Abstract
Shaziling pig, a Chinese indigenous breed, has been classified as a fatty pig model. However, the gut microbial development and role in lipid metabolism in Shaziling pigs has been rarely reported. Here, we compared the lipid metabolic and microbial profiles at 30, 60, 90, 150, 210, and 300 d of age between Shaziling and Yorkshire pigs. Predictably, there were marked differences in the liver lipids (i.e., cholesterol, glucose, and low-density lipoprotein) and the lipid related expressions (i.e., SREBP1/2, LXRα/β, DGAT1/2, and FABP1-3) between Shaziling and Yorkshire pigs. Bacteria sequencing in the ileal digesta and mucosa showed that Shaziling pigs had a higher α-diversity and higher abundances of probiotics, such as Lactobacillus johnsonii, Lactobacillus amylovorus, and Clostridium butyricum. Thirty-five differentiated metabolites were further identified in the mucosa between Shaziling and Yorkshire pigs, which were enriched in the carbohydrate, protein, glucose and amino acid metabolism and bile acid biosynthesis. Furthermore, 7 differentiated microbial species were markedly correlated with metabolites, indicating the role of gut microbiota in the host metabolism. Next, the role of differentiated L. johnsonii in lipid metabolism was validated in Duroc × Landrace × Yorkshire (DLY) pigs and the results showed that L. johnsonii mono-colonization promoted lipid deposition and metabolism by altering gut microbiota (i.e., Megasphaera elsdenii and L. johnsonii) and DGAT1/DGAT2/CD36-PPARγ gene expressions. In conclusion, Shaziling pigs exhibited different metabolic and microbial profiles compared with Yorkshire pigs, which might have contributed to the diverse metabolic phenotypes, and the significant enrichment of L. johnsonii in Shaziling pigs promoted lipid metabolism and obesity of DLY pigs, which provided a novel idea to improve the fat content of lean pigs.
Keywords: Shaziling pig, Gut microbiota, Lipid metabolism, Lactobacillus johnsonii, Fatty acid
1. Introduction
The Shaziling pig, a famous indigenous breed in China, is characterized by higher intramuscular fat and meat quality compared with imported pig breeds. Previous studies have mainly discussed the differences in genetics, involving epigenetics, quantitative genetics, and metagenomics between Shaziling and introduced pig breeds (Xu et al., 2015; Yang et al., 2016). However, there are few studies on the regulation of Shaziling pork quality from the perspective of nutrition, which hinders the breeding of Shaziling pigs and the promotion of meat products. Meanwhile, indigenous breeds are rich in microbial resources, which play an important role in host metabolism (Li et al., 2020; Fan and Pedersen, 2021; Yang et al., 2021), meat quality (Khanal et al., 2020), feed efficiency (Li et al., 2021; Wen et al., 2021), and gut development (Guevarra et al., 2018; Hu et al., 2018; Y. Li et al., 2021) in farming animals. Thus, analyzing and screening gut microbiota in Shaziling pigs may enrich the probiotic library and guide a more healthy pig industry (Azad et al., 2020).
In recent years, with the development of germ-free animals and microbiome analysis (16S rDNA sequencing, metagenomics), it has been widely confirmed that gut microbiota are involved in the regulation of host lipid metabolism (Yin et al., 2018; Aron-Wisnewsky et al., 2020, 2021; Li et al., 2020; Zhou et al., 2021). In addition, fecal microbial transplantation and probiotics treatment has further confirmed that remodeling gut microbial compositions improves host lipid metabolism (Korpela et al., 2020; Yin et al., 2020b), which may further target fat deposition in the muscle. However, microbial compositions have not been analyzed in Shaziling pigs, thus the current study aimed to compare the microbial and metabolic differences between Shaziling and Yorkshire pigs (an imported lean pig breed) to provide a microbial perspective for the research on meat quality of indigenous pig breeds in China.
2. Materials and methods
The animal model and experimental procedures used in this experiment were approved by the Hunan Agricultural University Institutional Animal Care and Use Committee (202005).
2.1. Bacterial strains and reagents
The Lactobacillus johnsonii (BNCC135265) strains used in this study were purchased by Beijing Beina Chuanglian Biotechnology Institute (Beijing, China). Unless otherwise stated, bacterial strains were grown in MRS broth (MRSB, Qingdao Hope Bio-technology) or on MRS agar (MRSA) plates at 37 °C, pH = 6.2.
2.2. Animals and diets
Shaziling pig (a local breed in Hunan province, China) was selected as the fatty pig model, and Yorkshire pig as the lean animal model. All pigs were given free access to basic diets (satisfying the nutritional requirements) and drinking water and slaughtered at 30, 60, 90, 150, 210, 300 d of age. Then, 12 male pigs (Duroc × Landrace × Yorkshire, 28.6 ± 2.0 kg) with a terminal ileal fistula were divided into 2 groups, including a control group and a L. johnsonii group, in which pigs received L. johnsonii mono-colonization for continuous 14 d. Samples of blood, liver, ileal chyme (at 0, 1, 3, 5, 7, 10 and 14 d) and mucosa, abdominal adipose tissue and muscle were collected and stored at −80 °C (n = 6).
2.3. Biochemical parameter analysis
The liver samples (0.1 g each) were homogenized in a centrifuge tube with 0.9 mL normal saline and the blood was collected at slaughter, then the supernatant was extracted and centrifuged at 1,500 × g, 4 °C for 10 min. Triglyceride (TG), total cholesterol (TC), total bile acid (TBA), glucose (GLU), high density lipoprotein (HDL), and low-density lipoprotein (LDL) were detected by an automatic biochemical instrument (KHB 450, Shanghai Kehua bio-engineering co., Ltd).
2.4. Reverse transcription-PCR
Mucosa, muscle, adipose tissue, and liver samples were frozen in liquid nitrogen and ground, and total RNA was isolated by using TRIzol reagent (Invitrogen, USA) and then treated with DNase I (Invitrogen, USA). Reverse transcription was conducted at 37 °C for 15 min at 95 °C for 5 s. The primers used in this study were designed according to the pig sequence (Table 1). PCR cycling and relative expression determination were performed according to previous studies (Yin et al., 2018).
Table 1.
Primers used for gene expression analysis by real-time PCR.
| Gene | Forward sequence (5′–3′) | Reverse sequence (5′–3′) |
|---|---|---|
| β-actin | CTACGCCAACACGGTGCTGTC | CTCCTGCTTGCTGATCCACATCTG |
| GAPDH | TCGGAGTGAACGGATTTGGC | TGACAAGCTTCCCGTTCTCC |
| ACC | CGGAATATCCAGAAGGCCGA | CCAGTCCGATTCTTGCTCCA |
| PPARα | CAGCAATAACCCGCCTTTCG | CTCCTTGTTCTGGATGCCGT |
| PPARγ | GCAGGAGCAGAGCAAAGAGGTG | GCCAGGTCGCTGTCATCTAATTCC |
| SREBP1 | CACGGAGGCGAAGCTGAATA | CTGGTTGCTCTGCTGAAGGA |
| SREBP2 | TGCTCTTGTGGCTGGTAAATGGTG | CGCTGCTCTTGGCTTCATCCTC |
| LXRα | GACAAGGGACTGCACCATCC | CCTCCACCCACAAGGACATC |
| LXRβ | GAACAAGGGGACGAAAGCAG | TGAAGGGGACCATTACCACC |
| MOGAT2 | GGCGCTCTGAGGCCATAA | AAGGGGACCCGAAACCAC |
| DGAT1 | AACCTGACCTACCGCGATCT | GGAAGCGGGAAAAGTTGAGC |
| DGAT2 | GCGGGAGTACCTGATGTCTG | AACCAGGTCGGCTCCGT |
| CD36 | GCATCACAGCCTACACCACAGC | CGAGCCAGAGATTGAACCCACATC |
| FABP1 | ATCACTACCGGGTCCAAGGT | CAACTGAACCACTGTCTTGACC |
| FABP2 | CGGAACTGAACTCACTGGGAA | CTGGACCATTTCATCCCCGA |
| FABP3 | GATGACAGGAAGGTCAAGTCCA | TAAGTGCGAGTGCAAACTGC |
| FABP4 | AAGAAGTGGGAGTGGGCTTTGC | ATTCTGGTAGCCGTGACACCTTTC |
ACC = acetyl-CoA carboxylase; PPARα = peroxisome proliferator activated receptor α; PPARγ = peroxisome proliferator activated receptor γ; SREBP1 = cholesterol regulatory element binding protein 1; SREBP2 = cholesterol regulatory element binding protein 1; LXRα = liver X receptor α; LXRβ = liver X receptor β; MOGAT2 = monoacylglycerol-O-acyltransferase 2; DGAT1 = diacylglycerol Acyltransferase 1; DGAT2 = diacylglycerol acyltransferase 2; CD36 = cluster of differentiation 36; FABP1 = fatty acid-binding protein 1; FABP2 = fatty acid-binding protein 2; FABP3 = fatty acid-binding protein 3; FABP4 = fatty acid-binding protein 4.
