Abstract
Carotenoid esterification plays indispensable roles in preventing degradation and maintaining the stability of carotenoids. Although the carotenoid biosynthetic pathway has been well characterized, the molecular mechanisms underlying carotenoid esterification, especially in floral organs, remain poorly understood. In this study, we identified a natural mutant flowering Chinese cabbage (Caixin, Brassica rapa L. subsp. chinensis var. parachinensis) with visually distinguishable pale-yellow petals controlled by a single recessive gene. Transmission electron microscopy (TEM) demonstrated that the chromoplasts in the yellow petals were surrounded by more fully developed plastoglobules compared to the pale-yellow mutant. Carotenoid analyses further revealed that, compared to the pale-yellow petals, the yellow petals contained high levels of esterified carotenoids, including lutein caprate, violaxanthin dilaurate, violaxanthin-myristate-laurate, 5,6epoxy-luttein dilaurate, lutein dilaurate, and lutein laurate. Based on bulked segregation analysis and fine mapping, we subsequently identified the critical role of a phytyl ester synthase 2 protein (PALE YELLOW PETAL, BrPYP) in regulating carotenoid pigmentation in flowering Chinese cabbage petals. Compared to the yellow wild-type, a 1,148 bp deletion was identified in the promoter region of BrPYP in the pale-yellow mutant, resulting in down-regulated expression. Transgenic Arabidopsis plants harboring beta-glucuronidase (GUS) driven by yellow (BrPYPY::GUS) and pale-yellow type (BrPYPPY::GUS) promoters were subsequently constructed, revealing stronger expression of BrPYPY::GUS both in the leaves and petals. Furthermore, virus-induced gene silencing of BrPYP significantly altered petal color from yellow to pale yellow. These findings demonstrate the molecular mechanism of carotenoid esterification, suggesting a role of phytyl ester synthase in carotenoid biosynthesis of flowering Chinese cabbage.
Keywords: carotenoid esterification, pale yellow petals, phytyl ester synthase, plastoglobules, flowering Chinese cabbage
Introduction
Carotenoids play a number of vital roles in higher plant growth and development, harvesting light for photosynthesis (Hermanns et al., 2020) and protecting photosynthetic apparatus from photooxidation (Niyogi and Truong, 2013), as well as providing precursor metabolites for the biosynthesis of abscisic acids (ABA) and strigolactones (SLs) (Jia et al., 2018). The presence of carotenoids is also an evolutionary strategy that results in colorful petals and fruit, thereby acting to attract pollinators (Yuan et al., 2015).
Flower color, which is narrowly defined as petal color, is an essential trait of crops and vegetables with characteristics of phenotype variation, genetic stability and environmental adaptability (Rahman, 2001). It also plays an important role in hybrid seed purity by removing mixed non-target plant material and revealing the reliability of interspecific trait transfer (Zhang X. et al., 2018). Carotenoids range in color from yellow to red, resulting in a variety of eye-catching flower colors (Nisar et al., 2015). Carotenoid pigments are typically synthesized and stored in chromoplasts, a type of non-photosynthetic plastid (Sadali et al., 2019). Chromoplasts are characterized by the accumulation of plastoglobules (PGs), which contain a large quantity of xanthophyll esters (Mulisch and Krupinska, 2013). However, the role of chromoplasts in petal coloration is yet to be explored.
The concentration of carotenoid is co-regulated by its biosynthesis, degradation, and sequestration (Galpaz et al., 2006; Tanaka and Ohmiya, 2008; Yoshioka et al., 2012). Biosynthesis begins with isoprene precursors generated by the methylerythritol phosphate (MEP) pathway, with the subsequent reaction producing isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP) (Rodriguez-Concepcion and Boronat, 2015). Phytoene is the first carotenoid product, and is catalyzed from geranylgeranyl pyrophosphate (GGPP) by phytoene synthase (PSY) then converted into red lycopene via a series of desaturation and isomerization reactions (Cazzonelli and Pogson, 2010). Cyclization of lycopene by lycopene ε-cyclase (LycE) and/or lycopene β-cyclase (LycB) produces α-carotene and β-carotene, which are further hydroxylated to produce xanthophylls, such as lutein and zeaxanthin (Sun et al., 2018; Hermanns et al., 2020). Xanthophyll esterification effectively prevents carotenoid degradation and subsequently increases total levels within the chromoplasts (Ariizumi et al., 2014). However, little is known about the factors underlying xanthophyll esterification in chromoplasts and other higher plant plastids.
Brassica rapa crops belong to the Cruciferae family, which includes a number of economically important vegetables and oil crops, especially in East Asia. Flower color plays a significant role in seed purity, removing miscellaneous plants, and aiding the selection and breeding of Brassica varieties (Li et al., 2015). Recently, a number of genes controlling white and orange flower color were reported in Brassica crops. For example, the carotenoid isomerase gene CRTISO and plastid-lipid associated gene PAP were found to be related to orange and white flower color in Chinese cabbage, respectively (Li et al., 2015; Zhang et al., 2020). Furthermore, the carotenoid cleavage dioxygenase 4 gene (CCD4) was found to be responsible for the white flower color in several B. oleracea subspecies, including cabbage (Han et al., 2019; Zhang et al., 2019) and kale (Xu et al., 2019). Moreover, BjuA008406 and BjuB027334 were identified as candidate genes of white flower color in B. juncea (Zhang X. et al., 2018). Recently, the white flower gene BrWF3, which might associate with the production of xanthophyll esters, was identified based on the fine mapping and carotenoid analysis (Yang et al., 2021). However, the underlying molecular mechanism in B. rapa remains poorly understood.
In this study, we identified a natural mutant flowering Chinese cabbage plant (Caixin, B. rapa L. subsp. Chinensis var. parachinensis) with visually distinguishable pale-yellow petals. Inheritance pattern analysis followed by transmission electron microscopy (TEM) assay and carotenoid profiling were subsequently performed. Based on fine mapping and virus-induced gene silencing, we revealed that a phytyl ester synthase 2 protein [PALE YELLOW PETAL (BrPYP)] is responsible for the mutant pale-yellow petal color. Staining of transgenic Arabidopsis plants harboring beta-glucuronidase further revealed the function of the BrPYP promoter in regulating petal carotenoid pigmentation in flowering Chinese cabbage. Overall, our findings demonstrate the molecular mechanism underlying BrPYP-mediated regulation of petal carotenoid accumulation, and suggest a role of phytyl ester synthase in carotenoid esterification in B. rapa.
Materials and Methods
Plant Materials
Inbred lines of the pale-yellow parent (20M-293, P1) and yellow parent (20M-294, P2) were used in all experiments. P1 was crossed with P2 to produce the hybrid F1 then the F1 was self-pollinated to obtain the F2 population. The F1 plants were also backcrossed with 20M-293 to produce a BC1 population. All materials were grown in a greenhouse at the Beijing Vegetable Research Center.
