Abstract
Precision antisense antibacterial agents may be developed into novel antibiotics in the fight against multidrug-resistant Gram-negative bacteria. In this study, a series of diaminobutanoic acid (DAB) dendrons are presented as novel carriers for the delivery of antisense antibacterial peptide nucleic acids (PNAs). The dendron–PNA conjugates targeting the essential acpP gene exhibit specific antisense antimicrobial bactericidal activity against Escherichia coli and Klebsiella pneumoniae at one-digit micromolar concentrations, while showing low toxicity to human cells. One compound selected from a structure–activity relationship series showed high stability in mouse and human serum (t1/2 ≫ 24 h) as well as in vivo activity against a multidrug-resistant, extended spectrum beta-lactamase-producing E. coli in a murine peritonitis model. The compound was also well tolerated in mice upon i.v. administration up to a dose of 20 mg/kg, and in vivo fluorescence imaging indicated clearance via renal excretion with slight accumulation in the kidneys and liver. Thus, DAB-based dendrons constitute a promising new chemistry platform for development of effective delivery agents for antibacterial drugs with possible in vivo use.
Keywords: antisense, antibiotics, PNA, dendron, Gram-negative bacteria
The World Health Organization has identified bacterial infections as one of the biggest threats to public health1 because of increasing and widespread multidrug resistance (MDR).2 The most commonly reported resistant bacteria are Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus.2,3 These pathogens pose a severe burden to the society, prolonging hospitalization and increasing treatment costs and mortality rates.2 Only two mechanistically new classes of antibiotics (oxazolidinones and the cyclic lipopeptide daptomycin) have been approved since the golden era of antibiotics (1940–1970),4 and most resources have been directed toward the improvement of existing “best-in-class” drugs rather than for search of novel “first-in-class” compounds. Thus, the pipeline for development of new antibacterial drugs is very limited.5 Antisense agents hold great promise as antimicrobials as they in principle can be designed to target any specific mRNA, and therefore, any expressed protein coding gene is potentially druggable using this approach. Therefore, new antibiotic targets are accessible in order to effectively circumvent existing resistance, and in addition, the new antibiotics should not be affected by resistance to existing antibiotic drugs. Furthermore, antisense agents may be modified and adapted to new genetic variants of resistant strains by simple modification of the nucleobase sequence.6 Peptide nucleic acid (PNA) is a charge-neutral nucleic acid mimic exhibiting exquisite biological stability and excellent sequence-specific RNA affinity, for which the potential as antimicrobial agents was first demonstrated by targeting the mRNA of the essential acpP (acyl carrier protein) gene in the Gram-negative E. coli.7,8 The presence of an outer lipopolysaccharide (LPS)-coated membrane acts as a highly impermeable barrier, impeding cellular uptake of many antibiotics, and is one of the challenges concerning Gram-negative pathogens.9−11 Poor cellular uptake of unmodified PNA also constitutes a major limitation for the antibacterial activity, which initially could only be demonstrated in the hyper-permeable E. coli mutant AS19.7 Subsequently, covalent attachment of bacterial penetrating peptide (BPP) carriers was shown to facilitate the envelope transport of antibacterial PNAs, allowing low micromolar activity in wild-type Gram-negative bacteria.8,12−14 Commonly, such cationic amphipathic linear carrier peptides show some intrinsic antibacterial activity as well as toxicity to mammalian cells, and they are prone to proteolytic degradation.15,16 In addition, it was later demonstrated that the uptake of many PNA and PMO (phosphorodiamidate morpholino oligomer) BPP conjugates is dependent on the non-essential ABC inner membrane transporter SbmA, thereby being prone to fast resistance development.17,18 Thus, as a minimum, effective antibacterial PNA-conjugate antibiotics require biologically stable BPPs capable of effective SbmA-independent bacterial delivery.
We recently synthesized and characterized a series of cationic peptide dendrons based on non-natural amino acids [primarily diaminobutanoic acid (DAB)] as cell-penetrating peptides (CPPs) for PNA delivery to eukaryotic cells.19 We now demonstrate that similar dendrons are also effective BPPs for SbmA-independent cellular delivery of antisense PNAs to Gram-negative bacteria both in vitro and in vivo.
Results and Discussion
Structure–Activity Relationship
The robust synthesis of cationic, amphipathic dendrons consisting of three generations of DAB functionalized with terminal guanidinylated ligands was recently reported.19 This novel dendrimeric moiety effectively delivers covalently conjugated PNA to eukaryotic cells, and in analogy to certain arginine peptides, these dendrons were anticipated to also interact with Gram-negative membranes and potentially facilitate uptake of antisense PNAs in bacteria.
