Abstract
A multiplex PCR was developed for the rapid detection of genes encoding Shiga toxins 1 and 2 (stx1 and stx2), intimin (eaeA), and enterohemolysin A (hlyA) in 444 fecal samples derived from healthy and clinically affected cattle, sheep, pigs, and goats. The method involved non-solvent-based extraction of nucleic acid from an aliquot of an overnight culture of feces in EC (modified) broth. The detection limit of the assay for both fecal samples and pure cultures was between 18 and 37 genome equivalents. stx1 and hlyA were the most commonly encountered virulence factors.
Enterohemorrhagic Escherichia coli (EHEC) is the most important recently emerged group of food-borne pathogens. It can cause severe gastrointestinal disease, including fatal infections, and is being detected more frequently worldwide. More investigations regarding the laboratory diagnosis of these organisms have been carried out in recent years than with any other group of food-borne pathogens, yet this group remains the most difficult to detect. EHEC strains not only produce potent cytotoxins (verotoxins) but have also acquired the ability to adhere to the intestinal mucosa in an intimate fashion (4, 15, 21). They are also defined by the presence of specific virulence factors; all strains produce hemolysin (most producing an EHEC-specific plasmid-encoded hemolysin, encoded by hlyA) (26) and at least one Shiga-like toxin (encoded by stx1 or stx2) (21), and many produce intimin, a 97-kDa attachment-and-effacement protein (encoded by eaeA) (19). Although E. coli O157:H− is currently the most common EHEC strain in many regions of the world (3), serotypes O5, O26, O91, O111, and O113 are also recognized as a serious threat to public health and have been recovered from infected patients (4). Strains of E. coli O157:H− are comparatively easy to isolate because of unique biochemical characteristics; however, the other serotypes can be differentiated from commensal E. coli only by specialized techniques, such as those described in this report (reference 11 and references therein).
Paton et al. (22) described a PCR for the amplification of stx1 and stx2 sequences in primary human fecal cultures. However, oligonucleotide hybridization probes were required to distinguish between the two toxins in a separate test. The presence of stx-positive fecal cultures in asymptomatic individuals (9, 22) suggested that other virulence factors besides stx are required to cause serious disease in humans. Fratamico et al. (12) described a multiplex PCR capable of detecting stx1, stx2, eaeA, and EHEC hlyA sequences. However, this PCR was not tested with fecal samples; primers for each target gene sequence showed differential sensitivities, and stx primers were unable to distinguish stx1 from stx2 by agarose gel electrophoresis. Ideally, PCR-based detection methods should be rapid and sensitive without requiring extensive sample preparation. More recently Paton and Paton (23) developed a multiplex PCR utilizing four PCR primer pairs for the detection of stx1, stx2, eaeA, and EHEC hlyA in human feces and foodstuffs. However, the relatively lengthy PCR template preparation protocol used was considered inappropriate for testing large numbers of samples.
Ruminants, particularly cattle (5, 7, 30) and sheep (7, 17), are natural reservoirs of EHEC, although other domestic animals, including goats, pigs, poultry, cats and dogs, can also harbor these bacteria (1, 5, 6). However, methodologies which provide comparatively rapid (24-h) and sensitive detection of stx1, stx2, eaeA, and hlyA gene sequences in animal feces have not been reported. The aim of this study was to develop and evaluate a multiplex PCR for this purpose.
EHEC reference strains O111:H8, O157:H7, O128:H2, O91:H−, O113:H21, and O5:H− were provided by the Victorian Infectious Diseases Reference Laboratory (Fairfield, Australia). A positive EHEC control, E. coli O111:H−, was provided by the Victorian Institute of Animal Science (Fairfield, Australia), and the negative control strain E. coli JM109 was provided by Mark Walker (Wollongong University, Wollongong, Australia). Two hundred thirty-five diagnostic fecal samples from sheep, cattle, and pigs submitted to the Regional Veterinary Laboratory, Elizabeth Macarthur Agricultural Institute (Menangle, New South Wales, Australia), for microbiological analysis and a further nine bovine fecal specimens from cattle from a dairy farm with an EHEC history were used for this study. Two hundred fecal samples were also collected from apparently healthy animals from four sheep flocks and four bovine herds (25 samples from each herd or flock).
