Abstract
The COVID‐19 pandemic poses a challenge in coming up with quick and effective means to counter its cause, the SARS‐CoV‐2. Here, we show how the key factors governing cysteine reactivity in proteins derived from combined quantum mechanical/continuum calculations led to a novel multi‐targeting strategy against SARS‐CoV‐2, in contrast to developing potent drugs/vaccines against a single viral target such as the spike protein. Specifically, they led to the discovery of reactive cysteines in evolutionary conserved Zn2+‐sites in several SARS‐CoV‐2 proteins that are crucial for viral polypeptide proteolysis as well as viral RNA synthesis, proofreading, and modification. These conserved, reactive cysteines, both free and Zn2+‐bound, can be targeted using the same Zn‐ejector drug (disulfiram/ebselen), which enables the use of broad‐spectrum anti‐virals that would otherwise be removed by the virus's proofreading mechanism. Our strategy of targeting multiple, conserved viral proteins that operate at different stages of the virus life cycle using a Zn‐ejector drug combined with other broad‐spectrum anti‐viral drug(s) could enhance the barrier to drug resistance and antiviral effects, as compared to each drug alone. Since these functionally important nonstructural proteins containing reactive cysteines are highly conserved among coronaviruses, our proposed strategy has the potential to tackle future coronaviruses.
This article is categorized under:
Structure and Mechanism > Reaction Mechanisms and Catalysis
Structure and Mechanism > Computational Biochemistry and Biophysics
Electronic Structure Theory > Density Functional Theory
Keywords: COVID‐19, cysteine reactivity, labile Zn‐sites, SARS‐CoV‐2, structural Zn
We reveal the key factors controlling the reactivity of free and Zn‐bound Cys in proteins and apply them to identify reactive Cys in multiple, conserved SARS‐CoV‐2 domains that can be targeted by clinically safe drugs, disulfiram or ebselen, to inhibit viral polyprotein cleavage, and viral RNA replication/transcription.
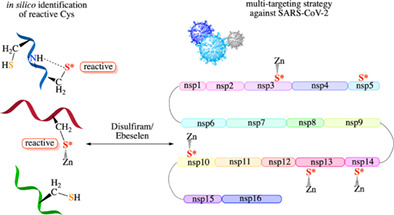
1. INTRODUCTION
The cell in all the complexity of its constituents and physiological processes can be thought of as a microcosm populated by various biological entities including organelles, proteins, lipids, and nucleic acids where the information flows as chemical signals. 1 Experimental methods generate a plethora of quantitative data on DNA/RNA/protein sequences, structures, and dynamics, revealing intricate patterns of the genome and protein universe. 2 The vast and sheer complexity of such data is quite overwhelming. Computational approaches can complement experimental data on complex biological processes by revealing their key underlying physical principles and elucidating recognition/reaction mechanisms. For example, experimental methods are limited in describing short‐lived transition states/intermediates in enzyme‐catalyzed reactions, whereas QM/MM methods can reveal how enzymes stabilize transition states/intermediates and help inhibitor design as well as test hypotheses/mechanisms and identify new reaction pathways. 3 , 4 , 5 , 6 , 7 As another example, computational approaches have elucidated the physical origins of noncovalent aromatic (π) interactions, 8 and the relationship between the sequences, structural properties, and functions of intrinsically disordered proteins. 9
At the atomic level, covalent, ionic, and metallic bonds as well as hydrogen‐bonding and van der Waals interactions in various dielectric environments form the “physicochemical language” of the cell. 1 Dynamical/conformational changes of large systems over long‐time scales can be probed using Markov state models, 10 , 11 coarse‐grained, 12 , 13 or ultra‐coarse‐grained 14 models, whereas those for smaller systems in the microsecond time scale are accessible by molecular dynamics (MD) simulations. 15 , 16 However, changes in the electron distribution during a chemical reaction, charge transfer, and/or polarization effects are best described by quantum theory. 17 , 18 , 19 Among the various quantum mechanical methods, density functional theory (DFT) has become the method of choice because of its good trade‐off between accuracy and computational cost. 20 , 21 DFT combined with molecular mechanics 3 or continuum methods 22 have proven to be successful in revealing biophysical trends behind the secondary structure formation in proteins, 23 and predicting the ionization free energy (pK a) of metal‐bound water molecules 24 as well as amino acid (aa) residues and substrate/drug‐like molecules in proteins. 25 , 26 , 27 They have helped to elucidate enzyme mechanisms, 4 , 6 , 28 , 29 and the physicochemical principles underlying metal‐binding affinity/selectivity in metalloproteins, 30 , 31 including cation selectivity of ion channel selectivity filters. 32 , 33 They have also been used to investigate the effects of macromolecular crowding on protein aggregation and stability. 34
In this focus article, we describe how studies of the physicochemical factors modulating cysteine (Cys) reactivity in proteins by combined DFT and continuum dielectric approach led to a novel strategy to battle the ongoing COVID‐19 pandemic caused by SARS‐CoV‐2. We first present an overview of Cys: (i) its importance, (ii) its various states, (iii) its biological roles, and (iv) its applications in research and biotechnology. Next, we delineate the key factors governing the reactivity of free and metal‐bound Cys in proteins. We then present a multi‐targeting strategy exploiting the key factors controlling the reactivity of Zn2+‐bound Cys in combination with evolutionary principles to identify novel drug targets in the SARS‐CoV‐2. Finally, we discuss how this strategy may be beneficial for tackling new coronaviruses that cause future epidemics/pandemics.
1.1. Importance of Cys
Despite being the second least abundant aa residue (only ∼1.9% of all aa residues in proteins), 35 Cys is very versatile with a myriad of functions. Cysteine reactivity has long been recognized as a key factor in the activity of many proteins. 36 Roughly 80% of Cys in proteins possess some functional importance. 1 Furthermore, Cys point mutations in proteins are associated with genetic diseases more often than expected, based on its occurrence frequency in protein sequences. 37 , 38 Conversely, point mutations to Cys in certain proteins can also lead to disease, as exemplified by the cancer‐causing mutation of the native Gly‐12 to Cys in the K‐Ras enzyme that catalyzes the hydrolysis of guanosine triphosphate. 39
1.2. The diverse nature of cysteine: Free, bound, and derivatized
Cysteine is the most versatile aa building block in proteins, as it can exist as free, bound or derivatized in vivo. 40 , 41 Being a “soft” ligand, Cys prefers binding to “borderline” (e.g., Fe2+, Co2+, Ni2+, Zn2+) or “soft” (e.g., Cu+, Cd2+, Hg2+) metal ions. 42 , 43 It can bind monodentately to a single cation or bridge two cations, as found in metallothioneins or FeS proteins. 44 The most common biogenic metal ion ligated by the Cys side chain is Zn2+, 45 the second most abundant transition metal ion in organisms. 46 Binding to Zn2+ lowers the Cys pK a, 47 so Zn2+‐bound Cys is generally deprotonated under physiological conditions. 48 Because S has an electron configuration of [Ne]3s 23p 4 and d‐orbitals for bonding, Cys can be readily oxidized to form derivatives with oxidation states ranging from −2 to +5 in vivo (Figure 1). 42 As evident from Figure 1, the Cys S atom has a different oxidation state/number, depending on its surrounding atoms, unlike its charge, which only depends on the number of electrons and protons in the S atom.
FIGURE 1.