Identification of Candidate Genes Using Bulk Segregation Analysis and Kompetitive Allele-Specific PCR Assay
Candidate genes were identified according to the BSA method (Takagi et al., 2013). Fifty yellow and 50 pale-yellow individuals from the F2 population were selected for each pool. Equal amounts of DNA from the selected individuals were then combined and DNA libraries were prepared using a TruSeq Library Construction Kit before sequencing on the HiSeq™ PE150 platform (Illumina, San Diego, United States).
The raw reads were filtered by removing reads with adapters, reads with an N value greater than 10%, and low-quality reads with a quality value Q ≤ 10. The clean reads were then aligned to the Brassica rapa reference genome V3.0 (Zhang L. et al., 2018) using Burrows-Wheeler Aligner (BWA) (Li and Durbin, 2010). Detection of SNPs and InDels was carried out using a GATK software toolkit (McKenna et al., 2010) with default parameters. The average ΔSNP-index across 10 chromosomes in the B. rapa genome was measured individually using the sliding-window approach, with a 2 Mb window size and a 10 kb sliding-window step.
High−quality SNP candidates between two parents were selected for KASP marker development. For each SNP, two allele-specific forward primers and one common reverse primer were designed by LGC Genomics (Supplementary Table 1). The amplifications and analysis methods were performed based on the previously published guidelines (Li et al., 2019).
Gene Cloning and Sequence Analysis
Total genomic DNA was extracted from fresh petals using the cetyl trimethylammonium bromide (CTAB) method (Chen and Ronald, 1999). The reference sequences of BraA02g037160.3C and BraA02g037170.3C were obtained from the B. rapa database, and used to design gene-specific and promoter primers (Supplementary Table 2). The cloning and sequencing of candidate genes were performed as described previously (Li et al., 2021).
The sequences of BraA02g037170.3C were then aligned using ClustalW1 and displayed using ESPript2 The cis- and trans-acting elements of the BraA02g037170.3C promoter region were also predicted using the PlantCARE3 online analysis software program.
RNA Extraction and Expression Analysis
Total RNA was extracted from petals of the two parent lines using the RNAprep Pure Plant Plus Kit (TIANGEN, Beijing, China). First-strand cDNA, which was used for real-time PCR (RT-PCR) and quantitative real-time PCR (qRT-PCR), was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Japan). Specific primers of BraA02g037160.3C, BraA02g037170.3C, and two phytoene desaturase (PDS) genes for RT-PCR and qRT-PCR were designed using the Primer Premier 6.0 software. PCR amplification and conditions were as described previously (Li et al., 2020). Gene expression was normalized using the housekeeping gene GAPDH. Reactions were performed with three biological replicates and three technical replicates, and transcript levels in different colored flowers were analyzed using the 2–Δ Δ Ct method (Livak and Schmittgen, 2001). Primer sequences are listed in Supplementary Table 3.
Carotenoid Detection
We detected carotenoid contents by Metware4 based on the AB SCIEX QTRAP 6500 liquid chromatography with tandem mass spectrometry (LCMS/MS) platform. Three biological replicates of each parent were assessed. In brief, samples from pale-yellow and yellow petals were freeze-dried then stored at -80°C until use. The dried samples were then homogenized in a grinder before extracting 50 mg of dried powder with a solution of n-hexane: acetone: ethanol. An internal standard was then added and the extract was centrifuged for 20 min at room temperature. The supernatants were then collected and the above steps were repeated to re-extract the residue. The extract was then evaporated to dryness under nitrogen flow and reconstituted in methanol: MTBE solution then filtered through a 0.22 μm filter for LC-MS analysis.
The analytical conditions were as follows: column, YMC C30 (3 μm, 100 mm × 2.0 mm); solvent system, methanol: acetonitrile (1:3, V/V) with 0.01% BHT and 0.1% formic acid (A), methyl tert-butyl ether with 0.01% BHT (B); gradient program, started at 0% B (0–3 min), increased to 70% B (3–5 min), then increased to 95% B (5–9 min), finally ramped back to 0% B (10–11 min); flow rate, 0.8 mL/min; temperature, 28°C; injection volume: 2μL.
AB 6500 Q TRAP LC/MS/MS System, equipped with an APCI Heated Nebulizer interface, operating in a positive ion mode, and controlled by Analyst 1.6.3 software (AB Sciex). The APCI source operation parameters were as follow: ion source, APCI +; source temperature 350°C; curtain gas (CUR) was set at 25.0 psi. DP and CE for individual MRM (Multiple Reaction Monitoring) transition was done with further DP and CE optimization. A specific set of MRM transitions were monitored for each period according to the carotenoids eluted within this period.
Virus-Induced Gene Silencing Assay
To determine the efficiency of the VIGS technique in B. rapa, the PDS gene, which can be easily used to score the photobleached phenotype, was used as a visual marker. The hairpin sequence and plasmid construction of BrPDS were performed based on the previously published protocol (Yu et al., 2018). An appropriate fragment of 40 nt of BrPYP was then selected and a 80-nt palindromic DNA fragment of with SnaBI restriction sites and homologous arms on both sides was designed (TTAAACTACTTAGCTCCGTCTTCGGGCTCTCTCCTGTCT CGAGACAGGAGAGAGCCCGAAGACGGAGCTAAGTAGTT TAA). The palindromic DNA fragment was produced by Nanjing Genscript Technology Company (Nanjing, China) and linked to pTYs using the ClonExpress Entry One Step Cloning Kit (Vazyme) (pTYsBrPDS and pTYsBrPYP). The plasmid was then transformed into competent Super Stbl3 (Beijing, Huayueyang) on an Ampicillin (50 μg/ml) resistant plate then cultured overnight before selecting single clones. The clones were then placed in 1 L LB liquid medium and shaken at 225 rmp at a temperature of 30°C for 16 h. The plasmid sample was extracted using a Plasmid Giga Kit (OMEGA, United States) then the working concentration was adjusted to 300 μg/μl. A 5 μl leaf sample was then selected, rubbed with quartz sand, left to stand for 1 min then washed with water. The second vaccination was carried out 4–5 days later. Plants used for the VIGS assay were grown in a greenhouse with a temperature regimen of 25°C day/18°C night. Bacterial solution was then infiltrated into the leaves to obtain the silencing effect.
