Abstract
Nonsyndromic orofacial clefts belong to a class of congenital malformations characterized by a complex and multifactorial etiology. During early facial development, multiple factors can disrupt fusion leading to a cleft; this includes the shape of the embryonic face. The face shape hypothesis (FSH) of orofacial clefting emerged in the 1960s, influenced by morphological differences observed within affected families, comparative studies of mouse models, and advances in modeling genetic liability for complex traits in populations. For the past five decades, studies have documented changes in the shape or spatial arrangement of facial prominences in embryonic mice and altered post-natal facial shape in individuals at elevated risk for orofacial clefting due to their family history. Moreover, recent studies showing how genes that impact facial shape in humans and mice are providing clues about the genetic basis of orofacial clefting. In this review, I discuss the origins of the FSH, provide an overview of the supporting evidence, and discuss ways in which the FSH can inform our understanding of orofacial clefting.
Keywords: face shape, cleft lip, cleft palate, craniofacial anomalies, multifactoral threshold
Introduction
It is well established that nonsyndromic orofacial clefts have a complex multifactorial etiology (Garland et al., 2020; Leslie, 2021). Genes, endogenous and exogenous environmental factors, interactions among genes and/or environmental agents, and epigenetic modifications are all believed to play a role. These factors can lead to clefts though different mechanisms (Jiang et al., 2006). For example, the primary deficiency may involve the fusion process. In such cases, the facial prominences may grow in a normal manner and make contact, but something interferes with the molecular machinery that drives tissue fusion (reviewed in Ji et al., 2020). However, another possibility is that the nascent facial prominences are not able to contact one another or meet too late for proper fusion to take place. This implies that the tissues are in principle capable of fusing, but never get the opportunity. One way this could happen is an intrinsic deficiency in the growth of one or more of the facial prominences (e.g., due to hypoplasia). Another is that the potential for cellular proliferation is normal, but the trajectory of facial prominence growth may be abnormal. Yet another possibility is that an extrinsic force (e.g., brain growth) displaces the facial prominences in a way that interferes with their positions relative to one another. The key principle uniting all the above scenarios is that the precise spatial arrangement required for the facial prominences to meet and fuse is disrupted leading to a cleft. Put another way, the above scenarios point to alterations in biological shape, either of individual facial prominences or of the entire developing facial complex. The idea that shape can act as a precipitating factor in the emergence of an orofacial cleft is the fundamental premise of the face shape hypothesis (FSH). In this review I discuss the origins of, lines of supporting evidence for, and future perspectives on the FSH of orofacial clefting.
Origins of the Face Shape Hypothesis
Three lines of converging evidence led to the development of the FSH: the first is observational and relates to the presence of distinctive orofacial traits in individuals whose baseline genetic risk for clefting is higher than average; the second is experimental and relates to work done using embryos of mouse strains with varying natural susceptibility to clefting; and the third is theoretical and relates to the establishment of models to conceptualize disease liability in populations.
The familial pattern of orofacial clefts was recognized at least as early as the 18th century (Trew, 1757). With no formal knowledge of genetics, 19th century surgeons suspected that parents with cleft affected children were passing on this hereditary tendency and claimed they could see the manifestation of this tendency in their faces (Fergusson, 1870; Rose, 1891). Studies documenting minor defects or asymmetries in the oronasal region of otherwise-healthy relatives (parents and/or siblings) from families with a history of orofacial clefting first surfaced in the 1930’s (Mengel, 1939) and again in the 1960’s (Fukuhara and Saito, 1962; Fukuhara and Saito, 1963; Rusconi and Brusati, 1966; Mills et al., 1968; Niswander, 1968). Likewise, Dixon (1966) noted a strong tendency toward a class III skeletal relationship (relative maxillary retrusion) in the parents of children with orofacial clefts. Many of these studies lacked proper controls and methods of phenotypic assessment were crude, but they established an important principle: those at genetically higher risk for orofacial clefting, by means of their familial connections, tend to exhibit a pattern of facial traits that seem to be in some way related to cleft predisposition.
As early as the late 1950’s, Fraser and colleagues noted that the rate of cortisone-induced cleft palate differed according to mouse strain and that several factors, including dimensions of the developing murine palate and head, can influence risk of spontaneous cleft palate (Fraser et al., 1957). A decade later, Trasler directly explored the possibility of facial shape as a predisposing factor (Trasler, 1968). In this work, Trasler qualitatively compared the faces of embryos from mouse strains with high and low rates of spontaneous clefting of the primary palate prior to fusion. More susceptible mice were characterized by differences in the relative position and shape of the medial nasal processes, such that the probability of contact with the adjacent lateral nasal and maxillary process was reduced. Since Trasler published her seminal findings, a number of subsequent studies have corroborated and expanded upon these observations by comparing the facial morphology and development of mouse strains with varying degrees of susceptibility to spontaneous clefting (Smiley et al., 1971; Juriloff and Trasler, 1976; Sulik et al., 1979; Trasler and Machado, 1979; Diewert, 1982; Millicovsky et al., 1982; Trasler and Ohannessian, 1983; Ciriani and Diewert, 1986; Jacobson and Trasler, 1992; Wang and Diewert, 1992). These studies point to alterations in the size/shape of individual facial prominences and/or their spatial arrangement relative to one another as being a factor in the lead up to failed fusion. More recently, studies applying modern morphometric methods and advanced imaging have found that susceptible mouse strains exhibit differences in facial shape (both in embryos and as adults), have less morphologically integrated faces, and show excess facial shape variation (Hallgrímsson et al., 2004; Young et al., 2007; Parsons et al., 2008).