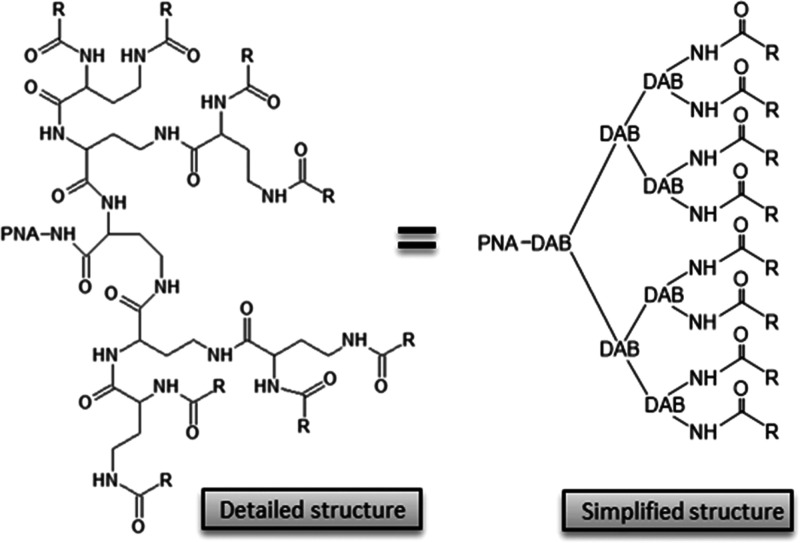
To this end, a series of DAB dendrons were conjugated to a PNA oligomer (Figure 1) targeting the translation start of the acpP mRNA, coding for an essential gene required for fatty acid synthesis7,12,17 (Tables S1 and S2). For each compound, a corresponding mismatch control (mm) was tested to validate whether the activity originates from specific antisense targeting of acpP mRNA rather than a non-specific cytotoxic growth inhibitory effect (e.g., membrane interaction/disruption or intracellular off-targeting). By exploiting the chemical versatility of the dendron structure and the robustness of divergent solid phase synthesis, we investigated the structure–activity relationship (SAR) focusing on the effect of terminal guanidinylation as well as the length and hydrophobicity of the terminal dendron ligands (Figure 1). The conjugates were initially tested for antibacterial activity [by minimal inhibitory concentration (MIC) determination] in the wild-type (wt) E. coli laboratory strain MG1655 and a corresponding deletion ΔsbmA mutant strain as well as a wt K. pneumoniae strain (ATCC 13883) (Table 1). Previously, effective BBPs have been identified among polycationic linear peptides such as (KFF)3K, (RXR)4, and selected antimicrobial peptides (AMPs).8,13,20,21 Thus, conjugates having eight terminal positive charges were designed to compare the activity of amino versus guanidino derivatives as well as the effect of varying terminal carbon linker chains (Figure 1 and Table 1). None of the compounds with terminal amino groups [i.e., (Apr)8-DAB-PNA (PNA4883), (Abu)8-DAB-PNA (PNA4887), and (Apn)8-DAB-PNA (PNA4842)] inhibited growth of E. coli at concentrations <16 μM, while the corresponding guanidinylated derivatives [i.e., (Gbu)8-DAB-PNA (PNA4883) and (Gpn)8-DAB-PNA (PNA4887)] exhibited MIC values in the low micromolar range and showed a significant difference between match and mismatch compounds (Table 1). These results demonstrate that DAB dendrons can deliver PNAs to bacteria and that terminal guanidinylation is necessary for effective uptake. This conclusion corroborates analogous findings for antibacterial antisense PMOs, showing that arginine-containing BPPs are more active than similar ligands based on lysine or ornithine.20 In order to further validate the delivery properties of guanidinylated DAB dendrons independent of the targeted gene, we also synthesized a guanidinobutanoic acid (Gbu)8-DAB-PNA derivative targeting the essential FtsZ gene (PNA4986), and this conjugate showed activity in E.coli comparable to the analogous acpP compound (Table 1). Previously, it was demonstrated that the antibacterial activity of antisense PNAs exhibits an optimum around 10 nucleobases in terms of oligomer length,7 and this was ascribed to size-limited bacterial uptake of the compounds.22 The present PNA dendron conjugates exhibit a similar behavior, showing the highest antibacterial activity for the decamer PNA4897 (Table S3). Next, we investigated the effect of the length of the terminal carbon chain of the dendron–PNA conjugates, comparing the activity of derivatives terminally functionalized with guanidinobutanoic acid (Gbu)8-DAB-PNA (PNA4897), guanidinopentanoic acid (Gpn)8-DAB-PNA (PNA4856), guanidinohexanoic acid (Ghx)8-DAB-PNA (PNA4737), guanidinoheptanoic acid (Ghp)8-DAB PNA4857, and guanidinooctanoic acid (Goc)8-DAB PNA4850. The C8 linker produced the most active compound ((Goc)8-DAB, PNA4850) with MIC = 0.25 μM, while decreasing the terminal carbon chain length resulted in up to 8-fold loss of activity for (Ghx)8-DAB (PNA4737). Interestingly, we observed a tendency of inversion in this trend with terminal carbon chains shorter than 6 atoms, and indeed, (Gbu)8-DAB-PNA (PNA4897) inhibited bacterial growth at a concentration of 0.5 μM. The mismatch version of the conjugates with long terminal carbon chains had a minor inhibitory activity with MIC values of 16 and 8 μM for (Ghp)8-DAB-PNA (PNA4852) and (Goc)8-DAB-(PNA4853), respectively. This non-acpP target growth inhibition effect seems to be directly correlated with the terminal carbon chain length and thus to the overall hydrophobicity of the conjugates. This could be linked to compromising the integrity of the bacterial envelope in analogy to the mechanism observed for AMPs, for example, colistin, but in contrast to colistin, PNA4853 did not induce envelope disruption as measured by Sytox staining (Figure S1A). However, most interestingly, both (Goc)8-DAB PNAs 4850 and 4853 significantly increased the colistin-induced uptake of SYTOX (Figure S1B,C), thereby indicating a contributing membrane destabilization by these PNAs. Furthermore, such an effect was not observed for the less hydrophobic and less toxic PNA4897 (and the corresponding mismatch PNA4898) SYTOX (Figure S1D,E), thereby assigning the effect to the (Goc)8-DAB peptide part. Thus, the acpP-targeting conjugates may inhibit bacterial growth by at least two independent mechanisms: (i) the specific antisense targeting of the acpP gene by the PNA moiety (low concentrations) and (ii) a (much weaker) non-antisense, membrane-associated activity by the cationic amphipathic carrier that may be analogous to the action of some AMPs (usually at higher concentrations than the antisense effect21). Therefore, the antisense specificity (estimated as MICmismatch/MICmatch) appears to be higher for the dendrons with shorter terminal carbon chains, as in the case of the sub-micromolar activity of the short (Gbu)8-DAB-PNA (PNA4897) (MIC = 0.5 μM) compared to its mismatch (Gbu)8-DAB-PNA-(PNA4898) (MIC >16 μM).
Figure 1.