Glucuronidase Staining and Activity
A sequence approximately 2 kb upstream of the gene start codon was used as a template for the cloned gene promoter sequence. Specific PCR primers were designed, and the target fragment was amplified then connected to the PBI121 (HindIII/BamhI) vector via homologous recombination. After transforming DH5α, a single clone was selected then sequenced before extracting the plasmid and transforming it into Agrobacterium GV3101. After plaque detection, A. thaliana was transformed using the floral dipping method (Clough and Bent, 1998). Seeds from the transformed Arabidopsis plants were then screened on 1/2 MS medium supplemented with 1.5% (w/v) sucrose and 0.8% (w/v) agar containing 25 mg/L of kanamycin (Yue et al., 2019). GUS staining of positive plants was performed using GUS staining solution at 37°C for 24 h with a GUSBlue KIT (Beijing Huayueyang, China). They were then decolorized with 75% ethanol and observed under a stereoscope (Olympus, Japan).
Results
Phenotypic Characterization of Pale-Yellow and Yellow Petals in Flowering Chinese Cabbage
Natural mutant flowering Chinese cabbage plants with visually distinguishable pale-yellow flowers were found in the field (Figures 1A,B). To clarify the inheritance pattern, we constructed two populations using a pale-yellow parent (P1) and yellow parent (P2) (Figures 1A,B). As expected, the flowers of F1 plants derived from a reciprocal cross between the two parents were all yellow. Of 442 F2 individuals, 343 had yellow flowers and 99 had pale yellow flowers. A χ2-test subsequently confirmed the F2 segregation ratio to be 3:1 (χ20.05 = 3.84; P > 0.05; Table 1). Furthermore, flower color was also examined in a BC1 population derived from F1 individuals backcrossed with the pale yellow-flowered parent, revealing an expected Mendelian inheritance ratio of 1 yellow: 1 pale yellow (χ20.05 = 3.84; P > 0.05; Table 1). These results indicated that the pale-yellow flower trait is controlled by a single recessive BrPYP gene (Table 1).
FIGURE 1.

Phenotypes of the yellow and pale-yellow flowering Chinese cabbage petals. Yellow (A) and pale-yellow flower petals (B). Plastid morphology in the yellow (C,E) and pale-yellow petals (D,F) was compared by TEM analysis. Enlarged images are also shown (E,F). PG: plastoglobules. Scale bars: (C,D) 2 μm, (E,F) 200 nm.
TABLE 1.
The segregation of flower colors in the F2 and BC1 population.
| Generation | Total plants | Yellow flower plants | White flower plants | Goodness of fit X2 |
| F2 | 442 | 343 | 99 | 1.46 |
| BC1 | 1,002 | 521 | 481 | 1.51 |
X2 > X20.05 = 3.84 is considered significant.
Since carotenoids are biosynthesized and stored in chromoplasts, TEM was used to determine whether the pale-yellow color was due to changes in the morphology of the chromoplasts. Accordingly, chromoplasts in the yellow petals were well developed and surrounded by numerous fully developed PGs (Figures 1C,E). However, in contrast, chromoplasts in the pale-yellow petals only contains far fewer well-developed PGs (Figures 1D,F).
Carotenoid Analysis of Pale-Yellow and Yellow Flowers Using LC-MS/MS
To examine whether the petal coloration was due to a reduction in carotenoid accumulation, carotenoid components were analyzed in three pale-yellow and three yellow flowers. Ultra-performance liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was used to identify non-esterified and esterified carotenoids (Table 2). As a result, a total of 34 carotenoid compounds were differently accumulated between yellow and pale-yellow petals (Table 2), 28 and 25 of which were found in the pale-yellow and yellow petals, respectively. Of these, 22 compounds were less abundant in the pale-yellow petals, including nine compounds that were not detected (Table 2).
TABLE 2.
Difference of carotenoids accumulation and their derivative esters present in yellow and pale-yellow petals.
| Compounds | Class | Yellow (μ g/g) | Pale-yellow (μ g/g) | Log2FC | Type |
| Lutein caprate | Carotenoid esters | 266 ± 5 | 1.17 ± 0.09 | -7.83 | Down |
| (E/Z)-Phytoene | Carotenes | 89.7 ± 7.5 | 32.27 ± 5.81 | -1.47 | Down |
| Violaxanthin dilaurate | Carotenoid esters | 46.97 ± 5.99 | 0.41 ± 0.1 | -6.84 | Down |
| Violaxanthin-myristate-laurate | Carotenoid esters | 17.03 ± 3.35 | NA | NA | Down |
| 5,6epoxy-luttein dilaurate | Carotenoid esters | 15.87 ± 2.76 | 0.16 ± 0.06 | -6.63 | Down |
| Lutein dilaurate | Carotenoid esters | 8.89 ± 1.63 | 1.09 ± 0.05 | -3.03 | Down |
| Lutein laurate | Carotenoid esters | 6.61 ± 0.31 | NA | NA | Down |
| β-Cryptoxanthin | Xanthophylls | 5.96 ± 0.39 | 2.83 ± 0.11 | -1.07 | Down |
| Rubixanthin caprate | Carotenoid esters | 3.35 ± 0.64 | NA | NA | Down |
| α-Carotene | Carotenes | 2.25 ± 0.27 | NA | NA | Down |
| α-Cryptoxanthin | Xanthophylls | 2.24 ± 0.15 | 0.84 ± 0.15 | -1.42 | Down |
| Neochrome palmitate | Carotenoid esters | 1.9 ± 0.28 | 0.84 ± 0.33 | -1.18 | Down |
| β-Cryptoxanthin laurate | Carotenoid esters | 1.59 ± 0.28 | 0.11 ± 0.02 | -3.85 | Down |
| 5,6-epoxy-lutein-caprate-palmitate | Carotenoid esters | 1.48 ± 0.44 | NA | NA | Down |
| Violaxanthin palmitoleate | Carotenoid esters | 1.09 ± 0.22 | NA | NA | Down |
| γ-Carotene | Carotenes | 0.7 ± 0.17 | NA | NA | Down |
| Rubixanthin palmitate | Carotenoid esters | 0.54 ± 0.08 | 0.19 ± 0.01 | -1.51 | Down |
| β-Cryptoxanthin palmitate | Carotenoid esters | 0.52 ± 0.1 | 0.23 ± 0.02 | -1.18 | Down |
| Zeaxanthin dilaurate | Carotenoid esters | 0.19 ± 0.04 | 0.01 ± 0 | -4.25 | Down |
| β-Cryptoxanthin oleate | Carotenoid esters | 0.11 ± 0 | NA | NA | Down |
| Echinenone | Xanthophylls | 0.04 ± 0 | 0.01 ± 0 | -2.00 | Down |
| Canthaxanthin | Xanthophylls | 0.01 ± 0 | NA | NA | Down |
| Apocarotenal | Xanthophylls | NA | 0.03 ± 0 | NA | Up |
| ε-Carotene | Carotenes | NA | 0.15 ± 0.02 | NA | Up |
| Antheraxanthin dipalmitate | Carotenoid esters | NA | 0.33 ± 0.1 | NA | Up |
| Violaxanthin laurate | Carotenoid esters | NA | 0.36 ± 0.03 | NA | Up |
| Violaxanthin dipalmitate | Carotenoid esters | NA | 0.47 ± 0.09 | NA | Up |
| Zeaxanthin dipalmitate | Carotenoid esters | NA | 0.58 ± 0.15 | NA | Up |
| Lutein dimyristate | Carotenoid esters | 12.67 ± 0.9 | 24.25 ± 1.35 | 0.94 | Up |
| Violaxanthin myristate | Carotenoid esters | 1.56 ± 0.52 | 4.73 ± 0.96 | 1.60 | Up |
| Violaxanthin palmitate | Carotenoid esters | 1.02 ± 0.09 | 3.59 ± 0.8 | 1.82 | Up |
| Violaxanthin-myristate-palmitate | Carotenoid esters | 0.46 ± 0.03 | 7.85 ± 1.42 | 4.09 | Up |
| Violaxanthin dimyristate | Carotenoid esters | 0.4 ± 0.13 | 16.03 ± 2.84 | 5.32 | Up |
| β-Citraurin | Xanthophylls | 0.03 ± 0 | 0.08 ± 0.01 | 1.42 | Up |
NA means that the substance is not detected in this assay.
