Yao et al. describe GIMAP6 deficiency in humans and mice showing immune dysfunction and susceptibility to bacterial infections. They find impaired autophagy in GIMAP6-deficient immune cells and define a functional complex composed of GIMAP6, GIMAP7, and GABARAPL2.
Abstract
Inborn errors of immunity (IEIs) unveil regulatory pathways of human immunity. We describe a new IEI caused by mutations in the GTPase of the immune-associated protein 6 (GIMAP6) gene in patients with infections, lymphoproliferation, autoimmunity, and multiorgan vasculitis. Patients and Gimap6−/− mice show defects in autophagy, redox regulation, and polyunsaturated fatty acid (PUFA)–containing lipids. We find that GIMAP6 complexes with GABARAPL2 and GIMAP7 to regulate GTPase activity. Also, GIMAP6 is induced by IFN-γ and plays a critical role in antibacterial immunity. Finally, we observed that Gimap6−/− mice died prematurely from microangiopathic glomerulosclerosis most likely due to GIMAP6 deficiency in kidney endothelial cells.
Introduction
GTPase of immunity-associated proteins (GIMAPs) are highly conserved gene families whose molecular functions have been elusive (Krucken et al., 2004; Liu et al., 2008; Nitta and Takahama, 2007; Poirier et al., 1999). The genes are highly expressed and necessary in immune cells for development, survival, and function (Barnes et al., 2010; MacMurray et al., 2002; Pascall et al., 2018; Saunders et al., 2010; Schulteis et al., 2008; Yano et al., 2014). Gimaps are also expressed in mouse kidney endothelial cells and human blood-vessel endothelial and lung septal cells (Hellquist et al., 2007). GIMAP proteins occur as soluble forms in the cytoplasm or are localized to distinct organelles, suggesting they control diverse intracellular functions (Limoges et al., 2021). GIMAP1, GIMAP2, and GIMAP5 contain C-terminal anchors for the Golgi complex, lipid droplets, and lysosomes, respectively (Schwefel et al., 2010; Wong et al., 2010). GIMAP4, GIMAP6, GIMAP7, and GIMAP8 are cytosolic. GIMAPs are phylogenetically related to the septin/dynamin GTP-binding proteins that control cytoskeleton and membrane dynamics (Limoges et al., 2021). Similar to septin/dynamins, GIMAPs undergo GTP-dependent homo- and hetero-dimerization via a conserved interface that regulates GTPase activity (Schwefel et al., 2010). For example, GIMAP7 can activate an otherwise catalytically silent GIMAP2 (Schwefel et al., 2013). The molecular functions of GIMAPs and whether they are inducible during infection are largely unknown.
GIMAP6 is a 32-kD protein expressed in lymphocytes and endothelial cells (Bhasin et al., 2010; Gay et al., 2013; Pascall et al., 2013; Yue et al., 2017). Polymorphisms in GIMAP6 have been linked to pulmonary disease (Lee et al., 2014), lymphocyte number (Astle et al., 2016), cholesterol levels (Hoffmann et al., 2018), fibrinogen (de Vries et al., 2017), and C-reactive protein (Sakaue et al., 2021). GIMAP6 also controls cell survival and autophagy (Ho and Tsai, 2017; Pascall et al., 2013; Pascall et al., 2018). A human case of GIMAP6 deficiency has been reported with recurrent infections, but the molecular pathogenesis was unclear (Shadur et al., 2020).
Autophagy packages and transports damaged organelles and proteins for degradation and recycling in the lysosomes (Zhao and Zhang, 2019). Autophagy contributes to immune cell function and cytokine and immunoglobulin release (Ho and Tsai, 2017; Pascall et al., 2018). Gamma-amino butyric acid receptor-associated protein-like 2 (GABARAPL2) is an autophagy gene 8 (ATG8) homolog co-recruited with GIMAP6 to autophagosomes during autophagy (Pascall et al., 2013; Pascall et al., 2018). GABARAPL2 regulates lysosome–autophagosome fusion and IFN-γ–induced clearance of pathogens (Lee and Lee, 2016; Sasai et al., 2017). Whether the GIMAP6/GABARAPL2 complex involves other GIMAP proteins is uncertain (Pascall et al., 2013).
New genomics technologies have increased gene discovery in inborn errors of immunity (IEIs) leading to exciting novel therapies (Casanova and Abel, 2021; Lenardo et al., 2016; Lenardo and Holland, 2019; Tangye et al., 2021). Here, we describe a new IEI in which autophagic flux, metabolism, redox state, and antibacterial activity are affected in patients with germline GIMAP6 mutations, who present with an autoimmune lymphoproliferative syndrome–like disease, recurrent infections, and vasculitis. We also report a new germline Gimap6−/− mouse showing early mortality due to progressive kidney disease.
Results
Clinical phenotype of GIMAP6 deficiency patients
Timelines of clinical phenotypes are shown in Fig. S1 A. Patient 1 (Pt 1) is a 30-yr-old Turkish female who presented at 6 mo old with Coombs-positive hemolytic anemia and hepatosplenomegaly and was treated with steroids and intravenous immunoglobulins (Table S1). She had recurrent pneumonia and underwent a splenectomy at 18 yr of age for refractory thrombocytopenia. At age 21, Pt 1 was hospitalized for meningitis, otitis media, and pneumonia. Cranial magnetic resonance imaging (MRI) showed bilateral effusions, sulcal hyperintensity, and lateral parietal subcortical acute focal ischemic lesions (Fig. 1, A–C; and Table S1). Vasculitis occurred in the central nervous system (Fig. 1 C), lungs (Fig. 1, F and G), and skin. Recurrent pneumonia caused bronchiectasis and atelectasis, and she required a lobectomy at age 21 (Fig. 1 D). She gradually developed pulmonary hypertension with a pulmonary artery pressure of 40 mm Hg and an enlarged pulmonary artery (3.5 cm; Fig. 1 E). Lung biopsy showed luminal narrowing, intimal thickening, damage in the elastic layer of the artery wall, mononuclear cell infiltrate, and histiocytic giant cell reaction in the tunica adventitia of one of the pulmonary artery branches, indicating vasculitis (Fig. 1, F and G). Transient lymphopenia was observed. Liver enzyme abnormalities were attributed to pulmonary hypertension. Antiphospholipid and anticardiolipin antibodies (IgM+, IgG−) were detected in the serum, and skin vasculitis was diagnosed by biopsy from livedo reticularis lesions. She suffered a herpes zoster infection when she was 22 yr old. Purpuric lesions localized on fingers resembled erythema multiforme, possibly herpetic, and were treated with acyclovir. Over time, she developed elevated serum IgM and β-2 microglobulin but reduced IgA and IgG levels (Table S1). Pt 1 had persistent lymphadenopathy during infections (Fig. 1, H and I). Currently, her hemoglobin is low, but kidney function is normal (Table S1). She is stable on plaquenil, bosentan, coraspin, vitamin D, and intravenous immunoglobulin (Fig. S1 A).
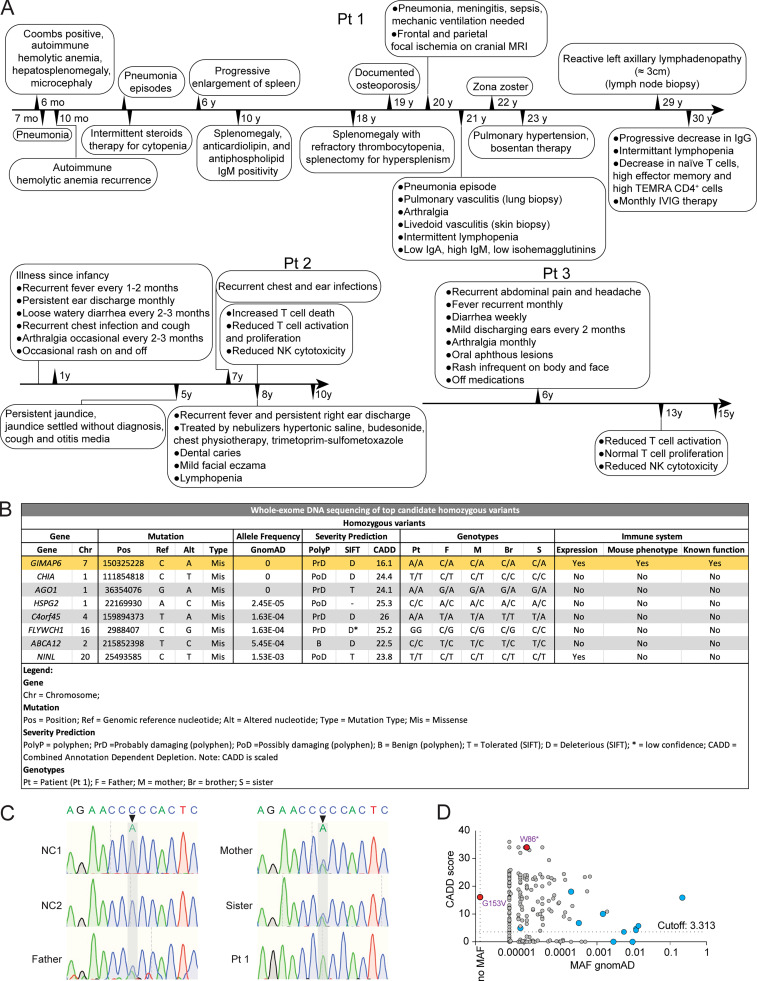
Figure S1.
Gene variants from whole-exome DNA sequencing. (A) Medical chronological summary of patients. (B) Table of homozygous mutations upon ranking by severity using PolyP, Sift, and CADD scores. Table shows information of gene, mutation, allele frequency, severity prediction, genotypes, and immune system expression, phenotype, and function. After additional prioritization by MAF, gene expression pattern, mutation severity, and known biological functions, the novel homozygous mutation in GIMAP6 was confirmed. (C) Sanger sequencing showing the mutations of GIMAP6 in Pt 1 and family members. (D) CADD/MAF diagram indicating the variant in the index (red) and all homozygous missense (blue) and deleterious variants from the public databases GnomAD.
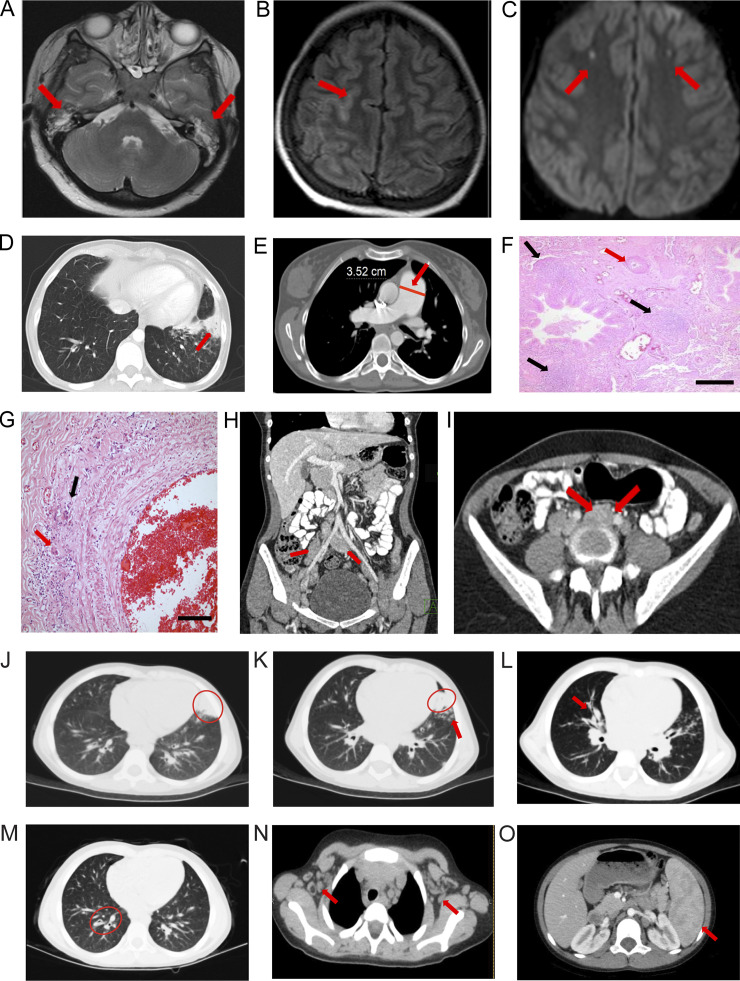
Figure 1.
Radiological and pathological findings. (A–I) Pt 1. (A) T2-weighted axial MRI slice; bilateral effusions in mastoid air cells (red arrows). (B) Axial FLAIR image and sulcal hyperintensity (red arrow). (C) Axial diffusion-weighted MRI shows diffusion-restricted lesions in both frontal lobes (red arrows). (D) Chest CT scan slices show bronchiectasis, atelectasis, and consolidation (red arrow). (E) Chest CT scan slices show pulmonary artery diameter in centimeters (red line and arrow; E). (F and G) H&E-stained lung biopsy showing mononuclear cell reaction with follicle formation around bronchioles (black arrows; F) and intimal thickening causing lumen narrowing of the pulmonary artery (red arrow; F); scale bar = 200 µm (F). Higher magnification of inflammatory infiltrate with histiocytic giant cells in the tunica adventitia (red arrow, G) and destruction of the external elastic layer (black arrow; G); scale bar = 50 µm (G). (H) Coronal abdominal CT showing para-aortic and iliac nodes (red arrows). (I) Abdominal MRI illustrating a para-aortic nodes (red arrows). (J–O) Pt. 2. Chest CT shows (J) a lingular consolidation (red oval), (K) bibasilar bronchial wall thickening and bronchiectasis (red oval and red arrow), (L) right peribronchial consolidation (red arrow), (M) mild bronchiectasis (red oval), (N) bilateral axillary adenopathy (red arrows), and (O) splenomegaly (red arrow).
Patient 2 (Pt 2) is a 10-yr-old female from Gaza, Palestine (Shadur et al., 2020). She was well until age 5 and then developed recurrent purulent otitis media and a chronic wet cough (Table S1). She also developed persistent jaundice and was hospitalized. At age 7, she was hospitalized for recurrent chest and ear infections. Chest computed tomography (CT) showed a lingular consolidation, mild bronchiectasis, bibasilar bronchial wall thickening (Fig. 1, J and K), right peribronchial consolidation (Fig. 1 L), right lower lobe bronchiectasis (Fig. 1 M), bilateral axillary lymphadenopathy (Fig. 1 N), and splenomegaly (Fig. 1 O). Patient 3 (Pt 3) is the older brother of Pt 2 and has suffered headaches, abdomen pain, mouth ulcers, and recurrent infections (Table S1).
GIMAP6 genetic defect causes protein abnormality
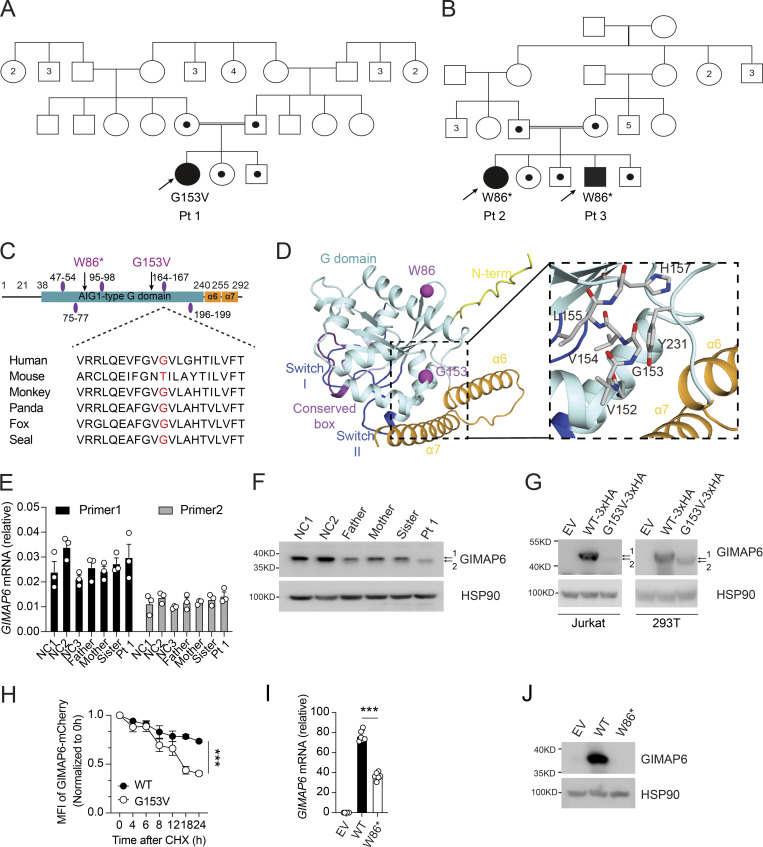
We suspected an IEI and performed whole-exome DNA sequencing. We focused on homozygous recessive variants since family 1 was consanguineous (Figs. 2 A and S1 A). We prioritized eight homozygous variants by minor allele frequency (MAF), expression pattern, mutation severity, and known immune functions, yielding a nonsynonymous mutation in GIMAP6 at position c.458G>T (p.153Gly>Val) as a strong candidate (Fig. S1, B and C). In kindred 2, Pts 2 and 3 had a homozygous stop-gain variant in GIMAP6 at position c.257G>A (p.Trp86Ter; Fig. 2, B–D; Shadur et al., 2020). The Pt 1 mutation site, Gly-153, is conserved among species (Fig. 2 C). The AlphaFold 2 model predicts a Ras-like GTPase (G) domain (Fig. 2 D; Jumper et al., 2021), with the two C-terminal helices, α6 and α7, folding back on the G domain, as reported for GIMAP2 and GIMAP7 (Schwefel et al., 2013). Gly-153 locates to a short helix in the G domain (Fig. 2 D). The Val substitution creates a hydrophobic stretch (VVVL) that may disrupt folding and the interaction of the G domain with the C-terminal helical region. This contact may control the GTP-dependent release of the helical region from the G domain (Schwefel et al., 2010). The G153V variant was predicted to be pathogenic (Combined Annotation Dependent Depletion [CADD] score of 16.1), unique in the ExAC, 1000 Genome, and GnomAD databases, and was validated by the mutation significance cutoff test (Itan et al., 2016; Kircher et al., 2014; Fig. S1, B and D). Both parents and siblings were heterozygous (Fig. S1 C). Although the mRNA level was comparable among Pt 1, family members, and healthy controls (Fig. 2 E), we found that Pt 1’s T cell blasts had a reduced amount of a faster-migrating GIMAP6 protein (Fig. 2 F, band 2). The overexpression of the GIMAP6G153V allele in Jurkat and HEK293T cells produced a similar protein (Fig. 2 G). Using a cycloheximide (CHX) chase assay, we found that the GIMAP6G153V protein had a shorter half-life (Fig. 2 H). Thus, GIMAP6G153V is a loss of function variant.
