Abstract
The amylomaltase gene of the thermophilic bacterium Thermus aquaticus ATCC 33923 was cloned and sequenced. The open reading frame of this gene consisted of 1,503 nucleotides and encoded a polypeptide that was 500 amino acids long and had a calculated molecular mass of 57,221 Da. The deduced amino acid sequence of the amylomaltase exhibited a high level of homology with the amino acid sequence of potato disproportionating enzyme (D-enzyme) (41%) but a low level of homology with the amino acid sequence of the Escherichia coli amylomaltase (19%). The amylomaltase gene was overexpressed in E. coli, and the enzyme was purified. This enzyme exhibited maximum activity at 75°C in a 10-min reaction with maltotriose and was stable at temperatures up to 85°C. When the enzyme acted on amylose, it catalyzed an intramolecular transglycosylation (cyclization) reaction which produced cyclic α-1,4-glucan (cycloamylose), like potato D-enzyme. The yield of cycloamylose produced from synthetic amylose with an average molecular mass of 110 kDa was 84%. However, the minimum degree of polymerization (DP) of the cycloamylose produced by T. aquaticus enzyme was 22, whereas the minimum DP of the cycloamylose produced by potato D-enzyme was 17. The T. aquaticus enzyme also catalyzed intermolecular transglycosylation of maltooligosaccharides. A detailed analysis of the activity of T. aquaticus ATCC 33923 amylomaltase with maltooligosaccharides indicated that the catalytic properties of this enzyme differ from those of E. coli amylomaltase and the plant D-enzyme.
Amylomaltase (4-α-glucanotransferase; EC 2.4.1.25) catalyzes glucan transfer from one α-1,4-glucan to another α-1,4-glucan or to glucose. This enzyme was first found in Escherichia coli (18) but has since been found in many bacterial species. The gene that encodes the enzyme has been cloned from E. coli (20), Streptococcus pneumoniae (14), Clostridium butyricum (6), and Chlamydia psittaci (7), and putative genes have been identified in the genomes of Haemophilus influenzae (4), Aquifex aeolicus (3), Synechosystis sp. (10), Mycobacterium tuberculosis (2), and Borrelia burgdorferi (5). A similar enzyme, 4-α-glucanotransferase, is also present in plants and is called disproportionating enzyme (D-enzyme) (EC 2.4.1.25). Analyses of the activity of the E. coli enzyme (19) and the activities of the potato (9) and barley (27) enzymes indicated that amylomaltase and D-enzyme catalyze similar reactions. cDNA for D-enzyme has been isolated only from potato tubers, but the deduced amino acid sequence of D-enzyme exhibited significant homology to the amino acid sequences of bacterial amylomaltases (23).
It has been thought that the substrates of amylomaltase and D-enzyme are maltooligosaccharides, and most of the work on amylomaltase and D-enzyme has been carried out with maltooligosaccharides. However, it was recently revealed that potato D-enzyme can act not only on maltooligosaccharides but also on amylose (24). When potato D-enzyme was incubated with amylose, it catalyzed the amylose cyclization reaction, which produced cyclic α-1,4-glucans (cycloamyloses) with degrees of polymerization (DP) ranging from 17 to a few hundred. This finding has caused amylomaltase and D-enzyme to receive much attention. No cyclization reaction has been reported with any amylomaltase from a bacterial source, but the structural and catalytic similarities between the two enzymes strongly suggest that such activity is possible. Here we describe isolation of amylomaltase from the thermophilic bacterium Thermus aquaticus ATCC 33923. The amylomaltase obtained from recombinant E. coli exhibited significant thermal stability and efficiently produced cycloamylose from amylose.
MATERIALS AND METHODS
Materials.
Synthetic amylose AS-110 was purchased from Nakano Vinegar Co. (Aichi, Japan). Maltose (G2), maltotriose (G3), and maltotetraose (G4) were purchased from Hayashibara Biochemical Laboratories Inc. (Okayama, Japan), and maltopentaose (G5) was purchased from Ensuiko Sugar Refining Co., Ltd. (Yokohama, Japan). Rhizopus sp. glucoamylase was purchased from Toyobo Co., Ltd. (Osaka, Japan), and porcine pancreas α-amylase was purchased from Sigma. Unless otherwise specified, all chemicals were purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan). A cycloamylose standard with DP ranging from 10 to 31 prepared from synthetic amylose AS-1000 (Nakano Vinegar Co.) with cyclodextrin glucanotransferase (CGTase) (EC 2.4.1.19) was kindly donated by K. Koizumi (Mukogawa Women’s University, Hyogo, Japan).
Purification of T. aquaticus amylomaltase.
