Abstract
The cornerstone of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) detection is reverse‐transcription polymerase chain reaction (RT‐PCR) of viral RNA. As a surrogate assay SARS‐CoV‐2 RNA detection does not necessarily imply infectivity. Only virus isolation in permissive cell culture systems can indicate infectivity. Here, we review the evidence on RT‐PCR performance in detecting infectious SARS‐CoV‐2. We searched for any studies that used RT‐PCR and cell culture to determine infectious SARS‐CoV‐2 in respiratory samples. We assessed (i) diagnostic accuracy of RT‐PCR compared to cell culture as reference test, (ii) performed meta‐analysis of positive predictive values (PPV) and (iii) determined the virus isolation probabilities depending on cycle threshold (Ct) or log10 genome copies/ml using logistic regression. We included 55 studies. There is substantial statistical and clinical heterogeneity. Seven studies were included for diagnostic accuracy. Sensitivity ranged from 90% to 99% and specificity from 29% to 92%. In meta‐analysis, the PPVs varied across subgroups with different sampling times after symptom onset, with 1% (95% confidence interval [CI], 0%–7%) in sampling beyond 10 days and 27% (CI, 19%–36%) to 46% (CI, 33%–60%) in subgroups that also included earlier samples. Estimates of virus isolation probability varied between 6% (CI, 0%–100%) and 50% (CI, 0%–100%) at a Ct value of 30 and between 0% (CI, 0%–22%) and 63% (CI, 0%–100%) at 5 log10 genome copies/ml. Evidence on RT‐PCR performance in detecting infectious SARS‐CoV‐2 in respiratory samples was limited. Major limitations were heterogeneity and poor reporting. RT‐PCR and cell culture protocols need further standardisation.
Keywords: cell culture, infectivity, real‐time polymerase chain reaction, SARS‐CoV‐2, systematic review
Abbreviations
- COVID‐19
coronavirus disease 2019
- Ct
cycle threshold
- DTA
diagnostic test accuracy
- FN
false negative
- FP
false positive
- PPV
positive predictive value
- PRISMA
preferred reporting items for a systematic review and meta‐analysis
- PROSERO
iternational prospective register of systematic reviews
- QUADAS‐2
quality assessment of diagnostic accuracy studies 2
- ROC
receiver operating characteristics
- RT‐PCR
reverse‐transcription polymerase chain reaction
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- TN
true negative
- TP
true positive
1. INTRODUCTION
Since the outbreak of the coronavirus disease 2019 (COVID‐19) pandemic the reverse‐transcription polymerase chain reaction (RT‐PCR) has been the mainstay of the laboratory diagnostics of acute severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection. 1
The detection of SARS‐CoV‐2 RNA by real‐time RT‐PCR is based on the amplification of specific gene targets. 1 In each amplification cycle, gene target regions are reversely transcribed until their fluorescence signals become detectable above a threshold level, the cycle threshold (Ct). The Ct value is inversely proportional to the amount of amplified DNA gene targets in the samples. However, the conversion of Ct values to genome copy number is not directly possible since for the interpolation of viral load the generation of a standard curve through amplification of a known concentration of a target is required. 2
As a nucleic acid‐based detection method, RT‐PCR detects RNA molecules derived from infectious and non‐infectious virus particles, unpackaged RNA from infected cells and non‐replicating RNA degradation products. Therefore, the duration of detectability of SARS‐CoV‐2 RNA in respiratory samples may differ substantially from the duration of virus isolation 3 and can thereby delay hospital discharge management or quarantine duration. Attempts have been made to overcome this limitation by inferring a relationship between Ct values and cell culture results to draw conclusions about infectivity of COVID‐19‐patients.
Virus isolation in cell culture is considered as the best available laboratory‐based method for assessing infectivity. The inoculation of permissive cell lines with infectious samples may elicit cytopathic effects. Confirmatory techniques such as RT‐PCR or immunofluorescence allow differentiation between nonspecific cytotoxic effects and virus‐induced cytopathic effects as well as molecular identification of the causative virus. 4
However, virus isolation by cell cultures is laborious and time consuming, explaining why RT‐PCR‐negative samples are most commonly not confirmed by independent cell culture in clinical practice. Therefore, data on sensitivity and specificity may be limited and other measurements must be used to evaluate the performance of RT‐PCR compared to cell culture. Cell culture results based on RT‐PCR‐positive samples allow the classification into true positive (TP) and false positive (FP), based on which only the determination of the positive predictive value (PPV) is possible.
We aimed to summarise available study data (i) to determine the performance of RT‐PCR compared to cell culture in detecting infectious respiratory samples by assessing its diagnostic accuracy, (ii) to provide a meta‐analysis of PPV, and (iii) to estimate virus isolation probability in relationship to Ct or log10 genome copies/ml.
2. MATERIALS AND METHODS
2.1. Design and registration
This study was conducted according to Preferred Reporting Items for a Systematic Review and Meta‐analysis guidelines and was registered in PROSPERO (International Prospective Register of Systematic Reviews, CRD42021239149).
2.2. Information sources and search strategy
The literature was searched by an experienced information specialist (MIM) up to 19 April 2021 using three bibliographic databases: Cochrane COVID‐19 Study Register, Web of Science, and COVID‐19 Open Access Project Living Evidence on COVID‐19. Details of the search strategies are available as supplementary material S1. References of included studies were screened to identify additional records. No language restrictions were applied.
2.3. Eligibility criteria
Studies that met the following criteria were included in this systematic review:
Types of studies: Any study design (diagnostic test accuracy (DTA) studies, case series, cohort studies, cross‐sectional studies) was eligible for inclusion, provided that the study performed simultaneous RT‐PCR and cell culture on respiratory samples to assess infectivity. In‐vitro, in‐silico, medical intervention, animal studies and studies with less than 5 participants were excluded. Peer‐reviewed and preprint studies not otherwise published were included.
Participants: Participants of any gender, ethnicity, age with suspected or known SARS‐CoV‐2 infection.
Index test: RT‐PCR
Reference standard: Virus isolation with any permissive cell line was regarded as reference test.
Target condition: Detection of infectious respiratory specimens.
Outcomes: Sensitivity and specificity, PPV, means of Ct values or log10 genome copies/ml in cell culture positive and negative samples, cell culture result and corresponding Ct value or log10 genome copies/ml.
2.4. Selection of studies
Search results were deduplicated in EndNoteTM X8 and imported into the web‐based screening software Rayyan (www.rayyan.ai/).
Screening of titles and abstracts of retrieved records was performed to assess eligibility, and relevant articles were read in full text.
2.4.1. Data extraction and quality evaluation
The following information from each included study was extracted: first author name, study design, publication date, country, RT‐PCR assay details, cell culture details, TP, FP, false negative (FN), and true negative (TN) values, specimen types, symptom onset to test time, gene target, means of Ct and log10 genome copies/ml and their upper bound of virus isolation. Genome copies/ml were converted to log10 genome copies/ml (S2 represents a full list of extracted data items). In the case of multiple or overlapping publications, the study with the most recent date or the largest data set was extracted.