Figure 2.
Two kindreds with deleterious mutations in GIMAP6. (A and B) Pedigrees of GIMAP6 genotypes. Open symbols: unknown or WT genotype, dots: heterozygotes, and solid symbols: homozygous genotypes in affected individuals; double line indicates consanguinity. Numbers in symbols show additional same sex siblings. Arrows indicate probands. (C) GIMAP6 protein showing the avrRpt2 induced gene 1 (AIG1)–type guanine nucleotide-binding (G) domain (cyan box) with amino acid changes (arrows) and GTP-binding motifs (purple ovals; Krucken et al., 2004). Below is a sequence alignment showing glycine residue (red) conservation. (D) AlphaFold 2 model of human GIMAP6, featuring the G domain, the P-loop (light blue), switches I and II (dark blue), residues of the conserved box involved in dimerization (magenta), helices α6 and α7 (orange), and part of the disordered N-terminus (yellow). The G153V and W86* mutations are indicated (purple). Magnified area shows the amino acids surrounding G153, which locates to a short connecting helix. (E) GIMAP6 mRNA levels in Pt 1, family members, and healthy donors (NC) using two primer sets. (F) WB of T cell lysates from Pt 1, family members, and healthy donors (NC) showing GIMAP6 and HSP90 (loading control). 1 = WT; 2 = mutant. (G) WBs of Jurkat cells transduced with empty vector (EV), GIMAP6WT (WT-3xHA), or GIMAP6G153V (G153V-3xHA) and HEK293T cells transfected with empty vector (EV), HA-tagged GIMAP6WT (WT-3xHA), or HA-tagged GIMAP6G153V (G153V-3xHA) and probed for GIMAP6 and HSP90 (loading control). 1 = WT; 2 = mutant. (H) CHX assay using HEK293T cells stably expressing mCherry-tagged GIMAP6WT (WT) or mCherry-tagged GIMAP6G153V (G153V). The mCherry signal was normalized to 0 h to calculate the protein degradation ratio. ***, P < 0.001 (two-way ANOVA with the Geisser-Greenhouse correction). (I) GIMAP6 mRNA in HEK293T cells transduced with empty vector (EV), GIMAP6WT (WT), or GIMAP6W86* (W86*). ***, P < 0.001 (unpaired Student’s t test). (J) WBs of GIMAP6 and HSP90 (loading control) of HEK293T cells transduced with EV, WT, or W86*. Bars (E, H, and I) represent mean ± SEM. Data represent two (E) or three experiments (F–J).
Kindred 2 patients with p.Trp86Ter (W86*) had decreased GIMAP6 mRNA stability with no GIMAP6 protein expression in both HEK293T cells and the previous case report (Fig. 2, B–D, I, and J; Shadur et al., 2020).
GIMAP6 deficiency results in defective autophagy in T cells and interruption of GIMAP7 and GABARAPL2 interactions
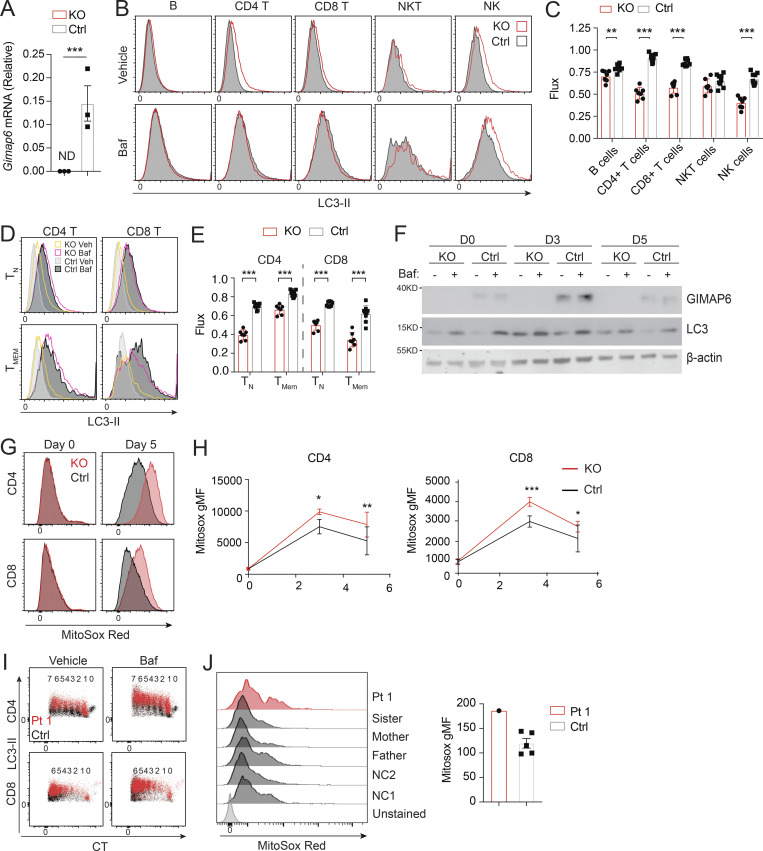
Previously, GIMAP6 was shown to localize to autophagosomes upon autophagy induction in mouse T cells, and Gimap6 conditional KO mouse lymphocytes had an autophagic defect (Pascall et al., 2013; Pascall et al., 2018). We, therefore, characterized a germline Gimap6 KO mouse strain (Gimap6−/−) newly available from the Knockout Mouse Project (KOMP; Austin et al., 2004; Dickinson et al., 2016). We first examined autophagic flux. Resting T cells have low basal autophagy that is upregulated by activation, so we assayed after TCR activation (Hubbard et al., 2010; Li et al., 2006). Activated Gimap6−/− CD4+ T cells showed increased LC3-II, indicating an accumulation of autophagosomes, mainly on day 5 after activation (Fig. 3, A and B). The accumulation of LC3-II+ autophagosomes could result from either increased biogenesis of autophagosomes or defective degradation of autophagosomes and LC3-II in the lysosomes. We, therefore, used bafilomycin A1 (Baf), which prevents lysosomal acidification and autophagosome–lysosome fusion, to measure autophagic flux (Mauvezin et al., 2015; Yamamoto et al., 1998). As expected, Baf treatment increased LC3-II in activated control cells (Fig. 3, A and B). In both Gimap6−/− CD4+ and CD8+ T cells, LC3-II+ autophagosomes accumulated prior to Baf treatment and did not further increase, indicating that autophagic flux was blocked perhaps during autophagosome maturation or autophagosome–lysosome fusion. Reduced autophagic flux was cumulative in Gimap6−/− T cells, peaking at 5 d after stimulation (Fig. 3 C). We also observed that ex vivo autophagic flux was significantly reduced in Gimap6−/− CD4+ and CD8+ T cells, B cells, and NK (natural killer) cells (Fig. S2, A–C). Since autophagy is important for memory T cell maintenance, we also examined autophagic flux in CD44− naive and CD44+ T cells and found that the flux was reduced in both naive and memory populations of Gimap6−/− T cells (Fig. S2, D and E). Given the prominent effects in CD4+ Gimap6−/− T cells, we carried out a time course of stimulation followed by a 2-h incubation with vehicle or 10 nM Baf and found that the GIMAP6 protein was induced and peaked at 3 d after stimulation, but was not affected by Baf treatment (Fig. S2 F). LC3 was present before Baf treatment in Gimap6−/− cells, whereas in WT cells, as expected, Baf caused LC3 protein accumulation. Also, this autophagy defect was intrinsic to hematopoietic cells using bone marrow chimeras (data not shown). Defects in autophagy can lead to the accumulation of mitochondrial reactive oxygen species (Mitosox; Filomeni et al., 2015), and indeed we found that Gimap6 KO CD4+ and CD8+ T cells showed increased Mitosox production (Fig. S2, G and H).
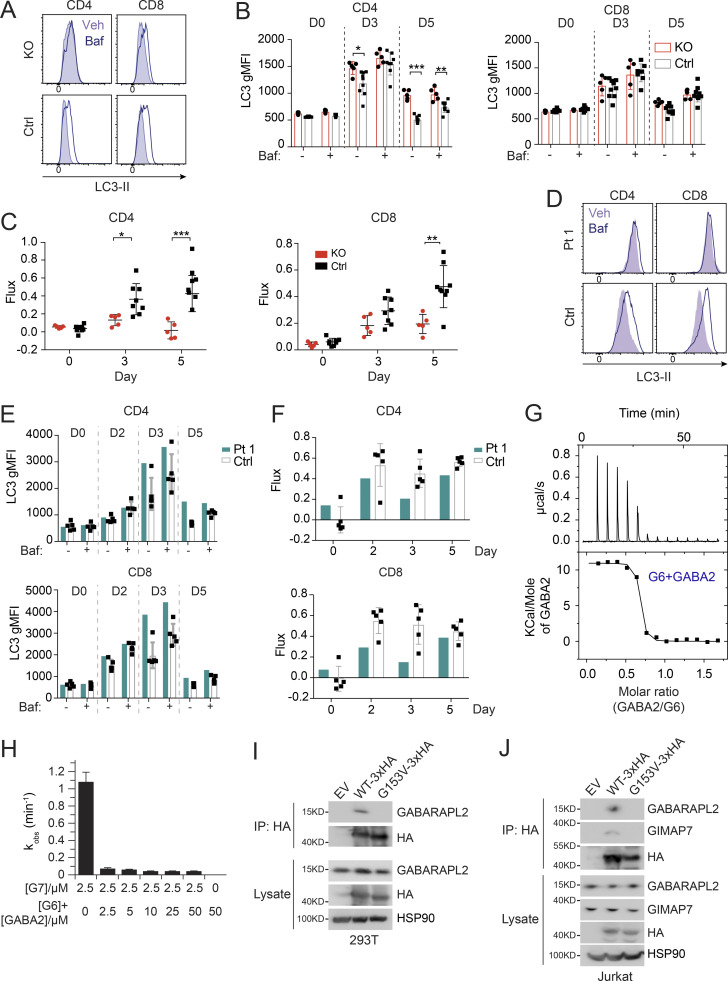
Figure 3.
Defective autophagy in T cells from Pt 1 and Gimap6−/− mice. (A) Whole splenocytes from Gimap6−/− (KO) or Gimap6+/+ (WT control, Ctrl) mice were treated with 1 μg/ml each of anti-CD3/CD28 for 3 d and then with 10 nM Baf or vehicle (Veh) for 2 h. Flow cytometry of LC3 expression (LC3-II) in CD4+ and CD8+ T cells. (B) Quantification of A for the indicated days. **, P < 0.01 (unpaired Student’s t test). nKO = 5; ncontrol = 8. (C) Autophagic flux of A, calculated as (gMFI LC3Baf − gMFI LC3Veh)/gMFI LC3Veh. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired Student’s t test). nKO = 5; ncontrol = 8. Shown is one of two independent repeats. gMFI, geometric MFI. (D) Flow cytometry histograms of LC3-II in human PBMCs from Pt 1 and healthy controls (Ctrl) stimulated with 1 μg/ml each of anti-CD3/CD28 for 3 d before treatment with Baf or vehicle (Veh) followed by flow cytometry and gating on CD4+ and CD8+ cells. (E) Quantification of gMFI of LC3-II of D as in B for the indicated days. (F) Quantification of autophagic flux of D as in C. Shown is one of two independent repeats and each dot indicates an individual donor in E and F. (G) 600 µM GABARAPL2 (GABA2) was titrated into 50 µM GIMAP6 (R134D; G6) and the resulting heat change was monitored in an ITC device. Data were fitted to a KD of 40 + −10 nM (n = 0.63 + −0.01). Shown is one of three experiments. (H) GTPase inhibition of 2.5 μM GIMAP7 (G7) by various concentrations of a 1:1 M GIMAP6 (G6)-GABARAPL2 (GABA2) complex. Shown is one of three experiments. (I) WB of HEK293T cells transfected with empty vector (EV), HA-tagged GIMAP6WT (WT-HA), or HA-tagged GIMAP6G153V (G153V-HA) for 1 d. GIMAP6WT and GIMAP6G153V were immunoprecipitated with HA tag antibody. The cell lysate and IP were probed for GABARAPL2, HA, and HSP90. Shown is one of three experiments. (J) WB of Jurkat cells transduced with lentivirus of empty vector (EV), HA-tagged GIMAP6WT (WT-HA), or HA-tagged GIMAP6G153V (G153V-HA) and selected with puromycin (1 µg/ml) for 4 d. GIMAP6WT and GIMAP6G153V were immunoprecipitated with HA tag antibody. The cell lysate and IP were probed for GABARAPL2, GIMAP7, HA, and HSP90. Shown is one of three experiments. Bars (B, C, E, F, and H) represent mean ± SEM.
Figure S2.
Defective autophagy in Gimap6−/− lymphocytes. (A) Quantitative PCR analysis of Gimap6 mRNA expression in T cells isolated from WT (Ctrl) and Gimap6−/− (KO) mice. ND, not detectable. Data represent three experiments. (B and C) Whole splenocytes from WT (Ctrl) and Gimap6−/− (KO) mice were treated with 100 nM Baf or an equal volume of vehicle (100% ethanol, Veh) for 2 h and then intracytoplasmically stained with antibody against LC3. (B) Representative flow plots of splenocytes gating on B cells (B220+), CD4 T cells (CD3+CD4+), CD8 T cells (CD3+CD8+), NK cells (NK.1+CD3−), and NKT cells (NK1.1+CD3+). (C) Quantification of autophagic flux (gMFI LC3Baf − gMFI LC3Veh)/gMFI LC3Veh. nKO = 7; nCtrl = 9. Data represent three experiments. (D) Representative flow plots showing LC3-II levels in naive (TN; CD44−) or memory (TMEM; CD44+) CD4 or CD8 T cells. (E) Autophagic flux as calculated in D. nKO = 7; nCtrl = 9. Data represent three experiments. (F) WB of enriched CD4 T cells activated for the indicated number of days (D) with 1 µg/ml of plate bound anti-CD3 and anti-CD28. On the day of harvest, cells were incubated for 2 h with vehicle (100% ethanol) or 10 nM Baf and then lysed in radioimmunoprecipitation assay buffer. Lysates were run on Bis-Tris gels and transferred onto PVDF membranes before immunoblotting with antibodies against mouse GIMAP6, LC3, and β-actin. Cells from one to three mice were pooled for each experiment. Shown is one representative experiment of three. (G and H) Whole mouse splenocytes were activated for 3 or 5 d with 1 µg/ml plate bound anti-CD3 and soluble anti-CD28 before staining with MitoSox Red for 15 min. (G) Representative flow plots showing MitoSox Red staining of CD4 and CD8 T cells. (H) Quantification of G. nKO = 4; nCtrl = 11. Shown is one of two experiments. (I) Representative flow plot showing LC3-II staining in proliferating Pt 1 and control (Ctrl) T cells activated for 3 d. The number of divisions each population has undergone is shown above, as indicated by dilution of CTV. Data represent three experiments. (J) Purified T cells from controls (Ctrl, including two NC and family members) and Pt 1 were activated for 15 d with Dynabeads Human T-Activator CD3/CD28 before staining with MitoSox Red for 15 min. Flow plots showing MitoSox Red staining (left) and quantification (right). Data represent three experiments. An unpaired t-test was used for A, C, E, and H (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bars (A, C, E, H, and J) represent mean ± SEM.
We next examined Pt 1 carrying the G153V mutation and found that the T cells of Pt 1 had an elevated basal LC3-II (Fig. 3, D and E). Similar to Gimap6−/− mice, G153V Pt 1 T lymphocytes had reduced autophagic flux (Fig. 3 F). LC3 turnover was decreased in Pt 1 T cells at all stages of proliferation (Fig. S2 I). Moreover, similar to Gimap6−/− mice, Pt 1 T cells had increased mitochondrial ROS production, suggesting oxidative stress (Fig. S2 J).
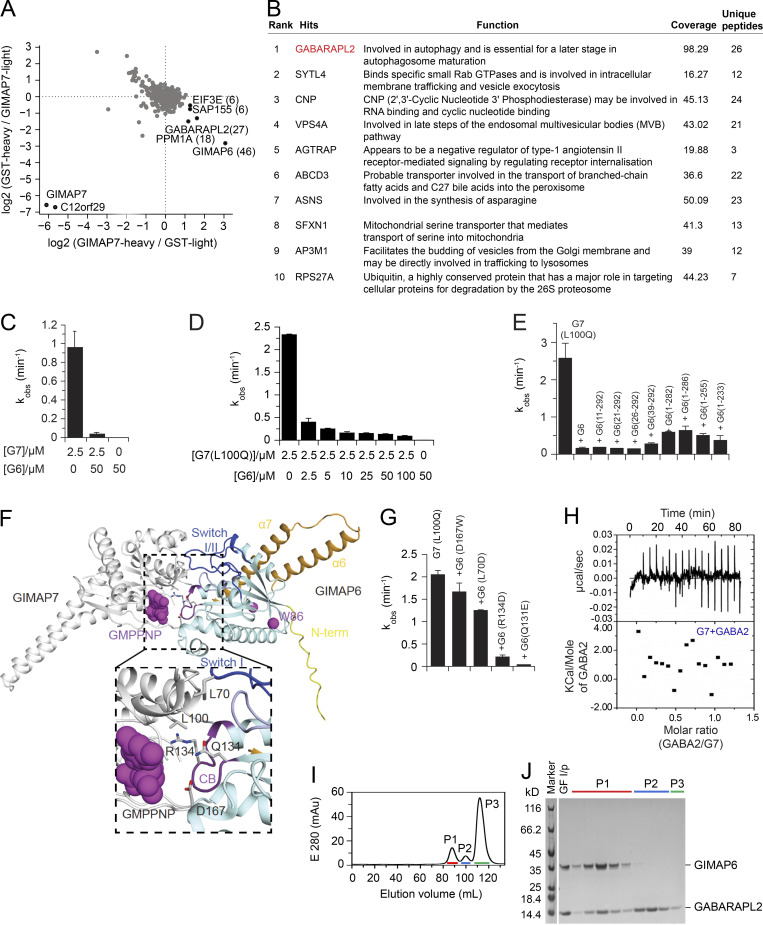
GIMAPs have been reported to form functional homo- and hetero-oligomers (Limoges et al., 2021; Schwefel et al., 2013). To understand how GIMAP6 could bind and regulate other GIMAPs, we screened for interaction partners through pull-down experiments using Jurkat cell lysates and stable isotope labeling using amino acids in cell culture (SILAC)–based mass spectrometry (MS) using different GIMAPs as bait (Ong and Mann, 2006; Paul et al., 2017). In this way, we identified GIMAP6 and GABARAPL2 as interactors of GIMAP7 (Fig. S3 A). A previous study showed that GIMAP6 interacts with GABARAPL2 (Pascall et al., 2013). Using a non-biased tandem affinity, co-immunoprecipitation (IP)/MS with GIMAP6 as bait in HEK293T cells yielded GABARAPL2 as the top hit (Fig. S3 B; Torres et al., 2009).