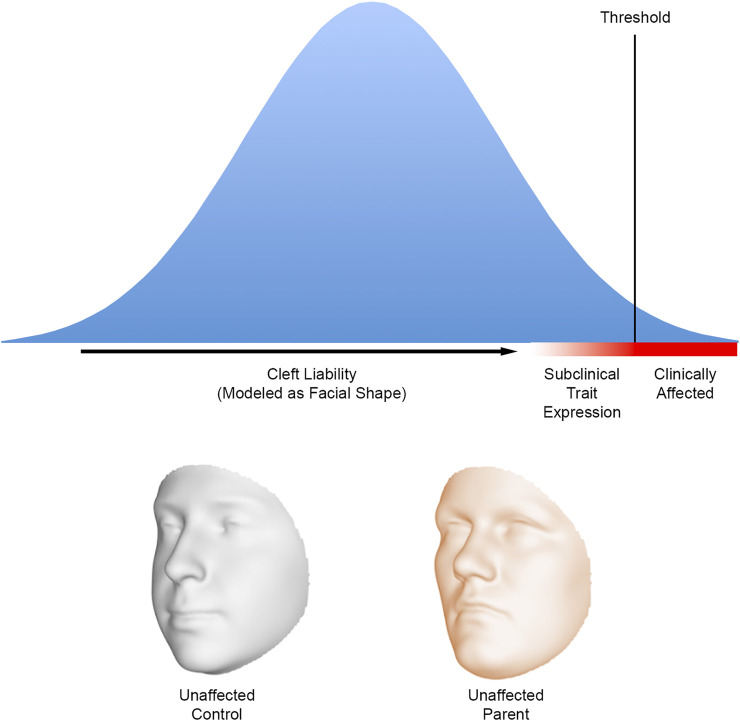
The multifactorial threshold (MFT) concept emerged in the 1930’s as a way to model the inheritance of discrete traits in mammals that did not follow strict Mendelian patterns (Wright, 1934a; 1934b). Application of the MFT model to human diseases and malformations, including orofacial clefting, began in the 1960’s with the work of Falconer, Carter, and Fraser (Curnow and Smith, 1975). The MFT model offered a way to conceptualize liability as a continuum and make predictions about recurrence. For clefting, the nature of what might underlie the liability, and threshold has been a matter of considerable debate (Fraser, 1976). In the context of the FSH, the MFT model provided a conceptual framework for prior research showing altered facial features in at-risk mouse strains and families with a history of orofacial clefting. Facial shape could now be considered one of the factors underlying cleft liability. Because seemingly unaffected family members possess at least some the same genetic risk factors as their affected relatives, they should be on average closer to the threshold than the rest of the population (Figure 1). As a result of their position on the liability continuum, we then expect these relatives to manifest craniofacial differences compared to the general population (Fraser, 1970).
FIGURE 1.
A hypothetical liability threshold model for orofacial clefting where the liability is modelled as facial shape. The facial shape of individuals near the defect threshold is expected to differ on average from the general population; the altered facial shape can be considered a subclinical expression resulting from the presence of cleft risk factors. The biological relatives of those affected with orofacial clefts are expected to be closer to the threshold—and therefore exhibit facial differences—compared with an individual chosen at random from the population. The faces below the graph show how the 3D facial surface changes shape from a hypothetical control to a hypothetical unaffected parent of a child affected by nonsyndromic CL/P. These 3D morphs are based on a comparison of 264 parental faces to 3,171 control faces (Indencleef et al., 2021). Note the relative midfacial flattening and upper facial broadening in the parental face. Faces adapted from Indencleef et al. (2021), CC BY 4.0.
Over 50 Years of Measuring Faces
By the start of the 1970’s, the notion that certain aspects of facial shape may contribute to cleft liability was beginning to gain acceptance. Fraser and Pashayan (1970) published a widely cited study comparing the facial dimensions of non-cleft parents with affected children to controls. Using several measurement approaches, they described a suite of facial differences in parents, including increased upper facial breadth, increased facial length, and reduced maxillary projection. The notion that quantitative facial differences will be present in the unaffected members of families with a history of clefting is based on a few assumptions: 1) that facial morphology can be reliability quantified; 2) that the relevant facial phenotypes are at least partly heritable; and 3) that alterations in embryonic facial shape are retained and can be detected postnatally. The first two assumptions are well supported, while the third is not plausible to test in humans, but is supported in mice (Boughner et al., 2008).
Since these early efforts, there have been over 30 quantitative studies comparing facial morphology between the ostensibly unaffected parents and/or siblings of individuals with clefts and controls with no family history of the defect. These studies employ a variety of methods including cephalometry (Coccaro et al., 1972; Kurisu et al., 1974; Shibasaki et al., 1978; Nakasima and Ichinose, 1983, 1984; Procházková and Tolarová, 1986; Sato, 1989; Ward et al., 1989, 1994; Raghavan et al., 1994; Procházková and Vinšová, 1995; Laatikainen et al., 1996; Mossey et al., 1998; AlEmran et al., 1999; Suzuki et al., 1999; McIntyre and Mossey, 2003; Perkiomaki et al., 2003; Chatzistavrou et al., 2004; McIntyre and Mossey, 2004; Yoon et al., 2004), direct anthropometry (Figalová and Šmahel, 1974; Blanco et al., 1992), 2D photogrammetry (Erickson, 1974) and 3D surface imaging (Weinberg et al., 2008, 2009; Miller et al., 2014; Roosenboom et al., 2017; Indencleef et al., 2021). A distinct subset of these studies is also focused on measuring patterns of craniofacial asymmetry in at-risk relatives (Pashayan and Fraser, 1971; Farkas and Cheung, 1979; Sigler and Ontiveros et al., 1999; McIntyre and Mossey, 2002a, 2010; Yoon et al., 2003; Kumar et al., 2010; Miller et al., 2014; Zhang et al., 2018). These studies cover a diverse array of populations, including individuals of East Asian, European, and Latin American Admixed ancestry. It is neither practical, nor worthwhile, to review every one of these studies. One reason is that they are so heterogeneous in terms of morphometric approach, sampling, and inclusion criteria that it is difficult to draw meaningful comparisons.
What we can state is that every study conducted thus far has reported at least some significant differences in facial morphology between unaffected relatives and controls. Moreover, these differences have been reported for every part of the craniofacial complex and many appear to be sex-specific. However, sorting out which specific facial features are reliable indicators of cleft-risk has been a challenge. There have been a few narrative reviews on the topic (McIntyre and Mossey, 2002b; Ward et al., 2002; Maulina et al., 2006), but it is difficult to avoid biased conclusions when the literature is rife with conflicting results. In attempt to get at something of a consensus, Weinberg et al. (2006) performed a meta-analysis of the parent versus control cephalometric literature available up to that point. After analyzing 28 measures, they concluded that the following suite of facial differences best characterized unaffected parents of children with CL/P: wider interorbital, nasal cavity and upper facial dimensions; narrower cranial vaults; longer cranial bases; longer and more protrusive mandibles; vertically shorter upper faces; and vertically longer lower faces. While these results still need to be considered with caution due to the small number of studies that met the inclusion criteria, it is worth noting that some of the traits identified—most notably a tendency toward increased midface retrusion and broader mid and upper facial dimensions—have now been validated by more recent studies using advanced 3D surface imaging and geometric morphometric techniques to assess biological shape (Weinberg et al., 2008, 2009; Indencleef et al., 2021). These differences are shown in Figure 1. Perhaps ironically, this morphological pattern echoes the original findings of Fraser and Pashayan from over 50 years ago.
