AutoDock Vina. |
| El‐Demerdash et al., 2021 | United Kingdom | Crambescidin 786; Norcrambescidic acid; Crambescidin 826; | Mpro (6LU7); spike glycoprotein (6VYB); Nucleocapsid (6VYO); membrane glycoprotein (6M17); NSP10 (6W4H) | Crambescidin 786 showed better binding energy for Mpro (−8.05 kcal/mol) and for NSP10 (−9.06 kcal/mol). Crambescidin 826 showed better binding energy for spike glycoprotein (−6.95 kcal/mol) and for nucleocapsid (−8.01 kcal/mol). Norcrambescidic acid shower better binding energy for membrane glycoprotein (−7.34 kcal/mol). | Molecular operating environment (MOE). |
| Sumitha et al., 2020 | India | Quinine | NSP12 (6NUR) | Quinine showed binding energy of −6.14 kcal/mol to NSP12 target. | AutoDock |
| M, Reddy, Hema, Dodoala, & Koganti, 2021 | India |
Jatrorrhizine Nafamostat a Camostat a |
TMPRSS2 (Genbank 7,113, UniProt O15393). | Jatrorrhizine showed the highest binding energy with TMPRSS2 (−7.5 kcal/mol). Nafamostat a and camostat a showed binding energies of −7.8 and −8.4 kcal/mol with TMPRSS2 target, respectively. | AutoDock vina |
Abbreviations: 2‐O‐MTase, 2′‐O‐ribose methyltransferase; ACE2, Angiotensin converting enzyme 2; CLpro, chymotrypsin‐like protease; CTB‐ACE2, C‐terminal domain of S1 protein in complex with ACE2; Mpro, main protease; NSP10, Nonstructural protein 10; NSP12, Nonstructural protein 12; NSP15, Nonstructural protein 15; NSP16, Nonstructural protein 16; NSP2, Nonstructural protein 2; PDB, Protein Data Bank; PLpro, Papain‐like protease; RBD‐ACE2, Complex between the SARS‐CoV‐2S protein receptor binding domain and ACE2 receptor; RdRp, RNA‐dependent RNA polymerase; sACE2‐INS1, soluble angiotensin‐converting enzyme II receptor in complex with spike (S) protein of Indian SARS‐CoV‐2; TMPRSS2, Human transmembrane protease; USA, United States of America.
Standard endo/exogenous compound/molecule also analyzed by the authors for the comparison of values.
Overall, 11 studies (25.58%) approached the spike glycoprotein as target protein, being the alkaloids analyzed the following: crambescidin 826, cryptospirolepine, deoxynortryptoquivaline, ergotamine, hyoscyamine, nicotine docking on soluble angiotensin‐converting enzyme II receptor in complex with spike (S) protein of Indian SARS‐CoV‐2 (sACE2‐INS1), nigellidine, norbolidine, pallidine, piperine and quinadoline B (Borquaye et al., 2020; El‐Demerdash et al., 2021; Firdiana et al., 2021; Gyebi et al., 2020; Ismail et al., 2021; Selvaa Kumar, Kumar, & Wei, 2020; Maiti, Banerjee, & Kanwar, 2021; Maiti, Banerjee, Nazmeen, Kanwar, & Das, 2020; Mao, Bie, Xu, Wang, & Gao, 2021; Ogunyemi et al., 2020; Skariyachan, Gopal, Muddebihalkar, & Uttarkar, 2021).
The alkaloids nicotine, nigellidine, norboldine, piperine, quinadoline A and trytoquivaline showed binding capacity to the angiotensin‐converting enzyme II (ACE2) (Firdiana et al., 2021; Ismail et al., 2021; Maiti et al., 2021; Maurya, Kumar, Prasad, Bhatt, & Saxena, 2020; Selvaa Kumar, Kumar, & Wei, 2020).
Caffeine plus ribavirin binding to the complex between SARS‐CoV‐2 S protein receptor binding domain and ACE2 receptor (RBD‐ACE2), quinine, cryptospirolepine binding on RBD‐ACE2 and on ACE2, nicotine binding on C‐terminal domain of S1 protein bound angiotensin‐converting enzyme II receptor (CTD‐ACE2) plus favipiravir were described in three studies (Gyebi, Adegunloye, et al., 2020; Lestari, Sitorus, Instiaty, Megantara, & Levita, 2020; Mohammadi et al., 2020).
The alkaloid 10‐hydroxyusambarensine showed binding capacity to human in transmembrane protease serine 2 (TMPRSS2) with high binding energy (−10.40 kcal/mol) (Gyebi, Adegunloye, et al., 2020). The jatrorrhizine alkaloid, obtained from Tinospora cordifolia (Willd.) Miers or Mahonia aquifolium (Pursh) Nutt. or Enantia chlorantha Oliv., was also evaluated for TMPRSSS target showing a binding energy of −7.50 kcal/mol (M, Reddy, Hema, Dodoala, & Koganti, 2021). One study showed that ergotamine has high binding energy with 2′‐O‐ribose methyltransferase (2′‐O‐MTase) (Sharma, Morla, Goyal, & Kumar, 2020).
In nine studies the inhibition of RdRp protein was evaluated, being the most promising alkaloids: caffeine, camptothecin, cryptomisrine, cryptospirolepine, emetine, ergotamine, protopine, michellamine B, and quinadoline B (Abd El‐Aziz, Eldin Awad, Shehata, & El‐Sohaimy, 2021; Borquaye et al., 2020; de Leon et al., 2021; Devasia, Mohammad, Alrefaei, & Manoharadas, 2021; Gul et al., 2020; Gurung, Ali, Lee, Farah, & Al‐Anazi, 2021; Gyebi, Adegunloye, et al., 2020; Ogunyemi et al., 2020; Pandeya, Ganeshpurkar, & Mishra, 2020; Quimque et al., 2020).
Alkaloids norquinadoline A, scedapin C, schizanthin Z, ergotamine, and norboldine demonstrated significant bindings with the PLpro of SARS‐CoV‐2 (Alfaro, Alfaro, & Angel, 2020; Firdiana et al., 2021; Jade et al., 2021; Quimque et al., 2020). The alkaloid nigellidine showed possible anti‐SARS‐CoV‐2 activity through a good binding energy (−6.60 kcal/mol) with nonstructural protein 2 (NSP2), as well as quinine presented similar energy of −6.14 kcal/mol for nonstructural protein 12 (NSP12) target (Maiti et al., 2020; Sumitha, Devi, Hari, & Dhanasekaran, 2020). The nonstructural protein 10 (NSP10) was evaluated by El‐Demerdash et al. (2021) and de Leon et al. (2021), who found good values for crambescidin 786 and michellamine B, respectively.
Michellamine B showed high affinity with the helicase and nonstructural protein 16 (NSP16) target (de Leon et al., 2021). Only one in silico study used the nonstructural protein 15 (NSP15) as one of the targets, reporting a high binding energy of quinadoline B with this target protein (Quimque et al., 2020). The nucleocapsid N was one of the targets that showed good binding energy with nigellidine and high binding energy with crambescidin 826 (El‐Demerdash et al., 2021; Maiti et al., 2020).
Mpro was used as a protein target in 58.53% (n = 24) of the studies. Among the alkaloids tested for this target protein, the following stood out: emetine deoxynortryptoquivaline, tubocurarine, 10‐Hydroxyusambarensine, noscapine plus hydroxychloroquine, cryptomisrine, 18‐hidroxy‐3‐epi‐alpha‐yohimbine, escholtzine, (S)‐stylopine, scequinadoline A, ergotamine; caffeine (Borquaye et al., 2020; Das, Sarmah, Lyndem, & Singha Roy, 2020; Elzupir, 2020; Singh & Florez, 2020; Garg & Roy, 2020; Gurung, Ali, Lee, Abul Farah, & Al‐Anazi, 2020; Gyebi, Adegunloye, et al., 2020; Gyebi, Ogunro, Adegunloye, Ogunyemi, & Afolabi, 2020; Ismail et al., 2021; N. Kumar, Awasthi, et al., 2020; Qiao, Zhang, Ji, & Chen, 2020; Quimque et al., 2020; Rahman et al., 2020; Sisakht, Mahmoodzadeh, & Darabian, 2021).
Noscapine, leuconicine F, nigellidine, epiisopiloturine, camptothecin, anisotine, norboldine were also analyzed through molecular docking for Mpro target (de Sá et al., 2021; Devasia et al., 2021; Firdiana et al., 2021; Ghosh, Chakraborty, Biswas, & Chowdhuri, 2020; Jade et al., 2021; D. Kumar, Kumari et al., 2020; N. Kumar, Sood, van der Spek, Sharma, & Chandra, 2020; Maiti et al., 2020).
A docking study using the main protease (Mpro) enzymes active sites (5R7Y, 5R7Z, 5R80, 5R81) showed that papaverine had the highest binding energies with 5R7Y and with 5R7Z. The same authors also analyzed ergotamine and theobromine for those complexes (Mostafa et al., 2021).
3.3. In vitro studies
The 13 available in vitro studies quantitatively evaluated the impact of seven alkaloids (complete table with alkaloids chemical structure classification is in Appendix D provided in the supplementary material) on the viability of Vero, Callu‐3 or Caco‐2 cells through the CellTiter = Glo® Luminescent Cell Viability Assay (CTG) spectroscopy assay and Cell Counting kit‐8 assay; colorimetric methods of 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) or 3‐(4,5‐dimethylthiazol‐2‐yl)‐5‐(3‐carboxymethoxyphenyl)‐2‐(4‐sulfophenyl)‐2H‐tetrazolium (MTS) or water‐soluble tetrazolium salt (WTS‐1).
Cell replication was estimated in two studies using the quantitative real‐time polymerase chain reaction (qRT‐PCR) assay (Gendrot et al., 2020; Pizzorno et al., 2020) and in one study (Choy et al., 2020) the counting of viral titers was performed using the median tissue culture infectious dose (TCID50). The assays varied in relation to the value of multiplicity of infection (MOI) (0.001 to 3.00) and incubation time (24 to 72 h) (see Table 2).
TABLE 2.
