Abstract
CasRx, a recently discovered member of the type VI CRISPR system with minimum size, offers a new approach for RNA manipulation with high efficiency and specificity in prokaryotes and eukaryotes. However, in vivo studies of functional recovery using the CasRx system have not been well characterized. Here, we sought to establish an adeno-associated virus (AAV)-CasRx-guide RNA (gRNA) system for the specific knockdown of Htra2 transcript to protect mice from aminoglycosides-induced hearing loss. For the study, we verified an optimized gRNA in vitro, which was packaged into a single AAV with CasRx, and injected the packaged AAV into mice with hearing loss induced by neomycin and auditory functions investigated by auditory brainstem response tests. Upon using the AAV-CasRx-gRNA system, we found the knockdown of Htra2 transcript led to less cochlear hair cell loss and improved auditory function, with low off-target and adverse side effects. Additionally, the decrease in Htra2 significantly inhibits mRNA expression of Casp3 and Casp9. In conclusion, the AAV-CasRx-gRNA-mediated knockdown of Htra2 transcript in mice has been proved effective and safe for preventing hearing loss induced by aminoglycosides and, thus, represents a promising genetic approach for the future clinical applications for treating non-inherited hearing loss.
Key words: RNA editing, AAV delivery, CRISPR-CasRx, Htra2, aminoglycosides, acquired sensorineural hearing loss
Graphical abstract
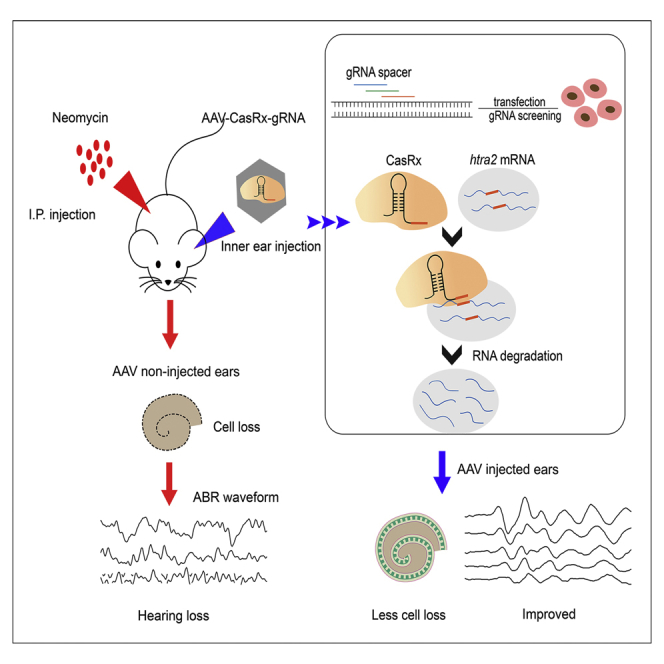
The study addressed the efficacy and safety of AAV-CasRx-mediated knockdown in preventing drug-induced ototoxicity in vivo and in vitro, which provide the novel approach for the treatment of non-inherited hearing loss in future clinical use.
Introduction
CRISPR-Cas systems were originally discovered in bacteria and archaea as a mechanism for defending against exogenic elements,1 which can be divided into the class I and the class II systems.2 The class II CRISPR system, which relies on a single effector enzyme, includes types II (CRISPR-Cas9), V (CRISPR-Cas12), and VI (CRISPR-Cas13), among which the types II and V mediate the targeting of DNA, while the type VI mediates the targeting and cleavage of single-strand RNA.3, 4, 5
Currently, six subtypes in the Cas13 family have been identified, including Cas13a, Cas13b, Cas13c, Cas13d, CasX, and CasY.6, 7, 8, 9, 10, 11 Since the first report of a Cas13 orthologue (CRISPR/LshCas13a), more Cas13 variants with various catalytic activities and specificities have been discovered.4,6,7 Among these subtypes, LwaCas13a, PspCas13, RfxCas13d, and Cas13X.1 have been widely applied for the knockdown experiments in mammalian cells, which exhibits higher efficiency and specificity than traditional RNA interference.6,7,10 Notably, Cas13d, a VI-D effector from Ruminococcus flavefaciens XPD3002 (CasRx) with the small size has shown robust catalytic activity for transcriptome targeting and engineering in mammals, and no side effects have been reported.8,10,12 In addition, CasRx-mediated processing of RNA is temporary and effective without inducing permanent and heritable changes.6 Thus, CasRx-mediated RNA engineering highlights the accessibility of AAV-mediated delivery for future clinical therapy.10,13 However, the validity of the CasRx system in in vivo function-restoration study remains unknown.
Acquired sensorineural hearing loss affects approximately 1.3 billion people worldwide and is mainly caused by noise, aging, infection, ototoxic drugs, and genetic defects.14 Notably, drug-induced ototoxicity is one of the major preventable factors that contribute to hearing loss.15 More than 150 drugs are known to be ototoxic, including aminoglycosides, macrolide antibiotics, quinine, and salicylic analgesics.16 Aminoglycosides are widely used owing to their low cost and high effectiveness, but the drugs can induce irreversible ototoxicity to auditory or vestibular components, leading to functional impairment or permanent hearing loss.17,18 Despite extensive researches, the iatrogenic ototoxicity remains a pressing issue.15
Omi/Htra2 (GenBank: NC_000072.7) encodes high-temperature requirement protein A2, which is a mitochondrial serine proteinase that participates in cell apoptosis, and the inhibitors of apoptosis proteins (IAPs) serve as endogenous inhibitors of apoptosis by inhibiting the activation of Caspase-3 and Caspase-9.19 Htra2 promotes apoptosis by directly binding with IAPs and inhibiting the protease activity of IAPs.20,21 Recent studies have demonstrated that Htra2 is highly expressed in the inner ears of mice with neomycin-induced ototoxicity,22,23 and knockout of Htra2 using CRISPR/Cas9 can effectively prevent the hearing loss caused by neomycin.23 However, the unintended off-target effects and irreversibility associated with CRISPR/Cas9 may limit its potential for clinical applications.24
Studies on CasRx-mediated knockdown have been reported in bacteria,25 plants,26 and various cell types.27,28 For example, Zhou et al.13 alleviated neurological disease in mice by knocking down Ptbp1 using CasRx, and Jiang et al.28 successfully applied the CasRx system to control pancreatic cancer progression by silencing mutant KrasG12D. However, the application of CasRx in acquired sensorineural hearing loss has not been well studied until now. In this study, we examined the protective effects of CasRx-mediated knockdown of Htra2 against inner ear damage and auditory function in mice with neomycin-induced hearing loss and evaluated the safety and potential of CasRx technology for future clinical applications.
