Abstract
Immunocompromised patients may experience prolonged viral shedding after their initial SARS-CoV-2 infection, however, symptomatic relapses after remission currently remain rare. We herein describe a severe COVID-19 relapse case of a kidney transplant recipient (KTR) following rituximab therapy, 3 months after a moderate COVID-19 infection, despite viral clearance after recovery of the first episode. During the clinical relapse, the diagnosis was established on a broncho-alveolar lavage specimen (BAL) by RT-PCR. The infectivity of the BAL sample was confirmed on a cell culture assay. Whole genome sequencing confirmed the presence of an identical stain (Clade 20A). However, it had an acquired G142D mutation and a larger deletion of 3-amino-acids at position 143–145. These mutations located within the N-terminal domain are suggested to play a role in viral entry. The diagnosis of a COVID-19 relapse should be considered in the setting of unexplained persistent fever and/or respiratory symptoms in KTRs (especially for those after rituximab therapy), even in patients with previous negative naso-pharyngeal SARS-CoV-2 PCR.
KEYWORDS: clinical research/practice, infection and infectious agents—viral, infectious disease, kidney transplantation/nephrology, lung disease: infectious, translational research/science
Abbreviations: ATG, antithymocyte globulin; AU, arbitrary units; CNI, calcineurin inhibitors; Ct, cycle threshold; eGFR, estimated glomerular filtration rate; IQR, interquartile range; KT, kidney transplantation; KTR, kidney transplant recipient; MPA, mycophenolic acid; mTOR, mammalian target of rapamycin; RT-PCR, reverse transcriptase-polymerase chain reaction; S Protein, spike protein; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SCr, serum creatinine
1. INTRODUCTION
Immunocompromised patients, including kidney transplant recipients (KTRs), may experience prolonged viral shedding for weeks after initial COVID-19 infection.1 , 2
Despite persistence of a replication competent virus,3 , 4 most immunocompromised patients remain asymptomatic. Moreover, only rare cases of COVID-19 relapses have been reported to date. We herein describe a severe COVID-19 relapse in a KTR without evidence of prolonged naso-pharyngeal (NP) SARS-CoV-2 shedding.
2. CASE REPORT
A 74-year-old man underwent a kidney transplantation in 2014 for diabetic nephropathy. Immunosuppressive treatment consisted of anti-interleukin 2 receptor for induction, and a maintenance regimen included tacrolimus, mycophenolic acid and steroids. The patient experienced immune thrombocytopenic purpura (ITP) in 2019. Rituximab was administered to the patient on September 2020 with two-1000 mg IV infusions separated by 2 weeks because of an ITP relapse.
On November 2020, he was admitted in our unit after a 10-day history of fever associated with confusion, weakness and a positive diagnosis for SARS-CoV-2 by RT-PCR on an NP sample ( Figure 1).
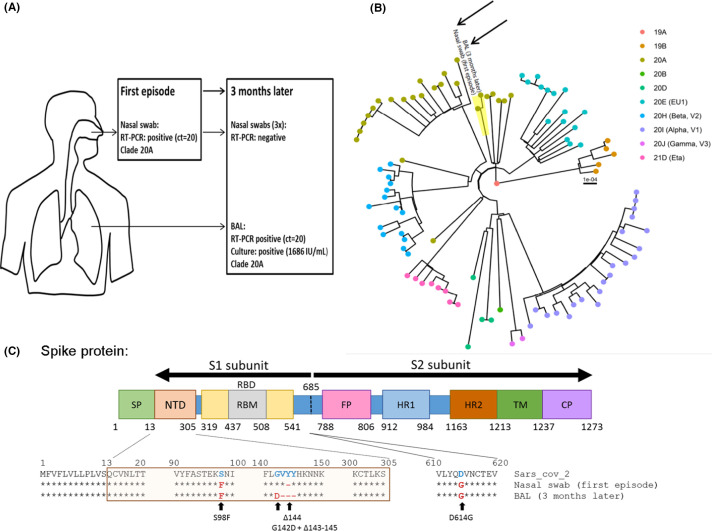
FIGURE 1.
