Abstract
Background
The highly contagious SARS‐CoV‐2 is mainly transmitted by respiratory droplets and aerosols. Consequently, people are required to wear masks and maintain a social distance to avoid spreading of the virus. Despite the success of the commercially available vaccines, the virus is still uncontained globally. Given the tropism of SARS‐CoV‐2, a mucosal immune reaction would help to reduce viral shedding and transmission locally. Only seven out of hundreds of ongoing clinical trials are testing the intranasal delivery of a vaccine against COVID‐19.
Methods
In the current study, we evaluated the immunogenicity of a traditional vaccine platform based on virus‐like particles (VLPs) displaying RBD of SARS‐CoV‐2 for intranasal administration in a murine model. The candidate vaccine platform, CuMVTT‐RBD, has been optimized to incorporate a universal T helper cell epitope derived from tetanus‐toxin and is self‐adjuvanted with TLR7/8 ligands.
Results
CuMVTT‐RBD vaccine elicited a strong systemic RBD‐ and spike‐IgG and IgA antibodies of high avidity. Local immune response was assessed, and our results demonstrate a strong mucosal antibody and plasma cell production in lung tissue. Furthermore, the induced systemic antibodies could efficiently recognize and neutralize different variants of concern (VOCs).
Conclusion
Our data demonstrate that intranasal administration of CuMVTT‐RBD induces a protective systemic and local specific antibody response against SARS‐CoV‐2 and its VOCs.
Keywords: COVID‐19, intranasal, SARS‐CoV‐2, vaccine, virus‐like particles
In this study, we describe a COVID‐19 vaccine based on virus‐like particles (VLPs) for intranasal administration. We demonstrate that the vaccine candidate (CuMVTT‐RBD) is highly immunogenic in mice and is capable of inducing mucosal and systemic RBD as well as spike specific antibody responses. The induced antibodies are capable of neutralizing SARS‐CoV‐2 and variants of concern (VOCs).

1. INTRODUCTION
Till date, COVID‐19 caused by SARS‐CoV‐2 is still considered a global pandemic that has wreaked havoc globally and put a heavy toll on public health and economy. The marketed vaccines such as mRNA, viral vector, and inactivated viruses have greatly reduced the number of COVID‐19 mortality and hospitalization and continue to provide different levels of protections against the emerging variants of concern (VOCs). 1
Viral tropism depends, among other factors, on the susceptibility of a specific host cell. COVID‐19 patients often present with respiratory illness that can progress to severe pneumonia. 2 These observations suggested that the lung is the primary organ infected by SARS‐CoV‐2. The fact that lung epithelial cells express the angiotensin converting enzyme 2 (ACE2), the viral receptor, substantiates this observation. 3 The primary port of entry to the body is alveolar epithelial cells, but vascular endothelial cells also express ACE2 and are a prominent place of viral replication. 4 , 5 These cells may be considered the base for early infection and viral replication as well as long‐term viral persistence in some cases. 6
The currently available marketed vaccines are administered intramuscularly (i.m.) producing systemic spike and RBD‐specific antibodies (Abs) that can recognize and neutralize the virus. 7 Given the tropism of SARS‐CoV‐2, recent research efforts have also been devoted toward the development of an intranasal (i.n.) COVID‐19 vaccine. Seven intranasal COVID‐19 vaccines candidates are currently in clinical trials. 8 Intranasal vaccination route may offer several advantages over i.m. route including: needle‐free administration, direct delivery to the site of infection, and most importantly, the induction of mucosal immunity in the respiratory tract. 9 Secretory IgA (sIgA) is of major importance in the respiratory tract where it presents an efficient line of defence against respiratory infections. 10 Furthermore, mucosal vaccination can result in resident B and T cell priming leading to long‐lived Ab secreting cells or tissue‐resident memory cells, which add in clearing the viral infection. 11 This locally induced immune reaction has been shown to reduce viral replication and shedding in lungs and nasal passages leading to lower infection and transmission. 12 The concept of i.n. vaccination goes back to 1960s based on observations with live‐attenuated influenza vaccines (LAIV) that mimic a natural influenza infection and have shown to elicit a protective local and systemic antibody as well as cellular responses. 13 Live‐attenuated virus or viral‐vector based vaccines need to infect cells for replication. Moreover, attenuated viruses may pose a small risk of retaining their replication ability, especially in people with weaker immune systems.
Efficient induction of mucosal immunity can be best achieved by vaccines that mimic mucosal pathogens. Virus‐like particles (VLPs) constitute an efficient and safe vaccine platform as they lack genetic material for replication in vivo. Their particulate and repetitive surface structure enables them to stimulate innate and adaptive immune response and target the mucosa as well as the underlying dendritic cells (DCs). 14 We have previously assessed the efficacy of Qß‐VLPs as an i.n. vaccine platform. Our results indicated efficient induction of specific‐IgGs in serum and lungs besides robust local IgA production. 14 Here, we provide a proof‐of‐concept (PoC) in murine model proving the immunogenicity and efficacy of i.n. administration of a SARS‐CoV‐2 vaccine based on VLPs. The developed vaccine candidate is based on our optimized plant‐derived VLPs (CuMVTT) displaying the receptor‐binding domain (RBD) of SARS‐CoV‐2. Our results demonstrate that CuMVTT‐RBD induces strong systemic and local B cell response including high levels of IgG and IgA, plasma cell (PC) formation as well as broad viral neutralization. Taken together, our vaccine constitutes an efficient candidate for the generation of Ab‐based vaccine that can be administered mucosally in a needle‐free manner.
2. METHODS
2.1. Mice
All in vivo experiments were performed using (8–12‐week‐old) wild‐type (wt) female BALB/cOlaHsd mice purchased from Harlan. All animal procedures were conducted in accordance with the Swiss Animal Act (455.109.1‐5 September 2008) of University of Bern. All animals were treated for experimentation according to the protocols approved by the Swiss Federal Veterinary Office.
2.2. Protein expression and purification
RBDwt of SARS‐CoV‐2 and mutant RBDs (RBDK417N, RBDE484K, RBDN501Y, RBDK417N/E484K/N501Y, and RBDL452R/E484Q) were expressed using Expi293F cells (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). The amino acid (a.a.) sequence of each RBD was inserted into a pTWIST‐CMV‐BetaGlobin‐WPRE‐Neo vector (Twist Bioscience, San Fransico, CA, USA). RBD‐His Tag construct was further transformed into competent XL‐1 Blue bacterial cells. After plasmid purification, 50 µg of the plasmid was then transfected into Expi293F cells at a density of 3 × 106 cells/ml in a 250 ml shaking flask using the ExpiFectamine 293 Transfection Kit (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). Ninety‐six hours later, the supernatant containing RBD was harvested and dialyzed with PBS. RBD protein was then captured using His‐Trap HP column (GE Healthcare, Wauwatosa, WI, USA) or HiTrap TALON crude column (Cytiva, Uppsala, Sweden). Fractions were collected and concentrated. Buffer‐exchanged to PBS was carried on using Vivaspin 20 5KDMWCO spin column (Sartorius Stedim Switzerland AG, Tagelswangen, Switzerland). Human ACE2 protein His Tag and SARS‐CoV‐2 spike were purchased from Sino Biological, Beijing, China.
2.3. CuMVTT expression and production
Expression and production of CuMVTT was described in detail in Zeltins et al. 15 The level of LPS is 10 endotoxin per mg of CuMVTT measured using LAL test (pierce).