A comparison of carotenoid profiles between the pale-yellow and yellow petals demonstrated abundant esterified carotenoids in the yellow petals. Moreover, carotenoid esters, such as lutein caprate, violaxanthin dilaurate, violaxanthin-myristate-laurate, 5,6epoxy-luttein dilaurate, lutein dilaurate, and lutein laurate, were highly abundant or found only in the yellow petals (Table 2). Taken together, these results suggest that yellow petals accumulate more carotenoid esters (namely, lutein caprate, and violaxanthin dilaurate) than pale-yellow petals, resulting in greater carotenoid accumulation and subsequent yellow pigmentation.
Identification of BrPYP Using Bulk Segregation Analysis and Fine Mapping
To identify candidates of the BrPYP gene, 50 pale yellow and 50 yellow individuals were selected from the F2 population and used to construct extreme pools for bulk segregation analysis (BSA). Genome resequencing of the pale-yellow parent (P1), yellow parent (P2) and two DNA pools (pale-yellow and yellow bulk) resulted in 202,531,626, 208,423,334, 209,006,868, and 207,592,868 high-quality clean reads, respectively (Supplementary Table 4). Meanwhile, 98.12, 98.35, 98.52, and 98.57% of the clean reads (Supplementary Table 5) were, respectively, aligned to the reference genome. Using a single-nucleotide polymorphism (SNP) index of 0.99 as the threshold, we were able to preliminarily map BrPYP to a candidate region of 2.76Mb (24,960,000–27,720,000) on chromosome A02 (Figure 2A).
FIGURE 2.
Fine mapping of the pale-yellow petal gene BrPYP. (A) SNP index plot of 10 chromosomes produced using BSA analysis. The x-axis represents the position of the 10 chromosomes, while the y-axis represents the SNP index. The dashed line represents the index threshold (0.99). (B) F2 and BC1 populations were applied to the BrPYP fine map, narrowing down the area to 101.79 kb. The SNP markers used for mapping are labeled in the schematic diagram of the chromosome. The number of recombinants within the same genotype are written on the left, with phenotypes on the right. “Y” and “PY” refer to yellow and pale-yellow phenotypes, respectively. White and gray segments indicate the same genotype as the yellow parent and F1, respectively, while the black segment represents the same genotype as the pale-yellow parent. (C) The 13 genes in the candidate region.
To fine map BrPYP, we used both BC1 and F2 individuals to conduct Kompetitive Allele-Specific PCR (KASP) genotyping analysis. SNPs were acquired based on the resequencing results with 50 SNP markers in the candidate region initially used to identify recombinant individuals (Figure 2B). Subsequently, 15 SNP markers exhibiting polymorphisms were detected between the pale-yellow bulk and yellow bulk (Supplementary Table 1). Finally, BrPYP was localized to a 101.79 kb region between SNP markers A02_25962798 and A02_26064584 (Figure 2B). Specifically, the SNP marker within the BraA02g037170.3C, A02_26057775 gene co-segregated with the pale-yellow flower trait.
Candidate Gene Cloning, Sequencing and Expression Analysis
To identify BrPYP in flowering Chinese cabbage, comparative gene annotation was carried out in Arabidopsis thaliana. Overall, 13 genes were identified in the candidate region, of which nine possessed functional annotations suggestive of Arabidopsis homologs (Table 3). However, among these, no carotenoid biosynthesis genes were identified. We therefore screened each gene according to previously published data, revealing two homologs of Phytyl ester synthase 2 (PES2) as candidates. PES2 encodes a carotenoid modifying protein, which plays a vital role in the production of xanthophyll esters in anthers and petals (Ariizumi et al., 2014).
TABLE 3.
Thirteen annotated genes in the candidate region of BrPYP.