THE Genetic Intersection Between Face Shape and Clefting
One expectation of the FSH is that the genes that drive face shape variation in humans will also be implicated in orofacial clefting (and vice-versa). Boehringer et al. (2011) tested this claim in two non-cleft European cohorts by evaluating the relationship between facial measures shown previously to differ between at-risk relatives and controls (nasal width and upper facial width) and 11 SNPs previously identified from orofacial clefting association studies. The result: two SNPs showed significant but weak genotype-phenotype relationships. A year later, in one of the first genome-wide association studies (GWAS) of normal-range facial morphology, Liu et al. (2012) performed targeted tests involving these same 11 candidate SNPs and 48 different facial measures; they reported significant associations at four loci involving horizontal upper facial dimensions. More recently, Indencleef et al. (2018) tested a set of 65 SNPs previously implicated in human studies of nonsyndromic clefting to determine their effects on normal-range 3D facial shape. They reported positive evidence for six SNPs including those near NOG (associated with philtrum shape), THADA (associated with supraorbital ridge morphology) and PAX7, MSX1, and PTCH1 (all associated with nose shape).
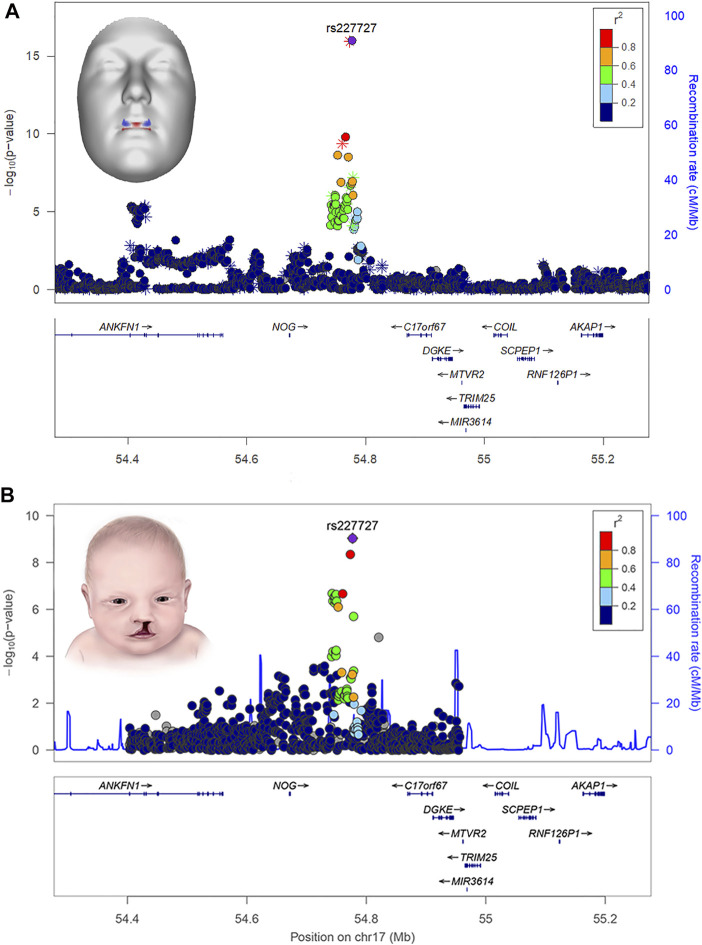
Several human face shape GWASs have now been published (Naqvi et al., in press) and many of the chromosomal regions identified have been found to harbor genes implicated in orofacial clefting and/or palate formation. To illustrate, in one of the most comprehensive facial GWAS to date, White et al. (2021) scanned the genomes of over 8,000 individuals and identified 203 regions across the genome associated with aspects of normal-range 3D facial shape. Bioinformatic analysis of the implicated genes revealed significant enrichment (i.e., statistical over-representation) for phenotypes and developmental processes directly related to clefting; some of the implicated cleft candidate genes included PRDM16, PAX7, PKDCC, SATB2, COL8A1, MSX1, FAT4, FGFR2, PAX9, BMP4, GREM1, NOG, BMP2, PAX1, and BMP7. Moreover, in many cases, phenotypic effects at the cleft-related GWAS signals were localized to anatomical regions directly relevant to clefting, like the central midface. For example, the rs227727 SNP near NOG was strongly associated with normal-range upper lip shape (Figure 2). Notably, this same variant was previously identified as functionally relevant for nonsyndromic CL/P (Leslie et al., 2015). In another example, PAX1 was associated with normal-range morphology of the nasal alae and philtrum and was recently shown to impact risk of cleft subtypes involving only of the lip (Curtis et al., 2021).
FIGURE 2.
LocusZoom plots showing the statistical association of SNP rs227727 near the gene NOG with lip shape (A) and nonsyndromic orofacial clefting (B). The 3D face inset for panel A (top) shows the effects of the SNP as color-coded heat map where red indicates regions of the face moving in an outward direction and blue indicates regions of the face moving in an inward direction. The lip shape association was reported originally in White et al. (2021). The cleft association was reported originally in Leslie et al. (2015). Attribution: the inset for panel A (top) was adapted from White and Indencleef, Insights into the genetic architecture of the human face, via Figshare, CC BY 4.0, https://doi.org/10.6084/m9.figshare.c.4667261.v6. The inset for panel B (bottom) is from the Centers for Disease Control and Prevention and is available under the Creative Commons CC0 1.0 Universal Public Domain Dedication, via Wikimedia Commons, https://commons.wikimedia.org/wiki/File:Cleftlip.jpg.