2.5. Bacterial profiling
Total genome DNA from ileal chyme and mucosa were extracted using the cetyltriethylammnonium bromide (CTAB) method, and DNA concentration and purity were monitored on 1% agarose gels. According to the concentration, DNA was diluted to 1 ng/μL using sterile water. 16S rDNA genes of distinct regions (16S V3–V4) were amplified using specific primer. All PCR reactions were carried out with 15 μL of Phusion High-Fidelity PCR Master Mix (New England Biolabs); 2 μmol/L of forward and reverse primers, and about 10 ng template DNA. Sequencing libraries were generated using TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, USA) following the manufacturer's recommendations and index codes were added according to our previous study (Yin et al., 2020a). Microbial communities were investigated by iTag sequencing of 16S rDNA genes (Coffey et al., 2019).
2.6. Mucosal metabolomics analysis
Thirty milligrams ileal mucosa was transferred to a 1.5-mL tube to perform metabolomics analysis via a LC–MS system. A volume of 20 μL of L-2-chlorophenylalanine (0.3 mg/mL) dissolved in methanol as an internal standard and a 0.4-mL mixture of methanol and water (4:1, vol:vol) were added to each sample. Samples were ground and then extracted by ultrasonic wave for 10 min in ice-water bath, stored at −20 °C for 30 min. The extract was centrifuged at 15,871 × g at 4 °C for 10 min (Eppendorf, 5452R). A volume of 300 μL supernatant in a glass vial was dried in a freeze concentration centrifugal dryer. A volume of 300 μL mixture of methanol and water (1:4, vol:vol) was added to each sample, samples were vortexed for 30 s, extracted by ultrasonic wave for 3 min in an ice-water bath, then placed at −20 °C for 2 h. Samples were centrifuged at 15,871 × g at 4 °C for 10 min. The supernatants (150 μL) from each tube were collected using crystal syringes, filtered through 0.22-μm microfilters and transferred to LC vials. Then the samples were analyzed by LC–MS system to detect the mucosal metabolic profiles.
2.7. Medium and long chain fatty acids analysis
Medium and long chain fatty acids were determined based on the GC–MS system. Muscle and chyme samples (50 mg) were accurately weighed into a 2-mL tube and 200 μL serum was extracted to a 1.5-mL tube. Then, 1-mL chloroform and methanol (1:1, vol:vol) was added, and the samples were freeze ground for 3 min (50 Hz, twice), subjected to low temperature ultrasound for 15 min, stood at −20 °C for 15 min, and centrifugated at 13,000 × g at 4 °C for 10 min (Eppendorf, 5452R). Next, the supernatant was absorbed into a 1.5-mL EP (Eppendorf) tube and dried with nitrogen. After nitrogen drying, 0.5 mL methylation reagent (0.5 mol/L sodium hydroxide methanol solution) was added and eddied for 30 s, and then water bath was carried out at 60 °C for 0.5 h. After the water bath had cooled, 0.5 mL n-hexane was added, and the vortex was rotated for 30 s at 4 °C and centrifugated at 13,000 × g at 4 °C for 10 min. Then the supernatant (n-hexane layer) was absorbed into a 1.5-mL tube, dried with nitrogen, and 100 μL n-hexane was added to re-dissolve. Vortex and low temperature ultrasound were performed for 10 min, then centrifugation at 13,000 × g at 4 °C for 10 min, and then finally, the supernatant was taken into the vial for GC–MS system.
2.8. Statistical analysis
All statistical analyses were performed using t-test and Pearson correlation analysis in SPSS 20.0 software (SPSS Inc., Chicago, Illinois, USA). The data are expressed as the means ± standard errors of the means (SEM). P < 0.05 was considered significant. All figures in this study were drawn by using GraphPad Prism 8.0.
3. Results
3.1. Lipid metabolism between Shaziling and Yorkshire pigs
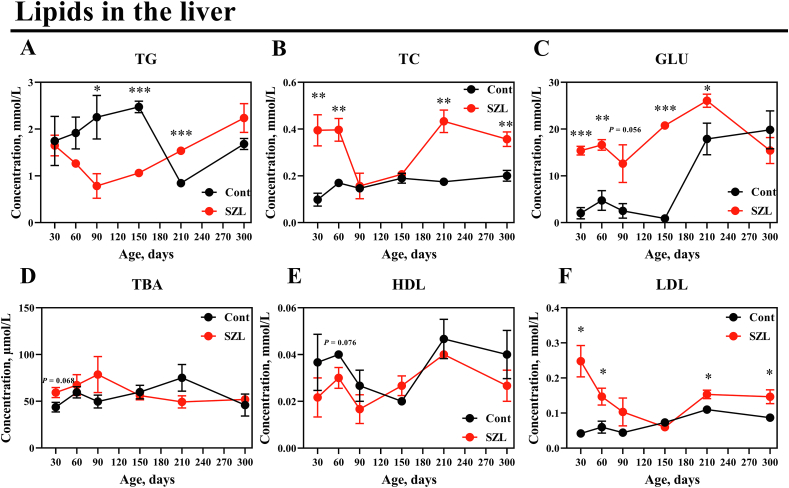
Exogenous intake and free lipids are assembled in the liver and distributed to the tissues of the body (Ko et al., 2020). Therefore, lipid levels were analyzed between Shaziling and Yorkshire pigs and the concentrations of TC and LDL were distinctly higher in Shaziling pigs at 30, 60, 210 and 300 d. GLU was increased in Shaziling pigs at 30, 60, 150, and 210 d, whereas TG was lowered at 90 and 150 d and higher at 210 d in Shaziling pigs (Fig. 1).
Fig. 1.
Lipids were altered between Shaziling and Yorkshire pigs. (A) TG, (B) TC, (C) GLU, (D) TBA, (E) HDL and (F) LDL were detected using t-test between Shaziling and Yorkshire pigs using SPSS 20.0. Figures were drawn using GraphPad Prism 8.0. Values are presented as the means ± SEM. ∗, P < 0.05, ∗∗, P < 0.01, ∗∗∗, P < 0.001. SZL = Shaziling pigs, Cont = Yorkshire pigs; TG = triglycerides; TC = total cholesterol; GLU = glucose; TBA = total bile acid; HDL = high-density lipoprotein; LDL = low-density lipoprotein.
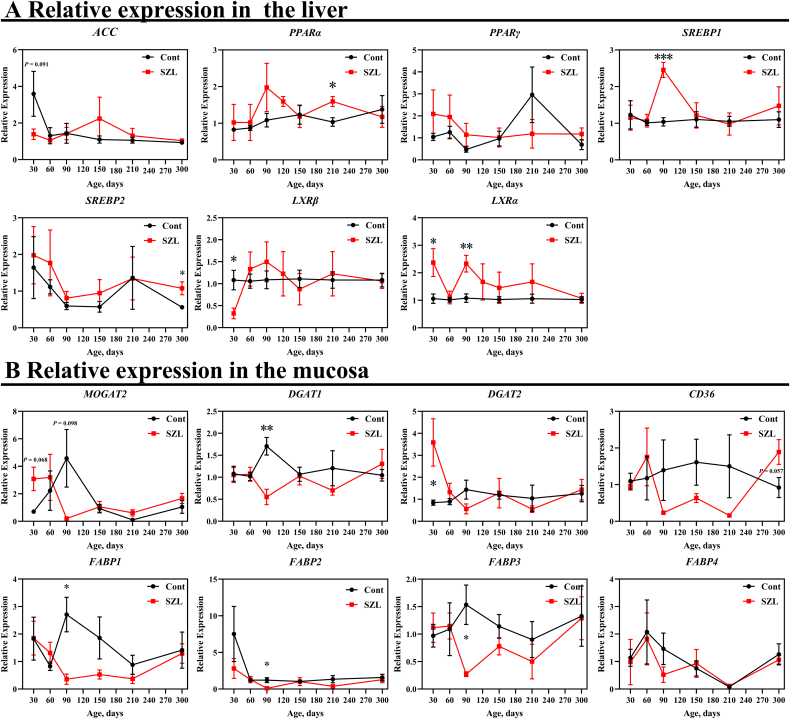
Then, we analyzed the expression of several genes associated with lipid metabolism in the mucosa, including monoacylglycerol-O-acyltransferase 2 (MOGAT2), diacylglycerol acyltransferase 1 (DGAT1), diacylglycerol Acyltransferase 2 (DGAT2), cluster of differentiation 36 (CD36), fatty acid-binding protein 1 (FABP1), fatty acid-binding protein 2 (FABP2), fatty acid-binding protein 3 (FABP3) and fatty acid-binding protein 4 (FABP4)) and liver (i.e., acetyl-CoA carboxylase (ACC), peroxisome proliferator activated receptor α (PPARα), peroxisome proliferator activated receptor γ (PPARγ), cholesterol regulatory element binding protein 1 (SREBP1), cholesterol regulatory element binding protein 2 (SREBP2), liver X receptor α (LXRα), liver X receptor β (LXRβ). Compared with the Yorkshire pigs, the expressions of PPARα, SREBP1, SREBP2, and LXRα tended to be higher in Shaziling pigs, respectively (Fig. 2A), whereas LXRβ mRNA abundance was lower at 30 d (Fig. 2A). In the mucosa, DGAT1, FABP1, FABP2, and FABP3 expressions were lower at 90 d in Shaziling pigs (Fig. 2B), whereas, DGAT2 was higher at 30 d (Fig. 2B). In summary, lipid metabolism and absorption related genes were differentiated between Shaziling and Yorkshire pigs, which might explain the different metabolic phenotypes.