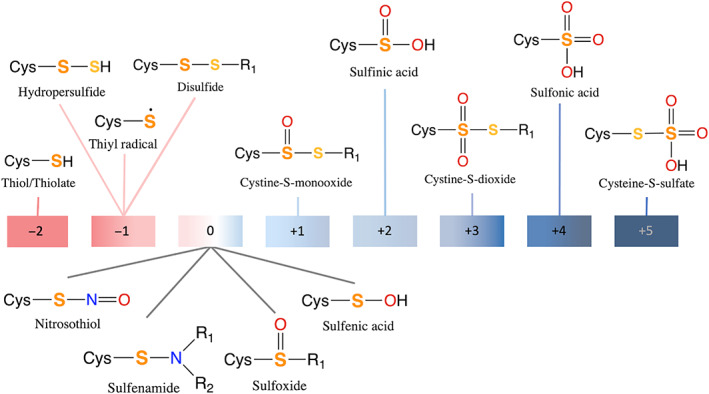
The various Cys(S) oxidation states found in vivo. The S corresponding to the indicated oxidation state is in bold (the oxidation state of the second S, if present, is −1)
1.3. Biological functions of cysteine
The Cys thiol shows a vast range of measured pK a values in proteins spanning from ~3 to 13, 49 , 50 reflecting its various biological functions, which fall into three main roles: (i) catalytic, (ii) regulatory, and (iii) structure‐stabilizing (Figure 2). 35 In enzymatic catalysis, Cys plays an essential catalytic role in nucleophilic substitution reactions catalyzed by diverse enzymes including Cys proteases and thioredoxin reductase. 43 Cysteine also participates in electron transfer catalyzed by glutathione reductase as well as in oxygen‐transfer, hydride‐transfer, or thiol/thiyl hydrogen radical transfer catalyzed by enzymes such as human peroxiredoxins, glyceraldehyde 3‐phosphate dehydrogenase, and ribonucleotide reductase, respectively. 43 In most of these reactions, the Cys thiolate serves as a catalytic nucleophile and can stabilize charges in the rate‐limiting transition state of a reaction. 51 , 52 Zn2+‐bound Cys can facilitate alkyl transfer reactions in enzymes (e.g., the DNA repair protein ADA, 53 whose Zn2+‐bound Cys removes methyl groups from the DNA backbone) or promote conversion of ribonucleotides to deoxyribonucleotides catalyzed by ribonucleotide reductase. 54
FIGURE 2.
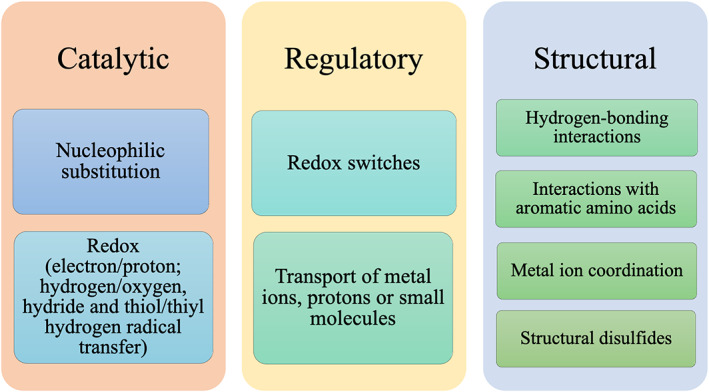
Catalytic, regulatory, or structural role of free, metal‐bound, or modified Cys
Apart from serving a direct catalytic role, Cys also plays a regulatory role in enzyme catalysis by changing its S oxidation state (Figure 1). For example, Cys oxidation, resulting in disulfide bond formation, can modulate protein structure and function, 41 and serve as regulatory functional switches. 55 It deactivates redox‐sensitive enzymes including caspases‐3/9, glyceraldehyde 3‐phosphate dehydrogenase, or protein tyrosine phosphatase that employ a catalytic Cys. 40 Zn2+‐bound Cys oxidation inhibits alcohol dehydrogenase by releasing the catalytic Zn2+, but activates matrix metalloproteases by freeing a coordination site for a nucleophilic water molecule/substrate to bind. 40
Cysteines, as part of nonprotein redox pairs such as glutathione disulfide/glutathione, regulate the reduction potential of the cell. 50 , 56 , 57 Post‐translationally modified Cys can regulate cellular signaling pathways: Cys palmitoylation regulates protein localization and trafficking. 58 Cysteine lipoxidation (modification by lipid‐derived electrophiles) triggers signaling pathways leading to stress adaptation. 59 Methylation of Zn2+‐bound Cys in the zinc finger domains of human TAB2 and TAB3 during bacterial infection disrupts the NF‐κB pathway, which mediates innate immune defense against microbial infection. 60 Another regulatory role of Cys is in ligand or cation transport: Cysteine thiols present on the cell surface mediate cell entry of molecules or proteins via thiol/disulfide exchange reactions followed by membrane fusion. 61 , 62 , 63 In metallothioneins, Cys thiolates regulate intracellular Zn2+ homeostasis, as their oxidation triggers release and transfer of Zn2+ ions to other proteins. 64 , 65
Cysteines are often employed to stabilize protein structure by forming disulfide bonds. 66 , 67 , 68 , 69 Notably, cysteine‐rich peptide toxins from animal venom secretions possess multiple disulfide bonds, resulting in exceptional thermal and chemical stability. 70 Thermophilic prokaryotes also employ structural disulfides in their proteins to protect them from thermal denaturation in high‐temperature environments. 71 , 72 Apart from forming covalent disulfide bonds, cysteines can form bonds with metal cations and stabilize specific folds in proteins such as Zn•Cys4, Zn•Cys3His, Zn•Cys2His2, and Zn2•Cys6 zinc fingers, zinc‐sensing proteins, zinc transport proteins, and metallothioneins. 73 , 74 , 75 By coordinating the catalytic metal cofactor in enzymes such as oxidoreductases 76 and zinc‐β‐lactamases II, 77 Cys helps to ensure the correct positioning of the active‐site residues and maintain enzyme catalytic efficiency/stability. By itself, Cys can also stabilize protein structure by interactions with aromatic amino acids, as evidenced by the Cys52–Phe65 interaction in SUMO‐1 where mutation of Cys52 to Ala significantly perturbed the SUMO‐1 secondary structure and thermal stability. 78
1.4. Applications of reactive cysteines
Due to its unique physicochemical properties, Cys is utilized in biochemical research. Based on the Cys thiolate reactivity, Cys scanning mutagenesis has been used to probe the structure and function of ion channels. 79 Cys thiol reactivity has also been exploited to design specific probes to investigate protein structure and functionality. 80 , 81 Cys, incorporated into lysine to form γ‐thialysine, has been proposed for studying methylation processes on intact histones and the nucleosome assembly, 82 as histone peptides containing native lysine and the unnatural γ‐thialysine are equally good substrates for methylation catalyzed by histone lysine methyltransferases. The ability of thiols to undergo via thiol/disulfide exchange on the cell surface (Section 1.3) has been exploited to enhance cellular uptake of a wide variety of cargos (e.g., small molecules, oligonucleotides, peptides/proteins, and synthetic constructs). 61 Structural motifs containing two Cys; for example, CXC where X denotes any aa, can enhance cellular uptake efficiency of cationic peptides, as the two Cys in the motif can form disulfide bonds with cell‐surface components. 83 , 84
Cysteines have also been used in biotechnology and in drug design. They have been used to conjugate antigen‐specific antibodies to potent small‐molecule drugs, creating antibody–drug conjugates. 85 Importantly, reactive Cys have become important drug targets for developing specific covalent inhibitors to treat various human diseases including cancers, arthritis, and bacterial/viral infections. 86 , 87 , 88 , 89 This is exemplified by one of the most frequent cancer‐causing mutations of the GTPase KRas enzyme, whereby the wild‐type G12 is mutated to Cys (G12C). By designing an inhibitor that covalently bonded specifically to the mutant C12 sulfur, KRasG12C is locked in the inactive GDP‐bound state and cannot switch to the active GTP‐bound state, hence oncogenic KRasG12C cell proliferation is blocked. 90 This strategy for inhibiting KRasG12C has been used successfully to develop the new anti‐cancer drug, Sotorasib (brand name: Lumakras), which was approved in May 2021 to treat metastatic nonsmall cell lung cancer patients with the KRas G12C mutation. As another example, reactive cysteines have also been chosen as targets in the battle against the COVID‐19 pandemic. Specific covalent inhibitors targeting the catalytic cysteines of the SARS‐CoV‐2 proteases (Mpro or PLpro) have been developed in silico 91 and in vitro, 92 , 93 , 94 and the inhibition mechanisms of several inhibitors have been investigated. 95 , 96
Apart from free Cys, reactive Cys bound to Zn2+ ions that play a structural role in stabilizing the protein (termed labile Zn2+‐site) can also serve as drug targets for retroviral or cancer therapy. 97 , 98 , 99 , 100 They react with electrophilic agents, resulting in the loss of structural Zn2+ cations and thus protein structure and function (Scheme 1). Such reactive Zn2+‐bound Cys has been found in the Zn fingers of several viruses including the HIV nucleocapsid p7, 101 herpes simplex virus, 102 and Junín virus, 103 as well as the human estrogen receptor DNA‐binding domain, which is essential for breast cancer growth. 104
SCHEME 1.