Results
Screening of gRNA for specific knockdown of Htra2 transcript
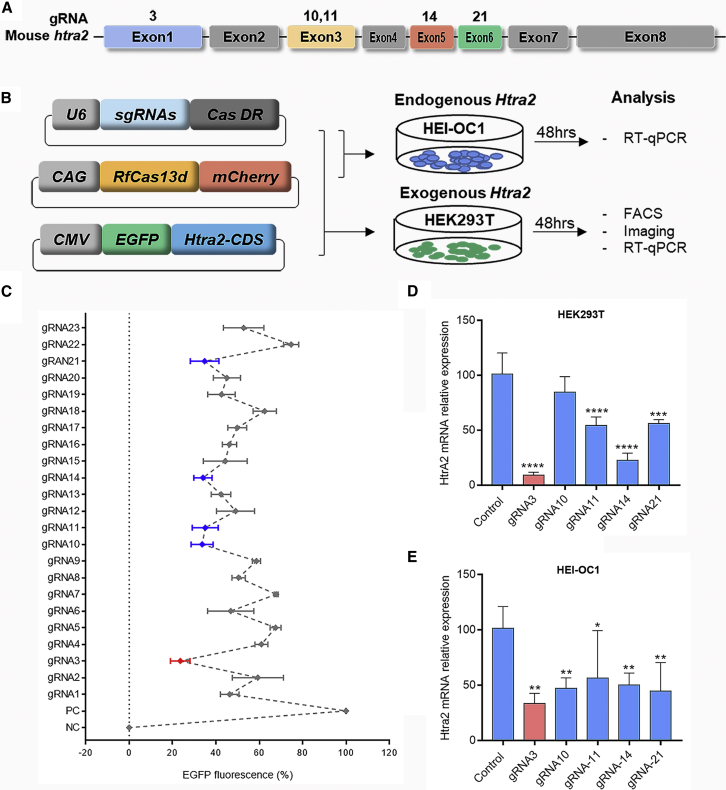
To achieve the specific knockdown of mouse Htra2 mRNA with high efficiency, we constructed a three-plasmid system, including an mCherry reporter-infused CasRx-expressing plasmid (CasRx-mCherry), an EGFP reporter-infused Htra2-expressing plasmid (Htra2-EGFP), and a gRNA-backbone plasmid that were co-transfected into HEK293T cells (Figure 1B). To optimize the gRNA, we designed 23 gRNAs targeting different exons, some of which were designed according to a guide-RNA design website (https://cas13design.nygenome.org)29 (Figure 1A and Table S1). Owing to the degradation of the exogenous Htra2 transcript by the CasRx system, we concluded that gRNA3 (g3)-transfected group was the most effective one based on the EGFP fluorescence among the 23 gRNA-transfected groups. A fluorescence-activated cell sorting (FACS) analysis showed that the average proportion of EGFP-positive cells in the g3-transfected group decreased by 76.4% compared with the control group (Figure 1C). In addition, the fluorescent images for the top five groups with low EGFP fluorescence were shown in Figure S1, and the sporadic fluorescent cells were captured in the g3-transfected group (Figure S1). To further verify the editing efficiency of g3 in vitro, we examined the mRNA expression of exogenous Htra2 by transfecting HEK293T cells with three-plasmid system and the mRNA expression of endogenous Htra2 by transfecting HEI-OC1 cells with CasRx-mCherry and g3 two-plasmid system (Figure 1B). The Htra2 mRNA expression in HEK293T and HEI-OC1 cells were dramatically decrease by 90.8% (Figure 1D) and 66.4% (Figure 1E), respectively, indicating that CasRx-g3 effectively knocked down the mouse Htra2 transcript.
Figure 1.
The screening of gRNA for specifically targeting Htra2 transcripts
(A) Design of five gRNAs targeting exons of the Htra2 gene. (B) Schematic illustration of the gRNA screening mediated by the CasRx system. Three plasmids, including the gRNA expression vector, RfxCas13d-mCherry fluorescence vector, and Htra2-EGFP fluorescence vector, were constructed. RfxCas13d-mCherry vector and g3 expression vector were transfected into HEI-OC1 cells, and the three plasmids mentioned above were transfected into HEK293T cells. (C) The percentage of HEK293T cells with EGFP fluorescence when transfected with different gRNAs. The graph shows the mean ± SD (n ≥ 3 biologically independent samples). (D and E) The relative mRNA expression was assessed by RT-qPCR after transfection with the top five gRNAs in HEK293T (D) and HEI-OC1 cells (E). Data are shown as the mean ± SD. The statistical analyses are performed by ordinary one-way ANOVA with Bonferroni’s multiple comparisons test, ∗, ∗∗,∗∗∗, ∗∗∗∗p < 0.05, 0.01, 0.001, 0.0001 vs. control.
Validation of AAV-PHP.eB transduction efficacy in the inner ear
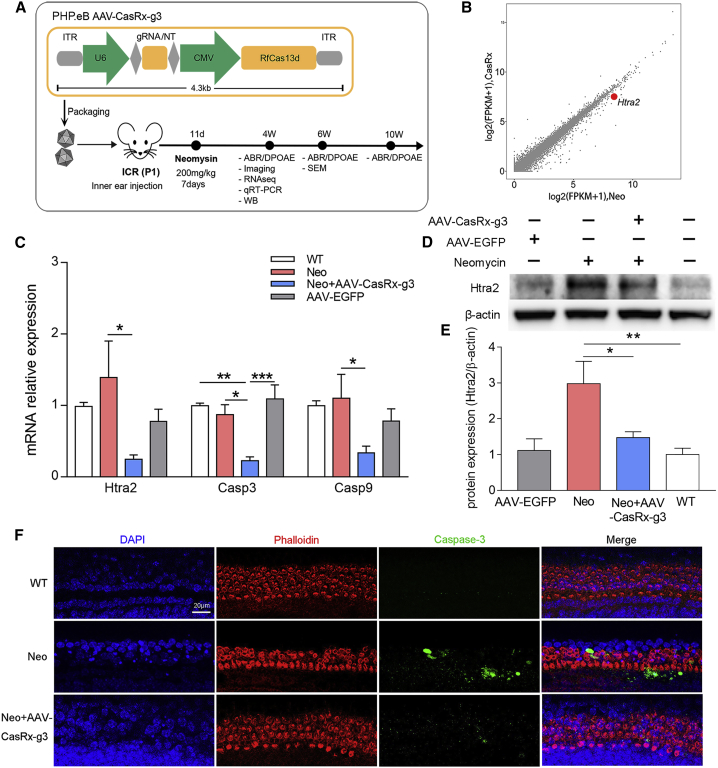
AAV-PHP.eB, which is an AAV serotype that has extremely high transduction efficacy in both outer hair cells (OHCs) and inner hair cells (IHCs),30 was used as the delivery vector for the following experiments. CasRx and gRNA3 were packaged into a single AAV-PHP.eB, which was then injected into the inner ears at P1 mice. The hearing loss model was constructed at P11 in mice by neomycin exposure for 7 days in a row. Next, the restoration of hearing function was examined at week 4 and 6 through functional studies and molecular experiments (Figure 2A).
Figure 2.
AAV-PHP.eB-CasRx-mediated Htra2 knockdown
(A) The experimental outline of AAV-CasRx-g3-mediated therapy in mice. (B) The fragments per kilobase of transcript per million fragments mapped reads (FPKM) values based on RNA-seq analysis were compared between Neo and AAV-CasRx-g3 groups, which showed the specific knockdown of Htra2 in the CasRx-mediated therapeutic system. (C) The relative mRNA expression of Htra2, Casp3, and Casp9 in using RT-qPCR. Data are presented as the mean ± SEM (n = 4). Statistical analysis was performed by one-way ANOVA with Bonferroni’s multiple comparisons test for each gene, ∗, ∗∗, ∗∗∗p < 0.05, 0.01, 0.001. (D) The western blot results of Htra2 in the WT, neomycin-treated (Neo), AAV-CasRx-g3-treated (AAV-CasRx-g3) and AAV-EGFP-treated (AAV-EGFP) groups at 4 weeks. (E) The statistical graph of western blot. The relative expression of Htra2 protein were analyzed by comparing it with β-actin expression based on their gray value using ImageJ software. Data are shown as the mean ± SEM (n = 3). Statistical analysis was performed by One-way ANOVA with Bonferroni’s multiple comparisons test, ∗p < 0.05, ∗∗p < 0.01. (F) The immunofluorescent staining of phalloidin and Caspase-3 in cochlear explants after AAV-CasRx-g3 treatment for 24 h before neomycin exposure. Samples were stained with phalloidin (red), Caspase-3 (green), and DAPI (blue). Scale bar: 20 μm. WT, control mice; Neo, neomycin treated mice; Neo+AAV-CasRx-g3, neomycin treated mice with AAV-CasRx-g3 transfection.
To verify its high transduction efficiency in hair cells, we injected 500 nL of AAV-PHP.eB-EGFP into the scala media of the inner ear and collected the cochleae for immunostaining after 2 weeks. The transduction efficiency of AAV-PHP.eB-EGFP in injected ears were nearly 100% for IHCs and 90.6 ± 0.8%, 94.3 ± 3.2%, and 89.1 ± 1.4% for the apical, middle, and basal turns of OHCs, respectively (Figures S2A and S2B). In addition, the low EGFP fluorescence were also detected in the apex, middle, and base turns of non-injected ears (Figures S2A and S2B), indicating a minimal spreading out of AAV from injected ears to the contralateral side. The results demonstrate that AAV-PHP.eB is an effective delivery vector with a high transduction efficacy for cochlear hair cells.