Fig. 2.
Gene expression in the liver and mucosa. Relative expressions in the (A) liver and (B) mucosa were detected using t-test between Shaziling and Yorkshire pigs using SPSS 20.0. Figures were drawn in GraphPad Prism 8.0. Values are presented as the means ± SEM. ∗, P < 0.05, ∗∗, P < 0.01, ∗∗∗, P < 0.001. ACC = acetyl-CoA carboxylase; PPARα = peroxisome proliferator activated receptor α; PPARγ = peroxisome proliferator activated receptor γ; SREBP1 = cholesterol regulatory element binding protein 1; SREBP2 = cholesterol regulatory element binding protein 1; LXRα = liver X receptor α; LXRβ = liver X receptor β; MOGAT2 = monoacylglycerol-O-acyltransferase 2; DGAT1 = diacylglycerol acyltransferase 1; DGAT2 = diacylglycerol acyltransferase 2; CD36 = cluster of differentiation 36; FABP1 = fatty acid-binding protein 1; FABP2 = fatty acid-binding protein 2; FABP3 = fatty acid-binding protein 3; FABP4 = fatty acid-binding protein 4.
3.2. Bacterial development in the ileal cavity
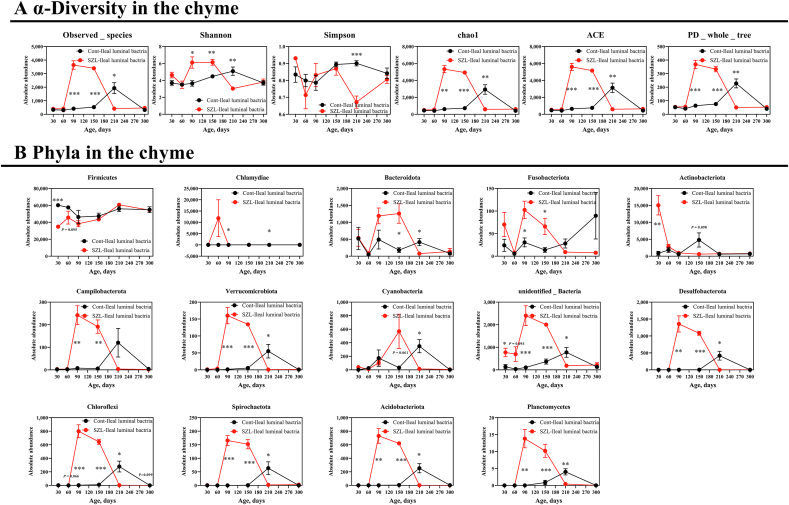
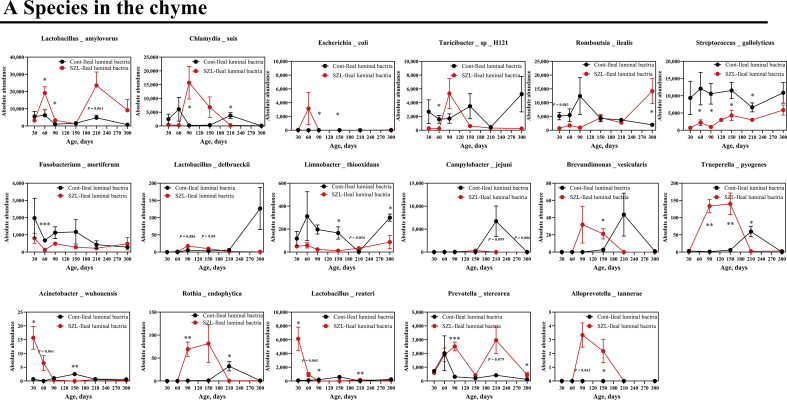
Next, we aimed to compare bacterial alterations at different ages between Shaziling and Yorkshire pigs. The results showed that the alpha diversity indices, including observed_species, Shannon, Chao1, abundance-based coverage estimator (ACE), and phylogenetic diversity (PD) whole tree of Shaziling pigs were significantly higher at d 90 and 150 compared with the lean subjects (Fig. 3A), whereas the alpha diversity was lowered at 210 d (Fig. 3A). Next, we analyzed the bacterial changes at the phylum level and found that 14 out of 15 phyla were altered and exhibited a time-dependent pattern, including Firmicutes, Chlamydiae, Bacteroidota, Fusobacteriota, Actinobacteriota, Campilobacterota, Verrucomicrobiota, Cyanobacteria, unidentified Bacteria, Desulfbacterota, Choroflexi, Spirochaetota, Acidobacteriota, and Planctomycetes (Fig. 3B), and 17 out of 20 species were identified to be differentiated between 2 datasets, such as Lactobacillus amylovorus, Chlamydia suis, Escherichia coli, Turicibacter sp. H121, Romboutsia ilealis, Streptococcus gallolyticus, Fusobacterium mortiferum, Lactobacillus delbrueckii, Limnobacter thiooxidans, Campylobacter jejuni, Brevundimonas vesicularis, Trueperella pyogenes, Acinetobacter wuhouensis, Rothia endophytica, Lactobacillus teuteri, Prevotella stercorea, and Alloprevotella tannerae (Fig. S1A).
Fig. 3.
Microbial development in the chyme. (A) α-Diversity and (B) phyla were assessed using t-test between Shaziling and Yorkshire pigs using SPSS 20.0. Figures were drawn in GraphPad Prism 8.0. Values are presented as the means ± SEM. ∗, P < 0.05, ∗∗, P < 0.01, ∗∗∗, P < 0.001.
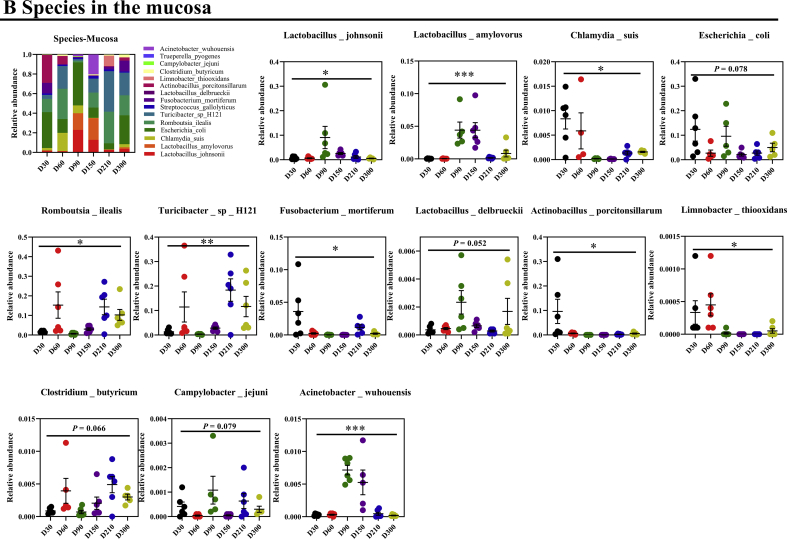
3.3. Commensal bacteria profiles between Shaziling and Yorkshire pigs
A previous study has indicated that the host metabolism is mainly governed by mucosal commensal bacteria (Cai et al., 2020), which has not been well-studied compared with chymous and fecal microbiota. Thus, we further explored the commensal bacteria in the mucosa between Shaziling and Yorkshire pigs. Similarly, the results showed that the α-diversity of Shaziling pigs was significantly higher at 90 and 150 d (Fig. 4A) and the principal component was markedly differentiated between the 2 datasets (Fig. 4A). In addition, the top 10 phyla were markedly differentiated between Shaziling and Yorkshire pigs, including Firmicutes, Proteobacteria, Chlamydiae, Fusobacteriota, Bacteroidota, Actinobacteriota, Campilobacterota, Verrucomicrobiota, Cyanobacteria, and unidentified Bacteria (Fig. 4B). At the species level, we found that 13 out of 20 species were markedly changed throughout the entire stage, including L. johnsonii, L. amylovorus, C. suis, E. coli, R. ilealis, Turicibacter sp. H121, F. mortiferum, L. delbrueckii, Actinobacillus porcitonsillarum, L. thiooxidans, Clostridium butyricum, C. jejuni, and A. wuhouensis (Fig. S1B). At 300 d, Shaziling pigs colonized higher abundances of L. johnsonii, L. amylovorus, C. suis, Turicibacter sp. H121, and C. butyricum, but lowered F. mortiferum and C. jejuni compared with the Yorkshire pigs (Fig. 4C). Together, there were marked differences in commensal microbiota between Shaziling and Yorkshire pigs, which might affect host lipid metabolism.