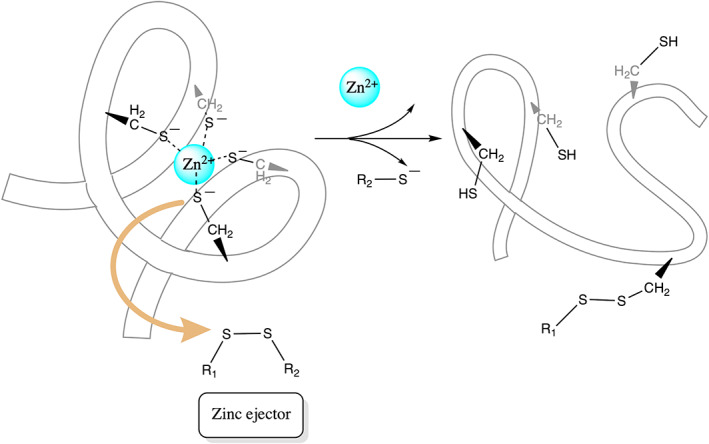
Proposed reaction mechanism for a reactive Zn2+‐bound Cys in a labile Zn2+‐finger to react with a Zn‐ejecting agent, causing loss of Zn2+ and protein structure
2. PHYSICAL PRINCIPLES UNDERLYING REACTIVE CYSTEINES
How can a simple amino acid such as Cys fulfill such diverse functional roles in proteins? The answer lies in the high reactivity of its side chain and its ability to interconvert between various oxidation states in vivo (Section 1.2). 43 Whether a Cys in a protein is reactive or inert depends on its environment, which modulates its pK a and consequently its reactivity. 105 For example, hydrogen bonds to metal‐bound Cys help to stabilize/protect the metal complex and enhance metal‐binding affinity/specificity, enzyme–substrate recognition, and enzyme activation. 106 Since anionic thiolates (S−) are more reactive/nucleophilic than neutral thiols (SH), a protein microenvironment that reduces the Cys pK a would enhance its reactivity. 49 , 107 Below, we summarize the key factors determining the reactivity of free or metal‐bound Cys in proteins from our previous studies, relying on the original references to provide details of the methodology. 35 , 51
2.1. Factors governing reactivity of free cysteine
Free Cys can be reactive as a catalytic or noncatalytic nucleophile in the Cys‐dependent enzymes. The reactivity of the free Cys side chain is dictated by its (i) solvent accessibility and (ii) hydrogen‐bonding interactions. 35 This is because the Cys thiolate is stabilized/destabilized to varying degrees depending on its hydrogen‐bonding partner and dielectric environment. 35 , 49 , 107 , 108 In peroxiredoxin enzymes, for example, Cys is stabilized in its reactive thiolate form by hydrogen‐bonding interactions with conserved Thr and Arg residues. 109 In thioredoxins, the number of hydrogen bonds to the catalytic Cys correlates with the decrease in the Cys pK a. 49 However, hydrogen‐bonding contacts to Cys alone do not suffice to determine the reactivity of free Cys. Protein conformational changes that position different hydrogen‐bonding partners to the Cys side chain and/or alter solvent access can modulate the nucleophilicity/reactivity of free Cys. Even two slightly different conformations of the same protein may differ greatly in their Cys reactivities due to differences in the Cys hydrogen‐bonding partners. 108 Along the same vein, mutations that alter the Cys hydrogen‐bonding partner and dielectric environment could change the degree of thiolate stabilization and thus Cys pK a. For example, in the disulfide‐binding protein A, mutation of His‐32 to Gly results in a loss of a strong hydrogen bond with the catalytic Cys‐30, whose unusually low pK a of 3.5 becomes elevated to 4.9, thus decreasing its reactivity. 110
2.2. Factors governing reactivity of Zn‐bound cysteines
In contrast to free Cys, which can exist as a neutral thiol (SH) or anionic thiolate depending on its pK a in the protein, Cys is deprotonated when bound to Zn2+, 48 thus Zn2+‐bound Cys serves as a nucleophile. The reactivity of a Zn2+‐bound thiolate towards an electrophile that can access the Zn2+‐site depends on whether (1) the thiolate S− can keep its negative charge, and (2) the positive charge on the cation is attenuated to free the Cys to undergo reaction. The negative charge on the Zn2+‐bound thiolate would be maintained if it is (i) not shared with a second Zn2+ or (ii) not withdrawn to the more electronegative carbonyl O via backbone hydrogen bonds to the Zn2+‐bound thiolate. Thus, whereas hydrogen bonds to free Cys stabilize the thiolate form, enabling it to serve as a nucleophile, hydrogen bonds to the Zn2+‐bound thiolate suppress nucleophilicity of the metal‐bound Cys (Figure 3). As for free Cys, conformational changes can affect the reactivity of the Zn2+‐bound thiolate due to changes in the hydrogen‐bonding interactions and/or solvent exposure of the structural Zn2+‐site. As negatively charged Cys− transfers much more charge to Zn2+ than neutral histidine ligands, 111 the positive charge on Zn2+ in Zn‐finger cores with >2 Cys would be reduced compared to that in Zn•Cys2His2 sites, where the higher positive charge on the cation would prohibit the two Cys from undergoing reaction. 51
FIGURE 3.
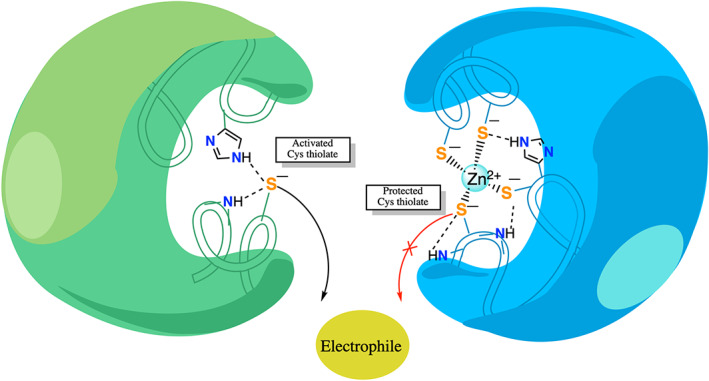
NH—S hydrogen bonds enhance nucleophilicity of free Cys (left), but suppress the nucleophilicity of Zn2+‐bound Cys (right)
3. APPLICATION OF PHYSICAL PRINCIPLES IN THE COVID‐19 PANDEMIC
3.1. Guidelines to identify labile Zn2+‐sites given the protein structure
The above factors controlling the Zn2+‐bound Cys reactivity in structural Zn2+‐sites have helped to establish guidelines to identify labile (druggable) Zn2+‐sites given the protein structure. Since neutral Zn2+‐bound ligands, or a second Zn2+, or hydrogen bonds to Zn2+‐bound Cys would suppress the reactivity of the Zn2+‐bound Cys, structural Zn‐Cys4 or Zn‐Cys3His, denoted collectively as Zn‐Cys4/(Cys3His) sites, with no hydrogen bond to any of the Zn2+‐bound Cys were predicted to be labile. 51 These guidelines were first used to identify putative labile Zn‐sites in human proteins that are promising drug targets, but whose Zn2+‐sites have not been considered to be drug targets. 112 Subsequently, they were used to predict labile Zn‐sites in the hepatitis C virus—the structural Zn2+‐Cys4 sites in NS5A, a multifunctional nonstructural protein (nsp), was predicted and subsequently verified to be labile. 113
3.2. How to target a viral labile Zn2+‐site without deadly cytotoxic effects
Since putative labile Zn‐sites are found in both human and viral Zn‐finger proteins, how can a Zn‐ejector selectively target the labile Zn2+‐site of a viral protein without affecting cellular Zn‐finger proteins? Instead of screening/designing a Zn‐ejecting compound that ejects Zn2+ from only the drug target viral protein, but not from essential human proteins, we had proposed using Zn‐ejecting agents that have passed safety tests in clinical trials or have been approved by the Food and Drug Administration. Although such Zn‐ejecting agents are generally not highly specific unlike antibody drugs that target a specific protein, they have been found to be clinically safe when used according to their recommended dosage. 114 The non‐specificity of clinically safe Zn‐ejector drugs can be exploited to target multiple viral proteins containing reactive Cys (see below).