| Gene | Chr | Start | End | Arabidopsis ID | Annotation |
| BraA02g037050.3C | A02 | 25,961,578 | 25,965,034 | AT3G26730 | E3 ubiquitin-protein ligase RMA1H1-like |
| BraA02g037060.3C | A02 | 25,965,421 | 25,965,848 | - | Light-regulated protein 1, LIR1 |
| BraA02g037070.3C | A02 | 25,968,852 | 25,969,076 | - | tRNA (guanine (10)-N2)-methyltransferase homolog |
| BraA02g037080.3C | A02 | 25,972,710 | 25,973,315 | - | Glycine-rich RNA-binding protein GRP1A-like isoform X1 |
| BraA02g037090.3C | A02 | 25,977,283 | 25,978,878 | AT3G26744 | Transcription factor ICE1, SCRM |
| BraA02g037100.3C | A02 | 25,984,959 | 25,986,267 | AT3G26760 | Short-chain dehydrogenase reductase 2a, SDR2a |
| BraA02g037110.3C | A02 | 26,003,742 | 26,005,011 | AT3G26770 | Short-chain dehydrogenase reductase 2a, SDR2a |
| BraA02g037120.3C | A02 | 26,009,406 | 26,011,108 | AT3G26780 | Aquaporin TIP1-2 |
| BraA02g037130.3C | A02 | 26,014,958 | 26,016,857 | AT3G26790 | B3 domain-containing transcription factor FUS3, FUS3 |
| BraA02g037140.3C | A02 | 26,029,608 | 26,031,989 | AT3G26810 | AUXIN SIGNALING F-BOX 2, AFB2 |
| BraA02g037150.3C | A02 | 26,033,422 | 26,034,954 | - | NIPA-like protein 2 |
| BraA02g037160.3C | A02 | 26,044,622 | 26,048,832 | AT3G26840 | Acyltransferase-like protein |
| BraA02g037170.3C | A02 | 26,053,073 | 26,057,985 | AT3G26840 | Acyltransferase-like protein |
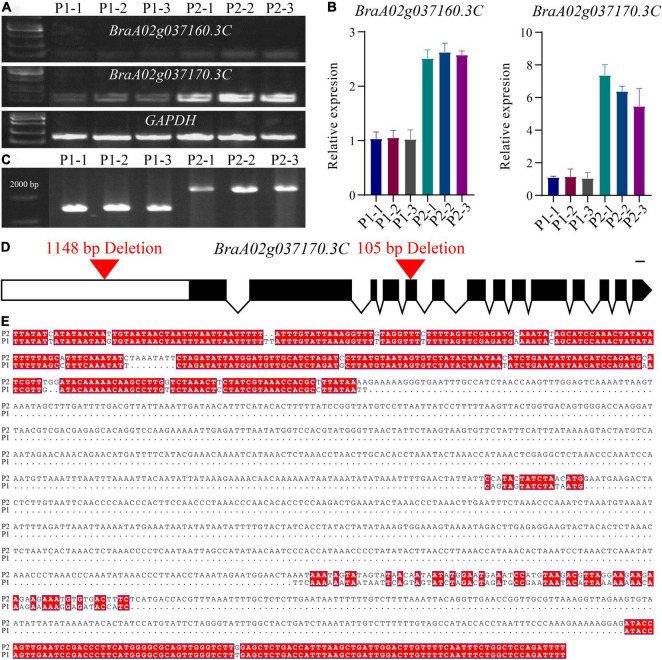
Quantitative real-time PCR (qRT-PCR) was subsequently performed to determine the expression level of BraA02g037160.3C and BraA02g037170.3C in the petals of the pale-yellow and yellow parent. Expression of both genes was lower in the pale-yellow mutant than the yellow parent (Figures 3A,B). However, in comparison, expression of BraA02g037170.3C was very high in the yellow parent and six-fold higher than in the pale-yellow parent. Meanwhile, expression of BraA02g037160.3C was only two-fold higher in the yellow parent than the pale-yellow parent, and was very low in both (Figure 3). To further characterize the difference between the two parents, the promoter of BraA02g037170.3C was subsequently amplified and sequenced. Sequence alignment revealed a 1,148 bp deletion starting 1,078 bp upstream of the promoter in the pale-yellow parent, with several other SNPs and InDels (Figures 3C,D). Analysis of Cis-acting regulatory elements further revealed that the deleted sequence contained multiple light-responsive motifs, including the TCT-motif, chs-CMA1a, MRE, GT1-motif, G-box, and ACE (Supplementary Table 6).
FIGURE 3.
Molecular cloning and confirmation of the BrPYP gene. (A) RT-PCR showing transcript abundance of BraA02g037160.3C and BraA02g037170.3C in the pale-yellow (P1) and yellow petals (P2). GAPDH was used as a control. (B) qRT-PCR showing transcript levels of BraA02g037160.3C and BraA02g037170.3C in the P1 and P2 petals. (C) Promoter amplification showing a large deletion in BraA02g037170.3C of P1. (D) Schematic representation of the gene structure of BrPYP showing the deletion in the promoter and the fifth exon of the pale-yellow parent. Introns are represented by a line, and exons are represented by black boxes. (E) The promoter was then sequenced and aligned (from 978 bp upstream to 2,500 bp) between P1 and P2.
Analysis of cDNA sequences of BraA02g037170.3C from both parents indicated a 105-bp deletion in the fifth exon of the pale-yellow parent, resulting in a 35-amino acid deletion (from 219 to 253) (Supplementary Figure 1a). Furthermore, protein analysis demonstrated two domains based on the NCBI Conserved Domain Database (Supplementary Figure 1b): alpha/beta hydrolases and lysophospholipid acyltransferases (LPLATs), both of which are involved in glycerophospholipid biosynthesis. Alpha/beta hydrolases are a functionally diverse superfamily consisting of proteases, lipases, peroxidases, epoxide hydrolases, and esterases. Interestingly, the 35-amino acid deletion in pale-yellow allele was in alpha/beta hydrolases domain (Supplementary Figure 1), which may also affect the function of BrPYP protein.
Silencing of BrPYP Causes a Pale-Yellow Petal Phenotype
Virus-induced gene silencing (VIGS) is a transient gene knockdown system that has successfully been used to characterize genes with different functions in B. rapa. We therefore used the VIGS approach to determine whether BrPYP can induce pale-yellow petals. Loss of chlorophyll pigmentation, caused by silencing of the PDS gene, was used as a visual marker to demonstrate the usefulness of this technique in efficient silencing of B. rapa genes (Figure 4A). Notably, both pTYsBrPDS and pTYs showed the same yellow petals as the yellow petal parent (Figures 4A,B). BrPYP gene silencing was subsequently monitored during flowering, revealing that successful silencing resulted in the pale-yellow phenotype (Figure 4C). The enlarged images of these petals were shown in Figures 4D–F.
FIGURE 4.

Transient silencing of BrPYP resulted in pale-yellow petals. (A) The evirus-induced gene silencing (VIGS)-mediated BrPDS gene silencing phenotype of flowering Chinese cabbage leaves. (B) Flowering Chinese cabbage plants were agro-drenched with empty pTYs vectors. (C) The VIGS-mediated BrPYP gene silencing phenotype of flowering Chinese cabbage petals. (D–F) Enlarged images of (A–C), respectively. (G) qRT-PCR analysis of VIGS-mediated BrPYP in flowering Chinese cabbage leaves and petals. The letters above the bars indicate significant differences at P < 0.05 (Student’s t-test).
qRT-PCR was also performed to further confirm the function of PYP at the transcriptional expression level mediated by VIGS (Figure 4G). The relative expression demonstrated that the BrPDS genes were successfully reduced in the leaves of plants infected with pTYsBrPDS (Supplementary Figure 2). Accordingly, transcript levels of BrPYP were substantially reduced both in the leaves and petals of pTYsBrPYP compared with pTYsBrPDS, pTYs and the yellow parent control (Figure 4G). Meanwhile, transcript levels of the BrPYP gene in BrPYP-silenced plants, which were similar to the pale-yellow parent, showed an approximate 70% reduction compared with the PDS-inoculated plants. This further confirms that BraA02g037170.3C represents the BrPYP gene responsible for the pale-yellow petal trait in flowering Chinese cabbage.