Study quality was evaluated using the Quality Assessment of Diagnostic Accuracy Studies tool (QUADAS‐2). 5 The QUADAS 2 tool was used for studies reporting sufficient data to create a 2 × 2 table (to derive diagnostic accuracy). As most of the studies did not intend to assess diagnostic accuracy of RT‐PCR compared to cell culture as reference standard a modified QUADAS 2 tool was created (S3 and S4).
At least two reviewers (AF, HM, KM) independently selected the literature using Rayyan's blind‐mode, extracted data and evaluated study quality.
Any disagreement was resolved by consultation with the third investigator (HH). Where necessary we contacted study authors for additional information.
2.5. Data synthesis and statistical analysis
All statistical analyses were performed using the open‐source software R (Version 4.1.0). 6
2.5.1. Diagnostic accuracy evaluation
We calculated the accuracy measurements using TP, FP, FN, and TN values. Chi‐square test was performed to test for equality of accuracy measurements. Heterogeneity was assessed visually by forest plots and receiver operating characteristics (ROC) ellipses. Plots were generated using the mada package (version 0.5.10). 7
2.5.2. Meta‐analysis of PPV
Studies providing the cell culture results amongst RT‐PCR positive respiratory samples were extracted to analyse the PPVs. The R‐package meta (version 4.18–2) was used. 8 A random effects model was applied to calculate the pooled PPV with their 95% confidence interval (CI) (95%‐CI). We estimated heterogeneity using 2 and Higgins inconsistency I2 statistic and considered I2 above 50% as high heterogeneity. 9 Subgroup analysis based on sampling time post‐symptom onset was performed to explain heterogeneity. Based on the maximum duration of virus isolation in non‐severe cases, which was reported as 9 days in the meta‐analysis by Cevik et al., 3 following categories were used in sampling time subgroup analysis: early mixed (presymptomatic, 0–10 days, >10 days), within 10 days, late mixed (0–10 days, >10 days), beyond 10 days and not reported sampling time post‐symptom‐onset.
2.5.3. Virus isolation probability estimation
Studies with extractable Ct values or log10 genome copies/ml and corresponding cell culture results were used to perform a logistic regression to estimate the probability of a positive culture depending on Ct value or log10 genome copies/ml. To allow visual comparability of the plots, the extracted raw data from individual studies were plotted in a common coordinate. Virus isolation probabilities were estimated at Ct values and log10 genome copies/ml which have been reported to be associated with nonviable virus (Ct value of 30 and 5 log10). For clarity reasons CI band of each study is provided separately in the supplementary material (S13‐32).
3. RESULTS
3.1. Literature search and selection
The database search identified 8939 references and 2 additional records from additional sources. After deduplication, 6219 records remained, of which 6074 were excluded during title and abstract screening (Figure 1). One hundred and fourty five full texts were screened for eligibility and 55 studies were eligible, 53 peer‐reviewed articles 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 and 2 preprints. 66 , 67
FIGURE 1.
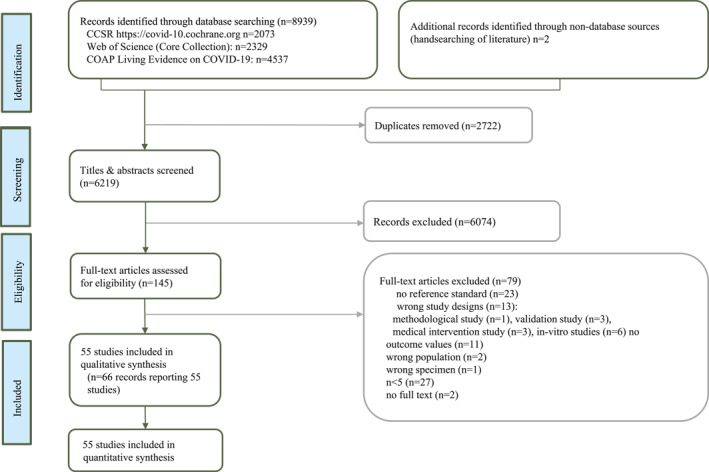
PRISMA flow diagram demonstrating the literature selection process
3.2. Study characteristics
A summary of characteristics of included studies is provided in Table 1 and detailed information in table S8 and S9. The studies analysed cover the period January 2020 to January 2021 of the pandemic. The majority of the studies consisted of case series (56.4%) and DTA studies (25.5%) with median sample size of 73.5 (IQR: 35–121.2). The objective of all but one of the DTA studies was to assess accuracy of rapid antigen test compared to RT‐PCR and cell culture. Most frequently non‐commercial RT‐PCR assays (54.5%) were used. The type of specimens varied across studies, with nasopharyngeal swabs (29.1%) being the most frequently collected. Most samples were collected in the late mixed time interval (43.6%). Vero E6 cells (43.6%) were most commonly used for cell culture and RT‐PCR (56.4%) was most commonly used as confirmation method of the cytopathic effect, while passaging of cells and the definition of cell culture positive result were poorly reported (72.3% not reported) and the definitions varied. Ct and log10 genome copies/ml cut‐offs of virus isolation were inconsistent across the studies, ranging from 20.2 10 to 37.3 11 for the upper limits of Ct values and from 2.34 12 to 6.01 37 for the lower limits of log10.
TABLE 1.