3.3. Multi‐targeting of conserved, vital SARS‐CoV‐2 nonstructural proteins
The SARS‐CoV‐2 that is responsible for the current pandemic employs multiple conserved Cys and Zn2+, which play crucial roles in the virus life cycle. Its main protease (Mpro) and papain‐like protease (PLpro) employ catalytic Cys to cleave the large viral polyproteins into its constituent nonstructural proteins. In addition to the catalytic Cys, structures of SARS‐CoV‐2 and the closely related SARS‐CoV viral proteins reveal conserved Zn2+‐bound Cys in (i) the PLpro enzyme of nsp3, (ii) the nsp10 zinc‐finger domain, (iii) the nsp12 RNA‐dependent RNA polymerase (RdRp), (iv) the nsp13 helicase, and (v) the nsp14 N‐terminal 3′→5′ exoribonuclease (ExoN) domain (see Table 1). Using the guidelines outlined in Section 3.1, each structure was checked to see if the Zn‐Cys4/(Cys3His) site lacks hydrogen bonds to the Zn‐bound thiolates. However, the cryo‐electron microscopy structure of SARS‐CoV‐2 nsp12 and the ≥3.2 Å crystal structures of SARS‐CoV nsp14 (5c8s, 5c8t, and 5c8u) have poor resolution, which prohibited reliable hydrogen‐bond analyses of these Zn‐sites. The other SARS‐CoV or SARS‐CoV‐2 structures in Table 1 show no hydrogen bonds to the Zn‐Cys4/(Cys3His) sites. Subsequently, the Zn‐sites in the SARS‐CoV‐2 PLpro, the nsp10 zinc‐finger, the nsp13 helicase, and the nsp14 ExoN domains were experimentally verified to be labile: clinically safe Zn‐ejector drugs, disulfiram/ebselen, can release Zn2+ from these four viral proteins and decrease their functional activities in vitro as well as inhibit SARS‐CoV‐2 replication in Vero E6 cells. 115 , 116
TABLE 1.
Labile Zn‐sites in SARS‐CoV and/or SARS‐CoV‐2 nsp proteins
| SARS‐CoV | SARS‐CoV‐2 | ||||
|---|---|---|---|---|---|
| Protein name | Structures a | Zn‐ligands | Structures a | Zn‐ligands | Experimentally confirmed labile Zn‐sites |
| PLpro subdomain of nsp3 | 4m0w, 3e9s, 5tl7 | C190, C193, C225, C227 | 6wrh, 6wrz, 6wuu, 6wx4, 6wzu, 7e35, 7jit, 7jn2, 7lbr, 7lbs, 7llf, 7llz, 7los | C189, C192, C224, C226 |
Lin, 2018 117 Sargsyan, 2020 115 |
| nsp10 |
2fyg, 2ga6, 2xyq, 2xyr, 2xyv, 3r24 5c8s, 5c8t, 5c8u, 5nfy |
C74, C77, C90, H83 C117, C120, C128, C130 |
6xkm, 6wjt, 6wvn, 6wrz, 6zpe, 7c2i, 7jib, 7jpe, 7jyy, 7jz0, 7krx |
C74, C77, C90, H83 C117, C120, C128, C130 |
Sargsyan, 2020 115 |
| nsp12 |
6nus, 6nur (EM) |
C487, C645, C646, H642 |
6xez, 6yyt, 7aap, 7b3b, 7b3c, 7b3d, 7btf, 7bv1, 7bv2, 7bw4, 7bzf, 7c2k, 7ctt, 7cxm, 7cyq (EM) |
C301, C306, C310 H295 C487, C645, C646, H642 |
None |
| nsp13 | 6jyt |
C50, C55, C72, H75 |
6zsl, 5rl7, 5rlb, 5rlc, 5rlg, 5rlz, 5rm1, 5rm4, 5rmc, 5rmd, 5rme, 5rml, 5rmm, 5rob, 7nio, 7nng |
C5, C8, C26, C29 C50, C55, C72, H75 |
Chen, 2021 116 |
| nsp14 |
5c8s, 5c8t, 5c8u, 5nfy |
C207, C210, C226, H229 C477, C452, C484, H487 |
7diy, 7mc5, 7mc6 (nsp14 ExoN domain) |
C207, C210, C226, H229 |
Chen, 2021 116 |
The Zn‐ligand residues are taken from the first PDB highlighted in bold; e.g., the Zn‐ligand numbers for SARS‐CoV‐2 nsp10 were taken from PDB 6xkm.
These labile Zn2+‐sites are attractive drug targets, as they play important functional roles in the SARS‐CoV‐2 life cycle: The Zn‐Cys4/(Cys3His) sites play important structural roles in nsp3 PLpro domain, 117 nsp10 zinc‐finger, 118 nsp12, 119 and nsp14, 120 whereas they play vital catalytic roles in the nsp13 helicase activity, 121 and the nsp14 3′→5′ exoribonuclease activity. 120 Furthermore, the viral proteins hosting these Zn2+‐sites are constituents of a large replication‐transcription complex that plays a critical role in viral (i) RNA synthesis, (ii) RNA proofreading, and (iii) RNA modification to evade the human immune response, as follows: 122 , 123 First, the nsp12 C‐terminal RdRp domain catalyzes viral RNA synthesis with the help of nsp7 and nsp8 cofactors. 119 , 124 , 125 Next, the nsp14 N‐terminal ExoN domain proofreads the viral RNA by recognizing erroneous nucleotides and catalyzing their excision, thereby maintaining the integrity of the SARS‐CoV‐2 genome. 122 Subsequently, the nsp13 helicase, as well as the nsp14 and nsp16 methyltransferase domains, are involved in modifying the newly synthesized viral RNA, enabling its efficient translation by host cell ribosomes. Without this modification, the viral RNA molecules would be degraded and may be detected as foreign, triggering innate immune responses. 126 The nsp10 zinc‐finger protein activates the nsp14 and nsp16 methyltransferase enzymatic activities and boosts the nsp14 ExoN nucleolytic activity. 118 Furthermore, it stabilizes the conserved domains involved in RNA proofreading (nsp14) and modification (nsp13–16). 122 , 126
3.4. Advantage of using Zn 2+‐ejecting drugs to target cysteines
Because the large nsp12–nsp13–nsp14–nsp10–nsp16 complex is indispensable for SARS‐CoV‐2 replication, using clinically safe Zn‐ejecting drugs such as disulfiram/ebselen to target labile Zn2+‐sites in the constituent proteins would reduce viral load, as shown in Vero E6 cells. By reacting with Zn2+‐bound Cys and ejecting “structural” Zn2+ cations from the multi‐functional nsp10 cofactor, 115 disulfiram/ebselen can destabilize the nsp10 zinc‐finger itself as well as its partner proteins, nsp14 and nsp16. The same Zn‐ejecting drug can not only affect protein stability, but also inhibit the enzyme activities of nsp3 PLpro, 115 nsp13, and nsp14, and probably nsp12 RdRp. In addition to Zn2+‐bound Cys, disulfiram/ebselen can also target catalytic Cys, 115 and thereby inhibit SARS‐CoV‐2 Mpro, 127 which does not possess a Zn2+‐site. By impeding Mpro‐ and PLpro‐catalyzed viral proteolysis, disulfiram/ebselen can prevent efficient cleavage of the replicase polyproteins into components.
Hence, these Zn‐ejecting drugs work against coronaviruses at various stages: First, they inhibit viral polypeptide proteolysis; then, they cripple the functions of several proteins that are crucial for viral RNA synthesis, proofreading, and modification. Targeting multiple viral proteins at different stages would create a high barrier to drug resistance. In contrast, drugs targeting a specific viral protein may lose their effectiveness if a lineage appears with mutations leading to drug resistance. For example, a popular SARS‐Cov‐2 drug target is the spike protein that mediates entry of the virus into the host cell. Numerous mutations are found in the genomic region corresponding to the spike protein that alters host cell entry. Some mutations lead to enhanced transmission, 128 and may, as a by‐product, evade neutralizing antibodies targeting the spike protein. 129 On the other hand, the number of mutations observed in the regions containing catalytic or reactive Zn2+‐bound Cys is much lower than that for the rest of the genome (https://nextstrain.org/ncov/global). Hence, the spontaneous occurrence of lineages with mutations conferring resistance to disulfiram/ebselen would be unlikely. Furthermore, mutations of the conserved catalytic or reactive Zn2+‐bound Cys would likely disrupt their vital catalytic/structural roles in the respective viral protein functions, incurring a destructive cost for the virus.