2.4. Development of CuMVTT‐RBD vaccine
RBDwt was conjugated to CuMVTT using the cross‐linker succinimidyl 6‐(beta‐maleimidopropionamido) hexanoate (SMPH) (Thermo Fisher Scientific, Waltham, MA, USA) at 7.5 molar excess to CuMVTT for 30 min at 25°C. The coupling reactions were performed with molar ratio RBD/CuMVTT (1:1) by shaking at 25°C for 3 h at 250 g on a DSG Titertek (Flow Laboratories, Irvine, UK). Unreacted SMPH and RBD proteins were removed using Amicon Ultra 0.5, 100 K (Merck Millipore, Burlington, MA, USA). VLP samples were centrifuged for 2 min at 12,000 g for measurement on ND‐1000. RBDwt. SDS‐PAGE was stained with InstantBlueTM Coomassie stain and image was obtained with Azure Biosystem using visible channel. Coupling efficiency was calculated by densitometry (as previously described for the IL17A‐CuMVTT vaccine 15 ), with a result of approximately 30%, meaning that there is about 60‐RBD per one VLP. RBDw coupling to CuMVTT was further analyzed by Western blot. For this purpose, RBDwt, CuMVTT, and coupled CuMVTT‐RBD were separated on a 12% SDS PAGE. Using the Trans‐Blot® Turbo™ Transfer System protein bands were transferred onto a 0.2 μM PVDF membrane (BIORAD, Hercules, USA). The membrane was further processed by using the iBindTM Flex Western Device (Invitrogen, USA) according to the manufacturer's protocol. As primary antibody SARS‐CoV‐2 Spike RBD antibody (R&DSystems, MAB10540) at (1:1000) and as detecting antibody goat anti‐mouse IgG conjugated to Horseradish Peroxidase (HRP) (Jackson ImmunoResearch, 115‐035‐071, West Grove, Pennsylvania) was added at (1:1000). Bound antibodies were detected by using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (ThermoScientific, 34579). WB image was obtained with Azure Biosystem c300 with exposure time of 2s using chemiluminescence channel. Packaging of ssRNA in CuMVTT‐VLPs and the developed vaccine CuMVTT‐RBD was confirmed by 1% Agarose gel run at 50 V for 40 min and visualized using Azure Biosystems c300 with exposure time of 10s using UV302 channel. A DNA Ladder (Thermo Scientific, Cat. Nr. SM0242) was included.
2.5. Electron microscopy
Physical stability and integrity of the candidate vaccine CuMVTT‐RBD were visualized by transmission electron microscopy (Philips CM12 EM). For imaging, sample‐grids were glow discharged and 10 μl of purified CuMVTT‐RBD (1.1 mg/ml) was added for 30s. Grids were washed 3× with ddH2O and negatively stained with 5 μl of 5% uranyl acetate for 30s. Excess uranyle acetate was removed by pipetting, and the grids were air dried for 10 min. Images were taken with 84,000× and 110,000× magnification.
2.6. Binding ELISA assay
To test if the vaccine can bind the relevant human receptor ACE2, ELISA plates were coated with 2 µg/ml of ACE2 in PBS at a volume of 50 µl/well. The plate was incubated at 4°C overnight. The plate was washed with PBS+Tween 0.01%. Added 50 µl/well of PBS‐Casein 0.15% and incubated for 1 h at RT on a shaker. Flicked off the blocking solution and added 50 µl of the CuMVTT‐RBD, CuMVTT, and SARS‐CoV‐2 RBDwt from 50 µg/ml to the first row of the plate followed by 1:3 dilution. The plate was incubated for 1 h at RT, washed with PBS+Tween 0.01%. 50 µl of mouse anti‐CuMVTT monoclonal antibody (clone 1‐1A8/ batch 2) at a concentration of 1 µg/ml was added to each well as a secondary antibody and incubated for 1 h at RT on a shaker. The plate was washed and 50 µl of the detection antibody; HRP labeled goat anti‐mouse IgG Fc gamma at a dilution of 1:1000 in PBS‐Casein 0.15% was added to each well. The plate was incubated for 1 h at RT. The plate was developed and read at OD450 nm (BioTek, USA).
2.7. Vaccination regimen
Wild‐type BALB/cOlaHsd mice (8–12 weeks, Harlan) were vaccinated intranasally (i.n.) with 40 µg with either CuMVTT‐RBD vaccine or CuMVTT as a control in a volume of 40 µl without any adjuvants (20 µg in each nostril). The mice were boosted with an equal dose at day 28 and bled until day 49. Serum was collected on a weekly basis via tail bleeding and the serum was isolated using Microtainer Tube (BD Biosciences, USA). For Fluorospot samples and Bronchoalveolar lavage (BAL), wt BALB/cOlaHsd mice (8–12 weeks, Harlan) were vaccinated i.n. as indicated above. Serum samples were collected on day 35.
2.8. Enzyme‐Linked Immunosorbent Assay (ELISA)
To determine the total IgG Abs against the candidate vaccine CuMVTT‐RBD in sera of vaccinated mice, ELISA plates were coated with SARS‐CoV‐2 RBDwt or spike protein (Sinobiological, Beijing, China) at concentrations of 1 μg/ml overnight. ELISA plates were washed with PBS‐0.01% Tween and blocked using 100 μl PBS‐Casein 0.15% for 2 h at RT. Sera from vaccinated mice serially diluted 1:3 starting with a dilution of 1:20 and incubated for 1 h at RT. After washing with PBS‐0.01%Tween, goat anti‐mouse IgG conjugated to Horseradish Peroxidase (HRP) (Jackson ImmunoResearch, 115‐035‐071, West Grove, Pennsylvania) was added at (1:2000) and incubated for 1 h at RT. ELISA was developed with tetramethylbenzidine (TMB), stopped by adding equal 1 M H2SO4 solution, and read at OD450 nm or expressed as Log10 OD50 which is the dilution of half‐maximal absorbance. Detecting RBD‐specific IgGs against mutated RBDs was carried out in a similar way.
To assess the subclass Ab response, the same procedure was performed. The following secondary Abs have been used: Rat anti‐mouse IgG1 (BD Pharmingen, Cat. 559626, 1:2000 dilution), biotinylated mouse anti‐mouse IgG2a (Clone R19‐15, BD Biosciences, Cat No 553391, USA, 1:2000 dilution), goat anti‐mouse IgG2b (Invitrogen, Ref. M32407, 1:2000 dilution), and goat anti‐mouse IgG3 (Southern BioTech, Cat No 1101‐05, 1:2000 dilution).
To detect IgA Abs, ELISA plates were coated with 1 µg/ml RBD protein and goat anti‐mouse IgA POX (ICN 55549, 1:1000 dilution) was used as a secondary Ab. IgG depletion was performed prior to serum incubation. 10 µl of Protein G beads (Invitrogen, USA) were transferred into a tube and placed into a magnet. The liquid was removed, and 75.6 µl diluted sera in PBS‐Casein 0.15% was added to the beads and mixed. The tube was incubated on a rotator at RT for 10 min. The tubes were placed back into the magnet, and ELISA was carried out as described above.
2.9. Avidity (ELISA)
To test the avidity of IgG and IgA Ab against RBD, the above‐described protocol was expanded by an additional step as previously described. 16 Following serum incubation at RT for 1 h, the plates were washed once in PBS/0.01% Tween, and then washed 3× with 7 M urea in PBS‐0.05%Tween or with PBS‐0.05% Tween for 5 min. After washing with PBS‐0.05%Tween, goat anti‐mouse IgG conjugated to Horseradish Peroxidase (HRP) (Jackson ImmunoResearch, West Grove, Pennsylvania) was added (1:2000) and incubated for 1 h at RT. IgA Abs were detected by using a goat anti‐mouse IgA POX (ICN 55549, 1:1000 dilution) detecting Ab. Plates were developed with TMB as described above and read at OD450 nm.
2.10. Bronchoalveolar Lavage
Bronchoalveolar Lavage (BAL) samples were collected as described in Sun et al. 17
2.11. Isolation of lymphocytes
2.11.1. From lung samples
Lungs were perfused with 10 ml of 1 mM EDTA in PBS via the right ventricle of the heart to remove blood cells from the lung vasculature. Lungs were dissected and digested for 30 min at 37°C using RPMI media (2% FBS+Pen/Strep, glutamine, 10 mM HEPES) containing 0.5 mg/ml Collagenase D (Roche). The digested fragments were passed through a 70 μm cell strainer (Greiner bio‐one, Art. Nr. 542070), and RBCs were lysed using ACK buffer. Lymphocytes were isolated using 35% Percoll gradient.
