Abstract
We have recently purified mammalian sterile 20 (STE20)–like kinase 3 (MST3) as a kinase for the multifunctional kinases, AMP-activated protein kinase–related kinases (ARKs). However, unresolved questions from this study, such as remaining phosphorylation activities following deletion of the Mst3 gene from human embryonic kidney cells and mice, led us to conclude that there were additional kinases for ARKs. Further purification recovered Ca2+/calmodulin-dependent protein kinase kinases 1 and 2 (CaMKK1 and 2), and a third round of purification revealed mitogen-activated protein kinase kinase kinase kinase 5 (MAP4K5) as potential kinases of ARKs. We then demonstrated that MST3 and MAP4K5, both belonging to the STE20-like kinase family, could phosphorylate all 14 ARKs both in vivo and in vitro. Further examination of all 28 STE20 kinases detected variable phosphorylation activity on AMP-activated protein kinase (AMPK) and the salt-inducible kinase 3 (SIK3). Taken together, our results have revealed novel relationships between STE20 kinases and ARKs, with potential physiological and pathological implications.
Keywords: AMPK, MAP4K5, STE20, phosphorylation, biochemical purification, signaling
Abbreviations: ACC1, acetyl-CoA carboxylase 1; AMPK, AMP-activated protein kinase; ARK, AMP-activated protein kinase–related kinase; BRSK1, BR serine/threonine kinase 1; CaMKK2, Ca2+/calmodulin-dependent protein kinase kinase 2; HEK, human embryonic kidney cell; LKB1, liver kinase B1; MAP, mitogen-activated protein; MAP4K5, mitogen-activated protein kinase kinase kinase kinase 5; MELK, maternal embryonic leucine zipper kinase; MS, mass spectrometry; MST3, mammalian sterile 20–like kinase 3; NUAK1, NUAK family SNF1-like kinase 1; PAK1, p21 protein-activated kinase 1; SIK3, salt-inducible kinase 3; SP HP, SP sepharose high performance column; STE20, sterile 20; TAOK1, thousand and one amino acid kinase 1; VRK1, vaccinia-related kinase 1
The AMP-activated protein kinase (AMPK) and AMP-activated protein kinase–related kinases (ARKs) play important roles in physiology and pathology (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13). They are known to be regulated by the liver kinase B1 (LKB1) (10, 14, 15, 16, 17, 18, 19, 20, 21), the Ca2+/calmodulin-dependent protein kinase kinase 2 (CaMKK2) (22, 23, 24, 25), and perhaps transforming growth factor-β–activated kinase 1 (TAK1) (26, 27, 28) (but see Ref. (29)).
Our first round of biochemical purification uncovered a mammalian sterile 20-like (MST) kinase 3 (MST3) (30, 31, 32). We then found that MST 1, 2, 3, 4, and 5 could phosphorylate salt-inducible kinase 3 (SIK3) at threonine (T) 221 and AMPKα at T172. However, removal of Mst3 by gene targeting in either mice or human embryonic kidney (HEK) cells did not eliminate LKB1-independent phosphorylation of SIK3-T221 and AMPK-T172. Our second round of purification led to the recovery of CaMKK2. Our third round of purification uncovered mitogen-activated protein (MAP) kinase kinase kinase kinase 5 (MAP4K5). When both Lkb1 and Camkk2 genes were deleted from HEK cells, MAP4K5 could still phosphorylate SIK3-T221 and AMPK-T172 in HEK cells, indicating that the effect of MAP4K5 was not dependent on LKB1 or CaMKK2 but rather directly. This suggestion was proven by using purified recombinant MAP4K5 on recombinant SIK3 and recombinant AMPK substrates. Furthermore, we found that MST3 and MAP4K5 could phosphorylate all 14 ARKs. Both MSTs and MAP4Ks belong to a mammalian subfamily of 28 serine/threonine kinases similar to the yeast sterile 20 (STE20) kinase (33). We examined all 28 mammalian STE20 kinases with each produced as recombinant proteins from bacteria and found that different STE20 kinases had different effects on AMPK and SIK3: some could phosphorylate both, some could phosphorylate one but not the other, and some could phosphorylate neither.
Our findings will stimulate laboratories in multiple fields to determine the exact endogenous signaling pathways mediated by one or more of the STE20 kinases together with one or more of the 14 ARKs in physiological and pathological processes.
Results
LKB1-independent and MST3-independent SIK3 and AMPKα1 phosphorylation shown by biochemistry and genetics
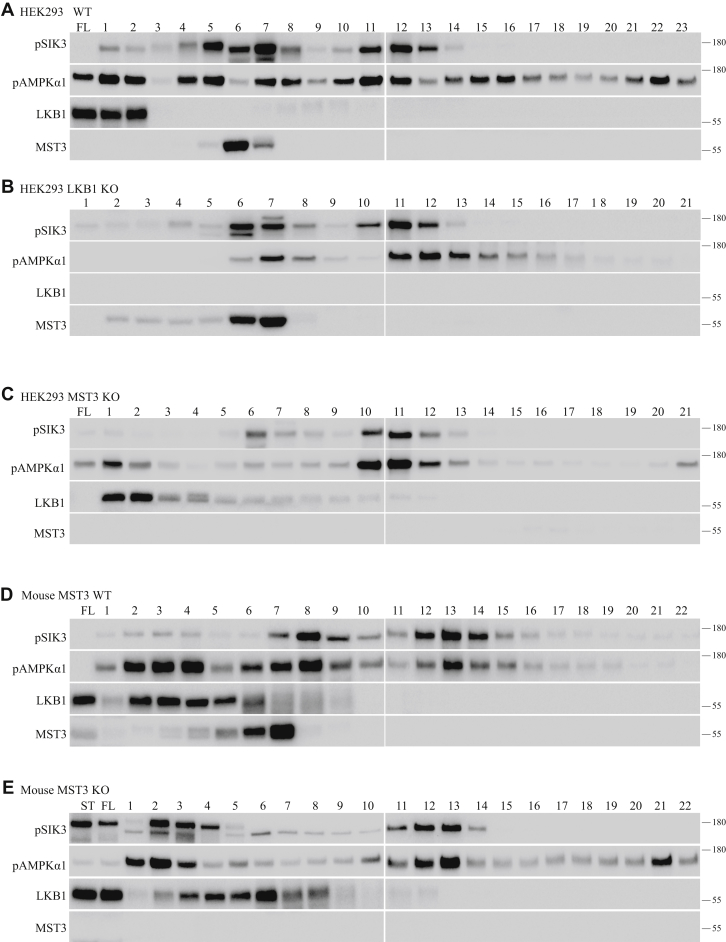
To investigate whether we have purified all SIK3 (and AMPKα1) phosphorylating activities from HEK cells in our first round of biochemical purification, we fractionated HEK lysates on an SP sepharose high performance column (SP HP) column again. SIK3-T221 phosphorylating activities partially overlapped with AMPKα1-T183 phosphorylating activities (Fig. 1A). Antibodies for LKB1 and MST3 showed that each of them correlated with some of the SIK3 and AMPKα1 phosphorylating activities. Notably, there were fractions such as 11 and 12 (300–330 mM NaCl), which contained strong SIK3 and AMPKα1 phosphorylating activities but were not positive for either LKB1 or MST3 (Fig. 1A).
Figure 1.
Phosphorylation of AMPKα1-T172 and SIK3 other than LKB1 and MST3. Lysates were prepared from HEK293T cells or mouse brains. They were fractionated on an SP HP column followed by NaCl gradient elution. Fractions were collected, and aliquots of 0.5 ml of each fraction were dialyzed against buffer A, followed by in vitro SIK3 and AMPK phosphorylation assays. A, fractions from WT HEK cells. AMPK phosphorylating activities were observed in more fractions than SIK3 phosphorylating activities. Both activities were present in more fractions than those containing LKB1 and MST3. For example, fractions 11 and 12 (300–330 mM NaCl) contained strong SIK3 and AMPK phosphorylation activities but no detectable LKB1 or MST3. Fraction 22 (1000 mM NaCl) had strong AMPK phosphorylation activity but no LKB1 or MST3. B, fractions from Lkb1 KO HEK cells. In the absence of LKB1, activities in fractions 6 and 7 (150–180 mM NaCl) and fractions 11 and 12 (300–330 mM NaCl) were still present. MST3 was present in fractions 5 and 6 (120–150 mM NaCl) but not fractions 10 to 13 (270–360 mM NaCl). C, fractions from Mst3 KO HEK cells. Phosphorylating activities in fractions 7 and 8 (180–210 mM NaCl) were reduced by not eliminating in Mst3 KO HEK cells. Phosphorylating activities existed in fractions 10 to 12 (270–330 mM NaCl). D, fractions from WT mouse brains. Fractions 12, 13, and 14 (330–390 mM NaCl) contained AMPK and SIK3 phosphorylating activities but neither LKB1 nor MST3. E, fractions from Mst3 KO mouse brains. Mst3 KO mice were generated. Their brains were isolated, and extracts were made. Brain lysates were fractionated on an SP HP column followed by NaCl gradient elution. Phosphorylating activities in fractions 7 and 8 (180–210 mM NaCl) were reduced by not eliminating. Phosphorylating activities in fractions 11 to 13 (300–360 mM NaCl) from Mst3 KO mouse brains contained strong AMPK and SIK3 phosphorylating activities. AMPK, AMP-activated protein kinase; HEK293T, human embryonic kidney 293T cell; LKB1, liver kinase B1; MST3, mammalian sterile 20–like kinase 3; SIK3, salt-inducible kinase 3.
After the Lkb1 gene was deleted in HEK cells (Fig. 1B), we fractionated HEK cell lysates on an SP HP column as described in the companion article and still detected strong SIK3 and AMPKα1 phosphorylating activities in fractions 6 and 7 (150–180 mM NaCl), with fractions 5 and 6 (120–150 mM NaCl) positive for MST3. Notably, fractions 11 and 12 (300–330 mM NaCl) from Lkb1 KO HEK cells were strongly positive for SIK3 and AMPKα1 phosphorylating activities but negative for MST3 and LKB1 (Fig. 1B).
After the Mst3 gene was deleted in HEK cells (Fig. 1C), we also fractionated HEK cell lysates on an SP HP column and found little AMPKα1 phosphorylating activities in fractions 6 and 7 (150–180 mM NaCl) and fractions 1 and 2 (0–30 mM NaCl) positive for LKB1. However, fractions 10, 11, and 12 (270–330 mM NaCl) from Mst3 KO HEK cells were strongly positive for SIK3 and AMPKα1 phosphorylating activities but were negative for LKB1 and MST3 (Fig. 1C). These results suggest that activities other than LKB1 and MST3 could phosphorylate SIK3-T221 and AMPKα1-T183 in HEK cells.
