Abstract
Because there is some controversy concerning the ligninolytic enzymes produced by Pleurotus species, ethylene release from α-keto-γ-thiomethylbutyric acid (KTBA), as described previously for Phanerochaete chrysosporium lignin peroxidase (LiP), was used to assess the oxidative power of Pleurotus eryngii cultures and extracellular proteins. Lignin model dimers were used to confirm the ligninolytic capabilities of enzymes isolated from liquid and solid-state fermentation (SSF) cultures. Three proteins that oxidized KTBA in the presence of veratryl alcohol and H2O2 were identified (two proteins were found in liquid cultures, and one protein was found in SSF cultures). These proteins are versatile peroxidases that act on Mn2+, as well as on simple phenols and veratryl alcohol. The two peroxidases obtained from the liquid culture were able to degrade a nonphenolic β-O-4 dimer, yielding veratraldehyde, as well as a phenolic dimer which is not efficiently oxidized by P. chrysosporium peroxidases. The former reaction is characteristic of LiP. The third KTBA-oxidizing peroxidase oxidized only the phenolic dimer (in the presence of Mn2+). Finally, a fourth Mn2+-oxidizing peroxidase was identified in the SSF cultures on the basis of its ability to oxidize KTBA in the presence of Mn2+. This enzyme is related to the Mn-dependent peroxidase of P. chrysosporium because it did not exhibit activity with veratryl alcohol and Mn-independent activity with dimers. These results show that P. eryngii produces three types of peroxidases that have the ability to oxidize lignin but lacks a typical LiP. Similar enzymes (in terms of N-terminal sequence and catalytic properties) are produced by other Pleurotus species. Some structural aspects of P. eryngii peroxidases related to the catalytic properties are discussed.
Lignin is a three-dimensional biopolymer composed of oxygenated phenylpropanoid units. The most common linkage between the units is an arylglycerol-β-aryl ether or β-O-4 bond (1). Degradation of recalcitrant lignin is an oxidative and nonspecific process carried out by white rot basidiomycetes, including the best-known ligninolytic organism, Phanerochaete chrysosporium (24). Some white rot fungi, such as Pleurotus eryngii and related species, have the capacity to remove lignin preferentially (i.e., with limited attack on cellulose) (30). This fact is relevant for environmentally friendly biotechnological delignification in paper pulp manufacture (7) and feed production (19).
The most prominent enzymes associated with lignin degradation are lignin peroxidase (LiP) and manganese-dependent peroxidase (MnP), as well as laccases and H2O2-producing oxidases (24). LiP appears to be the key enzyme in the oxidation of nonphenolic phenylpropanoid units which leads to polymer fragmentation. This enzyme oxidizes aromatic nuclei to aryl cation radicals which undergo several nonenzymatic reactions, including cleavage of C-C and C-O linkages (14, 24). MnP oxidizes Mn2+ to Mn3+, which in turn may attack phenolic structures in lignin (10) as long as it is stabilized by suitable metal chelators secreted by fungi (25, 27); and also may attack nonphenolic structures via lipid peroxidation (6).
Laccase and aryl-alcohol oxidase (AAO) have been purified from cultures of P. eryngii in glucose-ammonium medium and have been characterized (13, 34), and it has been suggested that these enzymes are involved in lignin degradation via oxygen activation and redox mediators (12, 31, 34). However, significant levels of LiP and MnP have not been detected in cultures of P. eryngii in glucose-ammonium medium. Recently, a high level of Mn2+-oxidizing peroxidase activity was observed in this fungus when peptone was used as an N source (32), and the possibility that LiP was present was suggested by the results of a Western blot hybridization analysis performed with antibodies against P. chrysosporium LiP (37). Nevertheless, LiP activity has not been detected in P. eryngii, and although a LiP type of peroxidase has been found in Pleurotus ostreatus (3), a Southern blot hybridization analysis performed with a DNA probe from the P. chrysosporium gene lpo demonstrated that P. eryngii does not have a similar gene (18).
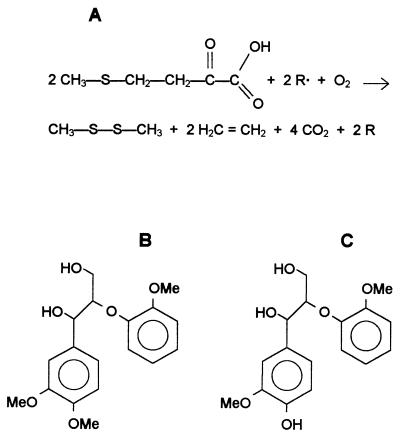
Veratryl alcohol oxidation, the most common test for LiP detection (42), can be inhibited by peptone constituents (40) or by aromatic fungal metabolites or lignin-derived products (43). Therefore, it would be useful to evaluate the production of ligninolytic enzymes in P. eryngii by using a different detection method. An assay which has been used to assess the potential for oxidative attack of the lignin macromolecule is the assay which measures ethylene release from α-keto-γ-thiomethylbutyric acid (KTBA) (21, 28). This assay is used to detect one-electron strong oxidants, such as LiP and the hydroxyl free radical (OH·) (Fig. 1A). A correlation between KTBA oxidation and ligninolytic activity has been observed in P. chrysosporium cultures (21). On the other hand, lignin model dimers with the most frequently found lignin intermonomer linkages (Fig. 1B and C) may be used to identify the specific degradation reactions of phenolic and nonphenolic phenylpropanoid units (the latter account for 80 to 90% of the total polymer) (9) produced by either ligninolytic organisms or enzymes (24).
FIG. 1.
Oxidation of KTBA to ethylene (4) (A) and transformation of nonphenolic (B) and phenolic (C) β-O-4 dimers were used to investigate the ligninolytic enzymes produced by P. eryngii. R· represents an aromatic radical from LiP or a hydroxyl radical from Fenton type reactions; generation of both of these chemical species requires H2O2, and generation of the hydroxyl radical also requires an Fe3+-reducing system. Me, methyl.
Both KTBA and lignin model dimers were used to study the mechanism of lignin degradation by P. eryngii. This study led to purification of KTBA-oxidizing enzymes from supernatants of liquid cultures and from wheat straw treated with the fungus under solid-state fermentation (SSF) conditions similar to the conditions found in the natural habitats of ligninolytic basidiomycetes. The ligninolytic potentials of these enzymes were evaluated by using different substrates, including phenolic and nonphenolic β-O-4 dimers.