2.11.2. From spleen samples
The spleen was collected from mice and transferred into 5 ml RPMI media (2% FBS+Pen/Strep, glutamine, 10 mM HEPES). A single cell suspension was prepared by passing the spleen through a 70 μm cell strainer. The suspension was collected and transferred into a falcon tube. The tube was centrifuged for 8 min at 4°C and 300×g. ACK lysis was performed, media added and centrifuged for 8 min at 4°C and 300×g. The pellet was resuspended in media.
2.11.3. From bone‐marrow (BM)
Tibia and femur were collected from mice and transferred into 5 ml RPMI media (2% FBS+Pen/Strep, glutamine, 10 mM HEPES). The BM cells were isolated using a syringe by rinsing the bones to flush out the cells. A single cell suspension was prepared by passing the spleen through a 70 μm cell strainer on a petri dish. The suspension was collected and transferred into a falcon tube. Petri dish was washed with 5 ml media and also added to the falcon tube. The tube was centrifuged for 8 min at 4°C and 300×g. ACK lysis was performed, media added and centrifuged for 8 min at 4°C and 300×g. The pellet was resuspended in media.
2.12. Fluorospot
Fluorospot assay was performed according to the manufacturer's protocol (FluoroSpot‐protocol.pdf (mabtech.com). Briefly, Fluorospot plate (Mabtech, Cat no. 3654‐FL) was coated with 100 μl RBD (50 μg/ml) per well, 4°C overnight. The next day, the plate was washed with PBS and blocked for 30 min at RT by the addition of 200 μl incubation medium (RPMI with 10% FBS, glutamine, pen/strep, 10 mm HEPES) per well. 2 × 106 cells from BM, spleen as well as 2 × 105 cells from lung were seeded in 120 μl medium per well. Plate was incubated at 37°C (5% CO2) for 20 h. Cells were removed and the plate was washed with PBS. For detecting IgG secreting plasma cells, a goat anti‐mouse IgG biotin primary Ab (SouthernBiotech, Cat no. 1030–08) at (1:1000) dilution in PBS‐0.1% BSA was used. For detecting IgA secreting plasma cells, goat anti‐mouse IgA biotin (Mabtech, Cat no. 3865‐6‐250) at (1:500) dilution in PBS‐0.1% BSA was used. 100 μl of Abs were added per well and plate incubated for 2 h at RT. Afterward, plate was washed with PBS and Streptavidin‐550 (Mabtech, Cat no. 3310‐11‐1000) diluted in PBS‐0.1% BSA 1:200 was added, 100 μl per well for 1 h at RT. Plate was washed with PBS and Fluorescence enhancer‐II (Mabtech, Cat no. 3641) was added, 50 μl per well for 15 min at RT. The plate was flicked and dried at RT. Finally, plate was read at 550 nm by using the Fluorospot reader (Mabtech IRIS).
2.13. BLI‐based assay
Antibody competitive binding activity was measured on an Octet RED96e (Fortébio) instrument which allows real‐time analysis due to the shift in the wavelength of the reflected light. Anti‐Penta‐HIS (HIS1K, Lot 2006292, FortéBio) biosensors were first loaded into a biosensor microplate and pre‐hydrated in BLI assay buffer (PBS, 0.1% BSA, 0.02% Tween 20) for 10 min. 96‐well microplates were loaded with 200 ml per well. The tips were immobilized with 15 μg/ml Sars‐Cov‐2 spike RBD containing a His‐Tag (Sino Biological, USA). After, they were loaded with sera from mice, diluted 1:20 in BLI assay buffer. Next, association with 50 nM of human receptor ACE2 (Sino Biological, USA) diluted in BLI assay buffer was measured. To regenerate the tips, two additional steps with regeneration buffer (0.1 M glycine, pH 1.5) and neutralization buffer (BLI assay buffer) were performed.
2.14. SARS‐COV‐2 wt and VOC live viruses
The SARS‐CoV‐2 2019‐nCov/Italy‐INMI1 clade V (Wuhan), the B.1.1.7 (UK VOC) named England/MIG457/2020, the B.1.351 (South Africa VOC) namedhCoV19/ Netherlands/NoordHolland_10159/2021, next strain clade 20H were all purchased from European Virus Archive (EVAg). SARS‐CoV‐2 strains were propagated in VERO E6 cells (ATCC—CRL 1586) in T175 Flasks using Dulbecco's Modified Eagle's‐high glucose medium (DMEM) (Euroclone, Pero, Italy) supplemented with 2 mM L‐glutamine (Lonza, Milano, Italy), 100 units/ml penicillin‐streptomycin (Lonza, Milano, Italy, and 2% fetal bovine serum (FBS) (Euroclo, Pero, Italy). All viral growth and neutralization assay with SARS‐CoV‐2 live viruses were performed inside the VisMederi Bisecurity Level 3 laboratories.
2.15. Neutralization Assay cytopathic‐effect‐based (CPE)
The neutralization assay was performed as previously reported by Manenti et al. 18 Briefly, 2‐fold serial dilutions of heat‐inactivated mice serum samples were mixed with an equal volume of viral solution containing between 25 TCID50 of SARS‐CoV‐2. 19 The serum‐virus mixture was incubated for 1 h at 37 °C in a humidified atmosphere with 5% CO2. After the incubation time, 100 μl of the mixture at each dilution point was passed to a 96‐well cell plate containing a sub‐confluent VERO E6 (ATCC—CRL 1586) monolayer. The plates were incubated for 3 days (Wuhan strain) and for 4 days (B.1.1.7 and B.1.351) at 37°C in a humidified atmosphere with 5% CO2. The day of the read‐out each well was inspected by means of an inverted optical microscope to evaluate the percentage of cytophatic effect (CPE) developed in each well. The neutralization titer has been reported as the reciprocal of the highest dilution of serum able to inhibit and prevent at least in 50% of cells the CPE.
2.16. Statistical analysis
Data were analyzed and presented as mean ± SEM using Student's t‐test or one‐way ANOVA as mentioned in the figure legend, with GraphPad PRISM 9. The value of p < .05 was considered statistically significant (*p < .05, **p < .01, ***p < .001, ****p < .0001).
3. RESULTS
3.1. CuMVTT constitute an efficient platform for vaccine development
In order to generate a vaccine‐candidate against SARS‐CoV‐2 for i.n. administration, we have utilized our optimized plant‐derived VLPs (CuMVTT) as a vaccine platform. 15 , 20 , 21 , 22 RBD amino acid sequence (a.a. Arg319‐Phe541) of SARS‐CoV‐2 was chemically coupled to CuMVTT using SMPH bifunctional cross‐linker (Figure 1A). The generated vaccine candidate CuMVTT‐RBD is self‐adjuvated with prokaryotic ssRNA (TLR7/8 agonist) which is packaged during expression and assembly in the bacterial E. coli system (Figure 1B). Efficiency of RBD coupling to CuMVTT was confirmed by SDS‐PAGE (Figure 1C) and Western blot (Figure 1D). The integrity of the VLPs following the coupling process was checked by electron microscopy and showed no signs of aggregation (Figure 1E). Finally, to confirm the antigenicity of CuMVTT‐RBD as well as the correct folding and confirmation of the RBD displayed on the particle`s surface, a receptor binding assay was performed. To this end, the human receptor ACE2 was coated on ELISA plate. CuMVTT‐RBD, CuMVTT, and RBD were added. Anti‐CuMVTT antibodies were used as a secondary antibody to detect receptor bound VLPs. The results revealed that CuMVTT‐RBD can bind to ACE2 receptor indicating correct folding of RBD on the surface of the VLP while the control did not show any binding (Figure 1F).
FIGURE 1.
