Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the causative agent of the coronavirus disease 2019 (COVID‐19) pandemic. This disease has currently affected more than 346 million people and resulted in more than 5.5 million deaths in many countries. Neutralising monoclonal antibodies (MAbs) against the SARS‐CoV‐2 virus could serve as prophylactic/therapeutic agents in COVID‐19 infection by providing passive protection against the virus in individuals. Until now, no Food and Drug Administration/European Medicines Agency‐approved neutralising MAb against SARS‐CoV‐2 virus exists in the market, though a number of MAbs have been authorised for emergency use. Therefore, there is an urgent need for development of efficient anti‐SARS‐CoV‐2 neutralising MAbs for use in the clinic. Moreover, neutralising anti‐SARS‐CoV‐2 MAbs could be used as beneficial tools for designing epitope‐based vaccines against the virus. Given that the target epitope of a MAb is a crucial feature influencing its neutralising potency, target epitopes of neutralising anti‐SARS‐CoV‐2 MAbs already reported in the literature and reactivity of these MAbs with SARS‐CoV‐2 variants are reviewed herein.
Keywords: COVID‐19, epitope mapping, immunotherapy, neutralising monoclonal antibody, RBD, SARS‐CoV‐2 virus
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- ADCC
antibody‐dependent cellular cytotoxicity
- ADCP
antibody‐dependent cellular phagocytosis
- COVID‐19
coronavirus disease 2019
- Cryo‐EM
cryo‐electron microscopy
- CTD
c‐terminal domain
- E
envelope
- EMA
European Medicines Agency
- EUA
emergency use authorisation
- Fab
fragment antigen‐binding
- Fc
fragment crystallisable
- FDA
food and drug administration
- FP
fusion peptide
- HR
hepta peptide repeat sequence
- IGHV
immunoglobulin heavy chain variable region
- IGLV
immunoglobulin light chain variable region
- M
membrane
- MAb
monoclonal antibody
- MERS‐CoV
middle east respiratory syndrome‐related coronavirus
- N
nucleocapsid
- nAb
neutralising antibody
- NK
natural killer cells
- nM
nanomolar
- NTD
n‐terminal domain
- RBD
receptor‐binding domain
- RBM
receptor‐binding motif
- RNA
ribonucleic acid
- S
spike
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- SH
stem helix
- TM
transmembrane
- TMPRSS2
transmembrane protease, serine 2
- VOC
variant of concern
- VOI
variant of interest
1. SARS‐CoV‐2 VIRUS AND CORONAVIRUS DISEASE 2019 INFECTION
Coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), was first identified in Wuhan, China in December 2019. 1 Until 23rd of January 2022, the disease affected more than 346 million people and resulted in more than 5.5 million deaths in many countries. 2 Currently, there are several effective drugs licenced based on the results of randomized clinical trials. The Food and Drug Administration (FDA) approved antiviral drugs including remdesivir, paxlovid, and molnupiravir for use in patients 12 years of age and older for the treatment of COVID‐19 requiring hospitalisation. The FDA and the World Health Organization (WHO) recommend several therapeutics for COVID‐19 such as IL‐6 receptor blockers (tocilizumab or sarilumab) as well as systemic corticosteroids in patients with severe or critical disease. 3 , 4 , 5 , 6 , 7 Meanwhile, many efforts have been made to produce SARS‐CoV‐2 neutralising monoclonal antibodies (MAbs). Sotrovimab, REGN‐CoV‐2, and the cocktail of bamlanivimab and etesevimab have been authorised for emergency use as post‐exposure prophylaxis for COVID‐19 in adults and children at high risk for progression to severe COVID‐19. 8 , 9 , 10 Considering the emergence of new variants and lack of efficacy of a number of the neutralising MAbs against the newly emerged Omicron variant, there is imperative need for MAbs able to efficiently cross‐neutralise various variants to be used as passive immunotherapy for the control SARS‐CoV‐2 infection and/or disease severity.
1.1. Structure of the SARS‐CoV‐2 virus
SARS‐CoV‐2 is a 29,881 bp single‐stranded ribonucleic acid (RNA)‐enveloped virus containing structural proteins including spike (S), envelope (E), membrane (M), and nucleocapsid (N) as well as non‐structural proteins including 3‐chymotrypsin‐like protease, papain‐like protease, and RNA‐dependent RNA polymerase (Figure 1a). Structural proteins are involved in virus attachment to the host cell membrane and subsequent entry into cells, viral assembly, and release from host cells. Among the structural proteins, the S protein is responsible for the virus binding to the host cell receptors and subsequent entrance into cells. Three other structural proteins including E, M, and N proteins contribute to viral assembly resulting in formation of the virus whole particle. N protein also facilitates packaging of the viral genome into a helical ribonucleocapsid. On the other hand, non‐structural proteins contribute to viral genome replication and transcription. 11
FIGURE 1.
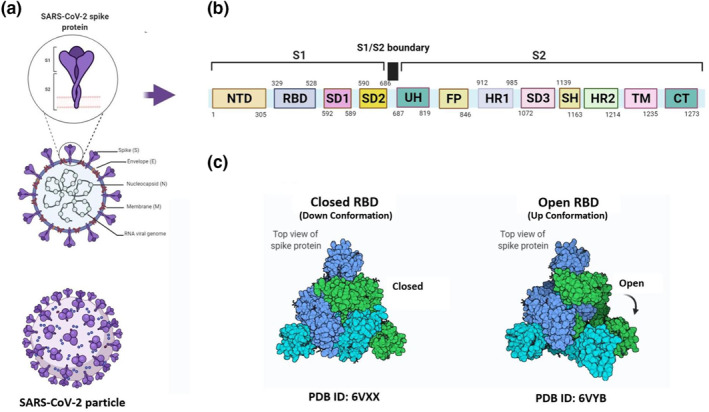
Structure of SARS‐CoV‐2 virus and spike Protein (a) Schematic representation of SARS‐CoV‐2 particle and structural proteins of the virus (b) Primary structure of spike protein (c) Diagram of S protein structure in the closed and open conformations (adapted from the Protein Database, 113 , 114 ). SARS‐CoV‐2: Severe acute respiratory syndrome coronavirus 2, S: Spike; NTD: N‐terminal domain; RBD: Receptor binding domain; SD: Subdomain; UH: Upstream helix; FP: Fusion peptide; HR: Heptad repeat; SH: Stem helix; TM: Transmembrane; CT: Cytoplasmic tail
1.2. S protein structure and function
The S protein of SARS‐CoV‐2 virus (1273 amino acids (aa)) is a clove‐shaped, type I Transmembrane (TM) protein and contains a large N‐terminal extracellular domain (aa: 1–1212), a TM domain (TM; aa: 1213–1237), and a short C‐terminal intracellular domain (aa: 1238–1273). It consists of a signal peptide (aa: 1–13), S1 subunit (aa: 14–685), and an S2 subunit (aa: 686–1273). 12 Moreover, the S1 subunit, responsible for binding the virus to host cell receptors, is composed of the N‐terminal domain (NTD; aa: 18–305), the C‐terminal receptor‐binding domain (RBD; aa: 329–528), subdomain‐1 (SD1; aa: 529–589), and SD2 (aa: 590–686; Figure 1b). 12 The Receptor binding domain (RBD) is composed of two sub‐domains including core sub‐domain composed of a β‐sheet with 5 anti‐parallel strands (β1, β2, β3, β4, and β7) in the inner side of the S protein and receptor‐binding motif (RBM) from the outer side that extends from the core sub‐domain and consists of β5 and β6 strands. 13 , 14
Angiotensin‐converting enzyme 2 (ACE2), a membrane‐bound zinc‐containing enzyme expressed on many tissues including lungs, arteries, heart, kidneys, and intestines, has been demonstrated to be the main receptor for virus attachment to target cells. 15 Moreover, transmembrane protease serine 2 (TMPRSS2) has been proposed for S protein priming. 16 The RBM sub‐domain of RBD, which forms a concave surface accommodating the N‐terminal α‐helix of the ACE2, is responsible for the virus binding to the ACE2. 12 The SARS‐CoV virus also employs a similar mechanism to bind to host cells. On the other hand, the S2 subunit mediates fusion of the viral membrane with the host cell membrane allowing virus entry into target cells. 12 The S2 subunit consists of upstream helix (UH; aa: 687–819), N‐terminal fusion peptide (FP; aa: 820–846), hepta peptide repeat sequence one (HR1; aa: 912–985), SD3 (aa: 1072–1139), stem helix (SH; aa: 1139–1163), HR2 (aa: 1163–1212), TM domain (aa: 1213–1237), and intracellular domain (aa: 1238–1273; Figure 1b). 12 , 17
The S protein possesses two distinct conformational states including prefusion and postfusion conformations. The prefusion state of the S protein, composed of three S1 subunits and three S2 subunits, exists in two conformations: (1) a closed conformation in which all three protomers of RBDs are hidden and thus preventing RBD‐ACE2 interaction (down conformation of RBD or receptor inaccessible state), (2) an open conformation in which one protomer of RBD is exposed allowing for RBD‐ACE2 interaction (up conformation of RBD or receptor accessible state; Figure 1c). Indeed, the up conformation of RBD provides the surface required for RBD interaction with the ACE2. 18 , 19 Upon RBD binding to the ACE2, a conformational change in S protein structure occurs allowing for proteolytic cleavage of the protein at the S1‐S2 boundary by host proteases. This converts S protein from the inactive prefusion state into the active postfusion state resulting in fusion of the viral membrane with the host cell membrane and entrance into the cell. 20 Though TMPRSS2 has been proven as the protease responsible for cleavage of the S protein, other host proteases such as trypsin were also recognized for their function in cleavage of the S protein. 21 Serum levels of antibodies specific for the spike RBD increase during the two weeks after onset of symptom. Higher levels of RBD‐specific IgM have been shown in deceased COVID‐19 patients rather than recovered patients. Also, a significant correlation was reported between RBD‐specific IgG and IgM in both groups of patients. 22 , 23 Furthermore, several studies found a positive correlation between serum neutralising capacity and disease severity in recovered patients with a wide range of disease severity (severe, moderate, mild, and asymptomatic). 24 , 25 , 26 , 27
2. NEUTRALISING ANTIBODIES
Passive protection against microbial agents, including viruses, could be achieved by neutralising antibodies (nAb). Neutralising antibodies bind to microbial structures responsible for binding to target cell receptors through their variable regions and consequently prevent cell entry and neutralise the toxic effects of bacterial toxins or any biological 28 (Figure 2).