PNA–dendron conjugate structure (DAB: diaminobutanoic acid). (Reproduced from Gabas, I. M.; Nielsen, P. E., Effective Cellular Delivery of Antisense PNA by Conjugation to Guanidinylated DAB-Based Peptide Dendrons. Biomacromolecules2020,21 (2), 472–483. Copyright 2020, ACS Publications).
Table 1. MIC Values for the Dendron–PNA Conjugatea.
| MIC μM |
||||||||
|---|---|---|---|---|---|---|---|---|
|
E. coli wt |
E. coli ΔSbmA |
K. pneumoniae |
||||||
| target gene | carrierb | PNAc | match | misM | match | misM | match | misM |
| acpP | (Gbu)8-(DAP) | 4990 | 1 | >16 | 0.5 | >16 | 4 | >16 |
| acpP | (Ghx)8-(DAP) | 4991 | 4 | >16 | 2 | >16 | 2 | >16 |
| acpP | (Gbu)8-(DAB) | 4897 | 0.5 | >16 | 0.5 | >16 | 8 | >16 |
| acpP | (Gpn)8-(DAB) | 4856 | 1 | >16 | 1 | >16 | 8 | >16 |
| acpP | (Ghx)8-(DAB) | 4737 | 2 | >16 | 2 | >16 | 2 | >16 |
| acpP | (Ghp)8-(DAB) | 4857 | 1 | 16 | 1 | 16 | 0.5 | 16 |
| acpP | (Goc)8-(DAB) | 4850 | 0.25 | 8 | 0.25 | 8 | 0.125 | 4 |
| acpP | (Apr)8-(DAB) | 4883 | >16 | >16 | >16 | >16 | >16 | >16 |
| acpP | (Abu)8-(DAB) | 4887 | >16 | >16 | 16 | >16 | >16 | >16 |
| acpP | (Apn)8-(DAB) | 4842 | >16 | >16 | >16 | >16 | >16 | >16 |
| ftsZ | (Gbu)8-(DAB) | 4986 | 2 | >16 | 4 | >16 | 16 | >16 |
MICs are reported as the lowest concentrations of the compound resulting in complete growth inhibition [measured as optical density (OD) at 595 nm] after 18 h of incubation at 37 °C.
Abbreviations: DAB: diaminobutanoic acid dendron; DAP: diaminopropanoic acid dendron; Gbu: guanidinobutanoyl Ghx: guanidinohexanoyl; Gpn: guanidinopentanoyl; Ghp: guanidinoheptanoyl; Goc: guanidinooctanoyl; Apr: aminopropanoyl; Abu: aminobutanoyl; Apn: aminopentanoyl.
PNA number for the E. coli match PNA.
In order to evaluate whether changes in the size of the core dendritic structure affect the antibacterial activity, we synthesized the conjugates (Gbu)8-DAP PNA4990 and (Ghx)8-DAP PNA4991 using the shorter diaminopropionic acid (DAP) [instead of DAB] as a monomer for the synthesis of the three-generation dendron. However, only slight differences in activity between the DAP- and DAB-based structures were observed, thereby suggesting that the outer shell of the dendron (i.e., terminal ligands) is more important than the core structure (and size) in determining the antibacterial properties of dendron-PNA conjugates (Table 1).
SmbA is a non-essential gene encoding an inner membrane transporter involved in the uptake of inter alia proline-rich AMPs23−25 and notably also (KFF)3K-PNA conjugates.16,17 Mutations in the sbmA gene do not affect bacterial growth under laboratory conditions but can confer resistance to (KFF)3K-PNA due to poor (inner membrane) cellular uptake. To study whether the antibacterial activity of DAB dendron–PNA conjugates is SbmA-dependent and therefore susceptible to the above-mentioned resistance mechanism, we tested a series of derivatives with different terminal ligands against a ΔsmbAE. coli mutant. The compounds showed virtually identical results for the wt and ΔsbmA strains. Importantly, this result indicates that the uptake of dendron–PNA conjugates is SbmA-independent and is consistent with the models, suggesting a carrier-independent, direct penetration of some arginine-rich peptides through both the outer and inner membranes,26−29 and most probably utilizing the electronegative potential over the inner membrane for crossing this.30 Interestingly, the D-form (kff)3k-PNA in contrast to the natural L-form (KFF)3K-PNA is active against the ΔsbmA mutant,16,17 inferring that the D-form is able to transverse the inner membrane by an sbmA-independent mechanism. It was also very recently found that the peptide part of the L-form (KFF)3K-PNA is rapidly degraded both in the medium and in the periplasm of the bacteria, thereby inferring that intact L-form (KFF)3K-PNA might indeed cross the inner membrane independently of sbmA in analogy to some arginine-based BPPs17,18,25 and that the resistance is due to insufficient stability and thus inability to actually reach the inner membrane.16
In the case of peptide–PMO conjugates (PPMOs), some arginine-rich peptides that could overcome ΔsbmA resistance have also been identified, such as (RX)6B and (RXR)4XB,18 analogous to the results with PNA. However, an (RFF)3RXB and even the corresponding D-form PPMO were reported inactive against a ΔsbmA strain.18 This apparent difference between BPP–PNA and PPMO behaviors is not clear at present, but the biological stability of these compounds is not available.