Characteristics of the thirteen in vitro studies
| Reference | Country | Alkaloid | Cell line | MOI | Incubation time (h) | Assay method | EC50 | IC50 | CC50 | Selectivity index (SI) |
|---|---|---|---|---|---|---|---|---|---|---|
| Pizzorno et al., 2020 | France | Berberine | Vero E6 | 0.01 | 48 | MTS assay and RT‐qPCR | ND | 10.577 μM | >400 e , b | >37.84 |
| Varghese et al., 2021 | The Netherlands | Vero E6 | 0.01 | 24 | CTG assay and plaque assay | 9.10 g μM | ND | >150 c μM | >16 | |
| Nasal epitelial | 10.00 | 72 | 10.70 g μM | ND | 87.3 μM | 8.15 | ||||
| Ren et al., 2021, ** | China | Cephaeline | Vero E6 | 0.05 | 24 | CCK8 assay kit and RT‐qPCR | 0.0123 ± 0.000503 b μM | ND | 49.048 ± 46.327 f μM | ND |
| Ianevski et al., 2020, ** | Norway | Emetine | Vero E6 | 0.10 | 72 | CTG assay | <0.01 c μM | ND | 0.7 ± 0.40 c μM | >700 |
| Choy et al., 2020, ** | China | Vero E6 | 0.02 | 48 | CTG assay and TCID50 assay. | 0.46 d μM | ND | 56.46 c μM | ND | |
| R. Kumar et al., 2021 | India | Vero E6 | 0.10 | 24 | Plaque assay | 0.147 g nM | ND | 1603.8nM | 10910.2 2 | |
| Ellinger et al., 2021, ** | Germany | Caco‐2 | 0.01 | 48 | CTG assay | ND | 0.52 ± 0.09 μM | 1.13 ± 0.5 μM | 2 | |
| Ren et al., 2021, ** | China | Vero E6 | 0.05 | 24 | CCK8 assay | 0.00771± 0.000117 b μM | ND | 2.170± 0.258 f μM | ND | |
| Huang et al., 2020, ** | Taiwan | Narciclasine | Vero E6 cells | NI | 72 | ACP assay and plaque assay | ND | 16.5± 2.20 nM g | 75.3 ± 0.946 nM i | 4.56 |
| Ianevski et al., 2020, ** | Norway | Homoharringt onine (HHT) | Vero E6 | 0.10 | 72 | CTG assay | 0.03 ± 0.02 c μM | ND | 0.36 ± 0.60 c μM c | 12.0 |
| Choy et al., 2020, ** | China | Vero E6 | 0.02 | 48 | CTG assay and TCID50 assay. | 2.55d, * μM | ND | 59.75 c μM c | ND | |
| Chen et al., 2021 | USA | Calu‐3 | 0.01 | 48 | CTG assay | 30 nM | ND | >10.00 b μM | ND | |
| Huang et al., 2020, ** | Taiwan | Vero cell | NI | 72 | ACP assay and plaque assay | ND | 165.7± 16.1 g nM | 1.110 ± 150 i nM | 6.70 | |
| Jin et al., 2021 | South Korea | Lycorine | Vero cell | 0.0125 | 24 | CTG assay | ND | 0.878 ± 0.022 μM | 10.00 μM | > 56.95 |
| Ren et al., 2021, ** | China | Vero E6 | 0.05 | 24 | CCK8 assay kit and qRT‐PCR | 0.439 ± 0.122 b μM | ND | >1000 f μM | ND | |
| Ellinger et al., 2021, ** | Germany | Papaverine | Caco‐2 cells | 0.01 | 48 | CTG assay | ND | 1.1 ± 0.39 μM | ND | ND |
| Gendrot et al., 2020 | France | Quinine | Vero E6 | 0.25 | 48 | MTT assay and RT‐qPCR | 10.7 ± 3.00 b μM | ND | >100 a μM | >9.00 |
| Große et al., 2021 | Germany | A549‐ACE2 | 0.20 | 48 | WST‐1 assay | ND | 5.98 μM | ND | ND | |
| A549ACE2/TMPRSS2_c1 | 0.20 | 48 | ND | 52.82 μM | ND | ND | ||||
| A549ACE2/TMPRSS2_c2 | 0.20 | 48 | ND | 3.75 μM | ND | ND | ||||
| Calu‐3 | 0.20 | 48 | ND | 27.00 μM | ND | ND | ||||
| Caco‐2 | 3.00 | 48 | ND | 50.00 μM | ND | ND | ||||
| Persoons et al., 2020 | Belgium | Quinine | Vero E6 | NI | 72 | MTS assay and CTG assay | ND | 61.60 h μM | >100 c μM | ND |
| Hul7 | NI | 72 | ND | >100 h μM | >100 c μM | ND |
Abbreviations: ACP, acid phosphatase assay; CC50, half‐maximal cytotoxic concentration; CTG, CellTiter‐Glo® Luminescent Cell Viability Assay (Promega); CCK8, Cell Counting kit‐8; EC50, half‐maximal effective concentration; IC50, half‐maximal inhibitory concentration; MOI, multiplicity of infection; MTS, 3‐(4,5‐dimethylthiazol‐2‐yl)‐5‐(3‐carboxymethoxyphenyl)‐2‐(4‐sulfophenyl)‐2H‐tetrazolium; MTT, 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide; ND, not determined; NI, not informed; qRT‐PCR, quantitative real‐time polymerase chain reaction; TCID50, median tissue culture infectious dose; WST, water‐soluble tetrazolium salt.
EC50 determined by cell viability assay trough MTS colorimetric method.
CC50 or EC50 Determined with serially‐diluted compounds in Vero E6 cells at 48/72 hr post‐infection by CTG assay.
CC50 determined by cell viability assay trough MTT colorimetric method.
Determined by collecting supernatants 48/72 hr post‐infection for subsequent viral titration by qRT‐PCR.
EC50 determined by infectious virus yield in culture supernatant at 48 hr post‐infection (log10 TCID50/ml).
CC50 determined by cell viability assay trough MTS colorimetric method.
CC50 determined by cell viability assay with CCK8 kit.
EC50 or IC50 determined by plaque assay.
EC50 determined by APC assay.
Reduction of infectious virus.
Studies reporting outcomes of more than one alkaloid.
A dose–response assay revealed that the association of berberine with remdesivir results in strong antagonism represented by the Loewe synergy score (−35.12) and by several negative peak values distributed along the synergy map (Pizzorno et al., 2020). One study performed a time‐of‐addition assay to decipher a putative mode of action for berberine, which indicated that it acts late in the viral infection cycle affecting the production of infectious virus particles (Varghese et al., 2021).
A synergism trial with emetine (0.195 μM) when associated to remdesivir (6.25 μM) provided an inhibition of up to 64.9% of viral production with a Loewe synergy score of 0.306, which indicates the need of clinical trials and observational studies to confirm these findings (Choy et al., 2020). Furthermore, an in vitro assay performed with Caco‐2 cells testing papaverine and emetine, elucidated that papaverine was eight‐fold more potent against SARS‐CoV‐2 in these cells than against Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) (Ellinger et al., 2021). On the other hand, a study demonstrated that emetine is effective at a low nanomolar range in Vero E6 cells, as well as performed an assessment of emetine's effect on SARS‐CoV‐2 life cycle showing a highly significant reduction in the total RNA virus (R. Kumar et al., 2021).
In another research, emetine, lycorine and cephaeline alkaloids were tested in vitro in a multi‐targeting‐based anti‐SARS‐CoV‐2 inhibitor designing strategy using RdRp complex. This study mentioned the three alkaloids as possible inhibitors of viral protein synthesis through interactions with human ribossome on distinct catalytic active sites. Furthermore, lycorine appeared to present a strong interaction with RdRp while RNA was presented, as well as emetine (KD = 25.7 μM) and cephaeline (KD = 19.6 μM) showed binding with RdRp (Ren et al., 2021).
The effects of natural compounds of emetine and HHT were assessed associated with nelfinavir, showing a possible synergistic action against SARS‐CoV‐2 virus. Conversely, amodiaquine, a synthetic aminoquinoline derivative of the quinine alkaloid, presented the highest synergy score in this study (Ianevski et al., 2020). Interestingly, emetine and HHT showed an inhibition of virus activity with an EC50 lower than 100 μM, among the compounds described in the literature with potential to inhibit SARS‐CoV‐2, such as the Food and Drug Administration (FDA) approved drug remdesivir (23.15 μM) (Choy et al., 2020; Lin et al., 2021).
It is believed that the concentration of HHT is achievable in human plasma demonstrating a marked resistance to SARS‐CoV‐2 infection in both live and pseudoviral in vitro models. Therefore, treatment of Caco‐2 and Calu‐3 cells with either HHT showed a reduction in endogenous TMPRSS2 protein expression (Chen et al., 2021). The protein synthesis inhibitors HHT and narciclasine were described as compounds capable of inducing a similar host response to IFN‐β (Huang et al., 2020).
An in vitro activity of the antimalarial drug quinine showed an inhibitory concentration (IC) values compatible with common oral treatment in the clinical management of malaria. When compared to the other antimalarial drug tested in the study, a mean antiviral activity for SARS‐CoV‐2 and a single oral dose of 600 mg of quinine sulfate represented a blood maximum concentration of 3.5 mg/L. Authors suggested that, if proven clinically effective, quinine would be a promising candidate for the treatment of hypercytokinaemia due to COVID‐19 (Gendrot et al., 2020).
Other assay treated five cell lines with quinine‐sulfate extracted from a tablet (Q‐L) and quinine‐sulfate as solid material (Q‐S). In this study, the median toxic dose (TD50) of Vero B4 cells was ~100 μM (Q‐S) and A549 cells presented a TD50 of ~150 μM (Q‐S)/~300 μM (Q‐L). Moreover, a reduction in viral replication by up to 90% was observed when10 μM of quinine was applied in Vero B4 cells (Große et al., 2021).
Caco‐2 cells are capable of expressing TMPRSS2 protein, and for this cell line doses above 50 μM can inhibit SARS‐CoV‐2 infection (MOI = 3.00), with a dose‐dependent effect up to 2 μM, as well as suppressing viral spread and replication of the virus. The authors concluded that quinine exhibits antiviral activity in A549 lung cancer cell lines, which can be modulated but not abrogated by TMPRSS2 expression (Große et al., 2021). Unlike what the two studies above exposed, Persoons et al. (2020) concludes that quinine only blocked coronavirus replication at higher concentrations.
3.4. Clinical trials and observational studies
Table 3 shows information from 5 clinical trials, designed as randomized controlled trials (Brazil, Canada, China, Greece and Russia) and 4 as observational studies (Italy and United States of America). Among the nine studies included, eight evaluated the use of colchicine and one evaluated the use of emetine.
TABLE 3.