E. coli isolates were each cultured on EC (modified) agar, which was prepared by adding 1.5% agar to EC (modified) broth (CM853; Oxoid, Basingstoke, United Kingdom) and incubated at 37°C for 18 to 20 h prior to nucleic acid extraction. Fecal broth cultures were prepared by inoculating 50 mg of feces into 10 ml of EC (modified) broth and incubated at 37°C for 18 to 20 h. For DNA sample preparation either a 15-μl aliquot of the overnight fecal culture or a single colony off EC agar was mixed in 1 ml of sterile water in a 1.7-ml microcentrifuge tube. Bacteria were pelleted by centrifugation at 11,000 rpm for 1 min in a Biofuge pico (Heraeus, Hanau, Germany). The supernatant was subsequently discarded, 200 μl of InstaGene matrix (Bio-Rad) was added to the pellet, and the mixture was incubated at 56°C for 30 min. After incubation, the mixture was vortexed for 10 s and then incubated at 100°C for 8 min, followed by vortexing and centrifugation at 11,000 rpm for 1 min prior to removal of the nucleic acid template for PCR.
Multiplex PCR for detection of stx1, stx2, eaeA, and EHEC hlyA gene sequences was performed with a PC-960 thermal cycler (Corbett Research). Oligonucleotide primers were manufactured commercially (GIBCO-BRL). Primers and the predicted lengths of PCR amplification products are listed in Table 1. These primers were chosen because they amplify conserved regions of the target genes and allow single-step identification of amplified DNA fragments by agarose gel electrophoresis. Each primer pair had been determined to be specific for E. coli and had been shown not to amplify products detectable by agarose gel electrophoresis using DNA templates derived from a range of gram-positive and gram-negative bacterial species from food and animal sources (12–14).
TABLE 1.
Primer sequences and predicted lengths of PCR amplification products
| Primer | Direc-tion | Primer sequence (5′-3′) | Fragment size (bases) | Refer-ence |
|---|---|---|---|---|
| EHEC hly | Forward | ACGATGTGGTTTATTCTGGA | 165 | 12 |
| Reverse | CTTCACGTGACCATACATAT | |||
| stx1 | Forward | ACACTGGATGATCTCAGTGG | 614 | 13 |
| Reverse | CTGAATCCCCCTCCATTATG | |||
| stx2 | Forward | CCATGACAACGGACAGCAGTT | 779 | 13 |
| Reverse | CCTGTCAACTGAGCAGCACTTTG | |||
| eaeA | Forward | GTGGCGAATACTGGCGAGACT | 890 | 14 |
| Reverse | CCCCATTCTTTTTCACCGTCG |
PCR assays were carried out in a 50-μl volume containing 2 μl of nucleic acid template prepared from fecal cultures (approximately 60 ng of DNA) or 1 μl of nucleic acid template prepared by using reference EHEC isolates (approximately 30 ng of DNA), 10 mM Tris-HCl (pH 8.4), 10 mM KCl, 3 mM MgCl2; 2 mM concentrations of each primer, 0.2 mM concentrations of each 2′-deoxynucleoside 5′-triphosphate, and 4 U of AmpliTaq DNA polymerase (Perkin-Elmer). Temperature conditions consisted of an initial 95°C denaturation step for 3 min followed by 35 cycles of 95°C for 20 s, 58°C for 40 s, and 72°C for 90 s. The final cycle was followed by a 72°C incubation for 5 min. Amplified DNA fragments were resolved by gel electrophoresis (25) using 2% (wt/vol) agarose. Gels were stained with 0.5 μg of ethidium bromide per ml, visualized with UV illumination, and imaged with a GelDoc 1000 fluorescent imaging system (Bio-Rad).
To determine the sensitivity of the multiplex PCR assay, the number of bacterial cells per milliliter in a stock suspension of E. coli O111:H− solution was determined with a hemocytometer. A 10-fold dilution series of the stock suspension was prepared, and from this the number of CFU was determined. Nucleic acid was extracted from a representative volume of each dilution of the titration by the InstaGene methodology. Preparation of bacterial template for PCR was carried out as described for fecal culture PCR; however, an initial 10-μl aliquot of each dilution series was washed in 1 ml of sterile water. Assay sensitivity was also determined by using the dilution series to inoculate a freshly cultured PCR-negative overnight fecal broth. Aliquots of the seeded enrichment broth were immediately prepared for multiplex PCR to determine what impact a complex coliform mixture might have on assay sensitivity. No further enrichment took place.