Effective knockdown of Htra2 transcripts in AAV-CasRx-g3-injected cochleae
To investigate the effect of Htra2 knockdown, cochleae were collected from neomycin-treated animals with or without AAV-CasRx-g3 at the age of 4 weeks for the analysis of gene and protein expression. The Htra2 mRNA level of all transgenomes was decreased after CasRx-mediated treatment through RNA sequencing (RNA-seq) analysis (Figure 2B) and was decreased by 82.4% in AAV-CasRx-injected ears compared with non-injected ears after neomycin exposure through quantitative RT-qPCR (Figure 2C). However, the mRNA expression of Htra2 in AAV-EGFP group were not significantly changed compared with the wild-type (WT) group. Each group included four independent samples. Moreover, the protein level of Htra2 was also evaluated by western blot. The relative expression of Htra2 compared with β-actin was significantly decreased in AAV-CasRx-g3 group, which indicated a much fainter band compared with the neomycin-treated group. However, the expression of the Htra2 had nosignificance between the AAV-EGFP group and the WT group, indicating that injection of AAV-EGFP had no effect on the expression of Htra2 (Figures 2D and 2E). The results suggest the specific knockdown of Htra2 transcripts by the AAV-CasRx-g3 system in vivo.
Inhibition of apoptosis by CasRx-mediated knockdown of Htra2
The role of Htra2 in apoptosis has been well studied.20 To determine the changes of apoptotic proteins after Htra2 knockdown, we evaluated the mRNA expression of Casp3 and Casp9 in vitro and in vivo. According to the RT-qPCR results in HEI-OC1 cells and HEK293T cells, the relative mRNA expression of Casp3 and Casp9 increased significantly in the neomycin-treated group, but decreased dramatically after the treatment of AAV-CasRx-g3 compared with the control group (Figure S3). Meanwhile, the relative mRNA expression of apoptotic proteins from different treatment groups were also analyzed. The mRNA level of Casp3 and Casp9 both showed a significant decrease in the AAV-CasRx-g3 group compared with the Neo group, but no significant variations between the WT group and the AAV-EGFP group were found (Figure 2C). Moreover, the basilar membranes from the postnatal day 1 (P1) mice were extracted and co-cultured with AAV-CasRx-g3 for 24 h followed by neomycin exposure, which were subjected to Caspase-3 staining for apoptotic measurements. Rhodamine-conjugated phalloidin, a bicyclic peptide used for actin filaments staining, was applied for labeling the hair cells. The confocal images showed the elevated Caspase-3 in neomycin-treated mice, and the minimum Caspase-3 in WT mice. In contrast, this increase was inhibited by AAV-CasRX-g3-treated mice which exhibited significantly lower Caspase-3 expression than the Neo group. These results are consistent with Casp3mRNA expression (Figure 2F).
Improvement of auditory function in mice with neomycin-induced hearing loss
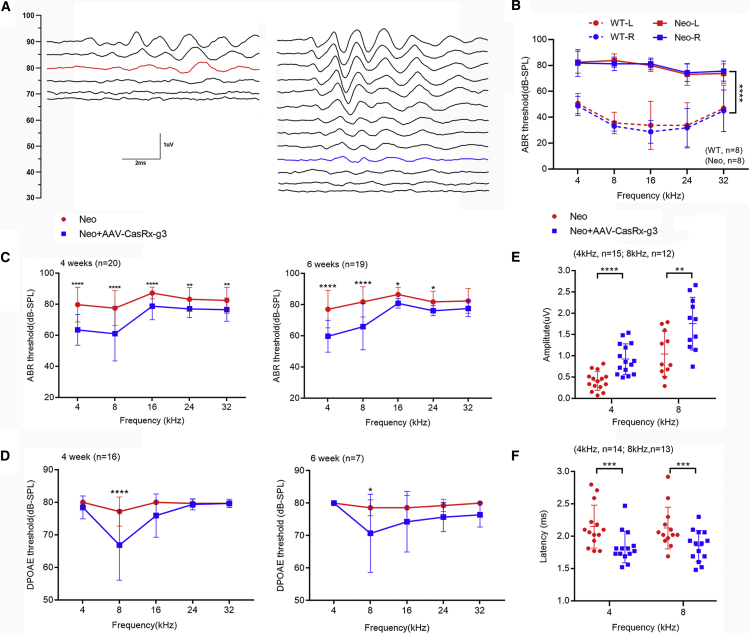
To confirm the neomycin-induced ototoxicity mouse model, we measured the frequency-specific tone-evoked auditory brainstem responses (ABRs). Frequency-specific ABRs are commonly used to study cochlear function in each region, including the apex, middle, and base turns.31,32 The ABR thresholds were elevated to 70–90 dB in the neomycin-treated mice compared with the ABR thresholds of 20–55 dB in WT mice, which demonstrated the hearing loss in mice after neomycin exposure (Figure 3B).
Figure 3.
The improved auditory function in AAV-CasRx-g3-injected mice after neomycin exposure
(A) The ABR waveform recorded at 2 weeks after the last dose of neomycin in AAV-CasRx-g3-injected and non-injected ears. The red line indicates the threshold for the non-injected ear, while the blue line indicates the threshold of the AAV-injected ear. (B) The comparison of ABR thresholds between the left and right ears in WT (n = 8) and neomycin-treated mice (n = 8). (C and D) The ABR (C) and DPOAE (D) thresholds of AAV-injected ears and non-injected ears were measured at 4 and 6 weeks after neomycin exposure. Statistical analysis was performed by two-way ANOVA with Bonferroni’s multiple comparisons test. (4-week ABR: n = 20; 6-week ABR: n = 19; 4-week DPOAE: n = 16; 6-week DPOAE: n = 7). (E and F) The peak amplitudes (E) and latencies (F) of ABR wave 1 evoked by 90 dB SPL in AAV-CasRx-g3 injected and non-injected ears after neomycin exposure. An unpaired two-tailed Student t-test was used for statistical analysis. All data are presented as the mean ± SD.∗, ∗∗, ∗∗∗, ∗∗∗∗p < 0.05, 0.01, 0.001, 0.0001 vs. each control.
To investigate the protective effects of the AAV-CasRx-g3 system on hearing function, ABR thresholds were measured in the bilateral ears in neomycin-treated mice at 4 and 6 weeks, respectively. The ears with AAV-CasRx-g3 injection showed significant restoration of hearing at low frequencies (4 kHz and 8 kHz) at all time points. At the age of 4 weeks, the ABR thresholds in the AAV-CasRx-g3-injected ears were significantly decreased at all frequencies (64 ± 10 dB, 61 ± 18 dB, 79 ± 9 dB, 77 ± 6 dB and 77 ± 7 dB at 4, 8, 16, 24, and 32 kHz, respectively) compared with non-injected ears (80 ± 11 dB, 78 ± 11 dB, 87 ± 4 dB, 83 ± 7 dB and 83 ± 8 dB at 4, 8, 16, 24 and 32 kHz, respectively) (Figure 3C). A representative waveform recorded at 45 dB in the AAV-CasRx-g3-injected ear and at 80 dB in the non-injected ear at a frequency of 8 kHz at 4 weeks is shown in Figure 3A. The ABR thresholds were also measured in neomycin-treated mice at 6 weeks, which showed a significant decrease at 4, 8, 16, and 24 kHz in the AAV-CasRx-g3-injected ear (Figure 3C). In contrast, the ABR thresholds of the AAV-control-injected ears with neomycin exposure had no significant changes compared with the non-injected ears at 4 weeks (Figure S4A). The ABR thresholds of mice injected with AAV-control without neomycin treatment showed no significant differences between the bilateral ears at any frequency, and their average values were close to those of WT mice (Figure S4B). The results indicated that AAV control injection had no treatment effects in neomycin-injured mice and no side effects. The evoked ABR peak 1 amplitude stimulated at 90 dB SPL at 4 kHz and 8 kHz were significantly higher in the AAV-CasRx-g3-injected group compared with the non-injected group at 4 and 6 weeks after neomycin exposure (Figure 3E). Similarly, the latencies of the evoked ABR peak 1 with 90 dB SPL stimulation were shorter in the AAV-CasRx-g3-injected group compared with the non-injected group (Figure 3F). In addition, we measured the distortion product otoacoustic emissions (DPOAEs) of AAV-CasRx-g3-injected mice to evaluate the function of OHCs. The DPOAE thresholds were significantly decreased in the ears injected with AAV-CasRx-g3 (67 ± 11 dB and 71 ± 12 dB at 4 and 6 weeks, respectively) at 8 kHz compared with the non-injected ears (77 ± 4 dB and 79 ± 2 dB at 4 and 6 weeks, respectively), which revealed the preservation of OHC function at a low frequency in the CasRx-mediated treatment of neomycin-injured mice (Figure 3D).