The targeted region of the acpP mRNA is relatively well conserved among Gram-negatives, and the PNA used in this work is also able to target acpP in K. pneumoniae. However, when testing the series against this pathogen, a slightly different trend than that seen in E. coli was observed, with a simple direct correlation between length of the terminal carbon linker and antimicrobial activity. As for E.coli, (Goc)8-DAB PNA4850 exhibits the highest antimicrobial activity (MIC = 0.125 μM), whereas the shorter (Gbu)8-DAB PNA4897 is much less active (MIC = 8 μM) (Table 1). However, as observed for E. coli, the activity of the mismatch controls also increases with the carbon-linker length. This behavior is similar to that previously observed in mammalian cells, showing that the gain in activity achieved by increasing hydrophobicity is commonly accompanied with an increase in non-specific cell toxicity.19
Finally, we also tested the activity of DAB dendron–PNA conjugates against Pseudomonas aeruginosa PAO1 and S. aureus ATCC 29213 using adequate antisense sequences to target the acpP mRNA in these species (Table S4). Only minor inhibitory activity was observed with the guanidinoheptanoic or guanidinoctanoic acid in P. aeruginosa. However, since the activities of the match and mismatch derivatives were not significantly different, the mechanism of action presumably is not antisense-mediated for this PNA in this pathogen. Also, the analogous PNA dendron did not show antimicrobial activity in the Gram-positive S. aureus. It is well established that the culture conditions, not least the growth medium (e.g., complex vs defined medium and cation contents), can have a dramatic influence on the antibacterial activity, especially for AMPs. The present experiments were conducted under standard MIC screening conditions in the complex Mueller Hinton Broth (MHB) medium, and no optimization by changing the medium was attempted. Similarly, the gene target position and length strongly influence activity. Furthermore, previous studies have shown that antisense BPP-PNA targeting of P. aeruginosa is quite challenging.13 Thus, it may be possible to optimize dendron structures to also target these bacteria.
Cell Toxicity of Dendron–PNAs
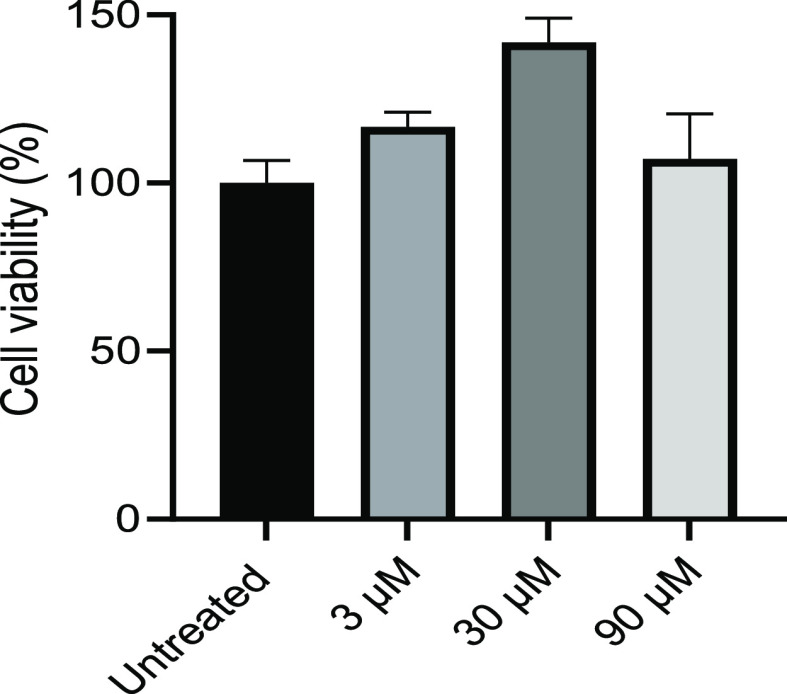
Based on the low MIC, combined with high MICmismatch/MICmatch ratios (>32) (Table 1), PNA4897 was chosen as the most interesting hit compound, and the cellular toxicity was determined in HepG2 cells, the standard cell line in antibiotic drug discovery. The results showed no significant toxicity at least up to 90 μM, which is more than 100-fold higher than the MIC (Figure 2). The cytotoxic effect of selected dendron–PNA conjugates was also determined in HeLa cells, which yielded higher toxicities than found for HepG2 cells (Figure S2). Furthermore, a direct correlation between the carbon chain length of the guanidinylated terminal group and reduction in cell viability was apparent in these cells. Specifically, (Gbu)8-DAB-PNA (PNA4897), carrying terminal guanidinium–butanoic acid, showed negligible toxicity at 20 μM (>10 × MIC), while the (Goc)8-DAB-PNA (PNA4850) with guanidinylated octanoic acid caused ∼35% reduction in viability at 20 μM. Although the substitution of DAB with DAP in the core dendrimer structure results in a reduction in size and molecular weight of the compound, reduced cell toxicity was not observed when comparing (Gbu)8-DAB-PNA (PNA4897) with (Gbu)8-DAP-PNA (PNA4990) or (Ghx)8-DAB-PNA (PNA4737) with (Ghx)8-DAP-PNA (PNA4991). This result would indicate that the outer shell of the dendron may also be more important than the core for cytotoxicity. Previously, it was demonstrated that long terminal groups are necessary for efficient delivery of DAB dendron conjugates to eukaryotic cells.19 Interestingly, identical dendron moieties conjugated to different PNA oligomers result in compounds with different cytotoxicities. Specifically, conjugates designed for eukaryotic antisense targeting (PNA length 18 nt)19 exhibit higher toxicity than the corresponding bacteria-targeting compounds (PNA length 10 nt), thereby indicating that the PNA oligomer increases the inherent cytotoxicity of the dendron CPP/BPP. It is also generally observed that increasing cationic charge as well as increased hydrophobicity of the delivery peptide increase cytotoxicity (e.g., ref (19)). These results supported the choice of (Gbu)8-DAB-PNA (PNA4897) as the most interesting candidate for further investigation.
Figure 2.
Cytotoxicity in human HepG2 cells after 48 h incubation with (Gbu)8-DAB-PNA (PNA4897). Cell viability was measured by ATP assay and expressed as mean with SD relative to the untreated control (n = 9).