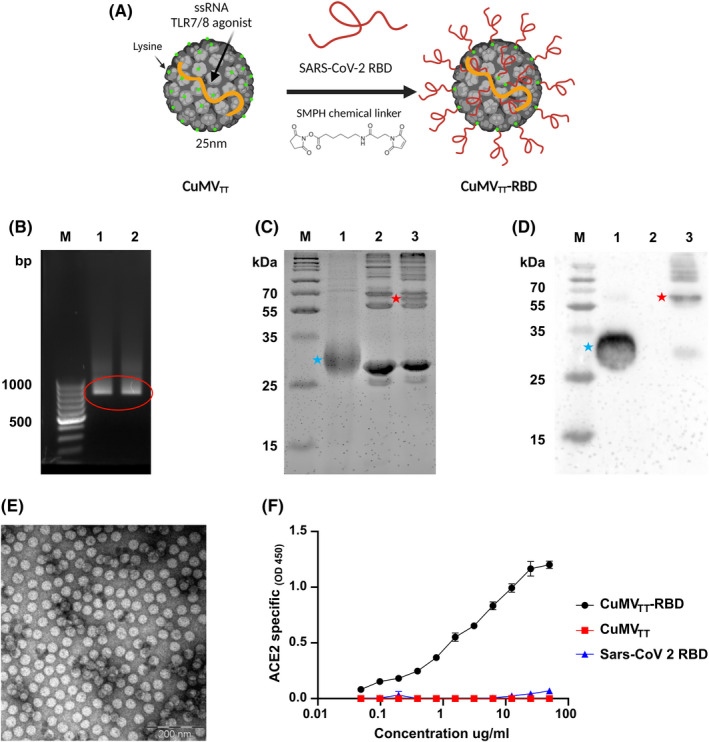
CuMVTT constitute an efficient platform for vaccine development. (A) Schematic representation of the chemical coupling of RBD to CuMVTT via SMPH bifunctional cross‐linker. (B) Agarose gel analysis of CuMVTT‐RBD and CuMVTT depicting nucleic acids packaged in CuMVTT. M. DNA Ladder, 1. CuMVTT‐RBD, 2. CuMVTT containing bands are labelled with red circle. (C) 12% SDS‐PAGE for CuMVTT‐RBD production. M. Protein marker, 1. RBD, 2. CuMVTT, 3. CuMVTT‐RBD post wash. Red star indicate the coupled CuMVTT‐RBD product and blue star indicate the RBD. Bands were visualized with InstantBlueTM Coomassie stain. (D) Western blot specific for RBD. M. Protein marker, 1. RBD, 2. CuMVTT, 3. CuMVTT‐RBD post‐wash. Red star indicate the coupled CuMVTT‐RBD product and blue star indicate the RBD. (E) Electron microscopy of CuMVTT‐RBD, scale bar 200 nm. (F) ACE2 binding of CuMVTT‐RBD, CuMVTT and RBD. Binding revealed with an anti‐CuMV mAb
3.2. Intranasal administration of CuMVTT‐RBD induces a systemic RBD‐ and spike‐specific IgG response of high avidity
To test the immunogenicity and the induction of a humoral immune response in murine models, BALB/c mice were i.n. primed on day 0 and boosted on day 28 with 40 µg of CuMVTT‐RBD vaccine or with 40 µg of CuMVTT as a control without addition of adjuvants. Vaccination and bleeding regimen are shown in Figure 2A. Total systemic RBD and spike‐specific IgG were measured by ELISA. Systemic RBD‐specific IgG response was detected in the group receiving CuMVTT‐RBD seven days after the priming dose. Furthermore, the induced response increased by about 1000‐fold following the booster dose on Day 35 (Figure 2B and C). Full‐length spike protein responses remained low after priming but increased significantly by 30‐folds upon a booster injection resulting in a stable IgG antibody titer (Figure 2D and E). ELISA plates were coated with 1 μg/ml of spike or RBD protein as mentioned in the method section, however, from a molar point of view, about 8‐fold more RBD is coated than spike. This is a likely explanation for the higher reactivity of RBD compared with spike. No IgG response has been detected in the control group which received CuMVTT only.
FIGURE 2.
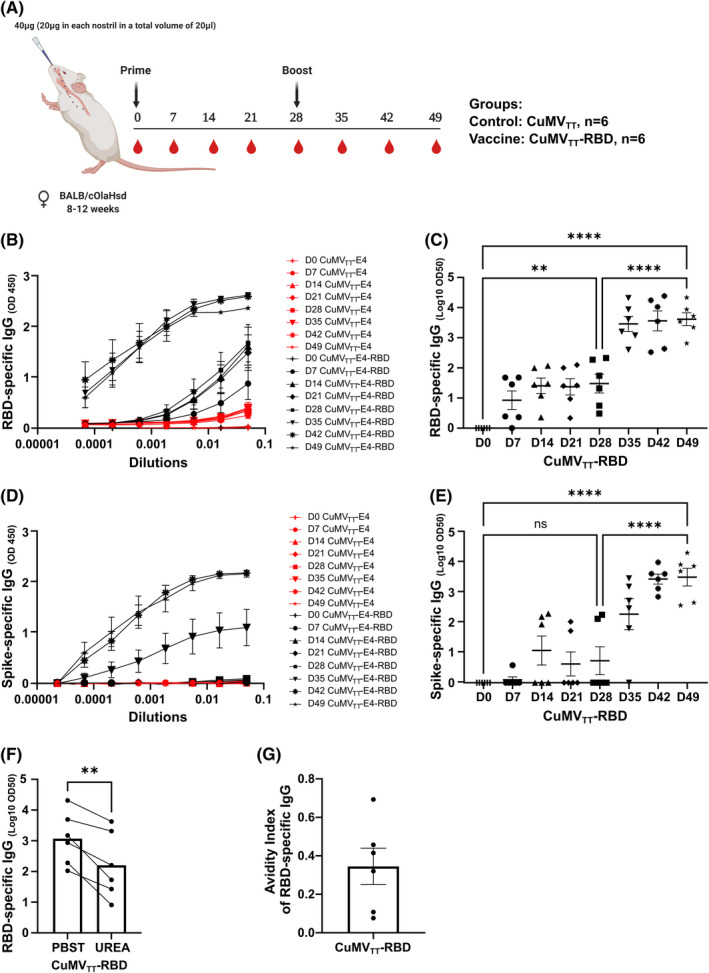
Intranasal administration of CuMVTT‐RBD induces a systemic RBD‐ and spike‐specific IgG response of high avidity. (A) Vaccination regimen and bleeding schedule. (B, C) RBD‐specific IgG titer on Days 0, 7, 14, 21, 35, 42, and 49 from mice immunized with CuMVTT‐RBD vaccine or CuMVTT control measured by ELISA, OD450 shown in B, LOG10 OD50 shown in C. (D, E) Spike‐specific IgG titer on Days 0, 7, 14, 21, 35, 42, and 49 from mice immunized with CuMVTT‐RBD vaccine or CuMVTT control measured by ELISA, OD450 shown in D, LOG10 OD50 shown in E. (F) RBD‐specific IgG titer at Day 49 from mice immunized with CuMVTT‐RBD vaccine, LOG10 OD50 shown. (G) Avidity index. Statistical analysis (mean ± SEM) using one‐way ANOVA (C and E) and Student's t‐test (F). Control group n = 6, vaccine group n = 6. One representative of 2 similar experiments is shown
The avidity of an Ab is defined as the binding strength through points of interaction. It can be quantified as the ratio of K d for the intrinsic affinity over the one for functional affinity of a multiple point interaction. 20 High avidity Abs are formed upon affinity maturation in germinal centers (GCs) and are associated with protective immunity against SARS‐CoV‐2 infection. 23 To assess the avidity of the induced IgG Abs against RBD, we carried out an avidity ELISA using day 49 sera. The obtained results indicated that about 40% of the systemically induced RBD‐specific IgGs are of high avidity following the i.n. vaccination (Figure 2F and G).
