Abstract
Introduction
Adverse drug reaction reports are usually manually assessed by pharmacovigilance experts to detect safety signals associated with drugs. With the recent extension of reporting to patients and the emergence of mass media-related sanitary crises, adverse drug reaction reports currently frequently overwhelm pharmacovigilance networks. Artificial intelligence could help support the work of pharmacovigilance experts during such crises, by automatically coding reports, allowing them to prioritise or accelerate their manual assessment. After a previous study showing first results, we developed and compared state-of-the-art machine learning models using a larger nationwide dataset, aiming to automatically pre-code patients’ adverse drug reaction reports.
Objectives
We aimed to determine the best artificial intelligence model identifying adverse drug reactions and assessing seriousness in patients reports from the French national pharmacovigilance web portal.
Methods
Reports coded by 27 Pharmacovigilance Centres between March 2017 and December 2020 were selected (n = 11,633). For each report, the Portable Document Format form containing free-text information filled by the patient, and the corresponding encodings of adverse event symptoms (in Medical Dictionary for Regulatory Activities Preferred Terms) and seriousness were obtained. This encoding by experts was used as the reference to train and evaluate models, which contained input data processing and machine-learning natural language processing to learn and predict encodings. We developed and compared different approaches for data processing and classifiers. Performance was evaluated using receiver operating characteristic area under the curve (AUC), F-measure, sensitivity, specificity and positive predictive value. We used data from 26 Pharmacovigilance Centres for training and internal validation. External validation was performed using data from the remaining Pharmacovigilance Centres during the same period.
Results
Internal validation: for adverse drug reaction identification, Term Frequency-Inverse Document Frequency (TF-IDF) + Light Gradient Boosted Machine (LGBM) achieved an AUC of 0.97 and an F-measure of 0.80. The Cross-lingual Language Model (XLM) [transformer] obtained an AUC of 0.97 and an F-measure of 0.78. For seriousness assessment, FastText + LGBM achieved an AUC of 0.85 and an F-measure of 0.63. CamemBERT (transformer) + Light Gradient Boosted Machine obtained an AUC of 0.84 and an F-measure of 0.63. External validation for both adverse drug reaction identification and seriousness assessment tasks yielded consistent and robust results.
Conclusions
Our artificial intelligence models showed promising performance to automatically code patient adverse drug reaction reports, with very similar results across approaches. Our system has been deployed by national health authorities in France since January 2021 to facilitate pharmacovigilance of COVID-19 vaccines. Further studies will be needed to validate the performance of the tool in real-life settings.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40264-022-01153-8.
Key Points
| Artificial intelligence models were successfully developed and showed good performance to automatically pre-code patient adverse drug reaction reports. |
| An artificial intelligence-based pharmacovigilance tool was thus nationally approved and deployed in France in January 2021, in particular to assist professionals with the monitoring of the COVID-19 vaccination campaign. |
| Further studies will be needed to validate the performance of the tool in real-life settings. |
Introduction
Pharmacovigilance (PV) is a medical field that has rapidly evolved over the last decades and most particularly in the context of sanitary crises. In the 1960s, the thalidomide tragedy, triggered by the publication of several case reports [1, 2], highlighted the importance of spontaneous reporting of adverse drug reactions (ADRs) by healthcare professionals. From the creation in 1964 of the “Yellow Card” system in the UK to the worldwide development of institutions managing and analysing ADR declarations [3], the thalidomide crisis gave birth to the pillars of the PV system of the late 20th century: signal detection through healthcare professional reports, followed by pharmacoepidemiological studies to properly assess the detected risks when necessary. While this approach proved efficient in many cases, limits quickly arose [4, 5]: new adverse reactions other than single cases are seldom seen by physicians, unlikely to recognise potential links, and ADRs are massively under-reported by professionals, even for new drugs or in countries where such a process is mandatory.
To cope with these limits, broader spontaneous reporting schemes were more recently developed, thanks to the rise of informatics and the appearance of the “expert patient” concept [6]. Noticeably, authorities throughout the world expanded to patients the possibility to declare ADRs for PV [7]. In France, such a system was first implemented in 2011, and further developed in 2017 with the creation of a national web portal collecting ADR reports from patients [8]. Globally, patient-reported ADRs proved to be an important and useful, yet heterogeneous, source of information [9, 10]. An unexpected downside occurred though: in an age of mass media and over communication about sanitary crises, patient reports may now frequently overwhelm PV systems. In 2008, New Zealand’s Centre for Adverse Reactions Monitoring experienced a dramatic surge of ADR reports following a change in the formulation of thyroxine and widespread web and media coverage on the subject [11]. Likewise, in 2017 in France, a similar issue concerning Levothyrox® prompted a 2000-fold increase of reports coming from patients [12]. In both cases, the tremendous flow of information, manually handled by experts, led to the saturation of PV activities for a few months. Specifically, the identification of ADRs and the evaluation of their seriousness in free text was found to be very time consuming. This activity mobilised important resources, possibly detrimental to other surveillance activities.
Overall, these sanitary crises, caused by the over-reporting of ADRs by patients, shaped the need to develop novel methods to help PV systems remain efficient and responsive in such situations. Experts could benefit from an automatic identification of symptoms and assessment of seriousness in reports, allowing them to focus on unexpected or serious reports by prioritising their manual assessment, and thus more rapidly detect safety signals or patients in need of special care. Over the last few years, the development of such tools has therefore been the object of research and efforts [13], helped with the recent advancements in artificial intelligence (AI) technologies, especially machine learning and natural language processing (NLP), though evidence of the application of computational linguistics applied to PV remains scarce.
A recent systematic review of the literature [14], focusing on machine learning to understand text in PV, found a total of 16 publications on the subject and concluded that the analysis of text had the potential to complement the traditional system, but it focused on social media or forum-related content. In other publications, Schmider et al. [15] found that it was feasible to use NLP to support data extraction from text-based ADR reports, but their study was restricted to PV documents coming from private industrial entities, and did not perform well in detecting ADRs. Other teams [16, 17] obtained better performance in more recent studies, but focused on published case report abstracts retrieved from PubMed, which do not share the same structure and style of patient-written reports, which are massively the cause of over-reporting.