Figure S3.
GIMAP6, GABARAPL2, and GIMAP7 exist as functional complex. (A) SILAC based pull-down assays with GST-GIMAP7 (L100Q) or GST (control). The log2 fold changes of heavy to light ratio from forward and reverse experiments are plotted on the x and y axes, respectively. Proteins that had at least six identified peptides are plotted on the graph. The total number of peptides from specific interaction partners used for quantification in both forward and reverse experiments are indicated in brackets. Specific GIMAP7-interactors found in both experiments are located in the right lower corner. (B) Rank table of proteins with the highest overall coverage and unique peptides from immunoprecipitation of tandem-tagged GIMAP6 overexpressed in HEK293T cells followed by MS protein identification. The top ranked protein (GABARAPL2) is in red. (C) kobs for GTP hydrolysis of GIMAP6 (G6), GIMAP7 (G7), and the GIMAP6:GIMAP7 complex at the indicated concentrations at 20°C. (D) GTP hydrolysis of 2.5 μM GIMAP7 (L100Q) (G7 [L100Q]) in the presence of different GIMAP6 (G6) concentrations at 20°C. (E) GTP hydrolysis of 2.5 µM GIMAP7 (L100Q) upon addition of 50 µM of the N- or C-terminal deletion constructs of GIMAP6. In these experiments, the L100Q variant of GIMAP7 was used, since it has more stable aqueous solubility and can be purified at 10-fold higher yields than WT; in addition, it shows a two-fold increased GTPase activity compared to WT (Schwefel et al., 2013). (F) The AlphaFold 2 model of GIMAP6 was aligned on the GIMAP7 homodimer (PDB: 3ZJC) to obtain a model of the GIMAP6-GIMAP7 hetero-dimeric complex. GIMAP6 colors are as in Fig. 2 D. Switches I and II (blue), the P-loop (light blue), and the conserved box (CB, magenta) are involved in hetero-dimerization. Interface residues of GIMAP6 that were probed for GTPase interference of GIMAP7 in G and Leu100 in GIMAP7 are indicated. (G) GTP hydrolysis of 2.5 µM GIMAP7 (L100Q) upon addition of 50 µM of the indicated GIMAP6 mutants. (H) Isothermal titration calorimetry measurement indicate no binding of GABARAPL2 (GABA2) to GIMAP7 (G7). (I) Gel filtration run of the GIMAP6:GABARAPL2 complex on a Superdex200 column. (J) SDS-PAGE with selected fractions of the gelfiltration run in I. Peak 1 (P1) fractions were used for the subsequent GTP hydrolysis assays. GF I/P, protein applied to gel filtration. Bars (C–E and G) represent mean ± SEM. Data represent three experiments (C–E and G–J).
As in previous experiments (Schwefel et al., 2013), 2.5 µM GIMAP7 efficiently hydrolyzed GTP with an apparent catalytic rate constant (kobs) of ∼0.9 min−1 (Fig. S3 C). By contrast, GIMAP6 displayed no GTPase activity, even at 20-fold higher enzyme concentrations. In fact, isothermal titration calorimetry experiments revealed no binding of GTP to GIMAP6 (data not shown). Strikingly, when co-incubated, GIMAP6 inhibited the GTPase activity of GIMAP7 in a dose-dependent manner (Fig. S3, C and D), suggesting a regulatory function of the interaction. To characterize the structural requirements of GIMAP6 for inhibition, truncation constructs of GIMAP6 were prepared. Most N- and C-terminal deletion constructs of GIMAP6, leaving the GTPase domain intact, inhibited GIMAP7 as efficiently as full-length GIMAP6 (Fig. S3 E). Thus, the core GTPase domain of GIMAP6 contributes to GIMAP7 GTPase inhibition.
Using the published GIMAP7 homodimer as a template, we modeled the GIMAP6–GIMAP7 heterodimer (Fig. S3 F). GIMAP6 amino acids L70, Q131, R134, and D167 localize to the dimerization interface (Fig. S3 F). To validate our model, we individually mutated these interface residues, finding that GIMAP6 mutants Q131E and R134D still inhibited the GIMAP7 GTPase activity, whereas L70D and D167W significantly reduced GIMAP7 inhibition (Fig. S3 G). This indicates that the G-interface of GIMAP6 mediates assembly with GIMAP7.
We hypothesized that GABARAPL2 might interact indirectly with GIMAP7 via GIMAP6 (Pascall et al., 2013). In agreement with the SILAC pull-down assays, isothermal titration calorimetry measurements showed that GABARAPL2 interacted in a 1:1 complex with GIMAP6 (R134D) with high affinity (KD of 40 ± 10 nM; Fig. 3 G), whereas no interaction was detected for GABARAPL2 and GIMAP7 (L100Q; Fig. S3 H). To support the heterotrimeric complex hypothesis, a GTP hydrolysis assay was performed using GABARAPL2, GIMAP6, and GIMAP7. GIMAP6 and GABARAPL2 were purified as a complex from Escherichia coli (Fig. S3, I and J). Similar to isolated GIMAP6, the GIMAP6:GABARAPL2 complex effectively inhibited GIMAP7 GTPase activity (Fig. 3 H). To verify our hypothesis of the GABARAPL2, GIMAP6, and GIMAP7 complex and analyze the effect of the G153V mutation on the complex, we carried out a co-IP assay in both HEK293T and Jurkat T cells. The result showed that GIMAP6WT bound GABARAPL2 in HEK293T cells (GIMAP7 is undetectable in HEK293T cells; Fig. 3 I), and GABARAPL2 and GIMAP7 in Jurkat cells (Fig. 3 J). However, the mutation in GIMAP6G153V abrogated binding to GABARAPL2 (Fig. 3, I and J). These results further strengthen the conclusion that GIMAP6, GABARAPL2, and GIMAP7 exist as a functional heterotrimeric regulatory complex that is disrupted in Pt 1.
GIMAP6 deficiency leads to imbalanced lymphocyte homeostasis
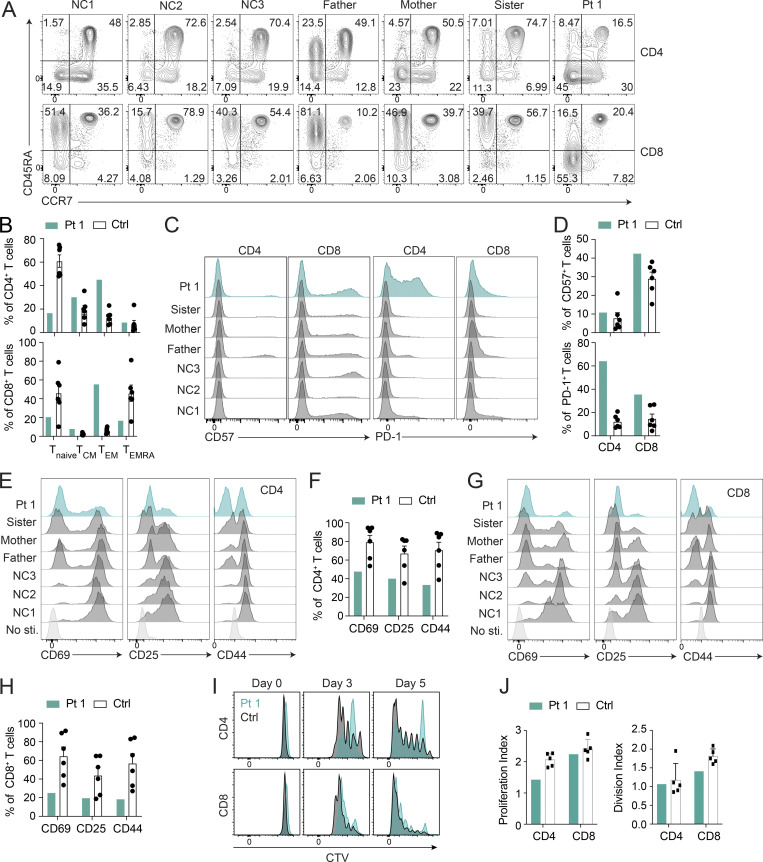
Autophagy is critical for T cell homeostasis (Botbol et al., 2016). Defective autophagy in T cells results in lymphopenia, compromised T cell memory, and reduced T cell proliferation (Pua et al., 2007; Puleston et al., 2014; Xu et al., 2014). Therefore, we examined Pt 1’s immunophenotype and found a dramatic skewing toward a CCR7−/CD45RA− memory-phenotype with a corresponding reduction of naive CCR7+/CD45RA+ CD4+ and CD8+ T cells (Fig. 4, A and B). Additionally, Pt 1 had increased levels of CD57 on CD8 T cells and a striking elevation of PD-1 on CD4 T cells, indicative of senescence and exhaustion, respectively, in those subsets (Agata et al., 1996; Brenchley et al., 2003; Palmer et al., 2005; Fig. 4, C and D). TCR activation, shown by the markers CD69, CD25, and CD44, was compromised in both CD4+ and CD8+ Pt 1 T cells (Fig. 4, E–H). Additionally, proliferation was moderately decreased, especially in CD4+ T cells (Fig. 4, I and J). These changes were consistent with the recent case report of immunodeficiency associated with a GIMAP6 loss of function mutation exhibiting an imbalance between naive and memory T cells along with no increase in CD4 and CD8 effector memory T (TEM) cells (Shadur et al., 2020). However, T cell exhaustion was not examined in that study (Shadur et al., 2020).
Figure 4.
Pt 1 lymphocyte surface phenotype and proliferation. (A) Flow cytometry plots of Pt 1, family, and NC samples showing naive (CCR7+CD45RA+), TCM (CCR7+CD45RA−), TEM (CCR7−CD45RA−), and effector memory re-expressing CD45RA (TEMRA; CCR7−CD45RA+) CD4+ and CD8+ T cells. (B) Quantification of A. Shown is one of two independent repeats and each dot is a different donor. (C) Flow cytometry showing CD57 and PD-1 expression on CD4+ and CD8+ T cells. (D) Quantification of C. Shown is one of two independent repeats and each dot is a different donor. (E and G) Flow cytometry showing CD69, CD25, and CD44 expression on CD4+ and CD8+ T cells after PBMCs were stimulated with 5 µg/ml each of plate-bound anti-CD3/CD28 for 24 h. (F) Quantification of E. (H) Quantification of G. Shown is one of two independent repeats. (I) Flow cytometry showing Cell Trace Violet (CTV) proliferation of CD4+ and CD8+ T cells stimulated with 1 µg/ml each of anti-CD3/CD28 for the indicated days. (J) Quantification of I. Bars (B, D, F, H, and J) represent mean ± SEM. Shown is one of two experiments, and each dot or square (n = 5−6) is a different donor in B, D, F, H, and J.
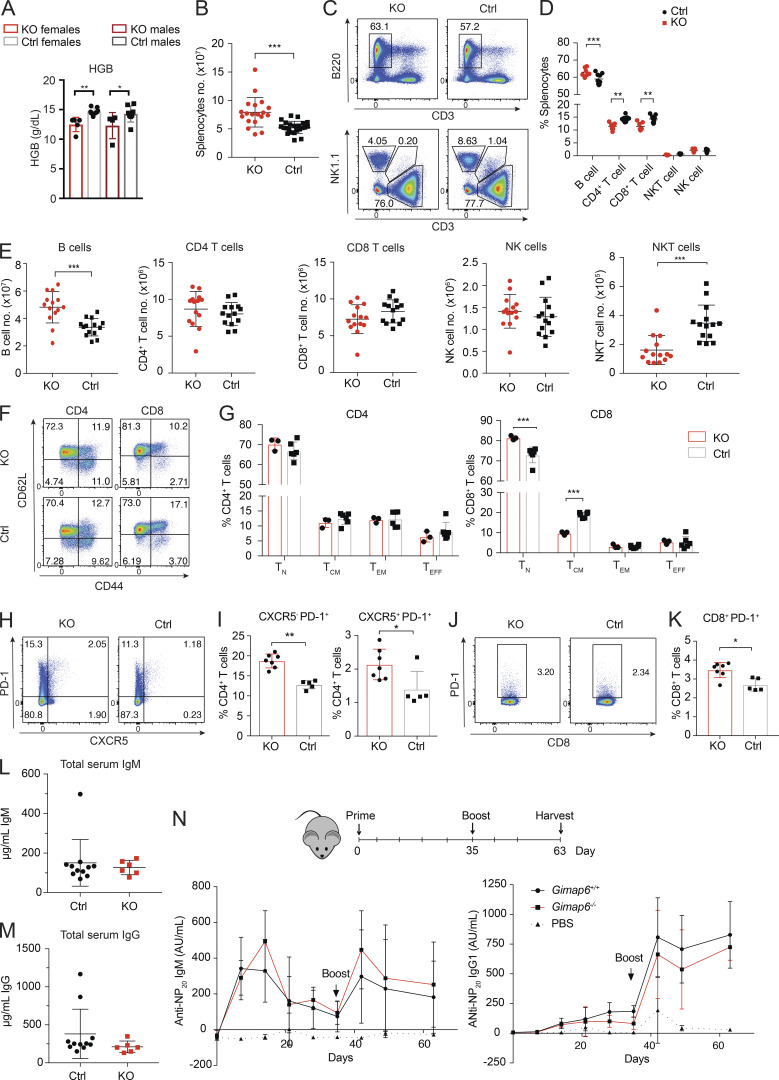
Except for Gimap4 and Gimap8, KO of individual Gimap genes in mice results in lymphopenia (Barnes et al., 2010; MacMurray et al., 2002; Pascall et al., 2018; Saunders et al., 2010; Schulteis et al., 2008), and a recently reported strain with a T and B cell ablation of Gimap6 (Gimap6fl/flCD2Cre) exhibited mildly decreased T cell numbers (Pascall et al., 2018). We observed that Gimap6−/− mice had anemia and greater splenic cellularity than littermates, with a slight increase in the B cell fraction and a corresponding reduction in the T cell fraction (Fig. S4, A–D). The absolute B cell numbers in the Gimap6−/− mice showed a mild increase in contrast to the reduced NKT and normal T cell numbers (Fig. S4 E). We found that naive and memory populations of CD4+ T cells were unaffected, but there was a mild expansion of naive CD8+ T cells and a reduction of CD8+ T central memory (TCM) cells in KO mice spleens (Fig. S4, F and G). PD-1 expression was increased on CD4+ T cells, irrespective of CXCR5 expression on CD8+ T cells (Fig. S4, H–K). IgM and IgG levels were normal (Fig. S4, L and M). We also tested T cell–dependent antibody responses to 4-hydroxy-3-nitrophenylacetyl-chicken γ globulin (NP-CGG) and found that KO mice responded equivalently to normal controls (Fig. S4 N). Thus, Gimap6−/− mice have mild derangements of immune phenotype but normal adaptive antibody responses under the conditions tested.
Figure S4.
Mouse immunophenotyping. (A) Whole blood was analyzed from 4-mo-old males and females using either the Pentraes 60 or Sysmex instruments and measured for hemoglobin content (HGB). nKO Female = 5; nControl Female = 7; nControl Male = 4; nKO male = 8. One of three experiments is shown. *, P < 0.05; **, P < 0.01. P values were calculated with an unpaired t-test. (B) Total splenocyte numbers of Gimap6−/− and control mouse. nKO = 19; nControl = 23. Three experiments were pooled. ***, P < 0.001. P values were calculated with an unpaired t-test. (C) Representative flow plots of B (B220+), NK (NK1.1+), T (CD3+), and NKT (NK1.1+/CD3+) splenocyte fractions from KO and control mice. nKO = 3; nControl = 6. One of three experiments is shown. (D and E) Quantitation of C for cell fraction (D) or for absolute cell count (E). Three experiments were pooled. (D) nKO = 7; nControl = 9. (E) nKO = 14; nControl = 14. **, P < 0.01; ***, P < 0.001. Mann-Whitney U test. (F) Flow cytometry dot plots of naive (TN; CD62L+CD44−), TCM (CD62L+CD44+), TEM (CD62L−CD44+), and effector (TEFF; CD62L−CD44+) CD4+ and CD8+ T cells. One of three experiments is shown. (G) Quantification of naive and memory populations in F. nKO = 3; nControl = 6. ***, P < 0.001. P values were calculated with an unpaired t-test. (H) CXCR5 and PD-1 expression in CD4+ T cells. One of three experiments is shown. (I) Quantification of H. nKO = 7; nControl = 5. *, P < 0.05; **, P < 0.01. Mann-Whitney U test. (J) PD-1 expression in CD8 T cells. One of three experiments is shown. (K) Quantification of J. nKO = 7; nControl = 5. *, P < 0.05. Mann-Whitney U test. (L and M) Total serum IgM (L) and total serum IgG (M) were analyzed by ELISA. nKO = 6; nControl = 11. (N) Mice were injected intraperitoneally with 50 µg NP-CGG in PBS 1:1 with Imject alum adjuvant and boosted 35 d later. PBS with alum was a negative control. Serum was analyzed for NP-specific IgM and IgG1 antibodies using an ELISA. Graphs show mean ± SD. AU, arbitrary units; NP, 4-Hydroxy-3-nitrophenylacetyl; NP-CGG, NP-chicken gamma globulin. nPBS = 3; nKO = 4; nControl = 8. One of two experiments is shown. Bars (A, B, D, E, G, I, K, and L–N) represent mean ± SD.