DNA probes for colony hybridizations (eaeA, stx1, stx2, and EHEC hlyA) were directly labelled with digoxigenin-11-dUTP (DIG) using the PCR as described by the manufacturer (Boehringer GmbH, Mannheim, Germany). PCR amplification was carried out as described in the previous section by using E. coli O111:H− as the template with the deoxynucleoside triphosphate mixture containing 10% DIG.
Overnight fecal cultures (determined to contain any combination of the four virulence factors by multiplex PCR) were diluted (to ensure single colonies), plated onto EC (modified) agar, and grown at 37°C for 18 to 20 h. Colonies (96) were picked and patched onto fresh EC (modified) agar plates prior to incubation at 37°C for 18 to 20 h. Colony lifting was subsequently used to transfer the bacteria onto nylon Hybond H+ membrane (Amersham, Little Chalfont, United Kingdom). Colony hybridization was carried out with a DIG chemiluminescence detection kit (Hyb; Boehringer Mannheim) per the manufacturer’s instructions. Hybridization was carried out in a minihybridization oven (Hybaid) at 58°C by using DIG Easy Hyb (Boehringer GmbH, Mannheim, Germany) with the denatured DIG-labelled DNA probe. After incubation, membranes were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) for 15 min at room temperature followed by two washes in 0.5× SSC–0.1% SDS at 68°C. All washes were performed in a minihybridization oven. Bound DIG-labelled probes were detected with CSPD (Boehringer GmbH, Mannheim, Germany) by following the manufacturer’s instructions. Membranes were subsequently exposed to X-ray film (Kodak) at room temperature for 10 to 60 min prior to development.
PCR products representing each of the four target EHEC virulence factors were amplified with E. coli O111:H− DNA template as a positive control (Fig. 1, lane 1). No amplification products were present in either the negative control (a fecal culture previously determined to lack sequences for any of the four virulence determinants by multiplex PCR) or a water control (no nucleic acid) after PCR (results not shown).
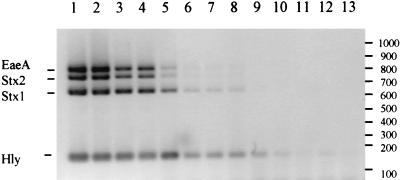
FIG. 1.
Sensitivity of multiplex PCR in detecting EHEC virulence factors using serial dilutions of E. coli O111:H−. DNA markers are indicated on the right (numbers are molecular weights, in base pairs). Lanes 1 through 13 contain 6,250, 2,500, 630, 372, 186, 93, 46, 37, 18, 9, 3, 1.5, and <1 genome equivalents, respectively.
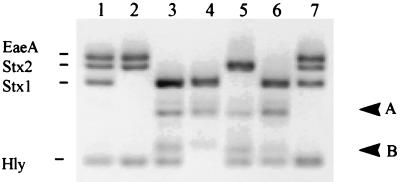
DNA extracted from strains representative of each of six EHEC serotypes was the template for a range of PCR amplification products (Fig. 2). stx gene sequences were detected in all six reference EHEC serotypes, and one strain contained both stx1 and stx2. Three strains contained only stx1, and the remaining two strains contained stx2. All reference strains except O91:H− contained the EHEC hlyA gene. The genotypic results obtained by the multiplex EHEC PCR assay were in agreement with phenotypic data provided by the Victorian Infectious Diseases Reference Laboratory for 17 of the known 18 factors. stx2, which had previously been detected in E. coli O91:H− by the Victorian Infectious Diseases Reference Laboratory, was not detected by multiplex PCR.
FIG. 2.
Multiplex PCR analysis of EHEC reference strains. Arrows A and B refer to nonspecific PCR bands (see the text). Lanes: 1, E. coli O111:H8; 2, E. coli O157:H7; 3, E. coli O128:H2; 4, E. coli O91:H−; 5, E. coli O113:H21; 6, E. coli O5:H−; 7, E. coli O111:H− positive control.
A stock suspension containing 6.3 × 107 E. coli O111:H− organisms per ml was determined. The sensitivity of the EHEC multiplex PCR assay was estimated to be between 37 and 18 genome equivalents. Amplified DNA bands were progressively lost in descending order from the largest (890-bp) eaeA product, which was lost between 37 and 18 genome equivalents, to the 165-bp EHEC hlyA product, which is detectable at less than 3 genome equivalents (Fig. 1). A similar level of sensitivity was achieved in fecal cultures which had previously been determined to be negative for the four virulence factors by multiplex PCR but which were seeded with titrations of stock suspension and tested without enrichment. All EHEC virulence factors were detectable between 37 and 18 genome equivalents (results not shown).