To examine whether activities independent of LKB1 and MST3 in the mouse brain could phosphorylate SIK3-T221 and AMPKα1-T183, we made extracts from 10 brains of 8-week-old WT mice and fractionated 50 mg of lysates on an SP HP column (Fig. 1D). We found that fewer fractions contained SIK3-T221 phosphorylating activities than those containing AMPKα1-T183 phosphorylating activities. Fractions 12, 13, and 14 (330–390 mM NaCl) contained activities phosphorylating both SIK3-T221 and AMPKα1-T183 but were negative for both LKB1 and MST3 (Fig. 1D).
We generated Mst3 KO mice by gene targeting (Fig. 1E). Fractionation with the SP HP column showed that activities phosphorylating SIK3-T221 and AMPKα1-T183 were significantly reduced in fractions 7 and 8 (180–210 mM NaCl) but were still strongly present in fractions 12 and 13 (330–360 mM NaCl, Fig. 1E). Reduction of activities in fractions 7 and 8 (180–210 mM NaCl) agreed with the results in Figure 1D, which detected MST3 in fractions 6 and 7 (150–180 mM NaCl). The activities remaining in fractions 11 to 13 (330–360 mM NaCl) could not be explained by LKB1 or MST3, which were absent from these fractions (Fig. 1E).
These results demonstrate the presence of SIK3-T221 and AMPKα1-T183 phosphorylating activities independent of LKB1 and MST3 both in HEK cells and in mouse brains.
Purification of CaMKKs as an AMPKα1 T183 phosphorylating activity from HEK cells
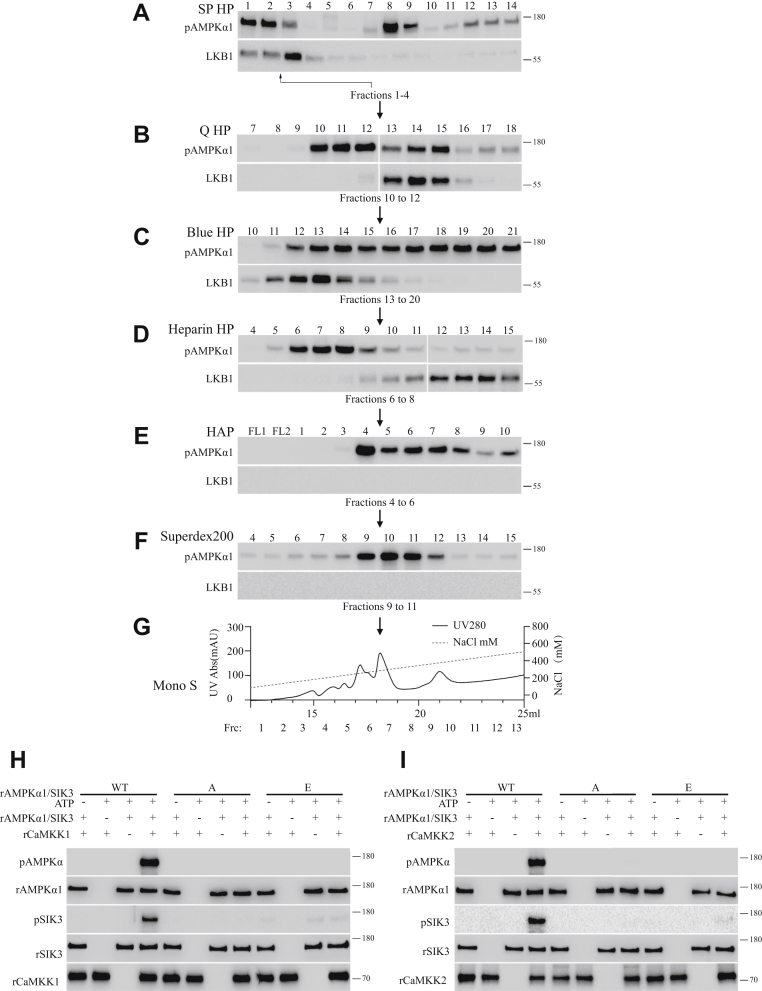
We carried out a second round of biochemical purification, in a scheme similar to that described for our purification of MST3, though we focused in this round on fractions 1 to 4 (0–90 mM NaCl) from the first SP HP cationic exchange column (Fig. 2).
Figure 2.
Purification of CaMKK2 from HEK cells.A–G, this was performed similarly as that described for MST3, with the exceptions that the focus was on fractions 1 to 4 (0–90 mM NaCl) of the SP HP column and that the phosphorylating activity for AMPKα1-T183, but not that for SIK3, was monitored. Active fractions from each column were pooled and loaded onto the next column: fractions 10 to 12 (270–330 mM NaCl) from the Q HP column, fractions 13 to 20 (360–570 mM NaCl) from the Blue HP column, fractions 6 to 8 (150–210 mM NaCl) from the heparin column, fractions 4 to 6 (45–75 mM K2PO4) from the HAP column and fractions 9 to 11 (240–300 mM NaCl) from the Superdex 200 column. The presence of LKB1 was also monitored. LKB1 was present in the fractions collected from the SP HP (A), Q HP (B), Blue HP (C), and heparin HP (D) columns but not in the fractions collected from the HAP (E), Supderdex 200 (F), and Mono S (G) columns. It indicates that we were successful in separating the kinase-active fractions (6–8, 150–210 mM NaCl) from the LKB1-containing fractions (9–15, 240–420 mM NaCl) of the heparin column (D). Results from the last (Mono S) column were shown in Supporting Appendix, Fig. S1. E, recombinant CaMKK1 could phosphorylate WT AMPK and WT SIK3 but could not phosphorylate AMPKα1 T183A or T183E, SIK3 T221A or T221E. F, recombinant CaMKK2 could phosphorylate WT AMPK and WT SIK3 but could not phosphorylate AMPKα1 T183A or T183E and SIK3 T221A or T221E. AMPK, AMP-activated protein kinase; CaMKK2, Ca2+/calmodulin-dependent protein kinase kinase-2; HEK, human embryonic kidney cell; LKB1, liver kinase B1; MST3, mammalian sterile 20–like kinase 3.
Because we noticed activities that were stronger in phosphorylating AMPKα1-T183 than SIK3-T221, we monitored and followed the AMPKα1-T183 phosphorylating activities and LKB1 in the first six columns (SP HP, Q HP, Blue HP, heparin, HAP, and Superdex 200) during this round of purification (Fig. 2). After the fourth (heparin) column, the phosphorylating activity was separated from the LKB1-positive fractions. After the last column (of Mono S), we examined phosphorylation of both AMPKα1-T183 and SIK3-T221 (Fig. S1B). The purified products appeared to phosphorylate AMPKα1-T183 but not SIK3-T221 (Fig. S1B).
Mass spectrometry (MS) analysis detected six kinases (CDK16, CaMKK1, serine/threonine-protein kinase 38 [STK38], CaMKK2, CaMK2D, and pantothenate kinase 1 [PANK1]). CaMKK2 had been previously found to phosphorylate AMPKα2 at T172 (22, 23, 24). With FLAG-tagged CaMKK2 expressed in and immunoprecipitated from HEK cells, the activities of CaMKK2 on AMPKα1-T183 were confirmed (Fig. S1D), and its activity on SIK3-T221 was weak (Fig. S1D). FLAG-tagged CaMKK1 immunoprecipitated from HEK cells had a very weak activity on AMPKα1-T183, whereas its activity on SIK3-T221 was even weaker (Fig. 3D).
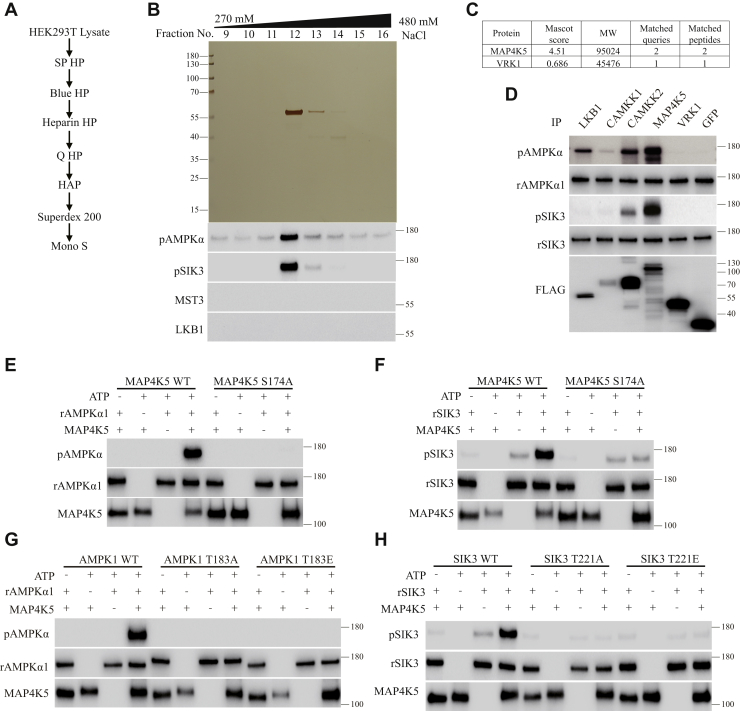
Figure 3.
Purification of MAP4K5 as an AMPKα1-T183 and SIK3-T221 phosphorylating activity from HEK cells.A, a diagram for our third round of purification. The focus was on fraction 12 (330 mM NaCl) from the first (SP HP) column (Fig. S2). B, silver staining and phosphorylation assays of fractions from the Mono S column. Fractions were dialyzed against buffer A and followed by silver staining and in vitro phosphorylation of AMPKα1-T183 and SIK3-T221. LKB1 and MST3 were examined but not detected in these fractions. The single visible bands in fractions 12 and 13 (330–360 mM NaCl) were isolated for MS analysis. C, two protein serine/threonine kinases (MAP4K5 and VRK1) were found by MS analysis. D, LKB1, CaMKK1, CaMKK2, MAP4K5, VRK1, and GFP were individually expressed as FLAG-tagged proteins in HEK cells and immunoprecipitated. They were tested for phosphorylation of recombinant AMPKα1 at T183 and recombinant SIK3 at T221. MAP4K5 phosphorylated both AMPKα1-T183 and SIK3-T221. LKB1 and CaMKK2 phosphorylated AMPKα1-T183 more strongly than SIK3-T221. VRK1 and GFP phosphorylated neither AMPKα1-T183 nor SIK3-T221. E, recombinant AMPKα1 was phosphorylated at T183 by MAP4K5 WT but not its S174A mutant immunoprecipitated from HEK cells. F, recombinant SIK3 was phosphorylated at T221 by MAP4K5 WT but not its S174A mutant immunoprecipitated from HEK cells. G, recombinant AMPKα1 WT, but not AMPKα1 T183A or AMPKα1 T183E, was phosphorylated by MAP4K5 WT immunoprecipitated from HEK cells. H, recombinant SIK3 T221, but not SIK3 T221A or SIK3 T221E, was phosphorylated by MAP4K5 WT immunoprecipitated from HEK cells. AMPK, AMP-activated protein kinase; CaMKK2, Ca2+/calmodulin-dependent protein kinase kinase 2; HEK, human embryonic kidney cell; LKB1, liver kinase B1; MAP4K5, mitogen-activated protein kinase kinase kinase kinase 5; MS, mass spectrometry; SIK3, salt-inducible kinase 3.