Combining disulfiram/ebselen with other broad‐spectrum anti‐viral drugs that target other viral regions/pathways could further enhance the barrier to drug resistance. It can also enhance antiviral effect compared to each drug alone. For example, disulfiram/ebselen combined with remdesivir exhibited synergistic inhibition of SARS‐CoV‐2 in cell‐based assays. 116 This is because remdesivir A stops viral RNA synthesis by the nsp12 RdRp domain, and can escape removal by the proofreading nsp14 ExoN, as disulfiram/ebselen inhibits nsp14 exoribonuclease activity and destabilizes its allosteric activator nsp10. Disulfiram/ebselen combined with the zinc ionophore, hydroxychloroquine, could also synergistically inhibit SARS‐Cov‐2, 115 as these drugs may increase the local Zn2+ concentration and inhibit nsp12 RNA‐dependent RNA polymerase. 130
Finally, the SARS‐CoV‐2 caused the current pandemic in less than two decades after the SARS‐CoV caused outbreaks in several countries in 2003. Hence, we should prepare for new emergent coronaviruses even after the current pandemic is over. Analysis of genomes for different bat and human coronaviruses belonging to the same family as SARS‐CoV‐2 shows the highest conservation in regions encoding the viral targets revealed herein. 131 This suggests the possibility of targeting the multiple conserved reactive free/Zn2+‐bound Cys in emergent coronavirus pathogens indispensable for the coronavirus replication using a cocktail of Zn2+‐ejecting drugs and other broad‐spectrum antivirals.
4. CONCLUSION AND FUTURE OUTLOOK
Reactive Cys is of widespread interest, as they can serve as important drug targets among many other applications (Section 1.4). Herein, we have delineated the key physicochemical principles underlying their reactivity in proteins: The strength of the hydrogen bond stabilizing the Cys thiolate, which depends on the solvent accessibility and hydrogen‐bonding partner of the Cys, dictate the reactivity of free Cys. On the other hand, the negative charge on the thiolate S− and the positive charge on the cation dictate the reactivity of Zn2+‐bound Cys. These principles have provided guidelines to identify labile Zn2+‐sites given the protein structure, which have been used to reveal novel druggable Zn2+‐sites in multiple SARS‐CoV‐2 proteins. These predicted SARS‐CoV‐2 druggable sites have been validated in in vitro and cell‐based experiments using clinically safe Zn2+‐ejector drugs, which target not only reactive Zn2+‐bound Cys, but also catalytic Cys. The importance of the multiple SARS‐CoV‐2 targets revealed herein is underscored by their evolutionary conservation and crucial functions in viral polypeptide proteolysis as well as viral RNA synthesis, proofreading, and modification (Figure 4).
FIGURE 4.
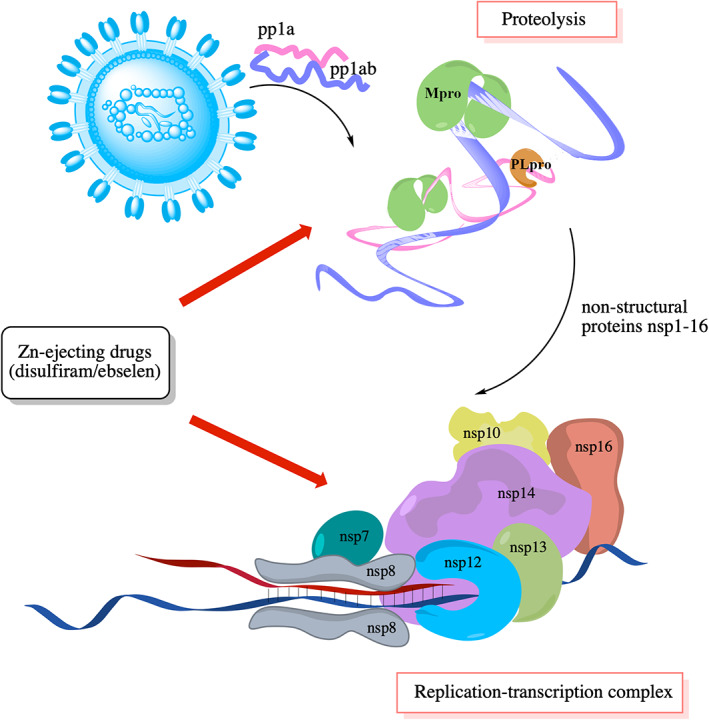
Viral drug targets and their functions
Because of the threat of another pandemic caused by a novel coronavirus, studying the best combination of Zn‐ejecting drugs and other broad‐spectrum antivirals targeting the multiple conserved viral regions/pathways in coronaviruses would be useful. Furthermore, since it takes time to solve structures of proteins for new infections, predicting which Cys are reactive and which are inert from sequence alone would also be useful.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
AUTHOR CONTRIBUTIONS
Karine Mazmanian: Visualization (lead); writing – original draft (equal). Ting Chen: Visualization (equal); writing – original draft (equal). Karen Sargsyan: Writing – original draft (equal). Carmay Lim: Conceptualization (equal); funding acquisition (equal); resources (equal); supervision (equal); writing – review and editing (equal).
RELATED WIREs ARTICLE
Mazmanian K, Chen T, Sargsyan K, Lim C. From quantum‐derived principles underlying cysteine reactivity to combating the COVID‐19 pandemic. WIREs Comput Mol Sci. 2022;12:e1607. 10.1002/wcms.1607
Ting Chen and Karen Sargsyan contributed equally to this study.
Edited by: Raghavan Sunoj, Associate Editor and Peter R. Schreiner, Editor‐in‐Chief
Funding information Academia Sinica, Taiwan, Grant/Award Number: AS‐IA‐107‐L03; Ministry of Science & Technology Taiwan, Grant/Award Number: MOST‐107‐2113‐M‐001‐018
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Parvez S, Long MJC, Poganik JR, Aye Y. Redox signaling by reactive electrophiles and oxidants. Chem Rev. 2018;118:8798–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeldovich KB, Chen P, Shakhnovich BE, Shakhnovich EI. A first‐principles model of early evolution: emergence of gene families, species, and preferred protein folds. PLoS Comp Biol. 2007;3:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mulholland AJ. Modelling enzyme reaction mechanisms, specificity and catalysis. Drug Discov Today. 2005;10:20–1402. [DOI] [PubMed] [Google Scholar]
- 4. Gao J, Ma S, Major DT, Nam K, Pu J, Truhlar DG. Mechanisms and free energies of enzymatic reactions. Chem Rev. 2006;106:3188–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sousa SF, Fernandes PA, Ramos MJ. Computational enzymatic catalysis ‐ clarifying enzymatic mechanisms with the help of computers. Phys Chem Chem Phys. 2012;14:12431–41. [DOI] [PubMed] [Google Scholar]
- 6. Blomberg MRA, Borowski T, Himo F, Liao R‐Z, Siegbahn PEM. Quantum chemical studies of mechanisms for metalloenzymes. Chem Rev. 2014;114:3601–58. [DOI] [PubMed] [Google Scholar]
- 7. Himo F. Recent trends in quantum chemical modeling of enzymatic reactions. J Am Chem Soc. 2017;139:6780–6. [DOI] [PubMed] [Google Scholar]
- 8. Neel AJ, Hilton MJ, Sigman MS, Toste FD. Exploiting non‐covalent π interactions for catalyst design. Nature. 2017;543:637–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nygaard M, Kragelund BB, Papaleo E, Lindorff‐Larsen K. An efficient method for estimating the hydrodynamic radius of disordered protein conformations. Biophys J. 2017;113:550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang H, Sheong FK, Zhu L, Gao X, Bernauer J, Huang X. Markov state models reveal a two‐step mechanism of miRNA loading into the human argonaute protein: selective binding followed by structural re‐arrangement. PLoS Comput Biol. 2015;11:e1004404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hempel T, Plattner N, Noé F. Coupling of conformational switches in calcium sensor unraveled with local Markov models and transfer entropy. J Chem Theory Comput. 2020;16:2584–93. [DOI] [PubMed] [Google Scholar]
- 12. Takada S. Coarse‐grained molecular simulations of large biomolecules. Curr Opin Struct Biol. 2012;22:130–7. [DOI] [PubMed] [Google Scholar]
- 13. Tai HC, Lim C. Gene silencing mechanisms revealed by dynamics of guide, target, and duplex binding to Argonaute. J Chem Theory Comput. 2020;16:688–99. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y, Cao Z, Zhang JZ, Xia F. Double‐well ultra‐coarse‐grained model to describe protein conformational transitions. J Chem Theory Comput. 2020;16:10. [DOI] [PubMed] [Google Scholar]
- 15. Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, et al. CHARMM‐GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 2016;12:405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan Y, Cembran A, Ma S, Gao J. Connecting protein conformational dynamics with catalytic function as illustrated in dihydrofolate reductase. Biochemistry. 2013;52:2036–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leopoldini M, Marino T, Michelini MD, Rivalta I, Russo N, Sicilia E, et al. The role of quantum chemistry in the elucidation of the elementary mechanisms of catalytic processes: from atoms, to surfaces, to enzymes. Theor Chem Acc. 2007;117:765–79. [Google Scholar]