Main characteristics from the nine clinical trials and observational studies
| Reference | Country | Alkaloid | Study design | Population (N°) | Treatment | Main outcomes |
|---|---|---|---|---|---|---|
| Clinical trials | ||||||
| Deftereos et al., 2020 | Greece | Colchicine | A small‐sized randomized, single‐center conventional treatment‐controlled trial | 105 patients were included. 50 adults received SoC and 55 received colchicine plus SoC. | 1.5 mg loading dose followed by 0.5 mg after one h on day 1. The maintenance daily dosage was 0.5 mg for a maximum of 20 days. | Patients who receive colchicine had statistically significant improved time to clinical deterioration (change in clinical condition requiring invasive or noninvasive mechanical respiratory support or death). Control group presented 83% in cumulative event‐free 10‐day survival and colchicine plus SoC group presented 97% (p = .03). There were four deaths in control group versus one death in colchicine plus SoC group. |
| Fan et al., 2021 | China | Emetine | A single‐center pragmatic randomized controlled clinical trial at Wuhan Fangcang shelter hospital and a multicenter real‐world research conducted at five hospitals in Anhui Province | At Wuhan Fangcang shelter hospital, 39 patients were included, being that 27 received emetine plus SoC and 12 received SoC. at five hospitals located in Anhui province, 24 patients were included. 10 received emetine plus SoC and 14 SoC. | Emetine was administered at a three times daily dose of 3.6 mg plus routine antiviral therapy (Arbidol) three times a day, for up to 10 days. | The outcomes of the patients in Wuhan Fangcang shelter hospital showed that low‐dose emetine can effectively improve percutaneous blood oxygen saturation and increase the blood oxygen concentration, which is conducive to treating the illness. The outcomes of the patients at hospitals in Anhui Province also showed that low‐dose emetine can improve percutaneous blood oxygen saturation and breathing difficulties. In both studies no, perceptible adverse effects and side effects of emetine were observed. |
| Lopes et al., 2021 | Brazil | Colchicine |
A randomized, double‐blinded, Placebo‐controlled Clinical trial |
72 patients were randomly assigned to either colchicine (36 patients) or placebo 36 patients) | 0.5 mg thrice daily for 5 days, then 0.5 mg twice daily for 5 days. |
It was observed that patients who received colchicine had a lower need of hospitalization maintenance at day 10 (9.0%, p = .002). At day 7, patients maintained the need for supplemental oxygen was 9% in the colchicine group and 42% in the placebo group (p = .001). There were two deaths in placebo group, but was an uncommon event, so it was not possible to ensure that colchicine reduced mortality in this clinical trial. |
| Mareev et al., 2021 | Russia | Colchicine | A prospective,comparative trial with patients randomized to four groups | 43 patients were included. 22 patients received SoC and 21 patients received colchicine plus SoC. | 1 mg colchicine during the first 3 days followed by 0.5 mg daily up to | It was observed a rapid and statistically significant decrease and normalization of CRP (from 99.4 to 4.2 mg / dl, p < .001), also a significantly LCR increased in both groups, but the intergroup difference was very significant. The delta in the colchicine group was 393 versus 54 in the control group (p = .003). The resulting median SaO2 of the colchicine group was 98.0% and 96.5% in the control group (p = .014). Two patients of the control group died. |
| Tardif et al., 2021–106 | Canada | Colchicine | It was a phase 3, randomized, double‐blind, adaptive, placebo‐controlled, multicenter, investigator‐initiated trial | A total of 4,488 patients were randomly assigned to either colchicine (2,235 patients) or placebo (2,253 patients). | 0.5 mg twice per day for the first 3 days and then once per day for 27 days with placebo in a 1:1 ratio. | Among COVID‐19 patients, colchicine led to a lower rate of the composite of death or hospital admission than placebo. The primary composite endpoint (composite of death or hospital admission because of COVID‐19 infection in the 30 days after randomization) was 96 (4.6%) in colchicine group and 126 (6.0%) in the placebo group, with p value of .042. It was observed nine deaths in placebo group versus five deaths in colchicine. |
| Observational studies | ||||||
| Brunetti et al., 2020 | United States of America | Colchicine | A single‐center cohort study | 66 patients were randomly assigned to colchicine plus SoC (n‐33) or SoC (n = 33). | 1.3 mg loading dose on day 1, followed by 0.6 mg twice a day for up to 27 additional days. | Colchicine was associated with a significant reduction (p = .012) of mortality in patients treated for 28 days. There were three deaths in colchicine group versus eleven deaths in the SoC group. |
| Manenti et al., 2021 | Italy | Colchicine | Observational, retrospective cohort study | 141 patients were included. 71 patients received SoC and 70 received colchicine plus SoC. | A daily dose of 1.0 mg for up until clinical improvement or up to a maximum of 21 days. | The cumulative mortality rate of patients treated with colchicine was 7.5% and 28.5% in the control group (p = .006). The clinical improvement at day 21 was 40.0% of the patients on colchicine treatment and 26.6% of control patients. |
| Scarsi et al., 2020 | Italy | Colchicine | A single‐center cohort study | 262 patients were included. 140 patients received SoC and 122 received colchicine plus SoC | A daily dose of 1.0 mg for up to 21 days. | The survival rate of patients treated with colchicine (84.3%) was significantly higher as compared with that of patients treated with standard of care (63.6%). There were 20 deaths in the colchicine plus SoC group versus 52 deaths in the SoC group related to COVID‐19 complications (p < .001). |
| Sandhu et al., 2020 | United States of America | Colchicine | A prospective comparative cohort study | 197 patients were included to comprehensive analysis. 144 patients received SoC and 53 patients received colchicine plus SoC. | A loading dose of 0.6 mg twice a day for 3 days and then 0.6 mg once a day for up to 12 days. | It was observed in the patients who received colchicine a lower mortality (49.1%, p = .002), a lower rate of intubations (52.8%, p = .006) and higher discharge rate (50.9%, p = 0.002). Patients in the control group had a mortality rate of 72.9%, a percentage of intubations of 73.6% and a discharge rate of 27.1%. Patients in the colchicine group also had significant decrease in inflammatory markers. |
Abbreviations: CRP, C‐reactive protein; HCQ, hydroxychloroquine; LCR, lymphocyte‐to‐CRP ratio; MP, methylprednisolone.
The alkaloids were administered to hospitalized patients in eight studies diagnosed with COVID‐19 (Brunetti et al., 2020; Deftereos et al., 2020; Lopes et al., 2021; Manenti et al., 2021; Mareev et al., 2021; Sandhu, Tieng, Chilimuri, & Franchin, 2020; Scarsi et al., 2020) and one clinical trial used patients that were not currently hospitalized and not under immediate consideration for hospitalization (Tardif et al., 2021).
The stage of disease progression varies between articles. Moreover, results found by all authors were based on alkaloids associated with other medications, with the control group receiving either standard‐of‐care (SoC) treatment (Brunetti et al., 2020; Deftereos et al., 2020; Manenti et al., 2021; Mareev et al., 2021; Sandhu et al., 2020; Scarsi et al., 2020) or placebo (Lopes et al., 2021; Tardif et al., 2021).
Among the studies that evaluated colchicine, the average age of patients included at intervention group was of 53–70 years and at control group was between 54 and 65 years. Overall, colchicine was administered orally in doses varying from 0.5 to 1.5 mg and within a time frame ranging from 10 to 28 days.
Five studies reported a decrease in the level of C‐ reactive protein (CRP), namely: Brunetti et al. (2020) (p = .021); Lopes et al. (2021) (p = .0001); Manenti et al. (2021) (p = .0009); Mareev et al. (2021) (0.66 μg/ml in the colchicine group and 1.14 μg/ml in the control group) and Sandhu et al. (2020) (p = .014).
Two colchicine studies reported a statistically significant reduction in D‐dimer level, an inflammatory marker, with p value of .037 (Sandhu et al., 2020) and 0.040 (Deftereos et al., 2020), but 62.5% of studies reported a lower level of D‐dimer in colchicine group.
A Chinese single‐center pragmatic randomized controlled clinical trial evaluated a total of 39 patients from Wuhan Shelter Hospital, and the same authors also performed a multicenter real‐world study assessing a total of 24 patients from five hospitals of Anhui Province, China. In both studies the low‐dose emetine intervention showed no significant effect on the viral nucleic acid conversion, apart from that, there was not a statistically significant difference in the axillary body temperature, oxygen saturation and respiratory rate of Wuhan and Anhui province patients (Fan et al., 2020).
Although, it was observed that the values of the cited parameters where different between treatment and control group, however the authors concluded it was related to time effect and not to the intervention measures. Nevertheless, the study concluded that the results should be cautiously interpreted and more comprehensive analysis is needed to demonstrate the clinical efficacy of emetine therapy for COVID‐19 (Fan et al., 2021).
4. DISCUSSION
To our knowledge, this is the first scoping review to gather evidence from more than 60 studies designed as in silico model, in vitro model, clinical trials and observational studies about the potential effects of alkaloids in different stages of the viral cycle of SARS‐CoV‐2 (Figure 2).
FIGURE 2.
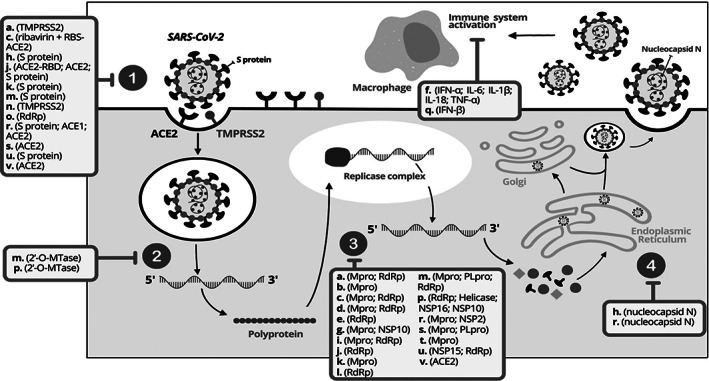
SARSCoV‐2 life cycle ith the main alkaloids discuss in the scoping review. The numbers 1, 2,3 and 4 dispose the viral cycle's phases: 1—Attachment and entry. 2—uncoating; 3—biosynthesis; 4—assembly and release. The main alkaloids of in silico, in vitro, clinical trials and observational studies are represented by letters and in parentheses their main action targets: (a). 10‐hydroxyysambarensine; (b). berberine; (c). caffeine; (d). camptothecin; (e). cephaeline; (f). colchicine; (g). crambescidin 786; (h). crambescidin 826; (i). cryptomisrine; (j). cryptospirolepine; (k). deoxynortryptoquivaline; (l). emetine; (m). ergotamine; (n). homoharringtonine; (o). lycorine; (p). michellamine B; (q). narciclasine; (r). nigellidine; (s). norboldine; (t). papaverine; (u). quinadoline B; (v). quinine. Adapted from Asselah et al., 2020
4.1. Replication of SARS‐CoV‐2
The viral cycle begins with the attachment and entry, wherein the coronavirus enters into human cells through glycoprotein spike of SARS‐CoV‐2 attachment to host‐cellular receptors (ACE2 and TMPRSS2) (Islam et al., 2020). The attachment forms a viral particle that fuses with the cell membrane or endosomal (Figure 2—1). Then, the uncoating of full‐length RNA genome subjects it to the immediate translation of two large open reading frames, ORF1a and ORF1b (primary translation and polyprotein processing) (Figure 2—2) (Asselah, Durantel, Pasmant, Lau, & Schinazi, 2020; V'Kovski, Kratzel, Steiner, Stalder, & Thiel, 2021).
The polyprotein formed is then processed by Mpro/ PLpro proteins in individual non‐structural proteins (NSP1 to NSP16). The viral RNA synthesis is formed by the viral replication and transcription complex (replicase complex around RdRp enzymatic activity) (Figure 2—3). The last phase occurs the formation of the viral nucleocapsid by packaging the genome into viral particles (virions), which are inserted into vesicles and secreted by exocytosis from the infected cell (Figure 2—4) (Asselah et al., 2020; V'kovski et al., 2021).
FIGURE 3.
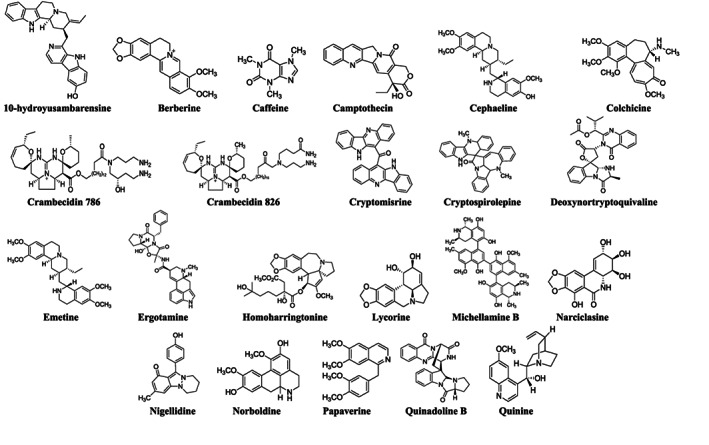
Chemical structures of the main alkaloids found in this scoping review for COVID‐19 treatment
The selection of the most promising alkaloids in the discussion section was guided by the previous viral cycle proteins presented. In summary, Figure 2 shows the possible phase of action of the 22 discussed alkaloids, in which the mechanisms of action are presented in a simplified way. The chemical structures of these main alkaloids are available in Figure 3.