Fig. 4.
Commensal bacteria in the Shaziling pigs. (A) Diversity and (B) phyla in the mucosa were assessed using one-way ANOVA by comparing at the different ages (30, 60, 90, 150, 210, 300 d) of Shaziling pigs using SPSS 20.0. (C) Species were altered in Shaziling pig. The data were assessed using t-test by comparing the Shaziling and Yorkshire pigs at 300 d using SPSS 20.0. Figures were drawn using GraphPad Prism 8.0. Values are presented as the means ± SEM. ∗, P < 0.05, ∗∗, P < 0.01, ∗∗∗, P < 0.001.
3.4. Ileal metabolic profiles between Shaziling and Yorkshire pigs
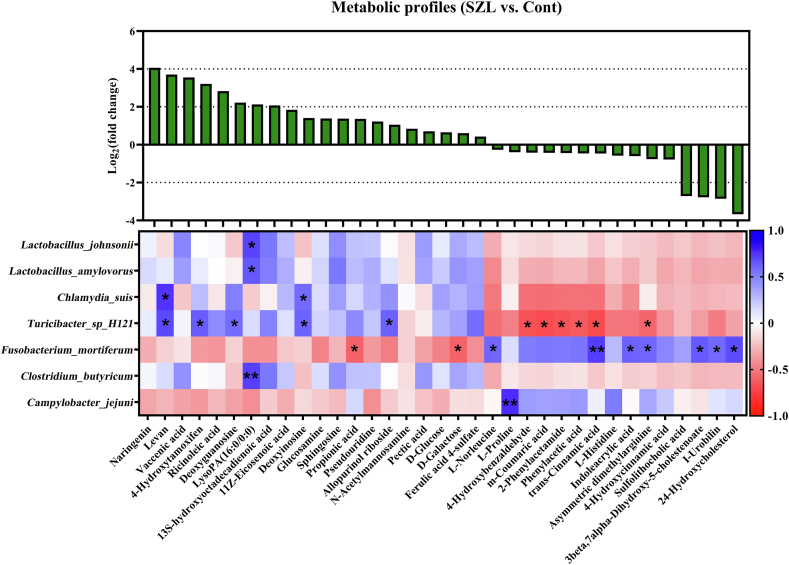
Microbial metabolites are one of the important factors affecting host lipid metabolism (Krautkramer et al., 2021). Thus, we further analyzed the metabolic profiles of the mucosa at 300 d of age between Shaziling and Yorkshire pigs (Fig. 5). First, we screened 86 different metabolites using variable importance in the projection (VIP) > 1, P < 0.05 as the standard (Table S1), and then we further identified 35 metabolites using VIP > 2, P < 0.05, including naringenin, levan, vaccenic acid, 4-hydroxytamoxifen, ricinoleic acid, deoxyguanosine LysoPA (16:0/0:0), 13S-hydroxyoctadecadienoic acid, 11Z-eicosenoic acid, deoxyinosine, glucosamine, sphingosine, propionic acid, pseudouridine, allopurinol riboside, N-acetylmannosamine, pectic acid, D-glucose, D-galactose, ferulic acid, 4-sulfate, L-norleucine, L-proline, 4-hydroxybenzaldehyde, m-coumaric acid, 2-phenylacetamide, phenylacetic acid, trans-cinnamic acid, L-histidine, indoleacrylic acid, asymmetric dimethylarginine, 4-hydroxycinnamic acid, sulfolithocholic acid, 3beta,7alpha-dihydroxy-5-cholestenoate, I-urobilin, and 24-hydroxycholesterol (Fig. 5). KEGG metabolic pathway enrichment showed that these differentiated metabolites were mainly related to carbohydrates, protein digestion and absorption, glucose and amino acid metabolism, and bile acid biosynthesis (Table S2).
Fig. 5.
Metabolic profile and correlation analysis. We screened 35 metabolites with VIP > 2 and P < 0.05 as standards, and then Pearson correlation analysis was performed with 7 different bacteria in the mucosa at 300 d. ∗, P < 0.05, ∗∗, P < 0.01. VIP = variable importance in the projection. SZL = Shaziling pig; Cont = Control.
Pearson correlation analysis showed that the significant positive correlations were identified between L. johnsonii and LsyoPA (16:0/0:0), L. amylovorus and LsyoPA (16:0/0:0), C. suis and levan, deoxyinosine, Turicibacter sp. H121 and levan, 4-hydroxytam deoxyguanosine, deoxyinosine, allopurinol riboside, F. mortiferum and L-norleucine, trans-cinnamic acid, indoleacrylic acid, asymmetric dimethylarginine, 3beta,7alpha-dihydroxy-5-cholestenoate, I-urobilin, 24-hydroxydium_sool, C. butyricum and LsyoPA (16:0/0:0), C. jejuni and L-proline. The negative correlations were noticed between Turicibacter sp. H121 and 4-hydroxybenzaldehyde, m-coumaric acid, 2-phenylacetamide, phenylacetic acid, trans-cinnamic acid, asymmetric dimethylarginine, F. mortiferum and propionic acid and D-galactose (Fig. 5).
3.5. L. johnsonii mono-colonization improves the lipids metabolism
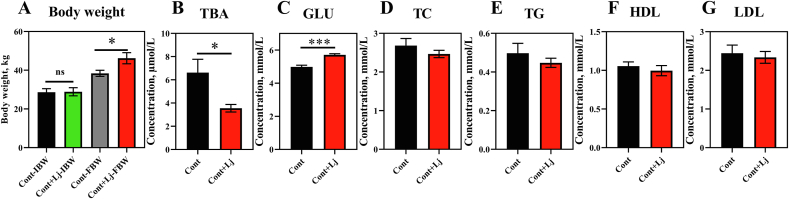
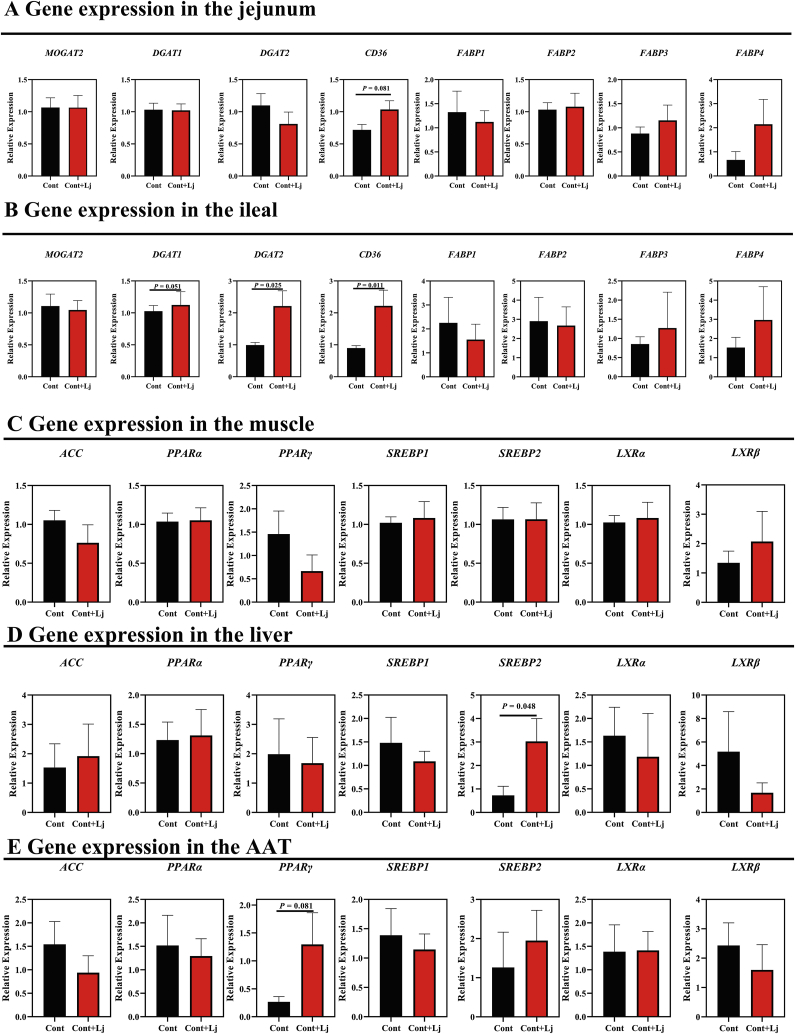
Mucosal symbiotic bacteria play an important role in lipid metabolism (Araujo et al., 2020; Huus et al., 2020; Paone and Cani, 2020), and we found that L. johnsonii, the most abundant bacteria in the mucous membrane, was significantly higher in Shaziling pigs (Fig. 4C). Thus, we speculated that L. johnsonii might help lipid absorption and accumulation in pigs. Interestingly, body weight was markedly increased by L. johnsonii mono-colonization (Fig. 6A). The serum TBA was reduced and Glu was markedly enhanced in L. johnsonii mono-colonized pigs (Fig. 6B–C). However, serum TC, TG, HDL, and LDL concentrations were not altered (Fig. 6D–G), which might be due to the short-term colonization, and the long-term effects of L. johnsonii are suggested in pigs. In addition, we next found that several genes associated with lipid transport and metabolism were significantly enhanced, such as CD36 in the jejunum (Fig. 7A), DGAT1/2 and CD36 in the ileum (Fig. 7B), SREBP2 in the liver (Fig. 7D), and PPARγ in the abdominal adipose tissue (Fig. 7E). These results indicated that L. johnsonii mono-colonization might promote lipid absorption and transport and fat accumulation.