Because of this lack of evidence and tools dedicated to the automatic processing of patient ADR reports, the French National Agency for the Safety of Medicines and Health Products (ANSM) and its associated Regional Pharmacovigilance Centres (CRPVs) partnered in 2020 with Synapse Medicine to develop an AI tool, automating the evaluation of patient ADR reports, with the ambition to help PV experts cope with the upcoming COVID-19 vaccination campaign and future sanitary crises. Following a recently published first study showing the feasibility of such a system [18], this partnership was strengthened by allowing Synapse Medicine to use a larger nationwide dataset from the French national PV web portal.
The aim of this study was thus to train and evaluate AI models and compare their predictive performance to identify ADRs and assess seriousness in patient reports, using a large nationwide PV dataset managed by French public institutions. The results will serve as supporting evidence for implementing AI to automatically pre-code text-based patient ADR reports.
Methods
Data Sources
We retrospectively collected all available cases of ADR reports filled by patients through the national ADR reporting web portal between March 2017 and December 2020 and transmitted by ANSM to Synapse Medicine. Each report form contained structured and unstructured free-text information filled by patients, together with the annotation and coding realised by PV experts from CRPVs. Cases were considered unusable when linkage between reports and coding by experts was impossible.
Coding of ADRs and Seriousness
The coding of ADRs by PV experts relied on the MedDRA standardised terminology [19]. The fourth level, “Preferred Terms (PTs)”, was used, as it is the internationally recommended standard for the coding of adverse events [20]. The PTs coded by experts according to the free-text ADR descriptions in reports were used as the reference standard to learn and evaluate the performance of our AI models for ADR identification.
The coding of seriousness by PV experts relied on the standards specified by the World Health Organization [21], considering ADRs as serious when corresponding to at least one of the following situations: death, life threatening, requiring or prolonging hospitalisation, resulting in persistent or significant disability or incapacity, provoking congenital anomalies or birth defects, or resulting in other significant medical events. We used this coding as the reference standard to learn and evaluate the performance of our AI models assessing reports’ seriousness.
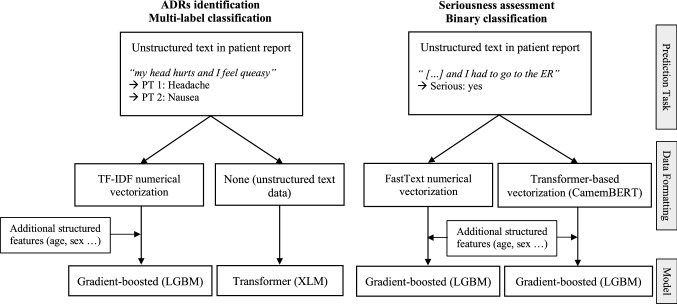
Experts could code multiple PTs for each ADR report, but assessed seriousness in a binary manner on each report as a whole. Identifying ADRs was therefore a multi-label classification task, where models can identify one or several distinct PTs. Assessing seriousness was a binary classification task, resulting from the integration of heterogeneous information regarding events, patient characteristics and outcomes. We therefore used different methods to identify ADRs and assess seriousness from reports, further discussed below and summarised in Fig. 1.
Fig. 1.
Summary of the methodological differences between tasks. ADRs adverse drug reactions, ER emergency room, LGBM Light Gradient Boosted Machine, PT Preferred Term, XLM Cross-lingual Language Model
Selection of Training, Internal Validation and External Validation Sets
Datasets from 27 CRPVs (total of 31) were available. We used data from 26 of them for the development set (train and internal validation set), aiming to represent approximately 90% of the data to maximise our learning abilities. The development set included the cases used in our previous study [18]. Data from the remaining CRPV were used as an external validation set to evaluate the robustness of our models on an unlearned sample and estimate the generalisability of their performance. We used a different CRPV for external validation to obtain a conservative estimate of our models’ performance, as practices or types of reports might be heterogeneous across teams, even if the coding of ADRs and their seriousness is supposedly standardised. The remaining CRPV dataset used for external validation was chosen so that it represented approximately 10% of all cases.
Data Extraction and Pre-Processing
Methods for data extraction and pre-processing were already described in our previous study [18]. Briefly, the datasets were provided by ANSM, and composed of tables in Portable Document Format containing fields of interest. We extracted the text from these Portable Document Formats using the Python library Camelot [22]. We performed basic text processing on the raw data (accents and punctuation removal, case lowering, stemming). We turned the following fields into structured features, to better take into account patient’s specificities and treatments: age, body mass index as a summary feature of weight and size, sex, a one-hot representation of outcomes and a one-hot representation of drugs. The final feature vectors were built concatenating these structured features and the free-text patient ADR descriptions (vectorised using approaches described below).
ADR Identification Task
Two different approaches were tested for the ADR identification task evaluated in this study. Considering the results of our previous study, we decided to compare a Light Gradient Boosting Machine (LGBM) [23] with a Cross-lingual Language Model (XLM) [24]. The first approach, a LGBM, is a gradient boosting framework that uses tree-based learning algorithms. Boosting is an ensemble learning method that aims to train many models sequentially, where each model learns from the errors of previous models. Successive iterations are found by applying a gradient descent in the direction of the average gradient of the previous (weaker) model leaf nodes, using error residuals of the loss function. A LGBM is known to provide generally good performance on various classification or regression machine learning problems [23]. It was selected as it was the best performing in our first study [18], which consisted of a similar study on a smaller dataset. The second approach, XLM, is a transformer model. Transformers, also called “BERT-like”, are deep learning attention-based neural networks [25], which process text sequence inputs simultaneously by forming direct connections between individual elements through an attention mechanism, unlike traditional methods that process each sequence element in turn. Unlike LGBMs, transformer models are already pre-trained on plain text corpus, and allow for a contextual bidirectional representation of words in sentences. Multi-class text classification in transformer-based models is then achieved using a classification layer on top of the transformer model, using n output neurons corresponding to each class. Transformers have recently been considered state of the art for NLP and subsequent classification tasks [26], but were not evaluated in our first study because the dataset was too small for deep learning approaches.
Regarding data formatting, distinct procedures were applied for both approaches. For the LGBM approach, we vectorised text in numerical vectors, as gradient-boosting models need numerical features as inputs. More specifically, we vectorised unstructured text data in numerical vectors using the Term Frequency-Inverse Document Frequency (TF-IDF) method [27]. We then used the TF-IDF vectors and the previously mentioned structured data (except the one-hot representation of outcomes) as model features. For the XLM approach, data formatting was not needed, as transformer models directly use unstructured text data as inputs. Additional structured features were not included in this approach, as valuable information regarding symptoms was considered to be in free text, and XLM does not support structured data as inputs.