- 18. Sakharov D, Lim C. Force fields including charge transfer and local polarization effects: application to proteins containing multi/heavy metal ions. J Comput Chem. 2009;30:191–202. [DOI] [PubMed] [Google Scholar]
- 19. Fox SJ, Pittock C, Fox T, Tautermann CS, Malcolm N, Skylaris C‐K. Electrostatic embedding in large‐scale first principles quantum mechanical calculations on biomolecules. J Chem Phys. 2011;135:224107. [DOI] [PubMed] [Google Scholar]
- 20. Verma P, Truhlar DG. Status and challenges of density functional theory. Trends Chem. 2020;2:302–18. [Google Scholar]
- 21. Rovira C. Study of ligand‐protein interactions by means of density functional theory and first‐principles molecular dynamics. In: Ulrich Nienhaus G, editor. Protein‐ligand interactions. Methods in molecular biology. Volume 305. Totowa, NJ: Humana Press; 2005. p. 517–54. [DOI] [PubMed] [Google Scholar]
- 22. Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chem Rev. 2005;105:2999–3093. [DOI] [PubMed] [Google Scholar]
- 23. Culka M, Rulíšek L. Interplay between conformational strain and intramolecular interaction in protein structures: which of them is evolutionarily conserved? J Phys Chem B. 2020;124:3252–60. [DOI] [PubMed] [Google Scholar]
- 24. Grauffel C, Lim C. Factors governing when a metal‐bound water is deprotonated in proteins. Phys Chem Chem Phys. 2018;20:29625–36. [DOI] [PubMed] [Google Scholar]
- 25. Gunner MR, Mao J, Song Y, Kim J. Factors influencing the energetics of electron and proton transfers in proteins. What can be learned from calculations. Biochim Biophys Acta. 2006;1757:942–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brás NF, Perez MA, Fernandes PA, Silva PJ, Ramos MJ. Accuracy of density functionals in the prediction of electronic proton affinities of amino acid side chains. J Chem Theory Comput. 2011;7:3898–908. [DOI] [PubMed] [Google Scholar]
- 27. Seybold PG, Shields GC. Computational estimation of pKa values. WIREs Comput Mol Sci. 2015;5:290–7. [Google Scholar]
- 28. Kazemia M, Himo F, Åqvist A. Enzyme catalysis by entropy without Circe effect. Proc Natl Acad Sci USA. 2016;113:2406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Glowacki DR, Harvey JN, Mulholland AJ. Taking Ockham's razor to enzyme dynamics and catalysis. Nat Chem. 2012;4:169–76. [DOI] [PubMed] [Google Scholar]
- 30. Dudev T, Lim C. Competition among metal ions for protein binding sites: determinants of metal ion selectivity in proteins. Chem Rev. 2014;114:538–56. [DOI] [PubMed] [Google Scholar]
- 31. Dudev T, Lim C. Metal binding and selectivity in metalloproteins: insights from computational studies. Annu Rev Biophys. 2008;37:97–116. [DOI] [PubMed] [Google Scholar]
- 32. Dudev T, Lim C. Ion selectivity strategies of sodium channel selectivity filters. Acc Chem Res. 2014;47:3580–7. [DOI] [PubMed] [Google Scholar]
- 33. Lim C, Dudev T. Potassium versus sodium selectivity in monovalent ion channel selectivity filters. In: Sigel A, Sigel H, Sigel RKO, editors. The alkali metal ions: their role for life in metal ions in life sciences. Cham, Switzerland: Springer International; 2016. [DOI] [PubMed] [Google Scholar]
- 34. Kinjo AR, Takada S. Effects of macromolecular crowding on protein folding and aggregation studied by density functional theory: statics. Phys Rev E. 2002;66:031911. [DOI] [PubMed] [Google Scholar]
- 35. Mazmanian K, Sargsyan K, Grauffel C, Dudev T, Lim C. Preferred hydrogen‐bonding partners of cysteine: implications for regulating Cys functions. J Phys Chem B. 2016;120:10288–96. [DOI] [PubMed] [Google Scholar]
- 36. Fowler NJ, Blanford CF, Visser SPD, Warwicker J. Features of reactive cysteines discovered through computation: from kinase inhibition to enrichment around protein degrons. Sci Rep. 2017;7:16338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu H, Ma BG, Zhao JT, Zhang HY. How similar are amino acid mutations in human genetic diseases and evolution. Biochem Biophys Res Commun. 2007;362:233–7. [DOI] [PubMed] [Google Scholar]
- 38. Marino SM, Gladyshev VN. Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J Mol Biol. 2010;404:902–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Giles NM, Watts AB, Giles GI, Fry FH, Littlechild JA, Jacob C. Metal and redox modulation of cysteine protein function. Chem Biol. 2003;10:677–93. [DOI] [PubMed] [Google Scholar]
- 41. Reddie KG, Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr Opin Chem Biol. 2008;12:746–54. [DOI] [PubMed] [Google Scholar]
- 42. Jacob C, Giles GI, Giles NM, Sies H. Sulfur and selenium: the role of oxidation state in protein structure and function. Angew Chem Int Ed. 2003;42:4742–58. [DOI] [PubMed] [Google Scholar]
- 43. Giles NM, Giles GI, Jacob C. Multiple roles of cysteine in biocatalysis. Biochem Biophys Res Commun. 2003;300:1–4. [DOI] [PubMed] [Google Scholar]
- 44. Palacios O, Atrian S, Capdevila M. Zn‐ and Cu‐thioneins: a functional classification for metallothioneins? J Biol Inorg Chem. 2011;16:991–1009. [DOI] [PubMed] [Google Scholar]
- 45. Dudev T, Lim C. Metal binding and selectivity in Zn proteins. J Chin Chem Soc. 2003;50:1093–102. [Google Scholar]
- 46. Sousa SF, Lopes AB, Fernandes PA, Ramos MJ. The zinc proteome: a tale of stability and functionality. Dalton Trans. 2009;14:7946–56. 10.1039/b904404c [DOI] [PubMed] [Google Scholar]
- 47. Hightower KE, Huang C‐C, Casey PJ, Fierke CA. H‐Ras peptide and protein substrates bind protein farnesyltransferase as an ionized thiolate. Biochemistry. 1998;37:15555–62. [DOI] [PubMed] [Google Scholar]
- 48. Dudev T, Lim C. Factors governing the protonation state of cysteines in proteins: an ab initio/CDM study. J Am Chem Soc. 2002;124:6759–66. [DOI] [PubMed] [Google Scholar]
- 49. Roos G, Foloppe N, Messens J. Understanding the pKa of redox cysteines: the key role of hydrogen bonding. Antioxid Redox Signal. 2013;18:94–127. [DOI] [PubMed] [Google Scholar]
- 50. Sousa SF, Neves RPP, Waheed SO, Fernandes PA, Ramos MJ. Structural and mechanistic aspects of S‐S bonds in the thioredoxin‐like family of proteins. Biol Chem. 2019;400:575–87. [DOI] [PubMed] [Google Scholar]
- 51. Lee Y‐M, Lim C. Factors controlling the reactivity of zinc finger cores. J Am Chem Soc. 2011;133:8691–703. [DOI] [PubMed] [Google Scholar]
- 52. Ferrer‐Sueta G, Manta B, Botti H, Radi R, Trujillo M, Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol. 2011;24:434–50. [DOI] [PubMed] [Google Scholar]
- 53. He C, Hus JC, Sun LJ, Zhou P, Norman DPG, Dötsch V, et al. A methylation‐dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli Ada. Mol Cell. 2005;20:117–29. [DOI] [PubMed] [Google Scholar]
- 54. Cerqueira NMFSA, Fernandes PA, Eriksson LA, Ramos MJ. Dehydration of ribonucleotides catalyzed by ribonucleotide reductase: the role of the enzyme. Biophys J. 2006;90:2109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fra A, Yoboue ED, Sitia R. Cysteines as redox molecular switches and targets of disease. Front Mol Neurosci. 2017;10:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–212. [DOI] [PubMed] [Google Scholar]