Fig. 6.
Lactobacillus johnsonii mono-colonization improves the body weight and serum lipids. (A) Body weight, (B) TBA, (C) GLU, (D) TC, (E) TG, (F) HDL and (G) LDL were detected using t-test between control and L. johnsonii colonization using SPSS 20.0. Figures were drawn using GraphPad Prism 8.0. Values are presented as the means ± SEM. ∗, P < 0.05, ∗∗∗, P < 0.001. Lj = L. johnsonii; Cont = control; TBA = total bile acid; GLU = glucose; TC = total cholesterol; TG = triglycerides; HDL = high-density lipoprotein; LDL = low-density lipoprotein.
Fig. 7.
L. johnsonii mono-colonization altered the gene expression. Lipid uptake and transport related genes in the (A) jejunum and (B) ileum and lipid metabolism related genes in (C) muscle, (D) Liver and (E) abdominal adipose tissue (AAT) were detected using t-test between control and L. johnsonii colonization using SPSS 20.0. Figures were drawn using GraphPad Prism 8.0. Values are presented as the means ± SEM. Lj = L. johnsonii; Cont = control; ACC = acetyl-CoA carboxylase; PPARα = peroxisome proliferator activated receptor α; PPARγ = peroxisome proliferator activated receptor γ; SREBP1 = cholesterol regulatory element binding protein 1; SREBP2 = cholesterol regulatory element binding protein 1; LXRα = liver X receptor α; LXRβ = liver X receptor β; MOGAT2 = monoacylglycerol-O-acyltransferase 2; DGAT1 = diacylglycerol Acyltransferase 1; DGAT2 = diacylglycerol Acyltransferase 2; CD36 = cluster of differentiation 36; FABP1 = fatty acid-binding protein 1; FABP2 = fatty acid-binding protein 2; FABP3 = fatty acid-binding protein 3; FABP4 = fatty acid-binding protein 4.
3.6. L. johnsonii mono-colonization affects gut microbiota compositions
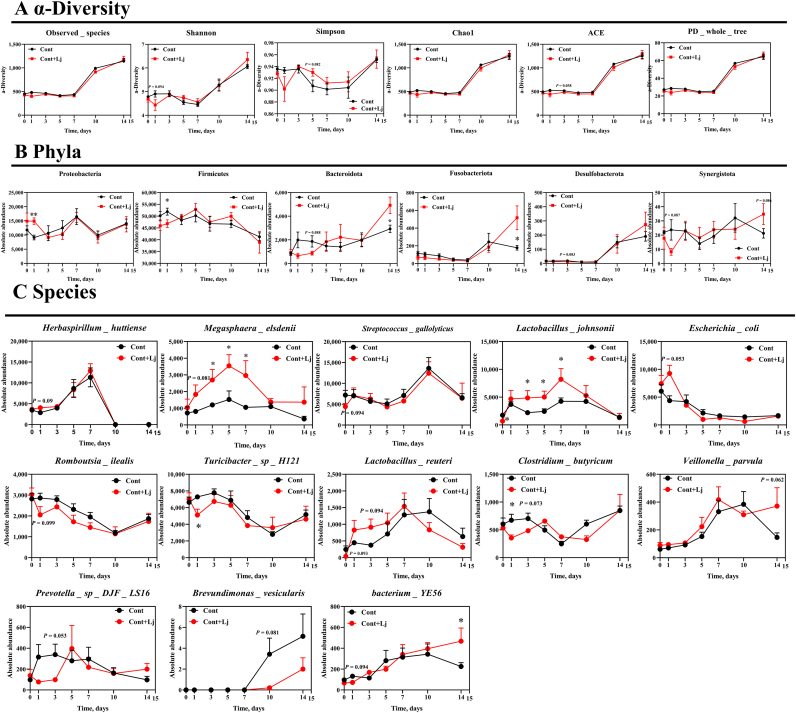
Next, we compared microbial alterations in the chyme in response to L. johnsonii mono-colonization (Fig. 8). Alpha diversity was not changed and the relative abundances of Proteobacteria, Bacteroidetes, and Fusobacteriota were significantly enhanced at d 1 and 14, while Firmicutes were decreased at d 1 after L. johnsonii mono-colonization (Fig. 8A and B). At the species level, L. johnsonii mono-colonization enhanced gut Megasphaera elsdenii, L. johnsonii and bacterium YE56 abundances at d 3, 5, 7, and 14 (Fig. 8C).
Fig. 8.
L. johnsonii mono-colonization improved the microbial development. (A) α-Diversity, (B) phyla, and (C) species in the chyme were assessed using t-test by comparing the control and control + L. johnsonii colonization using SPSS 20.0. Figures were drawn using GraphPad Prism 8.0. Values are presented as the means ± SEM. ∗, P < 0.05. ACE = abundance-based coverage estimator; PD whole tree = phylogenetic diversity whole tree.
3.7. L. johnsonii mono-colonization improves the composition of medium and long chain fatty acids
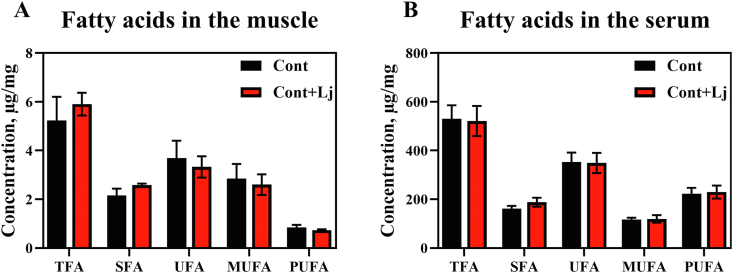
Medium long chain fatty acids are an essential intermediate product of fat metabolism, and play an important role in lipid metabolism whether lipolysis or fat synthesis (Kimura et al., 2020; Quan et al., 2020). To investigate L. johnsonii regulating the fat deposition, medium and long chain fatty acids were detected in the serum and muscle samples (Fig. 9 and Table S3). The results showed that total fatty acids (TFA) and saturated fatty acids (SFA) (i.e., methyl stearate, C18:0) tended to be increased in the muscle but not in the serum in L. johnsonii-fed pigs. These results indicated that L. johnsonii had a potential role in dietary lipid absorption and deposition in the muscle, but further investigation of the long-term effects of L. johnsonii are needed.
Fig. 9.