GIMAP6 deficiency changes global metabolism and lipid profiles
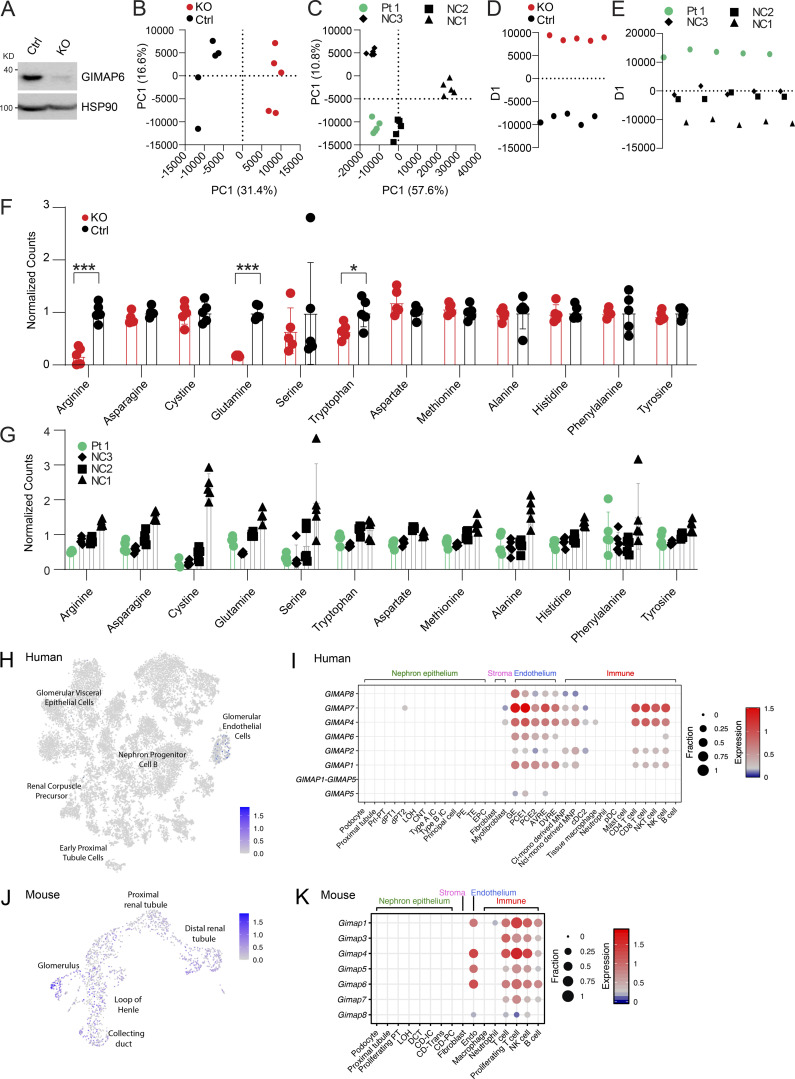
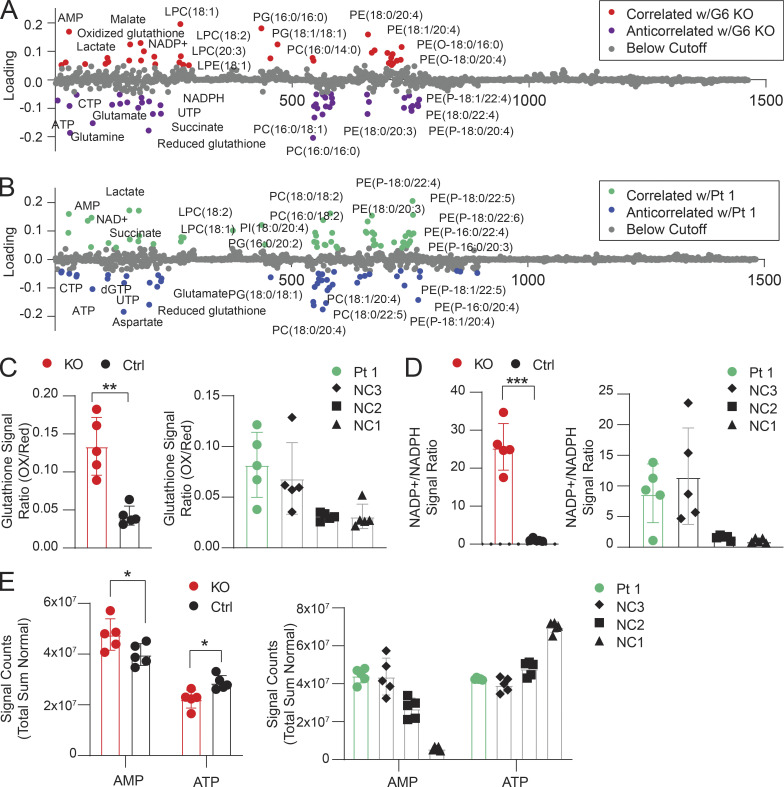
Lipids and metabolic processes regulate autophagy, and, conversely, autophagy defects affect lipid and basic metabolic circuits (Dall’Armi et al., 2013; Lahiri et al., 2019; Saito et al., 2019; Xie et al., 2020). Therefore, we carried out metabolomics and lipidomics analyses using multitargeted liquid chromatography MS (LCMS). By unbiased principal component analysis (PCA), Jurkat GIMAP6 KO (KO) samples were readily distinguishable from WT controls (Fig. S5, A and B). Healthy donor variability obscured patterns by PCA within the Pt 1 dataset (Fig. S5 C). To search for disease-associated patterns, especially given the changes in T cell subsets, patient datasets were analyzed using partial least squares discriminant analysis (PLS-DA) which revealed conserved changes in families of molecules that were correlated or anticorrelated with the primary axis of variance associated with GIMAP6 deficiency in both the Pt 1 and Jurkat datasets. These conserved patterns included positive correlations with lactate, malate, AMP, oxidized nicotinamide cofactors, lysophosphatidylcholine, and ether-linked phosphatidylethanolamine. Conserved negative correlates included nucleotide triphosphates, succinate, reduced glutathione, and phosphatidylcholine phospholipids (Figs. 5, A and B; and S5, D and E). In agreement with these conserved patterns, we found that KO cells exhibited a shift toward oxidation in redox metabolites including glutathione and nicotinamide adenine dinucleotide phosphate (NADPH; Fig. 5, C and D). Pt 1 T cells showed remarkably similar trends, although there was an overlap with the range for control (NC) cells (Fig. 5 D). GIMAP6 deficiency also disrupted energy homeostasis as reflected by a positive correlation in both datasets with AMP levels and a negative correlation with multiple nucleotide triphosphates (Fig. 5, A, B, and E). Amino acid pools were also affected, with GIMAP6-deficient Jurkat cells showing significant depletion of arginine, glutamine, and tryptophan (Fig. S5 F). In Pt 1 samples, amino acid levels trended uniformly lower, with arginine, cysteine, and serine being especially reduced (Fig. S5 G).
Figure S5.
Metabolite and lipid study, and kidney specific scRNA analyses. (A) WB analysis of GIMAP6 and HSP90 (control) protein expression in Cas9 stable expressing Jurkat cells treated with control (Ctrl) and GIMAP6 (KO) sgRNA. Shown is one of three experiments. (B–E) Multivariate analysis of metabolomic and lipidomic data separates Pt 1 and GIMAP6 KO samples from controls. (B) Unbiased PCA of the combined metabolomic and lipidomic datasets for Jurkat KO and Ctrl samples. PCA utilized nine principal components to account for variance and the first two components are shown. Percentage represents the percent variance accounted for by that component. (C) PCA analysis of Pt 1 and NC samples. 19 principal components were generated of which the first two are shown. (D) PLS-DA of the Jurkat dataset separated on a single axis of variance. (E) PLS-DA of the Pt 1 dataset with NC samples grouped together to identify features correlated with Pt 1 samples. All analysis was performed in MarkerView. (F and G) Signal levels of specific amino acids in both the KO Jurkat model and Pt 1 cells. (F) Normalized amino acid signals from targeted LCMS/MS analysis of metabolites from GIMAP6 KO and control (Ctrl) Jurkat cells. All levels are normalized to the mean of the control set for display. Bars represent mean ± SD. (G) Normalized amino acid signals for Pt 1 and NCs. All levels are normalized to the mean signal across all normal controls. Bars represent mean ± SD. (H) t-SNE showing GIMAP6 expression in 20,425 human kidney cells obtained from a previously described study (Hochane et al., 2019). (I) scRNA-seq of human kidney was obtained from a previously described study (Young et al., 2018). The expression of GIMAP family genes in the indicated cell populations are shown. Dot size (fraction) shows the percentage of GIMAP-expressing cells, and color scale bar (expression) shows the expression intensity. (J) UMAP projection showing Gimap6 expression in murine kidney cells obtained from 10x Genomics. (K) scRNA-seq of mouse kidney was obtained from a previously described study (Park et al., 2018). The expression of Gimap family genes in the indicated cell populations are shown. Dot size (fraction) shows the percentage of Gimap-expressing cells, and color scale bar (expression) shows the expression intensity. Data represent three experiments (B–G). P values were calculated with an unpaired t-test in F (*, P < 0.05; ***, P < 0.001).
Figure 5.
Metabolic and lipidomic changes in Pt 1 T cells and GIMAP6-deficient Jurkat cells. (A) PLS-DA of molecular correlates (red) and anticorrelates (purple) of GIMAP6-deficient (G6 KO) and WT Jurkat T cells. (B) Pt 1 T cells compared to control T cells. Features with loading scores >20% of maximum were colored as correlated (green) or anticorrelated (blue) with GIMAP6 deficiency. LPE, lysophosphatidylethanolamine; LPC, lysophosphatidylcholine; PG, phosphoglycerol; PC, phosphotidylcholine; PE, phosphotidylethanolamine. (C) Ratios of the signals of oxidized to reduced glutathione. (D) Ratio of the normalized signals of oxidized to reduced NADPH. (E) Total sum normalized signals for AMP and ATP. n = 5 for all groups. Bars (C–E) represent mean ± SD. Data represent three experiments (A–E). An unpaired t-test was used for C–E (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Correlations between the Jurkat KO model and Pt 1 samples extended into the lipidome. We found that GIMAP6 deficiency increased multiple lysophospholipids associated with cellular stress and T cell activation (Fig. 5, A and B; Asaoka et al., 1992). Furthermore, GIMAP6 loss disrupted families of both plasmenyl and plasmanyl ether–linked phosphatidylethanolamine, especially pools containing long-chain (≥C20) polyunsaturated fatty acids (PUFAs; Fig. 5, A and B). Although individual species were differentially affected in the Jurkat and Pt 1 datasets, we observed broad perturbations of both plasmenyl and plasmanyl. Because the LCMS methodology does not resolve fatty acid isomers, the exact identity of the PUFAs needs to be further defined, but the changes observed were likely limited to immune active long-chain ω3 and ω6 fatty acid metabolites.
Gimap6-deficient mice exhibit kidney disease and increased mortality
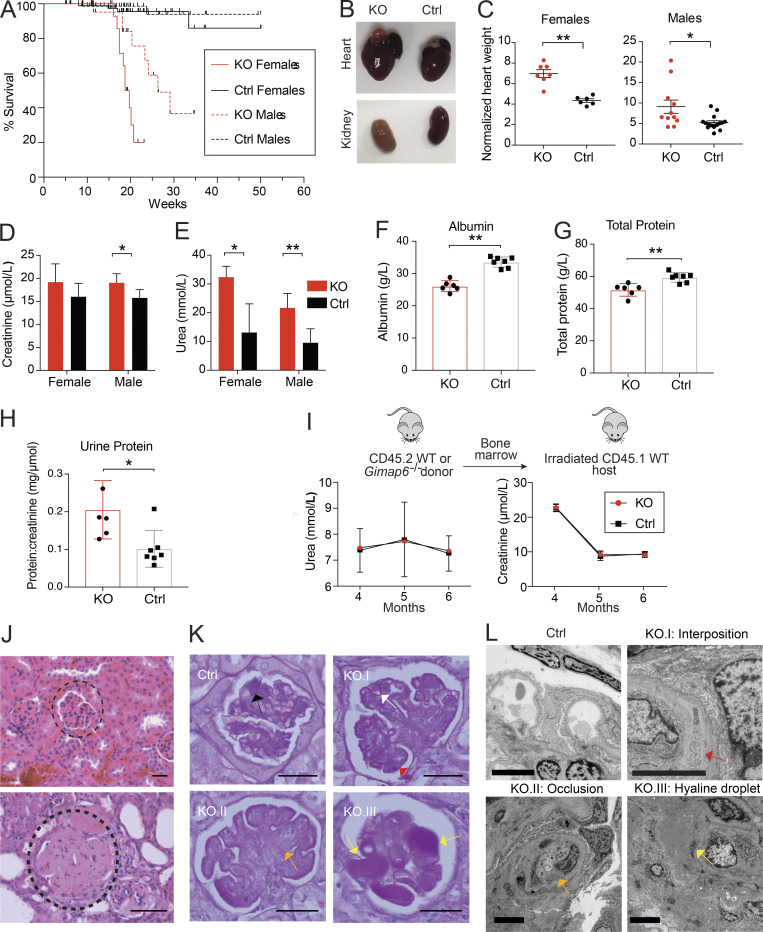
While carrying the Gimap6−/− (KO) mouse line, we unexpectedly observed a severely shortened lifespan, particularly in females (Fig. 6 A). This contrasted with the previous conditional, CD2 regulated, Gimap6 lymphocyte KO mouse strain that had normal longevity (Pascall et al., 2018). Gross pathology of the deceased KO mice showed enlarged hearts, pale, scarred kidneys, pulmonary edema, and splenomegaly, but only minor abnormalities in other organs (Fig. 6, B and C; and Table S2). KO mice had elevated plasma creatinine and urea, reduced serum albumin and total protein, and proteinuria indicative of renal failure (Fig. 6, D–H). Thus, we investigated the expression of GIMAP6 in kidney tissue. As previously found, GIMAPs are primarily expressed in immune tissues and lymphocytes, but further single-cell analysis of both mouse and human kidney cell subsets revealed selective expression of GIMAP6 in endothelial cells as well as in kidney-localized lymphocytes (Fig. S5, H–K; Park et al., 2018; Stewart et al., 2019; Young et al., 2018). To investigate the contribution of lymphocytes and endothelial cells to the kidney disorder, we performed a hematopoietic cell transplant experiment. We found the transplantation of KO hematopoietic cells into WT mice did not cause kidney disease (Fig. 6 I). Correspondingly, the transplantation of WT hematopoietic cells into KO hosts could not protect against kidney disease (data not shown). Conclusions from the latter data are provisional since irradiation can cause renal endothelial damage, but together, our data indicate that non-hematopoietic loss of GIMAP6 in the kidney itself caused sickness and death. H&E staining of select major organs confirmed that the most consistently and severely damaged organs were the kidneys with focal segmental glomerulosclerosis (Fig. 6 J and Table S2). Periodic acid–Schiff (PAS) staining showed that KO glomeruli had glomerular capillary wall thickening and duplication, mesangiolysis, and hyaline deposits (Fig. 6 K). Transmission electron microscopy of glomeruli revealed cellular interposition between basement membranes, occlusion of capillaries, and hyaline droplets, though fibrin thrombi were not observed (Fig. 6 L). Thus, glomerular microangiopathy causes renal disease and death in KO mice.
Figure 6.
Gimap6−/− mice have reduced survival due to microangiopathic renal disease. (A) Kaplan-Meier survival curve in Gimap6−/− (KO) and controls (Ctrl) pooled from WT and heterozygous mice. Culled mice were censored at death. nKO females = 33; nKO males = 35; nControl females = 99; nControl males = 80. Females: P < 0.0001; Males: P < 0.0005 (log-rank Mantel-Cox test). (B) Gross anatomy of hearts (upper) and kidneys (lower) in KO and Ctrl mice. (C) Normalized heart weight (mg heart/g body weight) in females (mean ± SEM, left, nKO = 6; nControl = 6) and males (right, nKO = 11; nControl = 15). (D) Plasma creatinine in 9–10-wk-old mice. mean ± SD, nKO Female = 5; nControl Female = 6; nControl Male = 8; nKO male = 8. (E) Plasma urea in 9–10-wk-old mice. mean ± SD, nKO Female = 5; nControl Female = 6; nControl Male = 8; nKO male = 8. (F) Serum albumin in 9–10-wk-old mice. mean ± SD, nKO = 6; nControl = 7. (G) Total protein in 9–10-wk-old mice. mean ± SD, nKO = 6; nControl = 7. (H) Urine protein in 9–10-wk-old mice (mean ± SD, nKO = 5; nControl = 7). (I) Bone marrow from CD45.2 Gimap6−/− (KO) or littermate controls was transferred to lethally irradiated CD45.1 WT mice. Top: Experimental scheme. Bottom: Plasma urea and creatinine after transfer (mean ± SD; n = 10 for both groups). (J) Photomicrographs (400×) of H&E-stained sections from Ctrl (top) and KO mouse (bottom) kidneys. Dotted curve encircles a glomerulus. Shown is one of three experiments. Scale bar = 20 μm. (K) Photomicrographs of glomeruli with PAS staining. Ctrl shows normal mesangium (black arrow). KO.I shows glomerular capillary wall thickening (white arrow) and duplication (red arrow). KO.II shows mesangiolysis (orange arrow). KO.III shows glomerular hyaline deposits (yellow arrows). nKO = 7; nControl = 5. Scale bar = 20 μm. (L) Representative transmission electron microscopy images of glomeruli from Ctrl and KO mice. Ctrl shows a normal glomerulus. KO.I shows cellular interposition (red arrow) between the basement membranes. KO.II shows capillary lumen occlusion (orange arrow). KO.III shows hyaline droplets (yellow arrows). nKO = 3; nControl = 1. Scale bar = 5 μm. An unpaired t-test was used for C–H (*, P < 0.05; **, P < 0.01).