MATERIALS AND METHODS
Organisms and cultures.
The fungal strains used were P. eryngii ATCC 90787 (= IJFM A169 = CBS 613.91) and P. chrysosporium ATCC 24725 (= VKM F-1767 = IJFM A547). Shaken cultures of P. eryngii in glucose-peptone medium were inoculated and grown as described elsewhere (32), and 5-ml samples were removed periodically for analysis. As indicated below, 0.1 mM MnSO4 was sometimes added to the medium. P. chrysosporium shallow cultures were grown as described by Tien and Kirk (42) by using Falcon 25-cm2 tissue culture flasks that contained 5 ml of medium inoculated with a spore suspension and were flushed with O2 through rubber caps.
Wheat (Triticum aestivum) straw from SAICA (Zaragoza, Spain) was sliced into 10- to 20-mm lengths and was sterilized at 120°C for 15 min. SSF was carried out in a horizontal rotary fermentor with six 2-liter bottles containing 125 g of straw and 375 ml of water (including inoculum) (7). No minerals or additional N sources were added. The inocula were obtained from 15-day-old stationary cultures grown in glucose-ammonium medium (13), which were homogenized, grown for additional 3 to 5 days at 180 rpm. After inoculation with the pellets obtained, the bottles were gently rotated for 24 h at 1 rpm and then incubated for 20 days at 28°C with rotation (1 rpm) for 1 h per day and a wet air flux of 166 ml/min per bottle.
Enzyme assays.
LiP activity was measured by monitoring the oxidation of 2 mM veratryl alcohol to veratraldehyde (ɛ310, 9.3 mM−1 cm−1) in 0.1 M sodium tartrate (pH 3) supplemented with 0.4 mM H2O2. Controls without H2O2 were included. AAO activity was measured in reaction mixtures containing 5 mM veratryl alcohol and 0.1 M phosphate buffer (pH 6). MnP activity was measured by monitoring the formation of the Mn3+-tartrate complex (ɛ238, 6.5 mM−1 cm−1) from 0.1 mM MnSO4 in 0.1 M sodium tartrate (pH 5) supplemented with 0.1 mM H2O2. Mn-mediated oxidation of 0.1 mM 2,6-dimethoxyphenol (DMP) to the dimeric product coerulignone (ɛ469, 27.5 mM−1 cm−1, referred to as the DMP concentration) (32) was estimated under the same conditions. Mn-independent peroxidase and laccase activities were measured by using 0.1 mM DMP in 0.1 M sodium tartrate (pH 5). In the case of Mn-independent peroxidase 0.1 mM H2O2 was added to the reaction mixture. One activity unit was defined as the amount of enzyme that oxidized 1 μmol of substrate per min. When purified enzymes were compared, the activities mentioned above (as well as the KTBA-oxidizing activity described below) were expressed as specific activities in units per milligram of protein, as estimated with the Bradford reagent obtained from Bio-Rad.
In vivo and in vitro ethylene release from KTBA.
Assays to determine oxidation of KTBA (final concentration, 3.3 mM) (Fig. 1A) were performed in triplicate by using flasks closed with rubber caps, and the reactions were stopped by adding 0.5 to 1 ml of 3 M H2SO4. In vivo assays were performed with 5-ml samples from P. eryngii liquid cultures or with 1-g straw samples from SSF (in 5 ml of reaction mixture) that were placed in sterile 25-cm2 tissue culture flasks and incubated for 5 to 8 h. In the case of P. chrysosporium, whole 5-ml cultures were used. As indicated below, 0.1 mM MnSO4 or 0.5 mM H2O2 was sometimes added. The in vitro assays to determine KTBA oxidation were performed in 12-ml flasks containing 1-ml reaction mixtures at room temperature for 30 min. For enzyme purification (see below), the KTBA assay was performed with 0.1-ml aliquots obtained from chromatographic fractions. The reaction mixtures contained 1 mM veratryl alcohol, 0.4 mM H2O2, and 0.1 M sodium tartrate (pH 3). The assay to determine KTBA oxidation by peroxidases purified from P. eryngii cultures was also performed with 0.1 M sodium tartrate (pH 5) supplemented with 0.1 mM MnSO4. The effects of H2O2 concentrations and pH were investigated. The possibility that KTBA oxidation could be increased by keeping the H2O2 concentration low was tested (the total H2O2 was added in six aliquots which were injected every 5 min). Oxidation of KTBA by LiP and MnP from P. chrysosporium was also analyzed. LiP was produced and purified as described previously (42), and MnP was a gift from B. Kurek. The LiP and MnP reactions were performed as described above for P. eryngii peroxidases in the presence of veratryl alcohol and Mn2+, respectively. KTBA oxidation by P. eryngii laccase (34) was examined by using up to 0.5 U of laccase per ml in 0.1 M sodium tartrate (pH 5) and adding 0.1 mM DMP or 10 mM 2,2-azinobis(3-ethylbenzothiazolin-6-sulfonate).
The ethylene in 0.5-ml samples obtained from the headspaces of in vivo and in vitro assay was analyzed by gas chromatography in the isothermic mode (95°C) by using a Porapack-N column and a flame ionization detector; the injector and detector temperatures were both 50°C, and the carrier gas was helium at a flow rate of 50 ml/min. Chromatographic measurements were made in triplicate, and the mean values are presented below. The KTBA-oxidizing activity of a culture was expressed as the amount of ethylene released per hour of reaction (in nanomoles per milliliter per hour). In the in vitro reactions, the activity corresponded to the total amount of ethylene released at the end of the KTBA oxidation reaction by chromatographic fractions (expressed in micromoles per milliliter) or purified enzymes (expressed in micromoles per milligram).
Purification of KTBA-oxidizing enzymes from P. eryngii.
The supernatants from liquid cultures were concentrated 15-fold and dialyzed (5-kDa cutoff membrane) against 10 mM sodium tartrate (pH 4.5). The contents of the SSF bottles were extracted with 3 liters of water by shaking the bottles at 200 rpm for 1 h, and the extracts were filtered, concentrated 20-fold, and dialyzed. The concentrates were loaded onto a Bio-Rad Q-cartridge (1 ml/min), and the retained fractions were eluted with 1 M NaCl. The fractions capable of oxidizing KTBA in the presence of veratryl alcohol or Mn2+ were applied to a Sephacryl S-200 HR (Pharmacia K16/100) column. The KTBA-oxidizing fractions were chromatographed with a Mono-Q column (Pharmacia HR 5/5) by using 10 mM sodium tartrate (pH 5) and a 0 to 0.25 M NaCl gradient (0.8 ml/min, 30 min).