SARS-CoV-2 compartmentalization in a kidney transplant recipient (KTR) with negative nasopharyngeal SARS-CoV-2 RT-PCR after rituximab. (A) Summary of SARS-CoV-2 virological results from the first and second COVID-19 infections including RT-PCR results in a nasal swab and BAL, infectious titer and clade according to Nextclade classification. (B) Phylogenetic tree from the first (nasal swab) and the second (BAL) infections (black arrows) in a representative group of other circulating SARS-CoV-2 strains (94 sequences) from the same geographical area at the time of sampling. Genomes were classified into clades using Nextclade. (C) Evolution of S protein sequences between the first and second infections compared to SARS-CoV-2 reference (SARS-CoV-2 MN908947.3). BAL, broncho-alveolar lavage; SP, signal peptide; NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor binding motif; FP, fusion peptide; HR1/2, heptad repeat 1 and 2; TM, transmembrane domain; CP, cytoplasmic domain
The diagnosis of a moderate COVID-19 was established with no additional diagnosis documented. Mycophenolic acid was suspended, and no further treatment was initiated. Fever progressively decreased and the patient recovered without needing specific therapy nor an oxygen support and was discharged at day 12. The patient was subsequently maintained on steroid and tacrolimus therapy.
Systematic SARS-CoV-2 RT-PCR on NP swabs performed 40 and 50 days after the COVID-19 diagnosis were reported negative. SARS-CoV-2-IgG directed against nucleocapsid antigen remained negative at 2 months after infection.
Three months after the first episode of COVID-19, the patient was readmitted for dyspnea and had a fever at 40°C. Three repeated NP-RT-PCR for SARS-CoV-2 remained negative using 3 different assays (Figure 1 and Supplemental Material 1). A thoracic-CT-scan depicted peri-broncho-vascular condensations with minimal ground glass opacities in the left pulmonary lobe. A broncho-alveolar lavage (BAL) was performed because the patient was immunocompromised and had hypoxemia (oxygen need = 4L/min). SARS-CoV-2 RT-PCR was positive while bacteriological and mycological cultures remained negative.
The multiplex PCR assay (BioFire® Respiratory 2.1 Panel, Biomérieux) which allows detection of 16 viruses and 4 intracellular bacteria, was performed on the BAL, and no other virus was amplified (Supplemental Material 1). In vitro culture of the BAL with SARS-CoV-2 susceptible cells revealed a high titer of infectious virus (1686 IU/ml).
Whole genome sequencing was performed on both the first positive nasal swab and the BAL specimen. Sequencing proved that the two stains were identical (Clade 20A), confirming the clinical relapse. Furthermore, a comparison with local circulating strains at the time of sampling ruled out re-infection (Figure 1 and Table S1). Viral genome sequencing was performed directly on the samples in duplicate. Compared to the first sample, we observed a complete change of sequence population (100%) with appearance of G142D mutation and 143–145 deletion in the N-terminal domain of the spike protein (S protein) (Figure 1). The patient progressively recovered without any specific treatment and was discharged at 10 days after his admission.
Over 7 months of follow-up, the patient remained free of respiratory symptoms and did not experience any other COVID-19 relapses. The NP RT-PCR for SARS-CoV-2 was negative 3 months after the relapse and no new BAL specimen was collected.
3. DISCUSSION
To our knowledge, this is the first description of a SARS-CoV-2 symptomatic relapse in a KTR without evidence of prolonged NP shedding after the first COVID-19 infection and negative NP SARS-CoV-2 PCR at relapse. The diagnosis of relapse was established on the BAL. Comparison of viral sequences suggests a bronchoalveolar persistence and evolution of the same SARS-CoV-2 strain that compartmentalized in the lower respiratory tract and mutated within the patient after the first infection rather than a re-infection. The number of observed mutations was in line with the described mutational rate of the virus. Moreover, the patient was not subsequently exposed to COVID-19 and nor were his close contacts diagnosed with COVID-19, between the two COVID-19 episodes. This finding supports the hypothesis of viral persistence in the lower respiratory tract.