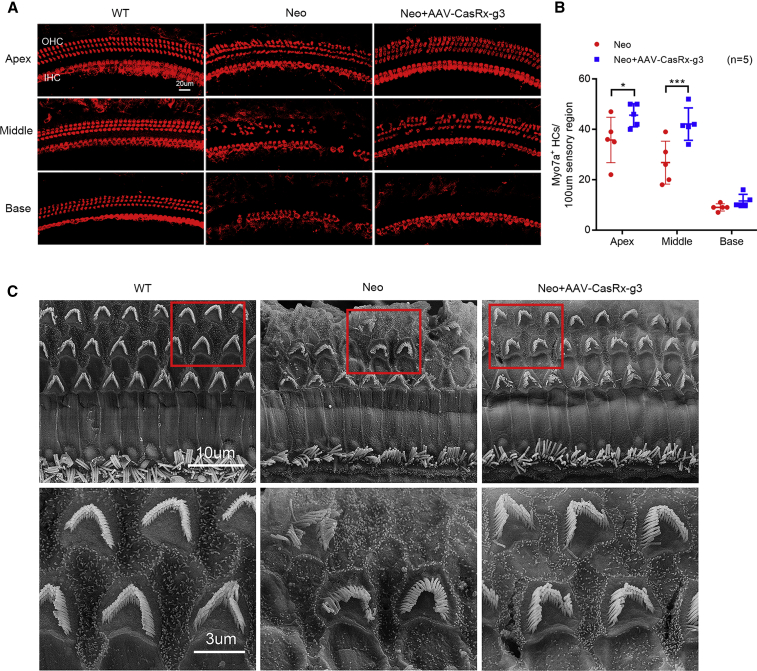
Protection of hair cells in mice with neomycin-induced hearing loss
To investigate hair cell survival in AAV-CasRx-g3-injected inner ears of mice after neomycin-induced ototoxicity, we collected the cochleae after measuring ABRs and DPOAEs. Surviving hair cells were identified with Myo7a antibody staining at 4 weeks (Figure 4A). IHCs and OHCs were present in all turns of the cochlea in WT mice, and the exposure to neomycin induced a great loss of hair cells. In the non-injected ears, a large number of OHCs were missing in the apical turn (8-kHz region) and middle turn (16-kHz region), while almost all OHCs were lost leaving only minimal IHCs in the basal turn (32-kHz region). In contrast, the AAV-CasRx-g3-injected ears showed significantly reduced hair cell loss in the apical and middle turns (Figure 4B). In the basal turn, IHCs survived but not OHCs (Figure 4A).
Figure 4.
The protective effects of AAV-CasRx-g3 in neomycin-injured hair cells
(A) Cochleae from WT, AAV-CasRx-g3-injected, and non-injected mice were immunostained with anti-Myo7a at 4 weeks after neomycin exposure. Representative images show the three functional segments, namely, the apex, middle, and base. Scale bar, 20 μm. (B) The total numbers of OHCs and IHCs per 100 μm sensory region of the cochleae were compared by paired two-tailed Student t-test. Data are shown as the mean ± SD, ∗p < 0.05, ∗∗∗p < 0.001. (C) The hair cell morphology from scanning electron microscopy images of middle-turn cochlear sensory epithelium was compared among WT, neomycin-treated, and neomycin + AAV-CasRx-g3 treated groups at 6 weeks. Scale bars, 10 μm (upper); 3 μm (lower).
We also examined the hair bundle morphology by scanning electron microscopy at 6 weeks to evaluate the protective effect of AAV-CasRx-g3 treatments against neomycin-induced ototoxicity. In WT mice, the classical staircase organization of hair bundles was observed in both IHCs and OHCs. Compared with WT mice, neomycin-treated ears displayed abnormal morphology, disorganized hair bundles, and loss of both IHCs and OHCs (Figure 4C). However, the hair cells from AAV-CasRx-g3-injected ears showed the decreased cell loss and the improved organization of IHCs and OHCs, comparable with normal hair bundle cells in WT mice (Figure 4C). These results were consistent with the ABR thresholds and demonstrated robust restoration at lower frequencies, but less so at a high frequency (32 kHz).
Safety evaluation of CasRx-mediated Htra2 knockdown in mice
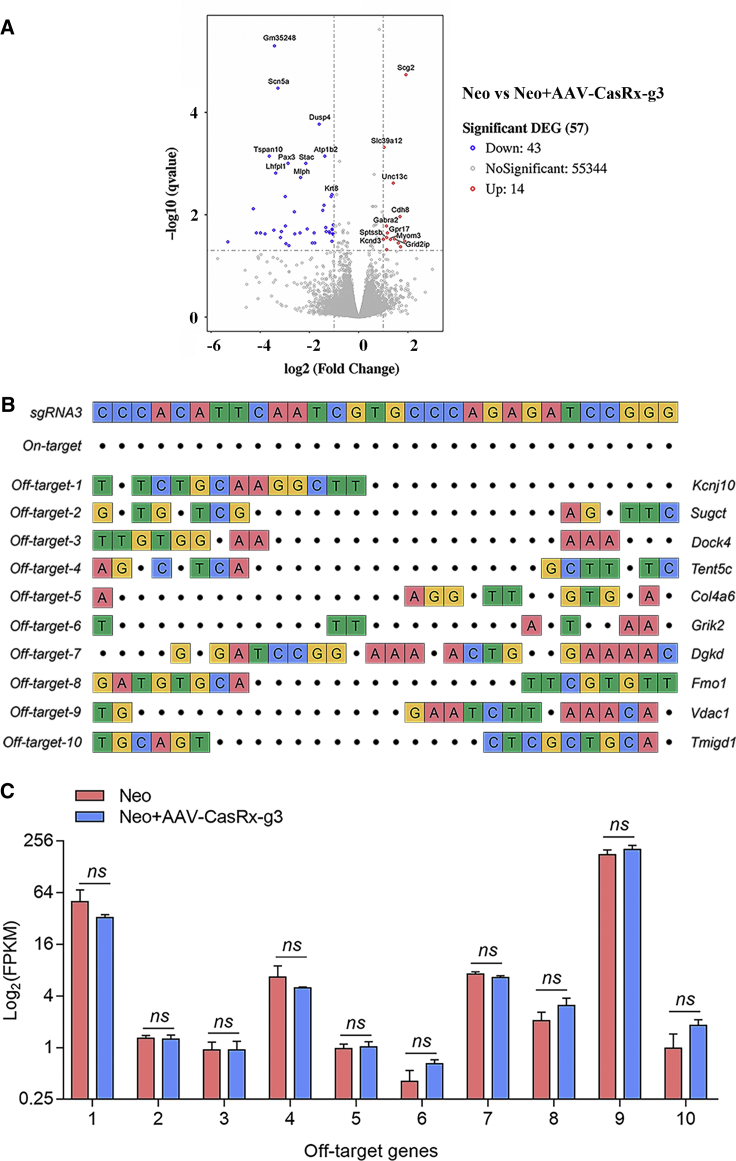
To determine the safety of CasRx-mediated knockdown in vivo, we collected the cochleae from AAV-CasRx-g3-treated mice for RNA-seq analysis. The significantly varied differentially expressed genes (DEGs) were analyzed and compared between neomycin-injured cochleae with or without AAV-CasRx-g3 treatment (Table S2). The percentage of DEGs was 0.1% among the whole transgenome based on RNA-seq data, which suggests limited off-target and side effects from the AAV-CasRx-g3 system in mice (Figure 5A). In addition, we aligned the mouse whole genome based on the 30 bp gRNA3 sequence to screen the top 10 most likely off-target genes that were not included in the 0.1% DEGs (Figure 5B). The mRNA expression of these off-target genes in the neomycin-exposed group and CasRx-treated group was compared, and no significant differences were detected in the top 10 genes (Figure 5C), suggesting the low off-target effects of CasRx-mediated knockdown in vivo.