To test for the possibility of direct phosphorylation of AMPK and SIK3 by CaMKKs, we expressed CaMKK1 and 2 in Escherichia coli and purified them. We examined their kinase activities on recombinant AMPKα1 and SIK3 purified from E. coli. WT AMPKα1 and SIK3 could be phosphorylated by CaMKK1 (Fig. 2H) and CaMKK2 (Fig. 2I), but neither AMPKα1 mutants (T183A and T183E) nor SIK3 mutants (T221A and T221E) could be phosphorylated by CaMKK1 (Fig. 2H) and CaMKK2 (Fig. 2I).
Purification of MAP4K5 as an AMPKα1 phosphorylating activity
We carried out a third round of purification, focusing on fraction 12 (330 mM NaCl) from the first SP HP column (Fig. S2), with the AMPKα1-T183 phosphorylating activities monitored (Fig. S2).
Similar to the purification of MST3 described previously, extracts were made from 50 l of HEK293T cells cultured in suspension, and lysates containing approximately 5000 mg of proteins were used for purification. About 10% were used in an initial run, with AMPKα1-T183 phosphorylating activities determined after each column from the second column (Blue HP) to the last (Mono S) (Fig. S2). Because we have used the first column (SP HP) at least eight times (e.g., Fig. 2A in this article and Figs. 1D and 2A in the companion article) and repeatedly detected kinase activities in fraction 12 (330 mM NaCl), we did not monitor the first column (SP HP) in this round and went ahead with fraction 12 from the SP HP column and loaded it on the second column (Blue HP) (Fig. S2, B–G). We tested the other six different columns before designing a purification scheme (Fig. 3A).
Both AMPKα1-T183 and SIK3-T221 phosphorylating activities were monitored in fractions from the second and third columns (Blue HP and heparin HP). Both LKB1 and MST3 were also monitored in those fractions (Fig. S2, B and C). The same fractions from the heparin column contained AMPKα1-T183 and SIK3-T221 phosphorylating activities but no LKB1 or MST3 (Fig. S2, B and C). From the fourth (Q HP), fifth (HAP), to the sixth (Superdex 200) column (Fig. S2, D–F), we only monitored AMPKα1-T183 phosphorylation. For fractions from the last (Mono S) column, we again monitored both AMPKα1-T183 and SIK3-T221 phosphorylating activities as well as both LKB1 and MST3 (Fig. 3B).
No LKB1 or MST3 was detected from any of the fractions from the Mono S column (Fig. 3B). Fractions 12 and 13 (330–360 mM NaCl) from the last column showed a single band upon silver staining and were positive in phosphorylating both AMPKα1-T183 and SIK3-T221 (Fig. 3B). These two bands were analyzed by MS, and two protein kinases were found: MAP4K5 and vaccinia-related kinase 1 (VRK1) (Fig. 3C).
We expressed FLAG-tagged LKB1, CaMKK1, CaMKK2, MAP4K5, VRK1, and GFP individually in HEK cells and immunoprecipitated them. Each was tested for phosphorylating activities on recombinant AMPKα1 and recombinant SIK3. Recombinant AMPKα1 was phosphorylated at T183 by LKB1, CaMKK2, MAP4K5, weakly by CaMKK1, but not by VRK1 or GFP (Fig. 3D). Recombinant SIK3 was phosphorylated at T221 strongly by MAP4K5, moderately by CaMKK2, but not by CaMKK1, VRK1, or GFP immunoprecipitated from HEK cells (Fig. 3D).
MAP4K5 WT but not MAP4K5 S174A immunoprecipitated from HEK cells phosphorylated recombinant AMPKα1-T183 (Fig. 3E) and SIK3-T221 (Fig. 3F). MAP4K5 immunoprecipitated from HEK cells did not phosphorylate the T183A mutant of AMPKα1 (Fig. 3G) or the T221 mutant of SIK3 (Fig. 3H).
In vivo and in vitro AMPKα phosphorylation and enhancement of AMPKα1 activity by MAP4K5
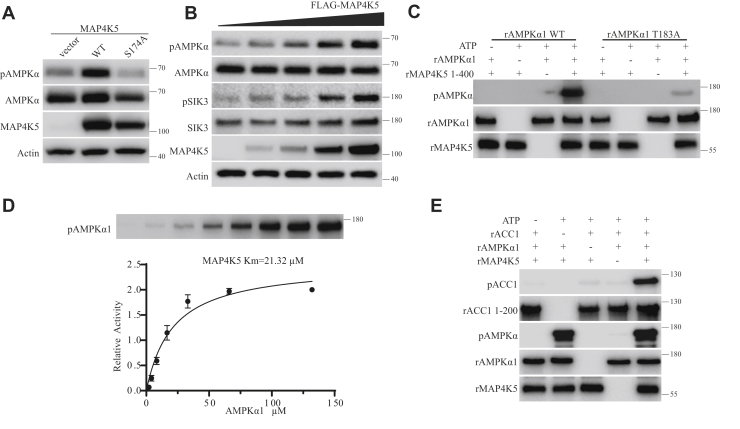
To investigate the effect of MAP4K5 on endogenous AMPKα at T172, we generated HEK293T cells from which both Lkb1 and Camkk2 genes were removed by gene targeting (“Lkb1 and Camkk2 double KO cells”). These cells were transfected with plasmids expressing MAP4K5 or MAP4K5 S174A mutant or a control plasmid. Phosphorylation of endogenous AMPKα-T172 was increased by MAP4K5 but not by MAP4K5 S174A or vector alone (Fig. 4A). We transfected an increasing dosage of the plasmid encoding MAP4K5 or its S174A mutant in Lkb1 and Camkk2 double KO cells. Higher levels of the MAP4K5 protein resulted in increased phosphorylation of AMPKα-T172 and SIK3-T221 (Fig. 4B). These results indicate that MAP4K5 could phosphorylate AMPKα-T172 and SIK3-T221 in HEK cells in a manner independent of LKB1 and CaMKK2.
Figure 4.
AMPKα and SIK3 phosphorylation by MAP4K5 in vivo and in vitro.A, control vector or plasmids encoding MAP4K5 WT or its S174A mutant were transfected into Lkb1 and Camkk2 double KO HEK293T cells. MAP4K5 WT but not its S174A mutant increased phosphorylation of endogenous AMPKα T172 in HEK cells. B, increasing concentrations of the plasmid expressing a FLAG-tagged MAP4K5 were transfected into Lkb1 and Camkk2 double KO HEK cells. After 24 h, cells were starved with DMEM without glucose and glutamine for 1 h before lysis and Western analysis. Increasing amounts of the plasmid led to increasing expression of FLAG-MAP4K5 protein and correspondingly increased phosphorylation of endogenous AMPKα-T172 and SIK3-T221 in HEK cells, in a manner independent of both LKB1 and CaMKK2. C, recombinant MAP4K5 expressed in Escherichia coli could directly phosphorylate recombinant AMPKα1 WT at T183 but not recombinant AMPKα1 T183A mutant. D, the Michaelis–Menten constant of recombinant MAP4K5 on recombinant AMPKα1 was 21.32 μM. E, recombinant MAP4K5 increased the catalytic activity of recombinant AMPKα1 on its substrate recombinant ACC1. The procedure was similar to that described in Figure 5J in the companion article. Prior treatment of recombinant AMPKα1 by recombinant MAP4K5 increased AMPKα1 catalyzed phosphorylation of recombinant ACC1. AMPK, AMP-activated protein kinase; CaMKK2, Ca2+/calmodulin-dependent protein kinase kinase 2; DMEM, Dulbecco's modified Eagle's medium; HEK293T, human embryonic kidney 293T cell; LKB1, liver kinase B1; MAP4K5, mitogen-activated protein kinase kinase kinase kinase 5; SIK3, salt-inducible kinase 3.
To test for direct phosphorylation of AMPKα1-T183 by MAP4K5, we expressed both as recombinant proteins in E. coli. Full-length MAP4K5 of 846 residues could not be generated from E. coli, and thus, its fragment containing amino acids 1 to 400 was generated and used as the MAP4K5 recombinant protein throughout this article (Figs. S6 and S7E). Recombinant AMPKα1 but not its T183A mutant could be phosphorylated by recombinant MAP4K5 (Fig. 4C).
To investigate the enzymatic kinetics of MAP4K5, we measured the Michaelis–Menten constant of MAP4K5 by using the same concentration of recombinant MAP4K5 with increasing concentrations of the recombinant AMPKα1 substrate (Fig. 4E). We found the Michaelis–Menten constant of MAP4K5 on AMPKα1-T183 to be 21.32 μM (Fig. 4D).
To examine whether recombinant MAP4K5 could increase the catalytic activity of recombinant AMPKα1, we pretreated AMPKα1 with MAP4K5 before assaying the activity of AMPKα1 on recombinant acetyl-CoA carboxylase 1 (ACC1). We found that recombinant MAP4K5 increased the activity of recombinant AMPKα1 on recombinant ACC1 (Fig. 4E). Recombinant MAP4K5 also phosphorylated recombinant AMPKα2 at T172 but not the T172A mutant of AMPKα2 (Fig. S3A). The Michaelis–Menten constant of MAP4K5 on AMPKα2-T172 was 20.10 μM (Fig. S3B). These results indicate that recombinant MAP4K5 phosphorylates both AMPKα1 and AMPKα2 directly in vitro.
Direct phosphorylation of ARKs by MST3 and MAP4K5
Fourteen ARKs have been reported (5, 6, 7, 8, 9). To test which ones could be phosphorylated by MST3 and MAP4K5, we expressed each of them as a recombinant protein in E. coli (Fig. S5) and tested them on each of the 14 recombinant ARKs as substrates (Fig. 5). Antibodies recognizing specific phosphorylation sites in other ARKs corresponding the T221 in SIK3 and T172 in AMPKα2 were used (10, 34, 35).
Figure 5.