FIGURE 2.
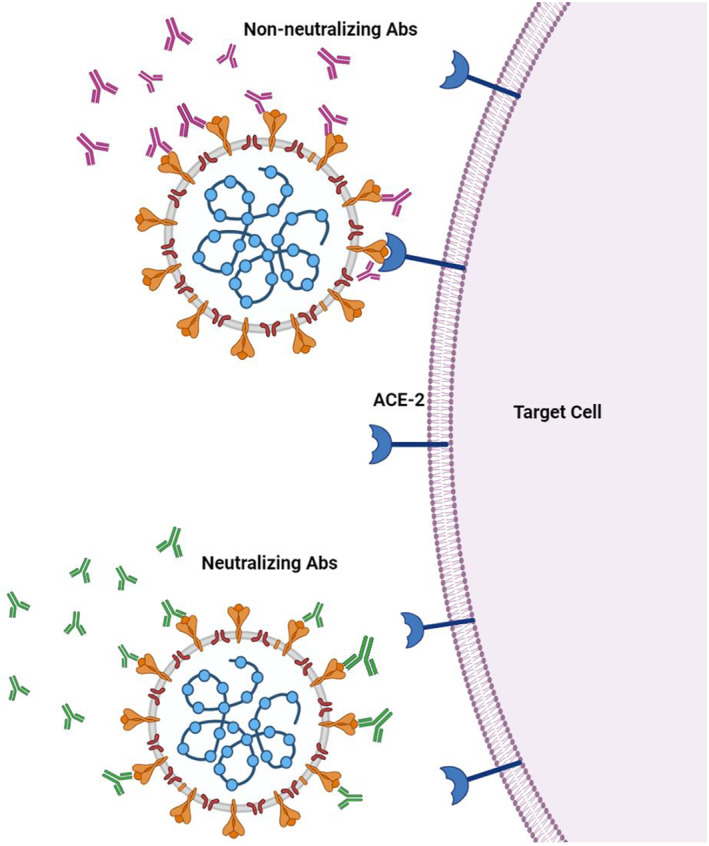
Prevention of virus binding to its receptor on target cell by neutralising antibodies. Binding of (a) non‐neutralising antibodies and (b) neutralising antibodies to spike protein. ACE2: Angiotensin‐converting enzyme two
Neutralising antibodies are a polyclonal pool of antibodies composed of a mixture of antibody clones recognising different epitopes of the corresponding antigen. These antibodies are prepared from human plasma of hyper‐immunised individuals or convalescent patients. 29 On 23 August 2020, FDA issued an emergency use authorisation (EUA) for COVID‐19 convalescent plasma as an investigational product for treatment of hospitalised patients with COVID‐19. 30 , 31 However, plasmas from infected patients with SARS‐CoV‐2 or convalescent patients have been shown to contain neutralising anti‐SARS‐CoV‐2 antibodies with varying neutralising capacity levels. 32 , 33 , 34 , 35 The nAb titre required for prevention of COVID‐19 infection in humans has not been determined yet. Using a non‐human primate model of COVID‐19 infection, prevention of clinical signs of the disease and reduced viral loads in bronchoalveolar lavage and nasal mucosa could be observed in re‐infected NHPs compared with post‐primary infection. The serum samples could potently neutralise the SARS‐CoV‐2 pseudovirus at a titre of 1:100 (ranging from 1:83‐1:197) and the authentic SARS‐CoV‐2 virus at a titre of 1:35‐1:326 on day 35 after rechallenge. 36
Polyclonal preparations of the antibodies have several limitations including insufficient level of neutralising potency in donor plasma, rapid decline of nAbs in convalescent patients, lot‐to‐lot heterogeneity, lack of plasma donors, and possibility of transmission of microbial agents and adverse reactions to plasma proteins. 29 , 37 , 38 Interestingly, neutralising MAbs targeting microbial antigens including COVID‐19 lack these limitations and could therefore be considered as prophylactic/therapeutic alternative for the passive immunotherapy. 29 , 37 Until now, no FDA/European Medicines Agency (EMA)‐approved neutralising MAb for COVID‐19 infection has entered in clinic, although a few number of the MAbs have been authorised for emergency use. 39 , 40 Therefore, there is an urgent need for development of efficient neutralising anti‐SARS‐CoV‐2 MAbs. Given that the target epitope of a MAb is a crucial feature influencing its neutralising potency, herein, epitope specificity of the neutralising anti‐SARS‐CoV‐2 MAbs already reported in the literature is delineated and discussed.
3. EPITOPE MAPPING OF NEUTRALISING ANTI‐SARS‐CoV‐2 MAbs
Identification of the target epitope of an antibody molecule is instrumental in development of effective prophylactic therapeutics and epitope‐based vaccines as well as molecular elucidation of MAb neutralising activities. 41 , 42 In a recent study, we performed epitope mapping of RBD in COVID‐19 patients' sera using a panel of linear epitopes spanning RBD. Our results demonstrated involvement of mostly conformational disulfide bond‐dependent epitopes in RBD‐specific IgG antibody 43 . Review of literature for neutralising anti‐SARS‐CoV‐2 MAbs indicates that all reported nAbs recognise epitopes within only the S protein and most of them are directed against the RBD (Figure 3, Table 1).
FIGURE 3.
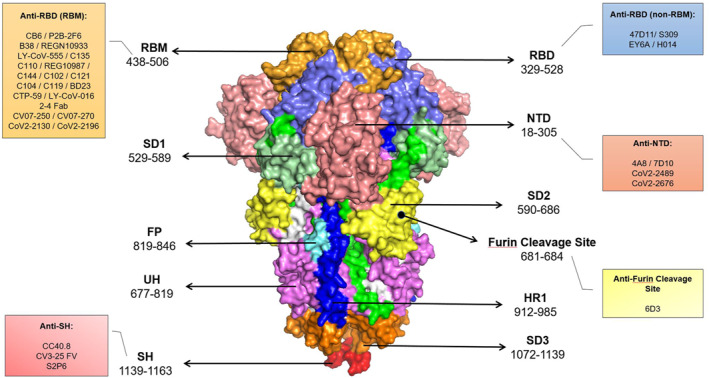
SARS‐CoV‐2 neutralising monoclonal antibodies (MAbs) grouped based on targeting regions of the spike protein. SARS‐CoV‐2 trimeric spike protein is illustrated showing S1 domain regions: the N‐terminal domain (NTD) (salmon), Receptor binding domain (RBD) (slate), receptor‐binding motif (RBM) (bright orange), SD1 (palegreen), and SD2 (yellow) and S2 domain regions: Upstream helix (UH) (violet), Fusion peptide (FP) (aquamarine), HR1 (blue), SD3 (orange) and Stem helix (SH) (red). SARS‐CoV‐2: Severe acute respiratory syndrome coronavirus 2, S: Spike; NTD: N‐terminal domain; RBD: Receptor binding domain; RBM: Receptor‐binding motif, SD: Subdomain; UH: Upstream helix; FP: Fusion peptide; HR: Heptad repeat; SH: Stem helix; TM: Transmembrane; CT: Cytoplasmic tail
TABLE 1.
Major characteristics of the reported neutralising anti‐SARS‐CoV‐2 monoclonal antibodies (MAbs)
| Ref. No. | MAb(s) designation | Target epitope | Inhibition of RBD‐ACE2 interaction | In vivo neutralisation activity | Affinity constant (nM) | IGHV and IGLV genes usage |
|---|---|---|---|---|---|---|
| 35 | 4A2, 4A12, 4D5, 4A10 | RBD | Yes | NI | 1.03–5.82 | NI |
| 32 | C101, C119, C121, C135, C145 | RBD | NI | NI | NI | IGHV: Multiple IGHVs |
| IGLV: Multiple IGLVs | ||||||
| 33 | ‐ | RBD | Yes | Prophylactic efficacy in syrian hamster (4.2 mg/kg) | NI | IGHV: IGHV1, IGHV3 |
| IGLV: NI | ||||||
| 45 | ‐ | RBD | NI | NI | NI | IGHV: Multiple IGHVs |
| IGLV: Multiple IGLVs | ||||||
| 44 | ‐ | RBD | NI | NI | NI | NI |
| 46 | BD23 | RBD | Yes | Prophylactic efficacy in hACE2 transgenic mice (20 mg/kg) | <15.9 | NI |
| Therapeutic efficacy in hACE2 transgenic mice (20 mg/kg) | ||||||
| 51 | COV2‐2196, COV2‐2130 | RBD | Yes | Prophylactic efficacy in hACE2 transgenic mice (10 mg/kg) | NI | NI |
| 52 | rRBD‐15 | RBD | Yes | NI | NI | NI |
| 49 | CV07–209, CV07–250 | RBD | Yes | Prophylactic efficacy in syrian hamster (18 mg/kg) | 0.006–1.1 | IGHV: IGHV1–2, IGHV3–53, IGHV3–66 |
| Therapeutic efficacy in syrian hamster (18 mg/kg) | IGLV: IGVK1–33, IGVK2–14 | |||||
| 60 | B38 | RBD | Yes | Therapeutic efficacy in hACE2 transgenic mice (25 mg/kg) | 1–100 | IGHV: IGHV1, IGHV3 |
| IGLV: IGVK1, IGVK2, IGVK3 | ||||||
| 67 | CB6 | RBD | Yes | Prophylactic efficacy in rhesus macaques monkey (50 mg/kg) | 2.49–68 | NI |
| Therapeutic efficacy in rhesus macaques monkey (50 mg/kg) | ||||||
| 65 | MW05 | RBD | Yes | Prophylactic efficacy in rhesus macaques monkey (40 mg/kg) | 0.40–0.46 | NI |
| Therapeutic efficacy in rhesus macaques monkey (40 mg/kg) | ||||||
| 61 | P2B‐2F6 | RBD | Yes | NI | 5.14 | NI |
| 70 | 47D11 | RBD | No | NI | NI | NI |
| 71 | S309 | RBD | No (Fc‐dependent effector functions, including ADCC and ADCP) | NI | NI | NI |
| 63 , 110 | REGN10933, REGN10987 | RBD | Yes | Prophylactic efficacy in rhesus macaques (25, 50 and 150 mg/kg) | NI | IGHV: IGHV3‐53, IGHV3‐66, IGHV2‐70 |
| Fc‐dependent effector functions, including ADCC and ADCP | Therapeutic efficacy in golden hamster (50, 5, 0.5 mg/kg) | IGLV: IGVK1‐9, IGVK1‐33, IGVK1‐39 | ||||
| 54 , 53 | LY‐CoV555 | RBD | Yes | Prophylactic efficacy in rhesus macaques (2.5 mg/kg) | NI | NI |
| Therapeutic efficacy in outpatients with mild or moderate COVID‐19 disease (2800 mg) | ||||||
| 111 | ‐ | RBD | Yes | NI | NI | NI |
| 112 | ‐ | RBD | Yes | NI | NI | IGHV:IGHV3‐53*01 |
| IGLV: IGKV3‐30*01 | ||||||
| 34 | MAb 2–4 | RBD | Yes | Prophylactic efficacy in syrian hamster (1.5 mg/kg) | NI | IGHV: IGHV3‐30 |
| IGLV:IGKV3‐20 | ||||||
| 50 | ‐ | RBD | Yes | NI | 1.8–15.6 | IGHV3‐64: IGKV1‐39 |
| IGHV3‐53: IGKV3‐20 | ||||||
| IGHV3‐53: IGKV1‐12 | ||||||
| IGHV3‐66: IGKV3‐20 | ||||||
| IGHV3‐23: IGLV3‐21 | ||||||
| 59 | CTP59 | RBD | Yes | Therapeutic efficacy in ferret, hamster, and rhesus monkey (3 and 30 mg/kg) | 0.027 | IGHV2‐70 |
| 77 | COV2‐2676, COV2‐4489 | NTD | Yes | Prophylactic and therapeutic efficacy in heterozygous K18‐hACE2 c57BL/6J mice (10 mg/kg) | NI |
|
| 33 | ‐ | Non‐RBD regions of S protein | NI | NI | NI | NI |
| 75 | ‐ | Non‐RBD regions of S protein | NI | NI | NI | NI |
| 74 | 4A8 | Non‐RBD regions of S1 subunit | NI | NI | 92.7 | NI |
Abbreviations: ACE2: Angiotensin‐converting enzyme 2, ADCC: Antibody‐dependent cellular cytotoxicity, ADCP: Antibody‐dependent cellular phagocytosis, IGHV: Immunoglobulin heavy chain variable region, IGLV: Immunoglobulin light chain variable region, NI: Not identified, nM: Nanomolar, NTD: N‐terminal domain, RBD: Receptor‐binding domain, S: spike.