The homogeneity of proteins was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% polyacrylamide gels and isoelectric focusing in 5% polyacrylamide gels by using a pH range of 2.5 to 5.5; protein bands were stained with AgNO3 or Coomassie blue R-250. In addition to SDS-PAGE including Bio-Rad standards, the Mr of proteins were estimated by matrix-assisted laser desorption and ionization-time of flight (MALDI-TOF) mass spectrometry by using a sinapic acid matrix. The N-terminal sequences were determined by automated Edman degradation of 5 μg of protein with an Applied Biosystems model 494 pulsed-liquid protein sequencer.
Transformation of lignin model dimers by the enzymes.
The two dimers used, veratrylglycerol-β-guaiacyl ether (nonphenolic β-O-4 dimer) and guaiacylglycerol-β-guaiacyl ether (phenolic β-O-4 dimer) (Fig. 1B and C) were synthesized by the methods described by Adler et al. (2) and Nakatsubo (35). The reaction mixtures used for degradation of the nonphenolic dimer contained peroxidase (0.5 U/ml, measured on the basis of the veratraldehyde formed), the dimer at a concentration of 0.25 mM, 0.1 M sodium tartrate (pH 3.5), and 0.1 mM H2O2 (added every 5 min). The effect of adding 0.25 mM veratryl alcohol or 0.1 mM MnSO4 was also determined. Phenolic dimer degradation was assayed like nonphenolic dimer degradation except that the pH was 4.5 (with or without 0.1 mM Mn2+) and the peroxidase concentration was 0.25 to 1 U/ml (measured on the basis of the Mn3+-tartrate formation). The reactions were monitored by high-performance liquid chromatography by using standard calibration curves for both the substrates (the phenolic and nonphenolic dimers) and the reaction product (veratraldehyde). Samples (20 μl) were injected into a Pharmacia system equipped with a Spherisorb S50DS2 column. The analyses were carried out at 30°C with a flow rate of 1 ml/min by using methanol-water as the eluent (starting with 20% methanol and ending with 40% methanol after 30 min). The UV detector was operated at 210 and 250 nm or at 280 and 310 nm. Fourier transform infrared spectra were obtained with a model IFS 28 Equinox spectrometer (Brucker) by using a DTGS-type detector and 100 scan accumulation.
RESULTS
Action of fungal cultures on KTBA.
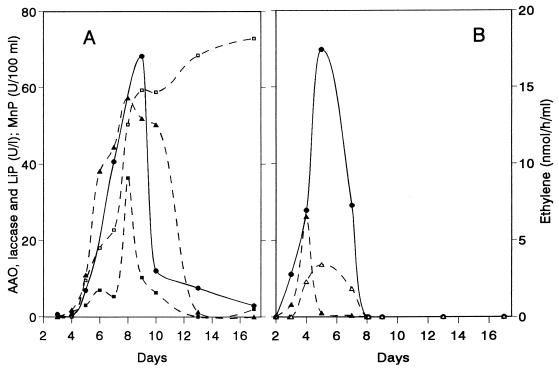
The ability to oxidize KTBA was investigated during growth of P. eryngii under both liquid and SSF culture conditions. The profiles obtained with liquid cultures were compared with the profiles obtained with P. chrysosporium (Fig. 2). Similar amounts of ethylene were released from KTBA by the two fungi despite the differences in the enzymatic activities produced. LiP was detected only in P. chrysosporium, and laccase and AAO were found only in P. eryngii. In both fungi the highest levels of activity were the levels of peroxidase that oxidize Mn2+ to Mn3+. The highest level of LiP activity appeared after the highest levels of MnP activity were observed, and the peak matched the KTBA oxidation activity in the P. chrysosporium culture (day 5). In P. eryngii, the broad peroxidase activity peak, the laccase activity peak, and the onset of AAO activity coincided roughly with the time when the maximum amount of ethylene was released from KTBA (day 9). The same enzymes were detected in SSF cultures of P. eryngii, and it was not possible to correlate ethylene release with an individual enzymatic activity.
FIG. 2.
Patterns of enzyme production and KTBA oxidation by P. eryngii (A) and P. chrysosporium (B) cultures. Symbols: ●, ethylene released from KTBA; ▵, LiP activity; ▴, MnP activity; ■, laccase activity; □, AAO activity.
When Mn2+ was included in the medium, KTBA oxidation was partially inhibited. However, addition of Mn2+ to culture samples before incubation with KTBA resulted in increases in the amounts of ethylene. Addition of H2O2 to the reaction mixture resulted in ethylene release similar to that of the control, which showed that H2O2 was not a limiting factor in KTBA oxidation as it could be generated by AAO during incubation.
Purification of KTBA-oxidizing enzymes.
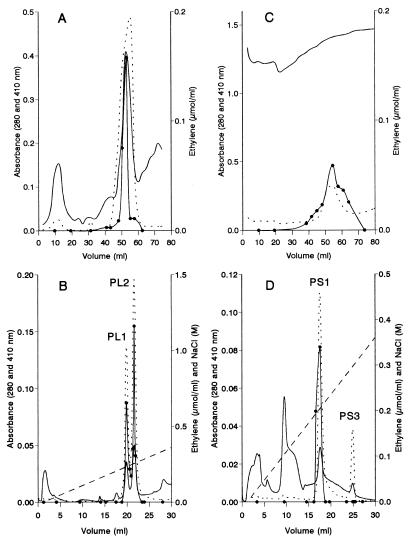
The enzymes responsible for KTBA oxidation were purified by using 8-day-old liquid cultures and 21-day-old SSF cultures of P. eryngii; these cultures exhibited maximal activity with KTBA. The ethylene release tests during protein purification were performed in the presence of H2O2 and veratryl alcohol (as described above for P. chrysosporium LiP detection), as well as in the absence of these compounds. Supernatants from liquid cultures and extracts from straw treated with the fungus were concentrated and dialyzed, and the colored compounds, which were especially abundant in the straw extracts, were removed by using a Q-cartridge. After molecular size exclusion gel permeation of samples from liquid cultures, a protein peak with strong KTBA-oxidizing peroxidase activity, as well as a high level of absorbance at 410 nm, was obtained (Fig. 3A). Mono-Q chromatography of this peak revealed two proteins (peroxidases PL1 and PL2) that were capable of oxidizing KTBA in the presence of H2O2 and veratryl alcohol (Fig. 3B). Despite the fact that all of the fractions obtained from Sephacryl chromatography of straw extracts had high levels of absorbance at 280 nm, a wide KTBA oxidation peak was identified, which corresponded to the peak in the 410-nm profile (Fig. 3C). Two 410-nm peaks were obtained by Mono-Q chromatography; however, only one of these peaks (peroxidase PS1) was able to oxidize KTBA, while the second peak (designated peroxidase PS3 because the peroxidase PS1 tail was designated peroxidase PS2) was not capable of oxidizing KTBA under the conditions described above (Fig. 3D). However, when KTBA oxidation was assayed in the presence of Mn2+ and H2O2, some ethylene was released with the four peroxidases obtained. These peroxidases, which were purified from liquid and SSF cultures, were different, as shown by their N-terminal sequences (Table 1) and the catalytic properties discussed below. Their molecular masses (42 to 45 kDa as determined by SDS-PAGE and 36 to 39 kDa as determined by MALDI-TOF) and isoelectric points (pI 3.65 to 3.80) differed only slightly.