4.2. In silico studies
Due to the large number of alkaloids found in in silico studies, being 45 different compounds, we chose to discuss those that showed higher binding value with more than two targets (host or viral proteins). The possibility of a compound acting simultaneously on more than two targets is interesting for the treatment of COVID‐19, owing to the emerging SARS‐CoV‐2 variants (Tao et al., 2021). Thus, we found 14 alkaloids (Figure 3—see complete table with alkaloids chemical structure classification is in Appendix D provided in the supplementary material) that fit in this requisition.
Almost 65.00% of the conducted studies in this scoping review refer to in silico assays. This approach gained strength in the past year given the increasing interest in drug repurposing (i.e., drug repositioning or rediscovery) to accelerate the identification of compounds that can cure or prevent COVID‐19 (Y. Kumar, Singh, & Patel, 2020).
In the context, we found that 10‐hydroxyusambarensine, a bisindole alkaloid from Strychnos usambarensis Gilg ex Engl. (Loganiaceae), showed high binding capacity to the host TMPRSS2 target and Mpro SARS‐CoV‐2 target (Gyebi, Adegunloye, et al., 2020; Gyebi, Ogunro, et al., 2020). The TMPRSS2 protein performs the proteolytic cleavage of the S1/S2 and S2 sites in the S protein, thus facilitating viral fusion with the host cell (adsorption) (Hoffmann et al., 2020). The main SARS‐CoV‐2 target protein, known as Mpro, cleaves polyproteins involved in the production of RNA that encodes structural viral proteins, so its blockage is capable of interrupting the viral cycle at its initial stages (Dai et al., 2020).
Some species of Loganiaceae family are used in folk African medicine to treat fever and malaria, as well as being known to have an alkaloid rich content (Asuzu & Nwosu, 2020). The indole alkaloids 10‐hydroxyusambarensine, chrysopentamine, strychnopentamine and isostrychnopentamine can be extracted from the species S. usambarensis and demonstrated ability to bind with coronaviruses' main protease (Gyebi, Ogunro, et al., 2020).
In addition, 10‐hydroxyusambarensine also showed high binding energy and binding affinity with RdRp target, which is responsible for the replication and transcription processes of the viral genome; besides having an active site in an accessible region (Aftab et al., 2020; Buonaguro, Tagliamonte, Tornesello, & Buonaguro, 2020; Ogunyemi et al., 2020).
Another compound with the ability to bind with two targets was, camptothecin, a pyrroloquinoline alkaloid, originally isolated from the Chinese tree, Camptotheca acuminata Decne. (Nyssaceae) that showed good value binds with Mpro and RdRp targets protein. Traditionally, it is used in Chinese medicine as a natural cancer drug and is currently an important alkaloid with proven anticancer properties acting as poison to the enzyme DNA topoisomerase 1 (TOP1). When infected by SARS‐CoV‐2 the human topoisomerase‐1 can activate transcriptional inflammatory genes, in view of that, camptothecin may eventually be studied for TOP1 inhibition activity, aiming to suppress inflammation by limitating the action of this enzyme (Devasia et al., 2021; Martino et al., 2017).
Caffeine is considered a purine alkaloid found in small amounts in fruit, seeds, and leaves of a variety of plants. This methylxanthine showed a significant binding energy with ACE2 when complexed to the active site of protein S plus ribavirin (Mohammadi et al., 2020), it either had good binding affinity to Mpro (Elzupir, 2020) and RdRp (Abd El‐Aziz et al., 2021). A review of possible beneficial actions of caffeine for COVID‐19 concluded that caffeine can be favoring bronchodilatation, immunomodulation and probably hindering viral intracellular transcription (Romero‐Martínez et al., 2021).
The African‐indigenous Cryptolepis sanguinolenta (Lindl.) Scheltr herb is traditionally used in the treatment of diverse diseases, such as malaria and bacterial respiratory disease. A review of C. sanguinolenta about its phytochemical composition explained that it is a species rich in alkaloids, tannins and flavones (Osafo, Mensah, & Yeboah, 2017). Three studies analyzed cryptomisrine (Borquaye et al., 2020) and cryptospirolepine (Gyebi, Adegunloye, et al., 2020; Ogunyemi et al., 2020), two secondary metabolites of C. sanguinlenta.
Cryptomisrine, an indolo[3,2‐b]quinoline dimeric alkaloid, showed good value binds with Mpro and RdRp targets proteins and Cryptospirolepine, an indolo‐quinoline alkaloid, demonstrated good binding value with ACE2, spike glycoprotein and RdRp targets. The Spike glycoprotein recognizes and binds to the ACE2 receptor performing the fusion enabling virus entry into the host cell (J. Yang et al., 2020). Therefore, these two compounds can potentially inhibit the virus' first interactions with the cell, its entry and replication (Elzupir, 2020; Mohammadi et al., 2020).
Alkaloids are one of the main secondary metabolites obtained from marine sponges with a wide range of biological activities. In this review, we found crambescidin 786 and crambescidin 826, classified as guanidinic alkaloids extracted from the marine sponge Monanchora arbuscula (Duchassaing & Michelotti, 1864).
Crambescidin 826 showed high binding energy with spike glycoprotein and nucleocapsid N (El‐Demerdash et al., 2021), whereas crambescidin 786 showed high binding value with Mpro and nonstructural protein 10 (NSP10), being the last a cofactor capable of attracting the replicase enzyme that implicates in the formation of replicase‐transcriptase complex (RTC) (Bouvet et al., 2014).
Another alkaloid found was deoxynortryptoquivaline, isolated from the mangrove‐derived fungus Cladosporium sp., showed high binding energy with the already mentioned main targets of SARS‐CoV‐2, Mpro and spike glycoprotein. This quinazoline alkaloid can also inhibit ACE2 target, in addition, this compound exhibits good pharmacokinetic and safety profiles. Nonetheless, more studies must be carried out to investigate it as possible natural multi‐target drug against COVID‐19 (Ismail et al., 2021).
The polypeptide ergot alkaloid, ergotamine, commonly used for migraine demonstrated high binding energy with 2′‐O‐MTase (Sharma et al., 2020), whose action is essential for viral replication and expression of coronaviruses in host cells, giving the virus the ability to escape from the immune system, ensuring less recognition of the host's immune response (Khan et al., 2021). It also has binding ability with more four targets showing high binding affinity with spike glycoprotein (Mao et al., 2021; Qiao et al., 2020), significant binding energy with RdRp (Gul et al., 2020), PLpro (Jade et al., 2021) and with Mpro complexes (Mostafa et al., 2021).
Michellamine B is an atropisomeric compound discovered in the leaves of the liana Ancistrocladus korupensis D.W. Thomas & Gereau (Ancistrocladaceae). This naphthylisoquinoline alkaloid showed the highest binding affinity for four SARS‐CoV‐2 targets namely, RdRp, NSP10, helicase and NSP16 (de Leon et al., 2021). It was found that several michellamine‐type alkaloids extracted from Congolese plants of the genus Ancistrocladus have antiviral activity due to its ability to inhibit the replication of the Human Immunodeficiency virus (HIV) reference strain, such as michellamine A1, A3, A4, and B (Shang et al., 2020).
Nigella sativa L., commonly known as black‐cumin, offers an indazole alkaloid named nigellidine. This herb is extensively employed traditional medicine like Ayurveda and Unani due to its extract safety, anti‐oxidant, anti‐bacterial, anti‐inflammatory, anti‐hypertensive activity and immunomodulatory effects. Therefore, this compound found in N. sativa has been associated with high energy binding with spike glycoprotein, NSP2, nucleocapsid N, ACE1, ACE2, AT1, AT2, in addition to having strong interactions with Mpro (Maiti et al., 2020, 2021). Renal and hepatic toxicities of the alkaloid nigellidine were evaluated in female Wistar rats, revealing no important safety concerns for the use of this substance (Maiti et al., 2020).
Norboldine, a compound classified as aporphine alkaloid that can be extract from Lindera aggregate (Sims) Kosterm., had binding potential with three target proteins, namely ACE2, Mpro and PLpro (Firdiana et al., 2021). The last target, PLpro acts as multifunctional protein with an essential role in the processing of viral polyproteins, maturation and assembly of RTC, it can also act on host cell proteins by interrupting the immune response and facilitating viral replication (Osipiuk et al., 2021). It has been an important target against several coronaviruses, such as MERS‐CoV (Kandeel et al., 2020; Maiti et al., 2020).
Quinadoline B, a fumiquinazoline alkaloid isolated from culture broth of Aspergillus sp., showed binding potential with three SARS‐CoV‐2 proteins, namely RdRp, NSP15 and protein S. A in silico profile of absorption, distribution, metabolism, excretion and toxicity (ADMET) showed a high gastrointestinal absorption, poor blood–brain barrier penetrability and high drug‐likeness. Also, it did not confer mutagenic, tumorigenic and reproductive toxicities, despite the high‐risk irritant (Quimque et al., 2020). As a side note, the NSP15 target has endoribonuclease activity, participates in viral RNA synthesis and plays a role in suppressing IFN‐α/INF‐β associated with the innate immune response of the host. For this reason, it limits the exposure of viral mRNA by the host's double‐stranded RNA (dsRNA) sensors, increasing its evasion capacity. Thus, quinadoline B may prevent virus evasion by inhibiting its replication and fusion to the host cells (Batool, Bibi, Amin, & Amjad, 2020; Deng et al., 2017; Quimque et al., 2020).
Finally, quinine, a quinoline alkaloid extracted from bark of Cinchona species that showed capability of binding with peptidase domain of the ACE2 receptor (Lestari et al., 2020) and to NSP12 (RdRp) target protein (Sumitha et al., 2020). The antiviral activities of several antimalarial drugs produced from quinine were investigated in an in vitro assay against SARS‐CoV‐2 (Gendrot et al., 2020).
A meta‐analysis of randomized trials showed that quinine analogue hydroxychloroquine (HCQ) is associated with increased mortality in COVID‐19 patients, and there is no benefit of using chloroquine (CQN). Currently, quinine alkaloids are the only effective treatment used against malaria and can be an option for further studies against COVID‐19, as their analogues HCQ and CQN have failed (Axfors et al., 2021).
In short, the therapeutic applications of these bioactive compounds must be explored in future pharmacological research. Overall, the information presented is a summary of the findings from the databases and we cannot support active recommendation of use, but evidence as a whole suggests that these 14 alkaloids can be analyzed more deeply about their simultaneous action on target proteins, of the other alkaloids represented in Table 1.
4.3. In vitro studies
Few in vitro studies were found in this scoping review. In vitro models provide a starting point for researchers to gather insights into how a cell responds to new substances in a controlled, isolated environment, but they often fail to recapitulate the complexity of human body systems (e.g., physiologically limited). Once a drug candidate demonstrates effectiveness through a series of in vitro essays, in vivo models can be used to advance drug development studies. Preclinical trials typically involve the use of animals or humans to further evaluate the safety, efficacy and delivery of a drug. However, this type of study is costly, and subject to strict regulations and compliance standards (Rosa et al., 2021).