T. aquaticus ATCC 33923 was grown at 70°C for 16 h in a medium containing 0.4% (wt/vol) yeast extract (Difco), 0.8% (wt/vol) Polypeptone (Wako), 0.2% (wt/vol) NaCl, and 1% (wt/vol) maltose (pH 7.5). After 18 h, the cells (141 g [wet weight] from a 28.8-liter culture) were harvested and washed twice with 1.8 liters of distilled water. The cells were suspended in 200 ml of 10 mM KH2PO4-Na2HPO4 (pH 7.0) (buffer A), disrupted by sonication at 4°C, and centrifuged (12,000 × g, 30 min) to remove cell debris. Solid ammonium sulfate was added slowly to the resulting supernatant to 20% saturation. The precipitate that formed was removed by centrifugation at 12,000 × g for 30 min. The supernatant was loaded onto a Phenyl-Toyopearl 650M (Tosoh Co., Ltd., Tokyo, Japan) column (2.6 by 15 cm) equilibrated with buffer A containing 1 M ammonium sulfate. After the column was washed with buffer A containing 0.3 M ammonium sulfate, the enzyme was eluted with a linear 0.3 to 0 M ammonium sulfate gradient in buffer A. The active fractions were pooled, dialyzed against buffer A, and loaded onto a Source 15Q (Pharmacia) column (1 by 10 cm) equilibrated with buffer A. After the column was washed with buffer A, the enzyme was eluted with a linear 0 to 0.4 M NaCl gradient in buffer A. The active fractions were pooled, dialyzed against buffer A, and loaded onto a Superdex 75pg (Pharmacia) column (1.6 by 60 cm) equilibrated with 50 mM KH2PO4-Na2HPO4 (pH 7.0) containing 0.15 M NaCl and eluted with the same buffer. The active fractions were pooled, dialyzed against buffer A, and stored at 4°C.
Determination of the amino acid sequence of purified amylomaltase.
Purified amylomaltase was subjected to reverse-phase high-performance liquid chromatography (HPLC) by using a C4 column (catalog no. 214TP54; 4.6 by 250 mm; Vydac, Hesperia, Calif.) equilibrated with 48.4% acetonitrile containing 0.1% trifluoroacetic acid. The enzyme was eluted with a linear 48.4 to 52.4% acetonitrile gradient supplemented with 0.1% trifluoroacetic acid, and the enzyme peak was collected and concentrated in vacuo. Part of the concentrate (80 μg of protein) was directly subjected to peptide sequencing, and the N-terminal amino acid sequence of the enzyme was determined. To obtain peptide fragments of the enzyme, another part of the concentrate (80 μg of protein) was digested with 3 × 10−3 U of lysyl endopeptidase (Wako) in the presence of 2 M urea, 0.1 M ammonium bicarbonate, 1.1 mM dithiothreitol, and 2.5 mM iodoacetoamide at 37°C for 24 h. The fragments generated were separated by reverse-phase HPLC by using a C18 column (catalog no. 218TP54; 4.6 by 250 mm; Vydac) and a 1.6 to 78.4% acetonitrile gradient containing 0.06% trifluoroacetic acid. Two peaks that were well separated from the other peaks were collected and lyophilized separately and then were subjected to an amino acid sequencing analysis.
Construction and screening of a T. aquaticus gene library.
Chromosomal DNA from T. aquaticus cells was prepared as described by Marmur (16). The chromosomal DNA was partially digested with Sau3AI and was size fractionated by NaCl density gradient centrifugation. The size-fractionated DNA fragments (5 to 10 kbp) were inserted into the BamHI site of the ZAP Express vector (Stratagene, La Jolla, Calif.). The gene library was plated onto E. coli VCS257 (Nippon Gene Co., Ltd., Toyama, Japan) and was transferred to Hybond N+ membrane filters (Amersham Pharmacia). Oligonucleotide hybridization was performed in 5× SSC (0.75 M NaCl plus 0.175 M sodium citrate) containing 5× Denhardt’s solution, 0.1% (wt/vol) sodium dodecyl sulfate (SDS), 0.05 mg of denatured salmon sperm DNA per ml, and a synthetic oligonucleotide probe that had been labeled with [γ-32P]ATP by using T4 polynucleotide kinase at 47°C overnight. The filters were washed twice in 5× SSC containing 0.1% (wt/vol) SDS at 47°C for 15 min and autoradiographed. DNA sequence was determined by the dideoxynucleotide chain termination method of Sanger et al. (21) by using a BigDye terminator cycle sequencing kit (Applied Biosystems).
Expression of the T. aquaticus amylomaltase gene in E. coli.