Summary of characteristics of included studies
| All studies (n = 55) | ||
|---|---|---|
| n | % | |
| Study design | ||
| Case series | 31 | 56.4 |
| Diagnostic test accuracy study | 14 | 25.5 |
| Cross‐sectional | 8 | 14.5 |
| Cohort | 2 | 3.6 |
| Continent | ||
| Europe | 26 | 47.3 |
| North America | 14 | 25.5 |
| Asia | 13 | 23.6 |
| South America | 1 | 1.8 |
| Australia | 1 | 1.8 |
| Period of the conduct of included studies | 01/20–01/21 | |
| Age groups | ||
| Mixed | 17 | 30.9 |
| Adults | 16 | 29.1 |
| Children | 1 | 1.8 |
| NR | 18 | 32.7 |
| Symptom status | ||
| Mixed (symptomatic‐asymptomatic‐postsymptomatic) | 22 | 40 |
| Symptomatic | 11 | 20 |
| Postsymptomatic | 3 | 5.5 |
| Asymptomatic | 1 | 1.8 |
| NR | 18 | 32.7 |
| Sample size | ||
| Range | 4‐3790 | |
| Median (IQR) | 73.5 (35–121.2) | |
| Sampling time post symptom onset | ||
| Early mixed (presymptomatic, 0‐10 d, >10 d) | 3 | 5.5 |
| Within 10 d | 6 | 10.9 |
| Late mixed (0‐10 d, >10 d) | 24 | 43.6 |
| Beyond 10d | 9 | 16.4 |
| NR | 13 | 23.6 |
| Sample types | ||
| NPS | 16 | 29.1 |
| Other mixed | 16 | 29.1 |
| Mixed NPS, OPS | 10 | 18 |
| Nasal | 3 | 5.5 |
| Mixed NPS, OPS, Sputum | 3 | 5.5 |
| Saliva | 2 | 3.6 |
| OPS | 2 | 3.6 |
| Sputum | 1 | 1.8 |
| NR | 2 | 3.6 |
| Cell line used for cell culture | ||
| Vero E6 cells | 24 | 43.6 |
| Vero CCL‐81 cells | 12 | 21.8 |
| Vero E6‐TMPRSS2 cells | 5 | 9 |
| Vero cells | 4 | 7.3 |
| Vero C1008 cells | 2 | 3.6 |
| Vero clone 118 cells | 2 | 3.6 |
| Caco‐2 cells | 2 | 3.6 |
| Vero B4 cells | 1 | 1.8 |
| Mixed Vero E6 and ML‐2 cells | 1 | 1.8 |
| Vero‐TMPRSS2 cells | 1 | 1.8 |
| NR | 1 | 1.8 |
| Confirmation method of the cytopathic effect | ||
| RT‐PCR | 31 | 56.4 |
| Transmission electron microscope | 1 | 1.8 |
| (scanning) EM and RT‐PCR | 2 | 3.6 |
| IF (Anti‐N‐Ab) | 4 | 7.3 |
| IF (Anti‐S‐Ab) | 1 | 1.8 |
| Plaque assay and IF (Anti‐N‐Ab | 1 | 1.8 |
| RT‐PCR or IF (Anti‐S‐Ab) | 1 | 1.8 |
| RT‐PCR or IF (Anti‐N‐Ab) | 1 | 1.8 |
| RT‐PCR and IF (Anti‐N‐Ab) | 1 | 1.8 |
| RT‐PCR and plaque assay | 1 | 1.8 |
| Plaque assay | 1 | 1.8 |
| Reinfection in new Vero monolayers | 1 | 1.8 |
| NR | 9 | 16.4 |
| Passaging | ||
| Performed | 13 | 23.6 |
| Not performed | 2 | 3.6 |
| NR | 40 | 72.3 |
| Cell culture positive definition | ||
| Reported | 13 | 23.6 |
| Not reported | 42 | 74.4 |
| RT‐PCR assay | ||
| In‐house | 30 | 54.5 |
| Commercial | 14 | 25.4 |
| Mixed | 9 | 16.4 |
| NR | 2 | 3.6 |
| Target genes reported for viral load quantification | ||
| N (1‐3, 1, 2) | 21 | 38.1 |
| E | 12 | 21.8 |
| ORF1ab | 2 | 3.6 |
| RdRP | 2 | 3.6 |
| S | 2 | 3.6 |
| E, RdRP, N | 2 | 3.6 |
| ORF1ab | 2 | 3.6 |
| E pp1ab | 1 | 1.8 |
| S, Nsp2 | 1 | 1.8 |
| N, S, ORF1ab | 1 | 1.8 |
| ORF1ab, N | 1 | 1.8 |
| E, Nsp12, N | 1 | 1.8 |
| E, S | 1 | 1.8 |
| ORF1ab, N, E, RdRP | 1 | 1.8 |
| E, N | 1 | 1.8 |
| NR | 4 | 7.3 |
Abbreviations: anti‐N‐Ab, anti‐nucleoprotein antibody; Anti‐S‐Ab, anti‐spike antibody; d, days; E, envelope protein gene; IF, immunofluorescence; N, nucleocapsid protein gene; NR, not reported; NPS, nasopharyngeal swab; Nsp2/12, nonstructural proteins 2/12 gene; OPS, oropharyngeal swab; ORF1ab, open reading frame 1ab gene; pp1ab, polyprotein 1ab; RdRP, RNA‐dependant RNA‐Polymerase gene; RT‐PCR, reverse transcription polymerase chain reaction; S, spike protein gene; SD, standard deviation; TMPRSS2, transmembrane protease; serine 2‐expressing Vero cells.
3.3. Methodological quality
Most studies providing data for diagnostic accuracy assessment showed high risk of bias (Summary table S5a, individual assessment S6). This judgement was mainly based on lack of blinding when conducting the ‘reference standard’ 14 , 15 , 16 , 17 , 18 , 19 , 20 and lack of inclusion of all samples when performing cell culture 14 , 17 , 20 or the studies did not report whether there was a ‘sample processing delay’ before inoculation of clinical specimen. 15 , 16 , 18 , 19 , 20
Studies which were included for PPV assessment (Summary table S5b, individual assessment S7) were most frequently assessed as high risk of bias in the domains ‘patients selection’ (22 out of 48 were scored high risk) and ‘flow and timing’ (32 out of 48 were scored high risk). Most of the studies were observational, thus patient selection was not random and inclusion and exclusion criteria were often unclear or not defined at all. ‘Flow and timing’ item was most commonly affected by the fact that not all patient samples underwent cell culture.
3.4. Accuracy of RT‐PCR
Sensitivity ranged from 90% to 99% and specificity ranged from 29% to 92%. Specificity varied across studies (Figure 2) and the chi‐square test suggested heterogeneity for specificities p < 0.001, while p = 0.794 was obtained for the test for equality of sensitivities. Forest plots (Figure 2) demonstrate greater variability of specificities compared to sensitivities and ROC ellipses (S11) showed a greater coverage of prediction regions of specificity than of sensitivity. As sample sizes of sensitivities were smaller than that of specificities (S8), the variability of specificities most likely arises from heterogeneity and not random error.
FIGURE 2.
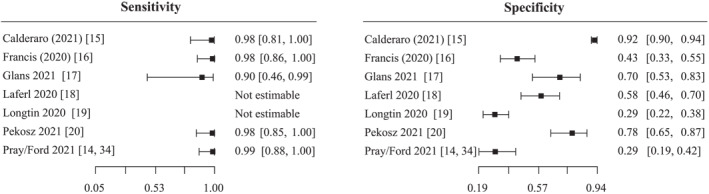
Forest plot of sensitivity and specificity estimates of reverse‐transcription polymerase chain reaction (RT‐PCR) for detection of infectious specimen compared to cell culture as reference standard. The squares and horizontal lines represent the point estimate and 95% confidence interval (CI) for each included study
Based on the paucity of studies for sensitivity (n = 5) and the limitation of the included studies with heterogeneity of index tests, reference standards, study population, no meta‐analysis of accuracy measurements could be performed.
3.5. Meta‐analysis of positive predictive values
A total of 9489 SARS‐CoV‐2 RT‐PCR‐positive respiratory samples were included.
Subgroup analyses were performed based on sampling time post‐symptom onset (Figure 3). There was significant between‐study heterogeneity (I 2 = 94%, 2 = 2.1; p < 0.01).
FIGURE 3.