HHT, a cytotoxic alkaloid isolated from the evergreen tree Cephalotaxus harringtonii (Knight ex J.Forbes) K.Koch, native to China, exhibits antiviral activity against a range of viruses, such as varicella‐zoster virus, hepatitis B virus and herpes simplex virus 1 (HSV1) (Fielding, da Filho, Ismail, & de Sousa, 2020). It is an anticancer approved by FDA, which demonstrates significant antiviral activity against diverse species of human and animal coronaviruses with the lowest IC50 (12 nM). Jointly, a review of natural products for COVID‐19 exposed HHT, lycorin and emetine as alkaloids with strong anti‐coronavirus effect (Boozari & Hosseinzadeh, 2021).
The effects of HHT and emetine were assessed in two studies, wherein different EC50 and 50% cytotoxicity concentration (CC50) data were obtained (Choy et al., 2020; Ianevski et al., 2020). This may be explained by the different used methodologies, MOI and incubation time, which prevent further comparison to be made for the use of these isolated alkaloids. HHT was identified as a reducer of the surface expression of TMPRSS2 protein target by altering its stability and modulating it through proteasomal‐mediated degradation, causing a reduction in endogenous TMPRSS2 protein expression. As a protein presents in host cells, there is less chance of mutations, making TMPRSS2 inhibition a potential therapeutic target (Chen et al., 2021; Rahman et al., 2020).
Futhermore, HHT and another alkaloid, narciclasine, demonstrated the ability to induce a similar host response to IFN‐β through a significant proportion of IFN‐b‐responsive genes and are intensively regulated by these alkaloids, which increase the expression of and IFNB1 (interferon beta 1). Narciclasine is a lycorine type alkaloid isolated for the first time from different varieties of Narcissus bulbs, which has a mechanism of action that involves inhibition of eukaryotic protein synthesis (Ceriotti, 1967; Huang et al., 2020).
On the other hand, we did not observe only analysis of isolated alkaloids, but also the result of the association between synthetic drugs and these compounds, like emetine and remdesivir, that revealed an important synergism with over 50% inhibition of SARS‐CoV‐2 production, which should be further investigated (Choy et al., 2020). Researchers suggested that emetine could affect viral genome synthesis in the target cells. Yet, the combination of drugs is a strategy to increase the effectiveness of the treatment, reducing the EC50 of the compounds and, thus, restricting the side effects (R. Kumar et al., 2021).
Conversely, an antagonism between the alkaloid berberine and remdesivir was demonstrated, suggesting that this combination should not be further explored (Pizzorno et al., 2020). A review concluded that berberine exerts anti‐inflammatory activity could be used to treat inflammation and other related diseases, such as downregulation of pro‐inflammatory cytokines and upregulation of anti‐inflammatory cytokines (Shang et al., 2020).
Berberine also has been associated with myocardial protective effects from ischemia/reperfusion injury. Moreover, it has shown very low toxicity and side effects in animal studies and in clinical trials only mild gastrointestinal reactions, including diarrhea and constipation were reported (Imenshahidi & Hosseinzadeh, 2019; Shang et al., 2020).
Lycorine, an indolizidine alkaloid isolated from the extract of the Chinese herb Lycoris radiata (L'Hér.) Herb., had a strong interaction with RdRp (Ren et al., 2021) showing a binding affinity (−6.20 kcal/mol) higher than the FDA approved drug remdesivir (−4.70 kcal/mol) (Jin et al., 2021). A review of compounds against coronavirus found that lycorine has antiviral activity in nanomolar concentration against SARS‐CoV with an EC50 value of 15.7 nM (Islam et al., 2020).
Isoquinoline is a subclass of alkaloids that can be extracted from plants of family Rubiaceae. Cephaeline is a terpenoid tetrahydroisoquinoline compound found in dried rhizome and roots of Cepahaelis ipecacuanha (Brot.) A. Rich (Rubiaceae), which may be more tolerable to patients when compared to its analogue, emetine, due to the similarly mechanism of action (inhibition of Zika and Ebola virus replication in vitro) (Ren et al., 2021; S. Yang et al., 2018).
Emetine is another isoquinoline alkaloid isolated from C. ipecacuanha, as well as plant families Alangiaceae and Icacinaceae, which displayed potent in vitro anti‐CoV and anti‐MERS‐CoV activity with EC50 value of 0.34 μM, moreover it demonstrated a strong interaction with SARS‐CoV‐2 replication protein, RdRp (Ren et al., 2021; Shen et al., 2019).
Among the articles that reported IC50, papaverine had the third lowest value. This alkaloid is extracted from opium poppy Papaver somniferum L. (Papaveraceae) and is classified as a benzylisoquinoline. It is a non‐narcotic alkaloid phosphodiesterase (PDE) inhibitor indicated for heart disease, impotence, pyschosis, and it also inhibits multiple strains of influenza virus (Ellinger et al., 2021; Kawase, Shirato, van der Hoek, Taguchi, & Matsuyama, 2012).
Finally, the in vitro inhibition of the virus replication caused by quinine, together with its strong interaction with hydrogen bond interaction and non‐polar interaction with active site of the protein RdRp (Sumitha et al., 2020), make this alkaloid a possible replacement drug for the treatment of COVID‐19 (Gendrot et al., 2020; Große et al., 2021).
4.4. Clinical trials and observational studies
Eight included studies assessed the anti‐inflammatory effects of colchicine for the reduction of cytokines release (e.g IL‐1β, IFNγ, IL‐18, and IL‐6) and decrease of TNF‐α receptor expression in the macrophages (Ribeiro, Lopes, Amaral, & Amaral, 2020), as these cytokines are responsible for several clinical manifestations of COVID‐19.
COLCORONA program assessed the use of colchicine in 4488 positive and non‐hospitalized patients with COVID‐19. Authors found significant (p = .040) reductions in hospitalizations between colchicine group and placebo group. In the view of this, early administration of colchicine may be paramount in the timely prevention of an acute hyperinflammatory state leading to deterioration (Tardif et al., 2021).
SARS‐CoV‐2 also stimulates the inflammasome by pathogen‐associated molecular patterns (PAMPs) and damage‐associated molecular Patterns (DAMPs) triggering numerous proinflammatory states that favor and contribute to cardiovascular process of a thrombotic nature (Páramo, 2020). A systematic review of the impact of colchicine on cardiovascular outcomes showed that results among clinical evidence have diverged, but they suggested a possible benefit from early initiation of colchicine when inflammation is more intense. In spite of this, they concluded that in post‐acute myocardial infarction (MI) patients, colchicine does not reduce cardiovascular or all‐cause mortality, recurrent MI, or other cardiovascular outcomes (Diaz‐Arocutipa et al., 2021).
Studies reported evidence that SARS‐CoV‐2 increase NLRP3 inflammasome activation (essential against viral infections, but linked to the inflammatory disorder when in excess, such as cytokine storm observed on COVID‐19) (Páramo, 2020). Colchicine suppresses NLRP3 inflammasome, by disrupting the microtubule‐dependent transport of mitochondria to the endoplasmatic reticulum, whose assembly with its adaptor and NLRP3 is required to be activated with inflammasomes (Lopes et al., 2021).
Another inflammatory marker observed by the researchers was CRP, whose raising levels have been observed in patients with COVID‐19. This non‐specific acute phase protein increases in acute infection or inflammation (Stringer et al., 2021). CRP is a considered a valid biomarker associated among severe and expired patients with COVID‐19 (p = .001) (Elshazli et al., 2020).
Like CRP, D‐dimer is a plasma biomarker used for the diagnostic investigation of pulmonary embolism. A meta‐analysis reported that D‐dimer and prothrombin time between severe and non‐severe cases (p‐value < .001) was statistically significant, in which severe cases presented a higher D‐dimer level, as well as CRP (Danwang et al., 2020).
The chronic use of colchicine, however, was associated with nausea, emesis, abdominal pain and diarrhea (Ribeiro et al., 2020) which can lead to drug discontinuation. In this context, gastrointestinal side‐effects were similar in colchicine studies, however, an incidence of diarrheal condition was observed in five studies (Deftereos et al., 2020; Lopes et al., 2021; Mareev et al., 2021; Tardif et al., 2021; Scarsi et al., 2020). On this basis, further studies on dose adjustments are needed to strength this evidence.
The greater limitations of emetine randomized controlled clinical trial was the exclusion of severe cases because of the limited conditions. However, in accordance with it emetine could be enriched in lung tissue for more than 12 h and still retain an effective concentration (Fan et al., 2021).
In view of that, it is suggested that the distinction observed in the clinical outcomes and values of the parameters analyzed was due to the difference among the severity of patients admitted to each study, ranging from patients with mild, moderate or severe course of COVID‐19. It is worth pointing out that the difference is also as a result of medicines associated with the alkaloids.
Overall, they were found as common clinical outcomes to the use of alkaloids colchicine and emetine: mortality rate reduction; decreased need for non‐invasive and invasive mechanical respiratory support (intubation); increased blood and percutaneous oxygenation; and reduction of inflammatory markers such as C‐reactive protein and D‐dimer. Despite of good outcomes, further studies are needed to confirm the isolated effects of alkaloids in patients with COVID‐19.
5. LIMITATIONS
Our study has some limitations. No statistical comparisons through meta‐analyses were possible given the high heterogeneity among studies (i.e., type of alkaloid, study designs, outcome assessment). This study focused on the antiviral potential of alkaloids, however, other natural compounds (alone or combined) may have some effect in this context and should be better evaluated in the future. Furthermore, well‐designed and high‐quality studies are needed to investigate the safety/efficacy of the alkaloids found and described in this review. In short, given the paramount importance of the COVID‐19 pandemic, as soon as more studies are published, this scoping review should be updated.
6. CONCLUSION
This scoping review summarized the available evidence from in silico, in vitro, clinical trials and observational studies on the potential effects of some alkaloids to be used alone or combined with other drugs. The compounds found with most promising antiviral effects against SARS‐CoV‐2 that can be investigated in additional in vitro assays, clinical trials and observational studies were: 10‐hydroxyusambarensine, berberine, caffeine, camptothecin, colchicine, crambescidin 786 and 826, cryptomisrine, cryptospirolepine, deoxynortryptoquivaline, emetine, ergotamine, HHT, lycorine, michellamine B, nigellidine, norboldine, papaverine, quinadoline B, and quinine. It is noteworthy, that further high quality studies are needed to firmly establish the possible clinical efficacy of the alkaloids compounds here described. In short, it is a hope that these findings can guide the design of other studies aiming at developing drugs with bioactive compounds and their derivatives to treat COVID‐19.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHOR CONTRIBUTION
Bárbara Longhini Gonzalez: Writing & Editing ‐ Original Draft (first reviewer). Natália Castelhano de Oliveira: Writing & Editing ‐ Original Draft (second reviewer). Mariane Roberta Ritter: Writing & Editing ‐ Original Draft (third reviewer). Fernanda Stumpf Tonin: Writing ‐ Review & Editing. Eduardo Borges Melo: Writing ‐ Review & Editing. Andreia Cristina Conegero Sanches: Writing ‐ Review & Editing. Fernando Fernandez‐Llimos: Writing ‐ Review & Editing. João Carlos Palazzo de Mello: Writing ‐ Review & Editing. Danielly Chierrito: Writing ‐ Review & Editing. Project administration: Daniela Cristina de Medeiros Araújo: Writing ‐ Review & Editing, Project administration, Supervision.
Supporting information
Appendix S1: Supporting Information
ACKNOWLEDGMENT
We truly thank all involved in researching treatment against COVID‐19.