All reports transmitted by the 26 CRPVs were included in the development set for both approaches. Likewise, the external validation set included all transmitted reports from the remaining CRPV. Models were trained and evaluated considering PTs with at least ten occurrences in the development set, owing to the size of data needed for this NLP task.
Seriousness Assessment Task
For the assessment of ADRs seriousness, we decided to compare two LGBM models with different data formatting procedures. At first, similar to the ADR identification task, a full transformer approach was tested. It was deemed unsatisfactory though, considering that seriousness is a task that sensibly relies on the structured data available in reports (such as age, sex and outcomes) and that transformers cannot directly use structured data as inputs. Therefore, we compared two gradient boosting approaches: one with classical word embedding for data formatting, vs one with a transformer model dedicated to data formatting (both structuring unstructured text data into inputs usable by LGBM). For the first approach, we used the FastText library [28] and its extremeText extension, pre-trained on a dataset composed of text dealing with medical topics extracted from French reference sources. For the second approach, we used CamemBERT [29], which is a French pre-trained version of BERT, the first transformer model developed on text from Wikipedia. As previously mentioned, after data formatting, both approaches included the additional structured data available from reports as model features, and we used the LGBM model for the classification part. Both approaches were trained using only a sub-part of the reports available: we excluded those concerning levothyroxine (which caused a sanitary crisis in France between 2017 and 2018), as codings regarding seriousness for this drug were too heterogeneous across experts for machine learning purposes.
Statistical Methods
We evaluated the performance of all AI models using multiples metrics: receiver operating characteristics area under the curve (ROC AUC), positive predictive value1, sensitivity2 (= true positive rate), specificity3 (= true negative rate) and F-measure4 (also called F1 score, the harmonic mean of precision1 and recall2). We aimed to improve the result of our previous study [18]: AUC 0.93/F-Measure 0.72 for ADR identification, and AUC 0.75/F-Measure 0.60 for seriousness assessment.
For the initial prototyping of approaches, k-fold cross validation [30] was used for training and internal validation, using k = 10 for the identification of ADRs and k = 5 for the assessment of seriousness. Records in the development dataset were randomly divided into k parts, where k-1 parts were used for training and the kth one left out for testing. A similar process was repeated k times across the dataset, using a decision threshold that maximised the F-measure, distinct for each task or approach.
Metrics reported in this paper were estimated using a bootstrap procedure in order to obtain better average estimates and 95% confidence intervals (95% CIs). For training and internal validation, we used n = 100 samples, randomly split using a 90/10 ratio for ADR identification and an 80/20 ratio for seriousness assessment. Such ratios were chosen so that they could maximise learning abilities for both tasks, yet leave enough cases for a precise evaluation of models’ performance through the estimation of confidence intervals. A narrower ratio was thus used for the seriousness assessment because of the lower prevalence of serious cases compared with PTs (multiple PTs can be coded for each report, while seriousness is assessed in a binary manner). Models were therefore internally trained using random samples containing 90% or 80% of the data, according to the task, and evaluated on the remaining 10% or 20%. As for all metrics reported in this paper, we used the decision threshold that maximised the F-measure. For external validation, fully internally trained models were also evaluated using a bootstrap procedure on 100 random samples, representing 80% of the external validation dataset for both tasks. For each task and validation sets, medians and 95% CIs of all n bootstrap metrics were estimated. Confidence intervals were estimated with the percentile bootstrap interval, using the definition recommended by Hyndman and Fan for quantiles [31], considered unbiased regardless of data distribution.
Hyperparameters were tuned using a classical grid search strategy on a dedicated train-test tuning split. Overfitting was controlled by performing the internal validation of models using the previously described k-fold cross validation, bootstrapping, as well as externally validating the models on independent data (external validation set). Concerning model implementations, we used Python 3 (Python Software Foundation, Beaverton, OR, USA) and the following libraries: scikit-learn, lgbm, pytorch, xlm, transformers. Analysis also involved the use of R 4.0.2 (R Foundation for Statistical Computing, Vienna, Austria) for descriptive statistics, predictive metrics and plots (ROCR package).
Results
Dataset Selection and General Characteristics
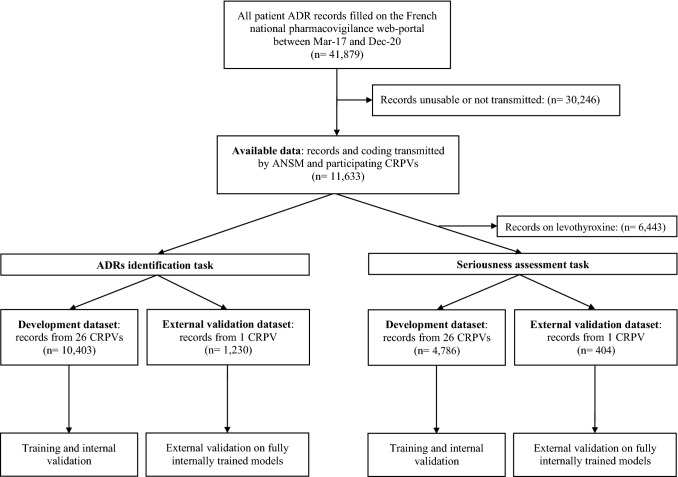
Figure 2 shows the selection process for datasets used for both tasks and both validations. A total of 11,633 usable ADR report forms were transmitted by ANSM and CRPVs to Synapse Medicine. These represented roughly one quarter of all patient reports filled on the national web portal during the study period. Among those, 10,403 reports (89.4% of the total), coded by 27 CRPVs, were included in the development dataset for ADR identification, and 4786 (41.1% of the total) in the development dataset for seriousness prediction. The external validation set for ADR identification included 1230 reports (10.6% of the total available), while the external validation set for seriousness assessment included 404 reports (3.5% of the total available). All reports in the external validation datasets came from Nantes’s CRPV.
Fig. 2.