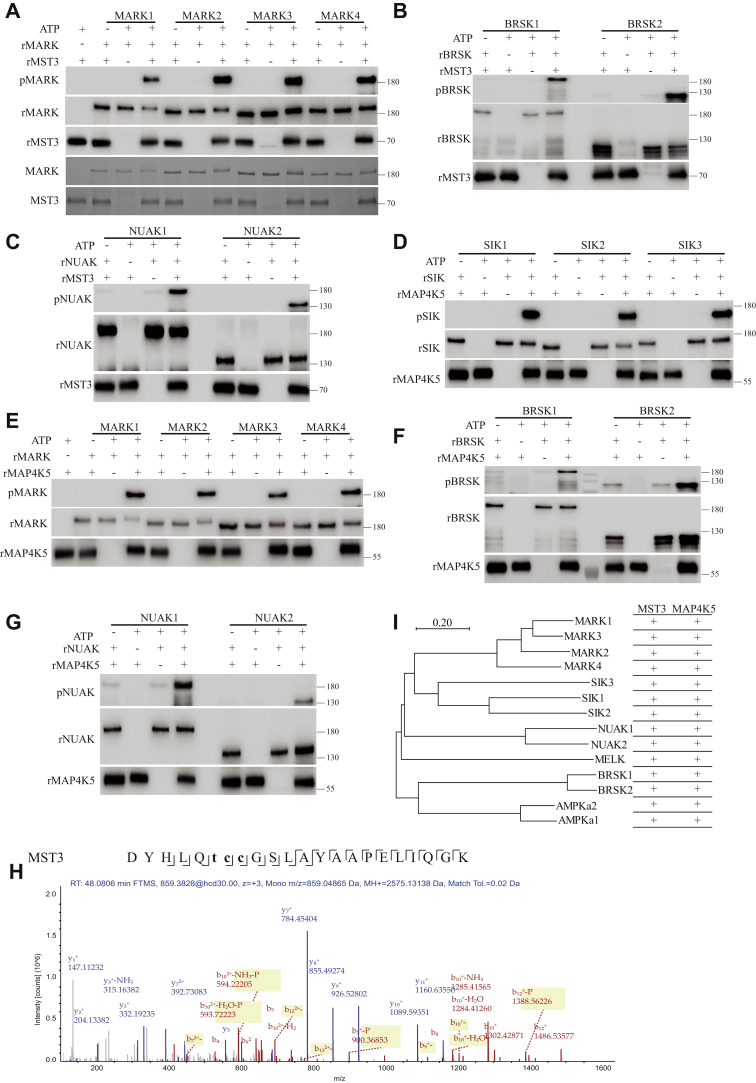
Phosphorylation of AMPK-related kinases by MST3 and MAP4K5 in vitro.A, recombinant MST3 from Escherichia coli phosphorylated recombinant MARK1 at T215, MARK2 at T208, MARK3 at T211, and MARK4 at T214 from E. coli (pMARK panel). The bottom two panels show Coomassie blue staining of recombinant proteins. B, recombinant MST3 phosphorylated recombinant BRSK1 at T189 and BRSK2 at T174 (pBRSK). rBRSK panel shows recombinant BRSK1 or 2 added to the reactions, rMST3 recombinant MST3 added to the reactions. C, recombinant MST3 phosphorylated recombinant NUAK1 at T211 and NUAK2 at T208. D, recombinant MAP4K5 phosphorylated recombinant SIK1 at T182, SIK2 at T175, and SIK3 at T221. E, recombinant MAP4K5 phosphorylated recombinant MARK1 at T215, MARK2 at T208, MARK3 at T211, and MARK4 at T214. F, recombinant MAP4K5 phosphorylated recombinant BRSK1 at T189 and BRSK2 at T174. G, recombinant MAP4K5 phosphorylated recombinant NUAK1 at T211 and NUAK2 at T208. H, MELK phosphorylation could not be analyzed by antibodies and was thus analyzed by MS. Phosphopeptides derived from MS chromatogram showing MELK T167 phosphorylation by MST3. Sequence: DYHLQTCCGSLAYAAPELIQGK, C7-carbamidomethyl (57.02146 Da), C8-carbamidomethyl (57.02146 Da), T6-phospho (79.96633 Da); charge: +3, monoisotopic m/z: 859.04865 Da (−1.44 mmu/−1.67 ppm), MH+: 2575.13138 Da, and RT: 48.0806 min; identified with: Sequest HT (version 1.17); XCorr: 5.65, percolator q value: 0.0e0, percolator PEP: 1.1e−5, ptmRS: best site probabilities: T6 (phospho): 100, and ions matched by search engine: 0/0. I, summary of MST3 and MAP4K5 phosphorylation of ARKs shown in the phylogeny tree of ARKs. AMPK, AMP-activated protein kinase; ARK, AMP-activated protein kinase–related kinase; MAP4K5, mitogen-activated protein kinase kinase kinase kinase 5; MELK, maternal embryonic leucine zipper kinase; MS, mass spectrometry; MST3, mammalian sterile 20–like kinase 3.
Recombinant MST3 phosphorylated recombinant MARK1 at T215, MARK2 at T208, MARK3 at T221, and MARK4 at T214 (Fig. 5A). Recombinant MST3 phosphorylated recombinant BR serine/threonine kinase 1 (BRSK1) at T189 and BRSK2 at T174 (Fig. 5B). Recombinant MST3 phosphorylated recombinant NUAK family SNF1-like kinase 1 (NUAK1) at T211 and NUAK2 at T208 (Fig. 5C). Taken together with our results that recombinant MST3 phosphorylated SIK3 at T221 (Fig. 4, A–C in the companion article), SIK1 at T182 and SIK2 at T175 (Fig. 4, F and G in the companion article), and AMPKα1 at T183 and α2 at T172 (Fig. 5D), these findings indicate that MST3 can directly phosphorylate all known ARKs.
Recombinant MAP4K5 phosphorylated recombinant SIK1-T182, SIK2-T175, and SIK3-T221 (Fig. 5D). Recombinant MAP4K5 phosphorylated recombinant MARK1-T215, MARK2-T208, MARK3-T221, and MARK4-T214 (Fig. 5E). Recombinant MAP4K5 phosphorylated recombinant BRSK1-T189 and BRSK2-T174 (Fig. 5F). Recombinant MAP4K5 phosphorylated recombinant NUAK1-T211 and NUAK2-T208 (Fig. 5G). No antibodies were available to recognize the corresponding phosphorylation of maternal embryonic leucine zipper kinase (MELK) at T167 (Fig. S5). We used MS to analyze phosphorylation of recombinant MELK expressed in and purified from E. coli by recombinant MST3 or recombinant MAP4K5. Phosphorylation of recombinant MELK at T167 by recombinant MST3 (Fig. 5H) and MAP4K5 was indeed detected by MS (Fig. S4).
Taken together with our results that recombinant MAP4K5 phosphorylated AMPKα1 and AMPKα2 (Fig. S3, A and B), these findings indicate that MAP4K5 can directly phosphorylate all 14 known ARKs. We summarize these results in Figure 5I by plotting the phosphorylating activities against the phylogenic tree of ARKs (Fig. 5I).
Phosphorylation of AMPKα1 and SIK3 by STE20-like kinases immunoprecipitated from HEK cells
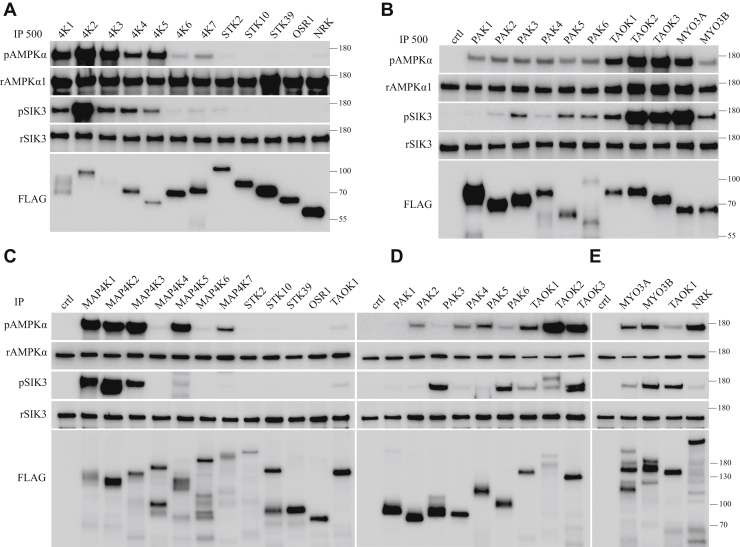
To study the effects of all STE20 kinases on AMPK and SIK3, we expressed each STE20 kinase as a FLAG-tagged fragment (containing the kinase domain) and in the full-length protein in HEK cells and immunoprecipitated them. Each was tested for phosphorylating activities on recombinant AMPKα1 and recombinant SIK3 (Fig. 6, A–E). For full-length STE20 kinases, thousand and one amino acid kinase 1 (TAOK1) was used in different panels to allow for comparison of signal strength (Fig. 6, C–E).
Figure 6.
Phosphorylation of AMPKα1 and SIK3 by STE20-like kinases immunoprecipitated from HEK cells.A–E, fragments (Figs. S6 and S7) or full-length forms of STE20-like kinases tagged with the FLAG epitope were individually expressed in HEK cells and immunoprecipitated. Each was tested for activities on recombinant AMPKα1 and recombinant SIK3. TAOK1 was used in different panels to allow references of signal strength (Fig. 6, C–E). AMPK, AMP-acivated protein kinase; HEK, human embryonic kidney cell; SIK3, salt-inducible kinase 3; STE20, sterile 20.
Recombinant AMPKα1 was phosphorylated at T183 strongly by immunoprecipitated fragments of MAP4K1, MAP4K2, MAP4K3, MAP4K5, TAOK2, TAOK3, and MYO3A (Fig. 6A), moderately by immunoprecipitated fragments of MAP4K4 and TAOK1 (Fig. 6, A and B), and weakly by MAP4K6, MAP4K7, MYO3B, p21 protein-activated kinase 1 (PAK1), PAK2, PAK3, PAK4, PAK5, and PAK6 (Fig. 6B) but not by immunoprecipitated fragments of STK2, STK10, STK39, OSR1, or NRK (Fig. 6A).
Recombinant SIK3 was phosphorylated at T221 strongly by immunoprecipitated fragments of MAP4K2 (Fig. 6A), TAOK3, TAOK2, and MYO3A (Fig. 6B), moderately by immunoprecipitated fragments of MAP4K1, MAP4K3, MAP4K4, and MAP4K5 (Fig. 6A), weakly by immunoprecipitated fragments of MAP4K7 (Fig. 6B), TAOK1, MYO3B, PAK3, PAK2, PAK6, PAK4, and PAK5 (Fig. 6B) but not by immunoprecipitated fragments of MAP4K6, STK2, STK10, OSR1, STK39, PAK1, and NRK (Fig. 6A).