Effect of Divalent Cations
Divalent cations such as Ca2+ and Mg2+ are known to significantly inhibit the activity of membrane-interacting cationic AMPs,31,32 and therefore, PNA delivery by DAB dendrons could be similarly affected. Thus, the MIC values of (Gbu)8-DAB-PNA (PNA4897) and the corresponding mismatch (Gbu)8-DAB PNA4898 were determined in MOPS minimal medium with different combinations of [Ca2+] and [Mg2+] (Table 2). The results show that both ions do indeed reduce the activity of the conjugates in E. coli: the MIC values for (Gbu)8-DAB-PNA (PNA4897) ranged from <0.5 μM at low cation concentrations (i.e. [Mg2+] = 50 μM, [Ca2+] < 200 μM) to >4 μM under high-cation conditions ([Ca2+] and [Mg2+] at 2.5 mM). Interestingly, the activity ratio between match and mismatch across the tested conditions is fairly constant (Figure S3), suggesting that the membrane stabilization effect of the divalent cations affects dendron–PNA uptake (implied by antisense activity) and membrane disruption [implied by non-antisense (mismatch) activity] in parallel and that these effects share a common or overlapping mechanism of outer LPS/membrane interactions.
Table 2. Effect of Calcium and Magnesium Ions on Apparent MIC of (Gbut)8-DAB-PNA (PNA4897) in MOPS Minimal Medium [MIC Values Relative to the Mismatch (Gbut)8-DAB-PNA (PNA4898) Are Shown in Parentheses]a.
| Ca2+ |
|||||||
|---|---|---|---|---|---|---|---|
| 0.5 μM | 5 μM | 50 μM | 200 μM | 500 μM | 2.5 mM | ||
| Mg2+ | 50 μM | <0.5 (4) | <0.5 (8) | <0.5 (16) | 1 (>16) | 2 | 4 |
| 200 μM | 1 (8) | 1 (8) | 1 (>16) | 1 (>16) | 2 | 4 | |
| 500 μM | 1-2 (16) | 1–2 (>16) | 1–2 (>16) | 2 (>16) | 2 | 4 | |
| 2.5 mM | 4 | 4 | 4 | 4 | 5 | >4 | |
MICs are reported as the lowest concentrations of the compound resulting in complete growth inhibition [measured as optical density (OD) at 595 nm] after 18 h of incubation at 37 °C.
As a further confirmation of the cation effect on DAB dendron activity, the effect on antibacterial activity (MIC) of (Gbu)8-DAB-PNA (PNA4897) was measured after addition of varying concentrations of the chelating agent ethylenediaminetetraacetic acid (EDTA) (from 0.2 μM to 2.5 mM) added to the (chemically undefined) MHB medium. Indeed, the results showed that addition of EDTA enhanced the antibacterial effect of the dendron–PNA in a concentration-dependent manner (Figure S4), supporting an inhibitory effect of divalent cations, presumably due to their binding to the outer LPS layer, thereby stabilizing the outer envelope of the Gram-negative bacteria or shielding ionic interactions with the cationic BPP.
Bactericidal Activity
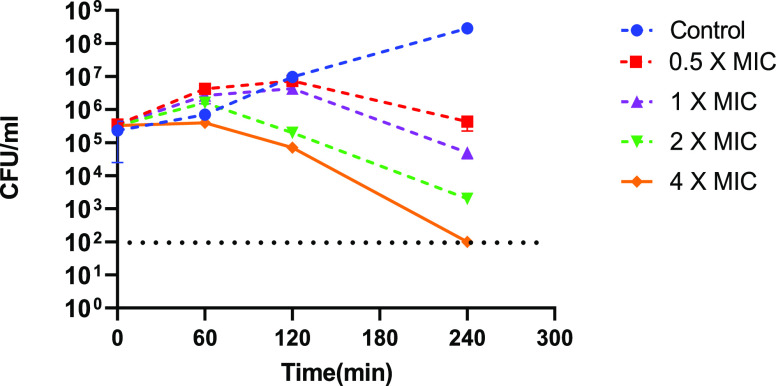
A time kill assay was employed to further characterize the antibacterial activity. The results of this clearly show that PNA4897 is bactericidal, causing more than 3 log reduction in CFU at 4 times MIC within 4 h (Figure 3).
Figure 3.
Representative time–kill graph for E. coli MG1655 grown in MHB in the presence of (Gbu)8-DAB-PNA (PNA4897) at 0.5 × MIC, 1 × MIC, 2 × MIC, and 4 × MIC. Samples were taken at 0, 1, 2, and 4 h to determine viable bacterial numbers (n = 3), and the experiments were performed at 2 independent days. Data are mean ± SD of two independent experiments. The dashed line indicates the detection limit.
Stability in Mouse and Human Serum
Prior to in vivo studies with (Gbu)8-DAB-PNA (PNA4897) (and mismatch PNA4898), the stability of this dendron–PNA conjugate was evaluated in mouse and human serum. As the compound is composed of non-natural amino acids, it is not expected to be a substrate for peptidases nor proteases and thus should exhibit good serum stability. Indeed, more than 95% of the compound remained intact after incubation for 24 h in both human and mouse serum (Figure S5).