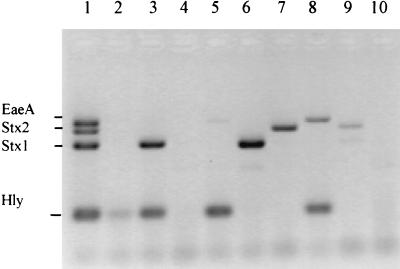
To further validate the utility of the multiplex PCR assay, 235 fecal samples were tested. Initial studies with 180 of these fecal samples used a multiplex PCR where primers for only three virulence factors, stx1, stx2, and eaeA, were tested (Table 2). stx1 was the most commonly encountered factor, being detected in 19.4% (35 of 180) of fecal samples, while stx2 and eaeA were each detected in 6.7% (12 of 180) of samples. Multiple EHEC factors were amplified in individual samples: stx1 plus stx2 5% (9 of 180), stx2 plus eaeA, 1.7% (3 of 180); and stx1 plus stx2 plus eaeA, 2.2% (4 of 180). Of particular interest is the prevalence of stx1 in the ovine samples (56.5%) compared to the almost equal distribution of stx1 (17.3%) and stx2 (21.8%) observed for cattle samples. Table 3 outlines the results for a further 64 fecal samples tested with the multiplex PCR, which included primers for the EHEC hlyA sequence. The EHEC hlyA factor was detected in 35.9% of all samples tested. The distribution of the stx1, stx2, and eaeA factors within these 64 samples was essentially the same (results not shown) as that within the 180 samples described in Table 2. Consequently, Table 3 displays EHEC factor combinations which include hlyA. A further 200 fecal samples from healthy cattle (four herds) and sheep (four flocks) were tested with the four-factor multiplex PCR assay (Table 4). Ovine samples from the four flocks also displayed a high prevalence of stx1 (54%), whereas bovine samples displayed a comparatively low prevalence of stx1 (17%) and stx2 (7%) (Table 4). Figure 3 shows amplification products of the multiplex PCR using DNA templates recovered from nine bovine fecal samples from a dairy farm with a history of bovine EHEC excretion. In summary, hlyA was detected in four samples, stx genes were detected in four samples, and eaeA was detected in two samples.
TABLE 2.
Multiplex PCR data derived from 180 livestock fecal samples
| Sample type | No. of samples positive for gene(s)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| None | stx1 | stx2 | eaeA | stx1 + stx2 | stx1 + eaeA | stx2 + eaeA | stx1 + stx2 + eaeA | Total | |
| Ovine | 14 | 25 | 1 | 4 | 1 | 1 | 46 | ||
| Bovine | 75 | 7 | 11 | 4 | 9 | 1 | 3 | 110 | |
| Caprine | 6 | 3 | 1 | 10 | |||||
| Porcine | 10 | 4 | 14 | ||||||
| Total | 180 | ||||||||
TABLE 3.
Multiplex PCR data derived from 64 livestock fecal samples
| Sample type | No. of samples positive fora:
|
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No hly | hly | hly + stx1 | hly + stx2 | hly + stx1 + stx2 | hly + eaeA | hly + stx2 + eaeA | hly + stx1 + stx2 + eaeA | ||
| Bovine | 36 | 8 | 2 | 1 | 2 | 1 | 1 | 51 | |
| Otherb | 5 | 3 | 2 | 1 | 1 | 1 | 13 | ||
| 64 | |||||||||
No samples were positive for hly plus stx1 plus eaeA.
Includes porcine, ovine, and caprine samples.
TABLE 4.
Multiplex PCR data derived from 200 fecal samples from four sheep flocks and four cattle herds
| Flock or herd | No. of samples positive for gene(s)a
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| None | stx1 | eaeA | stx1 + stx2 | stx1 + eaeA | hly | hly + stx1 | hly + stx2 | hly + stx1 + stx2 | hly + eaeA | hly + stx1 + eaeA | hly + stx1 + stx2 + eaeA | Total | |
| Flock 1 | 9 | 8 | 1 | 4 | 1 | 1 | 1 | 25 | |||||
| Flock 2 | 9 | 5 | 1 | 1 | 7 | 2 | 25 | ||||||
| Flock 3 | 6 | 5 | 5 | 3 | 2 | 1 | 1 | 2 | 25 | ||||
| Flock 4 | 10 | 13 | 2 | 25 | |||||||||
| Herd 1 | 12 | 1 | 9 | 1 | 2 | 25 | |||||||
| Herd 2 | 19 | 1 | 5 | 25 | |||||||||
| Herd 3 | 14 | 1 | 3 | 3 | 2 | 2 | 25 | ||||||
| Herd 4 | 2 | 1 | 14 | 5 | 25 | ||||||||
| 200 | |||||||||||||
No samples were positive for stx1 plus stx2 plus eaeA.