Leveraging existing data, Howe et al. (2018) investigated the shared genetics between facial shape and orofacial clefting by constructing a polygenic risk score (PRS) for clefting and testing whether that PRS was associated with selected facial dimensions previously shown to be relevant for cleft risk from family studies. They found that the cleft PRS was significantly associated with decreased philtrum width and, using Mendelian randomization, showed that this relationship was causal (i.e., genetic variants for clefting cause decreased philtrum width). Indencleef et al. (2021) also investigated the question of genetic overlap starting with an improved delineation of the cleft-relevant facial features. Using a highly innovative facial phenotyping approach that naturally breaks the 3D facial geometry into hieratically arranged regions (Claes et al., 2018), the authors explicitly measured aspects of 3D facial morphology that differed between at-risk relatives and controls. Once identified, the shape phenotypes indicative of elevated cleft risk were then measured in a sample of over 8,000 healthy individuals with available 3D facial and genomic data. A series of GWASs were then performed to better understand the genetic architecture of these cleft risk traits, resulting in the identification of 29 loci. Twenty-two of these loci were previously associated with normal facial variation and 18 were near genes known to play a role in orofacial clefting. Moreover, a PRS for nonsyndromic orofacial clefting was statically associated with several of the identified facial risk traits.
As noted earlier, strains of mice with high and low susceptibility to clefting show differences in facial shape. For example, Parsons et al. (2008) showed that cleft-susceptible A/WySn embryos had underdeveloped and more divergently spaced facial maxillary prominences compared with C57BL/6J embryos. In recent years, a picture has started to emerge regarding the molecular factors driving these and other shape changes associated with increased risk of clefting. For example, Lipinski et al. (2010) showed that interfering with sonic hedgehog expression early in facial formation results by gestational day (GD) 11.25 in mispositioned and misshapen medial nasal prominences that can no longer make contact to form an intact primary palate. When examined at a slightly later timepoint (GD 17), treated mice showed midface hypoplasia and broader upper facial dimensions, and many developed clefts (Lipinski et al., 2014). In another study, Tfap2a mutant mouse embryos that develop CL/P were shown at E10.5 and E11.5 to have misaligned and misshaped nasal and maxillary prominences with reduced cell proliferation, driven in part by aberrant Fgf signaling (Green et al., 2015). In a later study, Green et al. (2019) showed that the cleft-related shape changes evident in the A/WySn embryonic face are linked to the degree of Wnt9b methylation and expression. Taken together, these studies point to specific genes and pathways driving changes in facial shape and variation that can push individuals along the liability spectrum toward the threshold, leading to an increased risk of clefting.
Impact and Future Outlook
Our knowledge connecting facial shape to orofacial clefting has evolved from the clinical anecdotes of 19th century surgeons to modern experiments using the latest imaging, molecular and bioinformatic techniques. However, translating this knowledge into clinically meaningful and actionable information remains a significant challenge. As stated earlier, we know that different kinds of mechanisms can lead to clefts during early facial formation. Lumping all orofacial clefts together as a monolithic group ignores these differences, which can have implications for counseling, scientific discovery, and even treatment. The aspiration is that investigating patterns of facial shape in families can be used to help sort though some of this etiological heterogeneity and build a more complete picture of the genetic players involved in conferring orofacial cleft risk. Even a goal as crude as classifying individuals and their families into broad etiological classes based on whether the cleft was impacted by facial shape may prove useful. In the realm of genetic counseling, such knowledge could potentially lead to greater precision and personalization in cleft recurrence risk estimation. From the perspective of scientists invested in uncovering the genetic architecture of orofacial clefts, this type of phenotypic refinement has the potential to boost discovery power. Perhaps we can envision building a robust PRS for clefting, informed by genetic variants known to impact aspects of facial shape related to cleft risk. Studies investigating the genetic basis of facial risk phenotypes (e.g., Indencleef et al., 2021) can reveal variants that play a role in orofacial clefting, but whose effects may be too small to be detected in a standard cleft case-control GWAS. Such variants are likely to be modifiers of cleft risk, each with a small individual effect, but enough to nudge individuals toward the defect threshold or perhaps toward a more severe phenotypic expression. To illustrate this concept, Yu et al. (2022) showed that common genetic risk factors for clefting (modeled as a PRS) can modify the phenotypic penetrance of a rare damaging mutation in family with a history of nonsyndromic CL/P.
These problems are complicated and will require considerable effort to resolve. More immediately, though, there are still some basic questions about face shape and clefting that need to be investigated. For example, we have a poor understanding of whether the facial features associated with cleft predisposition differ according to the type of cleft(s) present in families (e.g., CL/P versus CPO), although at least one study has shown differences along these lines (Mossey et al., 1997). Similarly, we don’t know how the facial patterns observed in unaffected relatives compare across different ancestral backgrounds or whether any ancestry-specific patterns might help explain differences in cleft incidence among human populations. This was suggested by Chung and Kau (1985), who showed a relationship between specific facial dimensions and cleft predisposition by comparing individuals from various ancestral backgrounds (and therefore cleft prevalences) residing in a single location. Even the long-recognized sex biases we see in orofacial clefting may relate to sex differences in embryonic facial development, as initially suggested by Burdi and Silvey (1969). We know that sex differences in human facial morphology are present across the lifespan (Kesterke et al., 2016; Matthews et al., 2018) and that many of the facial differences we see in unaffected parents are sex-specific (Weinberg et al., 2008). Nevertheless, the data connecting sex, face shape, and cleft predisposition remains speculative. To answer these kinds of questions, we will undoubtedly need data from larger and more diverse samples, which should be a priority for future investigations.
Author Contributions
SW conceptualized and wrote the manuscript.