Figure 5.
The safety evaluation of RNA editing in vivo by RNA-seq
(A) The volcano plot of the DEGs between AAV-CasRx-g3-treated and non-treated cochleae at 4 weeks after neomycin exposure. Blue indicates downregulated genes and red indicates upregulated genes. The top 10 significantly up-regulated and down-regulated DEGs were labeled in the plot. (B) The off-target sites of the most likely off-targeted 10 genes were screened and listed. The 30 bp on-target sequence (gRNA3) is shown at the top. The mismatched bases compared with the on-target sites are highlighted in colors. (C) The FPKM values of the 10 off-target genes were compared between neomycin-treated and neomycin + AAV-CasRx-g3-treated groups. Data are shown as the mean ± SD. Statistical analysis was performed by the two-tailed Student t-test. ns, no significance.
In addition, to evaluate the adverse effects and the long-term safety of AAV-CasRx system to auditory function in mice, we measured the hearing level of mice at age of 10 weeks after the AAV-CasRx injection without neomycin exposure. The ABR thresholds of bilateral sides in AAV-CasRx injected mice had no significant difference from that of 10-week WT mice at all frequencies (Figure S5), which suggested the application of AAV-CasRx system had no detrimental effects to mice auditory function up to 10 weeks.
Discussion
Drug-induced ototoxicity refers to the degradation and dysfunction of cochlear hair cells by drugs or chemicals, ultimately resulting in hearing loss. Aminoglycosides are the most common ototoxicity-inducing drugs that cannot be replaced in the treatment of some diseases, among which amikacin, neomycin, dihydrostreptomycin, and kanamycin are the most cochleotoxic.18 Permanent hearing impairment can significantly affect the patient’s quality of life.33 Therefore, it is necessary to exploresuitable methods to prevent ototoxicity-induced hearing impairment.
In this study, we demonstrated that aminoglycoside-induced ototoxicity could be effectively prevented by the CasRx-mediated down-regulation of the over-expressed Htra2, which plays an important role in maintaining mitochondrial homeostasis and regulating cell apoptosis. The ABR results showed significant improvements of auditory function in the AAV-CasRx-g3-injected ears, especially at lower frequencies (4 kHz and 8 kHz), which presented less hair cell loss in apex and middle turns of cochleae compared with the non-injected ears after neomycin exposure. Notably, the AAV-CasRx-g3-injected ears showed the greatest increase of 40 dB at 8 kHz, which demonstrated the improved auditory function after the CasRx treatment. Consistent with these results, the inner ears treated with AAV-CasRx-g3 showed better hair cell bundle morphology and more surviving hair cells compared with the untreated ears.
Oxidative stress is the main cause of ototoxicity by inducing apoptosis and necrosis in cochlear hair cells.34 The IAPs are endogenous inhibitors of Caspase-3 and Caspase-9 activation.19,35 During apoptosis, the proapoptotic protein Htra2 is also released, which promotes caspase-dependent apoptosis by binding to X-linked inhibitor of apoptosis protein.19,36 In addition, it has been reported that the increased levels of Htra2 promote apoptosis and the knockdown of Htra2 transcripts protects cells from numerous apoptotic stimuli.37,38 According to a previous study,23 Htra2 transcript is significantly increased in neomycin-exposed inner ears, and thus Htra2 is a potential target for CasRx-mediated treatment of neomycin-induced ototoxicity.
CRISPR/Cas9 technology, the most powerful gene-editing tool currently available, has been widely used for gene therapy of various diseases.39 However, the changes in genomic DNA from CRISPR/Cas9 are permanent and can be inherited by the offspring.40,41 It has been demonstrated that the knockout of Htra2 in the mouse genome by CRISPR/Cas9 effectively protects hearing against ototoxic injuries.23 However, the deletion of Htra2 induces the generation of reactive oxygen species (ROS) and mutations, also may lead to mitochondrial damage and dysfunction.42 In addition, the permanent knockout of Htra2 by CRISPR/Cas9 technology might lead to unexpected side effects, which is not appropriate for the treatment of an acquired disease.43,44 Interestingly, our CasRx-mediated treatment had a comparable protective effect against neomycin-induced injury and hearing loss, but unlike heritable CRISPR/Cas9, CasRx targets RNA transcripts, which are temporary and nonpermanent. Moreover, the ROS intensity was detected in cochleae from the WT and AAV-CasRx-injected mice at the age of 10 weeks, which exhibited no significant difference (Figure S6). Thus, the CasRx system possesses the lower risk, suggesting its greater potential for clinical applications.
The Cas13 family is well known for RNA targeting and manipulating, the nuclease of which is composed of 64–66 nt crRNA/guide-RNA and protospacer-flanking site. The target specificity is encoded by a 24–30 nt spacer region, which is complementary to the target site.4,10 The discovery of Cas13 opens a new era for gene editing technology, which has been empowered of versatile applications in both mammalian cells and plants, including live imaging, RNA degradation, base editing, and nucleic acid detection.45 Recently, the newly identified Cas13 orthologs from the types VI–X and VI–Y families were originated from micro-organisms living in hypersaline environment, which showed comparable knockdown efficiency with RfxCas13d and a lower risk of pre-existing immunity found in Cas9 and Cas12 orthologs.46 The variety of Cas13 orthologs provide broad options for RNA manipulating in different models, highlighting the importance of RNA editing-based researches and therapeutic applications.
In this study, we applied CasRx ortholog which was reported of no side effects for in vivo study13,47 to reach the RNA knockdown with high efficiency in mice. As AAV delivery has been confirmed to be safe and efficient for inner ear treatments according to previous studies,30,48 we packaged CasRx together with the CRISPR array into a single AAV-PHP.eB, one of the serotypes of AAVs that has shown high transduction efficiency in cochlear hair cells in vivo.9,13,30 The AAV-CasRx-mediated treatment system is demonstrated a promising protective effect against neomycin-induced injury and hearing loss. In addition, the low proportion of DEGs caused by the CasRx system, and the low off-target efficiency reflected the safety and effectiveness of our AAV-CasRx-g3-mediated treatment in mice. This study provides a proof-of-concept that the CRISPR-CasRx system has great potential for clinical applications in treating non-inherited hearing loss.
Materials and methods
Animals
Institute of Cancer Research (ICR) WT mice were used for the whole study because of their high fertility. All mice were maintained with a 12 h light/12 h dark cycle in the Department of Laboratory Animal Science of Fudan University. Standard mouse chow and water were available ad libitum. All animal experiments were performed in accordance with institutional animal welfare guidelines and were approved by the Animal Care and Use Committee of Fudan University, China. For hearing loss model, experimental mice at P11 were treated with neomycin (200 mg/kg) through subcutaneous injection for 7 consecutive days.
Virus preparation
The CasRx system was packaged into PHP.eB AAV by OBIO Technology Corp., Ltd. (Shanghai, China). The therapeutic AAV virus (AAV-CasRx-g3) included a U6 promoter-driven gRNA targeting Htra2 and a CMV-driven RfxCas13d promoter. The viral titer was 1.04 × 1013 viral genomes (vg)/mL. The control AAV (AAV-control) was consistent with AAV-CasRx except for the gRNA sequence, which was replaced by a non-targeting sequence. The viral titer of AAV-control was 1.73 × 1012 vg/mL. Moreover, a PHP.eB AAV encoding an EGFP reporter was constructed for the determination of AAV delivery efficiency.
Inner ear injection
Newborn pups were injected at P1 with 500 nL AAV-CasRx-g3, AAV-control, or AAV-EGFP virus through the scala media. The pups were anesthetized using hypothermia by placing them in ice for 2–3 min. A tiny incision was made below the right ear to expose the otic bulla and the stapedial artery. Viruses were injected through a glass micropipette at a rate of 5 nL/s. After injection, the cut was sutured and pups were put on a 42°C heating pad for recovery.