FIG. 3.
Multiplex PCR analysis of bovine fecal samples. Lanes: 1, O111:H− positive control; 2 through 10, fecal samples collected on a farm previously identified as containing EHEC-positive animals.
DNA hybridization was carried out on the overnight fecal culture which possessed stx1 and EHEC hlyA (Fig. 3, lane 3). Four colonies from a total of 90 were identified as positive for both stx1 and EHEC hlyA (data not shown); none of the four virulence factors were observed among the other 86 colonies from this sample.
The multiplex PCR described in this study is an effective means of detecting EHEC virulence factors in bovine, ovine, porcine, and caprine feces. Although a number of different primer sequences which amplify EHEC virulence factors have been described, including those used in this study (12–14), the combination of primers reported here was chosen to generate a high level of sensitivity (37 to 18 genome equivalents) and to facilitate clear resolution of each of the four amplification products by size on a single 2% agarose gel. A multiplex PCR which amplified the four known EHEC virulence factors in human feces was recently described; however, the sensitivity levels for each primer pair were not reported, and preparation of PCR template required the use of lysozyme, proteinase K, boiling, and ethanol precipitation, a procedure requiring at least 3 to 4 h and significant cost (23). Sensitivity in an EHEC assay is essential, as the infectious dose for humans may be as low as 1 to 10 CFU. A combination of overnight culture in EC (modified) broth (which dilutes PCR inhibitors and increases the number of target bacteria) and the use of a comparatively rapid and inexpensive template preparation methodology not reliant on PCR-inhibiting solvent methodologies (8) facilitated the development of a multiplex PCR that is sensitive (between 37 and 18 genome equivalents), rapid, and amenable to high throughput sampling compared with similar PCR-based detection assays.
Multiplex PCR stx1 and eaeA profiles for the six reference E. coli strains were in agreement for the stx1 and eaeA marker results provided by the Victorian Infectious Diseases Reference Laboratory. The stx2 gene from E. coli O91:H− was the only factor not detected. This is not surprising, as Shiga-like toxins are readily lost during subculture (16); however, it should be noted that stx1 was detected in this particular strain. EHEC hlyA was detected in all strains except E. coli O91:H−; again, this result is not surprising, as the EHEC plasmid which carries the hlyA gene was not previously detected in this serotype (27). The gene encoding the attachment and effacement factor, eaeA, was detected only in E. coli O157 and O111; these strains are the most common EHEC strains cultured from patients with food-borne illness in Australia (11). With single-primer PCR, nonspecific amplification (labelled A and B in Fig. 2) appeared to be associated with amplification with eaeA and stx2 primers sets respectively. By altering annealing conditions in the multiplex PCR, these amplification products could be avoided; however, the sensitivity of detection of EHEC virulence factors was reduced (results not shown).
The multiplex PCR identified 26 of 46 (56.5%) ovine fecal samples containing stx1 gene sequences, compared to a comparatively equal distribution of stx1 (7 of 110 [6.4%]), stx2 (11 of 110 [10%]) and stx1 plus stx2 (9 of 110 [8.2%]) sequences in cattle feces. Similar finding were demonstrated in the samples collected from healthy sheep and cattle. A recent study (28) described the presence of stx sequences in feces of cattle, sheep and pigs in Queensland, Australia. This study identified 19 of 105 (18%), 70 of 101 (69%), and 27 of 129 (21%) bovine, ovine, and porcine fecal samples, respectively, to be positive for stx sequences, similar to the results presented in this study. Kudva et al. (18) reported that approximately 75% of isolates from a single sheep flock were stx1, stx2, and eaeA positive by colony hybridization while a further 22.9% of isolates were positive for only stx1 and eaeA. This high prevalence of the eaeA and stx2 was not observed among fecal samples collected from 46 sheep in our study, with only 2.2 and 8.7% of fecal samples being positive for stx2 and eaeA sequences, respectively. Beutin et al. 1997 (7) reported that the vast majority of bovine isolates were positive for stx2, all but one isolate were negative for eaeA, and only one isolate was positive for stx1. While these results vary considerably from our data, it has been shown that patterns of shedding of Shiga-toxin-producing E. coli are affected by diet, age, stress, and seasonal variation (17, 18).