Flowchart of the selection of datasets. ADR adverse drug reaction, ANSM French National Agency for the Safety of Medicines and Health Products, CRPV Regional Pharmacovigilance Centre
Table 1 presents detailed characteristics of the included datasets. Regarding data used for ADR identification, in the development set, patients were mostly women (84.5%), with a median age of 51 years (interquartile range [IQR] 37–62). Reports mentioned a median 1 drug per report (IQR 1–1), and 2.6% mentioned a vaccine. Experts coded a median number of four distinct PTs per report (IQR 2–6). Overall, 50,546 PT terms were coded in this dataset. Among these, 1465 were distinct PTs, and 311 (21.2%) were present in at least ten reports and thus learned by our ADR identification models. These 311 distinct PTs represented 94.7% of all PTs coded in the dataset (47,845/50,546). In the external validation dataset, patients were also mostly women (85.9%), with a median age of 51 years (IQR 39–63). Reports mentioned a median 1 drug per report (IQR 1–1), and 1.1% mentioned a vaccine. Experts coded a median number of five distinct PTs per report (IQR 3–7). Overall, 6636 PTs were coded in this dataset, representing 506 distinct terms. The 311 PTs considered during training corresponded to 95.0% (6302/6636) of all PTs coded in the external validation dataset. Regarding seriousness assessment, both development and external validation datasets were also very similar. Median age of patients was 41 years (30–58) and 39 years (31–56), respectively. Most patients were women in both datasets (78.0% vs 77.5%). In the development dataset, 23.6% of reports were coded as serious, compared with 20.8% in the external validation dataset.
Table 1.
Patient and report characteristics
| ADR identification task | Seriousness assessment task | |||
|---|---|---|---|---|
| Development dataset (n = 10,403) |
External validation dataset (n = 1230) |
Development dataset (n = 4786) |
External validation dataset (n = 404) |
|
| Patient age, years | ||||
| Median (IQR) | 51 (37–62) | 51 (39–63) | 41 (30–58) | 39 (31–56) |
| Patient sex, n (%) | ||||
| Female | 8792 (84.5) | 1056 (85.9) | 3732 (78.0) | 313 (77.5) |
| Encoded PTs per report, n | ||||
| Median (IQR) | 4 (2–6) | 5 (3–7) | 3 (1–5) | 3 (2–6) |
| Reported drugs per report, n | ||||
| Median (IQR) | 1 (1–1) | 1 (1–1) | 1 (1–1) | 1 (1–1) |
| Reports about vaccines, n (%) | ||||
| Yes | 275 (2.6) | 13 (1.1) | 275 (5.7) | 13 (3.2) |
| Reports considered serious, n (%) | ||||
| Yes | 2454 (23.6) | 136 (11.1) | 1128 (23.6) | 84 (20.8) |
ADRs adverse drug reactions, IQR interquartile range, PTs Preferred Terms
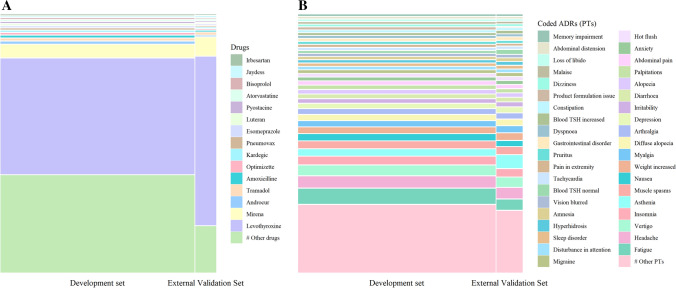
Figure 3A shows the most frequently coded drugs (top 15) and Fig. 3B the most frequently coded PTs (top 40) in both datasets used for ADR identification. Levothyroxine was by far the most frequently reported drug, accounting for around half of the reports in the development dataset and two thirds in the external validation dataset. Mirena® followed, accounting for around 5% of reports. Overall, across all datasets, 2836 distinct drug names were coded by experts. Regarding ADRs, fatigue was the most frequently coded PT, among 1529 distinct PTs in all datasets.
Fig. 3.
Repartition of the most frequently reported drugs and adverse drug reactions (ADRs) coded in Medical Dictionary for Regulatory Activities Preferred Terms (PTs) across complete datasets. A Mosaic plot of the most frequently reported drugs and B mosaic plot of the most frequently reported PTs
Comparison of Machine Learning Models
Table 2 shows the main median metrics and 95% CIs estimated for both tasks and validation sets, using a decision threshold maximising the F-Measure. Figures 4 and 5 show the ROC and precision-recall curves for the ADR identification and seriousness assessment, respectively. They allow for a more detailed view of the possible trade-offs between sensitivity, specificity and positive predictive value at various threshold settings.
Table 2.
Model comparison metrics, using the prediction threshold maximising F-measure
| Validation | Task | Models | AUC | F-measure | PPV | Sensibility | Specificity | TN | FP | FN | TP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Internal validation | ADR identification |