FIG. 3.
Purification of KTBA-oxidizing peroxidases from liquid (A and B) and SSF (C and D) cultures of P. eryngii: isolation of peroxidases PL1, PL2, PS1, and PS3 by Mono-Q chromatography (B and D) of the KTBA-oxidizing fractions (elution volumes, 50 to 55 ml in panel A and 50 to 60 ml in panel B) from Sephacryl S-200 gel permeation (A and C) of concentrated extracellular proteins. —, 280-nm profile; …, 410-nm profile; ●, amount of ethylene released from KTBA in the presence of 1 mM veratryl alcohol; ––––, NaCl gradient.
TABLE 1.
N-terminal sequences of Mn2+-oxidizing peroxidases (some of which also oxidize veratryl alcohol) produced by Pleurotus species in liquid cultures or on lignocellulosic substrates
| Organism | Culture | Peroxidase | Oxidation of:
|
N-terminal sequence | GenBank accession no. | Reference | |
|---|---|---|---|---|---|---|---|
| Veratryl alcohol | Mn2+ | ||||||
| P. eryngii | Wheat straw | PS1 | + | + | VTCATGQTT | This study | |
| PS2 | + | + | XTCATGQTT | This study | |||
| PS3 | − | + | VTCADGNTV | This study | |||
| Liquid | PL1a | + | + | ATCADGRTT | AF007223 | 32 | |
| PL2a | + | + | ATCDDGRTT | AF007224 | 32 | ||
| P. ostreatus | Sawdust | MnP1 | + | + | ATCADGRTT | 38 | |
| MnP2 | + | + | VTCATGQTT | 38 | |||
| Liquid | MnP | − | + | ATCAGGQVT | U21878 | 5 | |
| MnPL | + | + | ATCADGRTT | 39 | |||
| P. pulmonarius | Wheat straw | MnPS | + | + | VTCATGQTT | 8 | |
| Liquid | MnPL | + | + | ATCADGRTT | 8 | ||
Peroxidases PL1 and PL2 are two allelic variants previously described by Martínez et al. (32) as isoenzymes MP1 and MP2.
KTBA oxidation by P. eryngii enzymes.
Information on the mechanism of oxidation of KTBA by P. eryngii peroxidases was obtained by monitoring ethylene release by gas chromatography and veratryl alcohol oxidation by UV spectrophotometry (data not shown). In the absence of KTBA, veratryl alcohol oxidation to veratraldehyde by peroxidases PL1 and PL2 (and to some extent by peroxidase PS1) was revealed by increased absorbance at 310 nm. However, when KTBA was added to the reaction mixture, ethylene was obtained, but there was no increase in absorbance at 310 nm. This observation was attributed to the presence of an indirect oxidation mechanism. The mediator formed by the action of the enzyme, probably the veratryl alcohol cation radical, oxidized KTBA, and there was concomitant production of ethylene. In the absence of enzyme, mediators (veratryl alcohol or Mn2+), or cofactors (hydroperoxides), no KTBA oxidation was observed. Laccase from P. eryngii was not capable of releasing ethylene from KTBA, even in the presence of 2,2-azinobis(3-ethylbenzothiazolin-6-sulfonate) or DMP as a potential mediator.
The results of a comparison of KTBA oxidation by the four P. eryngii peroxidases in the presence of veratryl alcohol and Mn2+ are shown in Table 2. When phenols were used as mediators, no ethylene was produced. The activities obtained with Mn2+, DMP, and veratryl alcohol (as estimated directly with the fractions collected from Mono-Q chromatography) showed that peroxidase PS3 differed from the other peroxidases because of its higher specific activity with Mn2+ and its lack of activity with veratryl alcohol (Table 2). This explains the lack of KTBA oxidation in the presence of this compound and the release of high levels of ethylene in the presence of Mn2+.
TABLE 2.
Comparison of the four KTBA-oxidizing peroxidases isolated from liquid cultures (peroxidases PL1 and PL2) and SSF cultures (peroxidases PS1 and PS3) of P. eryngii: enzymatic activities, KTBA oxidation, and degradation of β-O-4 dimers (effects of Mn2+ and veratryl alcohol)
| Peroxidase | Enzymatic activities (U/mg) ona:
|
Ethylene release (μmol/mg)b
|
% Dimer transformationc
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phenolic dimer
|
Nonphenolic dimer
|
|||||||||
| Mn2+ | DMP | Veratryl alcohol | Direct | Veratryl alcohol | Mn2+ | Direct | Mn2+ | Direct | Veratryl alcohol | |
| PL2 | 125 | 44 | 10 | 0 | 54 | 22 | 65 | 100 | 19 | 19 |
| PL1 | 109 | 57 | 14 | 0 | 52 | 26 | +d | + | + | + |
| PS1 | 174 | 38 | 3 | 0 | 30 | 29 | 0 | 100 | 0 | 0 |
| PS3 | 356 | 65 | 0 | 0 | 0 | 88 | 0 | 100 | 0 | 0 |
Oxidation of 0.1 mM Mn2+ was determined at pH 5, and oxidation of 10 mM DMP or 10 mM veratryl alcohol was determined at pH 3.