Although PCR can detect EHEC virulence factors with a relatively high degree of sensitivity amongst fecal E. coli strains, the detection of a gene does not indicate whether that factor is being expressed. Kudva et al. (18) showed that 9 of 11 stx-positive isolates were capable of expressing Shiga-like toxin(s) in Vero cultures, indicating the presence of the appropriate gene regulatory sequences. While combinations of the four EHEC virulence factors typically are present in most EHEC strains which have been recovered from symptomatic patients, a small proportion of stx-positive E. coli isolates do not possess eaeA or EHEC hlyA and are still able to cause hemolytic-uremic syndrome (20). Furthermore, it has been suggested that stx-positive E. coli strains which lack eaeA sequences may be less virulent for humans than eaeA-positive EHEC isolates, although it is not known if all Shiga toxin-producing strains are equally pathogenic in this regard (6). These observations suggest that other factors may enable a small proportion of stx-positive E. coli isolates to induce symptoms associated with typical EHEC isolates. Several studies have demonstrated that EHEC virulence factors are mobile within bacterial populations (2, 24, 31), and the assortment of genes between E. coli organisms may lead to pathogenic strains. A multiplex PCR approach is advantageous in rapidly detecting EHEC pathogenicity factors in the natural environment while the concomitant use of colony hybridization enables the identification of specific EHEC isolates. Future studies will focus on applying the multiplex PCR in conjunction with colony hybridization to livestock fecal samples collected from different geographic locations within Australia and among herds and flocks utilizing different farm management practices.
Acknowledgments
This work was supported by funds from the Australian Meat Research Corporation.
REFERENCES
- 1.Acheson D W, Keusch G T. Which Shiga toxin-producing types of Escherichia coli are important? ASM News. 1996;62:302–306. [Google Scholar]
- 2.Agin T S, Cantey J R, Boedeker E C, Wolf M K. Characterization of the eaeA gene from rabbit enteropathogenic Escherichia coli strain RDEC-1 and comparison to other eaeA genes from bacteria that cause attaching-effacing lesions. FEMS Microbiol Lett. 1996;144:249–258. doi: 10.1111/j.1574-6968.1996.tb08538.x. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong G L, Hollingsworth J, Morris J G. Emerging food borne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol Rev. 1996;18:29–50. doi: 10.1093/oxfordjournals.epirev.a017914. [DOI] [PubMed] [Google Scholar]
- 4.Bettelheim K A. Enterohaemorrhagic Escherichia coli—a review. Int Food Hyg. 1996;7:5–9. [Google Scholar]
- 5.Beutin L, Geier D, Steinruck H, Zimmermann S, Scheutz F. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Clin Microbiol. 1993;31:2483–2488. doi: 10.1128/jcm.31.9.2483-2488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin L, Geier D, Zimmermann S, Karch H. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J Clin Microbiol. 1995;33:631–635. doi: 10.1128/jcm.33.3.631-635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutin L, Geier D, Zimmermann S, Aleksic S, Gillespie H A, Whittam T S. Epidemiological relatedness and clonal types of natural populations of Escherichia coli strains producing shiga toxins in separate populations of cattle and sheep. Appl Environ Microbiol. 1997;63:2175–2180. doi: 10.1128/aem.63.6.2175-2180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bretagne S, Guillou J P, Morand M, Houin R. Detection of Echinococcus multilocularis DNA in fox faeces using DNA amplification. Parasitology. 1993;106:193–199. doi: 10.1017/s0031182000074990. [DOI] [PubMed] [Google Scholar]
- 9.Brian M J, Frosolono M, Murray B E, Miranda A, Lopez E L, Gomez H F, Cleary T G. Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic uremic syndrome. J Clin Microbiol. 1992;30:1801–1806. doi: 10.1128/jcm.30.7.1801-1806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cubbon M D, Coia J E, Hanson M F, Thomson-Carter F M. A comparison of immunomagnetic separation, direct culture and polymerase chain reaction for the detection of verotoxin-producing Escherichia coli O157 in human feces. J Med Microbiol. 1996;44:219–222. doi: 10.1099/00222615-44-3-219. [DOI] [PubMed] [Google Scholar]