Limited cases of symptomatic relapses have been recently described in immunocompromised hosts with various conditions, such as anti-phospholipid syndrome or chronic lymphocytic leukemia (CLL).5, 6, 7 These patients usually presented a chronic viral shedding of SARS-CoV-2 before relapse and similarly to our patient, some of them received anti-CD20 therapy.
Rituximab therapy has been associated with severe forms of COVID-19 in patients with rheumatic diseases or hematologic malignancies.8, 9, 10, 11 Cases of prolonged SARS-COV-2 shedding or a COVID-19 relapse in patients treated with rituximab have also been reported12 , 13 in patients with hematologic malignancies. The patient failed to produce antibodies against SARS-CoV-2 supporting the conclusion drawn from previous studies that patients on rituximab are unable to produce neutralizing antibodies, resulting in more severe and more prolonged diseases. Therefore, on top of a chronic maintenance immunosuppressive regimen, rituximab may further increase the risk of long-term viral shedding and subsequent relapse in KTRs.
In addition, the discrepancy of RT-PCR results between the nasal swab (which is considered a highly sensitive technique for the diagnosis of COVID-19 in patients with acute pneumonia) and the BAL suggest a compartmentalized viral replication in immunocompromised patients and was previously demonstrated by Rueca et al.14 Moreover, no evidence of transmission from the patient to his close contacts was identified despite a well-established infectivity of the BAL specimen.
The G142D mutation and the 143–145 deletions observed in the BAL are located within the N-terminal domain of the viral S protein, which is suggested to play the primary role in viral entry as shown by the high neutralization potency of certain monoclonal antibodies targeting the NTD.15 , 16
Furthermore, the 143–145 deletions have been found in the Alpha and Omicron (B.1.1.529) variants and the G142D have been found in the Omicron and Delta variants (B.1.617.2) which were two variants that emerged later in the pandemic. This may indicate an epidemiological advantage of these mutations. It is also worth noting that the Alpha variant emerged from an immunocompromised patient17 and altogether, this may suggest that long-term shedding in immunocompromised hosts may favor the evolution of the virus and points out the potential need for close monitoring of these patients.
Notably, monoclonal antibodies were not available at our center when the patient experienced the first COVID-19 episode. Few cases of recovery after monoclonal antibodies18 , 19 or convalescent plasma have been reported.20 However, further studies are needed to assess the efficacy of these treatments to prevent COVID-19 relapses.
In conclusion, the diagnosis of COVID-19 relapse should be considered in the setting of unexplained persistent fever with respiratory symptoms and/or other characteristic symptoms of COVID-19 infection in KTRs. This is especially necessary for patients following rituximab therapy, including in cases in whom SARS-Cov-2 is cleared from the naso-pharyngeal compartment. Because of a possible viral broncho-alveolar compartmentalization, BAL should be systematically performed, and patients should be isolated until diagnosis.
ACKNOWLEDGMENTS
The authors would like to thank AcaciaTools for their proofreading services.
DISCLOSURE
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
DATA AVAILABILITY STATEMENT
Dataset can be made available by contacting the corresponding author via: Nathalie.chavarot@aphp.fr.
Footnotes
Antoine Morel and Sandrine Imbeaud contributed equally to this article.