Ethylene concentration resulting from KTBA oxidation in the presence of 1 mM veratryl alcohol or 3 mM Mn2+.
Level of transformation of 0.25 mM phenolic dimer (pH 4.5; 0.5 U/liter; estimated on the basis of Mn2+ oxidation) and nonphenolic dimer (pH 3.5; 0.5 U/liter estimated on the basis of veratryl alcohol oxidation) after 70 min.
+, Peroxidase PL1, an allelic variant of peroxidase PL2, has degradation abilities with model dimers similar to those of peroxidase PL2 (the transformation rates were not calculated).
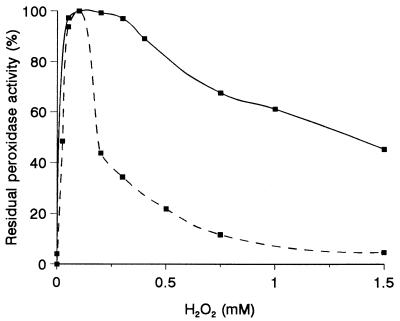
As with P. eryngii peroxidases, KTBA oxidation in the presence of Mn2+ was also observed with MnP from P. chrysosporium. However, this peroxidase was inhibited by H2O2 concentrations greater than 0.2 mM, which slightly affected P. eryngii peroxidases (Fig. 4) (P. chrysosporium LiP was more susceptible to H2O2 than the peroxidases discussed above). Moreover, when the concentration of H2O2 was greater than 0.5 mM, more ethylene was released by the P. eryngii peroxidases if the total dose was added in five aliquots during incubation.
FIG. 4.
Inhibition of P. eryngii peroxidase PL2 activity (—) by different amounts of H2O2, compared with P. chrysosporium MnP activity (––––).
Finally, we compared the amount of ethylene released from KTBA by P. eryngii PL2 with the amount of ethylene released by P. chrysosporium LiP in the presence of veratryl alcohol and the amount of ethylene released by the P. eryngii peroxidase with the amount of ethylene released by P. chrysosporium MnP in the presence of Mn2+. Under the reaction conditions used (we estimated the same activities by using veratryl alcohol or Mn2+ oxidation), the P. eryngii enzyme released more ethylene per activity unit (2,280 nmol/U in the presence of veratryl alcohol and 250 nmol/U in the presence of Mn2+) than P. chrysosporium LiP (1,730 nmol/U) and MnP (70 nmol/U) released. However, LiP produced more ethylene when the same amount of protein was used because of its higher specific activity.
Transformation of lignin model dimers by the P. eryngii peroxidases.
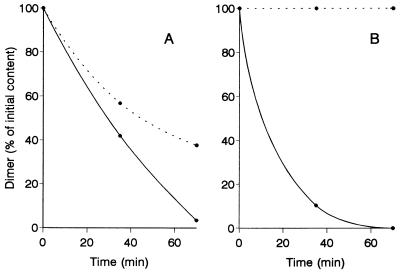
We investigated degradation of both phenolic and nonphenolic β-O-4 dimers by peroxidases from liquid and SSF cultures of P. eryngii. The phenolic dimer was degraded by peroxidases PL1 and PL2 in the absence of Mn2+ (Table 2). However, the reaction was enhanced by Mn2+ (Fig. 5A), and the highest degradation rate was observed at pH values around 4.5. On the other hand, peroxidases PS1 and PS3 from SSF cultures required Mn2+ in order to attack the phenolic dimer (Fig. 5B). Despite the fact that both of these enzymes exhibited Mn-independent activity with DMP, they differed from peroxidases obtained from liquid cultures by exhibiting lower levels of activity with guaiacol (the specific activities with 5 mM guaiacol were only 10 to 20% of the specific activities of the liquid culture peroxidases). During the peroxidase reaction with phenolic dimers, a pale brown precipitate was formed, which was characterized by Fourier transform infrared signals at 1,727, 1,628, 1,502, 1,464, 1,259, 1,215, and 1,125 cm−1 and probably resulted from condensation of the aromatic radicals formed.
FIG. 5.
Differences between P. eryngii peroxidases PL2 (A) and PS3 (B), as shown by oxidation of a phenolic β-O-4 dimer in the presence (—) and in the absence (…) of 0.1 mM Mn2+ (0.5 U of peroxidase per liter, estimated as Mn3+-tartrate, and 0.1 M tartrate [pH 4.5] were used).
As shown in Table 2, nearly 20% of the nonphenolic dimer was degraded with peroxidase PL2 from liquid cultures under the reaction conditions used. The dimer was stoichiometrically transformed into veratraldehyde, which was the only product detected by high-performance liquid chromatography. The highest reactions rates were observed at pH 3 to 3.5. Similar results were obtained with peroxidase PL1, but peroxidases PS1 and PS3 from SSF cultures (which exhibited a low level of activity and no activity with veratryl alcohol, respectively) were not able to catalyze this reaction. Transformation of the nonphenolic dimer was not affected by veratryl alcohol or Mn2+. Very similar degradation rates were obtained with P. chrysosporium LiP when the same enzyme activity and reaction pH were used.
Two different KTBA-oxidizing peroxidases have recently been purified from liquid (peptone medium) and SSF cultures of Pleurotus pulmonarius CBS 507.85 (data not shown). The peroxidase obtained from a liquid culture degraded lignin model dimers in the absence of Mn2+.
DISCUSSION
Detection of the ligninolytic activity of P. chrysosporium peroxidases was related to the use of KTBA and lignin model dimers as substrates. When Kuwahara et al. (28) purified the two ligninolytic peroxidases secreted by this fungus, LiP was detected on the basis of its ability to release ethylene from KTBA in the presence of veratryl alcohol. Its ability to degrade a nonphenolic β-O-4 dimer was considered a reflection of its ligninolytic capabilities (41). In the present study we found that P. eryngii, a fungus that was investigated because of its ability to degrade wheat lignin selectively (30), oxidizes KTBA to ethylene when it is grown in peptone-containing media, as well as during treatment of straw under the SSF conditions used for biomechanical and biosemichemical pulping (7, 11, 30). LiP activity was not detected in the cultures mentioned above, and the oxidation of veratryl alcohol observed was due to Pleurotus AAO. However, the KTBA oxidation observed was similar to that found in P. chrysosporium cultures grown under ligninolytic conditions. KTBA oxidation in the absence of a typical LiP has also been observed in some other fungi (22, 29).