3.3. Intranasal immunization with CuMVTT‐RBD promotes isotype switching to IgA and leads to balanced IgG subclass responses
IgG subclasses are of major importance in the immunological response against viruses because of enhancing opsonization as well as immune effector funcitons. 24 In additionally, IgA plays an important role in protection against respiratory viruses as it is found in mucosal tissue, the main entrance site for these kind of viruses. 10 The ability of the CuMVTT‐RBD vaccine to induce serum IgA and IgG subclasses was evaluated by performing ELISA against RBD with sera collected at day 42. All IgG subclasses were induced in vaccinated mice with IgG1, IgG2a, and IgG2b being the dominant ones. In contrast, no IgG subclasses were detected in the control group (Figure 3A). The vaccine was also able to induce isotype switching to IgA as shown in Figure 3B. Approximately, 20% of serum IgA Abs were of high avidity as shown in Figure 3C, D.
FIGURE 3.
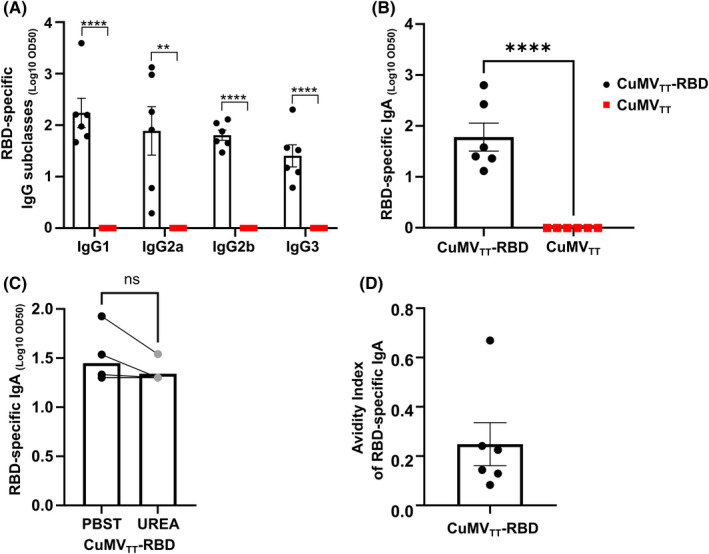
Intranasal immunization with CuMVTT‐RBD promotes isotype switching to IgA and leads to balanced IgG subclass responses. (A) RBD‐specific IgG1, IgG2a, IgG2b, and IgG3 titer for the groups vaccinated with CuMVTT‐RBD vaccine or CuMVTT control on Day 42 measured by ELISA, LOG10 OD50 shown. (B) RBD‐specific IgA titer for the groups vaccinated with CuMVTT‐RBD vaccine or CuMVTT control on Day 42 measured by ELISA, OD450 shown. (C) RBD‐specific IgA titer at Day 42 from mice immunized with CuMVTT‐RBD vaccine, LOG10 OD50 shown. Plates in duplicates: treated with PBSTween or 7 M urea. (D) Avidity index of RBD‐specific IgA titer. Statistical analysis (mean ± SEM) using Student's t‐test. Control group n = 6, vaccine group n = 6. One representative of 2 similar experiments is shown
3.4. CuMVTT‐RBD induced immune sera is able to recognize VOCs
The mutation potential of SARS‐CoV‐2 is considered a burden in vaccine design and development, especially in terms of prolonged protection. Accordingly, we studied the capability of the induced RBD‐specific IgG Abs in recognizing mutant RBDs of the different VOCs. Specifically, we have expressed and produced the following mutated RBDs: RBDK417N, RBDE484K, RBDN501Y, RBDK417N/E484K/N501Y, and RBDL452R/E484Q. 25 Compared with RBDwt, RBD VOCs specific IgG levels were slightly lower (Figure 4). However, the difference observed between RBD VOCs and RBDwt IgG levels was statistically not significant (p = .28). These findings indicate a broad potential binding capacity of Abs induced by i.n. vaccination with CuMVTT‐RBD.
FIGURE 4.
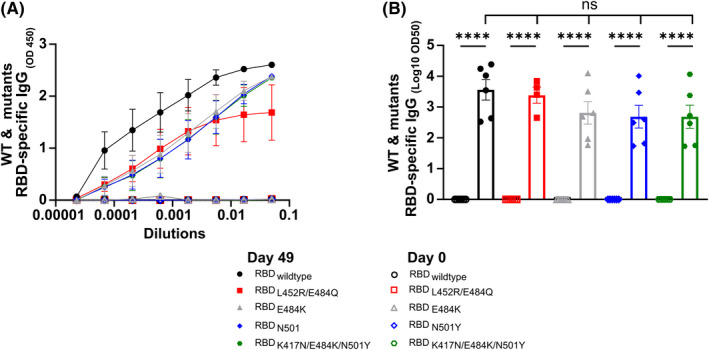
CuMVTT‐RBD induced immune sera is able to recognize VOCs. RBDwt and VOCs‐specific IgG titers on Day 0 and Day 49 for the group vaccinated with CuMVTT‐RBD measured by ELISA, OD450 in A, LOG10 OD50 in B. Statistical analysis (mean ± SEM) using one‐way ANOVA. Control group n = 6, vaccine group n = 6. One representative of 2 similar experiments is shown
3.5. RBD‐ and spike‐specific IgG and IgA were detected locally in BAL, with RBD‐specific IgG2b dominating the local subclass response
To test the ability of the CuMVTT‐RBD vaccine to induce a humoral immune response in the lung mucosa; BAL was collected two weeks after the booster injection (Day 42) and assessed for RBD and spike protein specific IgG and IgA (Figure 5A–F). RBD‐ and spike‐specific IgG Abs were detected at equal levels in the BAL (Figure 5A–C). However, IgA Abs in BAL were more abundant against RBD than against spike protein (Figure 5D–F). We have also assessed the induced RBD‐specific IgG subclasses in BAL. Interestingly, the local mucosal IgG response was less balanced than the serum response and dominated by IgG2b (Figure 5G). Next, we tested the quality of the induced RBD‐specific IgA in BAL samples, the results confirmed that about 60% of detected IgA Abs in BAL were of high avidity (Figure 5H–J).
FIGURE 5.