An oligonucleotide adapter,5′-GATCTAGATAGATGAAGGAGATATACATATGG ATCTATCTACTTCCTCTATATGTATACCCTAG-5′
which contained three stop codons in each frame, two additional restriction sites (XbaI and NdeI sites), and a ribosome-binding site (AGGA), was inserted into a BamHI site of plasmid pGEX-5X-3 (Amersham Pharmacia) in order to obtain expression plasmid pGEX-Nde. The amylomaltase gene was amplified by PCR by using two oligonucleotide primers, 5′-TTTCATATGGAGCTTCCCCGCGCTTTCGGTCTGCTT-3′ and 5′-TTTGAATTCGGGCTGGTCCACCTAGAGCCGTTCCGT-3′, which were designed to introduce additional NdeI and EcoRI sites before and after the coding sequence, respectively. The amplified fragment (length, 1.5 kbp) was digested with NdeI and EcoRI and then introduced into the NdeI-EcoRI site in pGEX-Nde in order to construct expression plasmid pFQG8.
E. coli MC1061 [hsdR mcrB araD139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi] carrying pFQG8 was grown at 37°C in Luria-Bertani medium (1% tryptone [Difco], 0.5% yeast extract [Difco], 1% NaCl; pH 7.0) containing 100 μg of ampicillin per ml. At the late log phase (optical density at 660 nm, 1.0), isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.01 mM. After 22 h, the cells (17 g [wet weight] from a 3-liter culture) were harvested and washed twice with 500 ml of buffer A. The cells were suspended in 50 ml of buffer A, disrupted by sonication at 4°C, and centrifuged (12,000 × g, 30 min) to remove the cell debris. The crude extract obtained in this way was heated at 70°C for 30 min and centrifuged (10,000 × g, 30 min). The recombinant amylomaltase was further purified from the supernatant by chromatography with Phenyl-Toyopearl 650M and Source 15Q columns as described above.
Enzyme activity assay.
T. aquaticus amylomaltase activity was assayed in a 120-μl reaction mixture containing 10% (wt/vol) maltotriose, 50 mM sodium acetate buffer (pH 5.5), and the enzyme. The mixture was incubated at 70°C for 10 min, and the reaction was terminated by heating the reaction tube at 100°C for 10 min. The amount of glucose released was measured by the glucose oxidase method (17). One unit of activity was defined as the amount of enzyme which produced 1 μmol of glucose per min under the assay conditions used.
TLC.
Three-microliter aliquots of reaction mixtures were spotted onto a thin-layer chromatography (TLC) plate (Silica Gel 60; Merck, Darmstadt, Germany), which was developed three times with 1-butanol–ethanol–water (5:5:3). The carbohydrates were detected by spraying the plate with sulfuric acid-methanol (1:1, vol/vol) and then baking it at 130°C for 5 min.
HPAEC.
High-performance anion-exchange chromatography (HPAEC) was carried out by using the DX-300 system (Dionex Corp., Sunnyvale, Calif.) with a pulsed electrochemical detector (model PED-II; Dionex) and a Carbopac PA-100 column (4 by 250 mm; Dionex). A 25-μl sample was injected and eluted with a gradient of sodium acetate (concentrations: zero time to 2 min, 50 mM; 2 to 37 min, increase from 50 to 350 mM with installed gradient program 3; 37 to 45 min, increase from 350 to 850 mM with installed gradient program 7; 45 to 47 min, 850 mM) in 150 mM NaOH by using a flow rate of 1 ml/min. To determine the DP of cycloamylose, 25 μl of a sample was injected and eluted with a gradient of sodium nitrate (concentrations: zero time to 2 min, 8 mM; 2 to 13 min, increase from 22 to 26 mM with installed gradient program 3; 13 to 35 min, increase from 26 to 70 mM with installed gradient program 4; 35 to 40 min, increase from 70 to 200 mM with installed gradient program 7; 40 to 42 min, 200 mM) in 150 mM NaOH by using a flow rate of 1 ml/min.
Analysis of amylomaltase activity with amylose.
Amylose AS-110 (0.2 g) was dissolved in 10 ml of 90% (vol/vol) dimethyl sulfoxide. A 3.5-ml reaction mixture containing 50 mU of T. aquaticus amylomaltase from recombinant E. coli, 0.35 ml of the amylose AS-110 solution, and 50 mM sodium acetate buffer (pH 5.5) was incubated at 70°C, and the reaction was terminated by heating the solution at 100°C for 30 min. Then 50 μl of the reaction mixture was incubated with glucoamylase (1.8 U) with or without α-amylase (0.26 U) at 40°C for 3 h. After the reaction was terminated by boiling the reaction mixture for 10 min, the amount of glucose released in each tube was measured by the glucose oxidase method (17). The amount of glucoamylase-resistant glucan was calculated by subtracting the amount of glucose released by glucoamylase and α-amylase from the amount of glucose released by glucoamylase alone. The glucoamylase-resistant glucan in the 50-μl reaction mixture was precipitated by adding 500 μl of ethanol. The pellet was dried in vacuo, redissolved in 50 μl of water, and analyzed by HPAEC.
Other procedures.