The search for ligninolytic enzymes in P. eryngii resulted in identification of four proteins that have the ability to release ethylene from KTBA (Table 2). Three of these proteins (one protein from SSF cultures and two proteins from liquid cultures) were detected under the conditions used to identify LiP in P. chrysosporium (20) (i.e., they degraded KTBA in the presence of H2O2 and veratryl alcohol, which in the absence of KTBA was oxidized to veratraldehyde). These peroxidases also oxidized Mn2+, as well as different phenols in the absence of Mn2+. Phenolic compounds inactivate LiP (15) and can interfere with veratryl alcohol oxidation by P. eryngii peroxidases because of competitive inhibition. This explains why peroxidase activity with veratryl alcohol was not detected in peptone-containing medium or in SSF extracts, which contained different phenols (40, 43). The KTBA-oxidizing enzymes obtained from liquid cultures were the same peroxidases as those described previously by Martínez et al. (32) as two MnP isoenzymes that exhibit Mn-independent activity with some aromatic substrates. In the present study we observed some of the LiP characteristics of these enzymes, such as the ability to degrade nonphenolic β-O-4 dimers (yielding veratraldehyde) and the ability to release ethylene from KTBA in the presence of veratryl alcohol. It is interesting that the P. eryngii peroxidases oxidized both dimers in the absence of mediators, Mn2+, or veratryl alcohol, whereas KTBA oxidation was dependent on the presence of these compounds. The third enzyme that could oxidize KTBA in the presence of veratryl alcohol and H2O2 (peroxidase PS1) was found in the SSF cultures. This peroxidase exhibited high levels of activity with Mn2+ and DMP, lower levels of activity with veratryl alcohol and guaiacol, and no activity with the dimers used. It differs from P. chrysosporium MnP mainly by its Mn-independent activity with DMP and veratryl alcohol. The lack of activity with the two model dimers revealed that this enzyme, isolated for the first time in this study, is different from the peroxidases obtained from liquid cultures (as confirmed by N-terminal sequencing). The three peroxidases discussed above were identified on the basis of their abilities to oxidize KTBA in the presence of veratryl alcohol. However, when assessing KTBA oxidation by P. eryngii cultures, we found that adding Mn2+ to the reaction mixtures enhanced ethylene release. A similar result suggested that MnP may be involved in KTBA oxidation by Trametes versicolor (4). This finding led to identification and purification of a fourth peroxidase in the SSF cultures of P. eryngii (peroxidase PS3), which oxidized KTBA in the presence of Mn2+. The catalytic properties of this enzyme, which was isolated for the first time in this study, are similar to those of previously described Mn-dependent peroxidases (it does not oxidize veratryl alcohol or nonphenolic dimers, and oxidation of the phenolic dimer is strictly dependent on Mn2+).
In this study we found that P. eryngii produces three types of extracellular peroxidases. One of these peroxidases, which is found only in SSF cultures, is related to the MnP of P. chrysosporium. The second peroxidase is a polyvalent peroxidase that is isolated from liquid cultures and exhibits both LiP and MnP types of catalytic properties, including Mn2+ oxidation and direct oxidation of phenolic and nonphenolic dimers. It differs from P. chrysosporium MnP and LiP with respect to oxidation of phenols (P. chrysosporium MnP requires Mn2+ to oxidize phenols [10], and LiP requires veratryl alcohol [26]). The third type of peroxidase, which is isolated from SSF cultures, exhibits activity with Mn2+ and phenols, a low level of activity with veratryl alcohol, and no activity with dimers, suggesting that the redox potential is intermediate between the redox potentials of the other two P. eryngii peroxidases. Similar Mn-oxidizing peroxidases are produced by other Pleurotus species grown in liquid media or on lignocellulosic substrates under SSF conditions (Table 1). Based on their catalytic properties and N-terminal sequences (or whole protein sequences when available), the peroxidases could be classified into the following four groups: (i) group i, including P. eryngii peroxidase PS1 from wheat straw cultures, P. ostreatus peroxidase MnP2 from sawdust (38), and P. pulmonarius peroxidase from wheat straw (8) (N terminus, VTCATGQTT); (ii) group ii, including P. eryngii peroxidase from liquid cultures (32) (including two allelic variants), P. ostreatus peroxidases from sawdust (MnP1) (38) and liquid cultures (39), and P. pulmonarius peroxidase from liquid cultures (8) (N terminus, ATCDDGRTT); (iii) group iii, including P. eryngii peroxidase PS3 from wheat straw (N terminus, VTCADGNTV); and (iv) group iv, including the P. ostreatus peroxidase described by Asada et al. (5) from liquid cultures (N terminus, ATCAGGQVT). The peroxidases in groups iii and iv can be considered close relatives of P. chrysosporium MnP, whereas the peroxidases in groups i and ii correspond to two different versatile peroxidases which have been found in the three Pleurotus species. Similar peroxidases have been found recently in Bjerkandera adusta (16) and Bjerkandera sp. (33); the latter was described as a LiP-MnP “hybrid” peroxidase. These Pleurotus and Bjerkandera Mn2-oxidizing peroxidases can oxidize typical LiP substrates, such as veratryl alcohol and dimethoxybenzenes, and inhibition studies have shown that they have different binding sites for Mn2+ and veratryl alcohol or dyes (17, 33).
Molecular models for the new polyvalent peroxidases produced by P. eryngii (peroxidases PL1 and PS1) have been constructed recently (Brookhaven Protein Database entries 1A20 and 1BQW), and these models reveal greater structural (and sequence) homology with P. chrysosporium LiP than with MnP. This could explain the LiP type properties observed and the crossed-reactivity with the anti-LiP antibodies used by Orth et al. (37). Moreover, these P. eryngii peroxidases have a putative Mn interaction site (which includes inner heme propionate and peroxidase PL1 Glu-36, Glu-40, and Asp-175 residues or peroxidase PS1 Glu-36, Glu-40, and Asp-181 residues) that is involved in Mn2+ oxidation (17). It is necessary to mention that Mn2+ oxidation by both wild-type and recombinant LiP isoenzyme H2 has been reported (23, 36), but this isoenzyme does not have an Mn-binding site (Brookhaven Protein Database entry 1QPA) and the mechanism involved is not known. The versatile catalytic properties of some Pleurotus peroxidases can explain why these enzymes have been reported by different authors to be either LiP (3) or MnP (32, 39). Additional studies on the regulation and structure-function relationships of these enzymes are under way in order to better understand their role in lignin degradation by P. eryngii and other Pleurotus species which do not produce typical LiP.