TF-IDF + LGBM |
0.97 (0.96–0.97) |
0.80 (0.78–0.81) |
0.85 (0.83–0.87) |
0.75 (0.73–0.78) |
1 (1–1) |
353,938 (345,353–360,293) |
703 (612–842) |
1329 (1194–1438) |
4028 (3857–4271) |
| XLM | 0.97 (0.96–0.97) |
0.78 (0.76–0.79) |
0.84 (0.82–0.86) |
0.73 (0.70–0.75) |
1 (1–1) |
353,854 (353,563–354,099) |
736 (609–883) |
1469 (1314–1592) |
3916 (3702–4131) |
||
| Seriousness assessment |
FastText + LGBM |
0.85 (0.82–0.87) |
0.63 (0.59–0.68) |
0.58 (0.52–0.69) |
0.69 (0.60–0.82) |
0.85 (0.77–0.91) |
629 (559–682) |
110 (62–166) |
69 (43–94) |
156 (129–194) |
|
| CamemBERT + LGBM |
0.84 (0.81–0.87) |
0.63 (0.57–0.67) |
0.56 (0.49–0.65) |
0.71 (0.57–0.81) |
0.83 (0.75–0.90) |
615 (542–672) |
126 (72–183) |
65 (44–96) |
160 (125–192) |
||
| External validation | ADR identification |
TF-IDF + LGBM |
0.97 (0.97–0.97) |
0.82 (0.81–0.82) |
0.88 (0.86–0.89) |
0.76 (0.75–0.78) |
1 (1–1) |
287,770 (287,640–287,896) |
502 (444–573) |
1128 (1054–1198) |
3631 (3530–3751) |
| XLM |
0.97 (0.97–0.97) |
0.80 (0.79–0.80) |
0.87 (0.86–0.88) |
0.74 (0.73–0.75) |
1 (1–1) |
288,717 (288,604–288,837) |
530 (476–558) |
1256 (1208–1310) |
3527 (3434–3602) |
||
| Seriousness assessment |
FastText + LGBM |
0.87 (0.85–0.89) |
0.65 (0.60–0.70) |
0.58 (0.49–0.69) |
0.77 (0.60–0.88) |
0.85 (0.75–0.92) |
274 (244–299) |
50 (25–80) |
21 (11–36) |
69 (54–79) |
|
| CamemBERT + LGBM |
0.86 (0.83–0.89) |
0.63 (0.59–0.68) |
0.56 (0.49–0.67) |
0.74 (0.59–0.84) |
0.84 (0.76–0.91) |
271 (246–294) |
53 (30–78) |
23 (14–37) |
67 (53–76) |
ADR adverse drug reactions, AUC area under the curve, FP false positive, FN false negative, LGBM Light Gradient Boosted Machine, PPV positive predictive value, TF-IDF Term Frequency-Inverse Document Frequency, TN true negative, TP true positive, XLM Cross-lingual Language Model
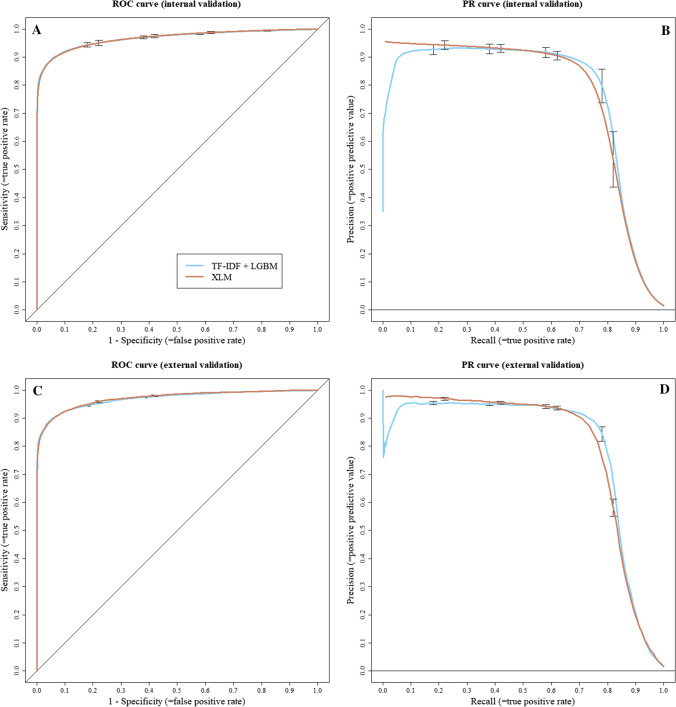
Fig. 4.
Adverse drug reaction identification: receiver operating characteristic (ROC) and precision-recall (PR) curves. A ROC curve of internal validation, B PR curve of internal validation, C ROC curve of external validation and D PR curve of external validation
Fig. 5.
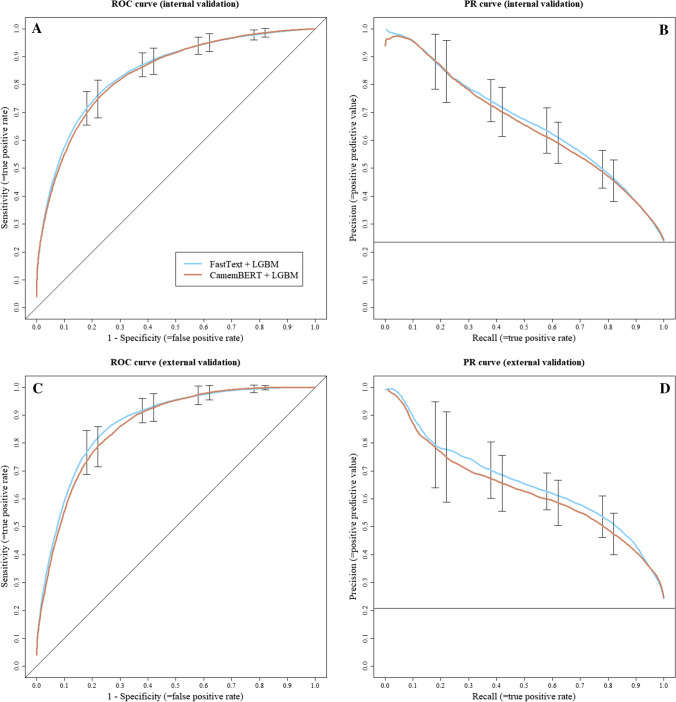
Seriousness assessment: receiver operating characteristic (ROC) and precision-recall (PR) curves. A ROC curve of internal validation, B PR curve of internal validation, with dataset seriousness prevalence at y = 0.24, C ROC curve of external validation and D PR curve of external validation, with dataset seriousness prevalence at y = 0.21
For the identification of ADRs, both approaches presented similar performances during internal validation, with TF-IDF + LGBM obtaining an AUC of 0.97 (95% CI 0.96–0.97) and an F-measure of 0.80 (95% CI 0.78–0.81), while XLM obtained an AUC of 0.97 (95% CI 0.96–0.97) and an F-measure of 0.78 (95% CI 0.76–0.79). Precision, sensitivity, specificity and true/false positives/negatives were also very close between models for internal validation (Table 2). Regarding external validation, both approaches yielded consistent results with the internal validation, with TF-IDF + LGBM obtaining an AUC of 0.97 (95% CI 0.97–0.97) and an F-measure of 0.82 (95% CI 0.81–0.82), while XLM obtained an AUC of 0.97 (95% CI 0.97–0.97) and an F-measure of 0.80 (95% CI 0.79– 0.80). External validation also yielded consistent and close results for other metrics (Table 2).
For the assessment of seriousness, both approaches also presented similar performance during internal validation. FastText + LGBM obtained an AUC of 0.85 (95% CI 0.85–0.87) compared with an AUC of 0.84 (95% CI 0.81–0.87) for CamemBERT + LGBM, and both approaches had a similar F-Measure of 0.63 (95% CI 0.59–0.68 and 0.57–0.67, respectively). Precision, sensitivity, specificity and true/false positives/negatives were also very close between models for internal validation (Table 2). Regarding external validation, performances were consistent with internal validation, with FastText + LGBM obtaining an AUC of 0.87 (95% CI 0.85–0.89) and an F-measure of 0.65 (95% CI 0.60–0.70), while CamemBERT + LGBM obtained an AUC of 0.86 (95% CI 0.83–0.89) and an F-measure of 0.63 (95% CI 0.59–0.68). External validation also yielded consistent and close results for other metrics (Table 2).