Gonzalez, B. L. , de Oliveira, N. C. , Ritter, M. R. , Tonin, F. S. , Melo, E. B. , Sanches, A. C. C. , Fernandez‐Llimos, F. , Petruco, M. V. , de Mello, J. C. P. , Chierrito, D. , & de Medeiros Araújo, D. C. (2022). The naturally‐derived alkaloids as a potential treatment for COVID‐19: A scoping review. Phytotherapy Research, 36(7), 2686–2709. 10.1002/ptr.7442
DATA AVAILABILITY STATEMENT
It is a review paper and this item is not applicable.
REFERENCES
- Abd El‐Aziz, N. M. , Eldin Awad, O. M. , Shehata, M. G. , & El‐Sohaimy, S. A. (2021). Inhibition of the SARS‐CoV‐2 RNA‐dependent RNA polymerase by natural bioactive compounds: Molecular docking analysis. Egyptian Journal of Chemistry, 64(4), 1989–2001. [Google Scholar]
- Aftab, S. O. , Ghouri, M. Z. , Masood, M. U. , Haider, Z. , Khan, Z. , Ahmad, A. , & Munawar, N. (2020). Analysis of SARS‐CoV‐2 RNA‐dependent RNA polymerase as a potential therapeutic drug target using a computational approach. Journal of Translational Medicine, 18(1), 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro, M. , Alfaro, I. , & Angel, C. (2020). Identification of potential inhibitors of SARS‐CoV‐2 papain‐like protease from tropane alkaloids from Schizanthus porrigens: A molecular docking study. Chemical Physics Letters, 761. 10.1016/j.cplett.2020.138068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselah, T. , Durantel, D. , Pasmant, E. , Lau, G. , & Schinazi, R. F. (2020). COVID‐19: Discovery, diagnostics and drug development. Journal of Hepatology, 74(1), 168–184. 10.1016/j.jhep.2020.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asuzu, C. U. , & Nwosu, M. O. (2020). Studies of the wood of some Nigerian alkaloid‐rich Strychnos species. Journal of Horticulture and Forestry, 12(2), 57–62. [Google Scholar]
- Axfors, C. , Schmitt, A. M. , Janiaud, P. , van't Hooft, J. , Abd‐Elsalam, S. , Abdo, E. F. , … Hemkens, L. G. (2021). Mortality outcomes with hydroxychloroquine and chloroquine in COVID‐19 from an international collaborative meta‐analysis of randomized trials. Nature Communications, 12(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batool, A. , Bibi, N. , Amin, F. , & Amjad, M. (2020). Drug designing against NSP15 of SARS‐COV2 via high throughput computational screening and structural dynamics approach. European Journal of Pharmacology, 892, 1–8. 10.1016/j.ejphar.2020.173779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boozari, M. , & Hosseinzadeh, H. (2021). Natural products for COVID‐19 prevention and treatment regarding to previous coronavirus infections and novel studies. Phytotherapy Research, 35(2), 864–876. [DOI] [PubMed] [Google Scholar]
- Borquaye, L. S. , Gasu, E. N. , Ampomah, G. B. , Kyei, L. K. , Amarh, M. A. , Mensah, C. N. , … Aboagye, C. I. (2020). Alkaloids from Cryptolepis sanguinolenta as potential inhibitors of SARS‐CoV‐2 viral proteins: An in silico study. BioMed Research International. 10.1155/2020/5324560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet, M. , Lugari, A. , Posthuma, C. C. , Zevenhoven, J. C. , Bernard, S. , Betzi, S. , … Morelli, X. (2014). Coronavirus Nsp10, a critical co‐factor for activation of multiple replicative enzymes. Journal of Biological Chemistry, 289(37), 25783–25796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bribi, N. (2018). Pharmacological activity of aporphinoid alkaloids: A review. Asian Journal of Botany, 1(5), 387–412. [Google Scholar]
- Brunetti, L. , Diawara, O. , Tsai, A. , Firestein, B. L. , Nahass, R. G. , Poiani, G. , & Schlesinger, N. (2020). Colchicine to weather the cytokine storm in hospitalized patients with COVID‐19. Journal of Clinical Medicine, 9(9), 2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaguro, L. , Tagliamonte, M. , Tornesello, M. L. , & Buonaguro, F. M. (2020, May). SARS‐CoV‐2 RNA polymerase as target for antiviral therapy. Journal of Translational Medicine. 10.1186/s12967-020-02355-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriotti, G. (1967). Narciclasine: An antimitotic substance from narcissus bulbs. Nature, 213(5076), 595–596. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Lear, T. B. , Evankovich, J. W. , Larsen, M. B. , Lin, B. , Alfaras, I. , … Chen, B. B. (2021). A high‐throughput screen for TMPRSS2 expression identifies FDA‐approved compounds that can limit SARS‐CoV‐2 entry. Nature Communications, 12(1). 10.1038/s41467-021-24156-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy, K. T. , Wong, A. Y. L. , Kaewpreedee, P. , Sia, S. F. , Chen, D. , Hui, K. P. Y. , … Yen, H. L. (2020). Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS‐CoV‐2 replication in vitro. Antiviral Research, 178, 104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, W. , Zhang, B. , Jiang, X. M. , Su, H. , Li, J. , Zhao, Y. , … Liu, H. (2020). Structure‐based design of antiviral drug candidates targeting the SARS‐CoV‐2 main protease. Science, 368(6497), 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danwang, C. , Endomba, F. T. , Nkeck, J. R. , Wouna, D. L. A. , Robert, A. , & Noubiap, J. J. (2020). A meta‐analysis of potential biomarkers associated with severity of coronavirus disease 2019 (COVID‐19). Biomarker Research, 8(1). 10.1186/s40364-020-00217-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S. , Sarmah, S. , Lyndem, S. , & Singha Roy, A. (2020). An investigation into the identification of potential inhibitors of SARS‐CoV‐2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 39(9), 3347–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon, V. N. O. , Manzano, J. A. H. , Pilapil, D. Y. H. , Fernandez, R. A. T. , Ching, J. K. A. R. , Quimque, M. T. J. , … Macabeo, A. P. G. (2021). Anti‐HIV reverse transcriptase plant polyphenolic natural products with in silico inhibitory properties on seven non‐structural proteins vital in SARS‐CoV‐2 pathogenesis. Journal of Genetic Engineering and Biotechnology, 19(1). 10.1186/s43141-021-00206-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sá, É. R. A. , Costa, A. N. , Costa, R. K. M. , Souza, J. L. , Ramos, R. M. , Lima, F. , & das, C. A. (2021). In silico study of the interactions of Pilocarpus microphyllus imidazolic alkaloids with the main protease (Mpro) of SARS‐CoV‐2. Molecular Simulation, 47(1), 74–87. 10.1080/08927022.2021.1873321 [DOI] [Google Scholar]
- Deftereos, S. G. , Giannopoulos, G. , Vrachatis, D. A. , Siasos, G. D. , Giotaki, S. G. , Gargalianos, P. , … Stefanadis, C. (2020). Effect of colchicine versus standard care on cardiac and inflammatory biomarkers and clinical outcomes in patients hospitalized with coronavirus disease 2019: The GRECCO‐19 randomized clinical trial. JAMA Network Open, 3(6), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X. , Hackbart, M. , Mettelman, R. C. , O'Brien, A. , Mielech, A. M. , Yi, G. , … Baker, S. C. (2017). Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proceedings of the National Academy of Sciences of the United States of America, 114(21), E4251–E4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devasia, R. M. , Mohammad, A. , Alrefaei, A. F. , & Manoharadas, S. (2021). Enhanced production of camptothecin by immobilized callus of Ophiorrhiza mungos and a bioinformatic insight into its potential antiviral effect against SARS‐CoV‐2. Journal of King Saud University—Science, 33, 1–8. 10.1016/j.jksus.2021.101344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz‐Arocutipa, C. , Benites‐Meza, J. K. , Chambergo‐Michilot, D. , Barboza, J. J. , Pasupuleti, V. , Bueno, H. , … Hernandez, A. V. (2021). Efficacy and safety of colchicine in post–acute myocardial infarction patients: A systematic review and meta‐analysis of randomized controlled trials. Frontiers in Cardiovascular Medicine, 8. 10.3389/fcvm.2021.676771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Demerdash, A. , Metwaly, A. M. , Hassan, A. , El‐Aziz, T. M. A. , Elkaeed, E. B. , Eissa, I. H. , … Stockand, J. D. (2021). Comprehensive virtual screening of the antiviral potentialities of marine polycyclic guanidine alkaloids against SARS‐CoV‐2 (COVID‐19). Biomolecules, 11(3), 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger, B. , Bojkova, D. , Zaliani, A. , Cinatl, J. , Claussen, C. , Westhaus, S. , … Ciesek, S. (2021). A SARS‐CoV‐2 cytopathicity dataset generated by high‐content screening of a large drug repurposing collection. Scientific Data, 8(1), 2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshazli, R. M. , Toraih, E. A. , Elgaml, A. , El‐Mowafy, M. , El‐Mesery, M. , Amin, M. N. , … Kandil, E. (2020). Diagnostic and prognostic value of hematological and immunological markers in COVID‐19 infection: A meta‐analysis of 6320 patients. PLoS One, 15(8), 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzupir, A. O. (2020). Caffeine and caffeine‐containing pharmaceuticals as promising inhibitors for 3‐chymotrypsin‐like protease of SARS‐CoV‐2. Journal of Biomolecular Structure and Dynamics, 40(5), 2113–2120. 10.1080/07391102.2020.1835732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, S. , Zhen, Q. , Chen, C. , Wang, W. , Wu, Q. , Ma, H. , … Zhang, L. (2021). Clinical efficacy of low‐dose emetine for patients with COVID‐19: A real‐world study. Journal of Bio‐X Research, 4, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding, B. C. , Filho, C. S. M. B. , Ismail, N. S. M. , & de Sousa, D. P. (2020). Alkaloids: Therapeutic potential against human coronaviruses. Molecules, 25(23). 10.3390/molecules25235496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firdiana, E. R. , Renjana, E. , Ningrum, L. W. , Angio, M. H. , Nikmatullah, M. , & Rizal, S. (2021). In Silico study of the active compounds of Lindera aggregata (Sims) Kosterm as anti‐coronavirus. Current Nutrition & Food Science, 17(4), 408–416. [Google Scholar]