The reducing power of glucan was determined by a modified Park-Johnson method (25). To investigate the ability of amylose to form a complex with iodine, 100 μl of a sample was mixed with 2 ml of an iodine solution (0.1% I2 and 1% KI in 3.8 mM HCl), and the absorbance at 660 nm was measured. SDS-polyacrylamide gel electrophoresis (PAGE) was carried out by using precast 8 to 16% acrylamide gradient gels (TEFCO Corp., Tokyo, Japan). Each gel was stained with a solution containing 0.1% Coomassie brilliant blue R-250, 40% methanol, and 10% acetic acid and destained with a solution containing 10% methanol and 7.5% acetic acid. Protein concentrations were determined by the method of Bradford (Bio-Rad, Hercules, Calif.) by using bovine gamma globulin as the standard.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession no. AB016244.
RESULTS AND DISCUSSION
Cloning of the T. aquaticus amylomaltase gene.
Amylomaltase was purified from a cell extract of T. aquaticus. The purified enzyme produced a single band on an SDS-PAGE gel at an estimated molecular mass of 57 kDa (data not shown). The purified enzyme was subjected to protein sequencing, and the N-terminal amino acid sequence, M-E-L-P-R-A, was determined. To determine the internal amino acid sequences of the enzyme, the purified amylomaltase was digested with lysyl endopeptidase, and two peptide fragments were purified by reverse-phase HPLC. The N-terminal amino acid sequences of these fragments were determined to be K-E-A-F-R-G-F and S-V-A-R-L-A-V-Y-P-V-Q-D-V. A mixed oligonucleotide probe, 5′-GCIGTITAYCCIGTICARGAYGT-3′ (Y = mixture of C and T; R = mixture of A and G), corresponding to the amino acid sequence A-V-Y-P-V-Q-D-V, was synthesized and used to screen a T. aquaticus gene library. Three positive plaques were found among the 40,000 plaques examined; one of these positive plaques had a 4.0-kbp DNA insert and contained the entire region encoding the T. aquaticus amylomaltase gene (designated malQ). The nucleotide sequence and deduced amino acid sequence data are available from the DDBJ/EMBL/GenBank databases (see Materials and Methods). malQ consisted of 1,503 nucleotides and encoded 500 amino acid residues (Mr, 57,221). The amino acid sequences obtained in the protein sequencing analysis were all found in the deduced amino acid sequence. The overall G+C content of malQ was 69 mol%, but the G+C content of the third positions of codons was 93 mol%. T. aquaticus amylomaltase exhibited high levels of homology with the amylomaltases of Synechocystis sp. (48% identity [10]), A. aeolicus (44% identity [3]), S. pneumoniae (43% identity [14]), C. butyricum (42% identity [6]), and B. burgdorferi (32% identity [5]), as well as potato D-enzyme (41% identity [23]), but low levels of homology with the amylomaltases of E. coli (19% identity [20]), H. influenzae (20% identity [4]), C. psittaci (21% identity [7]), and M. tuberculosis (20% identity [2]).
In E. coli, amylomaltase is a member of a maltooligosaccharide transport and utilization system, which includes maltodextrin phosphorylase and maltose transport proteins (22). The role of amylomaltase is to convert short maltooligosaccharides into longer chains upon which maltooligosaccharide phosphorylase can act. In the genome of E. coli, amylomaltase and maltodextrin phosphorylase are encoded by the genes of the malPQ operon and are transcribed as a single transcriptional unit. Similar operon structures have been found in S. pneumoniae (14), Klebsiella pneumoniae (1), and C. butyricum (6). On the other hand, the amylomaltase genes of H. influenzae (4) and A. aeolicus (3) are found in the glycogen operon, which includes genes for glycogen synthesis and degradation. In order to obtain information about the T. aquaticus amylomaltase, including its physiological role in vivo, we sequenced at least 500 bp of the 5′ upstream and 3′ downstream regions of the T. aquaticus amylomaltase gene. However, sequences homologous to genes encoding glycogen or maltooligosaccharide metabolism were not found.
Expression of T. aquaticus malQ in E. coli.