3.1. Neutralising monoclonal antibodies recognising the Receptor binding domain fragment of the S protein
Given the crucial role of the RBD fragment of the S protein in binding of the virus to its receptor on target cells, it is not surprizing that a major proportion of neutralising anti‐SARS‐CoV‐2 antibodies are directed against RBD (Figure 3, Table 1). Analysis of MAbs isolated from 25 COVID‐19‐infected patients showed that a majority of the nAbs recognized the S1 subunit of the virus. Removal of anti‐RBD antibodies from sera of patients abolished their neutralising activity, highlighting the dependency of the neutralising activity to anti‐RBD antibodies. 44 Accordingly, analysis of anti‐SARS‐CoV‐2 MAbs for their neutralising activities against the virus showed that a large number of neutralising MAbs (67/70) recognise the RBD fragment. 45 In line with these findings, none of the non‐RBD‐binding MAbs showed neutralising activities in a different study. 46 Therefore, RBD seems to be the most crucial domain of the S protein for eliciting nAbs against the virus.
3.1.1. Neutralising anti‐RBD monoclonal antibodies that interfere with RBD‐ACE2 interaction
Considering that the up conformation of RBD provides the surface required for RBD interaction with the ACE2, it is assumed that neutralising anti‐RBD MAbs should recognise S protein in the up conformation. 47 Barnes et al. classified neutralising anti‐RBD MAbs into 5 groups: (1) MAbs that recognise the up conformation of the S protein and prevent RBD‐ACE2 interaction, (2) MAbs that recognise both up and down conformations of the S protein and prevent RBD‐ACE2 interaction, (3) MAbs that recognise the up conformation of the S protein, but the epitopes are located outside of the ACE2‐binding site of RBD, (4) MAbs that recognise both up and down conformations of the S protein and do not bind to the ACE2‐binding site of RBD, 5) MAbs that recognise the down conformation of the S protein and prevent RBD‐ACE2 interaction. 19 , 48 Therefore, majority of neutralising anti‐RBD MAbs (4/5 groups) could recognise S protein in the up conformation. However, antibodies from group 5 unexpectedly bind to RBD epitopes that are solely available on the down conformation of the S protein. In efforts to explore neutralising mechanisms of these MAbs, the authors found MAbs binding to RBD epitopes on the down conformation locked S protein in the down conformation and consequently prevented accessibility of the ACE2‐binidng surface of RBD to the ACE2. 48 Liu et al. reported a similar finding. They isolated a neutralising anti‐RBD MAb (MAb 2–4) that bound to S protein in the down conformation and locked the protein in the receptor inaccessible state. 34
Robbiani et al. showed 54% of RBD‐binding MAbs neutralised the virus. 32 In accordance with this result, 46% of anti‐RBD MAbs isolated by Kreye et al. showed neutralising activity. 49 We have recently generated a panel of mouse MAbs against RBD and observed that less than half of these MAbs display neutralising activity in pseudovirus‐based neutralising assays, suggesting that recognising RBD is not necessarily sufficient for virus neutralisation (unpublished data). These findings indicate that RBD contains potent neutralising epitopes even if not all RBD epitopes contribute to virus neutralisation. Robbiani et al. identified three distinct neutralising epitopes on RBD including C144 and C101 in group 1; C121 and C119 in group 2 and C135 in group 3. They showed that groups 1 and 2 antibodies could bind to the RBD immunocomplexed with group 3 antibodies. Of note, groups 1 and 2 displayed different properties in binding specificity, so that group 1 could bind to the RBD immunocomplexed with group 2, but not vice versa. 32 Rogers et al. have also identified three distinct neutralising epitopes on RBD: the most potent neutralising MAbs were found to recognise the RBD‐A epitope. Further analysis showed that the RBD‐A epitope spans the ACE2‐binding site in RBD. These RBD‐A specific MAbs also efficiently neutralised the virus when administered prophylactically (antibody administration before virus challenge) in a Syrian hamster animal model of COVID‐19 infection. 33 Consistently, MAbs inhibiting RBD binding to ACE2 displayed the strongest neutralising activity. These MAbs also revealed in vivo efficiency when administered either prophylactically or therapeutically (the antibody administration after the virus challenge). BD23, another nAb in their panel that bound the “down” conformation of RBD also competed with ACE2. 46 Moreover, the MAbs that interrupted RBD‐ACE2 interaction imposed neutralising activity. Among the panel of human neutralising MAbs targeting the SARS‐CoV‐2 RBD isolated from patients at the acute phase, a subset of them inhibited binding to the human ACE2. 50 Also, a large number of neutralising anti‐RBD MAbs obtained by Zost et al. interfered with RBD‐ACE2 interaction. 51 The neutralising anti‐RBD MAb, rRBD‐15, inhibited binding of RBD to ACE2. 52 Neutralising anti‐RBD MAb LY‐CoV555, that prevented RBD‐ACE2 interaction, was successfully used in a phase two clinical trial conducted on outpatients with mild or moderate COVID‐19 disease. A single dose (2800 mg) administration of this MAb, also known as bamlanivimab, significantly improved clinical outcomes in patients by reducing severity of symptoms and viral load. 53 , 54 However, bamlanivimab was revoked by FDA because of increased risk for treatment failure due to continued development of SARS‐CoV‐2 escape variants. 55 Altogether, these findings indicate that the most potent neutralising epitopes are the epitopes involved in RBD binding to the ACE2. Based on this notion, Liu et al. used an innovative approach to isolate neutralising anti‐SARS‐CoV‐2 MAbs. In this approach, they initially isolated MAbs based on positive selection for RBD followed by negative selection of the isolated MAbs for a mutant RBD in which RBD residues that contribute to ACE2 binding were deleted. Indeed, these selections ensured isolation of neutralising MAbs including 4A2, 4A12, 4D5, and 4A10 accurately recognising RBD epitopes involved in ACE2 binding. 35
Cryo‐electron microscopy studies showed that RBD interact with ACE2 through hydrogen and ionic bonds. Residues A475, N487, E484, and Y453 in RBM interact with residues S19, Q24, K31, and H34 of ACE2, respectively. Moreover, the residues Q498, T500, and N501 form hydrogen bonds of RBD interact with Y41, Q42, K353, and R357 of ACE2. 13 , 14 In another study, RBD residues of Y505, Y449, G496, F497, and G502 bound to ACE2 residues including E37, D38, D38, K353, and G354, respectively. An ionic bond between P491 of RBD with K31 of ACE2 also contributed to RBD‐ACE2 interaction. 56 The other RBD residues, including T470, F486, Y489, and Q493, were also identified as crucial residues of RBD for interaction with the ACE2. 14 , 57 Furthermore, the SARS‐CoV‐2 RBM provides a larger and more favourable contact interface with ACE2 in comparison to the SARS‐CoV RBM. 58 In sum, RBD residues, including Y449, Y453, L455, F456, T470, A475, E484, F486, N487, Y489, F490, P491, Q493, G496, F497, Q498, T500, N501, G502 and Y505, might be considered as ACE2‐interacting residues of RBD, and all are located in SARS‐CoV‐2 RBM spanning from residue 438–506 of the S sequence (Figure 4). 58
FIGURE 4.