ACKNOWLEDGMENTS
We are indebted to Bernard Kurek (INRA, Reims, France) for a sample of P. chrysosporium MnP. Benjamín Rodríguez (IQOG, CSIC, Madrid, Spain) is acknowledged for the facilities used for organic synthesis, Jaime Amézaga is thanked for preparation of lignin model dimers, Alicia Prieto is thanked for help with the gas chromatography and MALDI-TOF analyses, Javier Varela is thanked for protein sequencing, and Angeles Guijarro and Teresa Raposo are thanked for technical assistance.
L.C. was supported by the Junta Nacional de Investigaçao Científica e Tecnológica (Portugal) and by the FAIR Programme of the European Union. This work was supported in part by European Project AIR2-CT93-1219 (Biological Delignification in Paper Manufacture) and by the Spanish Biotechnology Programme.
ADDENDUM IN PROOF
A molecular characterization of peroxidase PL from liquid cultures of Pleurotus eryngii (Brookhaven Protein Database entry 1A20) has been published recently (F. J. Ruiz-Dueñas, M. J. Martínez, and A. T. Martínez, Mol. Microbiol. 31:223–236, 1999).
REFERENCES
- 1.Adler E. Lignin chemistry—past, present and future. Wood Sci Technol. 1977;11:169–218. [Google Scholar]
- 2.Adler E, Lindgren B O, Saeden U. The β-guaiacyl ether of α-veratryl-glycerol as a lignin model. Sven Papperstidn. 1952;7:245–254. [Google Scholar]
- 3.Akhmedova Z R. Ligninolytic enzymes of basidiomycetes: lignin peroxidases from the fungus Pleurotus ostreatus UzBI-ZAX 108. 2. Isolation, purification, and characterization of isozymes. Biochemistry (Engl Transl Biokhimiya) 1996;61:981–987. [Google Scholar]
- 4.Archibald F S. Lignin peroxidase activity is not important in biological bleaching and delignification of unbleached Kraft pulp by Trametes versicolor. Appl Environ Microbiol. 1992;58:3101–3109. doi: 10.1128/aem.58.9.3101-3109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asada Y, Watanabe A, Irie T, Nakayama T, Kuwahara M. Structures of genomic and complementary DNAs coding for Pleurotus ostreatus manganese(II) peroxidase. Biochim Biophys Acta. 1995;1251:205–209. doi: 10.1016/0167-4838(95)00102-z. [DOI] [PubMed] [Google Scholar]
- 6.Bao W L, Fukushima Y, Jensen K A, Moen M A, Hammel K E. Oxidative degradation of non-phenolic lignin during lipid peroxidation by fungal manganese peroxidase. FEBS Lett. 1994;354:297–300. doi: 10.1016/0014-5793(94)01146-x. [DOI] [PubMed] [Google Scholar]
- 7.Camarero S, Barrasa J M, Pelayo M, Martínez A T. Evaluation of Pleurotus species for wheat-straw biopulping. J Pulp Paper Sci. 1998;24:197–203. [Google Scholar]
- 8.Camarero S, Böckle B, Martínez M J, Martínez A T. Manganese-mediated lignin degradation by Pleurotus pulmonarius. Appl Environ Microbiol. 1996;62:1070–1072. doi: 10.1128/aem.62.3.1070-1072.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camarero S, Galletti G C, Martínez A T. Preferential degradation of phenolic lignin units by two white rot fungi. Appl Environ Microbiol. 1994;60:4509–4516. doi: 10.1128/aem.60.12.4509-4516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenn J K, Akileswaran L, Gold M H. Mn(II) oxidation is the principal function of the extracellular Mn-peroxidase from Phanerochaete chrysosporium. Arch Biochem Biophys. 1986;251:688–696. doi: 10.1016/0003-9861(86)90378-4. [DOI] [PubMed] [Google Scholar]
- 11.Guadalix M E, Almendros G, Martínez A T, Camarero S, Barrasa J M, Pelayo M. Comparative analysis of wheat straw paperboards prepared after biomechanical and semichemical pulping. Bioresour Technol. 1996;57:217–227. [Google Scholar]
- 12.Guillén F, Evans C S. Anisaldehyde and veratraldehyde acting as redox cycling agents for H2O2 production by Pleurotus eryngii. Appl Environ Microbiol. 1994;60:2811–2817. doi: 10.1128/aem.60.8.2811-2817.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillén F, Martínez A T, Martínez M J. Substrate specificity and properties of the aryl-alcohol oxidase from the ligninolytic fungus Pleurotus eryngii. Eur J Biochem. 1992;209:603–611. doi: 10.1111/j.1432-1033.1992.tb17326.x. [DOI] [PubMed] [Google Scholar]
- 14.Hammel K E, Tien M, Kalyanaraman B, Kirk T K. Mechanism of oxidative Cα-Cβ cleavage of a lignin model dimer by Phanerochaete chrysosporium ligninase. J Biol Chem. 1985;260:8348–8353. [PubMed] [Google Scholar]
- 15.Harvey P J, Palmer J M. Oxidation of phenolic compounds by ligninase. J Biotechnol. 1990;13:169–179. [Google Scholar]
- 16.Heinfling A, Martínez M J, Martínez A T, Bergbauer M, Szewzyk U. Purification and characterization of peroxidases from the dye-decolorizing fungus Bjerkandera adusta. FEMS Microbiol Lett. 1998;165:43–50. doi: 10.1111/j.1574-6968.1998.tb13125.x. [DOI] [PubMed] [Google Scholar]
- 17.Heinfling A, Ruiz-Dueñas F J, Martínez M J, Bergbauer M, Szewzyk U, Martínez A T. A study on reducing substrates of manganese-oxidizing peroxidases from Pleurotus eryngii and Bjerkandera adusta. FEBS Lett. 1998;428:141–146. doi: 10.1016/s0014-5793(98)00512-2. [DOI] [PubMed] [Google Scholar]
- 18.Huoponen K, Ollikka P, Kälin M, Walther I, Mäntsälä P, Reiser J. Characterization of lignin peroxidase-encoding genes from lignin-degrading basidiomycetes. Gene. 1990;89:145–150. doi: 10.1016/0378-1119(90)90218-g. [DOI] [PubMed] [Google Scholar]
- 19.Kamra D N, Zadrazil F. Influence of gaseous phase, light and substrate pretreatment on fruit-body formation, lignin degradation and in vitro digestibility of wheat straw fermented with Pleurotus spp. Agric Wastes. 1986;18:1–17. [Google Scholar]