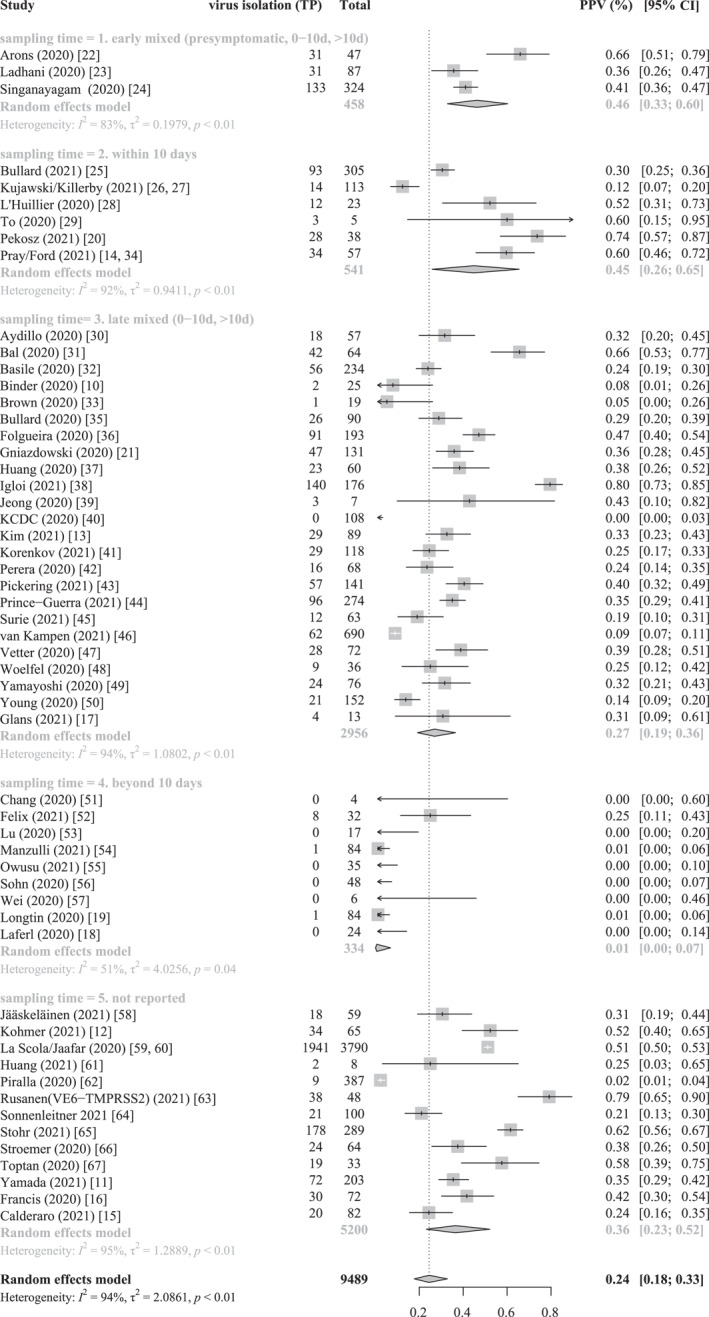
Forest plot shows random effect meta‐analysis of positive predictive values|positive predictive value (PPV) for virus isolation in total reverse‐transcription polymerase chain reaction (RT‐PCR) positive respiratory samples, subgrouped by sampling time post symptom onset. 5 Subgroups based on sampling time: (1). Early mixed sampling (presymptomatic, 0–10 days, >10 days), (2). Within 10 days, (3). Late mixed (0–10 days, >10 days), (4). Beyond 10 days, (5). Sampling time not reported. TP – true positive, FP – false positive, Total = TP + FP
The PPV was 45% (CI, 26%–65%, six studies) in the subgroup with sampling time post symptom onset of 0–10 days and 1% (CI, 0%–7%, nine studies) in the subgroup with symptom time beyond 10 days.
The estimated PPVs were 46% (CI, 33%–60%, three studies) for the subgroup of early mixed sampling time (presymptomatic, 0–10 days, >10 days), 27% (CI, 19%–36%, 24 studies) for the subgroup of late mixed sampling time (0–10 days, >10 days) and 36% (95% CI, 23%–52%, 13 studies) for the subgroup without reported sampling times. Due to poor reporting further subgroup analysis based on variables which we anticipated as potential sources of variation, such as symptom status, patients‘ age range, specimen type, cell line used for virus isolation could not be performed.
The sensitivity analysis excluding studies at high risk of bias included 10 studies with unclear risk of bias and provided results comparable to those obtained from all studies (S12).
3.6. Virus isolation probability in dependence on cycle threshold or log10 genome copies/ml
We included 13 studies for estimation of virus isolation probability depending on cycle threshold (N = 9) or log10 genome copies/ml (n = 4) with sampling size ranging from 33 65 to 3790 58 for studies providing Ct values and cell culture outcomes and from 12 28 to 690 46 for log10 genome copies/ml data. Five studies reported Ct values based on multiple gene targets with corresponding cell culture outcomes. Among studies reporting Ct values four studies reported performance of RT‐PCR based on standard curve. 12 , 13 , 37 , 64
Virus isolation probability estimation across the studies at Ct value of 30 and at 5 log10 genome copies/ml ranged for Ct from 6% (CI, 0%–100%) 13 to 50% (CI, 0%–100%) 12 and for log10 genome copies/ml from 0% (CI, 0%–22%) 45 to 63% (CI, 0%–100%) 12 (Figure 4a,b).
FIGURE 4.
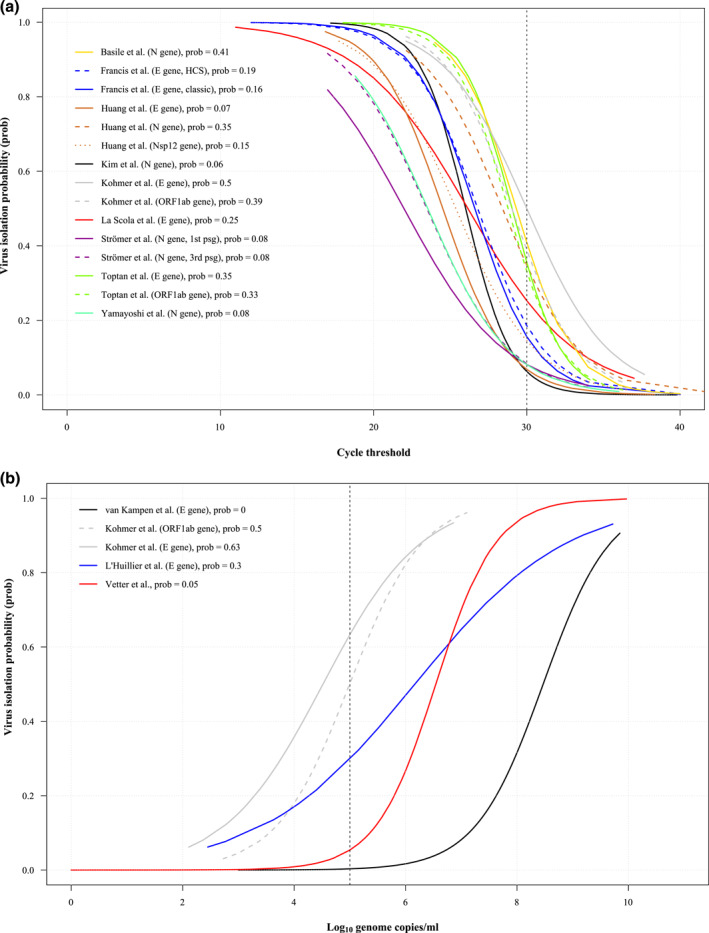
Overview of virus isolation probability curves assessed by logistic regression. Virus culture outcomes are plotted against Ct values (N = 9) (a) or log10 genome copies/ml (n = 5) (b) in a common coordinate to allow visual comparability of the curves. Estimation of virus isolation probability is represented by lines for each study. Colouration of the lines represents specific study, dashed or dotted lines have been used if studies provided Ct values for several target genes. For clearity reasons the 95% confidence intervals of the estimates are depicted for each study in supplementary material (S13‐32). (a) Overview of virus isolation probability versus cycle threshold. The dotted vertical line represents Ct of 30 and corresponding virus isolation probability estimates are depicted in the legend. (b) Overview of virus isolation probability versus log10 genome copies/ml. The dotted vertical line represents 5 log10 genome copies/ml and corresponding virus isolation probability estimates are depicted in the legend
At low virus isolation probability of 10% Ct values varied from 29.1 37 to 35.9 12 and from 2.6 12 to 7.1 46 log10 genome copies/ml (supplementary material S33, 34).