Expression Patterns of BrPYP
To determine whether the promoter of BrPYP functions in carotenoid esterification, we constructed transgenic Arabidopsis plants harboring beta-glucuronidase (GUS) driven by the synthetic promoter BrPYP (Figure 5). We first analyzed the expression pattern of BrPYP:GUS in the Arabidopsis seedlings, revealing strong GUS activity in the leaves under the yellow petal promoter (BrPYPY::GUS) compared with the pale-yellow petal promoter (BrPYPPY::GUS) (Figures 5A,B). As expected, GUS expression was also stronger in the petals of Arabidopsis with BrPYPY::GUS (Figures 5C,D), indicating that variation in the promoter of pale-yellow petals results in a decrease in BrPYP expression.
FIGURE 5.

Expression patterns of BrPYP in Arabidopsis. BrPYP promoter-GUS expression of the yellow (A) and pale-yellow (B) type in Arabidopsis leaves. BrPYP promoter-GUS expression of yellow (C) and pale-yellow (D) type in Arabidopsis flowers.
Discussion
Insufficient Plastoglobules in the Pale-Yellow Petals of Flowering Chinese Cabbage Result in the Poor Accumulation of Carotenoids
This study demonstrated that the chromoplasts in the pale-yellow petals of flowering Chinese cabbage only contains far fewer well-developed PGs compared to the yellow petals (Figure 1). Chromoplasts are a type of plastid that participate in carotenoid pigment biosynthesis and storage through multiple catalytic steps (Sun et al., 2018; Hermanns et al., 2020). An increase in the number and size of mature chromoplasts leads to an accumulation of carotenoids, resulting in colorful petals that then act to attract pollinators (Li and Yuan, 2013). The PGs in chromoplasts are much bigger than those in chloroplasts and contain more xanthophyll esters, which are formed during the degradation of carotenoids (Mulisch and Krupinska, 2013). However, irregularly shaped PGs, and disorganized and electron-lucent membrane structures lead to a decrease in carotenoid, as found in carnations, tomato, and B. juncea (Ariizumi et al., 2014; Zhang X. et al., 2018; Iijima et al., 2020). In this study, far fewer well-developed PGs were observed in the pale-yellow petals of flowering Chinese cabbage, suggesting that insufficient PGs reduces accumulation of carotenoid esters.
A Decrease in Carotenoid Esters Directly Affects Petal Color in Flowering Chinese Cabbage
(E/Z)-Phytoene and carotenoid esters, such as lutein caprate, violaxanthin dilaurate, violaxanthin-myristate-laurate, 5,6epoxy-luttein dilaurate, lutein dilaurate and lutein laurate, were highly accumulated or only found in the yellow flowering Chinese cabbage petals (Table 2). Carotenoids can be acylated by different fatty acids, generating a large number of possible carotenoid ester compounds (Schweiggert et al., 2016; Mariutti and Mercadante, 2018). Lutein esters, such as lutein caprate, 5,6epoxy-luttein dilaurate, lutein dilaurate and lutein laurate, are also the well-known predominant carotenoids in petals of marigold (Rodrigues et al., 2019) and tomato (Watanabe et al., 2021). Carotenoid esters are also generated by acylation of fatty acids (Fas), and esterification does not change the carotenoid chromophore; thus, all carotenoid esters present the same UV-VIS spectrum and color of free-carotenoid (Mercadante et al., 2017; Rodrigues et al., 2019). Interestingly, the pale-yellow flowering Chinese cabbage petals observed here had slightly more lutein dimyristate, violaxanthin myristate, violaxanthin-myristate-palmitate, and violaxanthin dimyristate than those of the yellow petals (Table 2). However, the absolute content of carotenoids in the pale-yellow petals was not very high, possibly resulting in the pale-yellow rather than pure white coloring. Based on the carotenoid profile detailed above, these findings suggest that the presence of lutein caprate and violaxanthin dilaurate are mainly responsible for the obvious color differences between yellow and pale-yellow flowering Chinese cabbage petals.
BrPYP Is the Key Factor Causing Pale-Yellow Petals in Flowering Chinese Cabbage
In many plants, yellow petals generally contain high concentrations of carotenoids, consisting largely of esterified lutein (Meng et al., 2019). A decease in carotenoids accumulation and subsequent alteration in petal color from yellow to white or pale yellow can result from carotenoid biosynthesis, degradation or sequestration (Ariizumi et al., 2014; Meng et al., 2019). In Medicago truncatula, disruption of the anthocyanin-related R2R3-MYB protein WP1, which functions as a transcriptional activator directly regulating expression of the carotenoid biosynthetic genes MtLYCe and MtLYCb, was found to cause a reduction in lutein accumulation (Meng et al., 2019). Meanwhile, the tetratricopeptide repeat protein RCP2 was found to regulate carotenoid biosynthesis and chromoplast compartment size in Monkeyflowers, suggesting that loss-of-function mutations in RCP2 damage the entire carotenoid biosynthetic pathway (Stanley et al., 2020). Plastid-localized CCDs are known to play significant roles in carotenoid degradation in many plants (Hai et al., 2012; Gonzalez-Jorge et al., 2013; Xu et al., 2019), which is negatively correlated with yellow pigment intensity, and is also part of the proteome of PGs. In addition to biosynthesis and degradation, carotenoid levels are also modulated by sequestration. Although the mechanisms involved in carotenoid sequestration remain elusive, several lines of evidence suggest the involvement of the phytyl ester synthase (PES) gene (Ariizumi et al., 2014; Yang et al., 2021). Disruption of PES caused a loss of xanthophyll esters, resulting in a decrease in total carotenoid levels and disruption of chromoplast development in the petals of plants (Ariizumi et al., 2014; Yang et al., 2021).
Our study strongly suggests that the BrPYP gene encodes an enzyme protein that controls the pale-yellow petal color in flowering Chinese cabbage (Figure 2). BrPYP is a homolog of Arabidopsis phytyl ester synthases 2 (PES2, At3g26840), a chloroplastic acyltransferase with phytyl ester synthesis and diacylglycerol acyltransferase activities (Lippold et al., 2012). Furthermore, BrPYP mapped to the same gene with BrWF3, which is a candidate gene responsible for white pigmentation in Chinese cabbage (Brassica rapa L. ssp. pekinensis) (Yang et al., 2021). However, different variations were identified between BrPYP and BrWF3. In the coding sequence of BrWF3, a SNP deletion was speculated to be key variant, causing a frameshift mutation and a premature stop codon (Yang et al., 2021). Meanwhile, we identified a functional 1,148 bp deletion in the promoter region of BrPYP that reduces promoter activity and expression level, and then verified by VIGS. As Caixin and Chinese cabbage are different subspecies of B. rapa, this could be different alleles between these two subspecies.