Assignment of epitope residues of spike protein for binding to Angiotensin‐converting enzyme 2 (ACE2) and SARS‐CoV‐2 neutralising monoclonal antibodies (MAbs). Epitope residues of (a) Receptor binding domain (RBD), (b) N‐terminal domain (NTD), (c) Furin cleavage site, and (d) SH‐targeting neutralising MAbs are highlighted in different colours. ACE2: Angiotensin‐converting enzyme 2, SARS‐CoV‐2: Severe acute respiratory syndrome coronavirus 2; NTD: N‐terminal domain; RBD: Receptor binding domain; SH: Stem helix
Crystallographic analysis showed that neutralising anti‐SARS‐CoV‐2 MAbs interfere with RBD‐ACE2 interaction to different extents. In a panel of neutralising MAbs, the two highest neutralising MAbs, CV07–209 and CV07–250, highly interfered with RBD‐ACE2 interaction. The RBD target epitope of MAb CV07–250 completely overlapped with the ACE2‐binding site. On the contrary, CV07‐270 binds from a different angle with partial overlapping with the ACE2 binding site. 49 Neutralising MAb CTP59 (also known as regdanvimab) showed direct recognition of RBM by binding to 12 (K417, Y449, Y453, L455, F456, E486, Y489, F490, Q493, G496, and Y505) of the 21 residues of the RBD/ACE2 interface without conformational changes. 59 Hence, the neutralising potencies of anti‐SARS‐CoV‐2 MAbs are affected by the extent of their abilities to interfere with RBD‐ACE2 interaction. This depends on the target residues of the epitope recognized by the corresponding MAb. For instance, MAb B38 completely abolished RBD‐ACE2 interaction by binding to 86% of RBD/ACE2 interacting residues. A single dose of this MAb (25 mg/kg) administrated to hACE2 transgenic mice after a viral challenge significantly reduced the viral load and inhibited pathologic damage in the lung tissue. 60
Although a few MAbs have been investigated for their paratope‐epitope interaction by cryo‐EM, crystallographic and/or mutagenesis experiments, current findings suggest paratope sites of some neutralising anti‐RBD MAbs interact with non‐RBM residues of RBD. Zost et al. found two of the most potent neutralising MAbs, including MAb COV2‐2196 (also known as tixagevimab or AZD8955) and MAb COV2‐2130 (also known as cilgavimab or AZD1061), recognise a linear peptide (60 aa) on RBD interacting with ACE2 binding. Crystal structure of a potent neutralising MAb, designated P2B‐2F6, recognising non‐ACE2‐interacting residues of RBD, including K444, G446, G447, N448, Y449, N450, L452, V483, E484, G485, F490, and S494, revealed interactions occurred between the MAb and the ACE2. 61 Also, CV07‐270 binds to a similar epitope as P2B‐2F6. 49 These interactions were detected between the light chain residues of the MAb, including R56, S58, G59, R63, S78, and G79 with D67, K68, A71, K74, E110, and K114 residues of ACE2, that prevented the efficient RBD‐ACE2 interaction (Figure 5). Thus, neutralising effect of an anti‐RBD MAb might be partially mediated via steric hindrance from the MAb on the ACE2 that consequently inhibits efficient ACE2 interaction with RBD. 61 Recently, a cocktail of two fully human non‐overlapping anti‐RBD MAbs (REGN‐COV2) received FDA EUA for treatment of mild to moderate COVID‐19 non‐hospitalised high‐risk patients. REGN10933 (casirivimab) and REGN10987 (imdevimab) were isolated from the Velocimmune mouse platform 62 and human B cells, respectively. Casirivimab binds to the spike‐like loop region of RBD on one side of the ACE2 interface from above, while imdevimab can only target RBD from the front or the lower left edge, providing the probability of simultaneous binding of two MAbs to distinct regions of the RBD. 63
FIGURE 5.
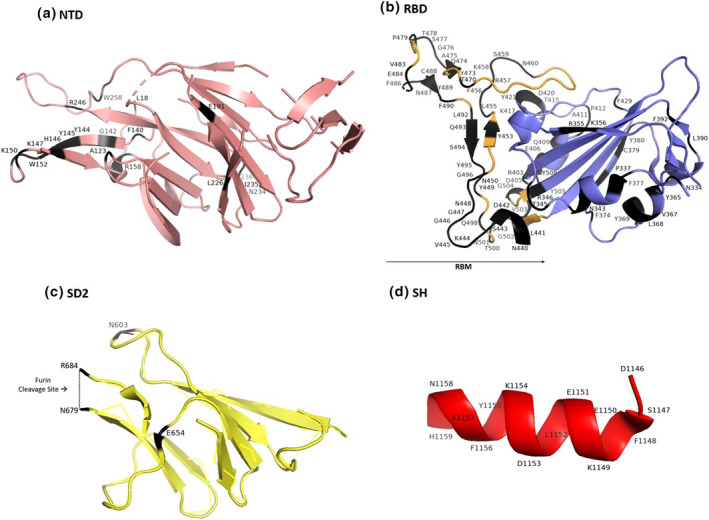
Assignment of epitope residues of (a) Receptor binding domain (RBD), (b) N‐terminal domain (NTD), (c) SD2, and (d) SH‐targeted by neutralising monoclonal antibodies (MAbs) illustrated in Figure 4. NTD: N‐terminal domain; RBD: Receptor binding domain; RBM: Receptor‐binding motif, SD2: Subdomain two; SH: Stem helix
Mutagenesis experiments allowed identifying hot spot residues. For example, F486 or N487 residues were defined as hot spots for MAb COV2‐2196 epitope binding. 51 , 64 Mutagenesis analysis for MW05 revealed E484 as the hot spot residue of RBD. In addition, the paratope of the MAb occupied a non‐ACE2‐interacting residue of RBD: F490. 65 MAb COV2‐2130 also recognized non‐ACE2‐interacting hot spot residues of RBD including K444 and G447. These findings indicate that mechanism(s) other than direct competition for RBD residues responsible for ACE2 interaction might mediate neutralising activity. Steric hindrance of the paratope site on the ACE2‐interacting surface of RBD may prevent efficient RBD binding to the ACE2 and, hence, partly explain the neutralising ability of the MAbs recognising non‐ACE2/RBD‐interacting domains. 51 , 66 Direct evidence supporting this notion comes from the crystallographic studies that investigated the epitope‐paratope interaction of a neutralising MAb, CB6. 67 Crystallography revealed steric hindrance of the paratope on the ACE2‐binding surface of RBD. In addition, great overlap in ACE2‐binding residues of RBD between the MAb and the ACE2 was identified indicating that the MAb also competitively prevented RBD‐ACE2 interaction. 67
Altogether, these studies indicated that anti‐RBD MAbs could neutralise the virus by preventing RBD‐ACE2 interaction. Monoclonal antibodies may hinder RBD‐ACE2 interaction through either direct competition of paratope for ACE2‐interacting residues of RBD and/or steric hindrance on the ACE2‐interacting surface of RBD. Alternatively, steric hindrance of the antibody paratope on the ACE2 could inhibit ACE2 interaction with RBD.
3.1.2. Neutralising anti‐RBD monoclonal antibodies unable to interfere with RBD‐ACE2 interaction
Evaluation of a panel of RBD‐specific anti‐SARS‐CoV‐2 MAbs for their abilities to inhibit RBD‐ACE2 interaction showed that only 26% of MAbs prevented RBD binding to ACE2. 33 Since RBD of SARS‐CoV and SARS‐CoV‐2 shares 73% homology, several studies have evaluated the neutralising potential of SARS‐CoV RBD‐specific MAbs to cross‐neutralise SARS‐CoV‐2 RBD. In this regard, Lindsley et al. reported cross reactivity of six SARS‐CoV RBD‐specific nAbs with SARS‐CoV‐2 RBD and showed that 18F3 and 7B11 cross‐neutralised SARS‐CoV‐2 infection. 18F3 recognized epitopes containing residues D392 and V394 in SARS‐CoV RBD which were conserved neutralising epitopes corresponding to residues D405 and V407 in SARS‐CoV‐2 RBD. 18F3 could not block binding between RBD and the ACE2 since its specific epitope did not overlap with the ACE2 binding site. 7B11 recognized epitopes containing I428, A430, and K439 in SARS‐CoV RBD which were not fully conserved in SARS‐CoV‐2 variants. Most epitopes recognized by 7B11 were in proximity of the ACE2 binding sites and resulted in blockade of RBD and ACE2 binding. 68 Moreover, a cross‐neutralising anti‐RBD MAb (47D11) did not inhibit RBD‐ACE2 interaction. 69 , 70 These findings suggest that neutralisation of SARS‐CoV‐2 can be achieved by Abs without interfering with ACE2 interaction. Interestingly, the human SARS‐CoV specific MAb 47D11 was able to cross‐neutralise the SARS‐CoV‐2 virus. Given that the core sub‐domain of RBD, rather than RBM, is more conserved between SARS‐CoV‐2 and SARS‐CoV viruses (aa identity of 86.3% for the core sub‐domain vs. 46.7% for RBM sub‐domain), the target epitope of MAb 47D11 is probably localised on the core sub‐domain of RBD of SARS‐CoV‐2. 70 S309 is another cross‐neutralising anti‐RBD MAb (also known as sotrovimab or Vir‐7831 which does not interfere with RBD‐ACE2 interaction) which recognises the non‐RBM region of RBD. 71 Therefore, it might be assumed that neutralising anti‐RBD MAbs, which are unable to inhibit RBD‐ACE2 interaction, recognise non‐RBM epitopes of RBD. It is not surprizing, as the RBM sub‐domain is responsible for the virus binding to ACE2. On the other hand, sotrovimab, which was recently authorised for emergency use by FDA, was found to enhance Fc‐dependent effector mechanisms including antibody‐dependent cellular cytotoxicity (ADCC) and antibody‐dependent cellular phagocytosis (ADCP) that could increase clearance of the virus as well as the infected cells. This suggests that activation of immune cells, including natural killer cells, monocytes, and macrophages by the neutralising anti‐RBD MAbs, could contribute to virus elimination. 71 EY6A is another RBD non‐RBM recognising nAb that cross‐reacts with SARS‐CoV. This MAb binds the highly conserved epitope, away from RBM. EY6A binds key residues involved in stabilizing the pre‐fusion spike. 72 H014 is a cross‐neutralising MAb which recognises an open RBD, non‐RBM conformational epitope. 73 In addition, comparison of neutralising activity of the intact antibody with its antigen‐binding fragment (Fab) showed higher neutralising activity by the intact IgG. This implies that cross‐linking and subsequent virus aggregation that facilitates virus clearance from circulation might partly mediate neutralisation by MAbs. 66 , 71 Intact forms of two other neutralising anti‐RBD MAbs possessed higher neutralising activity compared to their Fab fragments, and thus, the MAbs also caused Fc‐dependent effector functions including ADCC and ADCP. Interestingly, one of the MAbs recognized the ACE2‐binding surface of RBD suggesting that neutralising anti‐RBD MAbs that interfere with RBD‐ACE2 interaction may also contribute to virus elimination by increasing Fc‐mediated effector functions and virus cross‐linking. 63
3.2. Neutralising anti‐S monoclonal antibodies recognising non‐RBD epitopes