We succeeded in expressing the full-length form of every STE20 kinases. For NRK, it required approximately 100 times the number of cultured HEK cells to obtain the amount of protein equivalent to other STE20 kinases. Recombinant AMPKα1 was phosphorylated at T183 strongly by immunoprecipitated full-length forms of MAP4K1, MAP4K2, MAP4K3, MAP4K5 (Fig. 6C), TAOK2, and TAOK3 (Fig. 6D), moderately by NRK, MYO3A, and MYO3B (Fig. 6E), and weakly by MAP4K6, MAP4K4, MAP4K7, TAOK1 (Fig. 6C), PAK3, PAK2, PAK6, PAK4, and PAK5 (Fig. 6D), but not by immunoprecipitated full-length forms of STK2, STK10, OSR1, and STK39 (Fig. 6C). The major difference between the results from fragments and those from the full-length forms is that full-length NRK could, but its fragment could not, phosphorylate AMPKα1-T183.
Recombinant SIK3 was phosphorylated at T221 strongly by immunoprecipitated full-length forms of MAP4K1, MAP4K2, and MAP4K3 (Fig. 6C), moderately by MAP4K5 (Fig. 6C), TAOK3, PAK3, and PAK6 (Fig. 6D), weakly by NRK, MAP4K7 (Fig. 6E), TAOK1, TAOK2, MYO3A, MYO3B, PAK4, and PAK5 (Fig. 6D) but not by MAP4K6, STK2, STK10, OSR1, STK39 (Fig. 6C), and PAK1 (Fig. 6D). The major difference between the results from fragments and those from the full-length forms is that full-length NRK could weakly, but its fragment could not, phosphorylate SIK3-T221.
Figure 7I summarizes the aforementioned results under the “IP” columns, with “Frag” indicating a fragment of a STE20 kinase and “Full” as the full-length form of the same STE20 kinase (Fig. 7I, the left half).
Figure 7.
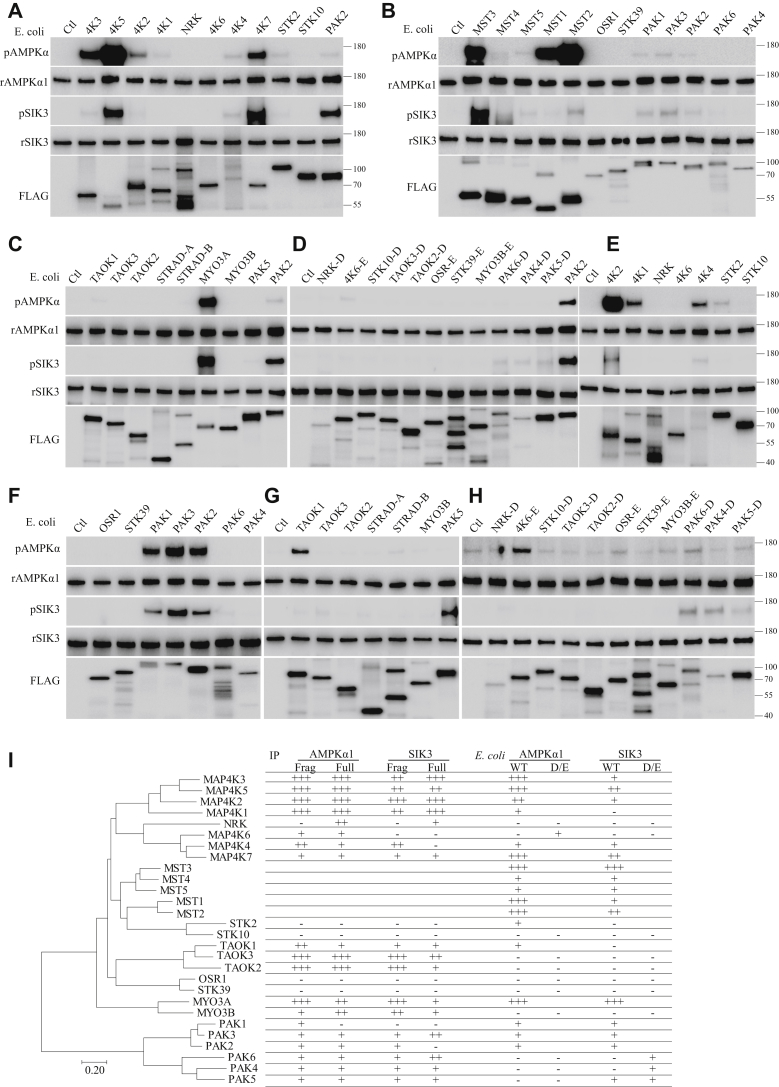
Phosphorylation of AMPKα1 and SIK3 by STE20-like kinases purified from Escherichia coli. A–C, full length or fragments of STE20-like kinases tagged with the FLAG epitope (Figs. S6 and S7) were individually expressed in and purified from E. coli before being assayed with recombinant SIK3 and AMPKα1. Kinases were aligned similar to the evolutionary tree (I). D and H, for STE20 kinases whose WT forms were unable to phosphorylate SIK3 and AMPKα1, recombinant proteins with S to D or T to E mutations were expressed in and purified from E. coli before being tested for their activities on SIK3 and AMPKα1. Signals in (D) can be compared with those in (A) by referencing to PAK2. E–H, exposure time in these panels was increased over those in (A–C) to allow for detection of weak signals. Signals in (E) can be compared with those in (A) by referencing MAP4K2. Signals in (F) can be compared with those in (A) by referencing PAK2. Signals in (G) can be compared with those in (C) by referencing TAOK1. I, summary of AMPKα1 and SIK3 phosphorylation by STE20 kinases, in the phylogeny tree of the latter. AMPK, AMP-activated protein kinase; SIK3, salt-inducible kinase 3; STE20, sterile 20.
Phosphorylation of AMPKα1 and SIK3 by STE20-like kinases immunoprecipitated from E. coli
To directly determine the catalytic activities of STE20 kinases, we also expressed every STE20 kinase as a recombinant protein in E. coli. If the full-length was too long to be expressed successfully in E. coli, we expressed fragments of them containing the entire kinase domain. If a recombinant kinase from E. coli was inactive on both AMPKα1 and SIK3, we further used E. coli to express its mutant carrying an S to D or T to E mutation in the activation loop (as described in Fig. S6) (36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49).
Recombinant STE20 kinases were assayed for phosphorylation of AMPKα1-T183 and SIK3-T221 (Fig. 7, A–G). The exposure time for Figure 7, A–C was similar and all shorter than that for Figure 7, E–G. The longer exposure time was used to allow weaker signals to show in the absence of glaringly strong signals. Usually one of the same samples was used in different panels to allow references of signal strength (PAK2 in Fig. 7, A–F; MAP4K2 and MAP4K1 for Fig. 7E comparison with Fig. 7A; TAOK1 for Fig. 7G comparison with Fig. 7C).
Recombinant AMPKα1-T183 was phosphorylated strongly by recombinant MAP4K3, MAP4K5, MAP4K7, MST3, MST1, MST2, and MYO3A, moderately by MAP4K2, and weakly by MAP4K1, MAP4K4, MST4, MST5, STK2, PAK1, PAK3, PAK2, and TAOK1. Recombinant AMPKα1-T183 was not phosphorylated by recombinant WT NRK, MAP4K6, STK10, STK39, TAOK3, TAOK2, OSR1, MYO3B, PAK6, PAK4, or PAK5. T187E mutation in the activation loop of MAP4K6 allowed it to phosphorylate AMPKα1-T183 (Fig. 7H). By contrast, similar mutations in the activation loops of NRK (S211D), MAP4K6 (T187E), STK10 (S191D), STK39 (T231E), TAOK3 (S177D), TAOK2 (S181D), OSR1 (T185E), STK39 (T231E), MYO3B (T190E), PAK6 (S560D), PAK4 (S474D), or PAK5 (S206D) did not allow any of the corresponding STE20 kinases to phosphorylate AMPKα1-T183 (Fig. 7, D and H).
SIK3-T221 was phosphorylated strongly by MST3 and MYO3A, moderately by MAP4K5, MAP4K7, and MST2, and weakly by MAP4K3, MAP4K4, MST4, MST5, MST1, PAK1, PAK3, PAK2, and PAK5. SIK3-T221 was not phosphorylated by MAP4K1, NRK, MAP4K6, STK2, STK10, TAOK1, TAOK3, TAOK2, OSR1, STK39, MYO3B, PAK6, or PAK4. Mutations in their activation loops (PAK6 S560D and PAK4 S474D) allowed PAK6 and PAK4 to phosphorylate SIK3-T221 weakly.
The right part in Figure 7I (under the “E. coli” columns) summarizes results of recombinant SIK3 phosphorylation at T221 and recombinant AMPKα1 phosphorylation at T183 by recombinant STE20 kinases (shown as WT in Fig. 6I) and recombinant D/E mutant versions of STE20 kinases (shown as D/E in Fig. 6I) (mutation sites listed in Supporting Appendix, Fig. S6). The phylogenic tree of human STE20 kinases is placed on the left side, and the substrates (AMPKα1 and SIK3) are placed on the top of the right part.
Of the 28 human STE20 kinases, 14 recombinant STE20 kinases could phosphorylate both AMPKα1-T183 and SIK3-T221 (Fig. 7I): MAP4K3, MAP4K5, MAP4K2, MAP4K7, MST3, MST4, MST5, MST1, MST2, STK2, MYO3A, PAK1, PAK3, and PAK2. Among these 14 recombinant STE kinases, MST3 and MYO3A phosphorylated both AMPK and SIK3 strongly, MAP4K4, MST4, MST5, PAK1, PAK2, and PAK3 phosphorylated both AMPK and SIK3 weakly, whereas MAP4K3, MAP4K5, MAP4K2, MAP4K1, MAP4K4, MAP4K7, MST1, MST2, STK2, and TAOK1 phosphorylated AMPKα1-T183 more strongly than SIK3-T221.
Ten recombinant STE20 kinases purified from E. coli could phosphorylate neither AMPKα1-T183 nor SIK3-T221 (Fig. 7, E–G): NRK, MAP4K6, STK10, TAOK3, TAOK2, OSR1, STK39, MYO3B, PAK6, and PAK4. Among these 10 STE20 kinases, T187E mutants of MAP4K6 and MAP4K7 could phosphorylate AMPKα1-T183 but still not SIK3-T221, whereas S560D or S474D mutant of PAK6 and PAK4 could phosphorylate SIK3 weakly but not AMPK. Among the 10 STE20 kinases, no phosphorylation activities on SIK3-T221 and AMPKα1-T183 were detected from either the WT or D/E mutant versions of seven recombinant kinases: NRK, STK10, TAOK3, TAOK2, OSR1, STK39, and MYO3B.