Funding
This work is supported by the following grants from the National Institute of Dental and Craniofacial Research: R01-DE016148 and R01-DE027023. The funder had no role in the conceptualization, writing or dissemination of the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- AlEmran S. E., Fatani E., Hassanain J. E. (1999). Craniofacial Variability in Parents of Children with Cleft Lip and Cleft Palate. J. Clin. Pediatr. Dent. 23, 337–341. [PubMed] [Google Scholar]
- Blanco R., Cifuentes L., Maldonado M. J., Rameau X., Muñoz M. A. (1992). Cleft Lip and Cleft Palate: Cephalometric Characteristics of Affected Individuals, Their Relatives and a Control Population. Rev. Med. Chil. 120, 13–19. [PubMed] [Google Scholar]
- Boehringer S., van der Lijn F., Liu F., Günther M., Sinigerova S., Nowak S., et al. (2011). Genetic Determination of Human Facial Morphology: Links between Cleft-Lips and Normal Variation. Eur. J. Hum. Genet. 19, 1192–1197. 10.1038/ejhg.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughner J. C., Wat S., Diewert V. M., Young N. M., Browder L. W., Hallgrímsson B. (2008). Short-faced Mice and Developmental Interactions between the Brain and the Face. J. Anat. 213, 646–662. 10.1111/j.1469-7580.2008.00999.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdi A. R., Silvey R. G. (1969). Sexual Differences in Closure of the Human Palatal Shelves. Cleft Palate J. 6, 1–7. [PubMed] [Google Scholar]
- Chatzistavrou E., Ross R. B., Tompson B. D., Johnston M. C. (2004). Predisposing Factors to Formation of Cleft Lip and Palate: Inherited Craniofacial Skeletal Morphology. Cleft Palate-Craniofacial J. 41, 613–621. 10.1597/03-090.1 [DOI] [PubMed] [Google Scholar]
- Chung C. S., Kau M. C. (1985). Racial Differences in Cephalometric Measurements and Incidence of Cleft Lip with or without Cleft Palate. J. Craniofac. Genet. Dev. Biol. 5, 341–349. [PubMed] [Google Scholar]
- Ciriani D., Diewert V. M. (1986). A Comparative Study of Development during Primary Palate Formation in A/WySn, C57BL/6, and Their F1 Crosses. J. Craniofac. Genet. Dev. Biol. 6, 369–377. [PubMed] [Google Scholar]
- Claes P., Roosenboom J., White J. D., Swigut T., Sero D., Li J., et al. (2018). Genome-wide Mapping of Global-To-Local Genetic Effects on Human Facial Shape. Nat. Genet. 50, 414–423. 10.1038/s41588-018-0057-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccaro P. J., D'Amico R., Chavoor A. (1972). Craniofacial Morphology of Parents with and without Cleft Lip and Palate Children. Cleft Palate J. 9, 28–38. [PubMed] [Google Scholar]
- Curnow R. N., Smith C. (1975). Multifactorial Models for Familial Diseases in Man. J. R. Stat. Soc. Ser. A General. 138, 131–156. 10.2307/2984646 [DOI] [Google Scholar]
- Curtis S. W., Chang D., Lee M. K., Shaffer J. R., Indencleef K., Epstein M. P., et al. (2021). The PAX1 Locus at 20p11 Is a Potential Genetic Modifier for Bilateral Cleft Lip. Hum. Genet. Genomics Adv. 2, 100025. 10.1016/j.xhgg.2021.100025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diewert V. M. (1982). A Comparative Study of Craniofacial Growth during Secondary Palate Development in Four Strains of Mice. J. Craniofac. Genet. Dev. Biol. 2, 247–263. [PubMed] [Google Scholar]
- Dixon D. A. (1966). “Abnormalities of the Teeth and Supporting Structures in Children with Clefts of Lip and Palate,” in The Causes and Natural History of Cleft Lip and Palate. Editors Drillien C., Ingram T., Wilkinson E. (Edinburgh: E & S Livingstone; ), 178–205. [Google Scholar]
- Erickson J. D. (1974). Facial and Oral Form in Sibs of Children with Cleft Lip with or without Cleft Palate. Ann. Hum. Genet. 38, 77–88. 10.1111/j.1469-1809.1974.tb01995.x [DOI] [PubMed] [Google Scholar]
- Farkas L. G., Cheung G. C. (1979). Nostril Asymmetry: Microform of Cleft Lip Palate? an Anthropometrical Study of Healthy North American Caucasians. Cleft Palate J. 16, 351–357. [PubMed] [Google Scholar]
- Fergusson W. (1870). A System of Practical Surgery. London: John Churchill & Sons. [Google Scholar]
- Figalová P., Smahel Z. (1974). Cephalometric Study of Families with Clefts. Acta Chir. Plast. 16, 247–255. [PubMed] [Google Scholar]
- Fraser F. C. (1970). The Genetics of Cleft Lip and Cleft Palate. Am. J. Hum. Genet. 22, 336–352. [PMC free article] [PubMed] [Google Scholar]
- Fraser F. C., Pashayan H. (1970). Relation of Face Shape to Susceptibility to Congenital Cleft Lip. A Preliminary Report. J. Med. Genet. 7, 112–117. 10.1136/jmg.7.2.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser F. C. (1976). The Multifactorial/threshold Concept-Uses and Misuses. Teratology 14, 267–280. 10.1002/tera.1420140302 [DOI] [PubMed] [Google Scholar]
- Fraser F. C., Walker B. E., Trasler D. G. (1957). Experimental Production of Congenital Cleft Palate: Genetic and Environmental Factors. Pediatrics 19, 782–787. 10.1542/peds.19.4.782 [DOI] [PubMed] [Google Scholar]
- Fukuhara T., Saito S. (1962). Genetic Consideration on the Dysplasia of the Nasopalatal Segments as a "Formes Frustes" Radiologically Found in Parents of Cleft Children: a Preliminary Report. Jpn. J. Hum. Genet. 7, 234–237. [Google Scholar]
- Fukuhara T., Saito S. (1963). Possible Carrier Status of Hereditary Cleft Palate with Cleft Lip: Report of Cases. Bull. Tokyo Med. Dent. Univ. 10, 333–345. [Google Scholar]