As demonstrated by the 95% CI bands (S13‐32) of the probability estimates the data has a high degree of uncertainty.
4. DISCUSSION
This systematic review assessed diagnostic accuracy of RT‐PCR assays compared to cell culture detection, synthesised data on the relationship of Ct or log10 genome copies/ml and virus isolation probability and collated data on PPV. The sensitivity to detect infectious virus was generally sufficient, while specificity varied across studies and PPV was variable across subgroups of sampling time after symptom onset. Ct cut‐off values indicating infectivity are currently not determinable with high reliability. However, the findings need to be interpreted in light of heterogeneity and quality limitations of available studies that were most commonly ad hoc initiated and performed under the unfavourable conditions of the ongoing SARS‐CoV‐2 pandemic.
The indispensable role of RT‐PCR in the COVID‐19 pandemic is undisputed; moreover, the evaluation of its performance compared to cell culture could provide important insights regarding the differentiation between infectious and non‐infectious samples. For pandemic management, both a highly specific and sensitive test is needed, as false negative results might lead to untoward transmission and false positive test results to unnecessary isolation and quarantining.
The sensitivity was generally adequate, with no false negative results obtained in any of the studies; the lowest estimate of 90% was due to the continuity correction and small sample size. 17 This emphasises the importance of RT‐PCR to rule out infectivity, provided that samples were collected and handled properly, and no gene dropout due to target gene mutations had occurred. 68
The specificity of RT‐PCR to detect infectious samples varied considerably amongst the studies. Optimization of specificity would have been possible by ROC curve analysis, but only one study provided raw data that could be used to generate sensitivity and specificity values per Ct cut‐off point. 16
The majority of the studies compared Ct or log10 genome copies/ml and cell culture results only from RT‐PCR positive samples. For this reason no valid ROC analysis based on sensitivity and specificity was possible. We performed a logistic regression and estimated the probability of virus isolation at a relatively high Ct value of 30, but the estimates obtained precluded this Ct value as a cut‐off for excluding infectivity.
In the meta‐analysis by Cevik et al. a maximum virus isolation time of 9 days after symptom onset was shown for non‐severe courses of the disease. 3 To quantify PPV, we performed subgroup analyses based on this sampling time after symptom onset.
Despite the large heterogeneity, the pooled PPV of 45% reflects that a substantial proportion of RT‐PCR‐positive samples do not result in detection of infectious virus within first 10 days of sampling. The PPV of 1% in samples beyond 10 days supports the observation of prolonged RNA excretion even when no virus can be isolated. 3 , 69
The high degree of heterogeneity among the studies might be due to different study designs, heterogenous populations studied at different time points, use of different respiratory specimen, variable permissiveness of cell lines and culture conditions and disparate RT‐PCR standards.
Considering cell culture as a biological, that is living detection system, we noted a wide range of methods used for the verification of cytopathic effects upon microscopic inspection. Poor reporting on storage conditions of samples before inoculation, the history of cell passaging, application of centrifugal enhancement, as well as the definition of cell culture positivity, was evident in most cases. If reported the factors applied varied between many studies. Further conditions that may influence the success of virus isolation include additional biological or engineered features of the cell line, inoculation volume, freeze‐thawing of the specimen, incubation environment and the density of cell layers varied amongst the studies. 4 , 70 , 71
Accordingly, there is a great need for consensus on standardisation of SARS‐CoV‐2 isolation by cell culture. To date, in the absence of universally accepted standardized cell culture guidelines, almost all included studies have followed their in‐house protocols. It must be noted that cell culture positivity only implies detection of viable virus within a body fluid of the patient representing a surrogate for the potential of virus transmission by an infected individual. Given the most relevant route of air‐borne transmission of SARS‐CoV‐2 infection, the extrapolation of virus isolation from swabs taken form the oro‐ or nasopharyngeal surfaces into the efficiency of virus transmission or even into a specific epidemiological context is currently not yet possible.
Several factors that influence RT‐PCR limits of detection and Ct values have been described, including pre‐analytic factors like sample type and matrix, its collection, storage and preparation, and analytic factors like reagents of RT‐PCR assay (primer sets, buffers, and enzymes, nucleic acid extraction efficiency, reference material of standard curve). 72 , 73 , 74
As shown in Figure 4, a high degree of variability in the correlation between Ct and virus isolation was observed between the studies. While low Ct values could be indicative of infectivity, high Ct values are difficult to interpret, as revealed by the variable virus isolation probabilities at a Ct of 30. Inter‐ and intralaboratory variations of RT‐PCR in conjunction with non‐standardized cell culture methodology might explain this variability. Due to pandemic‐related supply shortages, a locally uniform RT‐PCR methodology was not feasible everywhere. As a result, different test kits had to be used within the same laboratory. In addition, an international quantitative reference standard to improve interlaboratory agreement on viral load quantification has not yet been established. 75 , 76 Thus, no testing strategy for de‐isolation based solely on longitudinal Ct monitoring can be recommended at this time.
In addition to the difficulties inherent in detection methods, the constant mutations with the formation of new variants during the pandemic may pose another challenge for virus detection. Initial reports show lower virus recovery of Alpha compared to Delta variants without concomitant differences in the Ct means of cell culture positive and negative samples 77 and target area related mutations can cause gene drop out without altering virus recovery. 68 Mutations could therefore be considered as another factor of variability in the accuracy effect estimates.
There are important limitations in our systematic review. The included studies differ in terms of study design, sample size (ranging from 4 to 3790) and patient characteristics (such as age, disease severity, symptom status and sampling time) and methodology of RT‐PCR as well as cell culture. Most published studies were case series and not designed to determine diagnostic accuracy, thus performed poorly in methodological quality assessment, as patient selection was most commonly by convenience and patients eligibility criteria were often unclear or not defined at all.
The heterogeneity across studies could not be fully explained in the PPV meta‐analysis and it was not possible to account for disease prevalence and potentially relevant subgroup analyses due to underreporting. Bivariate analysis for the sensitivity and specificity could not be performed because of the limited number of suitable studies.