Hearing test
ABRs and DPOAEs were recorded with a TDT BioSigRP system (Tucker-Davis Technologies, Alachua, FL) in a sound-proof chamber at 2, 4, and 8 weeks after neomycin exposure. Mice of either sex were anesthetized with xylazine (10 mg/kg) and ketamine (100 mg/kg) through an intraperitoneal injection and were placed on a heating pad. Three needle electrodes were subcutaneously stuck into the back of the pinna (the recording electrode), the dorsum between the ears (the reference electrode), and the rump (the ground electrode). Before each recording session, the animals were examined for otitis media or the unusual formation of cerumen in the ear canal. All ears identified as such were excluded from the study. Tone burst acoustic stimuli was decreased in 5-dB decrements from a 90-dB threshold down to 20 dB sound pressure level (SPL) at frequencies 4, 8, 16, 24, and 32 kHz. The responses were amplified (10,000 times), filtered (0.3–3 kHz), and averaged (500 responses) at each SPL with an analog to digital board in a data acquisition system. Test parameters and fast Fourier transform outputs were presented on the monitor and stored on a disk for offline analysis. The ABR threshold was determined as the lowest SPL at which the wave of the ABR test relative to background noise could be visually detected and repeated. The acoustic stimuli thresholds were defined by two independent observers. At the beginning of the DPOAE measurement, the system was calibrated for each ear by sequentially applying a chirp signal in each of the two speakers. The chirp consisted of equi-amplitude sine waves with a specified phase distribution.49 The f1 and f2 primary tones (f2/f1 = 1.2) were presented with f2 varied between 4, 8, 16, 24, and 32 kHz and L1 − L2 = 10 dB SPL. At each f2, L2 was decreased from 80 dB SPL to 20 dB SPL in 5-dB SPL decrements. The DPOAE threshold was defined from the average spectra as the L2 level eliciting a DPOAE of magnitude 5 dB above the noise floor.50 Data were analyzed and plotted using GraphPad Prism 8. Thresholds averages ± standard deviations (SD) are presented unless otherwise stated.
Plasmids
To construct the CasRx plasmid, the human codon-optimized RxCas13d gene was synthesized and cloned into a mammalian expression vector with the CAG promoter. An mCherry reporter was then cloned into the CasRx plasmid as an indicator of successful transfection, the expression of which was separated from the RfxCas13d gene by a P2A (2A self-cleaving peptides) sequence.
Next, we constructed the CasRx-gRNA cloning backbone that contained two direct repeat sequences and the Bbs1 recognition site under the U6 promoter for cloning. We designed 23 gRNAs for targeting different exons of the Htra2 gene. The gRNA oligos were annealed and cloned into the gRNA cloning backbone plasmid through restriction enzyme ligation. To select the optimized gRNA in HEK293T cells, we designed an Htra2 and EGFP gene co-expression vector under the control of the CMV promoter. When Htra2 mRNA was cleaved after transcription, the EGFP expression was also interrupted. The CDS sequence of Htra2 was synthesized and cloned into the 3′ end of the EGFP gene without the stop codon.
Cell culture and transfection
HEK293T and HEI-OC1 cell lines were originally obtained from the National Collection of Authenticated Cell Cultures (Shanghai, China) and cultured in DMEM (Corning Inc, Corning, NY) supplemented with 10% fetal bovine serum, 100 IU/mL penicillin, and 100 mg/mL streptomycin (ThermoFisher Scientific, Waltham, MA) at 37°C in a humidified atmosphere containing 5% CO2. HEK293T cells were seeded into 24-well plates overnight and transfected with mixed plasmids using Lipofectamine 3,000 (ThermoFisher Scientific) according to the manufacturer’s protocol. For each well of the plate, 1.2 μg plasmids (gRNA: RfxCas13d: Htra2 vector at a 1:2:1 ratio) were mixed with 1.8 μL Lipofectamine 3000 and 2.4 μL P3000 in 100 μL Opti-MEM (Life Technologies, Carlsbad, CA). For HEI-OC1 cells, 300 ng gRNA and 600 ng RfxCas13d were transfected into each well using Lipofectamine 3000 according to the manufacturer’s protocol. The mixtures were co-cultured for 6–8 h and the media were replaced with fresh media. To study the ototoxicity induced by neomycin, HEI-OC1 cells were incubated with 10 mM neomycin for 24 h after the transfection of the above plasmids. About 72 h after transfection, the cells were examined by fluorescence microscopy and then harvested for FACS and RT-qPCR.
Cochlear explant culture
The cochleae from WT P1 ICR mice were dissected, and the organ of Corti was cultured in DMEM/F12 (ThermoFisher Scientific) with 1% N2 supplement (ThermoFisher Scientific), 2% B27 supplement (ThermoFisher Scientific), and 50 mg/mL ampicillin (Sigma-Aldrich, St. Louis, MO) at 37°C with 5% CO2. For the viral transduction, 2 h after cochlear explants, AAV-CasRx-g3 (3 × 1012 vg/mL) was co-cultured with cochlear for 24 h. The medium was then changed to fresh DMEM/F12 mixture with 1 mM neomycin (Sigma-Aldrich), and cultured for 6 h. After 24 h of recovery in DMEM/F12 medium without neomycin, the cochlear explants were harvested for immunohistochemical staining.
Flow cytometry
HEK293T cells were washed with PBS and harvested 72 h after transfection. EGFP signals were examined by a BD LSRFortessa flow cytometer (BD Biosciences, Palo Alto, CA). For each sample, a total of 20,000 cell events were collected. Each sample had three repeats, and the results were analyzed by FlowJo software.
RT-qPCR
Cells were briefly washed before harvesting. The total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) and reverse transcribed into cDNA using the PrimeScript RT reagent Kit (Takara, Kusatsu, Japan) according to the manufacturer’s protocol. Gene expression was quantified by RT-qPCR using the ABI 7500 Fast Real-Time PCR Detection System (Applied Biosystems, Waltham, MA) and TB Green Premix Ex Taq II (Tli RNaseH Plus; Takara). The expression of β-actin was used as the loading control. Primer sequences were as follows: β-actin forward: 5′-CCGCAGCTAGGAATAATGGA-3′ and reverse: 5′-CAAATGCTTTCGCTCTGGTC-3′; Htra2 forward: 5′-TCATCGCAGATGTGGTGGAG-3′ and reverse: 5′-GATGAGCCCATCTGAAGCCA-3′; Casp3 forward: 5′-CGGGGTACGGAGCTGGACTGT-3′ and reverse: 5′-ATGCTGCAAAGGGACTGGATGAAC-3′; Casp9 forward: 5′-AGGCCCGTGGACATTGGTTCT-3′ and reverse: 5′-AGTTGGAGCCCGTGCGTGTG-3′. For each group, the mRNA levels were measured three times, and the data are presented as the relative expression compared with the control group.
Western blot
Htra2 expression was assessed by western blot analysis. Cochleae from 4-week-old mice were collected and lysed in RIPA buffer (Beyotime Biotechnology, Haimen, China) supplemented with phenylmethanesulfonyl fluoride (Beyotime Biotechnology). Proteins were separated by SDS-PAGE (GenScript, Piscataway, NJ) and transferred onto polyvinylidene difluoride membranes (Millipore, Burlington, MA). The membranes were blocked in 5% skim milk powder (Beyotime Biotechnology) and probed with anti-Htra2 (Cell Signaling Technology, Danvers, MA) or anti-β-actin antibodies (Beyotime Biotechnology) at 4°C overnight. The membranes were labeled with HRP-conjugated goat anti-rabbit IgG (H + L) secondary antibodies (Beyotime Biotechnology) for 1 h at room temperature, and the bands were visualized using western Blot ECL Blotting Substrate (Bio-Rad, Hercules, CA). The images were analyzed through ImageJ software.