L. johnsonii mono-colonization improved the medium and long chain fatty acid concentration. Medium and long chain fatty acid concentration in the (A) muscle and (B) serum were analyzed using t-test between control and L. johnsonii colonization using SPSS 20.0. Figures were drawn using GraphPad Prism 8.0. Values are presented as the means ± SEM. Lj = L. johnsonii; Cont = control; TFA = total fatty acids; SFA = saturated fatty acid; UFA = unsaturated fatty acid; MUFA = monounsaturated fatty acids; PUFA = polyunsaturated fatty acids.
4. Discussion
Chinese indigenous breeds such as Shaziling pigs, have many excellent traits, such as strong stress resistance and high meat quality. However, there is little research on the mechanism of regulating meat quality by nutritional methods. In this trial, we focused on the relationship between the differences in microbes at different stages and the lipid metabolism of the host, in order to explore new ideas for nutritional regulation of lipid metabolism in Shaziling pigs.
The liver is one of the important organs for host lipid metabolism, including lipid assembly and transportation (Zhao et al., 2020). Meanwhile, the absorption and transport of lipids in the small intestine also determines the fate of lipid metabolism (Ko et al., 2020), and a two-way effect occurs with intestinal microbes and metabolites (Martinez-Guryn et al., 2018; Huang et al., 2019). Thus, the expression of genes related to lipid absorption and transport in the small intestine and genes related to liver lipid metabolism may reveal the molecular mechanism of lipid metabolism. Germ-free mice demonstrated that the changes of the gut microbiota community including decreased relative abundance of Lachnospiraceae and an enhanced occurrence of Desulfovibrionaceae, Clostridium lactatifermentans and Flintibacter butyricus were associated with impaired glucose metabolism, lowered counts of enteroendocrine cells, fatty liver, and elevated amounts of hepatic triglycerides, cholesteryl esters, and monounsaturated fatty acids (Just et al., 2018). Furthermore, gut microbiota-mediated cholesterol metabolism via a microbial cholesterol dehydrogenase played an important role in host cholesterol homeostasis (Kenny et al., 2020). Interestingly, there may be a bridge between gut microbes and metabolites and lipid levels, and several relevant genes regulated the concentration of lipids. Recent studies have found that theabrownin reduced liver cholesterol and decreased lipogenesis by the gut microbe-bile acid-FXR-FGF15 signaling pathway (Huang et al., 2019). In diet-induced obesity, the deletion of the MyD88 gene in hepatocytes altered the gut microbes and their metabolites, and the host phenotype was attributed to the gene expression and transcription factor activity (PPARα, FXR, LXR and ATAT3) and bile acid profiles involved in glucose, lipid metabolism and inflammation (Duparc et al., 2017). In this trial, we found that the variational tendency of lipid concentration was consistent with the gene expression of relevant lipid metabolism in the liver. While the tendency of the gene expression associated with lipid absorption and transport was different in the mucosa, Shaziling gene expression was lower than the control group before 210 d of age, and reached or exceeded the control group after 210 d. This result indicated that there were temporal and spatial differences in lipid metabolism, which is worth further exploration.
Recently, the relationship between gut microbes and host lipid metabolism has been extensively studied (Canibe et al., 2019; Kim et al., 2020; Ko et al., 2020). Although Shaziling pigs are a fatty model, they are well known for their high meat quality. Further, 2 distinct types of pigs that have been studied for their microbial variations are Yorkshire and Shaziling pig at different stages, which helps us understand and grasp the relationship between microbes and lipid metabolism. The organism establishes a huge microbiota from birth to coordinate the health of the body. The abundance of microorganisms also changes and plays different roles at different stages (Errington and Scazzocchio, 2003; Robertson et al., 2019). A study investigated the human microbiota in the first 1,000 d and found that the concurrent assembly of the microbiota and endocrine, immune and metabolic pathways indicate tight regulatory interdependence and underlying growth and development between microbiota and host, and it is intergenerational, thereby perpetuating growth impairments into successive generations (Robertson et al., 2019). In addition, piglets are prone to weaning emergency syndrome due to factors such as weaning, environment, drugs and feed, resulting in intestinal microorganism disturbance (Gresse et al., 2017). Furthermore, one of the important roles of intestinal microbes in adults and the elderly is to participate in the body's lipid metabolism (Lippert et al., 2017; Zhou et al., 2019). In this trial, we investigated the development of microorganisms in Yorkshire and Shaziling pigs' chyme at 30, 60, 90, 150, 210 and 300 d. We found that the abundance of gut microbes in Shaziling pigs was higher than in Yorkshire pigs. We further analyzed that there are significant differences including 14 phyla (such as Firmicutes, Chlamydiae, Bacteroidota, etc.), and 17 species (such as L. amylovorus, C. suis, E. coli, Turicibacter sp. H121, etc.).
Additionally, we have analyzed that the microbial abundance in the mucosa was higher than in the chyme. Therefore, we further explored the commensal microorganisms of Shaziling pigs in the mucosa. Importantly, several studies have reported that the commensal microorganisms in the mucosa and the mucus layer together maintain the gut barrier function (Cai et al., 2020). Comprehensive studies have found that the molecular mechanisms of commensal bacteria regulating host lipid metabolism were different, Lactobacillus paracasei promoted lipid storage through the L-lactic acid-malonyl-CoA-lipid β oxidation pathway; in contrast, E. coli improved lipids oxidation by the acetic acid-acetyl-CoA/AMP-AMPK/PGC-1α/PPARα pathway (Araujo et al., 2020). In this experiment, we screened several discrepant bacteria, including 10 phyla (Firmicutes, Proteobacteria, Chlamydiae, etc.) and 13 species (L. johnsonii, L. amylovorus, C. suis, etc.).
Metabolomics is extensively studied for the relationship between microbial metabolites and host metabolism (Shen et al., 2019). Most experiments revealed that gut microbes governed host lipid metabolism (Jia et al., 2021; Zhang et al., 2021). However, there are manifold intestinal microbes and complex interactions between microorganisms, so the mechanism by which microbes regulate host lipid metabolism is ambiguous. Microbial metabolites may be a bridge to reveal the relationship between microbes and host metabolism (Legoux et al., 2019; Wu et al., 2020), thus we investigated mucosal metabolites by non-targeted metabolomics, and then analyzed the correlation between mucosal microbes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that metabolites were mainly involved in the digestion and absorption of protein and carbohydrates, glucose metabolism and primary bile acids biosynthesis. In addition, module–trait associations indicated that microorganisms were related to plasma metabolites, including Clostridales, Tenericutes, Methanobrevibacter and Christensenellaceae, and were positively correlated with acetate, glutamine, and polyunsaturated fatty acids, whereas Blautia was negatively correlated with TG, monounsaturated fatty acids, pyruvate, glycerol, alanine, isoleucine, leucine, and GlycA (Org et al., 2017). Further research found that microbiota-derived inositol phosphate regulates histone deacetylase 3 (HDAC3) activity to promote epithelial repair (Wu et al., 2020). In this experiment, we revealed that L. johnsonii, L. amylovorus, C. suis, Turicibacter sp. H121, F. mortiferum, C. butyricum and C. jejuni were markedly correlated with host lipid metabolism, and suggested that gut microbes and metabolites are involved in lipid metabolism. However, the function of microorganisms and the mechanism of regulating lipid metabolism need further study.
We found that L. johnsonii plays an important role in promoting obesity. Mono-colonization is frequently used to investigate the relationship between bacteria and host metabolism (Barroso-Batista et al., 2020). Recent reports suggested that L-lactic acid produced by L. paracasei inhibits chylomicron secretion by enterocytes and promotes lipid storage through subsequent inhibition of lipid β-oxidation, but E. coli, that promoted lipid oxidation through secreting acetate, was absorbed by enterocytes and an up-regulated AMPK/PGC-1A/PPARα pathway (Araujo et al., 2020). Commensal bacteria regulated the immune response through adaptation to the host nutritional environment. For example, undernourished mice fail to develop IgA recognition of intestinal Lactobacillus, but Lactobacillus adaptations were a direct response to nutritional pressure, independently of host IgA, and are associated with reduced mucosal bacteria colonization and with bacterial gene mutations in the carbohydrate process (Huus et al., 2020). L. johnsonii showed a role in promoting lipid deposition, such as increasing the final body weight and raising the concentration of GLU. But the concentration of TBA was markedly decreased, and there was a tendency that other serum lipids (i.e., TC, TG, HDL and LDL) were lower. It suggested a potentially beneficial effect of L. johnsonii on serum lipids. Lipids are absorbed and transported in the gut and then distributed to tissues by the host for metabolism and deposition (Ko et al., 2020). An investigation showed that lactobacillus acidophilus improves glucolipid metabolism-related genes in mice with type 2 diabetes, such as down-regulating the expression of glycogen synthase kinase 3β (GSK-3β), fatty acid synthase (FAS), sterol regulatory element-binding transcription factor 1c (SREBP-1c), and up-regulating the expression of protein kinase B (Akt) (Yan et al., 2019), and further research showed that Lactobacilli reshape the gut microbes (Rodrigues et al., 2021). Lactobacillus rhamnosus ameliorated high fat diet-induced obesity in mice by altering gut microbes, promoting carbohydrate metabolism, and reducing glucose content. We found that L. johnsonii promoted adipose deposition by upregulating CD36, DGAT1/2 in intestinal mucosa and PPARγ in adipose tissue, altering the gut microbiota involving M. elsdenii, L. johnsonii and bacterium YE56.