The fact that the pandemic is a highly dynamic, discontinuous event and driven by virus variants with distinct virological properties makes it clear that the work begun here should be continued. It will be interesting to learn whether the two major virus detection methods compared here undergo further standardisation of methodology and if this allows more robust conclusions regarding their correlations.
5. CONCLUSIONS
This systematic review identified an evidence gap in well designed diagnostic accuracy studies comparing RT‐PCR with cell culture as a surrogate for infectivity of SARS‐CoV‐2 patients. Such studies would provide essential information for clinical practice, where the differentiation between infectious and not infectious patients is highly important for clinical management.
Furthermore, we found a high degree of between study heterogeneity and a lack of standardisation especially of cell culture methodology.
However, it was possible to make rough estimations on the correlation between viral load measured by RT‐PCR and infectivity based on these studies.
Taking into account that the studies were performed in different waves of the ongoing SARS‐CoV‐2 pandemic, these shortcomings are comprehensible, but have to be addressed in future research and pandemic management planing.
CONFLICT OF INTEREST
The authors have no relevant competing interest to disclose in relation to this work.
ETHICAL STATEMENT
No ethical approval is needed as systematic reviewing is based on published trial results.
AUTHOR CONTRIBUTIONS
Alexey Fomenko designed the study, conducted analyses, and wrote the first draft of manuscript. Stephanie Weibel, Gerta Rücker and Christine Schmucker contributed to study design and analysis. Maria‐Inti Metzendor and Edith Motschall created and implemented the search strategy. Alexey Fomenko, Hartmut Hengel and Kristina Menger conducted the screening of articles, data extraction and quality assessment. Hartmut Hengel, Marcus Panning, Valeria Falcone, Daniela Huzly, Stephanie Weibel, Gerta Rücker, Christine Schmucker, Helia Moez, Kristina Menger and Maria‐Inti Metzendor critically reviewed and approved the final manuscript.
Supporting information
Supplementary Material S1
ACKNOWLEDGEMENTS
Antonio Piralla and Kelvin To provided additional information for this systematic review. Johanna Pilz and Tarek Assali helped in screening and extraction process. Furthermore, we would like to thank the CEOsys network in supporting this study. This work was supported by COVID‐19 evidence eco‐system (‘COVID‐19 Evidenzökosystem’ [CEO‐sys]) under a funding scheme issued by the National Research Network of University Medical Centers on COVID‐19 (Nationales Forschungsnetzwerk der Universitätsmedizin zu COVID‐19, NaFoUniMedCovid19) by the Federal Ministry of Education and Research of Germany (Bundesministerium für Bildung und Forschung, BMBF), grant number [FKZ]: 01KX2021 to HH.
Open access funding enabled and organized by Projekt DEAL.
Fomenko A, Weibel S, Moezi H, et al. Assessing severe acute respiratory syndrome coronavirus 2 infectivity by reverse‐transcription polymerase chain reaction: a systematic review and meta‐analysis. Rev Med Virol. 2022;32(5):e2342. 10.1002/rmv.2342
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. World Health Organization (WHO) . Diagnostic Testing for SARS‐CoV‐2. 2020. WHO/2019‐nCoV/laboratory/2020.6:20. https://www.who.int/publications/i/item/diagnostic‐testing‐for‐sars‐cov‐2 [Google Scholar]
- 2. Miranda RL, Guterres A, de Azeredo Lima CH, Filho PN, Gadelha MR. Misinterpretation of viral load in COVID‐19 clinical outcomes. Virus Res. 2021;296:198340. 10.1016/j.virusres.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta‐analysis. Lancet Microbe. 2021;2(1):e13‐e22. 10.1016/S2666-5247(20)30172-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007;20(1):49‐78. 10.1128/CMR.00002-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529‐536. 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 6. R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2021. https://www.R‐project.org/ [Google Scholar]
- 7. Doebler P. Mada: Meta‐Analysis of Diagnostic Accuracy. R package version 0.5.10. 2020. [Google Scholar]
- 8. Balduzzi S, Rücker G, Schwarzer G. How to perform a meta‐analysis with R: a practical tutorial. Evid based Ment health. 2019;22(4):153‐160. 10.1136/ebmental-2019-300117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta‐analyses. BMJ. 2003;327(7414):557‐560. 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Binder RA, Alarja NA, Robie ER, et al. Environmental and aerosolized SARS‐CoV‐2 among hospitalized COVID‐19 patients. J infect dis. 2020;222(11):1798‐1806. 10.1093/infdis/jiaa575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamada S, Fukushi S, Kinoshita H, et al. Assessment of SARS‐CoV‐2 infectivity of upper respiratory specimens from COVID‐19 patients by virus isolation using VeroE6/TMPRSS2 cells. BMJ open Respir Res. 2021;8(1):e000830. 10.1136/bmjresp-2020-000830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kohmer N, Toptan T, Pallas C, et al. The comparative clinical performance of four SARS‐CoV‐2 rapid antigen tests and their correlation to infectivity in vitro. J Clin Med. 2021;10(2):328. 10.3390/jcm10020328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim M‐C, Cui C, Shin K‐R, et al. Duration of culturable SARS‐CoV‐2 in hospitalized patients with covid‐19. N. Engl J Med. 2021;384(7):671‐673. 10.1056/NEJMc2027040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pray IW, Ford L, Cole D, et al. Performance of an antigen‐based test for asymptomatic and symptomatic SARS‐CoV‐2 testing at two university campuses ‐ Wisconsin, September‐October 2020. MMWR Morb Mortal Wkly Rep. 2021;69(5152):1642‐1647. 10.15585/mmwr.mm695152a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Calderaro A, De Conto F, Buttrini M, et al. Human respiratory viruses, including SARS‐CoV‐2, circulating in the winter season 2019‐2020 in Parma, Northern Italy. Int J Infect Dis. 2020. 10.1016/j.ijid.2020.09.1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Francis R, Le Bideau M, Jardot P, et al. High‐speed large‐scale automated isolation of SARS‐CoV‐2 from clinical samples using miniaturized co‐culture coupled to high‐content screening. Clinical microbiology and infection. 2020(1):128.e1‐128.e7. 10.1016/j.cmi.2020.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glans H, Gredmark‐Russ S, Olausson M, et al. Shedding of infectious SARS‐CoV‐2 by hospitalized COVID‐19 patients in relation to serum antibody responses. BMC Infect Dis. 2021;21(1):494. 10.1186/s12879-021-06202-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laferl H, Kelani H, Seitz T, et al. An approach to lifting self‐isolation for health care workers with prolonged shedding of SARS‐CoV‐2 RNA. Infection. 2021;49(1):95‐101. 10.1007/s15010-020-01530-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Longtin Y, Charest H, Quach C, et al. Infectivity of healthcare workers diagnosed with SARS‐CoV‐2 infection approximately 2 weeks after onset of symptoms: a cross‐sectional study. Infect Control Hosp Epidemiol. 