Fatty acid phytyl esters are a type of neutral lipid, the main compound of the PG core (Besagni and Kessler, 2013). Furthermore, characterization of phytyl ester synthases previously confirmed that PES is the main contributor to thylakoid decomposition and PG enlargement during chloroplast senescence and nitrogen deprivation through their role in the biosynthesis of fatty acid phytate and triacylglycerol (Lippold et al., 2012). Moreover, a homolog of PES in tomato was found to be involved in the formation of abundant carotenoid esters in chromoplasts, also affecting petal color (Ariizumi et al., 2014). The reduction in accumulation of esterized carotenoids in the pyp mutant was therefore expected. Taken together, these findings suggest that BrPYP forms more xanthophyll esters during the breakdown of carotenoids.
Conclusion
In conclusion, the results of this study unequivocally demonstrate that normal yellow petals of B. rapa contain more well-developed chromoplasts and PGs, with more esterified carotenoids compared to the pale-yellow mutant. Furthermore, the fine mapping and VIGS findings further suggest that BrPYP is the key factor causing higher accumulation of esterified carotenoids in the yellow flowers. Meanwhile, mutation in the promoter of BrPYP resulted in down-regulated expression, preventing carotenoid esterification and decreasing the efficiency of carotenoid sequestration within the chromoplast. Overall, these findings highlight the molecular mechanism underlying BrPYP-mediated regulation of petal carotenoid pigmentation, and suggest a role of phytyl ester synthase in carotenoid esterification in B. rapa.
Data Availability Statement
Sequencing data has been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA776414.
Author Contributions
PL, SY, and FZ designed the research. PL carried out sequencing and data analyses, and wrote the manuscript. SL performed the GUS and VIGS experiments. DZ provided the sequencing material. TS, XZ, WW, XX, and YZ provided comments relating to the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This work was supported by the National Natural Science Foundation of China (No. 31872094), the Innovation Project of Beijing Vegetable Genebank Resource Breeding and Optimization (KJCX20200113), the Science and Technology Innovation Capacity Project (KJCX20170710), the Scientist Training Program of BAAFS (JKZX201906), the Key Program of Beijing Municipal Science and Technology Committee (Z191100004019010), and the Earmarked Fund for China Agriculture Research System (CARS-23-A-05).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.844140/full#supplementary-material
The protein sequence and predicted structure of BrBYP. (a) Sequence alignment of BrBYP protein between the pale-yellow mutant and yellow type. (b) The predicted protein structure of BrBYP.
The expression patterns of BrPDS1 (a) and BrPDS2 (b) determined by real-time qRT-PCR.
References
- Ariizumi T., Kishimoto S., Kakami R., Maoka T., Hirakawa H., Suzuki Y., et al. (2014). Identification of the carotenoid modifying gene PALE YELLOW PETAL 1 as an essential factor in xanthophyll esterification and yellow flower pigmentation in tomato (Solanum lycopersicum). Plant J. 79 453–465. 10.1111/tpj.12570 [DOI] [PubMed] [Google Scholar]
- Besagni C., Kessler F. (2013). A mechanism implicating plastoglobules in thylakoid disassembly during senescence and nitrogen starvation. Planta 237 463–470. 10.1007/s00425-012-1813-9 [DOI] [PubMed] [Google Scholar]
- Cazzonelli C. I., Pogson B. J. (2010). Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15 266–274. 10.1016/j.tplants.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Chen D. H., Ronald P. C. (1999). A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol. Biol. Rep. 17 53–57. 10.1023/A:1007585532036 [DOI] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Galpaz N., Ronen G., Khalfa Z., Zamir D., Hirschberg J. (2006). A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 18 1947–1960. 10.1105/tpc.105.039966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jorge S., Ha S. H., Magallanes-Lundback M., Gilliland L. U., Zhou A., Lipka A. E., et al. (2013). Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25 4812–4826. 10.1105/tpc.113.119677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai N., Masuda J.-I., Miyajima I., Nguyen T., Mojtahedi N., Hiramatsu M., et al. (2012). Involvement of carotenoid cleavage dioxygenase 4 gene in tepal color change in Lilium brownii var. colchesteri. J. JPN. Soc. Hortic. Sci. 81 366–373. 10.2503/jjshs1.81.366 [DOI] [Google Scholar]
- Han F. Q., Cui H. L., Zhang B., Liu X. P., Yang L. M., Zhuang M., et al. (2019). Map-based cloning and characterization of BoCCD4, a gene responsible for white/yellow petal color in B. oleracea. Bmc Genomics 20:11. 10.1186/s12864-019-5596-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanns A. S., Zhou X., Xu Q., Tadmor Y., Li L. (2020). Carotenoid pigment accumulation in horticultural plants. Hortic. Plant J. 6 343–360. 10.1016/j.hpj.2020.10.002 [DOI] [Google Scholar]
- Iijima L., Kishimoto S., Ohmiya A., Yagi M., Okamoto E., Miyahara T., et al. (2020). Esterified carotenoids are synthesized in petals of carnation (Dianthus caryophyllus) and accumulate in differentiated chromoplasts. Sci. Rep. 10:15256. 10.1038/s41598-020-72078-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K. P., Baz L., Al-Babili S. (2018). From carotenoids to strigolactones. J. Exp. Bot. 69 2189–2204. 10.1093/jxb/erx476 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yuan H. (2013). Chromoplast biogenesis and carotenoid accumulation. Arch. Biochem. Biophys. 539 102–109. 10.1016/j.abb.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Li P., Su T., Yu S., Wang H., Wang W., Yu Y., et al. (2019). Identification and development of a core set of informative genic SNP markers for assaying genetic diversity in Chinese cabbage. Hortic. Environ. Biotechnol. 60, 411–425. 10.1007/s13580-019-00138-4 [DOI] [Google Scholar]
- Li P., Su T., Zhang D., Wang W., Xin X., Yu Y., et al. (2021). Genome-wide analysis of changes in miRNA and target gene expression reveals key roles in heterosis for Chinese cabbage biomass. Hortic. Res. 8:39. 10.1038/s41438-021-00474-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Zhang D., Su T., Wang W., Yu Y., Zhao X., et al. (2020). Genome-wide analysis of mRNA and lncRNA expression and mitochondrial genome sequencing provide insights into the mechanisms underlying a novel cytoplasmic male sterility system. BVRC-CMS96, in Brassica rapa. Theor. Appl. Genet. 133 2157–2170. 10.1007/s00122-020-03587-z [DOI] [PubMed] [Google Scholar]
- Li P., Zhang S., Zhang S., Li F., Zhang H., Liu X., et al. (2015). Carotenoid identification and molecular analysis of carotenoid isomerase-encoding BrCRTISO, the candidate gene for inner leaf orange coloration in Chinese cabbage. Mol. Breed. 35:72. 10.1007/s11032-015-0190-z [DOI] [Google Scholar]