- 57. Ulrich K, Jakob U. The role of thiols in antioxidant systems. Free Radic Biol Med. 2019;140:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kathayat RS, Elvira PD, Dickinson BC. A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling. Nat Chem Biol. 2017;13:150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Patinen T, Adinolfi S, Cortés CC, Härkönen J, Deen AJ, Levonen A‐L. Regulation of stress signaling pathways by protein lipoxidation. Redox Biol. 2019;23:101114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang L, Ding X, Cui J, Xu H, Chen J, Gong YN, et al. Cysteine methylation disrupts ubiquitin‐chain sensing in NF‐κB activation. Nature. 2011;481:204–8. [DOI] [PubMed] [Google Scholar]
- 61. Laurent Q, Martinent R, Lim B, Pham AT, Kato T, López‐Andarias J, et al. Thiol‐mediated uptake. JACS Au. 2021;1:710–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Aubry S, Burlina F, Dupont E, Delaroche D, Joliot A, Lavielle S, et al. Cell‐surface thiols affect cell entry of disulfide‐conjugated peptides. FASEB J. 2009;23:2956–67. [DOI] [PubMed] [Google Scholar]
- 63. Ryser HJ‐P, Flückiger R. Progress in targeting HIV‐1 entry. Drug Discov Today. 2005;10:1085–94. [DOI] [PubMed] [Google Scholar]
- 64. Maret W. Redox biochemistry of mammalian metallothioneins. J Biol Inorg Chem. 2011;16:1079–86. [DOI] [PubMed] [Google Scholar]
- 65. Babu CS, Lee Y‐M, Dudev T, Lim C. Modeling Zn2+ release from metallothionein. J Phys Chem A. 2014;118:9244–52. [DOI] [PubMed] [Google Scholar]
- 66. Yang Y, Chen M, Kesterson RA Jr, Harmon CM. Structural insights into the role of the ACTH receptor cysteine residues on receptor function. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1120–6. [DOI] [PubMed] [Google Scholar]
- 67. Olivella M, Caltabiano G, Cordomí A. The role of cysteine 6.47 in class a GPCRs. BMC Struct Biol. 2013;13:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rana R, Carroll CE, Lee H‐J, Bao J, Marada S, Grace CRR, et al. Structural insights into the role of the smoothened cysteine‐rich domain in hedgehog signalling. Nat Commun. 2013;4:2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dias RDO, Franco OL. Cysteine‐stabilized αβ defensins: from a common fold to antibacterial activity. Peptides. 2015;72:64–72. [DOI] [PubMed] [Google Scholar]
- 70. Maxwell M, Undheim EAB, Mobli M. Secreted cysteine‐rich repeat proteins “SCREPs”: a novel multi‐domain architecture. Front Pharmacol. 2018;9:1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Landeta C, Boyd D, Beckwith J. Disulfide bond formation in prokaryotes. Nat Microbiol. 2018;3:270–80. [DOI] [PubMed] [Google Scholar]
- 72. Beeby M, O'Connor BD, Ryttersgaard C, Boutz DR, Perry J. The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 2005;3:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gomes CM, Stafshede PW. Protein folding and metal ions: mechanisms, biology and disease. Boca Raton, FL: Taylor & Francis, CRC Press; 2010. [Google Scholar]
- 74. Krishna S, Majumdar I, Grishin NV. Structural classification of zinc fingers. Nucleic Acids Res. 2003;31:532–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cox EH, McLendon GL. Zinc‐dependent protein folding. Curr Opin Chem Biol. 2000;4:162–5. [DOI] [PubMed] [Google Scholar]
- 76. Vidal LS, Kelly CL, Mordaka PM, Heap JT. Review of NAD(P)H‐dependent oxidoreductases: properties, engineering and application. Biochim Biophys Acta Proteins Proteomics. 2018;1866:327–47. [DOI] [PubMed] [Google Scholar]
- 77. Mojica MF, Bonomo RA, Fast W. B1‐Metallo‐β‐lactamases: where do we stand? Curr Drug Targets. 2016;17:1029–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Drobecq H, Boll E, Sénéchal M, Desmet R, Saliou JM, Lacapère JJ, et al. A central cysteine residue is essential for the thermal stability and function of SUMO‐1 protein and SUMO‐1 peptide‐protein conjugates. Bioconjug Chem. 2016;27:1540–6. [DOI] [PubMed] [Google Scholar]
- 79. Liu X, Alexander C, Serrano J, Borg E, Dawson DC. Variable reactivity of an engineered cysteine at position 338 in cystic fibrosis transmembrane conductance regulator reflects different chemical states of the thiol. J Biol Chem. 2006;281:8275–85. [DOI] [PubMed] [Google Scholar]
- 80. Manna S, Karmakar P, Ali SS, Guria UN, Sarkar R, Datta P, et al. A Michael addition–cyclization‐based switch‐on fluorescent chemodosimeter for cysteine and its application in live cell imaging. New J Chem. 2018;42:4951–8. [Google Scholar]
- 81. Iacobucci C, Piotrowski C, Rehkamp A, Ihling CH, Sinz A. The first MS‐cleavable, photo‐thiol‐reactive cross‐linker for protein structural studies. J Am Soc Mass Spectrom. 2019;30:139–48. [DOI] [PubMed] [Google Scholar]
- 82. Temimi AHKA, Bruijne RVDW‐D, Proietti G, Guo H, Qian P, Mecinović J. γ‐Thialysine versus lysine: an insight into the epigenetic methylation of histones. Bioconjug Chem. 2019;30:1798–804. [DOI] [PubMed] [Google Scholar]
- 83. Li T, Gao W, Liang J, Zha M, Chen Y, Zhao Y, Wu C. Biscysteine‐bearing peptide probes to reveal extracellular thiol–disulfide exchange reactions promoting cellular uptake. Anal Chem. 2017;89:8501–8. [DOI] [PubMed] [Google Scholar]
- 84. Meng X, Li T, Zhao Y, Wu C. CXC‐mediated cellular uptake of Miniproteins: forsaking “arginine magic”. ACS Chem Biol. 2018;13:3078–86. [DOI] [PubMed] [Google Scholar]
- 85. Cal PM, Bernardes GJ, Gois PM. Cysteine‐selective reactions for antibody conjugation. Angew Chem Int Ed Engl. 2014;53:10585–7. [DOI] [PubMed] [Google Scholar]
- 86. Visscher M, Arkin MR, Dansen TB. Covalent targeting of acquired cysteines in cancer. Curr Opin Chem Biol. 2016;30:61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lagoutte R, Patouret R, Winssinger N. Covalent inhibitors: an opportunity for rational target selectivity. Curr Opin Chem Biol. 2017;39:54–63. [DOI] [PubMed] [Google Scholar]
- 88. Büttner D, Kramer JS, Klingler F‐M, Wittmann SK, Hartmann MR, Kurz CG, et al. Challenges in the development of a thiol‐based broad‐spectrum inhibitor for metallo‐β‐lactamases. ACS Infect Dis. 2018;4:360–72. [DOI] [PubMed] [Google Scholar]
- 89. Maurais AJ, Weerapana E. Reactive‐cysteine profiling for drug discovery. Curr Opin Chem Biol. 2019;50:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Goebel L, Müller MP, Goody RS, Rauh D. KRasG12C inhibitors in clinical trials: a short historical perspective. RSC Med Chem. 2020;11:760–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Martí S, Arafet K, Lodola A, Mulholland AJ, Świderek K, Moliner V. Impact of warhead modulations on the covalent inhibition of SARS‐CoV‐2 M pro explored by QM/MM simulations. ACS Catal. 2022;12:698–708. [DOI] [PubMed] [Google Scholar]
- 92. Dai W, Zhang B, Jiang X‐M, Su H, Li J, Zhao Y, et al. Structure‐based design of antiviral drug candidates targeting the SARS‐CoV‐2 main protease. Science. 2020;368:1331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hoffman RL, Kania RS, Brothers MA, Davies JF, Ferre RA, Gajiwala KS, et al. Discovery of ketone‐based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID‐19. J Med Chem. 2020;63:12725–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, et al. Crystal structure of SARS‐CoV‐2 main protease provides a basis for design of improved α‐ketoamide inhibitors. Science. 2020;368:409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ramos‐Guzmán CA, Ruiz‐Pernía JJ, Tuñón I. Multiscale simulations of SARS‐CoV‐2 3CL protease inhibition with aldehyde derivatives: Role of protein and inhibitor conformational changes in the reaction mechanism. ACS Catal. 2021;11:4157–68. [DOI] [PubMed] [Google Scholar]