Fatty acids, as important intermediates of fat metabolism and deposition, are mainly medium- and long-chain fatty acids from dietary triglycerides, and short-chain fatty acids produced by gut microbiota fermentation of indigestible dietary fiber, and constitute the main source of free fatty acids in the metabolic network (Kimura et al., 2020). Saturated fatty acids promote inflammatory responses, obesity and related diseases, including cardiovascular disease, insulin resistance, type 2 diabetes, and certain chronic diseases such as asthma (Zacek et al., 2019; Choi et al., 2020; Heileson, 2020; Machate et al., 2020). Additionally, SFA such as methyl stearate activated inflammation by the Toll-like receptor 4 (TLR4) pathway (Rogero and Calder, 2018; Sergi and Williams, 2020); meanwhile obesity was a chronic inflammatory process. Further SFA increased growth differentiation factor-15 (GDF15, involved in the development of obesity and insulin resistance) expression to induce obesity by inducing endoplasmic reticulum stress and activating the PERK/eIF2/CHOP signaling pathway (L'Homme et al., 2020), especially in cell experiments, and stearic acid potentiated macrophage inflammatory protein (MIP)-1α production in monocytic cells via MyD88 Independent TLR4/TBK/IRF3 signaling pathway to promote inflammation and obesity (Kochumon et al., 2020). We specially noticed that TFA and SFA tended to be elevated in the muscle, but other fatty acids were not altered, indicating that short-term colonization and long-term effects of L. johnsonii were suggested.
5. Conclusion
In conclusion, our results indicated that the gut microbes of Shaziling pigs influenced their metabolic phenotype. The gut microbial abundance, lipid level and gene expression of Shaziling pigs were altered at different stages, and the abundance was higher in the test group than the control group. Correlation analysis showed that microorganisms and metabolites were involved in host lipid metabolism. Further we investigated the promoting effect of L. johnsonii on lipid metabolism, but the effect on serum lipid level and medium- and long-chain fatty acids were not impacted immensely, and even tended to decrease serum lipid level and fatty acid contents. Therefore, the impact of L. johnsonii in promoting lipid deposition may need to be studied over a longer period to determine a long-term effect. This mechanism needs to be further explored.
Author contributions
Jie Ma is the primary investigator in this study. Yehui Duan participated in the animal experiments. Xiaoxiao Liang and Rui Li performed statistical data analysis. Tiejun Li and Xingguo Huang participated in sample analysis. Yulong Yin designed this study and Jie Yin examined the manuscript as corresponding author.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
This study was funded by the National Natural Science Foundation (32172761, U19A2037, and U20A2055), Earmarked Fund for China Agriculture Research System (CARS-35), Young Elite Scientists Sponsorship Program by CAST (2019-2021QNRC001), and Hunan Science Foundation for Outstanding Young Scholars (2020JJ3023). We thank the College of Animal Science and Technology, Hunan Agricultural University, Animal Nutrition Genome and Germplasm Innovation Research Center for support. All data generated or analyzed during this study are available from the corresponding author upon reasonable request.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2021.10.012.
Appendix A. Supplementary data
The following is the supplementary data to this article:
Fig. S1.
Fig. S2.
References
- Araujo J.R., Tazi A., Burlen-Defranoux O., Vichier-Guerre S. Fermentation products of commensal bacteria alter enterocyte lipid metabolism. Cell Host Microbe. 2020;27:358–375. doi: 10.1016/j.chom.2020.01.028. e7. [DOI] [PubMed] [Google Scholar]
- Aron-Wisnewsky J., Vigliotti C., Witjes J., Le P. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol. 2020;17:279–297. doi: 10.1038/s41575-020-0269-9. [DOI] [PubMed] [Google Scholar]
- Aron-Wisnewsky J., Warmbrunn M.V., Nieuwdorp M., Clement K. Metabolism and metabolic disorders and the microbiome: the intestinal microbiota associated with obesity, lipid metabolism and metabolic health: pathophysiology and therapeutic strategies. Gastroenterology. 2021;160:573–599. doi: 10.1053/j.gastro.2020.10.057. [DOI] [PubMed] [Google Scholar]
- Azad M.A.K., Gao J., Ma J., Li T.J. Opportunities of prebiotics for the intestinal health of monogastric animals. Anim Nutr. 2020;6:379–388. doi: 10.1016/j.aninu.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-Batista J., Pedro M.F., Sales-Dias J., Pinto C.J.G. Specific eco-evolutionary contexts in the mouse gut reveal Escherichia coli metabolic versatility. Curr Biol. 2020;30:1049–1062. doi: 10.1016/j.cub.2020.01.050. e7. [DOI] [PubMed] [Google Scholar]
- Cai R., Cheng C., Chen J., Xu X. Interactions of commensal and pathogenic microorganisms with the mucus layer in the colon. Gut Microbes. 2020;11:680–690. doi: 10.1080/19490976.2020.1735606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canibe N., O'Dea M., Abraham S. Potential relevance of pig gut content transplantation for production and research. J Anim Sci Biotechnol. 2019;10:55. doi: 10.1186/s40104-019-0363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Kim M., Kim S.J., Yoo H.J. Metabolic shift favoring C18:0 ceramide accumulation in obese asthma. Allergy. 2020;75:2858–2866. doi: 10.1111/all.14366. [DOI] [PubMed] [Google Scholar]
- Coffey M.J., Nielsen S., Wemheuer B., Kaakoush N. Gut microbiota in children with cystic fibrosis: a taxonomic and functional dysbiosis. Sci Rep. 2019;9:18593. doi: 10.1038/s41598-019-55028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duparc T., Plovier H., Marrachelli V.G., Van Hul M. Hepatocyte MyD88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut. 2017;66:620–632. doi: 10.1136/gutjnl-2015-310904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J., Scazzocchio C. Growth and development. Microbial development--regulation in space and time. Curr Opin Microbiol. 2003;6:531–533. doi: 10.1016/j.mib.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Fan Y., Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- Gresse R., Chaucheyras-Durand F., Fleury M.A., Van de Wiele T. Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol. 2017;25:851–873. doi: 10.1016/j.tim.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Guevarra R.B., Hong S.H., Cho J.H., Kim B.R. The dynamics of the piglet gut microbiome during the weaning transition in association with health and nutrition. J Anim Sci Biotechnol. 2018;9:54. doi: 10.1186/s40104-018-0269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heileson J.L. Dietary saturated fat and heart disease: a narrative review. Nutr Rev. 2020;78:474–485. doi: 10.1093/nutrit/nuz091. [DOI] [PubMed] [Google Scholar]
- Hu J., Ma L., Nie Y., Chen J. A microbiota-derived bacteriocin targets the host to confer diarrhea resistance in early-weaned piglets. Cell Host Microbe. 2018;24:817–832. doi: 10.1016/j.chom.2018.11.006. e8. [DOI] [PubMed] [Google Scholar]
- Huang F., Zheng X., Ma X., Jiang R. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat Commun. 2019;10:4971. doi: 10.1038/s41467-019-12896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huus K.E., Bauer K.C., Brown E.M., Bozorgmehr T. Commensal bacteria modulate immunoglobulin A binding in response to host nutrition. Cell Host Microbe. 2020;27:909–921. doi: 10.1016/j.chom.2020.03.012. e5. [DOI] [PubMed] [Google Scholar]
- Jia W., Wei M., Rajani C., Zheng X. Targeting the alternative bile acid synthetic pathway for metabolic diseases. Protein Cell. 2021;12:411–425. doi: 10.1007/s13238-020-00804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just S., Mondot S., Ecker J., Wegner K. The gut microbiota drives the impact of bile acids and fat source in diet on mouse metabolism. Microbiome. 2018;6:134. doi: 10.1186/s40168-018-0510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny D.J., Plichta D.R., Shungin D., Koppel N. Cholesterol metabolism by uncultured human gut bacteria influences host cholesterol level. Cell Host Microbe. 2020;28:245–257. doi: 10.1016/j.chom.2020.05.013. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal P., Maltecca C., Schwab C., Fix J. Modeling host-microbiome interactions for the prediction of meat quality and carcass composition traits in swine. Genet Sel Evol. 2020;52:41. doi: 10.1186/s12711-020-00561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Hwang S.W., Kim S., Lee Y.S. Dietary cellulose prevents gut inflammation by modulating lipid metabolism and gut microbiota. Gut Microbes. 2020;11:944–961. doi: 10.1080/19490976.2020.1730149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Ichimura A., Ohue-Kitano R., Igarashi M. Free fatty acid receptors in health and disease. Physiol Rev. 2020;100:171–210. doi: 10.1152/physrev.00041.2018. [DOI] [PubMed] [Google Scholar]