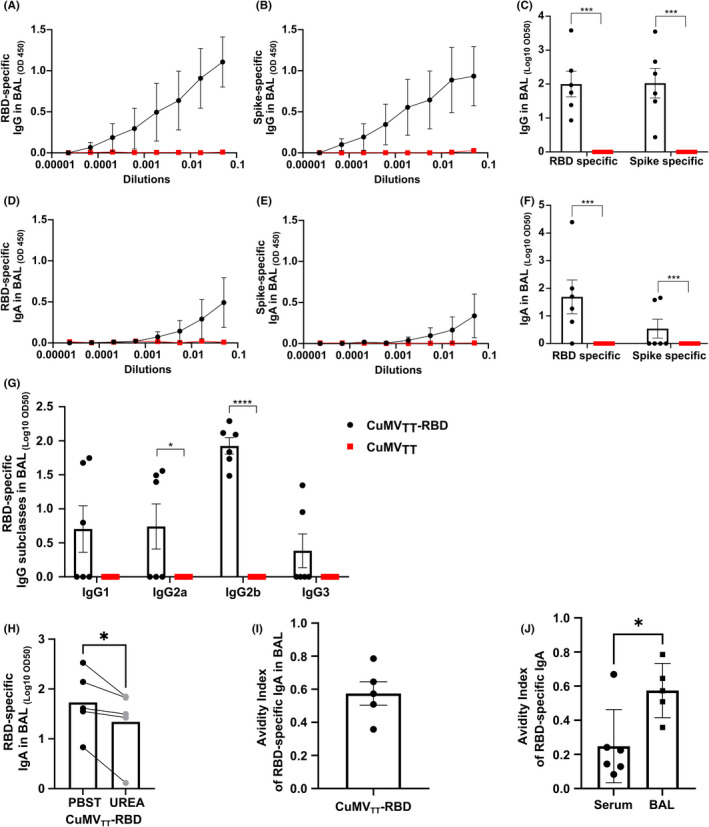
RBD‐ and spike‐specific IgG and IgA were detected locally in BAL, with RBD‐specific IgG2b dominating the local subclass response (A–C) RBD‐ (A, C) and spike‐ (B, C) specific IgG titer in BAL for the groups vaccinated with CuMVTT‐RBD vaccine or CuMVTT control, measured by ELISA, OD450 shown in A, B, LOG10 OD50 shown in C. (D–F) RBD‐ (D, F) and spike‐ (E, F) specific IgA titer in BAL for the groups vaccinated with CuMVTT‐RBD vaccine or CuMVTT control, measured by ELISA, OD450 shown in D, E, LOG10 OD50 shown in F. (G) RBD‐specific IgG1, IgG2a, IgG2b, and IgG3 titer in BAL for the groups vaccinated with CuMVTT‐RBD vaccine or CuMVTT control, measured by ELISA, LOG10 OD50 shown. (H) RBD‐specific IgA titer in BAL from mice immunized with CuMVTT‐RBD vaccine, LOG10 OD50 shown. Plates in duplicates: treated with PBSTween or 7 M urea. (I) Avidity index of RBD‐specific IgA titer. (J) Avidity indexes of RBD‐specific IgA Abs found in BAL and serum after CuMVTT‐RBD immunization. Statistical analysis (mean ± SEM) using Student's t‐test. Control group n = 6, vaccine group n = 6. One representative of 2 similar experiments is shown
3.6. Intranasal administration of CuMVTT‐RBD induced RBD‐specific IgG and IgA plasma cells locally and systemically
In order to characterize the humoral immune response upon i.n. CuMVTT‐RBD vaccination, specific plasma blasts were quantified in lymphoid organs and lung tissue. To this end, spleen, BM and lung were collected on day 42 and analyzed for the presence of RBD‐specific IgG and IgA secreting plasma cells. As shown in Figure 6 and Figure S1, IgG secreting plasma cells were detected in all investigated tissues. Around 25 IgG secreting plasma cells were found per two million cells in spleen and BM. In lung, this ratio was ten‐fold higher because the same amount of IgG plasma cells was observed while ten times less cells were seeded. IgA secreting cells were detected in all three tissues; however, at a lower level compared with IgG secreting plasma cells. RBD‐specific IgA producing plasma cells in lung were thereby about ten times more abundant compared with spleen or BM (Figure 6A–C). In overall term, i.n. vaccination with CuMVTT‐RBD induced a systemic humoral immune response which was accompanied by a potent local humoral immune response in the lung.
FIGURE 6.
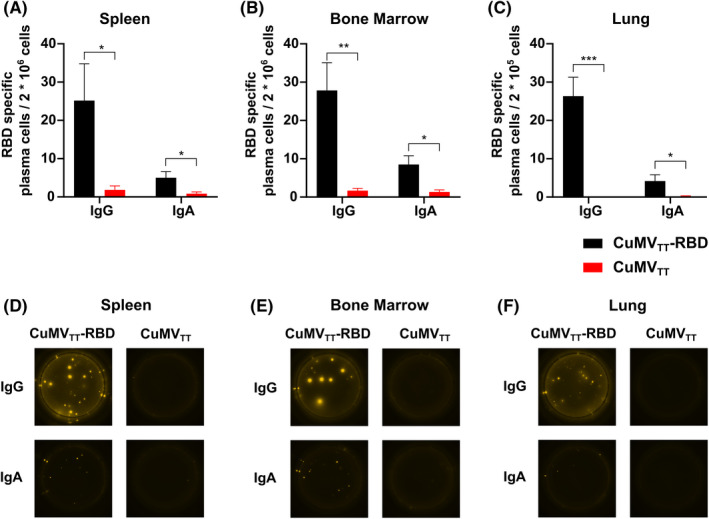
Intranasal administration of CuMVTT‐RBD induced RBD‐specific IgG and IgA plasma cells locally and systemically. (A–C) Number of RBD specific IgG and IgA secreting cells per 2 × 106 seeded cells in spleen (A), BM (B) or per 2 × 105 seeded cells in lung (C) after immunization with CuMVTT‐RBD vaccine or CuMVTT control, detected by Fluorospot. (D–F) Representative pictures of wells seeded with cells out of spleen (D), BM (E), lung (F). Statistical analysis (mean ± SEM) using Student's t‐test. Control group n = 6, vaccine group n = 6. One representative of 2 similar experiments is shown
3.7. Abs induced by intranasal vaccination are capable of neutralizing SARS‐CoV‐2 and its variants
To test the ability of the sera of immunized mice to inhibit binding of RBD to ACE2, a biolayer interferometry competition assay was performed. Accordingly, RBD was immobilized onto anti‐His biosensors and binding capacity of ACE2 to RBD in the presence of serum samples was quantified. As depicted in Figure 7A, the binding of ACE2 to RBD was reduced in the presence of sera from CuMVTT‐RBD vaccinated mice. In contrast, no binding inhibition was observed in the presence of sera from CuMVTT control mice. Percentage of ACE2 to RBD binding inhibition of individual mice is shown in Figure 7B. Interestingly, the binding inhibition correlated with RBD‐specific IgG titers in serum (R value = 0.78) (Figure 7C), indicating higher RBD specific IgG titer in serum are more efficient at blocking RBD‐ACE2 interaction.
FIGURE 7.
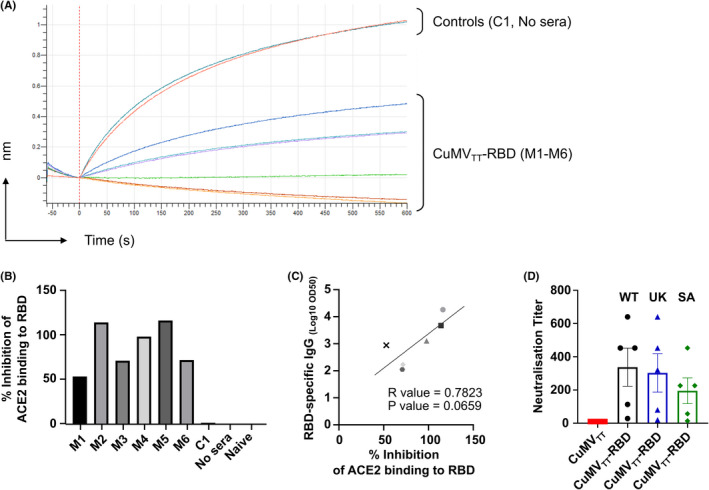
Antibodies induced by intranasal vaccination are capable of neutralizing SARS‐CoV‐2 and its variants. (A) BLI‐evaluation of ACE2 binding to RBD in the presence of vaccinated mice sera or controls. (B) Percentage of ACE2 to RBD binding inhibition of the individual mice vaccinated with CuMVTT‐RBD, control mouse vaccinated with CuMVTT (C1), with no serum and naïve mouse. (C) Correlation of RBD‐specific LOG10 OD50 IgG titer with the percentage of ACE2 to RBD binding inhibition from individual mice sera. Statistical analysis (mean ± SEM) using Pearson Correlation test. (D) Neutralization titer against SARS‐CoV‐2 wt, SA and UK isolates. Statistical analysis (mean ± SEM) using Student's t‐test. Control group n = 1 in a, b, c, n = 5 for D, vaccine group n = 6 for A, B, C, n = 5 for D. One representative of 2 similar experiments is shown
For the evaluation of viral neutralization capacity, sera from CuMVTT‐RBD and CuMVTT immunized mice were tested in a CPE‐based neutralization assay. To this end, sera from immunized mice were assessed for their ability to prevent cytopathic effects of wt SARS‐CoV‐2 as well as VOCs on Vero cells in vitro. As shown in Figure 7D, all sera from CuMVTT‐RBD vaccinated mice were able to neutralize the wt SARS‐CoV‐2 as well as SA and UK variants with high neutralization titers reaching to (1:600). In contrast, no neutralizing capacity was determined for sera from CuMVTT immunized mice.
4. DISCUSSION
The immune responses of the mucosal compartments are considered an early and essential line of defense against harmful pathogens such as SARS‐CoV‐2. 26 The majority of mucosal vaccines have been administered via oral or nasal routes, with the rectal, ocular, sublingual, or vaginal routes being less often used. 27 Ideally, an effective mucosal vaccine would induce both local and systemic responses including the distant mucosal tissues. Accordingly, i.n. administration of a vaccine appears to be a promising strategy.