Although a major proportion of the reported anti‐SARS‐CoV‐2 neutralising MAbs are directed against the RBD fragment of the S protein, neutralising anti‐S antibodies recognising non‐RBD epitopes of S protein have also been reported. 33 , 34 , 74 , 75 Sera from 40% of COVID‐19‐infected patients contained both neutralising anti‐S1 and neutralising anti‐S2 antibodies, and only 4% of patients with neutralising activity developed only anti‐S2 antibodies. 44 Rogers et al. isolated anti‐S MAbs that recognized non‐RBD epitopes. However, in sharp contrast with anti‐RBD MAbs, only a minority of these MAbs demonstrated neutralising activity. 33
On the contrary, Liu et al. identified that 52% of the neutralising MAbs recognise non‐RBD epitopes, 42% of them were directed against NTD and 10% recognise neither RBD nor NTD epitopes. Anti‐NTD MAbs possessed similar or slightly higher neutralising potency compared to anti‐RBD MAbs indicating that anti‐S MAbs recognising non‐RBD epitopes could neutralise the virus as efficient as anti‐RBD MAbs. 34 Although it is currently unclear how antibody binding to the NTD fragment neutralises the virus, it is possible that anti‐NTD MAbs prevent the prefusion‐to‐postfusion conversion of the S protein. It has been shown that coronaviruses NTD could bind to carbohydrate contents of target cells which facilitates conversion of the S protein from the prefusion state to the postfusion state. 74 Accordingly, an anti‐NTD MAb (designated 7D10) that recognized the MERS‐CoV virus (a type of coronaviruses) inhibited prefusion‐to‐postfusion conformational change of the S protein. 76 Given that the amino acid sequence of NTD is highly conserved between different coronaviruses, anti‐NTD SARS‐CoV‐2 MAbs may neutralise the virus by inhibiting fusion of the viral membrane to the target cell. Cryo‐electron microscopy analysis of the complex of an anti‐NTD MAb (4A8) with NTD revealed that the MAb could restrain the conformational change of the S protein from prefustion to postfusion state upon binding to NTD loops, including N3 and N5 loops. 74 Also, only two MAbs designated COV2‐2676 and COV2‐2489, belonging to a panel of human MAbs against different epitopes on the NTD of SARS‐CoV‐2 S protein, displayed neutralising activity via inhibiting a post‐attachment step in the infection cycle. 77 Interestingly, Cheng et al. found that there is a superantigen‐like motif in the proximity of S1/S2 cleavage site that is similar to a staphylococcal enterotoxin B segment in sequence and structure. They reported that an anti‐SEB MAb, designated 6D3, cross‐reacts with this viral motif, especially the polybasic PRRA insert (aa: 681–684). This interaction resulted in prevention of infection through interfering with the proteolytic activity of TMPRSS2/furin and blocking the access of host cell proteases to the cleavage site. 78
Stem helix (aa: 1134–1151) is a highly conserved sequence within the S2 fusion subunit of β‐coronaviruses. It forms a surface exposed membrane‐proximal helical bundle and is critical for membrane fusion in the prefusion conformation of trimeric spike. It has been shown that it induces the antibody response during natural infection. 79 , 80 , 81 A number of MAbs targeting SH demonstrated potent neutralising properties. CV3‐25 identified an epitope in the SH and blocked membrane fusion. 81 CC40.8 is another neutralising anti‐stem helix MAb with neutralising effects against SARS‐CoV‐2 in vivo. 80 Also, Pinto et al. reported that among the five nAbs recognising motif F1148KEELDKYF1156 of SH, S2P6 was the most broadly neutralising antibody against all β‐coronaviruses through blocking membrane fusion as well as Fc‐mediated effector functions. 79
Taken together, these findings indicate that non‐RBD epitopes of the S protein might induce nAbs with comparable neutralisation potency as anti‐RBD MAbs. This highlights importance of non‐RBD epitopes of the S protein as additional neutralising epitopes for vaccine design as well as passive immunotherapy purposes. Genome wide analysis of virus variants showed that the RBD sequence is the most variable region prone to mutations. 82 , 83 Currently, the exact mechanism(s) utilised by non‐RBD‐binding MAbs to neutralise the virus is not fully understood. Different neutralisation mechanisms have been proposed including inhibition of prefusion‐to‐postfusion conformational change of the S protein preventing virus membrane fusion with the host membrane, Fc‐mediated effector functions, steric hindrance of Fab as well as Fc regions on the ACE2‐binding site of the S protein and finally conformational changes in the ACE2 binding site leading to abrogation of the binding of the virus to its receptors on target cells. 47 , 66 , 84
Thus, vaccine designs based on the conserved regions in RBD and outside RBD are the favoured candidates for inducing protective immunity capable of neutralising the emerging pan‐coronavirus variants. 79
4. NEUTRALISING ACTIVITY OF MONOCLONAL ANTIBODIES AGAINST EMERGING SARS‐CoV‐2 VARIANTS
Since beginning of the COVID‐19 pandemic, ongoing evolution of SARS‐CoV‐2 has led to emergence and circulation of genetic lineages around the world. Emerging variants are classified either as variants of interest or as variants of concern (VOC) by the WHO Virus Evolution Working Group. VOIs are no longer circulating or are detected at very low levels and do not confer a significant or critical risk for public health. SARS‐CoV‐2 variants harbouring genetic changes predicted or known to affect transmissibility, disease severity, immune escape, and diagnostic or therapeutic escape are considered as VOIs. On the contrary, VOCs have been demonstrated to be associated with one or more of the following changes: increased transmissibility, deleterious change in COVID‐19 epidemiology, increased virulence and decreased effectiveness of public health measures or available diagnostics, vaccines, and therapeutics. Currently, the designated VOCs are as follows: Alpha variant (B.1.1.7), first identified in the United Kingdom, contains N501Y substitution in RBD. Beta variant (B.1.351), first detected in South Africa, contains three important mutations in RBD including N501Y, E484 K, and K417 N. Alpha and Beta variants are significantly more transmissible (43%–82% and 50%, respectively), 85 , 86 due to N501Y substitution that enhances the accessibility of RBD and binding affinity to ACE2. 85 , 87 , 88 , 89 Gamma variant (p.1) was first found in Brazil with biologically important mutations in the RBD region including N501Y, E484 K, and K417 N/T. Although K417 N/T substitutions found in Beta and Gamma variants decreased the binding affinity, N501Y and E484 K mutations enhanced the binding affinity of their RBDs to ACE2. 90 Delta variant (B.1.617.2), first documented in India, harbours two substitutions in the RBD, including L452 R and T478 K associated with its 97% transmissibility and higher affinity and stability of S protein conformation. 91 , 92 , 93 , 94 Omicron (B.1.1.529) which was first detected in South Africa, harbours 34 mutations, 15 of which are in the RBD region, leading to 4‐fold increased infectivity compared to wild‐type SARS‐CoV‐2. Lambda (C.37, Peru) and Mu (B.1.621, Colombia) variants are considered as VOIs. 95 Each variant has heavily mutated spike proteins. Continuous evolution of SARS‐CoV‐2 can reduce MAb effectiveness if any of the mutations change epitopes targeted by the antibodies. Figure 6 shows spike mutations in VOC variants.
FIGURE 6.
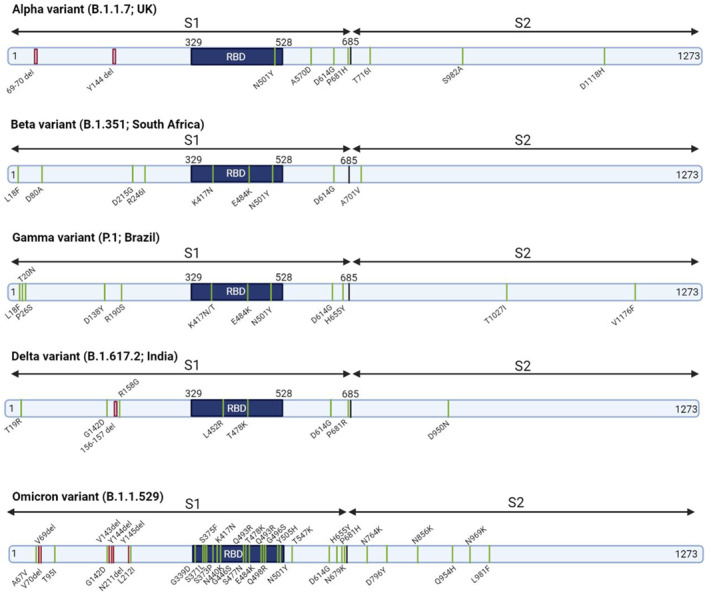
Spike amino acid mutations in Alpha, Beta, Gamma, Delta, and Omicron VOCs. VOCs: variants of concern (VOC)
Several studies have evaluated the neutralising activity of MAbs authorised for emergency use by the FDA‐emergency use (EU) in advanced clinical trials on new emerging variants in comparison with the prototype SARS‐CoV‐2 virus. Wang et al. performed an extensive study to assess the neutralising activity of several MAbs targeting outer side, RBM (including bamlanivimab and casirivimab), inner side of RBD (including imdevimab and sotrovimab) and NTD (including 4A8) against all current VOCs. Neutralising activity against B.1.1.7 was slightly reduced by a few MAbs such as sotrovimab because of N501Y substitution as well as NTD‐directed MAbs due to Δ144. The B.1.351 variant was resistant to the neutralising activity of most MAbs specific for NTD and RBM. Accordingly, the neutralising activity of bamlanivimab and casirivimab was completely or markedly abrogated against B.1.351 because of E484 K and K417 N mutations and the neutralising activity of 4A8 was abolished due to Δ242‐244 and/or R246I mutations. However, combination of casirivimab and imdevimab maintained much of the neutralisation activity against the B.1.351 variant. 96 They also reported that the p.1 variant is relatively refractory to neutralisation by the FDA‐EU authorised MAbs through adaptation of a conformation in trimer p.1 with one RBD in the ‘‘up’’ position, facilitating entry of the virus to target cells. 97 In contrast, while bamlanivimab efficiently neutralised the B.1.1.7 variant, it lost the neutralisation effect against B.1.135 carrying the E484 K substitution. 98 In another study, complete mapping of all mutations to RBD by bamlanivimab, and its cocktail combination with LY‐CoV016 (etesevimab) was conducted. The results indicated that the E484 K substitution escapes bamlanivimab and K417N/T escapes etesivimab. Both mutations are present in B.1.351 and p.1 variants. 99 In another study, casirivimab and imdevimab were tested against two VOCs including B.1.1.7 and B.1.351. Imdevimab maintains its neutralisation effectiveness against B.1.1.7 and B.1.351, but casirivimab lost reactivity against B.1.351 due to K417 N and E484 K mutations in RBD. 100 B.1.1.7 and B.1.351 variants reduced neutralisation activity of six out of eight MAbs obtained from blood samples of COVID‐19 convalescent patients. 101 In accordance with these findings, the majority of our RBD‐specific neutralising mouse hybridoma MAbs displayed significant neutralisation reduction to p.1 and B.1.135 variants in comparison with the wild‐type spike protein (unpublished data).