Discussion
In this validation study, we analysed the predictive performance of different AI models supporting the automatic coding of patient ADR reports. For ADR identification, both TF-IDF + LGBM and XLM achieved an AUC of 0.97 during internal validation, with an F-measure of 0.80 and 0.78, respectively. External validation yielded consistent and robust results. For the assessment of seriousness, during internal validation, FastText + LGBM achieved an AUC of 0.85, CamemBERT + LGBM an AUC of 0.84, and both approaches had an F-measure of 0.63. External validation also yielded consistent and robust results.
For both tasks, neither approach seemed to outweigh the other, with very similar performances. This could be explained by the balanced strengths and weaknesses of compared approaches. For ADR identification, the use of additional structured features on top of TF-IDF might help LGBM match XLM performances, which uses a more modern attention-based architecture based on neural networks but does not support additional structured features. For the seriousness assessment, the main difference was also related to classifiers’ inputs. While FastText word embedding is usually considered less efficient than the contextual bidirectional representation achieved by BERT, we used a specific version of FastText pre-trained on medical data, compared with the more general CamemBERT model, pre-trained on French Wikipedia data. Both approaches then used LGBM for the classification part, which might explain close results. In real-life settings, performances between approaches might diverge, as novel drugs, report styles or declarations emerge. We will therefore need to continue validating and comparing them using new data. In terms of computing, TF-IDF or FastText-based LGBM models are lighter and run faster than transformers, such as XLM and CamemBERT, but these might emerge as more accurate in the long term, as transformer-based deep learning models are currently considered state of the art for NLP [26]. They also have the advantage of being multilingual, potentially allowing for the transposability of our models in other languages than French.
Overall, while still perfectible, our AI models showed promising performance to support the automatic coding of ADR reports, though a more thorough interpretation of our results might be needed. In our study, reported precision (= positive predictive value), sensitivity ( = recall) and specificity were estimated using a predictive threshold aimed to maximise the F-measure. We decided to use such a threshold as the F-measure is the most common mathematical score used to compare machine learning classification models. As we aim to continue further improvement of our models, it allows for a good comparison over time or with novel approaches. The F-measure is a difficult statistic to interpret for the end user though, as its meaning varies with the prevalence of predicted classes, and it is not fit for tasks needing a particular trade-off between sensitivity, specificity or precision [32]. In a PV crisis use-case [1], experts might value sensitivity over precision for the automatic assessment of seriousness in ADR reports, in order to prioritise potentially severe yet rarer cases. In another use-case [2], experts might value precision when wishing to automatically pre-fill PT codings in reports. In this regard, ROC and PR curves are more informative, as they allow a better understanding of the possible trade-offs between measures. For a use-case [1], our seriousness assessment models allow for an 80% recall/50% precision trade-off, as shown in Fig. 4B and D. In our dataset, this translates to the manual assessment of roughly 32% of all cases to correctly capture four fifths of the 20% cases considered serious. For a use-case [2], experts may choose the 90% precision/70% recall trade-off of our models identifying ADRs, as shown in Fig. 5B and D. This could allow for a nearly automatic “fill-free” validation of 70% of the pre-coded PTs in all reports. As these estimates are only based on the reading of our internal and external validation ROC and PR curves, these will also need to be confirmed in further real-life studies. A study group composed of experts from different CRPVs has been established to arbitrate on clinical rules, such as the selected threshold for our distinct tasks, aiming to calibrate our models with the best parameters for PV professional end users.
These results are a major improvement in comparison with our first study [18], where regarding the identification of ADRs we reported an AUC of 0.93 and an F-Measure of 0.72 for TF-IDF + LGBM. Regarding the assessment of seriousness, the results of FastText + LGBM were an AUC of 0.75 and an F-measure of 0.60. This shows that having more quality data to train machine learning models is key to obtaining better performance, and that these novel AI approaches clearly outperform classical methods, such as regular expression (RegEx), which we tested in our first study by matching Medical Dictionary for Regulatory Activities terms to obtain a baseline for our benchmarks. RegEx only achieved an AUC of 0.69 and an F-Measure of 0.50.
Our study has several strengths compared with previous work on the subject. Our selection of data sources covered nearly a quarter of all ADR reports transmitted by patients to ANSM and CRPV between March 2017 and December 2020, providing a wide sample of reports over this period and across multiple teams, with a variety of suspected drugs and coded PTs. We used real-life expert-annotated data, in contrast with studies using case report abstracts or simulated data. We compared state-of-the art models for our classification tasks, and created in-house AI pipelines dedicated to processing Portable Document Format reports. In addition, we externally validated our models on a distinct CRPV to assess their generalisability in a different setting, which yielded consistent and robust results.
Our study also has some limitations. First, regarding selection, most reports focused on levothyroxine and Mirena®, two drugs involved in sanitary crises widely reported by the media in 2017–18. Most ADRs described in our datasets are therefore those associated with levothyroxine and levonorgestrel. Although these cover a wide variety of PTs, susceptible to be associated with other drugs, this could bias the predictions of our model identifying ADRs in other situations. Second, models identifying ADRs in reports were not trained on PTs coded in fewer than ten reports in the development dataset. They are therefore not appropriate for the identification of all possible PTs. Nevertheless, our ambition was not to be exhaustive of all potential PTs but to identify those representing the most frequent PTs, which are usually the issue with over-reporting. Our results showed that this only misses 5% of coded PTs. Finally, our models’ predictive performances, especially regarding seriousness, do not reach levels accurate enough to consistently fully replicate coding by PV experts. The goal of our project was not to create a tool replacing experts, but assist them in the time-consuming process of manually assessing ADR reports. Our seriousness assessment AI pipeline can allow them to prioritise reports according to the automatic coding, and focus their time on cases predicted as serious. Likewise, our ADR identification pipeline can allow them to save time in the coding of PTs frequently associated with reports, by quickly validating automatically predicted PTs.