Three recombinant STE20 kinases from E. coli (MAP4K1, STK2, and TAOK1) could phosphorylate AMPKα1-T183 weakly but not SIK3-T221. The S171D mutant of MAP4K1 (Fig. 6) phosphorylated AMPKα1-T183 more strongly but still not SIK3-T221 (Fig. 7E).
Three recombinant STE20 proteins from E. coli (PAK5, PAK6, and PAK4), either in WT or S to D mutant forms, could phosphorylate only SIK3-T221 but not AMPKα1-T183.
Discussion
Our biochemical purifications and characterizations have led to the discovery that multiple mammalian STE20 kinases can phosphorylate the ARKs.
The novelty of our discovery is supported by the findings that STE20 phosphorylation of ARKs is independent from LKB1 and CaMKK2 (Fig. 4, A and B in this article; Fig. 5B in the companion article; Fig. S4, A and B in the companion article), and that recombinant STE20 kinases can directly phosphorylate recombinant AMPK kinases (Figs. 4C, 5, D–H, 6, C–E, 7, A–H and S3; and Figs. 4, 5,E–G and I in the companion article; and Fig. S4, G and H in the companion article).
Because both ARKs and the STE20 kinases are functionally important, our discovery represents a major step in linking these two subfamilies biochemically and will stimulate further studies into the significance of their relationships in biologically interesting signaling as well as the physiology and the pathology of pairing between each of the STE20 kinases and each of the ARKs.
The phosphorylation of ARKs by STE20 kinases was specific. Although multiple fractions from each of the fractionation columns during our experiments were active, not all fractions were active, in phosphorylating SIK3-T221 and AMPKα2-T172 (Figs. 1, A–E, 2 and S2 in this article; Figs. 1, B–F and 2 in the companion article). During our experiments, we have tested other kinases and found them to be inactive in phosphorylating SIK3-T221 and AMPKα2-T172. FES (Fig. 3D in the companion article), STK39 (Fig. 3D in the companion article), ADCK2 (Fig. 3D in the companion article), STK38 (Fig. S1D), CDK16 (Fig. S1D), CaMKD2 (Fig. S1D), and VRK1 (Fig. 3D) were not STE20 kinases, and each of them expressed in and immunoprecipitated from HEK cells was inactive in phosphorylating SIK3-T221 or AMPKα2-T172. Seven recombinant STE20 kinases (NRK, STK10, TAOK3, TAOK2, OSR1, STK39, and MYO3B) (Fig. 7), either in their WT or S to D/T to E mutant forms, could not phosphorylate SIK3-T221 and AMPKα-T172 (Figs. 7 and 3D). Because the full-length NRK immunoprecipitated from HEK cells was active on AMPKα1 and SIK3, it is possible that the fragment expressed in E. coli was too short to be active.
The substrates for STE20 kinases were also specific. STE20 kinases tested here (MST3 and MAP4K5) could not phosphorylate HDAC4 (Fig. 4E) or ACC1 (Fig. 5J in the companion article and Fig. S4E and S4E in the companion article).
MSTs immunoprecipitated from HEK cells (Fig. 5,H and I in the companion article) showed more phosphorylation activities on AMPKα1-T183 and SIK3-T221 than MSTs purified from E. coli (Fig. 7). Again, it is possible that immunoprecipitation could bring down associated proteins (or other factors), which facilitated kinase activities.
When all 14 ARKs were tested as substrates of two STE20 kinases (MST3 and MAP4K5), they appeared to be phosphorylated by both. It should be noted that only two ARKs had been tested as substrates (AMPKα1-T183 and SIK3-T221) for all 28 STE20 kinases. It will be interesting to test for possible phosphorylation of all 14 ARKs by all 28 STE20 kinases.
While our biochemical reactions with recombinant STE20 kinases and ARKs from E. coli or with STE20 kinases immunoprecipitated from HEK and ARKs from E. coli have shown STE20 kinases can phosphorylate ARKs in vitro, we have no evidence whether any of the STE20 kinases identified here can phosphorylate any of the ARKs in vivo. We have no evidence that any STE20 kinase functions upstream of any ARKs in a physiologically significant signaling pathway. We also have no evidence whether any of the STE20 and ARK pairing is important in the pathogenesis of any diseases. But these are all interesting questions for future studies.
MST3 contributes to the endogenous activities phosphorylating AMPK and SIK3 present in HEK cells and in the mouse brain. When Lkb1 was removed by gene targeting in HEK cells (Fig. 1B, compared with Fig. 1A), the activities present in fractions 1 and 2 for phosphorylating SIK3 and AMPK1 were significantly reduced, and LKB1 protein was present in fractions 1 and 2 of the WT HEK cells (Fig. 1, A and C). When Mst3 was removed by gene targeting in HEK cells (Fig. 1C, compared with Fig. 1A), the activities present in fractions 5, 6, and 7 for phosphorylating SIK3 and AMPK1 were significantly reduced, and MST3 protein was present in fractions 5 and 6 of the WT HEK cells (Fig. 1, A and B). Furthermore, we have generated mouse KO for Mst3. When extracts from the brains of Mst3 KO mice were compared with extracts from WT mouse brains (Fig. 1, D and E), the activities present in fractions 6, 7, and 8 for phosphorylating SIK3 and AMPK1 were significantly reduced, and MST3 protein was present in fractions 6 and 7 of WT mouse brain extracts (Fig. 1D).
The exact in vivo relationships between each of the 28 mammalian STE20 kinases and the 14 ARKs will require time and efforts by many laboratories in different research fields to determine, with our discoveries regarding the biochemical relationship between two major subfamilies of kinases stimulating physiological and pathological studies because both subfamilies have been implicated in normal situations and in diseases ranging from (but not limited to) metabolic, neurological, and immunological disorders to cancer and longevity.
Experimental procedures
Generation of LKB1 and CaMKK2 KO HEK293 cells
Lkb1/Camkk2 double-deficient and Mst3-deficient HEK293T cells were generated by CRISPR–Cas9. Briefly, guide sequences targeting Lkb1 (CCACCGCATCGACTCCACCGAGG), Camkk2 (GAGACAGCTTGCGACCGGAGAGG), and Mst3 (GGCCATCTACCTAGCGGAGG) were inserted into the pX459 vector. The pX459 containing guide sequences were transfected into HEK293T cells, and the single clones were selected from 96-well plates by limited dilution. Success of gene targeting was confirmed by immunoblotting with appropriate antibodies.
Purification of CaMKK2 from HEK293T cells
The purification was similar to that of MST3. HEK293T cell lysates (500 ml at a concentration of 10 mg/ml) were purified by seven sequentially connected chromatography steps (SP HP, Q HP, Blue HP, heparin HP, HAP, Superdex 200, and Mono S) as diagramed in Fig. S6A. Results from each of the first six columns examined by Western analysis are shown in Figure 2, A–G. Results from the last column are shown in Fig. S1B.
Purification of MAP4K5 from HEK293T cells
The purification was similar to that of MST3. HEK293T cell lysates (500 ml at a concentration of 10 mg/ml) were purified by seven sequential connected chromatography steps as illustrated in Figure 3A and results of Western analysis shown in Fig. S2. Cell lysates were loaded onto two tandem-connected 5 ml HiTrap SP HP columns followed by in vitro SIK3 and AMPKα2 phosphorylation assays. Fraction 12 (330 mM NaCl) from the SP HP columns was pooled and loaded on two tandem-connected 5 ml HiTrap Blue HP columns and eluted with a linear gradient of 0 to 3000 mM NaCl in 30 column volume buffer A. Fractions were dialyzed and assayed of phosphorylating SIK3 and AMPKα1. Fractions 15 to 20 (1400–2000 mM NaCl) from the Blue columns (Fig. S2B) were pooled and loaded on Heparin HP. Fractions 13 to 16 (360–450 mM NaCl) from the heparin columns (Fig. S2C) were pooled and loaded on Q HP. Fractions 9 and 10 (240–270 mM NaCl) from the Q HP columns (Fig. S2D) were pooled and loaded to the HAP column. Fractions 13 to 15 (180–220 mM K2PO4) from the HAP columns (Fig. S2E) were pooled, concentrated to 0.5 ml, and subjected to Superdex 200 10/300 GL column, and eluted with 200 mM NaCl in buffer A. The resulting fractions were dialyzed and assayed for SIK3 and AMPKα1 phosphorylation activity. At the next step, the active fraction (fraction 17) from Superdex 200 (Fig. S2F) was loaded on a Mono S 5/50 GL column and eluted with a linear gradient of 20 column volume buffer A from 0 to 600 mM NaCl. The fractions were dialyzed against buffer A, and each was assayed for activity and silver staining (Fig. 3B).
Generation of Mst3 KO mice
Mst3/Stk24 KO mice were created with EGE system (CRISPR–Cas9) by Beijing Biocytogen. Different concentrations of Cas9 mRNA, single-guide RNA, and donor vector were mixed and coinjected into the cytoplasm of one-cell stage fertilized eggs. Founder 0 mice were mosaic, and mouse tail junction PCR and sequencing analysis were used to screen potential founder 0 mice with the correct genetic recombination, which were further bred with C57BL/6N mice. F1 mice were further screened by Southern blot, tail junction PCR with primers (WT-forward: TGTTTTCAGATGTCTCTAGCAC TGG; WT-reverse: GAAGAGGTATGAGGTTGCTTAGTGC with product size 425 bp and WT-forward: mutant-R CGAAAGGGGGTCTTACACTATCAC with product size 47,761 bp in WT and 486 bp in mutant mice) and sequencing to confirm the absence of random insertions as well as correct genetic recombination. Mutant lines were back-crossed to C57BL/6N for at least five generations to exclude possible off-targeting.
All procedures were approved by Center for Innovative Biomedical Resources animal research committee.
Data availability
All data are contained in the article.
Supporting information
This article contains supporting information (seven supporting figures).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful to Peking-Tsinghua Center for Life Sciences, Center for Innovative Biomedical Resources, Changping Laboratory, and Shenzhen Bay Laboratory for support, to Dr Dong Liu, Dr Qi Zhang, and Xin-dan Qiu at the National Center for Protein Sciences at Peking University for help with MS sample preparing and data analysis.