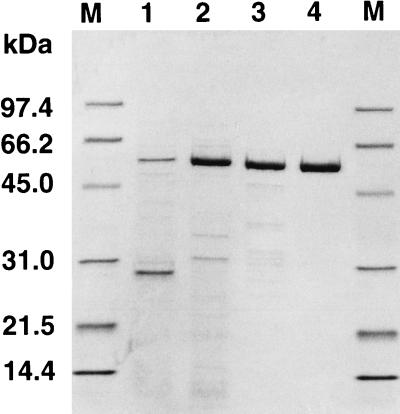
The coding sequence of malQ was amplified by PCR in order to create additional NdeI and EcoRI restriction sites at the translation initiation codon and 13 bp after the stop codon, respectively. The amplified fragment was digested with NdeI and EcoRI and ligated to the NdeI-EcoRI site of plasmid pGEX-Nde in order to construct plasmid pFQG8. When E. coli MC1061 carrying plasmid pFQG8 was grown with the inducer IPTG, thermostable amylomaltase activity was detected only in the soluble fraction of an extract of E. coli cells. Although activity was detected without IPTG, the activity was three times higher when the recombinant E. coli was grown with 0.01 mM IPTG. When the concentration of IPTG was increased to 0.1 mM, the activity was decreased because E. coli growth was suppressed. The amylomaltase was purified from E. coli cell extracts by the three purification steps summarized in Table 1. After each purification step samples were analyzed by SDS-PAGE (Fig. 1). The molecular mass of the recombinant enzyme was 57 kDa, and thus this enzyme was the same size as the enzyme purified from T. aquaticus cells (data not shown). As expected, heat treatment was a very effective method for purifying T. aquaticus amylomaltase from E. coli, because most of the endogenous proteins were denatured by this treatment and were easily removed by centrifugation.
TABLE 1.
Purification of T. aquaticus amylomaltase expressed in E. coli
| Purification step | Amt of total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) |
|---|---|---|---|---|
| Cell extract | 3,900 | 2,000 | 0.53 | 100 |
| Heat treatment | 590 | 1,200 | 2.1 | 60 |
| Phenyl-Toyopearl 650M | 330 | 860 | 2.7 | 42 |
| Source 15Q | 220 | 640 | 2.9 | 31 |
FIG. 1.
SDS-PAGE analysis performed at each purification step. A 10-μg portion of protein from each purification step was analyzed. Lane 1, cell extract; lane 2, cell extract after heat treatment; lane 3, Phenyl-Toyopearl 650 M pool; lane 4, Source 15Q pool; lanes M, marker proteins (Bio-Rad).
Enzymatic properties of amylomaltase.
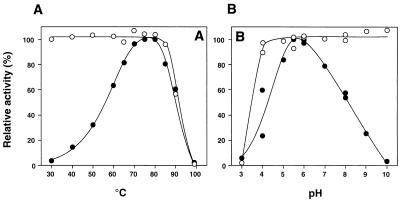
The optimum temperature for activity of the recombinant enzyme was around 75°C (Fig. 2A). This enzyme was stable after incubation at 85°C for 10 min (Fig. 2A), and about 50% of the enzyme activity was retained even after incubation at 80°C for 24 h (data not shown). However, almost all enzyme activity was lost after incubation at 100°C for 10 min. On the basis of these results, we decided to incubate the reaction mixture at 100°C for at least 10 min to stop the enzyme reaction. The optimum pH was 5.5 to 6.0 (Fig. 2B), and the enzyme was stable at pH 4.0 to 10.0 at 70°C for 10 min (Fig. 2B). These properties of the recombinant enzyme were the same as the properties of the enzyme purified from T. aquaticus cells (data not shown).
FIG. 2.
Effects of temperature (A) and pH (B) on activity (●) and stability (○) of the amylomaltase. (A) The enzyme activity was assayed at each temperature in 50 mM CH3COONa-CH3COOH (pH 5.5). To determine the thermal stability, the enzyme in 10 mM KH2PO4-Na2HPO4 (pH 7.0) was incubated at each temperature for 10 min and then immediately transferred to ice. The residual activity was assayed at 70°C in 50 mM CH3COONa-CH3COOH (pH 5.5). (B) The enzyme activity was assayed at 70°C in each buffer (50 mM). To determine the pH stability, the enzyme in each buffer (100 mM) was incubated at 70°C for 10 min. After the enzyme solution was diluted with 10 mM KH2PO4-Na2HPO4 (pH 7.0), the residual activity was assayed at 70°C in 50 mM CH3COONa-CH3COOH (pH 5.5). The buffers used were CH3COONa-HCl (pH 3.0 to 5.0), CH3COONa-CH3COOH (pH 4.0 to 6.0), KH2PO4-Na2HPO4 (pH 5.5 to 8.0), and H3BO4 · KCl-NaOH (pH 8.0 to 10.0).
Reaction with maltooligosaccharides.
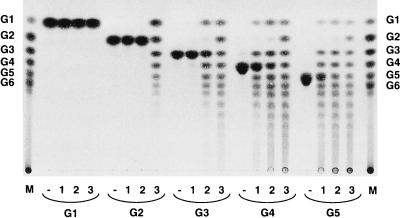
To confirm the identity of the amylomaltase expressed in E. coli, we investigated the activity of the purified enzyme with maltooligosaccharides. The enzyme was incubated with each maltooligosaccharide at a concentration of 1% in 20 mM sodium acetate buffer (pH 5.5) at 70°C for 6 h; three different enzyme concentrations (5, 10, and 50 mU/ml) were used. After each reaction solution was heated at 100°C for 10 min, the reaction products were analyzed by TLC (Fig. 3). At the highest enzyme concentration (50 mU/ml) (Fig. 3, lane 3), transglycosylation products (maltooligosaccharides and glucose) were produced from all of the maltooligosaccharides tested. However, at a lower enzyme concentration (10 mU/ml) (Fig. 3, lane 2), no transglycosylation products were obtained from G2, indicating that G2 was not an effective substrate for the enzyme under these conditions. When the concentration of the enzyme was decreased to 5 mU/ml (Fig. 3, lane 1), transglycosylation products were produced from G4 and G5 but not from G3 and G2. These results show that T. aquaticus amylomaltase catalyzes transglycosylation of maltooligosaccharides, but larger molecules (G4 and G5) are more effective substrates than smaller molecules (G2 and G3).