Our AI pipelines were judged accurate enough by ANSM for routine use. Since January 2021, a platform showing our models’ predictions for each new report filled by patients on the national web portal has been deployed in CRPVs, hoping to help experts cope with the expected surge of reports linked with the COVID-19 vaccination campaign [33]. As results were similar between approaches, we implemented TF-IDF + LGBM for ADRs identification and FastText + LGBM for the seriousness assessment, as they are lighter and quicker to run than transformers. This is, to our knowledge, the first AI-based platform nationally deployed for the automated coding of ADRs. British health authorities also launched a similar project in late 2020 with an industrial partner, to “ensure that no details from the ADRs’ reaction text are missed” [34], but no results or implementation have been communicated so far. The COVID-19 vaccination campaign is an interesting test for our system. Coding rules for ADRs indeed usually vary according to drugs and standards. For example, in the case of COVID-19 vaccines, CRPV experts received the instruction to prefer the use of the PT “flu syndrome”, rather than the multiple distinct PTs comprising the syndrome (“headache” + “fever”…), which our models may suggest. We already plan to alleviate this issue using online machine learning, which dynamically learns new patterns in reports, and adapt models to newly emerging situations, rules or drugs. We are currently working with ANSM to implement such a solution. Meanwhile, we had to manually tweak our models to better code flu syndrome ADRs associated with vaccines. In any case, it is important to remember that experts might code ADRs and seriousness heterogeneously across teams or individuals, even if standards or rules exist [35]. This might complicate the “true” validity of our models’ predictions. Because the reference standard (PV professional coding) used in this study might not be considered gold standard owing to possibly heterogeneous coding practices, further studies might more thoroughly examine and compare our model errors, as well as expert coding differences. This will be addressed by our CRPV experts committee. Further development and validation of our AI models over time will also be required to consolidate predictive performance and evaluate whether automatic coding was robust during the COVID-19 sanitary crisis.
Conclusions
In this study, we successfully trained and validated the performances of AI models identifying ADRs and assessing seriousness using unstructured text data from nationwide patient reports. Gradient boosting and transformer-based approaches yielded close results for both internal and external validation, probably in relation to the respective strengths and weaknesses of the distinct methods used for data formatting and classification.
Our system was considered accurate enough by ANSM for national deployment in France in January 2021, aiming to help PV experts cope with the COVID-19 vaccination campaign, during which authorities expected a massive reporting of ADRs. Further studies will be needed to validate the performance of our system in real-life settings and to continue identifying the best possible model, adding more evidence to the possible use of AI in the automatic pre-coding of PV reports.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Funding
Public funding was received by Synapse Medicine for this work, following a contract with the French Agency for the Safety of Health Products (Agence Nationale de Sécurité du Médicament et des Produits de Santé).
Conflict of interest
Guillaume Louis Martin, Julien Jouganous, Axel Bellec, Romain Savidan, Clément Goehrs and Louis Létinier were employed by Synapse Medicine at the time this research was conducted or hold stock/stock options therein. All other authors declared no competing interests.
Ethics approval
This study and the use of the French pharmacovigilance database have been validated by the French Agency for the Safety of Health Products (Agence Nationale de Sécurité du Médicament et des Produits de Santé). The data studied are the property of the Agence Nationale de Sécurité du Médicament et des Produits de Santé. Storage of the data is managed by service providers certified as “Hébergeur de Données de Santé” (Health Data Host).
Consent to participate
This study and the use of the French pharmacovigilance database have been validated by the French Agency for the Safety of Health Products (Agence Nationale de Sécurité du Médicament et des Produits de Santé).
Consent for publication
Not applicable.
Availability of data and material
Data are not shareable as they are the property of the Agence Nationale de Sécurité du Médicament et des Produits de Santé.
Code availability
The example code in Python for the AI pipelines is provided on the following git repository: https://github.com/louisletinier/MAITAI. For more information about data management or our algorithms, please contact the corresponding author.
Author contributions
G.L. Martin designed the research, analysed the data and wrote the manuscript. J. Jouganous, A. Bellec and R. Savidan designed the research, implemented models and critically reviewed the article. C. Goehrs critically reviewed the article. A. Pariente designed the research, critically reviewed the article, and was involved in the selection and acquisition of data. M. Benkebil, G. Miremont, J. Micallef and F. Salvo were involved in the selection and acquisition of data, and critically reviewed the article. L Létinier designed the research, analysed the data and critically reviewed the article. He is the guarantor; he had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors read and approved the final version of the manuscript and agreed to its publication.
Reporting guidelines
We used the STARD 2015 reporting guidelines [36], with its checklist available as Electronic Supplementary Material.
Footnotes
Also called precision in machine learning literature. PPV = TP/(TP+FP). If a prediction is made, it informs on its likelihood to be correct. In a multi-class problem, precision is the sum of true positives across all classes divided by the sum of true positives and false positives across all classes.
Also called recall in machine learning literature. Sensitivity = TP/(TP+FN). It informs on the likelihood to correctly capture positively predicted entities. In a multi-class problem, recall is calculated as the sum of true positives across all classes divided by the sum of true positives and false negatives across all classes.
Specificity = TN/(TN+FP). It informs on the likelihood to correctly capture negatively predicted entities. In a multi-class problem, specificity is calculated as the sum of true negatives across all classes divided by the sum of true negatives and false positives across all classes.
F-measure = 2×(precision × recall)/(precision + recall). The highest possible value of an F-score is 1.0, indicating perfect precision and recall, and the lowest possible value is 0.
References
- 1.Florence AL. Is thalidomide to blame? Br Med J. 1960;2:1954. doi: 10.1136/bmj.2.5217.1954. [DOI] [Google Scholar]
- 2.Mcbride WG. Thalidomide and congenital abnormalities. Lancet. 1961;278:1358. doi: 10.1016/S0140-6736(61)90927-8. [DOI] [Google Scholar]
- 3.Fornasier G, Francescon S, Leone R, Baldo P. An historical overview over pharmacovigilance. Int J Clin Pharm. 2018;40:744–747. doi: 10.1007/s11096-018-0657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crombie I. Inherent limitations of the Yellow Card system for the detection of unsuspected adverse drug reactions. Hum Toxicol. 1984;3:261–269. doi: 10.1177/096032718400300402. [DOI] [PubMed] [Google Scholar]
- 5.Hazell L, Shakir SAW. Under-reporting of adverse drug reactions: a systematic review. Drug Saf. 2006;29:385–396. doi: 10.2165/00002018-200629050-00003. [DOI] [PubMed] [Google Scholar]
- 6.Tattersall RL. The expert patient: a new approach to chronic disease management for the twenty-first century. Clin Med (Lond) 2002;2:227–229. doi: 10.7861/clinmedicine.2-3-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berrewaerts J, Delbecque L, Orban P, Desseilles M. Patient participation and the use of Ehealth tools for pharmacoviligance. Front Pharmacol. 2016;7:90. doi: 10.3389/fphar.2016.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ANSM. Déclarer un effet indésirable. 2021. https://ansm.sante.fr/documents/reference/declarer-un-effet-indesirable. Accessed 15 Jun 2021.