GIMAP6 is highly expressed in innate immune cells and is IFN-γ inducible
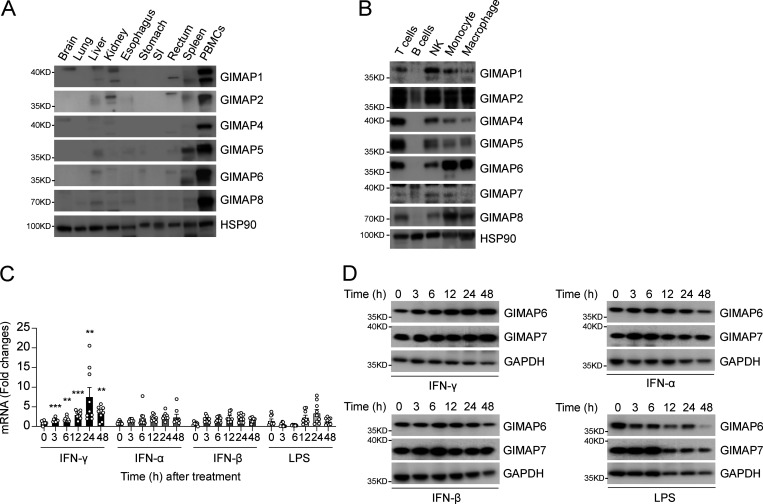
GIMAP family genes encode conserved homologous GTP-binding proteins mainly studied in the immune system (Ciucci and Bosselut, 2014). To confirm these previous results, we first examined individual GIMAPs in human tissues. All of the GIMAP family proteins showed high expression in peripheral blood mononuclear cells (PBMCs), with less consistent and significantly lower expression in the spleen or other tissues (Fig. 7 A). We also evaluated the different GIMAP family proteins in isolated immune cell types from PBMCs, which showed that all were highly expressed in T cells, NK cells, and innate immune cell populations such as monocytes and macrophages but were generally low or absent in B cells (Fig. 7 B). GIMAP7 appeared to have two isoforms: a lower molecular weight size that predominates in T and NK cells and a higher molecular weight form in B cells, monocytes, and macrophages. GIMAP6 was notably high in monocytes and macrophages, suggesting a role in innate immunity. Hence, we examined whether GIMAP6 was inducible by bacterial pathogen-associated molecular patterns or antibacterial/viral cytokines in PMA-induced macrophages derived from the THP-1 cell line. Unexpectedly, we found that GIMAP6 mRNA and protein, and to a lesser extent the slower migrating GIMAP7 protein, could be strongly induced by type II IFN-γ, but not type I IFN-α and IFN-β, or LPS, further implying that the GIMAP6:GIMAP7 complex may have critical functions in antibacterial activity in innate immune cells (Fig. 7, C and D).
Figure 7.
GIMAP6 is expressed in immune cells and is inducible by IFN-γ stimulation. (A) WB analysis of GIMAP family members and HSP90 (control) proteins in the indicated tissues from healthy donors. SI, small intestine. Shown is one of two experiments. (B) WB analysis of GIMAP family members and HSP90 (control) proteins in the indicated immune cell populations isolated from human PBMCs. Shown is one of four experiments. (C) Quantitative PCR analysis of fold-change of GIMAP6 mRNA expression over unstimulated condition in THP-1 differentiated macrophages after stimulating with IFNs (100 ng/ml) or LPS (1 µg/ml) for the indicated hours. Data are pooled from three experiments. Bars represent mean ± SEM. An unpaired t-test was used (**, P < 0.01; ***, P < 0.001). (D) WB analysis of GIMAP and GAPDH (control) proteins in THP-1 cells after stimulating with IFNs (100 ng/ml) or LPS (1 µg/ml) at the indicated times. Shown is one of three experiments.
Loss of Gimap6 results in increased host susceptibility to pathogen infections
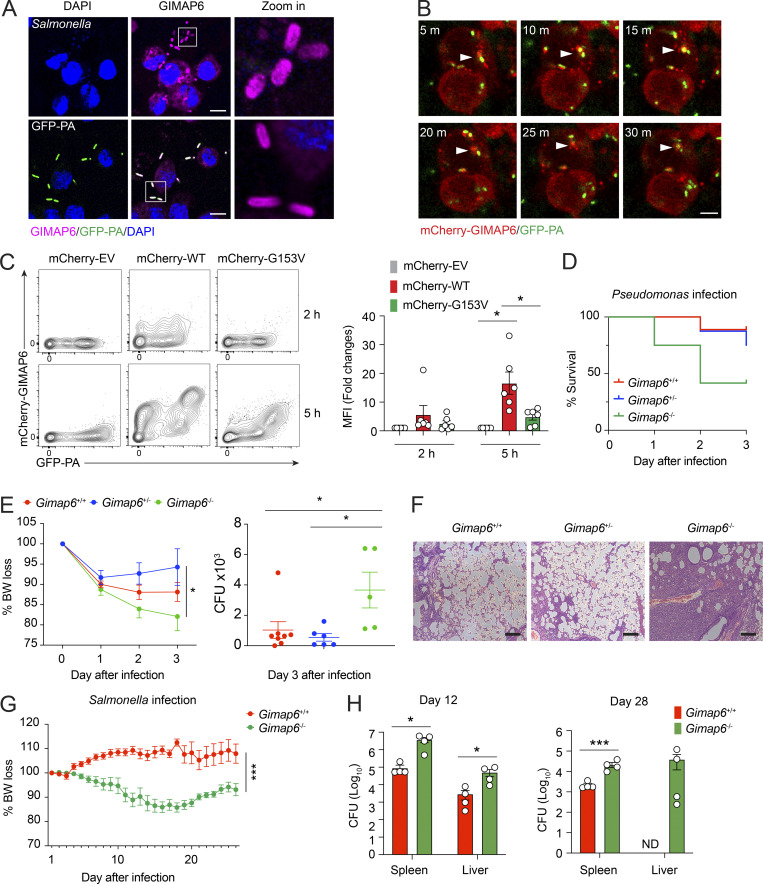
To investigate potential antibacterial activity, we examined whether GIMAP6 colocalized with pathogens as observed with other dynamin-related G proteins (Tretina et al., 2019). Pt 1 has experienced severe Pseudomonas aeruginosa pneumonia. Therefore, we used P. aeruginosa expressing the GF (GFP-PA) compared with the bacterial strain Salmonella typhimurium to infect THP-1–derived macrophages and found that both Salmonella and GFP-PA were coated with GIMAP6 (Fig. 8 A). Moreover, mCherry-tagged GIMAP6 was recruited to GFP-PA prior to bacterial degradation (Fig. 8 B and Video 1). Flow cytometry showed that GIMAP6G153V from Pt 1 bound poorly to GFP-PA compared with WT GIMAP6 (Fig. 8 C). These data reveal a potential defect in the antibacterial function of the GIMAP6G153V allele.
Figure 8.
GIMAP6 co-localizes with pathogenic bacteria and plays a role in antibacterial activity. (A) Confocal photomicrographs of immunostaining GIMAP6 and Salmonella (upper panel) or GFP-Pseudomonas (GFP-PA; lower panel) in differentiated THP-1 cells. Zoom in (right) is a 6× enlargement of the boxed region. Scale bar, 10 μm. Shown is one of three experiments. (B) Time-lapsed live cell confocal microscopy imaging of mCherry-tagged GIMAP6 (mCherry-GIMAP6) and GFP-PA in THP-1 differentiated macrophages. Images taken at 5 min (m) intervals. Arrow shows double-stained bacteria. Scale bar, 5 μm. Shown is one of two experiments. (C) Flow cytometry analysis of mCherry-GIMAP6 and GFP-PA in THP-1 differentiated macrophages transduced with WT or G153V mCherry-GIMAP6 lentivirus after infection with GFP-PA at 2 or 5 h. Statistics for mCherry-GIMAP6 MFI on GFP-PA (right). Data are pooled from three experiments. An unpaired t-test was used (*, P < 0.05). (D) Percentage of mouse survival after P. aeruginosa infection. Data are pooled from two out of three experiments. Gimap6+/−, n = 8; Gimap6+/+, n = 9; Gimap6−/−, n = 12. (E) Average body weight loss (BW loss %; left panel) and quantified CFUs of P. aeruginosa bacteria extracted from total lung tissue from surviving mice on day 3 after infection (right panel) of the same experiment. Gimap6+/−, n = 8; Gimap6+/+, n = 9; Gimap6−/−, n = 12. *, P < 0.05 (two-way ANOVA with the Geisser–Greenhouse correction). Data are pooled from two out of three experiments. (F) Photomicrographs of H&E-stained lung sections from WT and Gimap6−/− mice on day 3 following infection with 5 × 108 CFU of P. aeruginosa. Scale bar, 100 μm. Shown is one of three experiments. (G and H) Average body weight loss (BW loss %; G) and CFUs of Salmonella bacteria extracted from total spleen and liver tissues (H) of infected WT and Gimap6−/− mice. Gimap6+/−, n = 8; Gimap6−/−, n = 8. ***, P < 0.001 (two-way ANOVA with the Geisser-Greenhouse correction) in G. Gimap6+/−, n = 4; Gimap6−/−, n = 4. *, P < 0.05, ***, P < 0.001 (an unpaired t-test) in H. Bars (C, E, G, and H) represent mean ± SEM. Data represent three experiments.
Video 1.
Co-localization of mCherry-GIMAP6 and GFP-PA after infection. Time: 5 min. Scale bar: 4 µm.
We hypothesized that GIMAP6 surface binding plays a critical role in pathogenic bacteria clearance and tested the response of Gimap6 deficient mice (Gimap6−/−) to P. aeruginosa infection. WT and Gimap6−/− mice were inoculated with 5 × 108 CFUs of P. aeruginosa via nasal gavage. We observed an ∼10% mortality rate in WT (Gimap6+/+) and heterozygous (Gimap6+/−) mice, but KO (Gimap6−/−) mice had a 25% mortality rate one day after infection and a 60% mortality rate 3 d after infection (Fig. 8 D). Gimap6−/− mice showed increased weight loss and greater P. aeruginosa CFU load in the lung than WT mice, indicating defective bacterial clearance (Fig. 8 E). Microscopy indicated severe pathological changes with air sacs obliterated by lymphocyte infiltration in the lungs of Gimap6−/− mice (Fig. 8 F). We also infected Gimap6−/− mice with S. typhimurium, another widely used model pathogen. Following infection, Gimap6−/− mice exhibited more significant body weight loss and increased CFU load in the spleen and liver than WT mice (Fig. 8, G and H). Thus, our data illustrate that GIMAP6 binds to pathogenic bacteria, marking and licensing bacteria for clearance, thus participating in antibacterial innate immunity.
Discussion
GIMAPs are highly conserved septin-related GTP-binding proteins that regulate intracellular processes critical for cell function and survival (Krucken et al., 2004; Liu et al., 2008; Nitta and Takahama, 2007; Poirier et al., 1999). Septin/dynamin GTP-binding proteins have diverse roles in cytoskeletal organization, cell division, and membrane remodeling processes throughout phylogeny (Obar et al., 1990; Praefcke and McMahon, 2004). Like septin/dynamins, human and mouse GIMAPs function by forming higher-order structures (Sirajuddin et al., 2007). The GTPase activity is not regulated by GTPase-activating proteins and guanine nucleotide exchange factors but rather by homo- and hetero-oligomerization (Schwefel et al., 2013). Surprisingly, GIMAP6 alone had negligible GTPase activity under the conditions tested but was specifically associated with GIMAP7 and inhibited its catalytic activity. The relative composition of GIMAP7 complexes could shift from active to inactive as GIMAP6 is increased by IFN-γ or other inducers.
Autophagy is an evolutionarily conserved recycling pathway essential for immune cell responses (Clarke et al., 2018; Riffelmacher et al., 2018; Zhao and Zhang, 2019). We find that the complex of GIMAP6 and GIMAP7 binds GABARAPL2. Human GIMAP6 showed a stronger binding with GABARAPL2 than mouse GIMAP6, which is achieved through a C-terminal sequence of GIMAP6 present in most species but absent from mouse (Pascall et al., 2013). Together, GIMAP6:GABARAPL2 move to the autophagosome which could release active GIMAP7, which is mainly in the cytoplasm. Tethering to the autophagosome via GABARAPL2 may help clear GIMAP6-coated pathogens through xenophagy (Kimmey and Stallings, 2016).
Metabolomics and lipidomics showed that GIMAP6 loss caused energy imbalance, increased oxidative stress, and reduced pools of specific amino acids including glutamine, arginine, and tryptophan, possibly due to defective autophagy (Zhang et al., 2018). We detected lysophosphatidylcholine, phosphatidylcholine, and PUFA-containing ether-linked PE lipids imbalances, possibly reflecting the cellular response to stress and activation. Long-chain fatty acids can induce autophagy, so the changes in PUFA pools reflected in ether-linked PE lipids may be compensatory for the autophagy defect (Kim et al., 2018). PUFAs are also known to be sinks for intracellular ROS, and the absence of autophagy in T cells can lead to the accumulation of dysfunctional mitochondria, ROS overproduction, and cell death. Defective autophagy can cause lymphopenia followed by homeostatic proliferation with increased “virtual memory” T cells (Puleston et al., 2014). The observed shift in redox toward a more oxidized and energy-poor state in both Pt 1 and Jurkat models may derail ROS-triggered autophagy (Filomeni et al., 2015; Zhang et al., 2016). Hence, GIMAP6 loss broadly alters a metabolite and lipid network affecting autophagy, redox control, and energy metabolism. This comports with the dysregulated ROS and ROS byproducts in autoimmune conditions (Griffiths, 2005).
GIMAP6 has a tentative link to lipophagy and lipid droplets through its interaction with GIMAP7 (O’Neill and Pearce, 2016; Schwefel et al., 2013). Lysophosphatidylcholine (lyso-PC), a regulator of T cell activation and survival (Asaoka et al., 1992; Sakata-Kaneko et al., 1998), was elevated in both the KO cell lines and patient samples. In Jurkat cells, lyso-PC can stimulate ROS generation (Im et al., 2006). In addition, PUFA-containing ether-linked PE lipids are key precursor storage pools for generating eicosanoids and docosanoids, which could affect immunity (Lone and Tasken, 2013). The dysregulation of PUFAs and PUFA-derivatives has been associated with autoimmune diseases, and supplementation with the ω3 fatty acid eicosapentaenoic acid has been studied as a treatment for autoimmune vasculitis (Hirahashi et al., 2014). Thus, the phenotype of Pt 1 may be due, in part, to the metabolic and lipid defects we have discovered.
A potentially life-threatening aspect of Pt 1’s disease is pulmonary hypertension. This is observed in immunodeficiency diseases and other GIMAP deficiencies (Drzewiecki et al., 2021; Johnston et al., 2004). For GIMAP6, recurrent lung infections may cause aberrant lung vasculature leading to hypertension. However, for GIMAP5, portal hypertension is attributable to endothelial abnormalities. Furthermore, GIMAP6 is expressed in kidney endothelial cells perhaps causing hypertension (Corban et al., 2017). Thus, there may be key roles of GIMAP6 in selected cell-types outside of the immune system, suggesting an important new horizon for understanding GIMAPs. The new Gimap6 KO mice developed severe anemia, an enlarged heart, autophagy defects, and premature mortality due to kidney disease. Most striking was severe glomerulosclerosis and kidney failure due to an intrinsic kidney defect. Presently, human patients have a normal renal function, which may relate to the fact that human kidney endothelial cells have less GIMAP6 than mice.
Materials and methods
Whole-exome sequencing (WES)
Pt 1 provided written informed consent for enrollment in research protocol (06-I-0015) that was approved by the Institutional Review Board of the National Institute of Allergy and Infectious Diseases (NIAID) and National Institutes of Health (NIH). Patient and control specimens were obtained under approved protocols. WES was carried out as previously described (Zhang et al., 2014). In short, genomic DNA was isolated from whole blood. WES was performed using the SureSelect Human All Exon 50 Mb Kit (Agilent) followed by HiSeq next-generation sequencing (Illumina). An in-house custom analysis pipeline was then used to filter and prioritize candidates. Variants were first filtered based on the genetic model (de novo, homozygous recessive, and compound heterozygous). Variants that failed the quality call, had genotype quality <50, were non-synonymous, or had allele frequency >0.001 were excluded. The human exome and genome sequencing data of Pt 1 and Pt 1’s family has been deposited at dbGaP as phs002816.v1.p1.
Variants prioritization for WES analysis candidates
We performed WES for Pt 1, healthy parents, and two unaffected siblings. Since both parents are healthy and have a consanguineous relationship, Pt 1 most likely has a recessive inherited disease caused by a homozygous variant. Thus, we used variant filtering for rare homozygous variants in Pt 1, along with heterozygous genotypes in both parents and the two unaffected siblings. The autosomal recessive genetic model filtering eventually yielded eight homozygous variants in Pt 1. Among them, NINL, HSPG2, ABCA12, and FLYWCH1 were present with MAF around 0.1%, reflecting a reasonable number of heterozygous individuals in the public population. The other four variants are novel while C4orf45 is an open reading frame without any known biological function; CHIA is mainly expressed in internal tissues including rectum/prostate/stomach; AGO1 has a relatively homogeneous expression pattern across different tissues and a general biological function regarding association with small RNAs, RNA interference, and RNA silencing. GIMAP6 remained the top candidate based on these additional factors: (1) GIMAP6 has a specific high expression in the targeted immune-related tissues and cells; (2) Gimap6 KO mouse model has an immune system and a hematopoietic system phenotype similar to Pt 1’s phenotype; (3) GIMAP6 and GIMAP GTPase family genes, in general, are known to have immune-mediated biological functions from previous literature (see Fig. S1 B).