FIG. 3.
TLC of reaction products formed from the activity of amylomaltase with maltooligosaccharides. Reaction mixtures (300 μl) containing 1% (wt/vol) substrate in 20 mM sodium acetate buffer (pH 5.5) and 5 mU of enzyme per ml (lanes 1), 10 mU of enzyme per ml (lanes 2), 50 mU of enzyme per ml (lanes 3), or no enzyme (lanes −) were incubated at 70°C for 6 h. Three microliters of each reaction mixture was analyzed by TLC. Lanes M contained standard maltooligosaccharides. G1, glucose; G6, maltohexaose.
The major difference found so far between E. coli amylomaltase and plant D-enzyme is the minimum glucan unit that is transferred by the enzyme. E. coli amylomaltase can transfer glucose units and larger units (19), but plant D-enzyme cannot transfer glucose units (9, 27). In the case of T. aquaticus amylomaltase, G3 and G5 were produced from G4 and G4 and maltohexaose were produced from G5 at the lowest enzyme concentration used (Fig. 3, lane 1). These results strongly suggested that T. aquaticus amylomaltase can transfer glucose units and thus resembles E. coli amylomaltase.
E. coli amylomaltase and plant D-enzyme also differ in the ability to produce G2. E. coli amylomaltase can produce G2 (19) from maltooligosaccharides, but plant D-enzyme cannot (9, 27). There are two possible ways for these enzymes to produce G2. One is glucosyl transfer to glucose, and the other is cleavage of the linkage penultimate to the reducing end of the donor molecule. T. aquaticus amylomaltase did not produce G2 at the lowest enzyme concentration used (Fig. 3, lane 1), indicating that this enzyme cannot cleave the linkage penultimate to the reducing end like plant D-enzyme. However, when the enzyme concentration was increased, G2 was produced (Fig. 3, lanes 2 and 3). We think that the G2 produced at the higher enzyme concentration was due to glucosyl transfer to glucose, although this was not fully demonstrated.
Reaction with amylose.
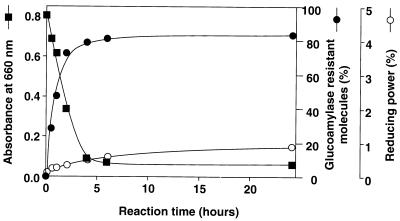
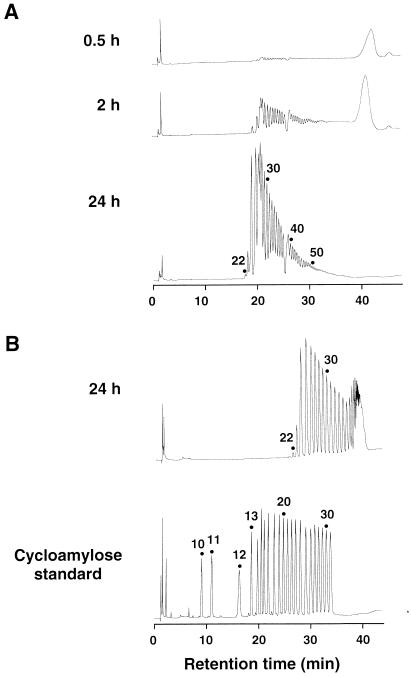
Recently, it was found that potato D-enzyme catalyzes not only the intermolecular transglycosylation (disproportionation) reaction but also the intramolecular transglycosylation (cyclization) reaction of amylose (24), but such activity has not been reported for a bacterial amylomaltase. The activity of T. aquaticus amylomaltase with amylose was investigated by using synthetic amylose AS-110 (average molecular mass, 110 kDa) as the substrate (Fig. 4). When 0.2% (wt/vol) amylose AS-110 was incubated with the enzyme, the ability of amylose to form an iodine complex, as determined by measuring the absorbance at 660 nm, decreased rapidly. However, the reducing power of the reaction mixture was not increased significantly and was less than 1% of the total sugar even after 24 h of reaction. These results strongly suggest that the enzyme catalyzed cyclization of amylose and produced cycloamylose, like potato D-enzyme. To demonstrate that cycloamylose was present, the reaction mixture was treated with glucoamylase since cycloamylose is resistant to this enzyme. The amount of glucoamylase-resistant glucans increased with time, and 84% of the total sugar was converted into glucoamylase-resistant glucans after 24 h of reaction. In order to confirm that the glucoamylase-resistant glucans were cycloamyloses, they were subjected to HPAEC. The glucoamylase-resistant glucans separated into many peaks (Fig. 5), whose retention times were the same as those of the cycloamylose standard with DP ranging from 22 to 31 (Fig. 5B). In addition to these results, the glucoamylase-resistant glucans were completely degraded to glucose by the combined action of α-amylase and glucoamylase (data not shown). On the basis of all of these results, we concluded that the glucoamylase-resistant glucans were cycloamyloses with DP of 22 or more.