- 9.McLernon DJ, Bond CM, Hannaford PC, Watson MC, Lee AJ, Hazell L, et al. Adverse drug reaction reporting in the UK: a retrospective observational comparison of yellow card reports submitted by patients and healthcare professionals. Drug Saf. 2010;33:775–788. doi: 10.2165/11536510-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Inch J, Watson MC, Anakwe-Umeh S. Patient versus healthcare professional spontaneous adverse drug reaction reporting: a systematic review. Drug Saf. 2012;35:807–818. doi: 10.1007/BF03261977. [DOI] [PubMed] [Google Scholar]
- 11.Faasse K, Cundy T, Petrie KJ. Thyroxine: anatomy of a health scare. BMJ. 2009;339:b5613. doi: 10.1136/bmj.b5613. [DOI] [PubMed] [Google Scholar]
- 12.Mouly S, Roustit M, Bagheri H, Perault-Pochat M-C, Molimard M, Bordet R. The French Levothyrox® crisis: we did the best we could but …. Therapie. 2019;74:431–435. doi: 10.1016/j.therap.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Basile AO, Yahi A, Tatonetti NP. Artificial intelligence for drug toxicity and safety. Trends Pharmacol Sci. 2019;40:624–635. doi: 10.1016/j.tips.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilipiec P, Liwicki M, Bota A. Using Machine Learning for Pharmacovigilance: a Systematic Review. Pharmaceutics. 2022;14(2):266. doi: 10.3390/pharmaceutics14020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmider J, Kumar K, LaForest C, Swankoski B, Naim K, Caubel PM. Innovation in pharmacovigilance: use of artificial intelligence in adverse event case processing. Clin Pharmacol Ther. 2019;105:954–961. doi: 10.1002/cpt.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negi K, Pavuri A, Patel L, Jain C. A novel method for drug-adverse event extraction using machine learning. Inform Med Unlocked. 2019;17:100190. doi: 10.1016/j.imu.2019.100190. [DOI] [Google Scholar]
- 17.Giorgi J, Wang X, Sahar N, Shin WY, Bader GD, Wang B. End-to-end named entity recognition and relation extraction using pre-trained language models. arXiv preprint. 2019;1912.13415. 10.48550/arXiv.1912.13415.
- 18.Létinier L, Jouganous J, Benkebil M, et al. Artificial intelligence for unstructured healthcare data: application to coding of patient reporting of adverse drug reactions. Clin Pharmacol Ther. 2021;110(2):392–400. doi: 10.1002/cpt.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA) Drug Saf. 1999;20:109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 20.Bousquet C, Lagier G, Louët AL-L, Le-Beller C, Venot A, Jaulent M-C. Appraisal of the MedDRA conceptual structure for describing and grouping adverse drug reactions. Drug Saf. 2005;28:19–34. doi: 10.2165/00002018-200528010-00002. [DOI] [PubMed] [Google Scholar]
- 21.Edwards IR, Aronson JK. Adverse drug reactions: definitions, diagnosis, and management. Lancet. 2000;356:1255–1259. doi: 10.1016/S0140-6736(00)02799-9. [DOI] [PubMed] [Google Scholar]
- 22.Camelot. PDF table extraction for humans: Camelot 0.10.1 documentation. 2021. https://camelot-py.readthedocs.io/en/master/. Accessed 24 Nov 2021.
- 23.Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, et al. LightGBM: a highly efficient gradient boosting decision tree. Proc Int Conf Neural Inf Process Syst. 2017;2017:3149–3157. [Google Scholar]
- 24.Conneau A, Khandelwal K, Goyal N, Chaudhary V, Wenzek G, Guzmán F, et al. Unsupervised cross-lingual representation learning at scale. arXiv preprint. 2019;1911.02116. 10.48550/arXiv.1911.02116.
- 25.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. Proc Conf North Am Chapter Assoc Comput Linguist Hum Lang Technol. 2019;1:4171–4186. [Google Scholar]
- 26.Wolf T, Debut L, Sanh V, et al. HuggingFace’s transformers: state-of-the-art natural language processing. arXiv preprint. 2019;1910.03771. 10.48550/arXiv.1910.03771.
- 27.Ramos J. Using TF-IDF to determine word relevance in document queries. Proceedings of the first instructional conference on machine learning, vol. 242, Issue 1. 2003. p. 29–48.
- 28.Bojanowski P, Grave E, Joulin A, Mikolov T. Enriching word vectors with subword information. Transactions of the association for computational linguistics, vol. 5; 2017. p. 135–146.
- 29.Martin L, Muller B, Suárez PJO, Dupont Y, Romary L, de la Clergerie ÉV, et al. CamemBERT: a tasty French language model. Proceedings of the 45th Annual Meeting of the Association of Computational Linguistics. 2020; p. 7203–19.
- 30.Jung Y, Hu J. A K-fold averaging cross-validation procedure. J Nonparametric Stat. 2015;27:167–179. doi: 10.1080/10485252.2015.1010532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyndman R, Fan Y. Sample quantiles in statistical packages. Am Stat. 1996;50:361–365. [Google Scholar]
- 32.Powers DMW. What the F-measure doesn’t measure: features, flaws, fallacies and fixes. arXiv preprint. 2015;1503.06410. 10.13140/RG.2.1.1571.5369
- 33.ANSM. Dossier thématique: COVID-19: dispositif de surveillance renf. 2021. https://ansm.sante.fr/dossiers-thematiques/covid-19-vaccins/covid-19-dispositif-de-surveillance-renforcee-des-vaccins. Accessed 24 Jun 2021.
- 34.Editorial. Can technology increase COVID-19 vaccination rates? Lancet Digit Health. 2021;3:e274. [DOI] [PMC free article] [PubMed]
- 35.MedDRA and pharmacovigilance a complex and little-evaluated tool. Prescrire Int. 2016;25:247–250. [PubMed] [Google Scholar]
- 36.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Cohen JF, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.