McCarthy et al. assessed reactivity of 4A8 as an NTD‐binding nAb and showed that it does not recognise the S protein with the following deletions: Δ69–70 + Δ144/145 (both found in the B.1.1.7 lineage and Δ69–70 which is key for increased infectivity of the B.1.1.7 lineage), Δ141–145, Δ144/145, Δ146, and Δ243–244 (found in the B.1.351 lineage). However, its binding to Δ210 and Δ69/70 alone remained unchanged, suggesting that the NTD deletions are not enough as the battalion of neutralising antibodies targeting different S epitopes. 102 B.1.1.7 mostly conferred resistance to neutralisation by the NTD‐directed nAbs, 103 suggesting that developing nAbs against subdominant epitopes need to be considered against emerging variants. Thus, emergence of mutations similar to B.1.1.7 and B.1.351 is considered a critical challenge for therapeutic MAbs. B.1.351 was the most resilient variant to COVID‐19 patient‐derived MAbs, followed by p.1 and B.1.1.7 variants. This resistance is largely mediated by Δ144 and Δ242–244 mutations in NTD and K417 N/T, E484 K, and N501Y mutations in RBD. 104 The Delta variant was refractory to neutralisation activity of bamlanivimab by impaired binding of the MAb to the spike protein. 105 An extensive in vitro and in vivo study of a panel of MAbs including COV2‐2196, COV2‐2130, sotrovimab, 47D11, casirivimab, imdevimab, bamlanivimab, and etesevimab was conducted against B.1.1.7, B.1.351, and B.1.617.1 variants. In vitro experiments showed no significant changes in neutralising activity of all MAbs against B.1.351, and B.1.617.1 variants, however, imdevimab and bamlanivimab displayed 10‐fold decrease and complete loss of reactivity, against B.1.617.1, respectively. Low prophylactic doses of MAbs inhibited SARS‐CoV‐2 infection by tested variants in K18‐hACE2 transgenic mice, 129S2 immunocompetent mice and hamsters, except for bamlanivimab monotherapy and bamlanivimab and eteseviamb combination therapy, which demonstrated complete loss of protective activity against B.1.351 and B.1.617.1. 106 In another study, while sotrovimab showed 3‐fold reduction and the combination of COV2‐2130 and COV2‐2196 demonstrated ∼200‐fold reduction in neutralisation activity against Omicron, other RBM‐specific MAbs, including casirivimab, imdevimab, bamlanivimab, etesevimab, and CT‐P59 completely lost antiviral activity. 17 Also, Planas et al. reported that bamlanivimab, etesevimab, casirivimab, imdevimab, tixagevimab, and regdanvimab completely lost the neutralising potency against B.1.617.2 and Omicron variants. Interestingly, sotrovimab was the only antibody maintaining the neutralising potency with a relatively similar activity against these two variants. 107 Although 85% of nAbs lost antiviral efficacy against Omicron, this variant showed less negative effect on nAbs with broad sarbecovirus (the viral subgenus containing SARS‐CoV and SARS‐CoV‐2) neutralising activity. 108 Gruell and colleagues have recently investigated the neutralising activity of a number of MAbs including bamlanvimab, etesevimab, casirivimab, imdevimab, P2B‐2F6, and sotrovimab against a variety of VOCs. They demonstrated that all antibodies maintain neutralising activity against B.1.1.7, B.1.351, and B.1.617.2, with the exception of bamlanivimab which lost its neutralising activity against B.1.351, and B.1.617.2. Notably, sotrovimab was the only antibody that maintained neutralising activity against the Omicron variant. 109 Therefore, Omicron variant exerted substantial humoral immune evasion and nAbs recognising the sarbecovirus conserved region remain most effective. Altogether, ongoing major antigenic shifts and drifts and increased transmissibility and affinity of new emergent variants confer serious challenges to current therapeutic antibodies.
5. CONCLUSION
Neutralising anti‐SARS‐CoV‐2 MAbs could serve as prophylactic/therapeutic agents in COVID‐19 infection. Epitope mapping of the reported neutralising anti‐SARS‐CoV‐2 MAbs has revealed that the neutralising epitopes of SARS‐CoV‐2 virus are mainly located on the RBD fragment of the S protein. Considering the crucial role of the RBD fragment in the virus binding to the ACE2, this is not a surprizing finding. Inhibition of RBD‐ACE2 interaction by MAbs might be either mediated through direct competition with RBD residues responsible for ACE2 interaction and/or steric hindrance on ACE2‐interacting RBD residues mediated by the antibody paratope. Moreover, neutralising anti‐RBD MAbs could enhance viral neutralisation by increasing antibody effector functions, including ADCC and ADCP as well as virus cross‐linking.
Current studies also highlight that non‐RBD epitopes of the S protein, including the NTD fragment, might elicit nAbs with neutralising potency comparable to anti‐RBD antibodies. This indicates that non‐RBD epitopes of the S protein could be considered as neutralising epitopes particularly with respect to the emerging SARS‐CoV‐2 variants. Although the exact mechanisms of virus neutralisation by these MAbs are not fully understood, several neutralisation mechanisms have been proposed and discussed in this review. Altogether, more studies are required to focus on neutralising MAbs directed against non‐RBD regions of the S protein in order to generate MAbs with broad neutralising activity and to elucidate their possible neutralising mechanisms.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
Somayeh Ghotloo and Faezeh Maghsood wrote the initial draft of manuscript. Faezeh Maghsood designed and illustrated the figures. Forough Golsaz Shirazi, Mohammad Mehdi Amiri, and Christiane Moog edited the initial draft. Fazel Shokri conceived the study, edited and revised the manuscript. All the authors approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We would like to thank Dr Mina Tabrizi for technical and linguistic revision of the manuscript. This study was partially supported by a grant from the National Institute for Medical Research Development (NIMAD) of Iran (Grant No. 993421) and ANRS COVID Sud project (ECTZ144757).
Ghotloo S, Maghsood F, Golsaz‐Shirazi F, Amiri MM, Moog C, Shokri F. Epitope mapping of neutralising anti‐SARS‐CoV‐2 monoclonal antibodies: implications for immunotherapy and vaccine design. Rev Med Virol. 2022;32(5):e2347. 10.1002/rmv.2347
Somayeh Ghotloo and Faezeh Maghsood contributed equally to this manuscript
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
REFERENCES
- 1. WHO. Coronavirus Disease (COVID‐19) Pandemic. https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019 [Google Scholar]
- 2. Weekly Epidemiological Update on COVID‐19 ‐ 25 January 2022. https://www.who.int/publications/m/item/weekly‐epidemiological‐update‐on‐covid‐19‐‐‐25‐january‐2022 [Google Scholar]
- 3. Actemra EUA Letter of Authorization . https://www.fda.gov/news‐events/press‐announcements/coronavirus‐covid‐19‐update‐fda‐authorizes‐drug‐treatment‐covid‐19
- 4. U.S. Food and Drug Administration . Center for Drug Evaluation and Research FDA Briefing Document Antimicrobial Drugs Advisory Committee Meeting November. 30; 2021. https://www.fda.gov/media/154418/download [Google Scholar]
- 5. Paxlovid EUA Letter of Authorization . https://www.fda.gov/media/155049/download
- 6. Therapeutics and COVID‐19: Living Guideline. https://www.who.int/publications/i/item/WHO‐2019‐nCoV‐therapeutics‐2021.2 [Google Scholar]
- 7. Baricitinib Letter of Authorization . https://www.fda.gov/media/143822/download
- 8. FDA Authorizes Bamlanivimab and Etesevimab Monoclonal Antibody Therapy for Post‐exposure Prophylaxis (Prevention) for COVID‐19. https://www.fda.gov/drugs/drug‐safety‐and‐availability/fda‐authorizes‐bamlanivimab‐and‐etesevimab‐monoclonal‐antibody‐therapy‐post‐exposure‐prophylaxis [Google Scholar]
- 9. Casirivimab, Imdevimab EUA Letter of Authorization . https://www.fda.gov/media/143891/download
- 10. Sotrovimab EUA Letter of Authorization . https://www.fda.gov/media/149532/download
- 11. Mittal A, Manjunath K, Ranjan RK, Kaushik S, Kumar S, Verma V. COVID‐19 pandemic: insights into structure, function, and hACE2 receptor recognition by SARS‐CoV‐2. PLoS Pathog. 2020;16(8):e1008762. 10.1371/journal.ppat.1008762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang Y, Yang C, Xu X‐f, Xu W, Liu S‐w. Structural and functional properties of SARS‐CoV‐2 spike protein: potential antivirus drug development for COVID‐19. Acta Pharmacol Sin. 2020;41(9):1141‐1149. 10.1038/s41401-020-0485-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215‐220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- 14. Yi C, Sun X, Ye J, et al. Key residues of the receptor binding motif in the spike protein of SARS‐CoV‐2 that interact with ACE2 and neutralizing antibodies. Cell Mol Immunol. 2020;17(6):621‐630. 10.1038/s41423-020-0458-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bhalla V, Blish CA, South AM. A historical perspective on ACE2 in the COVID‐19 era. J Hum Hypertens. 2020;35:935‐939. 10.1038/s41371-020-00459-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cameroni E, Bowen JE, Rosen LE, et al. Broadly neutralizing antibodies overcome SARS‐CoV‐2 Omicron antigenic shift. Nature. 2021;602:664‐670. 10.1038/s41586-021-04386-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barros EP, Casalino L, Gaieb Z, et al. The flexibility of ACE2 in the context of SARS‐CoV‐2 infection. Biophysical J. 2021;120(6):1072‐1084. 10.1016/j.bpj.2020.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gur M, Taka E, Yilmaz SZ, Kilinc C, Aktas U, Golcuk M. Conformational transition of SARS‐CoV‐2 spike glycoprotein between its closed and open states. J Chem Phys. 2020;153(7):075101. 10.1063/5.0011141 [DOI] [PubMed] [Google Scholar]
- 20. Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS‐CoV‐2 spike protein and the role of the novel S1/S2 site. iScience. 2020;23(6):101212. 10.1016/j.isci.2020.101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun. 2020;11(1):1620. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maghsood F, Hassani D, Salimi V, et al. Differential antibody response to SARS‐CoV‐2 antigens in recovered and deceased Iranian COVID‐19 patients. Viral Immunol 2021;34(10):708‐713. 10.1089/vim.2021.0061 [DOI] [PubMed] [Google Scholar]