FIG. 4.
Activity of amylomaltase with amylose AS-110. Reaction were stopped at different times. The absorbance at 660 nm (■) and the reducing power (○) of the reaction mixture were determined. The reducing power when all of the amylose was broken down to glucose was defined as 100%. The amount of glucoamylase-resistant glucans (●) was determined as described in the text.
FIG. 5.
HPAEC analysis of products formed from the activity of amylomaltase with amylose AS-110. (A) The glucoamylase-resistant glucans in reaction mixtures (50 μl) prepared as described in the legend to Fig. 4 were precipitated with 10 volumes of ethanol, dried in vacuo, and then redissolved in 50 μl of distilled water. Each 25-μl sample was analyzed by HPAEC by using a sodium acetate gradient. (B) To determine the DP of cycloamylose, the glucoamylase-resistant glucans obtained after 24 h and the cycloamylose standard were analyzed by HPAEC by using a sodium nitrate gradient. The DP of cycloamyloses are indicated above the peaks.
It has been thought for a long time that CGTase is the only enzyme which can catalyze the cyclization of amylose. However, similar cyclization reactions were observed in 1996 with potato D-enzyme (24), more recently with the novel 4-α-glucanotransferase of Thermococcus litoralisi (8), and in this study with bacterial amylomaltase (Fig. 5). These results suggest that the intramolecular transglycosylation (cyclization) reaction is not a special reaction that occurs only with CGTase but may be a common feature of all 4-α-glucanotransferases. Although all of the glucanotransferases catalyze similar reactions, they apparently can be distinguished on the basis of the cyclic glucans produced. T. aquaticus amylomaltase preferentially produced large cycloamyloses with DP of more than 60 (the large peaks that eluted around 42 min) in the initial stage of the reaction. These products were subsequently converted into smaller products with DP of 22 or more (Fig. 5A). The potato D-enzyme (24) and CGTase (26) reaction patterns were similar, but the lowest DPs of cycloamyloses were 17 and 6, respectively. The mechanism which determines the smallest cyclic glucan is not known but is of great interest. There is significant homology (40% identity) between the amino acid sequences of potato D-enzyme and T. aquaticus amylomaltase, but there is no similarity between D-enzyme (amylomaltase) and CGTase. Surprisingly, the novel 4-α-glucanotransferase of T. litoralis also exhibits no similarity to the other glucanotransferases described above (8). The tertiary structure of CGTase has been obtained by X-ray crystallographic studies (11–13, 15), but the tertiary structures of other 4-α-glucanotransferases are still not available.
Cycloamylose is highly soluble in water. It can form inclusion complexes with several guest molecules (24), and it is expected that cycloamylose will be used in the food, pharmaceutical, and chemical industries. Thermal stability is one of the most important properties of enzymes used for cycloamylose production. The amylomaltase of T. aquaticus is a good candidate, since it has high thermal stability and can efficiently convert amylose into cycloamylose.
ACKNOWLEDGMENTS
We especially thank K. Koizumi (Mukogawa Women’s University) for the generous gift of a cycloamylose standard.
This work was supported in part by a grant for the development of the next generation of bioreactor systems from the Society for Techno-Innovation of Agriculture, Forestry, and Fisheries (STAFF).