- Lippold F., vom Dorp K., Abraham M., Hölzl G., Wewer V., Yilmaz J. L., et al. (2012). Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant Cell 24 2001–2014. 10.1105/tpc.112.095588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mariutti L. R. B., Mercadante A. Z. (2018). Carotenoid esters analysis and occurrence: what do we know so far? Arch. Biochem. Biophys. 648 36–43. 10.1016/j.abb.2018.04.005 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Wang Z., Wang Y., Wang C., Zhu B., Liu H., et al. (2019). The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula. Plant Cell 31 2751–2767. 10.1105/tpc.19.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercadante A. Z., Rodrigues D. B., Petry F. C., Mariutti L. R. B. (2017). Carotenoid esters in foods-a review and practical directions on analysis and occurrence. Food Res. Int. 99 830–850. 10.1016/j.foodres.2016.12.018 [DOI] [PubMed] [Google Scholar]
- Mulisch M., Krupinska K. (2013). “Ultrastructural analyses of senescence associated dismantling of chloroplasts revisited,” in Plastid Development in Leaves during Growth and Senescence, eds Biswal B., Krupinska K., Biswal U. C. (Dordrecht: Springer Netherlands; ), 307–335. 10.1007/978-94-007-5724-0_14 [DOI] [Google Scholar]
- Nisar N., Li L., Lu S., Khin, Nay C., Barry J. (2015). Carotenoid metabolism in plants. Mol. Plant 8 68–82. 10.1016/j.molp.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Niyogi K. K., Truong T. B. (2013). Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr. Opin. Plant Biol. 16 307–314. 10.1016/j.pbi.2013.03.011 [DOI] [PubMed] [Google Scholar]
- Rahman M. H. (2001). Inheritance of petal colour and its independent segregation from seed colour in Brassica rapa. Plant Breed. 120 197–200. 10.1046/j.1439-0523.2001.00607.x [DOI] [Google Scholar]
- Rodrigues D. B., Mercadante A. Z., Mariutti L. R. B. (2019). Marigold carotenoids: much more than lutein esters. Food Res. Int. 119 653–664. 10.1016/j.foodres.2018.10.043 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M., Boronat A. (2015). Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr. Opin. Biol. 25 17–22. 10.1016/j.pbi.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Sadali N. M., Sowden R. G., Ling Q., Jarvis R. P. (2019). Differentiation of chromoplasts and other plastids in plants. Plant Cell Rep. 38 803–818. 10.1007/s00299-019-02420-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiggert R. M., Vargas E., Conrad J., Hempel J., Gras C. C., Ziegler J. U., et al. (2016). Carotenoids, carotenoid esters, and anthocyanins of yellow-, orange-, and red-peeled cashew apples (Anacardium occidentale L.). Food Chem. 200 274–282. 10.1016/j.foodchem.2016.01.038 [DOI] [PubMed] [Google Scholar]
- Stanley L. E., Ding B., Sun W., Mou F., Hill C., Chen S., et al. (2020). A tetratricopeptide repeat protein regulates carotenoid biosynthesis and chromoplast development in Monkeyflowers (Mimulus). Plant Cell 32 1536–1555. 10.1105/tpc.19.00755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T. H., Yuan H., Cao H. B., Yazdani M., Tadmor Y., Li L. (2018). Carotenoid metabolism in plants: the role of plastids. Mol. Plant 11 58–74. 10.1016/j.molp.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Takagi H., Abe A., Yoshida K., Kosugi S., Natsume S., Mitsuoka C., et al. (2013). QTL-Seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74 174–183. 10.1111/tpj.12105 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Ohmiya A. (2008). Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotech. 19 190–197. 10.1016/j.copbio.2008.02.015 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Yamagaki T., Azuma T., Horikawa M. (2021). Distinguishing between isomeric neoxanthin and violaxanthin esters in yellow flower petals using liquid chromatography/photodiode array atmospheric pressure chemical ionization mass spectrometry and tandem mass spectrometry. Mass Spectrom. 35:e9142. 10.1002/rcm.9142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Luo W., Guo J., Chen H., Akram W., Xie D., et al. (2019). Fine mapping and candidate gene analysis of the yellow petal gene ckpc in Chinese kale (Brassica oleracea L. var. alboglabra Bailey) by whole-genome resequencing. Mol. Breed. 39:96. 10.1007/s11032-019-1011-6 [DOI] [Google Scholar]
- Yang S., Tian X., Wang Z., Wei X., Zhao Y., Su H., et al. (2021). Fine mapping and candidate gene identification of a white flower gene BrWF3 in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 12:646222. 10.3389/fpls.2021.646222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka S., Aida R., Yamamizo C., Shibata M., Ohmiya A. (2012). The carotenoid cleavage dioxygenase 4 (CmCCD4a) gene family encodes a key regulator of petal color mutation in chrysanthemum. Euphytica 184 377–387. 10.1007/s10681-011-0602-z [DOI] [Google Scholar]
- Yu J., Yang X., Wang Q., Gao L., Yang Y., Xiao D., et al. (2018). Efficient virus-induced gene silencing in Brassica rapa using a turnip yellow mosaic virus vector. Biol. Plant 62 826–834. 10.1007/s10535-018-0803-6 [DOI] [Google Scholar]
- Yuan H., Zhang J. X., Nageswaran D., Li L. (2015). Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2:11. 10.1038/hortres.2015.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X., Guo Z., Shi T., Song L., Cheng Y. J. (2019). Arabidopsis AGC protein kinases IREH1 and IRE3 control root skewing. J. Genet. Genomics 46 259–267. 10.1016/j.jgg.2019.02.007 [DOI] [PubMed] [Google Scholar]
- Zhang B., Han F., Cui H., Li X., Ren W., Fang Z., et al. (2019). Insertion of a CACTA-like transposable element disrupts the function of the BoCCD4 gene in yellow-petal Chinese kale. Mol. Breed. 39 130–137. 10.1007/s11032-019-1008-1 [DOI] [Google Scholar]
- Zhang L., Cai X., Wu J., Liu M., Grob S., Cheng F., et al. (2018). Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 5:50. 10.1038/s41438-018-0071-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Chen L., Ma S., Wang R. F., He Q., Tian M., et al. (2020). Fine mapping and candidate gene analysis of the white flower gene Brwf in Chinese cabbage (Brassica rapa L.). Sci. Rep. 10:6080. 10.1038/s41598-020-63165-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li R., Chen L., Niu S., Chen L., Gao J., et al. (2018). Fine-mapping and candidate gene analysis of the Brassica juncea white-flowered mutant Bjpc2 using the whole-genome resequencing. Mol. Genet. Genomics 293 359–370. 10.1007/s00438-017-1390-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The protein sequence and predicted structure of BrBYP. (a) Sequence alignment of BrBYP protein between the pale-yellow mutant and yellow type. (b) The predicted protein structure of BrBYP.
The expression patterns of BrPDS1 (a) and BrPDS2 (b) determined by real-time qRT-PCR.
Data Availability Statement
Sequencing data has been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA776414.