In Vivo Efficacy on MDR E. coli of Dendron–PNAs
In view of the positive in vitro results, the antimicrobial effect of (Gbu)8-DAB-PNA (PNA4897) and the corresponding mismatch PNA4898 was evaluated against a small series of MDR E.coli clinical isolates including extended spectrum beta-lactamase (ESBL), carbapenem, and colistin-resistant strains, and except for one (AMA 817), the PNA exhibited good activity against all strains (Table S5). The multidrug-resistant E.coli EC-106-09 was chosen for an in vivo efficacy experiment in a neutrophenic murine peritonitis model. This clinical isolate carries the ESBL gene CTX-M-27 and is additionally resistant to quinolones, sulphonamides, and trimethoprim.33
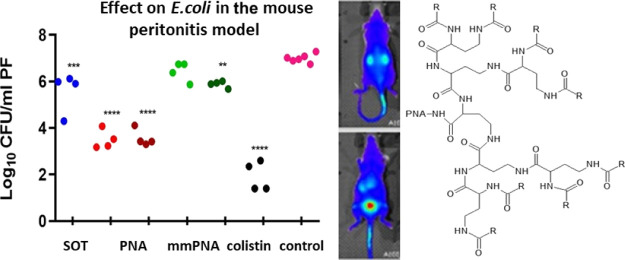
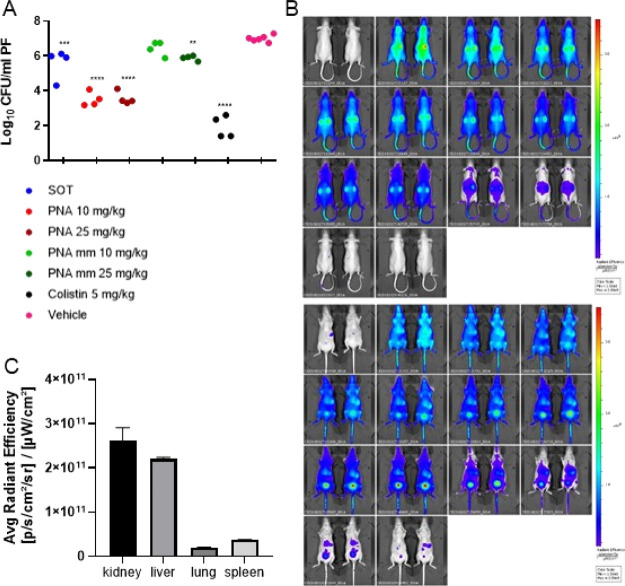
Initially, the acute toxicity of the compound was assessed in female NMRI mice, and it was found to be well tolerated up to 25 mg/kg upon i.p. and up to 20 mg/kg upon i.v. administration (Table S6). An intraperitoneal (IP) inoculum of E. coli (0.5 mL containing 6 × 1010 CFU/mL) was used to establish an IP infection. At t = 1 h post inoculum, the mice were treated with 10 mg/kg or 25 mg/kg of either PNA4897 or the mismatch control PNA4898 or colistin (5 mg/kg) as the positive control, while citrate buffer was used for the vehicle treatment group. A significant increase in CFUs of ∼1.5 log between t = 0 and the sampling at t = 5 h (Figure 4A) was observed in the vehicle treatment group. The CFU levels in the peritoneum of mice treated with PNA 4897 (10 and 25 mg/kg) were significantly lower (ca 3.5 log) compared to the vehicle-treated group, although no significant dose response was observed. For comparison, only the highest dose of the mismatch control PNA4898 exhibited a minor CFU count reduction (∼1 log). No signs of distress in the animals were observed during the experiment (with the exception of one mouse in the group treated with the highest dose of PNA4897 showing minor clinical signs). Overall, the match PNA4897 was significantly more effective than the mismatch PNA4898 in reducing the infection burden.
Figure 4.
(A) In vivo efficacy of dendron–PNA conjugates in a murine peritonitis infection. Evaluation of (Gbu)8-DABPNA (4897) and (Gbu)8-DAB MM-PNA (4898) antibacterial potential after 4 h IP treatment of a peritonitis NMRI mice infection model, inoculated with 2.5 × 106 CFU/mouse of E. coli (ESBL). Colistin was used as the positive control, and the vehicle (citrate buffer) was used as the negative control. **p < 0.01; ***p < 0.001; ****p < 0.0001. (B) In vivo biodistribution of AF680-PNA4897 after i.v. administration. Top (back) and bottom (front) scans; the time points from the top left corner are T = 0 (pre-injection), 5, 10, 15, 20, 25, 30, 40, 50 min, 1 h, 2 h, 4 h, 24 h, 48 h. (C) AF680-PNA4897 average organ accumulation (independent of organ size) 48 h post i.v. administration (n = 2).
In Vivo Biodistribution and Organ Accumulation of Dendron–PNAs
To further characterize the in vivo behavior of the dendron–PNA conjugates and obtain information about their pharmacokinetics behavior, PNA4897 was labeled with the near-IR fluorophore, AlexaFluor680 (AF680), and body distribution was followed by in vivo fluorescence imaging after intravenous administration in nude mice via the tail vein (Figure 4B). As expected, the compound distributed readily (t < 5 min) in the whole body after i.v. administration, and some initial accumulation was located in the kidneys. The compound was eliminated fast by renal excretion with an estimated body half-life of 40 min, which is slightly longer than previously observed for a 20mer unmodified PNA.34−36 Intact PNA4897 was detected in the urine by HPLC and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Figure S7) in accordance with the high serum stability of the (unlabeled) compound (Figure S4), and we have previously shown that a fully analogously AF680-labeled PNA can be recovered intact in the urine following i.v. administration,34−36 strongly supporting that the imaging fluorescence signal does indeed reflect intact AF680-PNA4897. At 20 min post administration, a significant amount of fluorescence was assigned to the bladder and to the liver, and after 2 h, most of the remaining fluorescence was concentrated in the liver, kidneys, and bladder (Figure 4B), whereas at 24 h, very little fluorescence was detected and at 48 h, practically no fluorescence was detected in the mouse. Scanning of the excised organs showed that the remaining fluorescence had accumulated predominantly in the kidney and in the liver (Figure 4C). While the relative accumulation was fast (0–2 h) in the kidneys, a more steady relative accumulation was seen in the liver (Figure S8). It remains to be seen whether the slight kidney accumulation is accompanied by nephrotoxicity as seen for, for example, colistin.37
Conclusions
The present dendron–PNA conjugates add a new chemical platform for BPPs that allow design and easy synthesis of (PNA) conjugates exhibiting specific antisense antimicrobial bactericidal activity against E. coli and K. pneumoniae at one-digit micromolar concentrations, while exhibiting low toxicity to human cells. One hit compound selected from an SAR series of such dendron–PNA conjugates showed high stability in mouse and human serum (t1/2 ≫ 24 h) as well as in vivo activity against a multidrug-resistant, ESBL-producing E. coli in a murine peritonitis model. The compound is well tolerated up to a dose of 20 mg/kg in mice upon i.v. administration, and in vivo fluorescence imaging indicated that it is cleared from the body (t1/2 ∼ 1 h) via renal excretion with very slight accumulation in the kidneys and liver. Thus, DAB-based dendrons are interesting and effective delivery agents for antibacterial drugs with possible in vivo use. Specifically, the dendrons provide a novel BPP chemical architecture for subsequent extended medicinal chemistry SAR and optimization of both antibacterial and not least in vivo properties in terms of pharmacokinetics, toxicity, and efficacy and thus therapeutic index of novel precision antisense antibiotics. In principle, the dendrons might also serve as delivery agents for other antimicrobials for which bacterial delivery is a limiting challenge.