Processing of human whole blood
PBMCs were isolated by Ficoll–Paque PLUS (GE Healthcare) density-gradient centrifugation, and RBCs were lysed with ammonium–chloride–potassium lysing buffer (Thermo Fisher Scientific). Cells were resuspended in complete RPMI (cRPMI) with 10% FBS, 2 mM glutaMAX (Gibco), 50 mM 2-mercaptoethanol, 20 mM HEPES, and 100 U/ml each of penicillin and streptomycin at 1 × 106 cells/ml.
Sanger sequencing
Genomic DNA was extracted from blasting T cells using QuickExtract DNA extraction solution (Epicentre). The region of interest of GIMAP6 was amplified using KOD Hot Start DNA Polymerase (EMD Millipore) and the primers: 5′-TGCTCCTGGTGACACAACTG-3′ (forward) and 5′-GATCTGGGACAGTCCTTCCA-3′ (reverse) by Applied Biosystems Genetic Analyzer.
Mouse strains and husbandry
The B6N(Cg)-Gimap6tm1b(KOMP)Wtsi/J were created by the KOMP (Abeler-Dorner et al., 2020; Austin et al., 2004; Dickinson et al., 2016; Skarnes et al., 2011). The mice were created by the insertion of a “knockout-first” trapping cassette and a promoter-driven selection cassette downstream of exon 1 of Gimap6 and flanking of exons 2 and 3 with loxP sites (Austin et al., 2004; Skarnes et al., 2011; Testa et al., 2004). CD45.1 B6.SJL was purchased from the University of Oxford Department of Biomedical Services or Charles River Laboratories. Mice were housed in pathogen-free facilities and given food and water ad libitum. Mice were used between 8 and 35 wk of age. All animal works performed at the NIH were carried out under a protocol approved by the NIAID Animal Care and Use Committee. All animal works performed at the University of Oxford were approved by the local ethical review committee and performed under UK project license 30/3388.
Mouse samples
Splenocytes were isolated by pressing spleens through a 70-μm cell strainer (Thermo Fischer Scientific) with a syringe plunger into cRPMI. After centrifugation at 300 g, RBCs were lysed using RBC lysis buffer (BioLegend) for 1–2 min. Cells were washed twice with PBS, resuspended in cRPMI, and quantitated using a TC20 Automated Cell Counter (Bio-Rad). Serum and plasma were collected either through lateral tail vein bleeding or cardiac puncture into uncoated microcentrifuge tubes (serum) or lithium heparin microvette CB300 capillary tubes (Sarstedt). Serum was allowed to coagulate for 1–2 h at 4°C and plasma was kept at 4°C and processed within 2 h. Samples were centrifuged at 5,000 g for 10 min at 4°C and the supernatant was carefully aspirated. Samples were then snap-frozen on dry ice and stored at −20°C until analysis.
Bone marrow chimera
Bone marrow was isolated from femur and tibia by crushing with a mortar and pestle in PBS with 0.1% BSA and 2 mM EDTA. Cells were then filtered through 70-μm cell strainers, spun down, and resuspended at 2 million cells/100 μl in PBS. For mixed bone marrow chimeras, CD45.1+ SJL bone marrow was mixed at a ratio of 1:1 with either CD45.2+ Gimap6−/− or Gimap6+/+ bone marrow. Adult female SJL mice hosts were lethally irradiated at 450 cGy twice, with 4 h in between doses. After 1–2 h of the final irradiation dose, mice were injected with 200 μl of donor bone marrow suspension in the tail vein. Host mice were given antibiotic water (Baytril) for 2 wk after irradiation.
T cell activation
Cells were stained with 5 μM of CellTrace Violet (Thermo Fisher Scientific) for 5 min at room temperature. A half volume of FBS was then added for 5 min at room temperature before washing with media. Mouse splenocytes were activated using 1 μg/ml of plate-bound anti-mouse CD3 (clone 17A2; BioLegend) and 1 μg/ml soluble anti-mouse CD28 (clone 37.51; BioLegend) in cRPMI. Human PBMCs were stimulated with 1 µg/ml Ultra-LEAF purified anti-CD3 (clone HIT3α; BioLegend) and 1 µg/ml Ultra-LEAF purified anti-CD28 (clone CD28.2; BioLegend) soluble antibodies in supplemented RPMI-1640 (10% FBS, 100 U/ml penicillin and streptomycin, 2 mM Glutamax, and 20 mM HEPES). To culture blasting T cells, activated T cells were washed and then cultured in a medium supplemented with 100 U/ml IL-2.
Flow cytometry
In general, cells were surface-stained with antibodies purchased from BD Biosciences and BioLegend against mouse (CD3, CD4, CD8, CD45R, CD69, CD11b, CD40L, CD19, and CD25), anti-human (CD3, CD4, CD8, and CD40L), or eBioscience anti-human (CD4 and CD69). Cells were stained with a viability dye and Fc block, as necessary, in PBS for 15–20 min at 4°C. Cells were then surface-stained for 20–30 min at 4°C with antibodies diluted in FACS buffer (5% FBS and 0.1% sodium azide in PBS). After washing, cells were fixed in 2–4% paraformaldehyde (PFA), washed, and resuspended in PBS before flow cytometric analysis. LC3-II staining of cells treated for 2 h with 10 nM Baf or vehicle was performed using the FlowCellect Autophagy LC3 Antibody-based Assay Kit (Merck Millipore) according to the manufacturer’s instructions. ROS was measured using cells treated with 5 μM MitoSOX Red Mitochondrial Superoxide Indicator (Thermo Fisher Scientific) diluted in PBS with 5% FCS at 37°C for 15 min before antibody staining and analyzed without fixation. We performed acquisition on a BD LSRFortessa (BD Biosciences) flow cytometer and analysis on FlowJo 10.4.
NP-CGG immunization
A total of 50 μg of 4-hydroxy-3-nitrophenyl-chicken γ globulin (NP-CGG; N-5055D-5; Biosearch Technologies) per mouse dissolved in PBS was mixed 1:1 with an equivalent volume of Inject Alum adjuvant (Thermo Fisher Scientific) and mixed on a rotator at room temperature for 30 min. Then 200 μl (50 μg NP-CGG) was injected i.p. per mouse on day 0. Similarly, a secondary immunization was performed on day 35. Peripheral blood samples were collected from the lateral tail vein on indicated days.
ELISAs
A microplate 96 PS half area high binding plate (Greiner Bio-one) was coated overnight at 4°C in 5 μg/ml of NP20-BSA (2B Scientific) in 100 mM bicarbonate/carbonate coating buffer (pH 9.6). The next day, plates were washed with PBS three times and blocked with 5% skimmed milk in PBS for 1–3 h at 37°C. Serum samples were thawed and diluted in 1% milk at 1:500 for the anti-NP20 IgM ELISA and 1:1,000 (primary response) or 1:5,000 (secondary response) for the anti-NP20 IgG1 ELISA. Pooled sera from day 7 after a secondary immunization from mice that were previously injected with NP-CGG in a previous experiment was serially diluted and used as a standard curve for all ELISA plates, starting with a 1:400 or 1:4,000 dilution for the IgM or IgG1 ELISA, respectively. After blocking, the plates were further washed and diluted sera was added to the plates for 1 h at 37°C. Plates were then washed with 0.05% Tween-20 in PBS before detection. Anti-NP20 IgM was detected with 1:1,000 AP-conjugated goat anti-mouse IgM, human adsorbed (1020-04; Southern Biotech); anti-NP20 IgG1 was detected with 1:2,000 AP-conjugated goat anti-mouse IgG1, human adsorbed (1070-04; Southern Biotech). These antibodies were diluted in 1% milk and incubated for 1 h at 37°C. Following further washing, plates were developed by incubating phosphatase substrate (S0942; Sigma-Aldrich) in p-nitrophenyl phosphate buffer for 10–15 min at room temperature in the dark. Absorbance was then measured at 405 nM by an ELISA plate reader (FLUOstar Omega, BMG Labtech). Serum was tested for total IgM and IgG levels using the IgG (Total) Mouse (88-50400-22; Thermo Fischer Scientific) and IgM Mouse (88-50470-22; Thermo Fisher Scientific) ELISA kits. All kits were used according to the manufacturer’s instructions.
Blood and plasma analysis
Plasma was typically diluted 1:3 and analyzed on the AU680 Analyser (Beckman-Coulter) by the Clinical Pathology Service Laboratory at MRC Harwell.
Mouse histology
Organs were fixed with 4% PFA at 4°C and then moved to 80% ethanol. Samples were processed using the Tissue Tek VIP Sakura (GMI Inc) and then sectioned. H&E staining was also done using the Tissue TEK VIP Sakura. PAS staining was performed using the NovaUltra PAS Stain kit (IW-3009; IHC World Online) according to the manufacturer’s directions.
Transmission electron microscopy
After harvesting kidneys, tissue pieces were fixed in 2.5% glutaraldehyde with +4% PFA in 0.1 M sodium cacodylate buffer (SC; pH 7.2) for 3–4 h at room temperature and then stored at 4°C until processing. Samples were washed in 0.1 M SC for ∼3 h, with several changes of buffer and the addition of 50 mM glycine for one of these wash steps. Secondary fixation was performed with 1% osmium tetroxide and 1.5% potassium ferrocyanide in 0.1 M SC for 2 h at 4°C. Samples were then washed 5–6 times in milli-Q water for 10 min each. Tertiary fixation was performed at 4°C in the dark with 0.5% Uranyl acetate overnight before washing three times with milli-Q water for 5–10 min each. The samples were then dehydrated in increasing concentrations of ice-cold ethanol). Samples were incubated with increasing concentrations of low viscosity epoxy resin (Agar Scientific) in pure ethanol. To embed samples, tissue pieces were placed in flat dish embedding molds filled with 100% resin and polymerized in a 60°C oven for 48 h. Blocks were sectioned using a Leica UC7 ultramicrotome. Semi-thin (500 nm) sections were transferred to glass slides and stained with toluidine blue to check for the presence of glomeruli. Ultra-thin (90 nm) sections were taken using a Diatome diamond knife and transferred to 50-mesh copper grids and then post-stained for 5 min with lead citrate. Sections were imaged on a FEI Tecnai 12 transmission electron microscope operated at 120 kV using a Gatan OneView camera.
Transfections
HEK293T cells were transfected at 50–80% confluence using GeneJuice (Merck Millipore) according to the manufacturer’s instructions. Empty vector (pCDNA3.1 zeo [+]) was added to keep the amounts of DNA transfected equal for all samples. After 24 h, cells were harvested and the protein lysates prepared for Western blots (WBs).
WB
Cells were lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 5 mM EDTA [pH 8.0], 50 mM Tris [pH 8.0], 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) complete with protease and phosphatase inhibitor cocktails, heated at 95°C for 5 min with reducing reagent, and run on a Bis-tris protein gel. Primary antibodies used were anti-β actin (clone 2D1D10; GenScript), anti-HA tag (clone 6E2; Cell Signaling Technologies), anti-LC3 rabbit polyclonal (cat no. L8919; Sigma-Aldrich), anti-human GIMAP1 (PA5-60858; Thermo Fischer Scientific), anti-human GIMAP4 (HPA019137; Sigma-Aldrich), anti-human GIMAP7 (HPA020268; Sigma-Aldrich), and anti-β tubulin Dylight 680 (clone BT7R; Life Technologies). Anti-human and anti-mouse GIMAP6 antibodies (MAC445 and MAC436, respectively), anti-human GIMAP2 antibody, and anti-human GIMAP8 antibody were sourced as previously described (Pascall et al., 2013). Secondary antibodies used were IRDye 800CW Donkey anti-Mouse (Murine) IgG, IRDye 800CW Donkey anti-Rabbit IgG, and IRDye 800CW Goat anti-Rat IgG (all from Li-COR Biosciences). Membranes were imaged using the Odyssey CLx Imaging System (LI-COR Biosciences) or PXi imager (Syngene). Data were analyzed using Image Studio Lite.
CHX chase assay
mCherry-tagged human WT and G153V GIMAP6 were cloned into pLV-EF1a-IRES-Puro (plasmid #85132; Addgene) to generate pLV-GIMAP6-mCherry-WT and pLV-GIMAP6-mCherry-G153V. The lentivirus was generated by transfecting Lenti-X 293T Cells (cat. no. 632180; TaKaRa) with pLV-GIMAP6-mCherry, psPAX2 (plasmid #12260; Addgene), and pMD2.G (plasmid #12259; Addgene) at a ratio of 10:10:1. The lentivirus-containing supernatant was collected after 48 h of transfection, and the HEK293T cells were infected with lentivirus to produce GIMAP6-mCherry stable expressing cells. Puromycin was added to remove the uninfected cells after 3 d of lentivirus transduction. After puromycin selection, the GIMAP6–mCherry–WT and GIMAP6–mCherry–G153V stable expressing cells were split into 12 wells and CHX (50 μg/ml) was added 24, 18, 12, 8, 6, and 2 h before collecting the cells. The mean fluorescence intensity (MFI) of mCherry at each time point was measured by flow cytometry and compared to 0-time points.
Metabolite and lipid sample preparation
Media was removed from cell culture samples and the cells were washed with 0.5 ml of 0.9% sodium chloride. Cells were immersed in 0.5 ml of ice-cold methanol for 5 min. Then 0.5 ml of ice-cold water followed by 0.5 ml of ice-cold chloroform were added to each sample. Samples were agitated for 30 min at 4°C and subsequently centrifuged at 16,000 g for 20 min. The top (aqueous) and bottom (organic) layers were collected separately. The organic layer was taken to dryness in a Savant DNA120 SpeedVac concentrator (Thermo Fisher Scientific). Aqueous metabolite and lipid samples were resuspended in 50% methanol or 5 µg/ml butylated hydroxytoluene in 6:1 isopropanol:methanol, respectively for analysis.
Metabolomics and lipidomics LCMS
Tributylamine and all synthetic standards were purchased from Millipore Sigma-Aldrich. LCMS grade water, methanol, isopropanol, and acetic acid were purchased through Thermo Fisher Scientific.
Aqueous metabolite and lipid samples were analyzed by targeted multiple-reaction monitoring (MRM) on a Sciex ExionLC AC system and a Sciex 5500 QTRAP mass spectrometer. Aqueous metabolites were analyzed using a previous protocol (McCloskey et al., 2015). Quality control samples were injected after every 10 injections. Samples were separated across a Waters Atlantis T3 column (100 Å, 3 µm, 3 × 100 mm) and eluted using a binary gradient from 5 mM tributylamine, 5 mM acetic acid in 2% isopropanol, 5% methanol, 93% water (vol/vol) to 100% isopropanol over 15 min. Analytes were detected in negative mode using two distinct MRM pairs for each metabolite. Heavy-labeled standards were not utilized, and relative quantification was performed. The fidelity of features was confirmed using a synthetic molecular reference. Lipid samples were analyzed using a previously established hydrophilic interaction chromatography method with modification (Mackenzie Pearson, 2018). Samples were separated on a Water XBridge Amide column (3.5 µm, 3 × 100 mm) and eluted using a 12-min binary gradient from 100% 5 mM ammonium acetate, 5% water in acetonitrile apparent pH 8.4–95% 5 mM ammonium acetate, and 50% water in acetonitrile apparent pH 8.0. Samples were analyzed using separate positive and negative mode MRM methods. All signals were integrated using MultiQuant Software 3.0.3. Molecules with >50% missing values were discarded and the remaining missing values were replaced with the lowest registered signal value. All signals were total sum normalized. Single and multivariate analysis were performed in MarkerView Software 1.3.1. For PLS-DA, samples were Pareto scaled. For Pt 1 cell extracts, all healthy donor samples were grouped together for PLS-DA analysis.
SILAC and Jurkat cell lysis
SILAC media was prepared as described in Ong and Mann (2006). Jurkat cells were cultured at 37°C with 5% CO2 in SILAC media supplemented with 10% (vol/vol) dialyzed FCS (Sigma-Aldrich), 4 mM L-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were split every third day for at least six passes for maximal incorporation of isotopes. A total of 10 × 106 cells were cultured per pull-down. Cells in the SILAC medium were centrifuged at 1,500 rpm at 20°C for 5 min. The pellet was washed twice with PBS and lysed using lysis buffer containing PBS, 2.5 mM dithiothreitol (DTT), 1 protease inhibitor cocktail tablet (Roche) per 10 ml buffer, 1 μM DNase mix, 1% NP-40 detergent, and 100 μM GTPγS. Cells were disrupted by gentle pipetting and the lysate was centrifuged at 13,200 rpm for 15 min. The cleared lysate was separated and used for pull-down assay.
SILAC pull down assay
SILAC experiments were conducted as label swap experiments similar to a previously published method but without GTP loading (Paul et al., 2017). In a forward experiment, GST-GIMAP7 (L100Q; bait) was incubated with a heavy and GST (control) was incubated with light lysate. The reverse experiment was performed with swapped lysates. Active N-hydroxy-succinimide sepharose beads were washed with 500 μl ice-cold equilibration buffer (1 mM HCl) and resuspended in 1 ml of washing buffer (PBS, 5 mM MgCl2) to which 2 mg of bait and control proteins were added separately and incubated for 2 h at room temperature. Non-bound protein was removed by centrifugation, and the beads were washed with 1 ml buffer A (0.5 M ethanolamine, 0.5 M NaCl, pH 8.3) followed by 1 ml buffer B (0.1 M Na-acetate, 0.5 M NaCl, pH 4.0). Subsequently, the beads were incubated in buffer A for 30 min and washed with buffer B, A, and again B followed by two-wash steps with washing buffer. Heavy and light cell lysates were added to the respective beads and incubated for 60 min at 4°C. The non-bound lysate was removed, and heavy and light beads were mixed in 1:1 ratio to obtain the pull-down pair. The mixed beads were washed with lysis buffer twice and the bound protein complexes were eluted with 200 μl denaturation buffer (6 M urea and 2 M thio-urea in 10 mM HEPES, pH 8) by shaking at 1,400 rpm on Thermo shaker for 15 min. The eluate was frozen at −80°C for subsequent mass spectrometric analysis.