- 20.Kelley R L. Ligninolytic activity of Phanerochaete chrysosporium measured as ethylene production from α-keto-γ-methylthiolbutyric acid. Methods Enzymol. 1988;161:79–82. [Google Scholar]
- 21.Kelley R L, Reddy C A. Ethylene production from α-oxo-g-methylthiobutyric acid is a sensitive measure of ligninolytic activity by Phanerochaete chrysosporium. Biochem J. 1982;206:423–425. doi: 10.1042/bj2060423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerem Z, Hadar Y. Effect of manganese on lignin degradation by Pleurotus ostreatus during solid-state fermentation. Appl Environ Microbiol. 1993;59:4115–4120. doi: 10.1128/aem.59.12.4115-4120.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khindaria A, Barr D P, Aust S D. Lignin peroxidases can also oxidize manganese. Biochemistry. 1995;34:7773–7779. doi: 10.1021/bi00023a025. [DOI] [PubMed] [Google Scholar]
- 24.Kirk T K, Farrell R L. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 25.Kishi K, Wariishi H, Marquez L, Dunford H B, Gold M H. Mechanism of manganese peroxidase compound II reduction. Effect of organic acid chelators and pH. Biochemistry. 1994;33:8694–8701. doi: 10.1021/bi00195a010. [DOI] [PubMed] [Google Scholar]
- 26.Koduri R S, Tien M. Oxidation of guaiacol by lignin peroxidase. Role of veratryl alcohol. J Biol Chem. 1995;270:22254–22258. doi: 10.1074/jbc.270.38.22254. [DOI] [PubMed] [Google Scholar]
- 27.Kuan I C, Tien M. Stimulation of Mn-peroxidase activity: a possible role for oxalate in lignin biodegradation. Proc Natl Acad Sci USA. 1993;90:1242–1246. doi: 10.1073/pnas.90.4.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwahara M, Glenn J K, Morgan M A, Gold M H. Separation and characterization of two extracellular H2O2-dependent oxidases from ligninolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 1984;169:247–250. [Google Scholar]
- 29.Leontievsky A A, Myasoedova N M, Golovleva L A. Production of ligninolytic enzymes of the white rot fungus Panus tigrinus. J Biotechnol. 1994;32:299–307. [Google Scholar]
- 30.Martínez A T, Camarero S, Guillén F, Gutiérrez A, Muñoz C, Varela E, Martínez M J, Barrasa J M, Ruel K, Pelayo M. Progress in biopulping of non-woody materials: chemical, enzymatic and ultrastructural aspects of wheat-straw delignification with ligninolytic fungi from the genus Pleurotus. FEMS Microbiol Rev. 1994;13:265–274. [Google Scholar]
- 31.Martínez M J, Muñoz C, Guillén F, Martínez A T. Studies on homoveratric acid transformation by the ligninolytic fungus Pleurotus eryngii. Appl Microbiol Biotechnol. 1994;41:500–504. [Google Scholar]
- 32.Martínez M J, Ruiz-Dueñas F J, Guillén F, Martínez A T. Purification and catalytic properties of two manganese-peroxidase isoenzymes from Pleurotus eryngii. Eur J Biochem. 1996;237:424–432. doi: 10.1111/j.1432-1033.1996.0424k.x. [DOI] [PubMed] [Google Scholar]
- 33.Mester T, Field J A. Characterization of a novel manganese peroxidase-lignin peroxidase hybrid isozyme produced by Bjerkandera species strain BOS55 in the absence of manganese. J Biol Chem. 1998;273:15412–15417. doi: 10.1074/jbc.273.25.15412. [DOI] [PubMed] [Google Scholar]
- 34.Muñoz C, Guillén F, Martínez A T, Martínez M J. Laccase isoenzymes of Pleurotus eryngii: characterization, catalytic properties, and participation in activation of molecular oxygen and Mn2+ oxidation. Appl Environ Microbiol. 1997;63:2166–2174. doi: 10.1128/aem.63.6.2166-2174.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakatsubo F. Synthesis of lignols and related compounds. Methods Enzymol. 1988;161B:57–65. [Google Scholar]
- 36.Nie G, Reading N S, Aust S D. Expression of the lignin peroxidase H2 gene from Phanerochaete chrysosporium in Escherichia coli. Biochem Biophys Res Commun. 1998;249:146–150. doi: 10.1006/bbrc.1998.9106. [DOI] [PubMed] [Google Scholar]
- 37.Orth A B, Royse D J, Tien M. Ubiquity of lignin-degrading peroxidases among various wood-degrading fungi. Appl Environ Microbiol. 1993;59:4017–4023. doi: 10.1128/aem.59.12.4017-4023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmieri G, Giardina P, Zocchi I, Sannia G. Proceedings of the 7th International Conference on Biotechnology in the Pulp and Paper Industry. Montreal, Canada: Canadian Pulp and Paper Association; 1998. Manganese peroxidase isoenzymes from Pleurotus ostreatus; pp. B253–B256. [Google Scholar]
- 39.Sarkar S, Martínez A T, Martínez M J. Biochemical and molecular characterization of a manganese peroxidase isoenzyme from Pleurotus ostreatus. Biochim Biophys Acta. 1997;1339:23–30. doi: 10.1016/s0167-4838(96)00201-4. [DOI] [PubMed] [Google Scholar]
- 40.Ten Have R, Hartmans S, Field J A. Interference of peptone and tyrosine with the lignin peroxidase assay. Appl Environ Microbiol. 1997;63:3301–3303. doi: 10.1128/aem.63.8.3301-3303.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tien M, Kirk T K. Lignin-degrading enzyme from the hymenomycete Phanerochaete chrysosporium Burds. Science. 1983;221:661–663. doi: 10.1126/science.221.4611.661. [DOI] [PubMed] [Google Scholar]
- 42.Tien M, Kirk T K. Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol. 1988;161:238–248. [Google Scholar]
- 43.Vares T, Kalsi M, Hatakka A. Lignin peroxidases, manganese peroxidases, and other ligninolytic enzymes produced by Phlebia radiata during solid-state fermentation of wheat straw. Appl Environ Microbiol. 1995;61:3515–3520. doi: 10.1128/aem.61.10.3515-3520.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]