- Ko C.W., Qu J., Black D.D., Tso P. Regulation of intestinal lipid metabolism: current concepts and relevance to disease. Nat Rev Gastroenterol Hepatol. 2020;17:169–183. doi: 10.1038/s41575-019-0250-7. [DOI] [PubMed] [Google Scholar]
- Kochumon S., Arefanian H., Azim R., Shenouda S. Stearic acid and TNF-alpha co-operatively potentiate MIP-1alpha production in monocytic cells via MyD88 independent TLR4/TBK/IRF3 signaling pathway. Biomedicines. 2020;8:403. doi: 10.3390/biomedicines8100403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpela K., Helve O., Kolho K.L., Saisto T. Maternal fecal microbiota transplantation in cesarean-born infants rapidly restores normal gut microbial development: a proof-of-concept study. Cell. 2020;183:324–334. doi: 10.1016/j.cell.2020.08.047. e5. [DOI] [PubMed] [Google Scholar]
- Krautkramer K.A., Fan J., Backhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat Rev Microbiol. 2021;19:77–94. doi: 10.1038/s41579-020-0438-4. [DOI] [PubMed] [Google Scholar]
- L'Homme L., Sermikli B.P., Staels B., Piette J. Saturated fatty acids promote GDF15 expression in human macrophages through the PERK/eIF2/CHOP signaling pathway. Nutrients. 2020;12:3771. doi: 10.3390/nu12123771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoux F., Bellet D., Daviaud C., Morr Y.E. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 2019;366:494–499. doi: 10.1126/science.aaw2719. [DOI] [PubMed] [Google Scholar]
- Li T., Huang S., Lei L., Tao S. Intrauterine growth restriction alters nutrient metabolism in the intestine of porcine offspring. J Anim Sci Biotechnol. 2021;12:15. doi: 10.1186/s40104-020-00538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ma J., Yao K., Su W. Circadian rhythms and obesity: timekeeping governs lipid metabolism. J Pineal Res. 2020;69:e12682. doi: 10.1111/jpi.12682. [DOI] [PubMed] [Google Scholar]
- Li Y., Xia S., Jiang X., Feng C. Gut microbiota and diarrhea: an updated review. Front Cell Infect Microbiol. 2021;11:625210. doi: 10.3389/fcimb.2021.625210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert K., Kedenko L., Antonielli L., Kedenko I. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef Microbes. 2017;8:545–556. doi: 10.3920/BM2016.0184. [DOI] [PubMed] [Google Scholar]
- Machate D.J., Figueiredo P.S., Marcelino G., Guimaraes R.C.A. Fatty acid diets: regulation of gut microbiota composition and obesity and its related metabolic dysbiosis. Int J Mol Sci. 2020;21:4093. doi: 10.3390/ijms21114093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Guryn K., Hubert N., Frazier K., Urlass S. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe. 2018;23:458–469. doi: 10.1016/j.chom.2018.03.011. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org E., Blum Y., Kasela S., Mehrabian M. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017;18:70. doi: 10.1186/s13059-017-1194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paone P., Cani P.D. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut. 2020;69:2232–2243. doi: 10.1136/gutjnl-2020-322260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan L.H., Zhang C., Dong M., Jiang J. Myristoleic acid produced by enterococci reduces obesity through brown adipose tissue activation. Gut. 2020;69:1239–1247. doi: 10.1136/gutjnl-2019-319114. [DOI] [PubMed] [Google Scholar]
- Robertson R.C., Manges A.R., Finlay B.B., Prendergast A.J. The human microbiome and child growth - first 1000 d and beyond. Trends Microbiol. 2019;27:131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- Rodrigues R.R., Gurung M., Li Z., Garcia-Jaramillo M. Transkingdom interactions between Lactobacilli and hepatic mitochondria attenuate western diet-induced diabetes. Nat Commun. 2021;12:101. doi: 10.1038/s41467-020-20313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogero M.M., Calder P.C. Obesity, inflammation, toll-like receptor 4 and fatty acids. Nutrients. 2018;10(4):432. doi: 10.3390/nu10040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergi D., Williams L. Potential relationship between dietary long-chain saturated fatty acids and hypothalamic dysfunction in obesity. Nutr Rev. 2020;78:261–277. doi: 10.1093/nutrit/nuz056. [DOI] [PubMed] [Google Scholar]
- Shen X., Wang R., Xiong X., Yin Y. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat Commun. 2019;10:1516. doi: 10.1038/s41467-019-09550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Yan W., Mai C., Duan Z. Joint contributions of the gut microbiota and host genetics to feed efficiency in chickens. Microbiome. 2021;9:126. doi: 10.1186/s40168-021-01040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.E., Hashimoto-Hill S., Woo V., Eshleman E.M. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature. 2020;586:108–112. doi: 10.1038/s41586-020-2604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Li Q.H., He C.Q., Wang L.Y. The complete mitochondrial genome of the Shaziling pig. Mitochondrial DNA. 2015;26:619–620. doi: 10.3109/19401736.2013.834431. [DOI] [PubMed] [Google Scholar]
- Yan F., Li N., Shi J., Li H. Lactobacillus acidophilus alleviates type 2 diabetes by regulating hepatic glucose, lipid metabolism and gut microbiota in mice. Food Funct. 2019;10:5804–5815. doi: 10.1039/c9fo01062a. [DOI] [PubMed] [Google Scholar]
- Yang H., Xu X.L., Ma H.M., Jiang J. Integrative analysis of transcriptomics and proteomics of skeletal muscles of the Chinese indigenous Shaziling pig compared with the Yorkshire breed. BMC Genet. 2016;17:80. doi: 10.1186/s12863-016-0389-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Zhang Y., Li W., You B. Gut microbiota composition affects procyanidin A2-attenuated atherosclerosis in ApoE–/– mice by modulating the bioavailability of its microbial metabolites. J Agric Food Chem. 2021;69(25):6989–6999. doi: 10.1021/acs.jafc.1c00430. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- Yin J., Li Y., Han H., Chen S. Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice. J Pineal Res. 2018;65:e12524. doi: 10.1111/jpi.12524. [DOI] [PubMed] [Google Scholar]
- Yin J., Li Y., Han H., Ma J. Administration of exogenous melatonin improves the diurnal rhythms of the gut microbiota in mice fed a high-fat diet. mSystems. 2020;5 doi: 10.1128/mSystems.00002-20. e00002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Ma J., Li Y., Ma X. Branched-chain amino acids, especially of leucine and valine, mediate the protein restricted response in a piglet model. Food Funct. 2020;11:1304–1311. doi: 10.1039/c9fo01757g. [DOI] [PubMed] [Google Scholar]
- Zacek P., Bukowski M., Mehus A., Johnson L. Dietary saturated fatty acid type impacts obesity-induced metabolic dysfunction and plasma lipidomic signatures in mice. J Nutr Biochem. 2019;64:32–44. doi: 10.1016/j.jnutbio.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu C., Jiang Q., Yin Y. Butyrate in energy metabolism: there is still more to learn. Trends Endocrinol Metab. 2021;32:159–169. doi: 10.1016/j.tem.2020.12.003. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhao M.F., Jiang S., Wu J. Liver governs adipose remodelling via extracellular vesicles in response to lipid overload. Nat Commun. 2020;11:719. doi: 10.1038/s41467-020-14450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Yu B., Sun J., Liu Z. Short-chain fatty acids can improve lipid and glucose metabolism independently of the pig gut microbiota. J Anim Sci Biotechnol. 2021;12:61. doi: 10.1186/s40104-021-00581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Xiao X., Zhang Q., Zheng J. Maternal genistein intake mitigates the deleterious effects of high-fat diet on glucose and lipid metabolism and modulates gut microbiota in adult life of male mice. Front Physiol. 2019;10:985. doi: 10.3389/fphys.2019.00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.