Seven ongoing clinical trials are currently testing the efficacy of i.n. vaccination against COVID‐19 and are based on live‐attenuated virus, viral‐vectors or protein subunits. Some drawbacks from using attenuated viruses or viral‐vectors includes: pre‐existing Abs that can impair the vaccine efficacy and the risk of reversion of the live‐attenuated viruses especially in newborns or immunocompromised people. In the current study, we have tested the immunogenicity and efficacy of a conventional vaccine based on VLPs displaying RBD of SARS‐CoV‐2 for i.n. administration. The multi‐protein VLP platform does not contain any genetic materials for replication and thus are considered a safe platform for vaccine development. Marketed vaccines against human‐papilloma virus (HPV), hepatitis‐B (HBV), hepatitis‐E virus (HEV) and malaria are based on VLPs. 20 , 28 Our immunologically optimized CuMVTT incorporates a universal TH cell epitope derived from tetanus toxin (TT) and are self‐adjuvanted with prokaryotic ssRNA, a potent TLR7/8 agonist. We have shown in previous studies the essential role of TLR7 signaling in licensing the generation of secondary plasma cells as well as the production of systemic IgA Abs. Such processes are usually dependent on TLR7 expression in B cells; however, in contrast to systemic IgA, a successful induction of mucosal IgA requires TLR signaling in DCs. 29 , 30 Our results here show effective induction of a systemic response of RBD‐specific IgGs one week after the priming dose which increased significantly (p < .001) following the booster dose on Day 28 and antibodies showed a high degree avidity maturation. Moreover, a significant increase in RBD‐specific IgA in serum was observed. Along with this, RBD‐specific IgA and IgG secreting plasma cells were detected in spleen, BM, and within lung tissues. Especially, IgG secreting cells showed a high Ab secretion rate. Consequently, i.n. vaccination with CuMVTT‐RBD is able to induce a strong systemic humoral immune response.
As COVID‐19 presents a respiratory disease and the virus is invading through the respiratory system, an enhanced immunological local protection in the lung should be in the focus when it comes to vaccine design. However, all vaccines currently licensed are applied intramuscularly, 8 thus ignoring this aspect. By applying CuMVTT‐RBD i.n. instead of subcutaneously, 31 we were able to induce RBD—as well as spike‐specific IgA and IgG Abs in the lung. IgA localized in lung mucosa has previously been shown to be of major importance for SARS‐CoV‐2 neutralization. 32 Furthermore, IgA may neutralize the virus in the lung without causing inflammation. 10 The fact that, around 60% of the RBD specific IgA Abs in the lung were of high avidity confirms the quality of the local humoral immune response upon i.n. vaccination with CuMVTT‐RBD. Interestingly, only 20% of the RBD specific IgA Abs were of high avidity in serum. This significant difference might be explained by the 2 different forms of IgA: While IgA Abs at mucosal sides are mostly found in a dimeric form, serum IgA is usually present monomeric form. 33 In addition to IgA, IgG in the lung might also mediate protection. 9 By passive transudation across alveolar epithelium, IgG can pass from blood into the lower lung and from there by the mucociliary escalator further be carried to the upper respiratory tract and nasal passages. However, only at high serum concentrations local protection through IgG is achieved in the lung. 9
While RBD‐specific IgG subclass response in serum was well balanced, IgG2b was the most abundant subclass in BAL. Besides IgG2a, IgG2b is the only subclass that binds all three activating Fc receptors (FcγRI, FcγRIII, and FcγRIV) and the only inhibitory receptor (FcγRIIB) in mice. 34 IgG2b Abs therefore mediate a wide variety of effector functions, which is of key importance in the maintenance of immune protection.
SARS‐CoV‐2 has the ability to mutate, albeit a proof‐reading system is in place to keep the large genome of almost 30 kD genetically stable. 35 Previously, we have described that a single N501Y mutation increased the binding affinity to ACE2, but could still be detected by convalescent sera. Contrary were the results with the E484K mutation, where no enhanced binding to ACE2 was shown but much lower recognition by convalescent sera. Triple mutant RBD (K417N/E484K/N501Y) exhibited both features: stronger affinity to ACE2 and much lower detection by convalescent sera. 36 Since vaccines optimally mediate protection for many years, vaccine induced Abs should therefore be able to recognize new virus variants as well. In the present study, we could show that i.n. applied CuMVTT‐RBD induced serum IgG Abs that are able to recognize wt RBD as well as numerous RBD VOCs.
A crucial milestone in vaccine development is effective neutralization of the virus. Sera induced after i.n. administration of CuMVTT‐RBD could completely inhibit the cytopathic effect of wt SARS‐CoV‐2 as well as other VOCs, specifically SA and UK variants. This may be explained by the highly repetitive, rigid antigenic surface array of VLPs which are spaced by 5 nm and displaying RBD domains at a spacing of 5–10 nm. 37 Such array of highly organized epitopes is considered a pathogen‐associated structural patterns (PASPs) which are recognized by the immune system. 38 In contrast, naturally induced Abs by SARS‐CoV‐2 are low in number and wane rapidly. 39
It has been shown that B and T cells are primed by mucosal vaccination or natural infection, express receptors which promote homing of these cells to mucosal sites as Ab‐secreting cells or effector or tissue‐resident T cells. 40 We have shown in our previous studies that VLP‐specific TH cell response mediate specific B cell isotype‐switch. Furthermore, the packaged RNA in VLPs can drive CD8+ T cells as well as TH1 responses. 41 The role of T cells after i.n. vaccination using VLPs is an area we are currently investigating.
Collectively, we have shown in this study that our COVID‐19 vaccine candidate CuMVTT‐RBD is highly immunogenic and capable of inducing both mucosal and systemic IgG and IgA response against SARS‐CoV‐2 upon i.n. administration. The induced Abs could effectively recognize and neutralize wt as well as the emerging VOCs. The ability of the vaccine candidate to stop nasal viral shedding and transmission is currently under investigation. As COVID‐19 pandemic continues to present a global threat to human health, it seems rational to further develop an i.n. vaccine based on conventional platform.
CONFLICT OF INTERESTS
M. F. Bachmann is a board member of Saiba AG and holds the patent of CuMVTT. Senta A. Walton works for Saiba AG. Mona O. Mohsen received payments by Saiba AG to work on the development of vaccines against Dengue and SARS‐CoV‐2. Martin F. Bachmann and Mona O. Mohsen are shareholder of Saiba AG.
AUTHOR CONTRIBUTIONS
MB, MM, PK, and DR involved in design of experiments. DR, PK, AN, IB, ACV, XC, AM, FV, GR, SW, and MM involved in methodology. DR, PK, MM, MB, ACV, AN, AM, FV, GR, EM, MV, and AZ involved in acquisition of data, and interpretation and analysis of data. DR, PK, MM, and MB involved in writing, revision, and editing of manuscript. EM and AZ involved in technical, material and tool support. MB, MM, and MV involved in study supervision. All authors read and approved the final manuscript.
Supporting information
Fig S1
ACKNOWLEDGEMENT
This publication was funded by Saiba AG, the Swiss National Science Foundation (SNF grants 31003_185114 and IZRPZO_194968). Open access funding provided by Inselspital Universitatsspital Bern.[Correction added on 14‐May‐2022, after first online publication: CSAL funding statement has been added.]
Rothen DA, Krenger PS, Nonic A, et al. Intranasal administration of a VLP‐based vaccine induces neutralizing antibodies against SARS‐CoV‐2 and variants of concern. Allergy. 2022;00:1–13. doi: 10.1111/all.15311
Rothen and Krenger contributed equally to this work
Contributor Information
Dominik A. Rothen, Email: dominik.rothen@dbmr.unibe.ch.
Pascal S. Krenger, Email: pascal.krenger@dbmr.unibe.ch.