- Singh, S. , & Florez, H. (2020). Bioinformatic study to discover natural molecules with activity against COVID‐19 [version 1; peer review: 2 approved]. F1000Research, 9, 1–15. 10.12688/f1000research.26731.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg, S. , & Roy, A. (2020). In silico analysis of selected alkaloids against main protease (Mpro) of SARS‐CoV‐2. Chemico‐Biological Interactions, 332, 109309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrot, M. , Andreani, J. , Boxberger, M. , Jardot, P. , Fonta, I. , Le Bideau, M. , … Pradines, B. (2020). Antimalarial drugs inhibit the replication of SARS‐CoV‐2: An in vitro evaluation. Travel Medicine and Infectious Disease, 37, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, R. , Chakraborty, A. , Biswas, A. , & Chowdhuri, S. (2020). Identification of alkaloids from Justicia adhatoda as potent SARS CoV‐2 main protease inhibitors: An in silico perspective. Journal of Molecular Structure, 1229, 129489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Große, M. , Ruetalo, N. , Layer, M. , Hu, D. , Businger, R. , Rheber, S. , … Schubert, U. (2021). Quinine inhibits infection of human cell lines with SARS‐CoV‐2. Viruses, 13(4), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul, S. , Ozcan, O. , Asar, S. , Okyar, A. , Barıs, I. , & Kavakli, I. H. (2020). In silico identification of widely used and well‐tolerated drugs as potential SARS‐CoV‐2 3C‐like protease and viral RNA‐dependent RNA polymerase inhibitors for direct use in clinical trials. Journal of Biomolecular Structure and Dynamics, 39(17), 6772–6791. 10.1080/07391102.2020.1802346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung, A. B. , Ali, M. A. , Lee, J. , Abul Farah, M. , & Al‐Anazi, K. M. (2020). Structure‐based virtual screening of phytochemicals and repurposing of FDA approved antiviral drugs unravels lead molecules as potential inhibitors of coronavirus 3C‐like protease enzyme. Journal of King Saud University—Science, 32, 2845–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung, A. B. , Ali, M. A. , Lee, J. , Farah, M. A. , & Al‐Anazi, K. M. (2021). The potential of Paritaprevir and emetine as inhibitors of SARS‐CoV‐2 RdRp. Saudi Journal of Biological Sciences, 28(2), 1426–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi, G. A. , Adegunloye, A. P. , Ibrahim, I. M. , Ogunyemi, O. M. , Afolabi, S. O. , & Ogunro, O. B. (2020). Prevention of SARS‐CoV‐2 cell entry: Insight from in silico interaction of drug‐like alkaloids with spike glycoprotein, human ACE2, and TMPRSS2. Journal of Biomolecular Structure and Dynamics, 40, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi, G. A. , Ogunro, O. B. , Adegunloye, A. P. , Ogunyemi, O. M. , & Afolabi, S. O. (2020). Potential inhibitors of coronavirus 3‐chymotrypsin‐like protease (3CLpro): An in silico screening of alkaloids and terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, 39(9), 3396–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, J. , & Thomas, J. (2021). Cochrane handbook for systematic reviews of interventions. Version 6.2 . https://training.cochrane.org/handbook/current
- Hoffmann, M. , Mösbauer, K. , Hofmann‐Winkler, H. , Kaul, A. , Kleine‐Weber, H. , Krüger, N. , … Pöhlmann, S. (2020). Chloroquine does not inhibit infection of human lung cells with SARS‐CoV‐2. Nature, 585(7826), 588–590. 10.1038/s41586-020-2575-3 [DOI] [PubMed] [Google Scholar]
- Huang, C. T. , Chao, T. L. , Kao, H. C. , Pang, Y. H. , Lee, W. H. , Hsieh, C. H. , … Juan, H. F. (2020). Enhancement of the IFN‐β‐induced host signature informs repurposed drugs for COVID‐19. Heliyon, 6(12), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianevski, A. , Yao, R. , Fenstad, M. H. , Biza, S. , Zusinaite, E. , Reisberg, T. , … Kainov, D. E. (2020). Potential antiviral options against SARS‐CoV‐2 infection. Viruses, 12(6), 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imenshahidi, M. , & Hosseinzadeh, H. (2019). Berberine and barberry (Berberis vulgaris): A clinical review. Phytotherapy Research, 33(3), 504–523. [DOI] [PubMed] [Google Scholar]
- Islam, M. T. , El‐kersh, C. S. D. M. , Jamaddar, S. , Uddin, S. J. , Shilpi, J. A. , & Mubarak, M. S. (2020). Natural products and their derivatives against coronavirus: A review of the non‐clinical and pre‐clinical data. Phytotherapy Research, 34, 2471–2492. [DOI] [PubMed] [Google Scholar]
- Ismail, E. M. O. A. , Shantier, S. W. , Mohammed, M. S. , Musa, H. H. , Osman, W. , & Mothana, R. A. (2021). Quinoline and quinazoline alkaloids against Covid‐19: An in silico multitarget approach. Journal of Chemistry. 10.1155/2021/3613268 [DOI] [Google Scholar]
- Jade, D. , Ayyamperumal, S. , Tallapaneni, V. , Joghee Nanjan, C. M. , Barge, S. , Mohan, S. , & Nanjan, M. J. (2021). Virtual high throughput screening: Potential inhibitors for SARS‐CoV‐2 PLPRO and 3CLPRO proteases. European Journal of Pharmacology, 901. 10.1016/j.ejphar.2021.174082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y. H. , Min, J. S. , Jeon, S. , Lee, J. , Kim, S. , Park, T. , … Kwon, S. (2021). Lycorine, a non‐nucleoside RNA dependent RNA polymerase inhibitor, as potential treatment for emerging coronavirus infections. Phytomedicine, 86, 153440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joanna Briggs Institute . (2015). Methodology for JBI scoping reviews Joanna Briggs Institute . Manual. https://joannabriggs.org/assets/docs/sumari/Reviewers-Manual_Methodology-for-JBI-Scoping-Reviews_2015_v2.pdf
- Kandeel, M. , Abdelrahman, A. H. M. , Oh‐Hashi, K. , Ibrahim, A. , Venugopala, K. N. , Morsy, M. A. , & Ibrahim, M. A. A. (2020). Repurposing of FDA‐approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS‐CoV‐2 papain‐like protease. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1784291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase, M. , Shirato, K. , van der Hoek, L. , Taguchi, F. , & Matsuyama, S. (2012). Simultaneous Treatment of Human Bronchial Epithelial Cells with Serine and Cysteine Protease Inhibitors Prevents Severe Acute Respiratory Syndrome Coronavirus Entry. Journal of Virology, 86(12), 6537–6545. 10.1128/jvi.00094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R. J. , Jha, R. K. , Amera, G. M. , Jain, M. , Singh, E. , Pathak, A. , … Singh, A. K. (2021). Targeting SARS‐CoV‐2: A systematic drug repurposing approach to identify promising inhibitors against 3C‐like proteinase and 2′‐O‐ribose methyltransferase. Journal of Biomolecular Structure & Dynamics, 39(8), 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D. , Kumari, K. , Jayaraj, A. , Kumar, V. , Kumar, R. V. , Dass, S. K. , … Singh, P. (2020). Understanding the binding affinity of noscapines with protease of SARS‐CoV‐2 for COVID‐19 using MD simulations at different temperatures. Journal of Biomolecular Structure and Dynamics, 39(7), 2659–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N. , Awasthi, A. , Kumari, A. , Sood, D. , Jain, P. , Singh, T. , … Chandra, R. (2020). Antitussive noscapine and antiviral drug conjugates as arsenal against COVID‐19: A comprehensive chemoinformatics analysis. Journal of Biomolecular Structure and Dynamics, 40, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, C. S. , Kumar, S. A. , & Wei, H. (2020). Comparative docking studies to understand the binding affinity of nicotine with soluble ACE2 (sACE2)‐SARS‐CoV‐2 complex over sACE2. Toxicology Reports, 7, 1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N. , Sood, D. , van der Spek, P. J. , Sharma, H. S. , & Chandra, R. (2020). Molecular binding mechanism and pharmacology comparative analysis of noscapine for repurposing against SARS‐CoV‐2 protease. Journal of Proteome Research, 19(11), 4678–4689. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Afsar, M. , Khandelwal, N. , Chander, Y. , Riyesh, T. , Dedar, R. K. , … Kumar, N. (2021). Emetine supresses SARS‐CoV‐2 replication by inhibiting interaction of viral mRNA with eIF4E. Antiviral Research, 189, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, Y. , Singh, H. , & Patel, C. N. (2020). In silico prediction of potential inhibitors for the main protease of SARS‐CoV‐2 using molecular docking and dynamics simulation based. Journal of Infection and Public Health, 12, 1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestari, K. , Sitorus, T. , Instiaty, Megantara, S. , & Levita, J. (2020). Molecular docking of quinine, chloroquine and hydroxychloroquine to angiotensin converting enzyme 2 (ACE2) receptor for discovering new potential COVID‐19 antidote. Journal of Advanced Pharmacy Education and Research, 10(2), 1–4. [Google Scholar]
- Li, J. , Han, T. , Woodward, M. , Anderson, C. S. , Zhou, H. , Chen, Y.‐D. , & Neal, B. (2020). Special article—The impact of 2019 novel coronavirus on heart injury: A systematic review and meta‐analysis. Progress in Cardiovascular Diseases, 63, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. X. J. , Cho, S. , Meyyur Aravamudan, V. , Sanda, H. Y. , Palraj, R. , Molton, J. S. , & Venkatachalam, I. (2021). Remdesivir in coronavirus disease 2019 (COVID‐19) treatment: A review of evidence. Infection, 49(3), 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M. I. , Bonjorno, L. P. , Giannini, M. C. , Amaral, N. B. , Menezes, P. I. , Dib, S. M. , … Oliveira, R. D. R. (2021). Beneficial effects of colchicine for moderate to severe COVID‐19: A randomised, double‐blinded, placebo‐controlled clinical trial. RMD Open, 7(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti, S. , Banerjee, A. , & Kanwar, M. (2021). In silico Nigellidine (N. sativa) bind to viral spike/active‐sites of ACE1/2, AT1/2 to prevent COVID‐19 induced vaso‐tumult/vascular‐damage/comorbidity. Vacular Pharmacology, 138, 106856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti, S. , Banerjee, A. , Nazmeen, A. , Kanwar, M. , & Das, S. (2020). Active‐site molecular docking of nigellidine with nucleocapsid‐NSP2‐MPro of COVID‐19 and to human IL1R‐IL6R and strong antioxidant role of Nigella‐sativa in experimental rats. Journal of Drug Targeting, 2330, 1–23. [DOI] [PubMed] [Google Scholar]
- Manenti, L. , Maggiore, U. , Fiaccadori, E. , Meschi, T. , Antoni, A. D. , Nouvenne, A. , … Peruzzi, L. (2021). Reduced mortality in COVID‐19 patients treated with colchicine: Results from a retrospective, observational study. PLoS One, 16, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, R. , Bie, L. , Xu, M. , Wang, X. , & Gao, J. (2021). Antiviral drug design based on the opening mechanism of spike glycoprotein in SARS‐CoV‐2. Physical Chemistry Chemical Physics, 23(22), 12549–12558. [DOI] [PubMed] [Google Scholar]
- Mareev, V. Y. , Orlova, Y. A. , Plisyk, A. G. , Pavlikova, E. P. , Akopyan, Z. A. , Matskeplishvili, S. , … Kamalov, A. A. (2021). Proactive anti‐inflammatory therapy with colchicine in the treatment of advanced stages of new coronavirus infection. The first results of the COLORIT study. Kardiologiya, 61(2), 15–27. [DOI] [PubMed] [Google Scholar]
- Martino, E. , Della Volpe, S. , Terribile, E. , Benetti, E. , Sakaj, M. , Centamore, A. , … Collina, S. (2017). The long story of camptothecin: From traditional medicine to drugs. Bioorganic and Medicinal Chemistry Letters, 27(4), 701–707. 10.1016/j.bmcl.2016.12.085 [DOI] [PubMed] [Google Scholar]
- Maurya, V. K. , Kumar, S. , Prasad, A. K. , Bhatt, M. L. B. , & Saxena, S. K. (2020). Structure‐based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS‐CoV‐2 spike glycoprotein and its cellular receptor. VirusDisease, 31(2), 179–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi, S. , Heidarizadeh, M. , Entesari, M. , Esmailpour, A. , Esmailpour, M. , Moradi, R. , … Doustkhah, E. (2020). In silico investigation on the inhibiting role of nicotine/caffeine by blocking the s protein of sars‐cov‐2 versus ace2 receptor. Microorganisms, 8(10), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa, E. M. , Gamal, M. , Ghoneim, M. M. , Hussein, S. , El‐Ghorab, A. H. , Abdelgawad, M. A. , & Musa, A. (2021). Repurposing of FDA approved alkaloids as COVID 19 inhibitors; in silico studies. Pharmacognosy Journal, 13(1), 110–123. [Google Scholar]