- Garland M. A., Reynolds K., Zhou C. J. (2020). Environmental Mechanisms of Orofacial Clefts. Birth Defects Res. 112, 1660–1698. 10.1002/bdr2.1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. M., Feng W., Phang T., Fish J. L., Li H., Spritz R. A., et al. (2015). Tfap2a-dependent Changes in Mouse Facial Morphology Result in Clefting that Can Be Ameliorated by a Reduction in Fgf8 Gene Dosage. Dis. Model. Mech. 8, 31–43. 10.1242/dmm.017616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. M., Leach C. L., Diewert V. M., Aponte J. D., Schmidt E. J., Cheverud J. M., et al. (2019). Nonlinear Gene Expression‐phenotype Relationships Contribute to Variation and Clefting in the A/WySn Mouse. Dev. Dyn. 248, 1232–1242. 10.1002/dvdy.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrimsson B., Dorval C. J., Zelditch M. L., German R. Z. (2004). Craniofacial Variability and Morphological Integration in Mice Susceptible to Cleft Lip and Palate. J. Anat. 205, 501–517. 10.1111/j.0021-8782.2004.00356.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L. J., Lee M. K., Sharp G. C., Davey Smith G., St Pourcain B., Shaffer J. R., et al. (2018). Investigating the Shared Genetics of Non-syndromic Cleft Lip/palate and Facial Morphology. PLoS Genet. 14, e1007501. 10.1371/journal.pgen.1007501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indencleef K., Hoskens H., Lee M. K., White J. D., Liu C., Eller R. J., et al. (2021). The Intersection of the Genetic Architectures of Orofacial Clefts and Normal Facial Variation. Front. Genet. 12, 626403. 10.3389/fgene.2021.626403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indencleef K., Roosenboom J., Hoskens H., White J. D., Shriver M. D., Richmond S., et al. (2018). Six NSCL/P Loci Show Associations with Normal-Range Craniofacial Variation. Front. Genet. 9, 502. 10.3389/fgene.2018.00502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson D., Trasler D. G. (1992). Morphometric Analysis of Heterozygote Dancer Mice Predisposed to 6-Aminonicotinamide-Induced Cleft Lip. Teratology 45, 393–400. 10.1002/tera.1420450410 [DOI] [PubMed] [Google Scholar]
- Ji Y., Garland M. A., Sun B., Zhang S., Reynolds K., McMahon M., et al. (2020). Cellular and Developmental Basis of Orofacial Clefts. Birth Defects Res. 112, 1558–1587. 10.1002/bdr2.1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Bush J. O., Lidral A. C. (2006). Development of the Upper Lip: Morphogenetic and Molecular Mechanisms. Dev. Dyn. 235, 1152–1166. 10.1002/dvdy.20646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juriloff D. M., Trasler D. G. (1976). Test of the Hypothesis that Embryonic Face Shape Is a Causal Factor in Genetic Predisposition to Cleft Lip in Mice. Teratology 14, 35–41. 10.1002/tera.1420140106 [DOI] [PubMed] [Google Scholar]
- Kesterke M. J., Raffensperger Z. D., Heike C. L., Cunningham M. L., Hecht J. T., Kau C. H., et al. (2016). Using the 3D Facial Norms Database to Investigate Craniofacial Sexual Dimorphism in Healthy Children, Adolescents, and Adults. Biol. Sex. Differ. 7, 23. 10.1186/s13293-016-0076-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S D., Gopalkrishnan K., Bhasker Rao C., V Ganeshkar S. (2010). Evaluation of Parental Nasomaxillary Asymmetry as a Risk Factor for Development of Palatal Clefts in Their Offsprings. Int. J. Clin. Pediatr. Dent. 3, 163–171. 10.5005/jp-journals-10005-1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu K., Niswander J. D., Johnston M. C., Mazaheri M. (1974). Facial Morphology as an Indicator of Genetic Predisposition to Cleft Lip and Palate. Am. J. Hum. Genet. 26, 702–714. [PMC free article] [PubMed] [Google Scholar]
- Laatikainen T., Ranta R., Nordström R. (1996). Craniofacial Morphology in Twins with Cleft Lip and Palate. Cleft Palate-Craniofacial J. 33, 96–103. 10.1597/1545-1569_1996_033_0096_cmitwc_2.3.co_2 [DOI] [PubMed] [Google Scholar]
- Leslie E. J. (2021). Genetic Models and Approaches to Study Orofacial Clefts. Oral Dis. (in press). 10.1111/odi.14109 [DOI] [PubMed] [Google Scholar]
- Leslie E. J., Taub M. A., Liu H., Steinberg K. M., Koboldt D. C., Zhang Q., et al. (2015). Identification of Functional Variants for Cleft Lip with or without Cleft Palate in or Near PAX7, FGFR2, and NOG by Targeted Sequencing of GWAS Loci. Am. J. Hum. Genet. 96, 397–411. 10.1016/j.ajhg.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski R. J., Song C., Sulik K. K., Everson J. L., Gipp J. J., Yan D., et al. (2010). Cleft Lip and Palate Results from Hedgehog Signaling Antagonism in the Mouse: Phenotypic Characterization and Clinical Implications. Birth Defects Res. A Clin. Mol. Teratol. 88, 232–240. 10.1002/bdra.20656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski R. J., Holloway H. T., O'Leary-Moore S. K., Ament J. J., Pecevich S. J., Cofer G. P., et al. (2014). Characterization of Subtle Brain Abnormalities in a Mouse Model of Hedgehog Pathway Antagonist-Induced Cleft Lip and Palate. PLoS One 9, e102603. 10.1371/journal.pone.0102603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., van der Lijn F., Schurmann C., Zhu G., Chakravarty M. M., Hysi P. G., et al. (2012). A Genome-wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans. PLoS Genet. 8, e1002932. 10.1371/journal.pgen.1002932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews H. S., Penington A. J., Hardiman R., Fan Y., Clement J. G., Kilpatrick N. M., et al. (2018). Modelling 3D Craniofacial Growth Trajectories for Population Comparison and Classification Illustrated Using Sex-Differences. Sci. Rep. 8, 4771. 10.1038/s41598-018-22752-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulina I., Urtane I., Jakobsone G. (2006). The Craniofacial Morphology of the Parents of Children with Cleft Lip And/or Palate: a Review of Cephalometric Studies. Stomatologija 8, 16–20. [PubMed] [Google Scholar]
- McIntyre G. T., Mossey P. A. (2010). Asymmetry of the Craniofacial Skeleton in the Parents of Children with a Cleft Lip, with or without a Cleft Palate, or an Isolated Cleft Palate. Eur. J. Orthod. 32, 177–185. 10.1093/ejo/cjp067 [DOI] [PubMed] [Google Scholar]