2021;43:1‐104. 10.1017/ice.2020.1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pekosz A, Parvu V, Li M, et al. Antigen‐based testing but not real‐time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. Clin Infect Dis. 2021;73(9):e2861‐e2866. 10.1093/cid/ciaa1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gniazdowski V, Paul Morris C, Wohl S, et al. Repeat COVID‐19 molecular testing: correlation of SARS‐CoV‐2 culture with molecular assays and cycle thresholds. Clin Infect Dis. 2021;73(4):e860‐e869. 10.1093/cid/ciaa1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arons MM, Hatfield KM, Reddy SC, et al. Presymptomatic SARS‐CoV‐2 infections and transmission in a skilled nursing facility. N. Engl J Med. 2020;382(22):2081‐2090. 10.1056/NEJMoa2008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ladhani SN, Chow JY, Janarthanan R, et al. Investigation of SARS‐CoV‐2 outbreaks in six care homes in London, April 2020. Eclinicalmedicine. 2020;26:100533. 10.1016/j.eclinm.2020.100533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT‐PCR cycle threshold values in cases of COVID‐19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. 10.2807/1560-7917.ES.2020.25.32.2001483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bullard J, Funk D, Dust K, et al. Infectivity of severe acute respiratory syndrome coronavirus 2 in children compared with adults. CMAJ. 2021;193(17):E601‐E606. 10.1503/cmaj.210263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Killerby ME, Ata Ur Rasheed M, Tamin A, et al. Shedding of culturable virus, seroconversion, and 6‐month follow‐up antibody responses in the first 14 confirmed cases of COVID‐19 in the United States. Journal of infectious diseases. 2021;224(5):771‐776. 10.1093/infdis/jiab125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. COVID‐19 Investigation Team . Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID‐19) in the United States. Nat Med. 2020;26(6):861‐868. 10.1038/s41591-020-0877-5 [DOI] [PubMed] [Google Scholar]
- 28. L'Huillier AG, Torriani G, Pigny F, Kaiser L, Eckerle I. Culture‐competent SARS‐CoV‐2 in nasopharynx of symptomatic neonates, children, and adolescents. Emerg Infect Dis. 2020;26(10):2494‐2497. 10.3201/eid2610.202403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. To KK, Tsang OT, Yip CC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841‐843. 10.1093/cid/ciaa149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aydillo T, Gonzalez‐Reiche AS, Aslam S, et al. Shedding of viable SARS‐CoV‐2 after immunosuppressive therapy for cancer. N. Engl J Med. 2020;383(26):2586‐2588. 10.1056/NEJMc2031670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bal A, Brengel‐Pesce K, Gaymard A, et al. Clinical and laboratory characteristics of symptomatic healthcare workers with suspected COVID‐19: a prospective cohort study. Sci Rep. 2021;11(1):14977. 10.1038/s41598-021-93828-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Basile K, McPhie K, Carter I, et al. Cell‐based culture of SARS‐CoV‐2 informs infectivity and safe de‐isolation assessments during COVID‐19. Clin Infect Dis. 2021;73(9):e2952‐e2959. 10.1093/cid/ciaa1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown CS, Clare K, Chand M, et al. Snapshot PCR surveillance for SARS‐CoV‐2 in hospital staff in England. J Infect. 2020;81(3):427‐434. 10.1016/j.jinf.2020.06.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ford L, Lee C, Pray IW, et al. Epidemiologic characteristics associated with SARS‐CoV‐2 antigen‐based test results, rRT‐PCR cycle threshold values, subgenomic RNA, and viral culture results from university testing. Clin Infect Dis. 2021;73(6):e1348‐e1355. 10.1093/cid/ciab303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bullard J, Dust K, Funk D, et al. Predicting infectious SARS‐CoV‐2 from diagnostic samples. Clin Infect Dis. 2020;71(10):2663‐2666. 10.1093/cid/ciaa638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Folgueira MD, Luczkowiak J, Lasala F, Perez‐Rivilla A, Delgado R. Persistent SARS‐CoV‐2 replication in severe COVID‐19. MedRxiv. 2020. 10.1101/2020.06.10.20127837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang CG, Lee KM, Hsiao MJ, et al. Culture‐based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID‐19. J Clin Microbiol. 2020;58(8):e01068‐20. 10.1128/JCM.01068-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iglοi Z, Velzing J, van Beek J, et al. Clinical evaluation of Roche SD biosensor rapid antigen test for SARS‐CoV‐2 in municipal health service testing site, The Netherlands. Emerg Infect Dis. 2021;27(5):1323‐1329. 10.3201/eid2705.204688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jeong HW, Kim SM, Kim HS, et al. Viable SARS‐CoV‐2 in various specimens from COVID‐19 patients. Clin Microbiol Infect. 2020;26(11):1520‐1524. 10.1016/j.cmi.2020.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Korea Central Disaster Control and Prevention (KCDC) . Finding from Investigation and Analysis of Re‐positive Cases; 2020. Accessed 19 May 2020. https://www.mofa.go.kr/eng/brd/m_22743/view.do?seq=3&srchFr=&srchTo=&srchWord=&srchTp=&multi_itm_seq=0&itm_seq_1=0&itm_seq_2=0&company_cd=&company_nm=&page=1&titleNm= [Google Scholar]
- 41. Korenkov M, Poopalasingam N, Madler M, et al. Evaluation of a rapid antigen test to detect SARS‐CoV‐2 infection and identify potentially infectious individuals. J Clin Microbiol. 2021;59(9):e00896‐21. 10.1128/JCM.00896-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perera RAPM, Tso E, Tsang OTY, et al. SARS‐CoV‐2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26(11):2701‐2704. 10.3201/eid2611.203219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pickering S, Batra R, Merrick B, et al. Comparative performance of SARS‐CoV‐2 lateral flow antigen tests and association with detection of infectious virus in clinical specimens: a single‐centre laboratory evaluation study. Lancet Microbe. 2021;2(9):e461‐e471. 10.1016/S2666-5247(21)00143-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prince‐Guerra JL, Almendares O, Nolen LD, et al. Evaluation of abbott BinaxNOW rapid antigen test for SARS‐CoV‐2 infection at two community‐based testing sites ‐ Pima county, Arizona, November 3‐17, 2020. Mmwr‐Morbidity Mortal Wkly Rep. 2021;70(3):100‐105. 10.15585/mmwr.mm7003e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Surie D, Huang JY, Brown AC, et al. Infectious period of severe acute respiratory syndrome coronavirus 2 in 17 nursing home residents‐Arkansas, June‐August 2020. Open Forum Infect Dis. 2021;8(3):ofab048. 10.1093/ofid/ofab048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Kampen JJA JJA, van de Vijver D, Fraaij PLA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease‐2019 (COVID‐19). Nat Commun. 2021;12(1):267. 10.1038/s41467-020-20568-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vetter P, Eberhardt CS, Meyer B, et al. Daily viral kinetics and innate and adaptive immune response assessment in COVID‐19: a case series. mSphere. 2020;5(6):e00827‐20. 