- Newman, D. J. , & Cragg, G. M. (2020). Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. Journal of Natural Products, 83(3), 770–803. [DOI] [PubMed] [Google Scholar]
- Ogunyemi, O. M. , Gyebi, G. A. , Elfiky, A. A. , Afolabi, S. O. , Ogunro, O. B. , Adegunloye, A. P. , & Ibrahim, I. M. (2020). Alkaloids and flavonoids from African phytochemicals as potential inhibitors of SARS‐Cov‐2 RNA‐dependent RNA polymerase: An in silico perspective. Antiviral Chemistry and Chemotherapy, 28, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafo, N. , Mensah, K. B. , & Yeboah, O. K. (2017). Phytochemical and pharmacological review of Cryptolepis sanguinolenta (Lindl.) Schlechter. Advances in Pharmacological Sciences. 10.1155/2017/3026370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk, J. , Azizi, S.‐A. , Dvorkin, S. , Endres, M. , Jedrzejczak, R. , Jones, K. A. , … Joachimiak, A. (2021). Structure of papain‐like protease from SARS‐CoV‐2 and its complexes with non‐covalent inhibitors. Nature Communications, 12(1), 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandeya, K. B. , Ganeshpurkar, A. , & Mishra, M. K. (2020). Natural RNA dependent RNA polymerase inhibitors: Molecular docking studies of some biologically active alkaloids of Argemone mexicana . Medical Hypotheses, 144, 109905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Páramo, J. A. (2020). Respuesta inflamatoria en relación con COVID‐19 y otros fenotipos protombóticos. Reumatología Clínica, 1(1), 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persoons, L. , Vanderlinden, E. , Vangeel, L. , Wang, X. , Dan, N. , Do, T. , … Jonghe, S. D. (2020). Broad spectrum anti‐coronavirus activity of a series of anti‐malaria quinoline analogues. Antiviral Research, 193, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno, A. , Padey, B. , Dubois, J. , Julien, T. , Traversier, A. , Dulière, V. , … Terrier, O. (2020). In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS‐CoV‐2. Antiviral Research, 181. 10.1016/j.antiviral.2020.104878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooja, M. , Reddy, G. J. , Hema, K. , Dodoala, S. , & Koganti, B. (2021). Unravelling high‐affinity binding compounds towards transmembrane protease serine 2 enzyme in treating SARS‐CoV‐2 infection using molecular modelling and docking studies. European Journal of Pharmacology, 890, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Z. , Zhang, H. , Ji, H. F. , & Chen, Q. (2020). Computational view toward the inhibition of SARS‐CoV‐2 spike glycoprotein and the 3CL protease. Computation, 8(2), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimque, M. T. J. , Notarte, K. I. R. , Fernandez, R. A. T. , Mendoza, M. A. O. , Liman, R. A. D. , Lim, J. A. K. , … Macabeo, A. P. G. (2020). Virtual screening‐driven drug discovery of SARS‐CoV2 enzyme inhibitors targeting viral attachment, replication, post‐translational modification and host immunity evasion infection mechanisms. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1776639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, N. , Basharat, Z. , Yousuf, M. , Castaldo, G. , Rastrelli, L. , & Khan, H. (2020). Virtual screening of natural products against type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS‐CoV‐2). Molecules (Basel, Switzerland), 25(10). 10.3390/molecules25102271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, P. X. , Shang, W. J. , Yin, W. C. , Ge, H. , Wang, L. , Zhang, X. L. , … Bai, F. (2021). A multi‐targeting drug design strategy for identifying potent anti‐SARS‐CoV‐2 inhibitors. Acta Pharmacologica Sinica, 1–11. 10.1038/s41401-021-00668-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro, S. A. , Lopes, C. , Amaral, R. , & Amaral, A. (2020). The therapeutic potential of colchicine in the complications of COVID‐19. Could the immunometabolic properties of an old and cheap drug help? Metabolism Open, 7, 100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero‐Martínez, B. S. , Montaño, L. M. , Solís‐Chagoyán, H. , Sommer, B. , Ramírez‐Salinas, G. L. , Pérez‐Figueroa, G. E. , & Flores‐Soto, E. (2021). Possible beneficial actions of caffeine in sars‐cov‐2. International Journal of Molecular Sciences, 22(11), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa, R. B. , Dantas, W. M. , do Nascimento, J. C. F. , da Silva, M. V. , de Oliveira, R. N. , & Pena, L. J. (2021). In vitro and in vivo models for studying SARS‐CoV‐2, the etiological agent responsible for COVID‐19 pandemic. Viruses, 13(3). 10.3390/v13030379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu, T. , Tieng, A. , Chilimuri, S. , & Franchin, G. (2020). A case control study to evaluate the impact of colchicine on patients admitted to the hospital with moderate to severe COVID‐19 infection. Canadian Journal of Infectious Diseases and Medical Microbiology, 2020, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarsi, M. , Piantoni, S. , Colombo, E. , Airó, P. , Richini, D. , Miclini, M. , … Andreoli, L. (2020). Association between treatment with colchicine and improved survival in a single‐Centre cohort of adult hospitalised patients with COVID‐19 pneumonia and acute respiratory distress syndrome. Annals of the Rheumatic Diseases, 79(10), 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, X. F. , Yang, C. J. , Morris‐Natschke, S. L. , Li, J. C. , Yin, X. D. , Liu, Y. Q. , … Lee, K. H. (2020). Biologically active isoquinoline alkaloids covering 2014–2018. Medicinal Research Reviews, 40(6), 2212–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, K. , Morla, S. , Goyal, A. , & Kumar, S. (2020). Computational guided drug repurposing for targeting 2′‐O‐ribose methyltransferase of SARS‐CoV‐2. Life Sciences. 10.1016/j.lfs.2020.118169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L. , Niu, J. , Wang, C. , Huang, B. , Wang, W. , Zhu, N. , … Tan, W. (2019). High‐throughput screening and identification of potent broad‐spectrum inhibitors of coronaviruses. Journal of Virology, 93(12), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões, C. M. O. , Schenkel, E. P. , de Mello, J. C. P. , Mentz, L. A. , & Petrovick, P. R. (2017). Farmacognosia: do produto natural ao medicamento. Porto Alegre, Brazil: Artmed. [Google Scholar]
- Sisakht, M. , Mahmoodzadeh, A. , & Darabian, M. (2021). Plant‐derived chemicals as potential inhibitors of SARS‐CoV‐2 main protease (6LU7), a virtual screening study. Phytotherapy Research, 35(6), 3262–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skariyachan, S. , Gopal, D. , Muddebihalkar, A. G. , & Uttarkar, A. (2021). Structural insights on the interaction potential of natural leads against major protein targets of SARS‐CoV‐2: Molecular modelling, docking and dynamic simulation studies. Computers in Biology and Medicine, 132, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi, C. , Alsafi, Z. , O'Neill, N. , Khan, M. , Kerwan, A. , Al‐Jabir, A. , … Agha, R. (2020). World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID‐19). International Journal of Surgery. 10.1016/j.ijsu.2020.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer, D. , Braude, P. , Myint, P. K. , Evans, L. , Collins, J. T. , Verduri, A. , … Carter, B. (2021). The role of C‐reactive protein as a prognostic marker in COVID‐19. International Journal of Epidemiology, 50(2), 420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana, J. , Crisafulli, S. , Gabbay, F. , Lynn, E. , Shakir, S. , & Trifirò, G. (2020). Challenges for drug repurposing in the COVID‐19 pandemic era. Frontiers in Pharmacology, 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitha, A. , Devi, P. B. , Hari, S. , & Dhanasekaran, R. (2020). COVID‐19—In silico structure prediction and molecular docking studies with doxycycline and quinine. Biomedical and Pharmacology Journal, 13(3), 1185–1193. [Google Scholar]
- Tao, K. , Tzou, P. L. , Nouhin, J. , Gupta, R. K. , de Oliveira, T. , Kosakovsky Pond, S. L. , … Shafer, R. W. (2021). The biological and clinical significance of emerging SARS‐CoV‐2 variants. Nature Reviews Genetics, 22(12), 757–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif, J. C. , Bouabdallaoui, N. , L'Allier, P. L. , Gaudet, D. , Shah, B. , Pillinger, M. H. , … Boivin, G. (2021). Colchicine for community‐treated patients with COVID‐19 (COLCORONA): A phase 3, randomised, double‐blinded, adaptive, placebo‐controlled, multicentre trial. The Lancet Respiratory Medicine, 9(8), 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricco, A. C. , Lillie, E. , Zarin, W. , O'Brien, K. K. , Colquhoun, H. , Levac, D. , … Hempel, S. (2018). PRISMA extension for scoping reviews (PRISMA‐ScR): Checklist and explanation. Annals of Internal Medicine, 169(7), 467–473. [DOI] [PubMed] [Google Scholar]
- V'Kovski, P. , Kratzel, A. , Steiner, S. , Stalder, H. , & Thiel, V. (2021). Coronavirus biology and replication: Implications for SARS‐CoV‐2. Nature Reviews Microbiology, 19(3), 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese, F. S. , van Woudenbergh, E. , Overheul, G. J. , Eleveld, M. J. , Kurver, L. , van Heerbeek, N. , … van Rij, R. P. (2021). Berberine and obatoclax inhibit SARS‐CoV‐2 replication in primary human nasal epithelial cells in vitro . Viruses, 12(282), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan, P. (2021). Repurposing drugs for treatment of COVID‐19. The Lancet. Respiratory Medicine. 10.1016/s2213-2600(21)00270-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, S. , Twilley, D. , Esmear, T. , Oosthuizen, C. B. , Reid, A. M. , Nel, M. , & Lall, N. (2020). Anti‐SARS‐CoV natural products with the potential to inhibit SARS‐CoV‐2 (COVID‐19). Frontiers in Pharmacology, 11. 10.3389/fphar.2020.561334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , Chen, X. , Cai, Y. , Xia, J. , Zhou, X. , Xu, S. , … Song, Y. (2020). Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Internal Medicine, 180(7), 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Petitjean, S. J. L. , Koehler, M. , Zhang, Q. , Dumitru, A. C. , Chen, W. , … Alsteens, D. (2020). Molecular interaction and inhibition of SARS‐CoV‐2 binding to the ACE2 receptor. Nature Communications, 11(1), 4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Xu, M. , Lee, E. M. , Gorshkov, K. , Shiryaev, S. A. , He, S. , … Zheng, W. (2018). Emetine inhibits Zika and Ebola virus infections through two molecular mechanisms: Inhibiting viral replication and decreasing viral entry. Cell Discovery, 4(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Data Availability Statement
It is a review paper and this item is not applicable.