- 23. Hassani D, Amiri MM, Maghsood F, et al. Does prior immunization with measles, mumps, and rubella vaccines contribute to the antibody response to COVID‐19 antigens? Iranian J Immunol. 2021;18(1):47‐53. 10.22034/iji.2021.87990.1843 [DOI] [PubMed] [Google Scholar]
- 24. Chen X, Pan Z, Yue S, et al. Disease severity dictates SARS‐CoV‐2‐specific neutralizing antibody responses in COVID‐19. Signal Transduct Target Ther. 2020;5(1):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ko J‐H, Joo E‐J, Park S‐J, et al. Neutralizing antibody production in asymptomatic and mild COVID‐19 patients, in comparison with pneumonic COVID‐19 patients. J Clin Med. 2020;9(7):2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong W‐H, Zhao R, Zhou J‐B, et al. Serologic response to SARS‐CoV‐2 in COVID‐19 patients with different severity. Virol Sin. 2020;35(6):752‐757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Long Q‐X, Tang X‐J, Shi Q‐L, et al. Clinical and immunological assessment of asymptomatic SARS‐CoV‐2 infections. Nat Med. 2020;26(8):1200‐1204. [DOI] [PubMed] [Google Scholar]
- 28. Abbas A, Lichtman A, Pillai S. Cellular and Molecular Immunology. 9 ed. Elsevier; 2017. [Google Scholar]
- 29. Pelletier JPR, Mukhtar F. Passive monoclonal and polyclonal antibody therapies. Immunologic Concepts in Transfusion Medicine; 2020:251‐348. 10.1016/B978-0-323-67509-3.00016-0 [DOI] [Google Scholar]
- 30. MyoClinic. Convalescent Plasma Therapy. https://www.mayoclinic.org/tests‐procedures/convalescent‐plasma‐therapy/about/pac‐20486440 [Google Scholar]
- 31. FDA. Recommendations for Investigational COVID‐19 Convalescent Plasma. 2021. [Google Scholar]
- 32. Robbiani DF, Gaebler C, Muecksch F, et al. Convergent antibody responses to SARS‐CoV‐2 in convalescent individuals. Nature. 2020;584(7821):437‐442. 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS‐CoV‐2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956‐963. 10.1126/science.abc7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu L, Wang P, Nair MS, et al. Potent neutralizing antibodies against multiple epitopes on SARS‐CoV‐2 spike. Nature. 2020;584(7821):450‐456. 10.1038/s41586-020-2571-7 [DOI] [PubMed] [Google Scholar]
- 35. Liu X, Gao F, Gou L, et al. Neutralizing antibodies isolated by a site‐directed screening have potent protection on SARS‐CoV‐2 infection. bioRxiv. 2020:2020.05.03.074914. 10.1101/2020.05.03.074914 [DOI] [Google Scholar]
- 36. Chandrashekar A, Liu J, Martinot AJ, et al. SARS‐CoV‐2 infection protects against rechallenge in rhesus macaques. Science. 2020;369(6505):812‐817. 10.1126/science.abc4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lang AB, Cryz SJ, Jr. , Schurch U, Ganss MT, Bruderer U. Immunotherapy with human monoclonal antibodies. Fragment A specificity of polyclonal and monoclonal antibodies is crucial for full protection against tetanus toxin. J Immunol. 1993;151(1):466‐472. [PubMed] [Google Scholar]
- 38. Kamei M, Hashizume S, Sugimoto N, Ozutsumi K, Matsuda M. Establishment of stable mouse/human‐human hybrid cell lines producing large amounts of anti‐tetanus human monoclonal antibodies with high neutralizing activity. Eur J Epidemiol. 1990;6(4):386‐397. 10.1007/bf00151713 [DOI] [PubMed] [Google Scholar]
- 39. Coronavirus FDA. COVID‐19 Update: FDA Authorizes Additional Monoclonal Antibody for Treatment of COVID‐19. https://www.fda.gov/news‐events/press‐announcements/coronavirus‐covid‐19‐update‐fda‐authorizes‐additional‐monoclonal‐antibody‐treatment‐covid‐19 [Google Scholar]
- 40. Coronavirus FDA. COVID‐19 Update: FDA Authorizes Monoclonal Antibodies for Treatment of COVID‐19; 2020. [Google Scholar]
- 41. Nilvebrant J, Rockberg J. An introduction to epitope mapping. Methods Mol Biol. 2018;1785:1‐10. 10.1007/978-1-4939-7841-0_1 [DOI] [PubMed] [Google Scholar]
- 42. Gershoni JM, Roitburd‐Berman A, Siman‐Tov DD, Tarnovitski Freund N, Weiss Y. Epitope mapping: the first step in developing epitope‐based vaccines. BioDrugs Clin Immunother Biopharm Gene Ther. 2007;21(3):145‐156. 10.2165/00063030-200721030-00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maghsood F, Shokri MR, Jeddi‐Tehrani M, et al. Identification of immunodominant epitopes on nucleocapsid and spike proteins of the SARS‐CoV‐2 in Iranian COVID‐19 patients. Pathog Dis. 2022;80. 10.1093/femspd/ftac001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen X, Pan Z, Yue S, et al. Disease severity dictates SARS‐CoV‐2‐specific neutralizing antibody responses in COVID‐19. Signal Transduct Target Ther. Sep 2 2020;5(1):180. 10.1038/s41392-020-00301-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zost SJ, Gilchuk P, Chen RE, et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS‐CoV‐2 spike protein. Nat Med. 2020;26(9):1422‐1427. 10.1038/s41591-020-0998-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS‐CoV‐2 identified by high‐throughput single‐cell sequencing of convalescent patients’ B cells. Cell. 2020;182(1):73‐84. e16. 10.1016/j.cell.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barnes CO, West AP, Jr. , Huey‐Tubman KE, et al. Structures of human antibodies bound to SARS‐CoV‐2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182(4):828‐842. e16. 10.1016/j.cell.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barnes CO, Jette CA, Abernathy ME, et al. SARS‐CoV‐2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682‐687. 10.1038/s41586-020-2852-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kreye J, Reincke SM, Kornau H‐C, et al. A therapeutic non‐self‐reactive SARS‐CoV‐2 antibody protects from lung Pathology in a COVID‐19 hamster model. Cell. 2020;183(4):1058‐1069. e19. 10.1016/j.cell.2020.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Noy‐Porat T, Makdasi E, Alcalay R, et al. A panel of human neutralizing mAbs targeting SARS‐CoV‐2 spike at multiple epitopes. Nat Commun. 2020;11(1):4303. 10.1038/s41467-020-18159-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zost SJ, Gilchuk P, Case JB, et al Potently neutralizing human antibodies that block SARS‐CoV‐2 receptor binding and protect animals. bioRxiv: The Preprint Server for Biology. 2020:2020.05.22.111005. 10.1101/2020.05.22.111005 [DOI] [Google Scholar]
- 52. Zeng X, Li L, Lin J, et al. Isolation of a human monoclonal antibody specific for the receptor binding domain of SARS‐CoV‐2 using a competitive phage biopanning strategy. Antibody Therapeutics. 2020;3(2):95‐100. 10.1093/abt/tbaa008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen P, Nirula A, Heller B, et al. SARS‐CoV‐2 neutralizing antibody LY‐CoV555 in outpatients with covid‐19. N. Engl J Med. 2020. 10.1056/NEJMoa2029849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jones BE, Brown‐Augsburger PL, Corbett KS, et al. The neutralizing antibody, LY‐CoV555, protects against SARS‐CoV‐2 infection in nonhuman primates. Sci Transl Med. 2021(593):13. 10.1126/scitranslmed.abf1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Coronavirus (COVID‐19) Update: FDA Revokes Emergency Use Authorization for Monoclonal Antibody Bamlanivimab. https://www.fda.gov/news‐events/press‐announcements/coronavirus‐covid‐19‐update‐fda‐revokes‐emergency‐use‐authorization‐monoclonal‐antibody‐bamlanivimab [Google Scholar]
- 56. Lim H, Baek A, Kim J, et al. Hot spot profiles of SARS‐CoV‐2 and human ACE2 receptor protein protein interaction obtained by density functional tight binding fragment molecular orbital method. Sci Rep. 2020;10(1):16862. 10.1038/s41598-020-73820-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Othman H, Bouslama Z, Brandenburg J‐T, et al. Interaction of the spike protein RBD from SARS‐CoV‐2 with ACE2: similarity with SARS‐CoV, hot‐spot analysis and effect of the receptor polymorphism. Biochem Biophys Res Commun. 2020;527(3):702‐708. 10.1016/j.bbrc.2020.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581(7807):221‐224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim C, Ryu D‐K, Lee J, et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS‐CoV‐2 spike protein. Nat Commun. 2021;12(1):288. 10.1038/s41467-020-20602-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu Y, Wang F, Shen C, et al. A noncompeting pair of human neutralizing antibodies block COVID‐19 virus binding to its receptor ACE2. Science. 2020;368(6496):1274‐1278. 10.1126/science.abc2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature. 2020;584(7819):115‐119. 10.1038/s41586-020-2380-z [DOI] [PubMed] [Google Scholar]
- 62. Murphy AJ, Macdonald LE, Stevens S, et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc Natl Acad Sci U S A. 2014;111(14):5153‐5158. 10.1073/pnas.1324022111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS‐CoV‐2 antibody cocktail. Science. 2020;369(6506):1010‐1014. 10.1126/science.abd0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dong J, Zost SJ, Greaney AJ, et al. Genetic and structural basis for SARS‐CoV‐2 variant neutralization by a two‐antibody cocktail. Nat Microbiol. 2021;6(10):1233‐1244. 10.1038/s41564-021-00972-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang S, Peng Y, Wang R, et al. Characterization of neutralizing antibody with prophylactic and therapeutic efficacy against SARS‐CoV‐2 in rhesus monkeys. Nat Commun. 2020;11(1):5752. 10.1038/s41467-020-19568-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ghotloo S, Amiri MM, Khoshnoodi J, et al. Contribution of Fc fragment of monoclonal antibodies to tetanus toxin neutralization. Neurotox Res. 2020;37(3):578‐586. 10.1007/s12640-019-00124-9 [DOI] [PubMed] [Google Scholar]
- 67. Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor‐binding site of SARS‐CoV‐2. Nature. 2020;584(7819):120‐124. 10.1038/s41586-020-2381-y [DOI] [PubMed] [Google Scholar]