Nathalie Chavarot and David Veyer contributed equally to this article.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
Supplementary Material
REFERENCES
- 1.Taramasso L, Sepulcri C, Mikulska M, et al. Duration of isolation and precautions in immunocompromised patients with COVID-19. J Hosp Infect. 2021;111:202–204. doi: 10.1016/j.jhin.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benotmane I, Risch S, Doderer-Lang C, Caillard S, Fafi-Kremer S. Long-term shedding of viable SARS-CoV-2 in kidney transplant recipients with COVID-19. Am J Transplant. 2021;21(8):2871–2875. doi: 10.1111/ajt.16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beran A, Zink E, Mhanna M, et al. Transmissibility and viral replication of SARS-COV-2 in immunocompromised patients. J Med Virol. 2021;93(7):4156–4160. doi: 10.1002/jmv.26970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tarhini H, Recoing A, Bridier-nahmias A, et al. Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectiousness among three immunocompromised patients: from prolonged viral shedding to SARS-CoV-2 superinfection. J Infect Dis. 2021;223(9):1522–1527. doi: 10.1093/infdis/jiab075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS-CoV-2 in an Immunocompromised Host. N Engl J Med. 2020;383(23):2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avanzato VA, Matson MJ, Seifert SN, et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183(7) doi: 10.1016/j.cell.2020.10.049. 1901-1912.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reuken PA, Stallmach A, Pletz MW, et al. Severe clinical relapse in an immunocompromised host with persistent SARS-CoV-2 infection. Leukemia. 2021;35(3):920–923. doi: 10.1038/s41375-021-01175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strangfeld A, Schäfer M, Gianfrancesco MA, et al. Factors associated with COVID-19-related death in people with rheumatic diseases: results from the COVID-19 global rheumatology alliance physician-reported registry. Ann Rheum Dis. 2021;80(7):930–942. doi: 10.1136/annrheumdis-2020-219498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.FAI2R/SFR/SNFMI/SOFREMIP/CRI/IMIDIATE Consortium and Contributors. Severity of COVID-19 and survival in patients with rheumatic and inflammatory diseases: data from the French RMD COVID-19 cohort of 694 patients. Ann Rheum Dis. 2020. 10.1136/annrheumdis-2020-218310 [DOI] [PMC free article] [PubMed]
- 10.Avouac J, Drumez E, Hachulla E, et al. COVID-19 outcomes in patients with inflammatory rheumatic and musculoskeletal diseases treated with rituximab: a cohort study. Lancet Rheumatol. 2021;3(6):e419–e426. doi: 10.1016/S2665-9913(21)00059-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duléry R, Lamure S, Delord M, et al. Prolonged in-hospital stay and higher mortality after Covid-19 among patients with non-Hodgkin lymphoma treated with B-cell depleting immunotherapy. Am J Hematol. 2021;96(8):934–944. doi: 10.1002/ajh.26209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monrad I, Sahlertz SR, Nielsen SSF, et al. Persistent severe acute respiratory syndrome Coronavirus 2 infection in immunocompromised host displaying treatment induced viral evolution. Open Forum Infect Dis. 2021;8(7):ofab295. doi: 10.1093/ofid/ofab295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderón-Parra J, Múñez-Rubio E, Fernández-Cruz A, et al. Incidence, clinical presentation, relapses and outcome of SARS-CoV-2 infection in patients treated with anti-CD20 monoclonal antibodies. Clin Infect Dis. 2021:ciab700. doi: 10.1093/cid/ciab700. [DOI] [PubMed] [Google Scholar]
- 14.Rueca M, Bartolini B, Gruber CEM, et al. Compartmentalized replication of SARS-Cov-2 in upper vs. lower respiratory tract assessed by whole genome quasispecies analysis. Microorganisms. 2020;8(9):1302. doi: 10.3390/microorganisms8091302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Planas D, Bruel T, Grzelak L, et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 17.Kemp SA, Collier DA, Datir RP, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592(7853):277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabascall CX, Lou BX, Navetta-Modrov B, Hahn SS. Effective use of monoclonal antibodies for treatment of persistent COVID-19 infection in a patient on rituximab. BMJ Case Rep. 2021;14(8) doi: 10.1136/bcr-2021-243469. e243469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailly B, Péré H, Veyer D, et al. Persistent COVID-19 in an immunocompromised host treated by SARS-CoV-2-specific monoclonal antibodies. Clin Infect Dis. 2021:ciab868. doi: 10.1093/cid/ciab868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark E, Guilpain P, Filip IL, et al. Convalescent plasma for persisting COVID-19 following therapeutic lymphocyte depletion: a report of rapid recovery. Br J Haematol. 2020;190(3):e154–e156. doi: 10.1111/bjh.16981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Dataset can be made available by contacting the corresponding author via: Nathalie.chavarot@aphp.fr.