REFERENCES
- 1. Swan DA, Bracis C, Janes H, et al. COVID‐19 vaccines that reduce symptoms but do not block infection need higher coverage and faster rollout to achieve population impact. Sci Rep. 2021;11:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peiris JSM, Chu CM, Cheng VCC, et al. Clinical progression and viral load in a community outbreak. Lancet. 2003;361:1767‐1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun K, Gu L, Ma L, Duan Y. Atlas of ACE2 gene expression reveals novel insights into transmission of SARS‐CoV‐2. Heliyon. 2021;7(1):e05850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jia HP, Look DC, Shi L, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79:14614‐14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harrison AG, Lin T, Wang P. Mechanisms of SARS‐CoV‐2 Transmission and Pathogenesis. Trends Immunol. 2020;41:1100‐1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS‐CoV‐2 receptor ace2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016‐1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krammer F. SARS‐CoV‐2 vaccines in development. Nature. 2020;586:516‐527. [DOI] [PubMed] [Google Scholar]
- 8. https://www.clinicaltrials.gov/
- 9. Renegar KB, Small PA, Boykins LG, Wright PF. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J Immunol. 2004;173:1978‐1986. [DOI] [PubMed] [Google Scholar]
- 10. Rodríguez A, Tjärnlund A, Ivanji J, et al. Role of IgA in the defense against respiratory infections: IgA deficient mice exhibited increased susceptibility to intranasal infection with Mycobacterium bovis BCG. Vaccine. 2005;23:2565‐2572. [DOI] [PubMed] [Google Scholar]
- 11. Allie SR, Bradley JE, Mudunuru U, et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat Immunol. 2019;20:97‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Doremalen N, et al. Intranasal ChAdOx1 nCoV‐19/AZD1222 vaccination reduces shedding of SARS‐CoV‐2 D614G in rhesus macaques. Serv Biol. 2021. doi: 10.1101/2021.01.09.426058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mohn KGI, Smith I, Sjursen H, Cox RJ. Immune responses after live attenuated influenza vaccination. Hum Vacc Immunother. 2018;14:571‐578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bessa J, et al. Efficient induction of mucosal and systemic immune responses by virus‐like particles administered intranasally: Implications for vaccine design. Eur J Immunol. 2008;38:114‐126. [DOI] [PubMed] [Google Scholar]
- 15. Zeltins A, West J, Zabel F, et al. Incorporation of tetanus‐epitope into virus‐like particles achieves vaccine responses even in older recipients in models of psoriasis, Alzheimer’s and Cat Allergy. NPJ Vaccines. 2017;2:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wiuff C, Thorberg BM, Engvall A, Lind P. Immunochemical analyses of serum antibodies from pig herds in a Salmonella non‐endemic region. Vet Microbiol. 2002;85:69‐82. [DOI] [PubMed] [Google Scholar]
- 17. Sun F, Xiao G, Qu Z. Murine bronchoalveolar lavage. Physiol Behav. 2017;176:139‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manenti A, et al. Evaluation of SARS‐CoV‐2 neutralizing antibodies using a CPE‐based colorimetric live virus micro‐neutralization assay in human serum samples. J Med Virol. 2020;92:2096‐2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manenti A, Molesti E, Maggetti M, et al. The theory and practice of the viral dose in neutralization assay: Insights on SARS‐CoV‐2 ‘doublethink’ effect. J Virol Methods. 2020;297:114261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mohsen MO, Augusto G, Bachmann MF. The 3Ds in virus‐like particle based‐vaccines: “Design, Delivery and Dynamics”. Immunol Rev. 2020;296:155‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mohsen MO, Balke I, Zinkhan S, et al. A scalable and highly immunogenic virus‐like particle‐based vaccine against SARS‐CoV‐2. Allergy Eur J Allergy Clin Immunol. 2022;77(1):243‐257. doi: 10.1111/all.15080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mohsen MO, Rothen D, Balke I, et al. Neutralization of MERS coronavirus through a scalable nanoparticle vaccine. NPJ Vaccines. 2021;6:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bauer G. The potential significance of high avidity immunoglobulin G (IgG) for protective immunity towards SARS‐CoV‐2. Int J Infect Dis. 2021;106:61‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hazenbos WL, et al. Murine IgG1 complexes trigger immune effector functions predominantly via Fc gamma RIII (CD16). J Immunol. 1998;161:3026‐3032. [PubMed] [Google Scholar]
- 25. Chang X, Augusto GS, Liu X, et al. BNT162b2 mRNA COVID‐19 vaccine induces antibodies of broader cross‐reactivity than natural infection, but recognition of mutant viruses is up to 10‐fold reduced. Allergy Eur J Allergy Clin Immunol. 2021;76(9):2895‐2998. doi: 10.1111/all.14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Russell MW, Moldoveanu Z, Ogra PL, Mestecky J. Mucosal immunity in COVID‐19: A neglected but critical aspect of SARS‐CoV‐2 infection. Front Immunol. 2020;11:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Czerkinsky C, Holmgren J. Mucosal delivery routes for optimal immunization. Curr Top Microbiol Immunol. 2012;354:1‐18. [DOI] [PubMed] [Google Scholar]
- 28. Mohsen MO, Zha L, Cabral‐Miranda G, Bachmann MF. Major findings and recent advances in virus–like particle (VLP)‐based vaccines. Semin Immunol. 2017;34:123‐132. [DOI] [PubMed] [Google Scholar]
- 29. Krueger CC, Zabel F, Keller E, et al. RNA and toll‐like receptor 7 license the generation of superior secondary plasma cells at multiple levels in a B cell intrinsic fashion. Front Immunol. 2019;10:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bessa J, Jegerlehner A, Hinton HJ, et al. Alveolar macrophages and lung dendritic cells sense RNA and drive mucosal IgA responses. J Immunol. 2009;183:3788‐3799. [DOI] [PubMed] [Google Scholar]
- 31. Zha L, Chang X, Zhao H, et al. Development of a vaccine against sars‐cov‐2 based on the receptor‐binding domain displayed on virus‐like particles. Vaccines. 2021;9:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Z, Lorenzi JCC, Muecksch F, et al. Enhanced SARS‐CoV‐2 neutralization by dimeric IgA. Sci Transl Med. 2021;13(577):eabf1555. doi: 10.1126/scitranslmed.abf1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Steffen U, Koeleman CA, Sokolova MV, et al. IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat Commun. 2020;11. doi: 10.1038/s41467-019-13992-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gosselin EJ, Bitsaktsis C, Li Y, Iglesias BV. Fc receptor‐targeted mucosal vaccination as a novel strategy for the generation of enhanced immunity against mucosal and non‐mucosal pathogens. Arch Immunol Ther Exp (Warsz). 2009;57:311‐323. [DOI] [PubMed] [Google Scholar]
- 35. Robson F, Khan KS, Le TK, et al. Coronavirus RNA proofreading: Molecular basis and therapeutic targeting. Mol Cell. 2020;79:710‐727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vogel M, Augusto G, Chang X, et al. Molecular definition of severe acute respiratory syndrome coronavirus 2 receptor‐binding domain mutations: Receptor affinity versus neutralization of receptor interaction. Allergy. 2022;77(1):143‐149. doi: 10.1111/all.15002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bachmann MF, Mohsen MO, Zha L, Vogel M, Speiser DE. SARS‐CoV‐2 structural features may explain limited neutralizing‐antibody responses. NPJ Vaccines. 2021;6:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bachmann MF, Jennings GT. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol. 2010;10:787‐796. [DOI] [PubMed] [Google Scholar]
- 39. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID‐19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363‐374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pizzolla A, Nguyen TH, Wakin LM, et al. Resident memory CD8+ T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci Immunol. 2017;2:1‐14. [DOI] [PubMed] [Google Scholar]
- 41. Skibinski DAG, Hanson BJ, Lin Y, et al. Enhanced Neutralizing Antibody Titers and Th1 Polarization from a Novel Escherichia coli Derived Pandemic Influenza Vaccine. PLoS One. 2013;8:e76571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1