- McIntyre G. T., Mossey P. A. (2002a). Asymmetry of the Parental Craniofacial Skeleton in Orofacial Clefting. J. Orthod. 29, 299–305. 10.1093/ortho/29.4.299 [DOI] [PubMed] [Google Scholar]
- McIntyre G. T., Mossey P. A. (2004). Parental Craniofacial Morphology in Orofacial Clefting. Eur. J. Orthod. 26, 375–384. 10.1093/ejo/26.4.375 [DOI] [PubMed] [Google Scholar]
- McIntyre G. T., Mossey P. A. (2003). Posteroanterior Cephalometric Analysis of the Parental Craniofacial Morphology in Orofacial Clefting. Cleft Palate-Craniofacial J. 40, 416–425. [DOI] [PubMed] [Google Scholar]
- McIntyre G. T., Mossey P. A. (2002b). The Craniofacial Morphology of the Parents of Children with Orofacial Clefting: a Systematic Review of Cephalometric Studies. J. Orthod. 29, 23–29. 10.1093/ortho/29.1.23 [DOI] [PubMed] [Google Scholar]
- Mengel W. (1939). Sippenuntersuchegen bei Lippen, Kiefer, und Gaumenspalte. Zeits. F. Konstlehre 23, 17–42. [Google Scholar]
- Miller S. F., Weinberg S. M., Nidey N. L., Defay D. K., Marazita M. L., Wehby G. L., et al. (2014). Exploratory Genotype-Phenotype Correlations of Facial Form and Asymmetry in Unaffected Relatives of Children with Non-syndromic Cleft Lip And/or Palate. J. Anat. 224, 688–709. 10.1111/joa.12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millicovsky G., Ambrose L. J. H., Johnston M. C. (1982). Developmental Alterations Associated with Spontaneous Cleft Lip and Palate in CL/Fr Mice. Am. J. Anat. 164, 29–44. 10.1002/aja.1001640104 [DOI] [PubMed] [Google Scholar]
- Mills L. F., Niswander J. D., Mazaheri M., Brunelle J. A. (1968). Minor Oral and Facial Defects in Relatives of Oral Cleft Patients. Angle Orthod. 38, 199–204. [DOI] [PubMed] [Google Scholar]
- Mossey P. A., McColl J. H., Stirrups D. R. (1997). Differentiation between Cleft Lip with or without Cleft Palate and Isolated Cleft Palate Using Parental Cephalometric Parameters. Cleft Palate-Craniofacial J. 34, 27–35. [DOI] [PubMed] [Google Scholar]
- Mossey P. A., McColl J., O'Hara M. (1998). Cephalometric Features in the Parents of Children with Orofacial Clefting. Br. J. Oral Maxillofac. Surg. 36, 202–212. 10.1016/s0266-4356(98)90498-3 [DOI] [PubMed] [Google Scholar]
- Nakasima A., Ichinose M. (1984). Size of the Cranium in Parents and Their Children with Cleft Lip. Cleft Palate J. 21, 193–203. [PubMed] [Google Scholar]
- Nakasima A., Ichinose M. (1983). Characteristics of Craniofacial Structures of Parents of Children with Cleft Lip And/or Palate. Am. J. Orthod. 84, 140–146. 10.1016/0002-9416(83)90178-1 [DOI] [PubMed] [Google Scholar]
- Naqvi S., Hoskens H., Wilke F., Weinberg S. M., Shaffer J. R., Walsh S., et al. (in press). Decoding the Human Face: Challenges and Progress in Understanding the Genetics of Craniofacial Morphology. Annu. Rev. Genomics Hum. Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswander J. D. (1968). Laminographic X-Ray Studies in Families with Cleft Lip and Cleft Palate. Archives Oral Biol. 13, 1019–1022. 10.1016/0003-9969(68)90017-4 [DOI] [PubMed] [Google Scholar]
- Parsons T. E., Kristensen E., Hornung L., Diewert V. M., Boyd S. K., German R. Z., et al. (2008). Phenotypic Variability and Craniofacial Dysmorphology: Increased Shape Variance in a Mouse Model for Cleft Lip. J. Anat. 212, 135–143. 10.1111/j.1469-7580.2007.00845.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashayan H., Fraser F. C. (1971). Nostril Asymmetry Not a Microform of Cleft Lip. Cleft Palate J. 8, 185–188. [PubMed] [Google Scholar]
- Perkiomaki M. R., Yoon Y.-J., Tallents R. H., Barillas I., Herrera-Guido R., Moss M. E., et al. (2003). Association of Distinct Craniofacial Features in Nonsyndromic Cleft Lip and Palate Family Members. Cleft Palate-Craniofacial J. 40, 397–402. 10.1597/1545-1569_2003_040_0397_aodcfi_2.0.co_2 [DOI] [PubMed] [Google Scholar]
- Procházková J., Tolarová M. (1986). Craniofacial Morphological Features in Parents of Children with Isolated Cleft Palate. Acta Chir. Plast. 28, 194–204. [PubMed] [Google Scholar]
- Procházková J., Vinsová J. (1995). Craniofacial Morphology as a Marker of Predisposition to Isolated Cleft Palate. J. Craniofac. Genet. Dev. Biol. 15, 162–168. [PubMed] [Google Scholar]
- Raghavan R., Sidhu S. S., Kharbanda O. P. (1994). Craniofacial Pattern of Parents of Children Having Cleft Lip And/or Cleft Palate Anomaly. Angle Orthod. 64, 137–144. [DOI] [PubMed] [Google Scholar]
- Roosenboom J., Indencleef K., Hens G., Peeters H., Christensen K., Marazita M. L., et al. (2017). Testing the Face Shape Hypothesis in Twins Discordant for Nonsyndromic Orofacial Clefting. Am. J. Med. Genet. 173, 2886–2892. 10.1002/ajmg.a.38471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose W. (1891). On Harelip and Cleft Palate. London: H.K. Lewis. [Google Scholar]
- Rusconi L., Brusati R. (1966). The Radiological Appearance of the Nasal Fossas in the Ancestors and Immediate Relations of Subjects with Cleft Palate. Preliminary Note. Minerva Stomatol. 15, 548–550. [PubMed] [Google Scholar]
- Sato T. (1989). Craniofacial Morphology of Parents with Cleft Lip and Palate Children. Shikwa Gakuho 89, 1479–1506. [PubMed] [Google Scholar]
- Shibasaki Y., Ohtsuka S., Hattori M., Nukatsuka S. (1978). A Cephalometric Study of Craniofacial Morphology of Parents of Children with Cleft Lip and Palate [in Japanese]. J. Jpn. Cleft Palate Assoc. 3, 31–43. [Google Scholar]
- Sigler A., Ontiveros D. S. (1999). Nasal Deformity and Microform Cleft Lip in Parents of Patients with Cleft Lip. Cleft Palate-Craniofacial J. 36, 139–143. 10.1597/1545-1569_1999_036_0139_ndamcl_2.3.co_2 [DOI] [PubMed] [Google Scholar]