10.1128/mSphere.00827-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581(7809):465‐469. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 49. Yamayoshi S, Sakai‐Tagawa Y, Koga M, et al. Comparison of rapid antigen tests for COVID‐19. Viruses. 2020;12(12):1420. 10.3390/v12121420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Young BE, Ong SWX, Ng LFP, et al. Viral dynamics and immune correlates of COVID‐19 disease severity. Clin Infect Dis. 2021;73(9):e2932‐e2942. 10.1093/cid/ciaa1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang D, Zhao P, Zhang D, et al. Persistent viral presence determines the clinical course of the disease in COVID‐19. J allergy Clin Immunol Pract. 2020;8(8):2585.e1‐2591.e1. 10.1016/j.jaip.2020.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lu J, Peng J, Xiong Q, et al. Clinical, immunological and virological characterization of COVID‐19 patients that test re‐positive for SARS‐CoV‐2 by RT‐PCR. EBioMedicine. 2020;59:102960. 10.1016/j.ebiom.2020.102960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Manzulli V, Scioscia G, Giganti G, et al. Real time PCR and culture‐based virus isolation test in clinically recovered patients: is the subject still infectious for SARS‐CoV2? J Clin Med. 2021;10(2):309. 10.3390/jcm10020309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Owusu D, Pomeroy MA, Lewis NM, et al. Persistent SARS‐CoV‐2 RNA shedding without evidence of infectiousness: a cohort study of individuals with COVID‐19. J Infect Dis. 2021;224(8):1362‐1371. 10.1093/infdis/jiab107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sohn Y, Jeong SJ, Chung WS, et al. Assessing viral shedding and infectivity of asymptomatic or mildly symptomatic patients with COVID‐19 in a later phase. J Clin Med. 2020;9(9):2924. 10.3390/jcm9092924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jaaskelainen AE, Ahava MJ, Jokela P, et al. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS‐CoV‐2 infection. J Clin Virology. 2021;137:104785. 10.1016/j.jcv.2021.104785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS‐CoV‐2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059‐1061. 10.1007/s10096-020-03913-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jaafar R, Aherfi S, Wurtz N, et al. Correlation between 3790 qPCR positives samples and positive cell cultures including 1941 SARS‐CoV‐2 isolates. Clin Infect Dis. 2020;72(11):e921. 10.1093/cid/ciaa1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang N, Perez P, Kato T, et al. SARS‐CoV‐2 infection of the oral cavity and saliva. Nat Med. 2021;27(5):892‐903. 10.1038/s41591-021-01296-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Piralla A, Ricchi M, Cusi MG, et al. Residual SARS‐CoV‐2 RNA in nasal swabs of convalescent COVID‐19: is prolonged quarantine always justified? Int J Infect Dis. 2021;102:299‐302. 10.1016/j.ijid.2020.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rusanen J, Kareinen L, Szirovicza L, et al. A generic, scalable, and rapid time‐resolved förster resonance energy transfer‐based assay for antigen detection—SARS‐CoV‐2 as a proof of concept. mBio. 2021;12(3):e00902‐e00921. 10.1128/mBio.00902-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sonnleitner ST, Julian D, Bianca J, et al. An in vitro model for assessment of SARS‐CoV‐2 infectivity by defining the correlation between virus isolation and quantitative PCR value: isolation success of SARS‐CoV‐2 from oropharyngeal swabs correlates negatively with Cq value. Virol J. 2021;18(1):71. 10.1186/s12985-021-01542-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stohr J, Zwart VF, Goderski G, et al. Self‐testing for the detection of SARS‐CoV‐2 infection with rapid antigen tests for people with suspected COVID‐19 in the community. Clin Microbiol Infect. 2021. 10.1016/j.cmi.2021.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Strömer A, Rose R, Schäfer M, et al. Performance of a point‐of‐care test for the rapid detection of SARS‐CoV‐2 antigen. Microorganisms. 2020;9(1):58. 10.3390/microorganisms9010058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Toptan T, Eckermann L, Pfeiffer AE, et al. Evaluation of a SARS‐CoV‐2 rapid antigen test: potential to help reduce community spread? J Clin Virol. 2020;135:104713. 10.1016/j.jcv.2020.104713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Felix AC, Vincente de Paula A, Ribeiro AC, et al. Iscontinuation of isolation for persons with COVID‐19: is 10 days really safe? MedRxiv. 2021;0129:21250753. 10.1101/2021.01.29.21250753 [DOI] [Google Scholar]
- 67. Wei S, Tong Y, Liu J, et al. Re‐positive discharged COVID‐19 patients are at low transmission risk for SARS‐CoV‐2 infection– a finding from recovered COVID‐19 patients in Wuhan, China. Research Square. 2020. 10.21203/rs.3.rs-93844/v1 [DOI] [Google Scholar]
- 68. Wollschläger P, Todt D, Gerlitz N, et al. SARS‐CoV‐2 N gene dropout and N gene Ct value shift as indicator for the presence of B.1.1.7 lineage in a commercial multiplex PCR assay. Clin Microbiol Infect. 2021;27(9):1353.e1‐1353.e5. 10.1016/j.cmi.2021.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jefferson T, Spencer EA, Brassey J, Heneghan C. Viral cultures for coronavirus disease 2019 infectivity assessment: a systematic review. Clin Infect Dis. 2021;73(11):e3884‐e3899. 10.1093/cid/ciaa1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ogando NS, Dalebout TJ, Zevenhoven‐Dobbe JC, et al. SARS‐coronavirus‐2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J General Virology. 2020;101(9):925‐940. 10.1099/jgv.0.001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS‐CoV‐2 by TMPRSS2‐expressing cells. Proc Natl Acad Sci. 2020;117(13):7001‐7003. 10.1073/pnas.2002589117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Basso D, Aita A, Navaglia F, et al. SARS‐CoV‐2 RNA identification in nasopharyngeal swabs: issues in pre‐analytics. Clin Chem Laboratory Med. 2020;58(9):1579‐1586. 10.1515/cclm-2020-0749 [DOI] [PubMed] [Google Scholar]
- 73. Han MS, Byun J‐H, Cho Y, Rim JH. RT‐PCR for SARS‐CoV‐2: quantitative versus qualitative. Lancet Infect Dis. 2021;21(2):165. 10.1016/S1473-3099(20)30424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Alcoba‐Florez J, Gil‐Campesino H, Artola DG‐MD, et al. Sensitivity of different RT‐qPCR solutions for SARS‐CoV‐2 detection. Int J Infect Dis. 2020;190‐192. 10.1016/j.ijid.2020.07.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mercer T, Almond N, Chain P, et al. A roadmap to better COVID‐19 testing from the coronavirus standards working group. Research Square; 2021. 10.21203/rs.3.rs-1066221/v1 [DOI] [Google Scholar]
- 76. Rhoads DD, Pinsky BA. The truth about SARS‐CoV‐2 cycle threshold values is rarely pure and never simple. Clin Chem. 2022;68(1):16‐18. 10.1093/clinchem/hvab146 [DOI] [PubMed] [Google Scholar]
- 77. Luo CH, Morris CP, Sachithanandham J, et al. Infection with the SARS‐CoV‐2 delta variant is associated with higher recovery of infectious virus compared to the alpha variant in both unvaccinated and vaccinated individuals. Clin Infect Dis. 2021;ciab986. 10.1093/cid/ciab986 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