- 68. Tai W, Zhang X, He Y, Jiang S, Du L. Identification of SARS‐CoV RBD‐targeting monoclonal antibodies with cross‐reactive or neutralizing activity against SARS‐CoV‐2. Antivir Res. 2020;179:104820. 10.1016/j.antiviral.2020.104820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fedry J, Hurdiss DL, Wang C, et al. Structural insights into the cross‐neutralization of SARS‐CoV and SARS‐CoV‐2 by the human monoclonal antibody 47D11. Sci Adv. 2021;7(23). 10.1126/sciadv.abf5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang C, Li W, Drabek D, et al. A human monoclonal antibody blocking SARS‐CoV‐2 infection. Nat Commun. 2020;11(1):2251. 10.1038/s41467-020-16256-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pinto D, Park Y‐J, Beltramello M, et al. Cross‐neutralization of SARS‐CoV‐2 by a human monoclonal SARS‐CoV antibody. Nature. 2020;583(7815):290‐295. 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 72. Zhou D, Duyvesteyn HME, Chen CP, et al. Structural basis for the neutralization of SARS‐CoV‐2 by an antibody from a convalescent patient. Nat Struct Mol Biol. 2020;27(10):950‐958. 10.1038/s41594-020-0480-y [DOI] [PubMed] [Google Scholar]
- 73. Lv Z, Deng YQ, Ye Q, et al. Structural basis for neutralization of SARS‐CoV‐2 and SARS‐CoV by a potent therapeutic antibody. Science. 2020;369(6510):1505‐1509. 10.1126/science.abc5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N‐terminal domain of the Spike protein of SARS‐CoV‐2. Science. 2020;369(6504):650‐655. 10.1126/science.abc6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brouwer PJM, Caniels TG, van der Straten K, et al. Potent neutralizing antibodies from COVID‐19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643‐650. 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhou H, Chen Y, Zhang S, et al. Structural definition of a neutralization epitope on the N‐terminal domain of MERS‐CoV spike glycoprotein. Nat Commun. 2019;10(1):3068. 10.1038/s41467-019-10897-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Suryadevara N, Shrihari S, Gilchuk P, et al. Neutralizing and protective human monoclonal antibodies recognizing the N‐terminal domain of the SARS‐CoV‐2 spike protein. Cell. 2021;184(9):2316‐2331. e15. 10.1016/j.cell.2021.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cheng MH, Porritt RA, Rivas MN, et al. A monoclonal antibody against staphylococcal enterotoxin B superantigen inhibits SARS‐CoV‐2 entry in vitro. Structure. 2021;29(9):951‐962. e3. 10.1016/j.str.2021.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pinto D, Sauer MM, Czudnochowski N, et al. Broad betacoronavirus neutralization by a stem helix–specific human antibody. Science. 2021;373(6559):1109‐1116. 10.1126/science.abj3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhou P, Yuan M, Song G, et al. A human antibody reveals a conserved site on beta‐coronavirus spike proteins and confers protection against SARS‐CoV‐2 infection. A Protective Broadly Cross‐Reactive Human Antibody Defines a Conserved Site of Vulnerability on Beta‐Coronavirus Spikes. bioRxiv; 2021. Mar 31. 10.1101/2021.03.30.437769 [DOI] [Google Scholar]
- 81. Li W, Chen Y, Prévost J, et al. Structural basis and mode of action for two broadly neutralizing antibodies against SARS‐CoV‐2 emerging variants of concern. Cell Rep. 2022;38(2):110210. 10.1016/j.celrep.2021.110210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Islam MR, Hoque MN, Rahman MS, et al. Genome‐wide analysis of SARS‐CoV‐2 virus strains circulating worldwide implicates heterogeneity. Sci Rep. 2020;10(1):14004. 10.1038/s41598-020-70812-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kaur N, Singh R, Dar Z, Bijarnia RK, Dhingra N, Kaur T. Genetic comparison among various coronavirus strains for the identification of potential vaccine targets of SARS‐CoV2. Infect Genet Evol. 2021;89:104490. 10.1016/j.meegid.2020.104490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ghotloo S, Golsaz‐Shirazi F, Amiri MM, Jeddi‐Tehrani M, Shokri F. Epitope mapping of tetanus toxin by monoclonal antibodies: implication for immunotherapy and vaccine design. Neurotox Res. 2020;37(2):239‐249. 10.1007/s12640-019-00096-w [DOI] [PubMed] [Google Scholar]
- 85. Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and Rapid Spread of a New Severe Acute Respiratory Syndrome‐Related Coronavirus 2 (SARS‐CoV‐2) Lineage with Multiple Spike Mutations in South Africa. MedRxiv; 2020. [Google Scholar]
- 86. Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS‐CoV‐2 lineage B. 1.1. 7 in England. Science. 2021(6538):372. eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Davies NG, Jarvis CI, Edmunds WJ, et al. Increased mortality in community‐tested cases of SARS‐CoV‐2 lineage B. 1.1. 7. Nature. 2021;593(7858):270‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Team EE. Updated rapid risk assessment from ECDC on the risk related to the spread of new SARS‐CoV‐2 variants of concern in the EU/EEA–first update. Euro Surveill. 2021;26(3):2101211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS‐CoV‐2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295‐1310. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Barton MI, MacGowan S, Kutuzov M, Dushek O, Barton GJ, van der Merwe PA. Effects of Common Mutations in the SARS‐CoV‐2 Spike RBD Domain and its Ligand the Human ACE2 Receptor on Binding Affinity and Kinetics; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Adam D. The rush to study fast spreading coronavirus variants. Nature. 2021;594:19‐20. [DOI] [PubMed] [Google Scholar]
- 92. Mahase E. Delta Variant: What Is Happening with Transmission, Hospital Admissions, and Restrictions? British Medical Journal Publishing Group; 2021. [DOI] [PubMed] [Google Scholar]
- 93. Campbell F, Archer B, Laurenson‐Schafer H, et al. Increased transmissibility and global spread of SARS‐CoV‐2 variants of concern as at June 2021. Euro Surveill. 2021;26(24):2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Motozono C, Toyoda M, Zahradnik J, et al. SARS‐CoV‐2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29(7):1124‐1136. e11. 10.1016/j.chom.2021.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tracking SARS‐CoV‐2 Variants. https://www.who.int/en/activities/tracking‐SARS‐CoV‐2‐variants/ [Google Scholar]
- 96. Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS‐CoV‐2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130‐135. 10.1038/s41586-021-03398-2 [DOI] [PubMed] [Google Scholar]
- 97. Wang P, Casner RG, Nair MS, et al. Increased resistance of SARS‐CoV‐2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747‐751. e4. 10.1016/j.chom.2021.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Widera M, Wilhelm A, Hoehl S, et al. Bamlanivimab Does Not Neutralize Two SARS‐CoV‐2 Variants Carrying E484K in Vitro. medRxiv; 2021:2021. 10.1101/2021.02.24.21252372 [DOI] [Google Scholar]
- 99. Starr TN, Greaney AJ, Dingens AS, Bloom JD. Complete map of SARS‐CoV‐2 RBD mutations that escape the monoclonal antibody LY‐CoV555 and its cocktail with LY‐CoV016. Cell Rep Med. 2021;2(4):100255. 10.1016/j.xcrm.2021.100255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tada T, Dcosta BM, Zhou H, Vaill A, Kazmierski W, Landau NR. Decreased Neutralization of SARS‐CoV‐2 Global Variants by Therapeutic Anti‐spike Protein Monoclonal Antibodies. bioRxiv; 2021. [Google Scholar]
- 101. Hu J, Peng P, Wang K, et al. Emerging SARS‐CoV‐2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell Mol Immunol. 2021;18(4):1061‐1063. 10.1038/s41423-021-00648-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. McCarthy KR, Rennick LJ, Nambulli S, et al. Recurrent deletions in the SARS‐CoV‐2 spike glycoprotein drive antibody escape. Science. 2021;371(6534):1139‐1142. 10.1126/science.abf6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Graham C, Seow J, Huettner I, et al. Impact of the B.1.1.7 Variant on Neutralizing Monoclonal Antibodies Recognizing Diverse Epitopes on SARS‐CoV‐2 Spike. bioRxiv; 2021. 10.1101/2021.02.03.429355 [DOI] [Google Scholar]
- 104. Wang R, Zhang Q, Ge J, et al. Analysis of SARS‐CoV‐2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54(7):1611‐1621. e5. 10.1016/j.immuni.2021.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS‐CoV‐2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276‐280. 10.1038/s41586-021-03777-9 [DOI] [PubMed] [Google Scholar]
- 106. Chen RE, Winkler ES, Case JB, et al. In vivo monoclonal antibody efficacy against SARS‐CoV‐2 variant strains. Nature. 2021;596(7870):103‐108. 10.1038/s41586-021-03720-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS‐CoV‐2 Omicron to antibody neutralization. Nature, 602, 671, 675. 2021/12/23 2021. 10.1038/s41586-021-04389-z [DOI] [PubMed] [Google Scholar]
- 108. Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS‐CoV‐2 neutralizing antibodies. Nature. 2021;602:657‐663. 10.1038/s41586-021-04385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gruell H, Vanshylla K, Tober‐Lau P, et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS‐CoV‐2 Omicron variant. Nat Med. 2022. 10.1038/s41591-021-01676-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Baum A, Ajithdoss D, Copin R, et al. REGN‐COV2 antibodies prevent and treat SARS‐CoV‐2 infection in rhesus macaques and hamsters. Science. 2020;370(6520):1110‐1115. 10.1126/science.abe2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chen X, Li R, Pan Z, et al. Human monoclonal antibodies block the binding of SARS‐CoV‐2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol. 2020;17(6):647‐649. 10.1038/s41423-020-0426-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Seydoux E, Homad LJ, MacCamy AJ, et al. Analysis of a SARS‐CoV‐2‐infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53(1):98‐105. e5. 10.1016/j.immuni.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Structure of the SARS‐CoV‐2 spike glycoprotein (closed state) . https://www.rcsb.org/structure/6VXX
- 114. SARS‐CoV‐2 spike ectodomain structure (open state) . https://www.rcsb.org/structure/6VYB
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.