Immunohistochemistry
The cochleae of both ears were collected from 4- or 6-week-old neomycin-injected mice after AAV injection. The temporal bones of the cochleae were perforated and perfused with 4% paraformaldehyde and incubated overnight at 4°C. After decalcification in 10% EDTA for 1–3 days, the cochleae were dissected into three pieces designated as the apical, middle, and basal turns for immunofluorescence staining. All of the tissue were permeabilized in PBS containing 1% Triton X-100 buffer (1% PBST) for 12–16 h at 4°C and blocked with 10% donkey serum for 2 h at room temperature. To check the EGFP fluorescence in the hair cells of the cochleae after the injection of AAV-EGFP, we incubated the tissue with rabbit anti-Myo7a (1:500 dilution, Proteus BioSciences, Waltham, MA) and chicken anti-eGFP (1:1,000 dilution, Abcam, Cambridge, UK) primary antibodies. For examining the apoptotic protein expression in the cochlea, the rabbit anti-cleaved-Caspase-3 primary antibody (1:1,000 dilution, Cell Signaling Technology) and rhodamine-conjugated phalloidin (1:500 dilution, Sigma-Aldrich) were used. To determine the expression of Htra2 in cochlear hair cells, rabbit anti-Htra2 primary antibody (1:200 dilution, Abcam, Waltham, MA) and rhodamine-conjugated phalloidin (1:500 dilution, Sigma-Aldrich) were added. The primary antibodies were incubated overnight at 4°C, and the corresponding luciferin-conjugated secondary antibody was added and incubated for 2 h in the dark, followed by rhodamine-conjugated phalloidin incubation for 1 h in the room temperature. DAPI was used to label the nuclei (1:1,000 dilution, Sigma-Aldrich). Specimens were mounted and fluorescent z stack confocal images were acquired with a Leica TCS SP8 laser scanning confocal microscope. Full cochlear images were reconstructed in Adobe Photoshop and ImageJ software.
Scanning electron microscopy
The temporal bones of 4-week-old adult mice were harvested and then perforated and perfused and fixed with 2.5% glutaraldehyde at 4°C overnight. The tissues were then transferred into 10% EDTA for decalcification at 4°C for 3 days. The decalcified cochleae were dissected into three pieces for whole mounts and fixed first in 2.5% glutaraldehyde and then in 1% osmium acid at 4°C for 2 h. The whole mount tissues were then dehydrated by ethanol gradient and dried in an HCP-2 critical point drier. The dried tissues were attached to the specimen tables and coated with platinum by an IB-3 ion sputtering instrument. Images were acquired with an emission scanning electron microscope (Hitachi 574 SU-8010, Tokyo, Japan) in a high vacuum field.
RNA-seq
Total RNA was extracted from the cochleae using TRIzol Reagent/RNeasy Mini Kit (Qiagen, Hilden, Germany) and quantified with an Agilent NanoDrop (ThermoFisher Scientific). A total of 1 μg RNA from each sample was used for the library preparations for next generation sequencing, and these were constructed according to the manufacturer’s protocol. The cDNA was synthesized by ProtoScript II Reverse Transcriptase and Strand Synthesis Enzyme Mix, and then treated with End Prep Enzyme Mix to repair both ends and to add tail-A, which followed by a ligation to add adaptors. Each sample was then amplified by PCR using P5 and P7 primers, with both primers carrying sequences that can anneal with the flow cell to perform bridge PCR and carrying indexes that allow for multiplexing. The libraries were then multiplexed and loaded onto an Illumina HiSeq/Novaseq instrument according to the manufacturer’s instructions (Illumina, San Diego, CA). The expression levels of all mapped genes were estimated by stringtie (v2.0), and the gene expression levels were determined by fragments per kilobase of transcript per million fragments mapped. Differential expression analysis was based on the DESeq2 Bioconductor package, and p values of less than 0.05 indicated DEGs. The analysis of pathways was based on the Kyoto Encyclopedia of Genes and Genomes database.
Off-target analysis
To analyze the off-target efficiency of CasRx-mediated knockdown across the whole transcriptome, the most likely off-target genes were screened and identified. The differential expression analysis of these genes between the CasRx injected and non-injected groups was performed using the DESeq2 R package based on RNA-seq data. The resulting p-values were adjusted using the Benjamini and Hochberg approach for controlling for the false discovery rate. Genes with an adjusted p value of less than 0.05 detected by DESeq2 were considered to be differentially expressed.
Statistical analysis
All data are presented as the mean ± SD. The statistical analysis was performed using GraphPad Prism version 7. A two-tailed Student t-test was used for comparing the differences between means. One-way or two-way ANOVA followed by Bonferroni’s test was performed for multiple comparisons. Any p values of less than 0.05 were considered significant.
Acknowledgments
Supported by National Key Research and Development Program of China grant 2020YFA0908201 (Y.S.), National Natural Science Foundation of China grant 81822011 (Y.S.), 82171148 (Y.S.), 81771013 (Y.S.), and the Science and Technology Commission of Shanghai Municipality 21S11905100 (Y.S). We thank Zhijiao Xu, Ziwen Zheng, Xi Gu, and Yu Zhao for giving suggestions and helping isolating basilar membranes from cochleae.
Author contributions
B.C., H.L., Y.S., and Y.G. jointly conceived the project and designed the experiments. Y.G. conducted the experiments and analyzed the data. L.H. and S.H. performed the in vivo experiments and part of the data analysis. Y.S., H.L., and B.C. developed and supervised the project. B.C., H.L., Y.S., and Y.G. wrote the manuscript. S.W., H.T., and C.C. reviewed and revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
All authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.04.014.
Contributor Information
Bing Chen, Email: bingchen@fudan.edu.cn.
Huawei Li, Email: hwli@shmu.edu.cn.
Yilai Shu, Email: yilai_shu@fudan.edu.cn.
Supplemental information
References
- 1.Barrangou R., Marraffini L.A. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J.J., Charpentier E., Horvath P., Moineau S., Mojica F.J.M., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., et al. Discovery and functional characterization of diverse Class 2 CRISPR-cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:f5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smargon A.A., Cox D.B., Pyzocha N.K., Zheng K., Slaymaker I.M., Gootenberg J.S., Abudayyeh O.A., Essletzbichler P., Shmakov S., Makarova K.S., et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell. 2017;65:618–630.e7. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., Scott D.A. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell. 2018;70:327–339.e5. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F., Wang L., Zou X., Duan S., Li Z., Deng Z., Luo J., Lee S.Y., Chen S. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol. Adv. 2019;37:708–729. doi: 10.1016/j.biotechadv.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C., Zhou Y., Xiao Q., He B., Geng G., Wang Z., Cao B., Dong X., Bai W., Wang Y., et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat. Methods. 2021;18:499–506. doi: 10.1038/s41592-021-01124-4. [DOI] [PubMed] [Google Scholar]
- 12.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H., Su J., Hu X., Zhou C., Li H., Chen Z., Xiao Q., Wang B., Wu W., Sun Y., et al. Glia-to-Neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell. 2020;181:590–603.e16. doi: 10.1016/j.cell.2020.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Eisenhut M. Evidence supporting the hypothesis that inflammation-induced vasospasm is involved in the pathogenesis of acquired sensorineural hearing loss. Int. J. Otolaryngol. 2019;2019:1–15. doi: 10.1155/2019/4367240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J., Chai R., Li H., Sun S. Protection of hair cells from ototoxic drug-induced hearing loss. Adv. Exp. Med. Biol. 2019;1130:17–36. doi: 10.1007/978-981-13-6123-4_2. [DOI] [PubMed] [Google Scholar]