Author contributions
Y. R. and Y. L. conceptualization; Y. L. methodology; Y. L., T. V. W., Y. C., C. L., and L. J. validation; Y. R., Y. L., T. V. W., C. L., and L. J. formal analysis; Y. L., T. V. W., Y. C., C. L., and L. J. investigation; Y. R. and Y. L. writing–original draft; Y. L., T. V. W., C. L., and L. J. writing–review and editing; Y. L., T. V. W., and Y. C. visualization; Y. R. supervision; Y. R. funding acquisition.
Edited by Henrik Dohlman
Supporting information
Purification of CaMKK2 from HEK293 T lysates. The scheme is similar to our purification of MST3, with the exception that the focus was on Fractions 1 to 4 (0–90 mM NaCl) from the first SP HP column (shown in Fig. 2). A, A diagram of purification with 7 chromatography columns. B, Silver staining (top panel), and AMPKα1 and SIK3 phosphorylating activities (middle panels) of fractions from the Mono S column. Bottom panel: Western analysis of LKB1. Bands 1, 2 and 3 from Fraction 9 were subjected to MS analysis. C, Kinases detected by MS analysis. CDK16, CaMKK1, STK38, CaMKK2, CaMK2D are protein serine/threonine kinases. PANK1 is pantothenate kinase 1. D, Recombinant AMPKα1 was strongly phosphorylated at T183 by FLAG-tagged CaMKK2, but not by FLAG-tagged CDK16, STK38, CaMK2D or PANK1 immunoprecipitated from HEK cells. Recombinant SIK3 was very weakly phosphorylated at T221 by CaMKK2, but not by CDK16, STK38, CaMK2D or PANK1 immunoprecipitated from HEK cells.
Purification of MAP4K5 from HEK293 T lysates. Purification was similar to that described for MST3, with the exception that, while MST3 was purified from Fraction 7 (180 mM NaCl) of the SP HP column, the present purification was focused on Fraction 12 (330 mM NaCl) from the SP HP column (cf. Fig. 2 of this paper, and Figs. 1 and 2 of the companion article). Results shown here came from purification of 500 mg lysates, passed onto SP HP, Blue HP, heparin HP, Q HP, HAP, Superdex 200 and Mono S columns. AMPKα1 T183 phosphorylating activities were monitored after each fractionation. LKB1 and MST3 were monitored by Western analysis after the Blue HP and the heparin HP columns. Active fractions from each column were pooled and loaded onto the next column: Fractions 15 to 20 (1400–1900 mM NaCl) from the Blue HP column, Fractions 13 to 16 (360–450 mM NaCl) from the heparin HP column, Fractions 9 and 10 (240–270 mM NaCl) from the Q HP column, Fractions 13 to 15 (180–210 mM K2PO4) from the HAP column, and Fraction 17 from the Superdex 200 column. Fractions from Mono S 5/50 Gl column were dialyzed against buffer A and each was assayed for kinase activity and by silver staining. After the initial run, all the rest of the 5000 mg lysates were purified through the same columns, with activities monitoring only after the last column (Mono S). Phosphorylating activities and silver staining of the fractions from Mono S column were shown in Figure 3.
Phosphorylation of AMPKα2-T172 by MAP4K5.A, Recombinant MAP4K5 directly phosphorylated recombinant AMPKα2 WT at T172 in vitro, but not T172A mutant of AMPKα2. B, The Michaelis-Menten constant of recombinant MAP4K5 on AMPKα2 T172 was 20.10 μM.
MELK Phosphorylation analyzed by mass spectrometry. Phosphopeptides derived from MS chromatogram showing MELK T167 phosphorylation by MAP4K5. Sequence: GNKDYHLQTCCGSLAYAAPELIQGK, C10-Carbamidomethyl (57.02146 Da), C11-Carbamidomethyl (57.02146 Da), T9-Phospho (79.96633 Da); Charge: +3, Monoisotopic m/z: 958.76752 Da (-2.35 mmu/-2.45 ppm), MH+: 2874.28800 Da, RT: 36.4501 min; Identified with: Sequest HT (v1.17); XCorr:3.56, Percolator q-Value:0.0e0, Percolator PEP:2.9e-6, ptmRS: Best Site Probabilities:T9(Phospho): 100, Ions matched by search engine: 0/0.
Recombinant AMPK related kinases: fragments and T to E mutants. Attempts to generate full length recombinant proteins in E. coli were successful for some ARKs but not for others (usually the larger ones). Thus, fragments for some ARKs were generated. “Truncated Protein” listed the length of each recombinant protein, and those not listed under “Truncated Protein” were produced as the full-length proteins. The T residue under “Activation Loop Phosphorylation Site” is equivalent to T221 of SIK3 and T172 of AMPKα2.
Fragments and S to D or T to E mutants of STE20 kinases. Attempts to generate full length recombinant STE20 proteins in E. coli were successful for some but not for others (usually the larger ones). We thus made fragments (as listed under “Truncated Protein” here). The S or T residue listed under “Activation Loop Phosphorylation Site” could be mutated to D or E for each STE20 kinases (as listed under “Mutant Protein).
Recombinant proteins expressed in E. coli. Coomassie blue staining of recombinant proteins used in this paper. 1 μg recombinant proteins were denatured by heating at 95 °C with the protein loading buffer and subjected to 12% SDS-PAGE followed by staining with Coomassie blue R250. MST1 to 5, MST3b, mMST3 and VRK1 were made as 3xFLAG tag-kinase-8xHis tag. The others were made as MBP-TEV site-3xFLAG tag-kinase-TEV site-GFP-8xHis tag. TEV site is the cleavage site for Tobacco Etch Virus protease. When these recombinant proteins were used as substrates, they were not cleaved. When they were used as kinases, they were first treated by the TEV protease to release the kinase part from the rest of the fusion protein.
References
- 1.Hardie D.G. AMP-activated protein kinase: Maintaining energy homeostasis at the cellular and whole-body levels. Annu. Rev. Nutr. 2014;34:31–55. doi: 10.1146/annurev-nutr-071812-161148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.López M., Nogueiras R., Tena-Sempere M., Diéguez C. Hypothalamic AMPK: A canonical regulator of whole-body energy balance. Nat. Rev. Endocrinol. 2016;12:421–432. doi: 10.1038/nrendo.2016.67. [DOI] [PubMed] [Google Scholar]
- 3.Hardie D.G., Schaffer B.E., Brunet A. Ampk: An energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 2016;26:190–201. doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herzig S., Shaw R.J. Ampk: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardie D.G. Ampk: A target for drugs and natural products with effects on both diabetes and cancer. Diabetes. 2013;62:2164–2172. doi: 10.2337/db13-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinberg G.R., Dandapani M., Hardie D.G. AMPK: Mediating the metabolic effects of salicylate-based drugs? Trends Endocrinol. Met. 2013;24:481–487. doi: 10.1016/j.tem.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carling D. AMPK signalling in health and disease. Curr. Opin. Cell Biol. 2017;45:31–37. doi: 10.1016/j.ceb.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Day E.A., Ford R.J., Steinberg G.R. AMPK as a therapeutic target for Treating metabolic diseases. Trends Endocrinol. Met. 2017;28:545–560. doi: 10.1016/j.tem.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Russell F.M., Hardie D.G. AMP-activated protein kinase: Do we need activators or inhibitors to treat or prevent cancer? Int. J. Mol. Sci. 2021;22:186. doi: 10.3390/ijms22010186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lizcano J.M., Göransson O., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Mäkelä T.P., Hardie G.G., Alessi D.R. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., Zhou E., Liu Y., Yu J., Yang J., Li C., Cui Y., Wang T., Li C., Liu Z., Rao Y. Sleep need, the key regulator of sleep homeostasis, is indicated and controlled by phosphorylation of threonine 221 in salt inducible kinase 3. bioRxiv. 2021 doi: 10.1101/2021.11.06.467421. [preprint] [DOI] [PubMed] [Google Scholar]
- 12.Funato H., Miyoshi C., Fujiyama T., Kanda T., Sato M., Wang Z., Ma J., Nakane S., Tomita J., Ikkyu A., Kakizaki M., Hotta-Hirashima N., Kanno S., Komiya H., Asano F., et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature. 2016;539:378–383. doi: 10.1038/nature20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park M., Miyoshi C., Fujiyama T., Kakizaki M., Ikkyu A., Honda T., Choi J., Asano F., Mizuno S., Takahashi S., Yanagisawa M., Funato H. Loss of the conserved PKA sites of SIK1 and SIK2 increases sleep need. Sci. Rep. 2020;10:8676. doi: 10.1038/s41598-020-65647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawley S.A., Boudeau J., Reid J.L., Mustard K.J., Udd L., Mäkelä T.P., Alessi D.R., Hardie D.G. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woods A., Johnstone S.R., Dickerson K., Leiper F.C., Fryer L.G.D., Neumann D., Schlattner U., Wallimann T., Carlson M., Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 16.Sutherland C.M., Hawley S.A., McCartney R.R., Leech A., Stark M.J.R., Schmidt M.C., Hardie D.G. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 2003;13:1299–1305. doi: 10.1016/s0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- 17.Hong S.-P., Leiper F.C., Woods A., Carling D., Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw R.J., Kosmatka M., Bardeesy N., Hurley R.L., Witters L.A., DePinho R.A., Cantley L.C. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto K., McCarthy A., Smith D., Green K.A., Hardie D.G., Ashworth A., Alessi D.R. Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J. 2005;24:1810–1820. doi: 10.1038/sj.emboj.7600667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw R.J., Lamia K.A., Vasquez D., Koo S.-H., Bardeesy N., DePinho R.A., Montminy M., Cantley L.C. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemminki A., Markie D., Tomlinson I., Avizienyte E., Roth S., Loukola A., Bignell G., Warren W., Aminoff M., Höglund P., Järvinen H., Kristo P., Pelin K., Ridanpää M., Salovaara R., et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 22.Hawley S.A., Pan D.A., Mustard K.J., Ross L., Bain J., Edelman A.M., Frenguelli B.G., Hardie D.G. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Hurley R.L., Anderson K.A., Franzone J.M., Kemp B.E., Means A.R., Witters L.A. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- 24.Woods A., Dickerson K., Heath R., Hong S.-P., Momcilovic M., Johnstone S.R., Carlson M., Carling D. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Anderson K.A., Ribar T.J., Lin F., Noeldner P.K., Green M.F., Muehlbauer M.J., Witters L.A., Kemp B.E., Means A.R. Hypothalamic CaMKK2 contributes to the regulation of energy balance. Cell Metab. 2008;7:377–388. doi: 10.1016/j.cmet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Momcilovic M., Hong S.P., Carlson M. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J. Biol. Chem. 2006;281:25336–25343. doi: 10.1074/jbc.M604399200. [DOI] [PubMed] [Google Scholar]