- 96. Parise A, Romeo I, Russo N, Marino T. The Se–S bond formation in the covalent inhibition mechanism of SARS‐CoV‐2 main protease by Ebselen‐like inhibitors: a computational study. Int J Mol Sci. 2021;22:9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rice WG, Schaeffer CA, Harten B, Villinger F, South TL, Summers MF, et al. Inhibition of HIV‐1 infectivity by zinc‐ejecting aromatic C‐nitroso compounds. Nature. 1993;361:473–5. [DOI] [PubMed] [Google Scholar]
- 98. Loo JA, Holler TP, Sanchez J, Gogliotti R, Maloney L, Reily MD. Biophysical characterization of zinc ejection from HIV nucleocapsid protein by anti‐HIV 2,2′‐dithiobis[benzamides] and benzisothiazolones. J Med Chem. 1996;39:4313–20. [DOI] [PubMed] [Google Scholar]
- 99. Jenkins LM, Durell SR, Maynard AT, Stahl SJ, Inman JK, Appella E, et al. Comparison of the specificity of interaction of cellular and viral zinc‐binding domains with 2‐mercaptobenzamide thioesters. J Am Chem Soc. 2006;128:11964–76. [DOI] [PubMed] [Google Scholar]
- 100. Miller Jenkins LM, Hara T, Durell SR, Hayashi R, Inman JK, Piquemal JP, et al. Specificity of acyl transfer from 2‐mercaptobenzamide thioesters to the HIV‐1 nucleocapsid protein. J Am Chem Soc. 2007;129:11067–78. [DOI] [PubMed] [Google Scholar]
- 101. Huang M, Maynard A, Turpin JA, Graham L, Janini GM, Covell DG, et al. Anti‐HIV agents that selectively target retroviral nucleocapsid protein zinc fingers without affecting cellular zinc finger proteins. J Med Chem. 1998;41:1371–81. [DOI] [PubMed] [Google Scholar]
- 102. Barlow PN, Luisi B, Milner A, Elliott M, Everett R. Structure of the C3HC4 domain by 1H‐nuclear magnetic resonance spectroscopy: a new structural class of zinc‐finger. J Mol Biol. 1994;237:201–11. [DOI] [PubMed] [Google Scholar]
- 103. Briknarova K, Thomas CJ, York J, Nunberg JH. Structure of a zinc‐binding domain in the Junin virus envelope glycoprotein. J Biol Chem. 2011;286:1528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schwabe JWR, Chapman L, Finch JT, Rhodes D, Neuhaus D. DNA recognition by the oestrogen receptor: from solution to the crystal. Structure. 1993;1:187–204. [DOI] [PubMed] [Google Scholar]
- 105. Mazmanian K, Sargsyan K, Lim C. How the local environment of functional sites regulates protein function. J Am Chem Soc. 2020;142:9861–71. [DOI] [PubMed] [Google Scholar]
- 106. Mazmanian K, Dudev T, Lim C. How first‐second shell interactions and metal substitution modulate protein function. Inorg Chem. 2018;57:14052–61. [DOI] [PubMed] [Google Scholar]
- 107. Wible RS, Sutter TR. Soft cysteine signaling network: the functional significance of cysteine in protein function and the soft acids/bases thiol chemistry that facilitates cysteine modification. Chem Res Toxicol. 2017;30:729–62. [DOI] [PubMed] [Google Scholar]
- 108. Marino SM. Protein flexibility and cysteine reactivity: influence of mobility on the H‐bond network and effects on pKa prediction. Protein J. 2014;33:323–36. [DOI] [PubMed] [Google Scholar]
- 109. Halls A, Nelson K, Poole LB, Karplus PA. Structure‐based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antiox Redox Signaling. 2011;15:795–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Karshikoff A, Nilsson L, Foloppe N. Understanding the −C−X1−X2−C− motif in the active site of the Thioredoxin superfamily: E. coli DsbA and its mutants as a model system. Biochemistry. 2013;52:5730–45. [DOI] [PubMed] [Google Scholar]
- 111. Lee YM, Lin YF, Lim C. Factors controlling the role of Zn and reactivity of Zn‐bound cysteines in proteins: application to drug target discovery. J Chin Chem Soc. 2014;61:142–50. [Google Scholar]
- 112. Lee Y‐M, Wang Y‐T, Duh Y, Yuan HS, Lim C. Identification of labile Zn‐sites in drug‐target proteins. J Am Chem Soc. 2013;135:14028–31. [DOI] [PubMed] [Google Scholar]
- 113. Lee YM, Duh Y, Wang ST, Lai MMC, Yuan HS, Lim C. Using an old drug to target a new drug site: application of disulfiram to target the Zn‐site in HCV NS5A protein. J Am Chem Soc. 2016;138:3856–62. [DOI] [PubMed] [Google Scholar]
- 114. Banys P. The clinical use of disulfiram (Antabuse®): a review. J Psychoactive Drugs. 1988;20:243–61. [DOI] [PubMed] [Google Scholar]
- 115. Sargsyan K, Lin C‐C, Chen T, Grauffel C, Chen YP, Yang WZ, et al. Multi‐targeting of functional cysteines in multiple conserved SARS‐CoV‐2 domains by clinically safe Zn‐ejectors. Chem Sci. 2020;11:6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Chen T, Fei CY, Chen YP, Sargsyan K, Chang CP, Yuan HS, et al. Synergistic inhibition of SARS‐CoV‐2 replication using disulfiram/ebselen and remdesivir. ACS Pharm Transl Sci. 2021;4(2):1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Lin MH, Moses DC, Hsieh CH, Cheng SC, Chen YH, Sun CY, et al. Disulfiram can inhibit MERS and SARS coronavirus papain‐like proteases via different modes. Antiviral Res. 2018;150:155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bouvet M, Lugari A, Posthuma CC, Zevenhoven JC, Bernard S, Betzi S, et al. Coronavirus Nsp10, a critical co‐factor for activation of multiple replicative enzymes. J Biol Chem. 2014;289:25783–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kirchdoerfer RN, Ward AB. Structure of the SARS‐CoV nsp12 polymerase bound to nsp7 and nsp8 co‐factors. Nat Commun. 2019;10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ma Y, Wu L, Shaw N, Gao Y, Wang J, Sun Y, et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci USA. 2015;112:9436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Jia Z, Yan L, Ren Z, Wu L, Wang J, Guo J, et al. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. A structural view of SARS‐CoV‐2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cell. 2020;9:1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS‐CoV‐2. Nat Rev Microbiol. 2021;19:155–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, et al. Structure of the RNA‐dependent RNA polymerase from COVID‐19 virus. Science. 2020;368:779–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yan L, Zhang Y, Ge J, Zheng L, Gao Y, Wang T, et al. Architecture of a SARS‐CoV‐2 mini replication and transcription complex. Nat Commun. 2020;11:5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Snijder EJ, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of Mpro from COVID‐19 virus and discovery of its inhibitors. Nature. 2020;582:289–93. [DOI] [PubMed] [Google Scholar]
- 128. Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS‐CoV‐2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JCC, et al. Escape from neutralizing antibodies by SARS‐CoV‐2 spike protein variants. Elife. 2020;9:e61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. te Velthuis AJW, van den Worm SHE, Sims AC, Baric RS, Snijder EJ, van Hemert MJ. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6:e1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Boni MF, Lemey P, Jiang X, Lam TTY, Perry BW, Castoe TA, et al. Evolutionary origins of the SARS‐CoV‐2 sarbecovirus lineage responsible for the COVID‐19 pandemic. Nat Microbiol. 2020;5:1408–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