- 16.Cianfrone G., Pentangelo D., Cianfrone F., Mazzei F., Turchetta R., Orlando M.P., Altissimi G. Pharmacological drugs inducing ototoxicity, vestibular symptoms and tinnitus: a reasoned and updated guide. Eur. Rev. Med. Pharmacol. Sci. 2011;15:601–636. [PubMed] [Google Scholar]
- 17.Schacht J., Talaska A.E., Rybak L.P. Cisplatin and aminoglycoside antibiotics: hearing loss and its prevention. Anat. Rec. (Hoboken) 2012;295:1837–1850. doi: 10.1002/ar.22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selimoglu E. Aminoglycoside-induced ototoxicity. Curr. Pharm. Des. 2007;13:119–126. doi: 10.2174/138161207779313731. [DOI] [PubMed] [Google Scholar]
- 19.Vaux D.L., Silke J. Mammalian mitochondrial IAP binding proteins. Biochem. Biophys. Res. Commun. 2003;304:499–504. doi: 10.1016/s0006-291x(03)00622-3. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasula S.M., Gupta S., Datta P., Zhang Z., Hegde R., Cheong N., Fernandes-Alnemri T., Alnemri E.S. Inhibitor of apoptosis proteins are substrates for the mitochondrial serine protease Omi/HtrA2. J. Biol. Chem. 2003;278:31469–31472. doi: 10.1074/jbc.C300240200. [DOI] [PubMed] [Google Scholar]
- 21.Yang Q.H., Church-Hajduk R., Ren J., Newton M.L., Du C. Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis. Genes Dev. 2003;17:1487–1496. doi: 10.1101/gad.1097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun S., Sun M., Zhang Y., Cheng C., Waqas M., Yu H., He Y., Xu B., Wang L., Wang J., et al. In vivo overexpression of X-linked inhibitor of apoptosis protein protects against neomycin-induced hair cell loss in the apical turn of the cochlea during the ototoxic-sensitive period. Front. Cell. Neurosci. 2014;8:248. doi: 10.3389/fncel.2014.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu X., Wang D., Xu Z., Wang J., Guo L., Chai R., Li G., Shu Y., Li H. Prevention of acquired sensorineural hearing loss in mice by in vivo Htra2 gene editing. Genome Biol. 2021;22:86. doi: 10.1186/s13059-021-02311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang K., Zhang Z., Kang J., Chen J., Liu J., Gao N., Fan L., Zheng P., Wang Y., Sun J. CRISPR/Cas13d-Mediated microbial RNA knockdown. Front. Bioeng. Biotechnol. 2020;8:856. doi: 10.3389/fbioe.2020.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahas A., Aman R., Mahfouz M. CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol. 2019;20:263. doi: 10.1186/s13059-019-1881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen C.C., Lin M.W., Nguyen B.K.T., Chang C.W., Shih J.R., Nguyen M.T.T., Chang Y.H., Hu Y.C. CRISPR-Cas13d for gene knockdown and engineering of CHO cells. ACS Synth. Biol. 2020;9:2808–2818. doi: 10.1021/acssynbio.0c00338. [DOI] [PubMed] [Google Scholar]
- 28.Jiang W., Li H., Liu X., Zhang J., Zhang W., Li T., Liu L., Yu X. Precise and efficient silencing of mutant Kras(G12D) by CRISPR-CasRx controls pancreatic cancer progression. Theranostics. 2020;10:11507–11519. doi: 10.7150/thno.46642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wessels H.H., Mendez-Mancilla A., Guo X., Legut M., Daniloski Z., Sanjana N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 2020;38:722–727. doi: 10.1038/s41587-020-0456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu X., Wang J., Yao X., Xiao Q., Xue Y., Wang S., Shi L., Shu Y., Li H., Yang H. Screened AAV variants permit efficient transduction access to supporting cells and hair cells. Cell Discov. 2019;5:49. doi: 10.1038/s41421-019-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X., Jen P.H., Seburn K.L., Frankel W.N., Zheng Q.Y. Auditory brainstem responses in 10 inbred strains of mice. Brain Res. 2006;1091:16–26. doi: 10.1016/j.brainres.2006.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akil O., Oursler A.E., Fan K., Lustig L.R. Mouse auditory brainstem response testing. Bio Protoc. 2016;6:e1768. doi: 10.21769/BioProtoc.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baguley D., Mcferran D., Hall D. Tinnitus. Lancet. 2013;382:1600–1607. doi: 10.1016/S0140-6736(13)60142-7. [DOI] [PubMed] [Google Scholar]
- 34.Ikeda K., Oshima T., Hidaka H., Takasaka T. Molecular and clinical implications of loop diuretic ototoxicity. Hear. Res. 1997;107:1–8. doi: 10.1016/s0378-5955(97)00009-9. [DOI] [PubMed] [Google Scholar]
- 35.Hu Y., Bi Y., Yao D., Wang P., Li Y. Omi/HtrA2 protease associated cell apoptosis participates in blood-brain barrier dysfunction. Front. Mol. Neurosci. 2019;12:48. doi: 10.3389/fnmol.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Loo G., van Gurp M., Depuydt B., Srinivasula S.M., Rodriguez I., Alnemri E.S., Gevaert K., Vandekerckhove J., Declercq W., Vandenabeele P. The serine protease Omi/HtrA2 is released from mitochondria during apoptosis. Omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity. Cell Death Differ. 2002;9:20–26. doi: 10.1038/sj.cdd.4400970. [DOI] [PubMed] [Google Scholar]
- 37.Martins L.M., Iaccarino I., Tenev T., Gschmeissner S., Totty N.F., Lemoine N.R., Savopoulos J., Gray C.W., Creasy C.L., Dingwall C., Downward J. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J. Biol. Chem. 2002;277:439–444. doi: 10.1074/jbc.M109784200. [DOI] [PubMed] [Google Scholar]
- 38.Hegde R., Srinivasula S.M., Zhang Z., Wassell R., Mukattash R., Cilenti L., Dubois G., Lazebnik Y., Zervos A.S., Fernandes-Alnemri T., et al. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J. Biol. Chem. 2002;277:432–438. doi: 10.1074/jbc.M109721200. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Zhang Y., Chen K.L., Shan Q.W., Wang Y.P., Liang Z., Gao C.X. [CRISPR/Cas: a novel way of RNA-guided genome editing] Yi Chuan. 2013;35:1265–1273. doi: 10.3724/sp.j.1005.2013.01265. [DOI] [PubMed] [Google Scholar]
- 40.Shin H.Y., Wang C., Lee H.K., Yoo K.H., Zeng X., Kuhns T., Yang C.M., Mohr T., Liu C., Hennighausen L. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 2017;8:15464. doi: 10.1038/ncomms15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goo H.G., Jung M.K., Han S.S., Rhim H., Kang S. HtrA2/Omi deficiency causes damage and mutation of mitochondrial DNA. Biochim. Biophys. Acta. 2013;1833:1866–1875. doi: 10.1016/j.bbamcr.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Goo H.G., Rhim H., Kang S. Pathogenic role of serine protease HtrA2/Omi in neurodegenerative diseases. Curr. Protein Pept. Sci. 2017;18:746–757. doi: 10.2174/1389203717666160311115750. [DOI] [PubMed] [Google Scholar]
- 44.Su X.J., Huang L., Qu Y., Mu D. Progress in research on the role of Omi/HtrA2 in neurological diseases. Rev. Neurosci. 2019;30:279–287. doi: 10.1515/revneuro-2018-0004. [DOI] [PubMed] [Google Scholar]
- 45.Smargon A.A., Shi Y.J., Yeo G.W. RNA-targeting CRISPR systems from metagenomic discovery to transcriptomic engineering. Nat. Cell Biol. 2020;22:143–150. doi: 10.1038/s41556-019-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He B., Peng W., Huang J., Zhang H., Zhou Y., Yang X., Liu J., Li Z., Xu C., Xue M., et al. Modulation of metabolic functions through Cas13d-mediated gene knockdown in liver. Protein Cell. 2020;11:518–524. doi: 10.1007/s13238-020-00700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isgrig K., Mcdougald D.S., Zhu J., Wang H.J., Bennett J., Chien W.W. AAV2.7m8 is a powerful viral vector for inner ear gene therapy. Nat. Commun. 2019;10:427. doi: 10.1038/s41467-018-08243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landegger L.D., Pan B., Askew C., Wassmer S.J., Gluck S.D., Galvin A., Taylor R., Forge A., Stankovic K.M., Holt J.R., et al. A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol. 2017;35:280–284. doi: 10.1038/nbt.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gyorgy B., Nist-Lund C., Pan B., Asai Y., Karavitaki K.D., Kleinstiver B.P., Garcia S.P., Zaborowski M.P., Solanes P., Spataro S., et al. Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nat. Med. 2019;25:1123–1130. doi: 10.1038/s41591-019-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parham K. Distortion product otoacoustic emissions in the C57BL/6J mouse model of age-related hearing loss. Hear. Res. 1997;112:216–234. doi: 10.1016/s0378-5955(97)00124-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.