- 27.Xie M., Zhang D., Dyck J.R., Li Y., Zhang H., Morishima M., Mann D.L., Taffet G.E., Baldini A., Khoury D.S., Schneider M.D. A pivotal role for endogenous TGF-beta-activated kinase-1 in the LKB1/AMP-activated protein kinase energy-sensor pathway. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17378–17383. doi: 10.1073/pnas.0604708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholz R., Sidler C.L., Thali R.F., Winssinger N., Cheung P.C., Neumann D. Autoactivation of transforming growth factor beta-activated kinase 1 is a sequential bimolecular process. J. Biol. Chem. 2010;285:25753–25766. doi: 10.1074/jbc.M109.093468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann D. Is TAK1 a direct upstream kinase of AMPK? Int. J. Mol. Sci. 2018;19:2412. doi: 10.3390/ijms19082412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dan I., Watanabe N.M., Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 31.Rawat S.J., Chernoff J. Regulation of mammalian Ste20 (Mst) kinases. Trends Biochem. Sci. 2015;40:149–156. doi: 10.1016/j.tibs.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pombo C.M., Iglesias C., Sartages M., Zalvide J.B. MST kinases and metabolism. Endocrinology. 2019;160:1111–1118. doi: 10.1210/en.2018-00898. [DOI] [PubMed] [Google Scholar]
- 33.Delpire E. The mammalian family of sterile 20p-like protein kinases. Pflug. Arch. Eur. J. Phy. 2009;458:953–967. doi: 10.1007/s00424-009-0674-y. [DOI] [PubMed] [Google Scholar]
- 34.Drewes G., Ebneth A., Preuss T., Mandelkow E.-M., Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 35.Brajenovic M., Joberty G., Küster B., Bouwmeester T., Drewes G. Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J. Biol. Chem. 2004;279:12804–12811. doi: 10.1074/jbc.M312171200. [DOI] [PubMed] [Google Scholar]
- 36.Abo A., Qu J., Cammarano M.S., Dan C., Fritsch A., Baud V., Belisle B., Minden A. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. Embo J. 1998;17:6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King C.C., Gardiner E.M.M., Zenke F.T., Bohl B.P., Newton A.C., Hemmings B.A., Bokoch G.M. p21-activated kinase (PAK1) is phosphorylated and activated by 3-phosphoinositide-dependent kinase-1 (PDK1) J. Biol. Chem. 2000;275:41201–41209. doi: 10.1074/jbc.M006553200. [DOI] [PubMed] [Google Scholar]
- 38.Parrini M.C., Lei M., Harrison S.C., Mayer B.J. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell. 2002;9:73–83. doi: 10.1016/s1097-2765(01)00428-2. [DOI] [PubMed] [Google Scholar]
- 39.Qu J., Cammarano M.S., Shi Q., Ha K.C., de Lanerolle P., Minden A. Activated PAK4 regulates cell adhesion and anchorage-independent growth. Mol. Cell Biol. 2001;21:3523–3533. doi: 10.1128/MCB.21.10.3523-3533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright J.H., Wang X., Manning G., LaMere B.J., Le P., Zhu S., Khatry D., Flanagan P.M., Buckley S.D., Whyte D.B., Howlett A.R., Bischoff J.R., Lipson K.E., Jallal B. The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. Mol. Cell Biol. 2003;23:2068–2082. doi: 10.1128/MCB.23.6.2068-2082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou T., Raman M., Gao Y., Earnest S., Chen Z., Machius M., Cobb M.H., Goldsmith E.J. Crystal structure of the TAO2 kinase domain: Activation and specificity of a Ste20p MAP3K. Structure. 2004;12:1891–1900. doi: 10.1016/j.str.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 42.Arnold R., Patzak I.M., Neuhaus B., Vancauwenbergh S., Veillette A., Van Lint J., Kiefer F. Activation of hematopoietic progenitor kinase 1 involves relocation, autophosphorylation, and transphosphorylation by protein kinase D1. Mol. Cell Biol. 2005;25:2364–2383. doi: 10.1128/MCB.25.6.2364-2383.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei M., Robinson M.A., Harrison S.C. The active conformation of the PAK1 kinase domain. Structure. 2005;13:769–778. doi: 10.1016/j.str.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Vitari A.C., Deak M., Morrice N.A., Alessi D.R. The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem. J. 2005;391:17–24. doi: 10.1042/BJ20051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pike A.C.W., Rellos P., Niesen F.H., Turnbull A., Oliver A.W., Parker S.A., Turk B.E., Pearl L.H., Knapp S. Activation segment dimerization: A mechanism for kinase autophosphorylation of non-consensus sites. Embo J. 2008;27:704–714. doi: 10.1038/emboj.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan L., Mieulet V., Burges D., Findlay G.M., Sully K., Procter J., Goris J., Janssens V., Morrice N.A., Lamb R.F. PP2A(T61 epsilon) is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol. Cell. 2010;37:633–642. doi: 10.1016/j.molcel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 47.Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucl. Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quintero O.A., Unrath W.C., Stevens S.M., Manor U., Kachar B., Yengo C.M. Myosin 3A kinase activity is regulated by phosphorylation of the kinase domain activation loop. J. Biol. Chem. 2013;288:37126–37137. doi: 10.1074/jbc.M113.511014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q., Nirala N.K., Nie Y., Chen H.-J., Ostroff G., Mao J., Wang Q., Xu L., Ip Y.T. Ingestion of food particles regulates the mechanosensing misshapen-yorkie pathway in Drosophila intestinal growth. Dev. Cell. 2018;45:433–449. doi: 10.1016/j.devcel.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Purification of CaMKK2 from HEK293 T lysates. The scheme is similar to our purification of MST3, with the exception that the focus was on Fractions 1 to 4 (0–90 mM NaCl) from the first SP HP column (shown in Fig. 2). A, A diagram of purification with 7 chromatography columns. B, Silver staining (top panel), and AMPKα1 and SIK3 phosphorylating activities (middle panels) of fractions from the Mono S column. Bottom panel: Western analysis of LKB1. Bands 1, 2 and 3 from Fraction 9 were subjected to MS analysis. C, Kinases detected by MS analysis. CDK16, CaMKK1, STK38, CaMKK2, CaMK2D are protein serine/threonine kinases. PANK1 is pantothenate kinase 1. D, Recombinant AMPKα1 was strongly phosphorylated at T183 by FLAG-tagged CaMKK2, but not by FLAG-tagged CDK16, STK38, CaMK2D or PANK1 immunoprecipitated from HEK cells. Recombinant SIK3 was very weakly phosphorylated at T221 by CaMKK2, but not by CDK16, STK38, CaMK2D or PANK1 immunoprecipitated from HEK cells.
Purification of MAP4K5 from HEK293 T lysates. Purification was similar to that described for MST3, with the exception that, while MST3 was purified from Fraction 7 (180 mM NaCl) of the SP HP column, the present purification was focused on Fraction 12 (330 mM NaCl) from the SP HP column (cf. Fig. 2 of this paper, and Figs. 1 and 2 of the companion article). Results shown here came from purification of 500 mg lysates, passed onto SP HP, Blue HP, heparin HP, Q HP, HAP, Superdex 200 and Mono S columns. AMPKα1 T183 phosphorylating activities were monitored after each fractionation. LKB1 and MST3 were monitored by Western analysis after the Blue HP and the heparin HP columns. Active fractions from each column were pooled and loaded onto the next column: Fractions 15 to 20 (1400–1900 mM NaCl) from the Blue HP column, Fractions 13 to 16 (360–450 mM NaCl) from the heparin HP column, Fractions 9 and 10 (240–270 mM NaCl) from the Q HP column, Fractions 13 to 15 (180–210 mM K2PO4) from the HAP column, and Fraction 17 from the Superdex 200 column. Fractions from Mono S 5/50 Gl column were dialyzed against buffer A and each was assayed for kinase activity and by silver staining. After the initial run, all the rest of the 5000 mg lysates were purified through the same columns, with activities monitoring only after the last column (Mono S). Phosphorylating activities and silver staining of the fractions from Mono S column were shown in Figure 3.
Phosphorylation of AMPKα2-T172 by MAP4K5.A, Recombinant MAP4K5 directly phosphorylated recombinant AMPKα2 WT at T172 in vitro, but not T172A mutant of AMPKα2. B, The Michaelis-Menten constant of recombinant MAP4K5 on AMPKα2 T172 was 20.10 μM.
MELK Phosphorylation analyzed by mass spectrometry. Phosphopeptides derived from MS chromatogram showing MELK T167 phosphorylation by MAP4K5. Sequence: GNKDYHLQTCCGSLAYAAPELIQGK, C10-Carbamidomethyl (57.02146 Da), C11-Carbamidomethyl (57.02146 Da), T9-Phospho (79.96633 Da); Charge: +3, Monoisotopic m/z: 958.76752 Da (-2.35 mmu/-2.45 ppm), MH+: 2874.28800 Da, RT: 36.4501 min; Identified with: Sequest HT (v1.17); XCorr:3.56, Percolator q-Value:0.0e0, Percolator PEP:2.9e-6, ptmRS: Best Site Probabilities:T9(Phospho): 100, Ions matched by search engine: 0/0.
Recombinant AMPK related kinases: fragments and T to E mutants. Attempts to generate full length recombinant proteins in E. coli were successful for some ARKs but not for others (usually the larger ones). Thus, fragments for some ARKs were generated. “Truncated Protein” listed the length of each recombinant protein, and those not listed under “Truncated Protein” were produced as the full-length proteins. The T residue under “Activation Loop Phosphorylation Site” is equivalent to T221 of SIK3 and T172 of AMPKα2.
Fragments and S to D or T to E mutants of STE20 kinases. Attempts to generate full length recombinant STE20 proteins in E. coli were successful for some but not for others (usually the larger ones). We thus made fragments (as listed under “Truncated Protein” here). The S or T residue listed under “Activation Loop Phosphorylation Site” could be mutated to D or E for each STE20 kinases (as listed under “Mutant Protein).
Recombinant proteins expressed in E. coli. Coomassie blue staining of recombinant proteins used in this paper. 1 μg recombinant proteins were denatured by heating at 95 °C with the protein loading buffer and subjected to 12% SDS-PAGE followed by staining with Coomassie blue R250. MST1 to 5, MST3b, mMST3 and VRK1 were made as 3xFLAG tag-kinase-8xHis tag. The others were made as MBP-TEV site-3xFLAG tag-kinase-TEV site-GFP-8xHis tag. TEV site is the cleavage site for Tobacco Etch Virus protease. When these recombinant proteins were used as substrates, they were not cleaved. When they were used as kinases, they were first treated by the TEV protease to release the kinase part from the rest of the fusion protein.
Data Availability Statement
All data are contained in the article.