REFERENCES
- 1.Bloch M A, Raibaud O. Comparison of the malA regions of Escherichia coli and Klebsiella pneumoniae. J Bacteriol. 1986;168:1220–1227. doi: 10.1128/jb.168.3.1220-1227.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, III C E B, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 3.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 4.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 5.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 6.Goda S K, Eissa O, Akhtar M, Minton N P. Molecular analysis of a Clostridium butyricum NCIMB 7423 gene encoding 4-α-glucanotransferase and characterization of the recombinant enzyme produced in Escherichia coli. Microbiology. 1997;143:3287–3294. doi: 10.1099/00221287-143-10-3287. [DOI] [PubMed] [Google Scholar]
- 7.Hsia R C, Pannekoek Y, Ingerowski E, Bavoil P M. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol Microbiol. 1997;25:351–359. doi: 10.1046/j.1365-2958.1997.4701834.x. [DOI] [PubMed] [Google Scholar]
- 8.Jeon B-S, Taguchi H, Sakai H, Ohshima T, Wakagi T, Matsuzawa H. 4-α-Glucanotransferase from the hyperthermophilic archaeon Thermococcus litoralis. Enzyme purification and characterization, and gene cloning, sequencing and expression in Escherichia coli. Eur J Biochem. 1997;248:171–178. doi: 10.1111/j.1432-1033.1997.00171.x. [DOI] [PubMed] [Google Scholar]
- 9.Jones G, Whelan W J. The action pattern of D-enzyme, a transmaltodextrinylase from potato. Carbohydr Res. 1969;9:483–490. [Google Scholar]
- 10.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 11.Klein C, Schulz G E. Structure of cyclodextrin glycosyltransferase refined at 2.0 Å resolution. J Mol Biol. 1991;217:737–750. doi: 10.1016/0022-2836(91)90530-j. [DOI] [PubMed] [Google Scholar]
- 12.Knegtel R M A, Wind R D, Rozeboom H J, Kalk K H, Buitelaar R M, Dijkhuizen L, Dijkstra B W. Crystal structure at 2.3 Å resolution and revised nucleotide sequence of the thermostable cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1. J Mol Biol. 1996;256:611–622. doi: 10.1006/jmbi.1996.0113. [DOI] [PubMed] [Google Scholar]
- 13.Kubota M, Matsuura Y, Sakai S, Katsube Y. Three-dimensional structure of cyclodextrin glucanotransferase and its reaction mechanism. J Appl Glycosci. 1994;41:245–253. [Google Scholar]
- 14.Lacks S A, Dunn J J, Greenberg B. Identification of base mismatches recognized by the heteroduplex-DNA-repair system of Streptococcus pneumoniae. Cell. 1982;31:327–336. doi: 10.1016/0092-8674(82)90126-x. [DOI] [PubMed] [Google Scholar]
- 15.Lawson C L, van Montfort R, Strokopytov B, Rozeboom H J, Kalk K H, de Vries G E, Penninga D, Dijkhuizen L, Dijkstra B W. Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose dependent crystal form. J Mol Biol. 1994;236:590–600. doi: 10.1006/jmbi.1994.1168. [DOI] [PubMed] [Google Scholar]
- 16.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 17.Miwa I, Okuda J, Maeda K, Okuda G. Mutarotase effect on colorimetric determination of blood glucose with β-d-glucose oxidase. Clin Chim Acta. 1972;37:538–540. doi: 10.1016/0009-8981(72)90483-4. [DOI] [PubMed] [Google Scholar]
- 18.Monod J, Torriani A M. Synthèse d’un polysaccharide de type amidon aux dèpens du maltose, en prèsence d’un extrait enzymatique d’origine bacterienne. C R Acad Sci. 1948;227:240–242. [Google Scholar]
- 19.Palmer T N, Ryman B E, Whelan W J. The action pattern of amylomaltase from Escherichia coli. Eur J Biochem. 1976;69:105–115. doi: 10.1111/j.1432-1033.1976.tb10863.x. [DOI] [PubMed] [Google Scholar]
- 20.Pugsley A P, Dubreuil C. Molecular characterization of malQ, the structural gene for the Escherichia coli enzyme amylomaltase. Mol Microbiol. 1988;2:473–479. doi: 10.1111/j.1365-2958.1988.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz M. The maltose regulon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1482–1502. [Google Scholar]
- 23.Takaha T, Yanase M, Okada S, Smith S M. Disproportionating enzyme (4-α-glucanotransferase; EC 2.4.1.25) of potato. J Biol Chem. 1993;268:1391–1396. [PubMed] [Google Scholar]
- 24.Takaha T, Yanase M, Takata H, Okada S, Smith S M. Potato D-enzyme catalyzes the cyclization of amylose to produce cycloamylose, a novel cyclic glucan. J Biol Chem. 1996;271:2902–2908. doi: 10.1074/jbc.271.6.2902. [DOI] [PubMed] [Google Scholar]
- 25.Takeda Y, Guan H-P, Preiss J. Branching of amylose by the branching isoenzymes of maize endosperm. Carbohydr Res. 1993;240:253–263. [Google Scholar]
- 26.Terada Y, Yanase M, Takata H, Takaha T, Okada S. Cyclodextrins are not the major cyclic α-1,4-glucans produced by the initial action of cyclodextrin glucanotransferase on amylose. J Biol Chem. 1997;272:15729–15733. doi: 10.1074/jbc.272.25.15729. [DOI] [PubMed] [Google Scholar]
- 27.Yoshio N, Maeda I, Taniguchi H, Nakamura M. Purification and properties of D-enzyme from malted barley. J Jpn Soc Starch Sci. 1986;33:244–252. [Google Scholar]