- Smiley G. R., Vanek R. J., Dixon A. D. (1971). Width of the Craniofacial Complex during Formation of the Secondary Palate. Cleft Palate J. 8, 371–378. [PubMed] [Google Scholar]
- Sulik K. K., Johnston M. C., Ambrose L. J. H., Dorgan D. (1979). Phenytoin (Dilantin)-induced Cleft Lip and Palate in A/J Mice: a Scanning and Transmission Electron Microscopic Study. Anat. Rec. 195, 243–255. 10.1002/ar.1091950201 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Takenoshita Y., Honda Y., Matsuura C. (1999). Dentocraniofacial Morphology in Parents of Children with Cleft Lip And/or Palate. Cleft Palate-Craniofacial J. 36, 131–138. 10.1597/1545-1569_1999_036_0131_dmipoc_2.3.co_2 [DOI] [PubMed] [Google Scholar]
- Trasler D. G., Machado M. (1979). Newborn and Adult Face Shapes Related to Mouse Cleft Lip Predisposition. Teratology 19, 197–206. 10.1002/tera.1420190210 [DOI] [PubMed] [Google Scholar]
- Trasler D. G., Ohannessian L. (1983). Ultrastructure of Initial Nasal Process Cell Fusion in Spontaneous and 6-Aminonicotinamide-Induced Mouse Embryo Cleft Lip. Teratology 28, 91–101. 10.1002/tera.1420280112 [DOI] [PubMed] [Google Scholar]
- Trasler D. G. (1968). Pathogenesis of Cleft Lip and its Relation to Embryonic Face Shape in A/J and C57BL Mice. Teratology 1, 33–49. 10.1002/tera.1420010106 [DOI] [PubMed] [Google Scholar]
- Trew C. (1757). Sistens Plura Exempla Palati Deficientis. Nova Acta Physico-Medica Acad. Caesareae Leopoldino-Carolinae Naturae Curiosorum 1, 445–447. [Google Scholar]
- Wang K. Y., Diewert V. M. (1992). A Morphometric Analysis of Craniofacial Growth in Cleft Lip and Noncleft Mice. J. Craniofac. Genet. Dev. Biol. 12, 141–154. [PubMed] [Google Scholar]
- Ward R. E., Bixler D., Raywood E. R. (1989). A Study of Cephalometric Features in Cleft Lip-Cleft Palate Families. I: Phenotypic Heterogeneity and Genetic Predisposition in Parents of Sporadic Cases. Cleft Palate J. 26, 318–325. [PubMed] [Google Scholar]
- Ward R. E., Moore E. S., Hartsfield J. K. (2002). “Morphometric Characteristics of Subjects with Oral Facial Clefts and Their Relatives,” in Cleft Lip and Palate: From Origin to Treatment. Editor Wyszynski D. (Oxford: Oxford University Press; ), 66–86. [Google Scholar]
- Ward R. E., Bixler D., Jamison P. L. (1994). Cephalometric Evidence for a Dominantly Inherited Predisposition to Cleft Lip-Cleft Palate in a Single Large Kindred. Am. J. Med. Genet. 50, 57–63. 10.1002/ajmg.1320500113 [DOI] [PubMed] [Google Scholar]
- Weinberg S., Maher B., Marazita M. (2006). Parental Craniofacial Morphology in Cleft Lip with or without Cleft Palate as Determined by Cephalometry: a Meta-Analysis. Orthod. Craniofac Res. 9, 18–30. 10.1111/j.1601-6343.2006.00339.x [DOI] [PubMed] [Google Scholar]
- Weinberg S. M., Neiswanger K., Richtsmeier J. T., Maher B. S., Mooney M. P., Siegel M. I., et al. (2008). Three-dimensional Morphometric Analysis of Craniofacial Shape in the Unaffected Relatives of Individuals with Nonsyndromic Orofacial Clefts: a Possible Marker for Genetic Susceptibility. Am. J. Med. Genet. 146A, 409–420. 10.1002/ajmg.a.32177 [DOI] [PubMed] [Google Scholar]
- Weinberg S., Naidoo S., Bardi K., Brandon C., Neiswanger K., Resick J., et al. (2009). Face Shape of Unaffected Parents with Cleft Affected Offspring: Combining Three-Dimensional Surface Imaging and Geometric Morphometrics. Orthod. Craniofacial Res. 12, 271–281. 10.1111/j.1601-6343.2009.01462.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. D., Indencleef K., Naqvi S., Eller R. J., Hoskens H., Roosenboom J., et al. (2021). Insights into the Genetic Architecture of the Human Face. Nat. Genet. 53, 45–53. 10.1038/s41588-020-00741-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. (1934a). An Analysis of Variability in Number of Digits in an Inbred Strain of Guinea Pigs. Genetics 19, 506–536. 10.1093/genetics/19.6.506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. (1934b). The Results of Crosses between Inbred Strains of guinea Pigs, Differing in Number of Digits. Genetics 19, 537–551. 10.1093/genetics/19.6.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y.-J., Perkiomaki M. R., Tallents R. H., Barillas I., Herrera-Guido R., Fong C.-T., et al. (2003). Association of Nasomaxillary Asymmetry in Children with Unilateral Cleft Lip and Palate and Their Parents. Cleft Palate-Craniofacial J. 40, 493–497. 10.1597/1545-1569_2003_040_0493_aonaic_2.0.co_2 [DOI] [PubMed] [Google Scholar]
- Yoon Y.-J., Perkiomaki M. R., Tallents R. H., Barillas I., Herrera-Guido R., Fong C.-T., et al. (2004). Transverse Craniofacial Features and Their Genetic Predisposition in Families with Nonsyndromic Unilateral Cleft Lip and Palate. Cleft Palate-Craniofacial J. 41, 256–261. 10.1597/02-134.1 [DOI] [PubMed] [Google Scholar]
- Young N. M., Wat S., Diewert V. M., Browder L. W., Hallgrímsson B. (2007). Comparative Morphometrics of Embryonic Facial Morphogenesis: Implications for Cleft-Lip Etiology. Anat. Rec. 290, 123–139. 10.1002/ar.20415 [DOI] [PubMed] [Google Scholar]
- Yu Y., Alvarado R., Petty L. E., Bohlender R. J., Shaw D. M., Below J. E., et al. (2022). Polygenic Risk Impacts PDGFRA Mutation Penetrance in Non-syndromic Cleft Lip and Palate. Hum. Mol. Genet. (in press). 10.1093/hmg/ddac037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Miller S. F., Roosenboom J., Wehby G. L., Moreno Uribe L. M., Hecht J. T., et al. (2018). Soft Tissue Nasal Asymmetry as an Indicator of Orofacial Cleft Predisposition. Am. J. Med. Genet. 176, 1296–1303. 10.1002/ajmg.a.38688 [DOI] [PMC free article] [PubMed] [Google Scholar]