Proteomics MS and sample preparation
Protein pellets were redissolved in a urea/thiourea buffer and reduced with dithiothreitol, alkylated with iodoacetamide, and digested with LysC and trypsin. The peptides were desalted off-line (Rappsilber et al., 2003) and analyzed by online LC-MS in an EASY-nLC system (Thermo Fisher Scientific) coupled to a Q Exactive Orbitrap (Thermo Fisher Scientific). Then 5 µl peptide samples were loaded onto a fritless microcolumn (75 µm inner diameter packed with ReproSil-Pur C18-AQ 3-µm resin, Dr. Maisch GmbH). Peptides were eluted with an 8–60% acetonitrile gradient and 0.5% formic acid. Runs were performed as 4 h gradients at a flow rate of 200 nl/min. Peptides were ionized at currents of 2–2.5 kV. The Q-Exactive Orbitrap device was operated in the data-dependent mode with a TOP10 method. One full scan (m/z range = 300–1,650, R = 70,000, target value: 106 ions, maximum injection time = 20 ms) was used to detect precursor ions. The 10 most intense ions with a charge state greater than one were selected for fragmentation (R = 17,500, target value 106 ions, isolation window = 3 m/z, maximum injection time = 60 ms). Dynamic exclusion time for fragmented precursor ions was set to 30 s.
Data analysis and label-free quantification with MaxQuant and Perseus
MS raw data files were analyzed with the MaxQuant software package (version 1.2.0.18) with standard settings (Cox and Mann, 2008). Proteins were searched against the IPI human database (version 3.84). Files produced by MaxQuant were further analyzed using the Perseus tool (version 1.3.0.4) available with the MaxQuant environment. Logarithmized normalized SILAC-ratios of protein intensities were plotted to distinguish interaction partners of GIMAP7 from background binders.
GIMAP6 protein expression and purification
GIMAP6 WT protein overexpressions were typically carried out in culture volumes varying between 10 and 20 liters of terrific broth medium. E. coli strain Rosetta 2 (DE3) carrying the pGEX-6P1-GIMAP6 plasmid was used for the expression of GIMAP6. The culture was grown at 37°C until the OD reached 0.5–0.6 while shaking. The cells were induced with 40 μM isopropyl β-D-1-thiogalactopyranoside and the temperature changed to 18°C for the overexpression of protein for 18–20 h. The bacterial cells were lysed by microfluidization at 8,000 psi. The lysate was cleared at 35,000 rpm at 4°C for 45 min. The supernatant was filtered using 0.2 μm filter and applied on chromatography column packed with 20 ml glutathione (GSH) sepharose beads pre-equilibrated with 5 column volume (CV) of equilibration buffer, EB (50 mM HEPES, pH 7.5, 800 mM NaCl, 2.5 mM DTT, 2 mM MgCl2, 10 mM KCl, 0.1 mM GDP, and 1 mM ATP). The column was extensively washed with 20 CV of EB. Subsequently, the column was washed with 10 CV of EB with 1% CHAPS followed by 5 CV of EB. For purification of GST-tag free GIMAP6, the column was unpacked and the protein-bound GSH sepharose was re-suspended in 40 ml of EB with 0.5 mg preScission protease and incubated overnight at 4°C. GSH sepharose was packed again into the chromatography column, and the cleaved protein was eluted with EB. The eluate was concentrated and injected into Superdex 200 column pre-equilibrated with 10 mM HEPES, pH 7.5, 300 mM NaCl, and 2.5 mM DTT. Fractions were analyzed on SDS-PAGE. Aliquots of GIMAP6 were flash-frozen and stored at −80°C.
GIMAP6:GABARAPL2 complex purification
pGEX-6P1 for GST-GABARAPL2 and pSKB-LNB for His-GIMAP6 expression were co-transformed in E. coli Rosetta 2(DE3). The expression and purification of the complex were carried out as described for GIMAP6. Eluate post-preScession-protease cleavage contained the GIMAP6:GABARAPL2 complex.
Isothermal titration calorimetry
Isothermal titration calorimetry experiments were carried out at 8°C in a buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 5 mM KCl in a microCal iTC200. 600 μM GABARAPL2 was titrated against 50 μM of GIMAP6(R134D) or GIMAP7(L100Q). The R134D mutant of GIMAP6 was used since it can be produced in ∼10-fold higher amounts than wt GIMAP6. Data were fitted with the Origin software.
GTP hydrolysis assays
GTPase assays were carried out using 20 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM KCl, and 5 mM MgCl2 at 20°C in the presence of 500 µM GTP using standard HPLC detection. The samples were diluted 10-fold and injected into the ODS-2 C18 HPLC column (250 × 4 mm). The flow rate was maintained at 1.3 ml/min. Nucleosil 100 C18 pre-column was used as a guard column to adsorb denatured protein. The running buffer contained 10 mM tetrabutylammonium bromide and 100 mM potassium phosphate (pH 6.5) with 7.5% acetonitrile. The eluting nucleotides were detected at a wavelength of 254 nm. A total of 50 μM of pure nucleotides were used as standards. Rates derived from a linear fit to the initial reaction rates (<40% GTP hydrolyzed) were plotted against the protein concentrations.
Single cell RNA sequencing (scRNA-seq)
scRNA-seq raw count data from a previously published dataset indicated in figure legends was analyzed using the Seurat package in R version 4.0.0. The data were log-normalized and scaled using the default settings in the Seurat package. We grouped the cells using k nearest neighbor on the top 12 dimensions from PCA. The cells were then clustered using the Louvain algorithm with a resolution of 0.5. The clusters were then projected into a two-dimensional space using UMAP and identified using canonical markers.
CRISPR GIMAP6 KO in Jurkat cells
GIMAP6 CRISPR RNAs (crRNAs) were purchased from IDT. crRNAs and trans-activating CRISPR RNAs (tracrRNAs; #1075928; IDT) from IDT were resuspended with Duplex buffer (#11-05-01-12; IDT) to 100 µM stock concentrations and combined with tracrRNA (#1072534; IDT; crRNA:tracrRNA) at a 1:1 ratio. The mixture of crRNA/tracrRNA complexes was heated to 95°C for 5 min in sterile PCR tubes in a thermocycler. After cooling down to room temperature, 9 µl of crRNA:tracrRNA complexes were mixed with 4.5 µl Cas9 protein (6.5 µg/µl from Berkeley Macrolab) and 3 µl of Duplex buffer. This mixture was incubated at room temperature for 10 min to form the Cas9:RNP complex. Meanwhile, 1–10 million Jurkat cells were spun down and resuspended in 20 µl P2 buffer (cat #V4XP-2024; Amaxa P2 4D kit from Lonza). Jurkat cells in P2 buffer were then mixed with Cas9:RNP complex and electroporation was carried out by following Amaxa 4D-Nucleofector Protocol for Jurkat clone E6.1 7 d after electroporation. Cell pellets were collected and lysed, and the GIMAP6 protein level was verified through WB.
Confocal microscopy and immunofluorescence staining
THP-1 cells in the log-phase were plated onto coverslips in 24-well plates and differentiated with 20 ng/ml PMA for 3 d to induce macrophages. GFP-P. aeruginosa and S. typhimurium were cultured in the log-phase and infected THP-1 differentiated macrophages at multiplicity of infection = 10. Cells on coverslips were washed twice with PBS and then permeabilized for 15 min in PBS buffer containing 1% Triton X-100 and 5% BSA. The cells were then blocked with 5% BSA in PBST (PBS + 0.05% Tween-20) for an additional 1 h. Primary antibodies diluted in dilution buffer (1:100; PBS buffer containing 1% Triton X-100 and 5% BSA) were added to the wells at 4°C overnight. After washing three times with PBS the following day, the cells were treated with fluorochrome-conjugated secondary antibodies (Invitrogen) in dilution buffer for 1 h at room temperature in the dark. Coverslips were mounted with DAPI Fluoromount-G (SouthernBiotech) after three washes with PBS. Confocal images were acquired on a Leica SP5 X849 WLL microscope, and analysis was performed using Imaris software. For the live images, THP-1 cells in the log-phase were plated into chamber slides (cat: 80841; ibidi) and differentiated with 20 ng/ml PMA for 3 d to induce macrophages. Cells were infected with GFP-P. aeruginosa at multiplicity of infection = 10 for 1 h and washed three times with PBS to remove extracellular bacteria. Live images were captured using Leica SP5 X849 WLL microscope and analysis was performed using Imaris software.
P. aeruginosa infectious pneumonia mouse model
P. aeruginosa (HER-1018) were seeded and cultured in LB Soy Agar Broth for 16–24 h at 37°C. Bacteria cultures were then spun down and resuspended in PBS to 1.25 × 1010 CFU per ml. Gimap6−/− and WT mice were inoculated with 40 µl of P. aeruginosa in PBS via a nasal infection under anesthesia conditions. After P. aeruginosa infection, the bodyweight of mice was recorded every day. On day 3, lungs from Gimap6−/− and WT were harvested and passed through a 70-µM cell strainer in a 6-well dish with 4 ml PBS. Each sample was diluted serially and 100 µl from each dilution was then plated on blood agar plates. CFUs were counted after 24 h.
Statistics
All statistical analysis was performed using Graphpad Prism 7.0. Data are shown as mean ± SD unless otherwise stated. Statistical tests (two-tailed Student’s t-test, Mann–Whitney U test, two-way ANOVA with Tukey’s multiple comparison test, repeat measures two-way ANOVA with Sidak’s multiple comparison test, or log-rank Mantel-Cox test) were used as appropriate. A difference was considered statistically significant when *, P < 0.05.
Study approval
All the patients, family members, and healthy controls provided written, informed consent in accordance with the Declaration of Helsinki under institutional review board-approved protocols from Hacettepe University or review board–approved protocols from Hadassah University Medical Center, the NIAID at the NIH (clinical trial identifier NCT00001355), and the University of Oxford.
Online supplemental material
Fig. S1 shows gene variants from whole-exome DNA sequencing. Fig. S2 shows defective autophagy in Gimap6−/− lymphocytes. Fig. S3 shows GIMAP6, GABARAPL2, and GIMAP7 exist as functional complex. Fig. S4 shows mouse immunophenotyping. Fig. S5 shows metabolite and lipid study, and kidney-specific scRNA analyses. Table S1 shows the clinical features of GIMAP6 deficiency patients. Table S2 shows histological findings of Gimap6−/− tissues. Video 1 shows the co-localization of mCherry-GIMAP6 and GFP-PA after infection.
Supplementary Material
shows clinical and laboratory findings of GIMAP6 patients.
shows histological findings of Gimap6−/− tissues.
Acknowledgments
We thank the participating clinicians, nurses, laboratory staff, and the patients and their families. We thank Dr. Helen Su, Nancie Ann Mooney, and the Kennedy Institute of Rheumatology. Dr. Bella Shadur was supported by the Australian Government Research Training Program Scholarship and Hadassah. We thank Hyun-Hee Lee, Emilio Flano, and Merck, Inc. for support and Ryan Kissinger for help with illustrations.We thank the Infection and Immunity Immunophenotyping (3I) consortium led by Adrian Hayday (King’s College) for the KOMP Gimap6 mouse and associated data.
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, the Oxford University McMichael Research Fund, and the German Research Foundation (Collaborative Research Grant 958, Project A12). Projects 013 D08103 001-341, 11/19-23, and 16/9087 were generously funded by Hacettepe University Coordination Unit for Scientific Research Projects.
Author contributions: Y. Yao, P.D. Jiang, B.N. Chao, and S. Gottlieb performed experiments, analyzed data, and co-wrote the manuscript. D. Cagdas provided clinical samples and data, analyzed data, and co-wrote the manuscript. S. Kubo, A. Balasubramaniyam, L. Zheng, M.A. Leney-Greene, A.Y. Park, F. Paul, H. Bulut, S. Clare, and S. Ganesan performed experiments and analyzed data. Y. Zhang performed bioinformatic analyses. B. Shadur, A.N. Eddin, I. Tezcan, and P. Stepensky provided clinical samples and data and analyzed data. L.R. Folio and S.D. Chauvin analyzed data and edited the manuscript. B. Schwarz and E. Bohrnsen performed metabolomics and lipidomics experiments and analyzed data. M. Lynberg performed scRNA analysis, analyzed data, and co-wrote the manuscript. A. Akdogan provided rheumatologic data and analyzed data. R. Gocmen provided radiological data and analyzed data. S. Onder provided pathological data and analyzed data. A. Rosenberg analyzed kidney samples and provided images. E.J. Soilleux analyzed mouse histological samples and provided images. E. Johnson performed the transmission electron microscopy and analyzed data. P.K. Jackson supervised IP/MS experiments and analyzed data. J. Demeter analyzed IP/MS experiments and data by network analysis. M. Selbach supervised IP/MS experiments. M.R. Clatworthy supervised scRNA analysis. Z.K. Tuong performed scRNA analysis and analyzed data. H. Zhang provided instruction and guidance on autophagy and mouse protocols. B. Stewart performed scRNA analysis and analyzed data. C.M. Bosio supervised metabolomics and lipidomics experiments and analyzed data. J.C. Pascall provided key reagents and guidance at early stages of the study. O. Daumke supervised biochemical and structural experiments and analyzed data. G.W. Butcher provided key reagents and guidance at early stages of the study. A.J. McMichael and A.K. Simon edited the manuscript and supervised the study. M.J. Lenardo co-wrote the manuscript and supervised the research.