- 11.Desmarchelier P M, Grau F H. Escherichia coli. In: Hocking A D, Arnold G, Jenson I, Newton K, Sutherland P, editors. Foodborne microorganisms of public health significance. 5th ed. Sydney, Australia: Australian Institute of Food Science and Technology; 1997. pp. 231–264. [Google Scholar]
- 12.Fratamico P M, Sackitey S K, Wiedmann M, Deng M Y. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–2191. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gannon V P J, King R K, Kim J Y, Golsteyn Thomas E J. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3809–3815. doi: 10.1128/aem.58.12.3809-3815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gannon V P J, Souza S D, Graham T, King R K, Rahn K, Read S. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J Clin Microbiol. 1997;35:656–662. doi: 10.1128/jcm.35.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman S L. Shiga-like toxins in hemolytic uremic syndrome and thrombotic thrombocytopenic purpura. JAMA. 1993;306:398–406. doi: 10.1097/00000441-199312000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Karch H, Meyer T, Russmann H, Heesemann J. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect Immun. 1992;60:3464–3467. doi: 10.1128/iai.60.8.3464-3467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kudva I T, Hatfield P G, Hovde C J. Escherichia coli O157:H7 in microbial flora of sheep. J Clin Microbiol. 1996;34:431–433. doi: 10.1128/jcm.34.2.431-433.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudva I T, Hatfield P G, Hovde C J. Characterization of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli serotypes isolated from sheep. J Clin Microbiol. 1997;35:892–899. doi: 10.1128/jcm.35.4.892-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louie M, de Azavedo J C S, Handelsman M Y C, Clark C G, Ally B, Dytoc M, Sherman P, Brunton J. Expression and characterization of the eaeA gene product of Escherichia coli serotype O157:H7. Infect Immun. 1993;61:4085–4092. doi: 10.1128/iai.61.10.4085-4092.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louie M, de Azavedo J C S, Clarke R, Borczyk A, Lior H, Richter M, Brunton J. Sequence heterogeneity of the eae gene and detection of verotoxin-producing Escherichia coli using serotype-specific primers. Epidemiol Infect. 1994;112:449–461. doi: 10.1017/s0950268800051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien A D, Holmes R K. Shiga and Shiga-like toxins. Microbiol Rev. 1987;51:206–220. doi: 10.1128/mr.51.2.206-220.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton A W, Paton J C, Goldwater P N, Manning P A. Direct detection of Escherichia coli Shiga-like toxin genes in primary fecal cultures by polymerase chain reaction. J Clin Microbiol. 1993;31:3063–3067. doi: 10.1128/jcm.31.11.3063-3067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton A W, Paton J C. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Schmidt H, Plaschke B, Franke S, Russman H, Schwarzkopf A, Heesemann J, Karch H. Differentiation in virulence patterns of Escherichia coli possessing eae genes. Med Microbiol Immunol. 1994;183:23–31. doi: 10.1007/BF00193628. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidjabat-Tambunan H, Bensink J C. Verotoxin-producing Escherichia coli from the faeces of sheep, calves and pigs. Aust Vet J. 1997;75:292–293. doi: 10.1111/j.1751-0813.1997.tb10100.x. [DOI] [PubMed] [Google Scholar]
- 29.Vernozy-Rozand C. Detection of Escherichia coli O157:H7 and other verocytotoxin-producing E. coli (VTEC) in food. J Appl Microbiol. 1997;82:537–551. doi: 10.1111/j.1365-2672.1997.tb03584.x. [DOI] [PubMed] [Google Scholar]
- 30.Wells J G, Shipman L D, Greene K D, Sowers E G, Green J H, Cameron D N, Downes F P, Martin M L, Griffin P M, Ostroff S M, Potter M E, Tauxe R V, Wachsmuth I K. Isolation of Escherichia coli O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J Clin Microbiol. 1991;29:985–989. doi: 10.1128/jcm.29.5.985-989.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto T, Honda T, Miwatani T, Yokota T. A virulence plasmid in Escherichia coli enterotoxigenic for humans: intergenetic transfer and expression. J Infect Dis. 1984;150:688–698. doi: 10.1093/infdis/150.5.688. [DOI] [PubMed] [Google Scholar]
- 32.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]