Methods
Synthesis of the Dendron–PNA Conjugate
The detailed procedure is described in ref (19). Briefly, the conjugate was synthesized by manual solid-phase synthesis using the Boc/Fmoc approach on MBHA resin LL (100–200 mesh, Novabiochem, loading 0.12 mmol/g). Fmoc-deprotection was performed with piperidine/DMF (1:4, v/v) two times for 10 min; Boc-deprotection was performed with TFA/anisol (95:5, v/v) two times for 4 min. Chain elongation was performed by using 0.2 M Boc-protected PNA monomer in DMF solution and Nα-Fmoc-Nγ-Boc-l-2,4-diaminobutanoic acid in DMF (4 equiv), in combination with an equal amount of 0.2 M hexafluorophosphate benzotriazole tetramethyl uronium as a coupling reagent and 4.5 equiv DIEA as an activator base. Guanidinylation of terminal amino groups was performed with 0.07 M of 1,3-di-Boc-2-(trifluoromethylsulfonyl)-guanidine (4 equiv) in DCM and 4.5 equiv DIPEA overnight (O/N). The conjugate was cleaved from the resin using TFA/TFMSA/m-cresol/thioanisole (6:2:1:1, v/v, 2 × 1 h), precipitated, and washed three times with cold Et2O. The conjugate was finally purified by C18 RP-HPLC and characterized by HPLC and MS-MALDI. PNA5512 used for bioimaging was synthesized via addition of a cysteine residue between the PNA and the dendrimer moiety of PNA4897, and the near-IR maleimide-functionalized AlexaFluor680 (Thermo Scientific, US) was coupled to the thiol group following the supplier’s instructions.
MIC Assay
The MIC values for the conjugates were determined using the broth microdilution protocol described in ref (38). MHB medium was purchased from Fluka/Sigma, and its content in calcium and magnesium cations (21.7 and 16.2 μM, respectively) was quantified by inductively coupled plasma atomic emission spectroscopy. For each compound, a 10 × 2-fold dilution series was prepared in a solution of 0.2% BSA, and a final volume of 10 μL was transferred in each well of a 96-well NUNC polystyrene plate. O/N cultures of the tested bacteria were grown in 5 mL of MHB, starting from a glycerol stock up to OD595 nm = 1. A dilution of ∼2.5 × 105 CFU/mL was prepared, and 90 μL was added to the compounds to a final volume of 100 μL. The plates were incubated in a rotatory shaker at 37 °C for 18 h, and finally, the OD595 was measured using a microplate reader (Tecan Sunrise, USA). MICs were defined as the lowest concentrations of the compound able to completely inhibit bacterial growth for 18 h at 37 °C. Each experiment was run in three biological replicates. For the evaluation of calcium and magnesium effects on the biological activity of the conjugates, growth inhibition experiments were carried out in MOPS minimal medium supplemented with 0.2% casamino acids (Gifco) following the same procedure as above for compound dilutions and plate assembly.
Cytotoxicity Determination
HepG2 cells were grown in DMEM with high glucose supplemented with 10% (v/v), fetal bovine serum, minimum essential medium non-essential amino acid solution (Gibco), and 1% (v/v) penicillin/streptomycin 10,000 U/mL (ThermoFisher). Cells were incubated for 48 h with the tested conjugates, and the viability was then evaluated measuring the amount of adenosine 5’ triphosphate (ATP): cells were lysed with 60 μL of passive lysis buffer (Promega), and ATP was quantified with a CellTiter-Glo luminescent cell viability assay kit (Promega) following the supplier’s protocol. The results are expressed as relative viability compared to the untreated control.
Time–Kill Assay
Time–kill curves were obtained by growing E. coli MG1655 in MHB (Oxoid) in the presence of PNA4897 at 0.5 × MIC, 1 × MIC, 2 × MIC, and 4 × MIC diluted in 2% BSA in H2O. The starting inoculum was 5 × 105 CFU/mL, and cells were grown with shaking at 37 °C for 4 h. Samples were taken at times 0, 1, 2, and 4 h; serially diluted; and plated onto agar plates to determine viable bacteria after O/N incubation at 37 °C. Experiments were performed in duplicates with the vehicle (solution containing 2% BSA in H2O) added to the broth as the growth control.
Serum Stability
Human or mouse serum was incubated with 60 μM of PNA4898 at 37 °C up to 24 h. At times 0, 2, 4, and 24 h, 30 μL of 3% TCA was added to 30 μL of serum incubated with PNA4898. Samples were kept on ice for 15 min and then centrifuged for 15 min at 14,000g. The supernatant was analyzed by HPLC and MALDI.
Acute Toxicity
A total of 15 female NMRI mice were weighted and dosed IP with 200 μL of 10 and 25 mg/kg conjugate solutions in isotonic citrate buffer (pH = 7.2) or citrate buffer alone. Mice were observed for signs of acute toxicity t = 0, 2, 15, 30, 60, and 120 min according to the score scheme (Table S6).