References
- Abeler-Dorner, L., Laing A.G., Lorenc A., Ushakov D.S., Clare S., Speak A.O., Duque-Correa M.A., White J.K., Ramirez-Solis R., Saran N., et al. 2020. High-throughput phenotyping reveals expansive genetic and structural underpinnings of immune variation. Nat. Immunol. 21:86–100. 10.1038/s41590-019-0549-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agata, Y., Kawasaki A., Nishimura H., Ishida Y., Tsubata T., Yagita H., and Honjo T.. 1996. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int. Immunol. 8:765–772. 10.1093/intimm/8.5.765 [DOI] [PubMed] [Google Scholar]
- Asaoka, Y., Oka M., Yoshida K., Sasaki Y., and Nishizuka Y.. 1992. Role of lysophosphatidylcholine in T-lymphocyte activation: Involvement of phospholipase A2 in signal transduction through protein kinase C. Proc. Natl. Acad. Sci. USA. 89:6447–6451. 10.1073/pnas.89.14.6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astle, W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A., et al. 2016. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 167:1415–1429.e19. 10.1016/j.cell.2016.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, C.P., Battey J.F., Bradley A., Bucan M., Capecchi M., Collins F.S., Dove W.F., Duyk G., Dymecki S., Eppig J.T., et al. 2004. The knockout mouse project. Nat. Genet. 36:921–924. 10.1038/ng0904-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, M.J., Aksoylar H., Krebs P., Bourdeau T., Arnold C.N., Xia Y., Khovananth K., Engel I., Sovath S., Lampe K., et al. 2010. Loss of T cell and B cell quiescence precedes the onset of microbial flora-dependent wasting disease and intestinal inflammation in Gimap5-deficient mice. J. Immunol. 184:3743–3754. 10.4049/jimmunol.0903164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin, M., Yuan L., Keskin D.B., Otu H.H., Libermann T.A., and Oettgen P.. 2010. Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genom. 11:342. 10.1186/1471-2164-11-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botbol, Y., Guerrero-Ros I., and Macian F.. 2016. Key roles of autophagy in regulating T-cell function. Eur. J. Immunol. 46:1326–1334. 10.1002/eji.201545955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley, J.M., Karandikar N.J., Betts M.R., Ambrozak D.R., Hill B.J., Crotty L.E., Casazza J.P., Kuruppu J., Migueles S.A., Connors M., et al. 2003. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 101:2711–2720. 10.1182/blood-2002-07-2103 [DOI] [PubMed] [Google Scholar]
- Casanova, J.L., and Abel L.. 2021. Lethal infectious diseases as inborn errors of immunity: Toward a synthesis of the germ and genetic theories. Annu. Rev. Pathol. 16:23–50. 10.1146/annurev-pathol-031920-101429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucci, T., and Bosselut R.. 2014. Gimap and T cells: A matter of life or death. Eur. J. Immunol. 44:348–351. 10.1002/eji.201344375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A.J., Riffelmacher T., Braas D., Cornall R.J., and Simon A.K.. 2018. B1a B cells require autophagy for metabolic homeostasis and self-renewal. J. Exp. Med. 215:399–413. 10.1084/jem.20170771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corban, M.T., Duarte-Garcia A., McBane R.D., Matteson E.L., Lerman L.O., and Lerman A.. 2017. Antiphospholipid syndrome: Role of vascular endothelial cells and implications for risk stratification and targeted therapeutics. J. Am. Coll. Cardiol. 69:2317–2330. 10.1016/j.jacc.2017.02.058 [DOI] [PubMed] [Google Scholar]
- Cox, J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Dall’Armi, C., Devereaux K.A., and Di Paolo G.. 2013. The role of lipids in the control of autophagy. Curr. Biol. 23:R33–R45. 10.1016/j.cub.2012.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, P.S., Sabater-Lleal M., Chasman D.I., Trompet S., Ahluwalia T.S., Teumer A., Kleber M.E., Chen M.H., Wang J.J., Attia J.R., et al. 2017. Comparison of HapMap and 1000 genomes reference panels in a large-scale genome-wide association study. PLoS One. 12:e0167742. 10.1371/journal.pone.0167742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., et al. 2016. . High-throughput discovery of novel developmental phenotypes. Nature. 537:508–514. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drzewiecki, K., Choi J., Brancale J., Leney-Greene M.A., Sari S., Dalgic B., Unlusoy Aksu A., Evirgen Sahin G., Ozen A., Baris S., et al. 2021. GIMAP5 maintains liver endothelial cell homeostasis and prevents portal hypertension. J. Exp. Med. 218:e20201745. 10.1084/jem.20201745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomeni, G., De Zio D., and Cecconi F.. 2015. Oxidative stress and autophagy: The clash between damage and metabolic needs. Cell Death Differ. 22:377–388. 10.1038/cdd.2014.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay, L., Miller M.R., Ventura P.B., Devasthali V., Vue Z., Thompson H.L., Temple S., Zong H., Cleary M.D., Stankunas K., and Doe C.Q.. 2013. Mouse TU tagging: A chemical/genetic intersectional method for purifying cell type-specific nascent RNA. Genes Dev. 27:98–115. 10.1101/gad.205278.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, H.R. 2005. ROS as signalling molecules in T cells-evidence for abnormal redox signalling in the autoimmune disease, rheumatoid arthritis. Redox Rep. 10:273–280. 10.1179/135100005X83680 [DOI] [PubMed] [Google Scholar]
- Hellquist, A., Zucchelli M., Kivinen K., Saarialho-Kere U., Koskenmies S., Widen E., Julkunen H., Wong A., Karjalainen-Lindsberg M.L., Skoog T., et al. 2007. The human GIMAP5 gene has a common polyadenylation polymorphism increasing risk to systemic lupus erythematosus. J. Med. Genet. 44:314–321. 10.1136/jmg.2006.046185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahashi, J., Kawahata K., Arita M., Iwamoto R., Hishikawa K., Honda M., Hamasaki Y., Tanaka M., Okubo K., Kurosawa M., et al. 2014. Immunomodulation with eicosapentaenoic acid supports the treatment of autoimmune small-vessel vasculitis. Sci. Rep. 4:6406. 10.1038/srep06406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, C.H., and Tsai S.F.. 2017. Functional and biochemical characterization of a T cell-associated anti-apoptotic protein, GIMAP6. J. Biol. Chem. 292:9305–9319. 10.1074/jbc.M116.768689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochane, M., van den Berg P.R., Fan X., Berenger-Currias N., Adegeest E., Bialecka M., Nieveen M., Menschaart M., Chuva de Sousa Lopes S.M., and Semrau S.. 2019. Single-cell transcriptomics reveals gene expression dynamics of human fetal kidney development. PLoS Biol. 17:e3000152. 10.1371/journal.pbio.3000152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, T.J., Theusch E., Haldar T., Ranatunga D.K., Jorgenson E., Medina M.W., Kvale M.N., Kwok P.Y., Schaefer C., Krauss R.M., et al. 2018. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 50:401–413. 10.1038/s41588-018-0064-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, V.M., Valdor R., Patel B., Singh R., Cuervo A.M., and Macian F.. 2010. Macroautophagy regulates energy metabolism during effector T cell activation. J. Immunol. 185:7349–7357. 10.4049/jimmunol.1000576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im, Y.J., Lee Y.K., Chung H.Y., and Im D.S.. 2006. Multiple actions of lysophosphatidylcholine in human Jurkat T cells. Acta Pharmacol. Sin. 27:700–707. 10.1111/j.1745-7254.2006.00339.x [DOI] [PubMed] [Google Scholar]
- Itan, Y., Shang L., Boisson B., Ciancanelli M.J., Markle J.G., Martinez-Barricarte R., Scott E., Shah I., Stenson P.D., Gleeson J., et al. 2016. The mutation significance cutoff: Gene-level thresholds for variant predictions. Nat. Methods. 13:109–110. 10.1038/nmeth.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, S.L., Hill S.J., Lock R.J., Dwight J.F., Unsworth D.J., and Gompels M.M.. 2004. Echocardiographic abnormalities in primary antibody deficiency. Postgrad. Med. J. 80:214–218. 10.1136/pgmj.2003.012443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper, J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature. 596:583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Jing K., Shin S., Jeong S., Han S.H., Oh H., Yoo Y.S., Han J., Jeon Y.J., Heo J.Y., et al. 2018. ω3-polyunsaturated fatty acids induce cell death through apoptosis and autophagy in glioblastoma cells: In vitro and in vivo. Oncol. Rep. 39:239–246. 10.3892/or.2017.6101 [DOI] [PubMed] [Google Scholar]
- Kimmey, J.M., and Stallings C.L.. 2016. Bacterial pathogens versus autophagy: Implications for therapeutic interventions. Trends Mol. Med. 22:1060–1076. 10.1016/j.molmed.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krucken, J., Schroetel R.M.U., Muller I.U., Saidani N., Marinovski P., Benten W.P.M., Stamm O., and Wunderlich F.. 2004. Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene. 341:291–304. 10.1016/j.gene.2004.07.005 [DOI] [PubMed] [Google Scholar]
- Lahiri, V., Hawkins W.D., and Klionsky D.J.. 2019. Watch what you (self-) eat: Autophagic mechanisms that modulate metabolism. Cell Metab. 29:803–826. 10.1016/j.cmet.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H., Cho M.H., Hersh C.P., McDonald M.L., Crapo J.D., Bakke P.S., Gulsvik A., Comellas A.P., Wendt C.H., Lomas D.A., et al. 2014. Genetic susceptibility for chronic bronchitis in chronic obstructive pulmonary disease. Respir. Res. 15:113. 10.1186/s12931-014-0113-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.K., and Lee J.A.. 2016. Role of the mammalian ATG8/LC3 family in autophagy: Differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 49:424–430. 10.5483/bmbrep.2016.49.8.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo, M., Lo B., and Lucas C.L.. 2016. Genomics of immune diseases and new therapies. Annu. Rev. Immunol. 34:121–149. 10.1146/annurev-immunol-041015-055620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo, M.J., and Holland S.M.. 2019. Introduction: Continuing insights into the healthy and diseased immune system through human genetic investigation. Immunol. Rev. 287:5–8. 10.1111/imr.12730 [DOI] [PubMed] [Google Scholar]
- Li, C., Capan E., Zhao Y., Zhao J., Stolz D., Watkins S.C., Jin S., and Lu B.. 2006. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J. Immunol. 177:5163–5168. 10.4049/jimmunol.177.8.5163 [DOI] [PubMed] [Google Scholar]
- Limoges, M.A., Cloutier M., Nandi M., Ilangumaran S., and Ramanathan S.. 2021. The GIMAP family proteins: An incomplete puzzle. Front. Immunol. 12:679739. 10.3389/fimmu.2021.679739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Wang T., Zhang W., and Li X.. 2008. Computational identification and analysis of immune-associated nucleotide gene family in Arabidopsis thaliana. J. Plant Physiol. 165:777–787. 10.1016/j.jplph.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Lone, A.M., and Tasken K.. 2013. Proinflammatory and immunoregulatory roles of eicosanoids in T cells. Front. Immunol. 4:130. 10.3389/fimmu.2013.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie Pearson, M., Kapil S.K., Wu S.J.. 2018. Achieve broad lipid quantitation using a high-throughput targeted lipidomics method. SCIEX. [Google Scholar]
- MacMurray, A.J., Moralejo D.H., Kwitek A.E., Rutledge E.A., Van Yserloo B., Gohlke P., Speros S.J., Snyder B., Schaefer J., Bieg S., et al. 2002. Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (Ian)-related gene. Genome Res. 12:1029–1039. 10.1101/gr.412702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin, C., Nagy P., Juhasz G., and Neufeld T.P.. 2015. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 6:7007. 10.1038/ncomms8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey, D., Gangoiti J.A., Palsson B.O., and Feist A.M.. 2015. A pH and solvent optimized reverse-phase ion-paring-LC-MS/MS method that leverages multiple scan-types for targeted absolute quantification of intracellular metabolites. Metabolomics. 11:1338–1350. 10.1007/s11306-015-0790-y [DOI] [Google Scholar]
- Nitta, T., and Takahama Y.. 2007. The lymphocyte guard-IANs: Regulation of lymphocyte survival by IAN/GIMAP family proteins. Trends Immunol. 28:58–65. 10.1016/j.it.2006.12.002 [DOI] [PubMed] [Google Scholar]
- O’Neill, L.A.J., and Pearce E.J.. 2016. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213:15–23. 10.1084/jem.20151570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar, R.A., Collins C.A., Hammarback J.A., Shpetner H.S., and Vallee R.B.. 1990. Molecular cloning of the microtubule-associated mechanochemical enzyme dynamin reveals homology with a new family of GTP-binding proteins. Nature. 347:256–261. 10.1038/347256a0 [DOI] [PubMed] [Google Scholar]
- Ong, S.E., and Mann M.. 2006. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1:2650–2660. 10.1038/nprot.2006.427 [DOI] [PubMed] [Google Scholar]
- Palmer, B.E., Blyveis N., Fontenot A.P., and Wilson C.C.. 2005. Functional and phenotypic characterization of CD57+CD4+ T cells and their association with HIV-1-induced T cell dysfunction. J. Immunol. 175:8415–8423. 10.4049/jimmunol.175.12.8415 [DOI] [PubMed] [Google Scholar]
- Park, J., Shrestha R., Qiu C., Kondo A., Huang S., Werth M., Li M., Barasch J., and Susztak K.. 2018. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 360:758–763. 10.1126/science.aar2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascall, J.C., Rotondo S., Mukadam A.S., Oxley D., Webster J., Walker S.A., Piron J., Carter C., Ktistakis N.T., and Butcher G.W.. 2013. The immune system GTPase GIMAP6 interacts with the Atg8 homologue GABARAPL2 and is recruited to autophagosomes. PLoS One. 8:e77782. 10.1371/journal.pone.0077782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascall, J.C., Webb L.M.C., Eskelinen E.L., Innocentin S., Attaf-Bouabdallah N., and Butcher G.W.. 2018. GIMAP6 is required for T cell maintenance and efficient autophagy in mice. PLoS One. 13:e0196504. 10.1371/journal.pone.0196504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, F., Zauber H., von Berg L., Rocks O., Daumke O., and Selbach M.. 2017. Quantitative GTPase affinity purification identifies rho family protein interaction partners. Mol. Cell. Proteomics. 16:73–85. 10.1074/mcp.M116.061531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier, G.M., Anderson G., Huvar A., Wagaman P.C., Shuttleworth J., Jenkinson E., Jackson M.R., Peterson P.A., and Erlander M.G.. 1999. Immune-associated nucleotide-1 (IAN-1) is a thymic selection marker and defines a novel gene family conserved in plants. J. Immunol. 163:4960–4969 [PubMed] [Google Scholar]
- Praefcke, G.J.K., and McMahon H.T.. 2004. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5:133–147. 10.1038/nrm1313 [DOI] [PubMed] [Google Scholar]
- Pua, H.H., Dzhagalov I., Chuck M., Mizushima N., and He Y.W.. 2007. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 204:25–31. 10.1084/jem.20061303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston, D.J., Zhang H., Powell T.J., Lipina E., Sims S., Panse I., Watson A.S., Cerundolo V., Townsend A.R., Klenerman P., and Simon A.K.. 2014. Autophagy is a critical regulator of memory CD8+ T cell formation. Elife. 3:e03706. 10.7554/eLife.03706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber, J., Ishihama Y., and Mann M.. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 75:663–670. 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- Riffelmacher, T., Richter F.C., and Simon A.K.. 2018. Autophagy dictates metabolism and differentiation of inflammatory immune cells. Autophagy. 14:199–206. 10.1080/15548627.2017.1362525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, T., Kuma A., Sugiura Y., Ichimura Y., Obata M., Kitamura H., Okuda S., Lee H.C., Ikeda K., Kanegae Y., et al. 2019. Autophagy regulates lipid metabolism through selective turnover of NCoR1. Nat. Commun. 10:1567. 10.1038/s41467-019-08829-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata-Kaneko, S., Wakatsuki Y., Usui T., Matsunaga Y., Itoh T., Nishi E., Kume N., and Kita T.. 1998. Lysophosphatidylcholine upregulates CD40 ligand expression in newly activated human CD4+ T cells. FEBS Lett. 433:161–165. 10.1016/s0014-5793(98)00898-9 [DOI] [PubMed] [Google Scholar]
- Sakaue, S., Kanai M., Tanigawa Y., Karjalainen J., Kurki M., Koshiba S., Narita A., Konuma T., Murakami Y., Akiyama M., et al. 2021. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53:1415–1424. 10.1038/s41588-021-00931-x [DOI] [PubMed] [Google Scholar]
- Sasai, M., Sakaguchi N., Ma J.S., Nakamura S., Kawabata T., Bando H., Lee Y., Saitoh T., Akira S., Iwasaki A., et al. 2017. Essential role for GABARAP autophagy proteins in interferon-inducible GTPase-mediated host defense. Nat. Immunol. 18:899–910. 10.1038/ni.3767 [DOI] [PubMed] [Google Scholar]
- Saunders, A., Webb L.M.C., Janas M.L., Hutchings A., Pascall J., Carter C., Pugh N., Morgan G., Turner M., and Butcher G.W.. 2010. Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood. 115:3249–3257. 10.1182/blood-2009-08-237586 [DOI] [PubMed] [Google Scholar]
- Schulteis, R.D., Chu H., Dai X., Chen Y., Edwards B., Haribhai D., Williams C.B., Malarkannan S., Hessner M.J., Glisic-Milosavljevic S., et al. 2008. Impaired survival of peripheral T cells, disrupted NK/NKT cell development, and liver failure in mice lacking Gimap5. Blood. 112:4905–4914. 10.1182/blood-2008-03-146555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwefel, D., Arasu B.S., Marino S.F., Lamprecht B., Kochert K., Rosenbaum E., Eichhorst J., Wiesner B., Behlke J., Rocks O., et al. 2013. Structural insights into the mechanism of GTPase activation in the GIMAP family. Structure. 21:550–559. 10.1016/j.str.2013.01.014 [DOI] [PubMed] [Google Scholar]
- Schwefel, D., Frohlich C., Eichhorst J., Wiesner B., Behlke J., Aravind L., and Daumke O.. 2010. Structural basis of oligomerization in septin-like GTPase of immunity-associated protein 2 (GIMAP2). Proc. Natl. Acad. Sci. USA. 107:20299–20304. 10.1073/pnas.1010322107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadur, B., Asherie N., Kfir-Erenfeld S., Dubnikov T., NaserEddin A., Schejter Y.D., Elpeleg O., Mor-Shaked H., and Stepensky P.. 2021. A human case of GIMAP6 deficiency: A novel primary immune deficiency. Eur. J. Hum. Genet. 29:657–662. 10.1038/s41431-020-00773-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin, M., Farkasovsky M., Hauer F., Kuhlmann D., Macara I.G., Weyand M., Stark H., and Wittinghofer A.. 2007. Structural insight into filament formation by mammalian septins. Nature. 449:311–315. 10.1038/nature06052 [DOI] [PubMed] [Google Scholar]
- Skarnes, W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T., et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 474:337–342. 10.1038/nature10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, B.J., Ferdinand J.R., Young M.D., Mitchell T.J., Loudon K.W., Riding A.M., Richoz N., Frazer G.L., Staniforth J.U.L., Vieira Braga F.A., et al. 2019. Spatiotemporal immune zonation of the human kidney. Science. 365:1461–1466. 10.1126/science.aat5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye, S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., Klein C., Morio T., Oksenhendler E., Picard C., et al. 2021. The ever-increasing array of novel inborn errors of immunity: An interim update by the IUIS committee. J. Clin. Immunol. 41:666–679. 10.1007/s10875-021-00980-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa, G., Schaft J., van der Hoeven F., Glaser S., Anastassiadis K., Zhang Y., Hermann T., Stremmel W., and Stewart A.F.. 2004. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 38:151–158. 10.1002/gene.20012 [DOI] [PubMed] [Google Scholar]
- Torres, J.Z., Miller J.J., and Jackson P.K.. 2009. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics. 9:2888–2891. 10.1002/pmic.200800873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretina, K., Park E.S., Maminska A., and MacMicking J.D.. 2019. Interferon-induced guanylate-binding proteins: Guardians of host defense in health and disease. J. Exp. Med. 216:482–500. 10.1084/jem.20182031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, V.W., Saunders A.E., Hutchings A., Pascall J.C., Carter C., Bright N.A., Walker S.A., Ktistakis N.T., and Butcher G.W.. 2010. The autoimmunity-related GIMAP5 GTPase is a lysosome-associated protein. Self Nonself. 1:259–268. 10.4161/self.1.3.12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y., Li J., Kang R., and Tang D.. 2020. Interplay between lipid metabolism and autophagy. Front. Cell Dev. Biol. 8:431. 10.3389/fcell.2020.00431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Araki K., Li S., Han J.H., Ye L., Tan W.G., Konieczny B.T., Bruinsma M.W., Martinez J., Pearce E.L., et al. 2014. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat. Immunol. 15:1152–1161. 10.1038/ni.3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., and Tashiro Y.. 1998. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23:33–42. 10.1247/csf.23.33 [DOI] [PubMed] [Google Scholar]
- Yano, K., Carter C., Yoshida N., Abe T., Yamada A., Nitta T., Ishimaru N., Takada K., Butcher G.W., and Takahama Y.. 2014. Gimap3 and Gimap5 cooperate to maintain T-cell numbers in the mouse. Eur. J. Immunol. 44:561–572. 10.1002/eji.201343750 [DOI] [PubMed] [Google Scholar]
- Young, M.D., Mitchell T.J., Vieira Braga F.A., Tran M.G.B., Stewart B.J., Ferdinand J.R., Collord G., Botting R.A., Popescu D.M., Loudon K.W., et al. 2018. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 361:594–599. 10.1126/science.aat1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, X., Acun A., and Zorlutuna P.. 2017. Transcriptome profiling of 3D co-cultured cardiomyocytes and endothelial cells under oxidative stress using a photocrosslinkable hydrogel system. Acta Biomater. 58:337–348. 10.1016/j.actbio.2017.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N., Yang X., Yuan F., Zhang L., Wang Y., Wang L., Mao Z., Luo J., Zhang H., Zhu W.G., and Zhao Y.. 2018. Increased amino acid uptake supports autophagy-deficient cell survival upon glutamine deprivation. Cell Rep. 23:3006–3020. 10.1016/j.celrep.2018.05.006 [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Dove C.G., Hor J.L., Murdock H.M., Strauss-Albee D.M., Garcia J.A., Mandl J.N., Grodick R.A., Jing H., Chandler-Brown D.B., et al. 2014. DOCK8 regulates lymphocyte shape integrity for skin antiviral immunity. J. Exp. Med. 211:2549–2566. 10.1084/jem.20141307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Cheng X., Yu L., Yang J., Calvo R., Patnaik S., Hu X., Gao Q., Yang M., Lawas M., et al. 2016. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat. Commun. 7:12109. 10.1038/ncomms12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.G., and Zhang H.. 2019. Core autophagy genes and human diseases. Curr. Opin. Cell Biol. 61:117–125. 10.1016/j.ceb.2019.08.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows clinical and laboratory findings of GIMAP6 patients.
shows histological findings of Gimap6−/− tissues.