IP Infection Model
A total of 30 outbred female NMRI mice weighing 25.9 to 31.4 g were rendered neutropenic with cyclophosphamide (Sendoxan). Briefly, 0.5 mL of the solution corresponding to a dose of 200 mg/kg cyclophosphamide was administered by i.p. injection 4 days before the experiment, and a dose corresponding to 100 mg/kg cyclophosphamide was administered by i.p. injection 1 day prior to inoculation. Fresh O/N colonies of E. coli EC-106-0933 from 5% horse blood agar were diluted to 2–5 × 106 CFU/mL in the sterile saline suspension, and mice were inoculated in the peritoneum with 0.5 mL of the bacterial suspension containing 6.6 log10 CFU/mL. 45 μL of Nurofen (20 mg ibuprofen/mL, 30 mg/kg approx.) was administered orally as pain relief. Test compounds were diluted to 1.4 and 3.5 mg/mL in isotonic citrate buffer (pH = 7), and mice were treated 1 h after the inoculation with 0.2 mL of the conjugate solution (citrate buffer was used in the vehicle treated control mice). A dose of 5 mg/kg of colistin administered s.c. in the neck region was used in the positive control group. The mice were sacrificed by cervical dislocation either at 1 h after inoculation (start of the treatment group) or after 4 h after the treatment. A total of 2 mL of sterile saline was injected i.p., and the abdomen was gently massaged before its opening to sample the fluid with a pipette. Each sample was 10-fold diluted in saline, and 20 μL spots were applied on blood agar plates for viable CFU counting. All agar plates were incubated 18–22 h at 35 °C in ambient air.
Bioimaging
Two NMRI-nu mice were treated systemically with 200 μL of 10 μM solution of Alexa Fluor 680 (AF680)-labeled PNA4897 (H-(Gbu)8-(DAB)4-(DAB)2-DAB-Cys(AF680)-CTCATACTCT-NH2) via tail vein injection. Mice were then anesthetized with 2% isoflurane at a flow rate of 1.5 L/min and placed in an IVIS spectrum CT (PerkinElmer) to run a near-IR epifluorescence acquisition. The 675/720 nm filters were used for excitation and emission, respectively. Images were acquired at t = 0 (pre-injection), 5, 10, 20, 25, 30, 60, 120, 240 min, 24 h, and 48 h. Mice were finally sacrificed to acquire an ex vivo organ scan. Living Image software was used for data analysis (PerkinElmer). Boundaries were drawn around the region of interest (ROI) to quantify the fluorescence signals from the body and kidneys. Using the radiant efficiency corresponding to the first three time points, liner regression analysis was performed with GraphPad Prism to determine the initial ROI values (Y0). Finally, all ROI values were converted into percentages based on the estimated initial signal and plotted into GraphPad Prism for the determination of the compound elimination “half-life” using nonlinear regression (one phase decay).
Statistical Analysis
GraphPad Prism was used for data collection, statistical analysis, and graph representation. Dunnett’s multiple comparison test was used in the i.p. infection model to evaluate the CFU levels of the treatment groups compared to the vehicle group. OriginPro9 was used to plot of the OD595 as a function of [Ca2+] and [Mg2+].
Acknowledgments
This work was supported by the NovoNordisk Foundation Challenge Program (grant no. NNF16OC0021700), the European Union JPIAMR program [SENBIOTAR through Innovation Fund Denmark (4209-000018)], and the Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 642738. The authors would like to thank Carina Vingsbo-Lundberg for assistance in the experiment design and planning and also to the technicians at the Contract research group at Statens Serum Institut for technical assistance in the animal experiments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.2c00089.
HPLC and MALDI-TOF data for the compounds and experimental data (PDF)
Author Contributions
M.I., I.M.G., L.M.C., E.H.M., E.L., F.B., N.Y., and C.B. performed experiments. M.I., I.M.G., L.M.C., E.H.M., E.L., F.B, N.Y., C.B., and P.E.N. designed experiments. M.I., C.B., and P.E.N. wrote the first draft of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Who Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. https://www.aidsdatahub.org/resource/who-global-priority-list-antibiotic-resistant-bacteria. (Accessed April 17, 2022).
- Collaborators A. R. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399, 629. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC; U.S. Department of Health and Human Services, C . Antibiotic Resistance Threats in the United States. 2019https://stacks.cdc.gov/view/cdc/82532. (Accessed April 17, 2022).
- Roemer T.; Boone C. Systems-level antimicrobial drug and drug synergy discovery. Nat. Chem. Biol. 2013, 9, 222–231. 10.1038/nchembio.1205. [DOI] [PubMed] [Google Scholar]
- Miethke M.; Pieroni M.; Weber T.; Brönstrup M.; Hammann P.; Halby L.; Arimondo P. B.; Glaser P.; Aigle B.; Bode H. B.; Moreira R.; Li Y.; Luzhetskyy A.; Medema M. H.; Pernodet J.-L.; Stadler M.; Tormo J. R.; Genilloud O.; Truman A. W.; Weissman K. J.; Takano E.; Sabatini S.; Stegmann E.; Brötz-Oesterhelt H.; Wohlleben W.; Seemann M.; Empting M.; Hirsch A. K. H.; Loretz B.; Lehr C.-M.; Titz A.; Herrmann J.; Jaeger T.; Alt S.; Hesterkamp T.; Winterhalter M.; Schiefer A.; Pfarr K.; Hoerauf A.; Graz H.; Graz M.; Lindvall M.; Ramurthy S.; Karlén A.; van Dongen M.; Petkovic H.; Keller A.; Peyrane F.; Donadio S.; Fraisse L.; Piddock L. J. V.; Gilbert I. H.; Moser H. E.; Müller R. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. 10.1038/s41570-021-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty J. P.; Stewart D. B. Advances in therapeutic bacterial antisense biotechnology. Appl. Microbiol. Biotechnol. 2018, 102, 1055–1065. 10.1007/s00253-017-8671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good L.; Nielsen P. E. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 1998, 16, 355–358. 10.1038/nbt0498-355. [DOI] [PubMed] [Google Scholar]
- Good L.; Awasthi S. K.; Dryselius R.; Larsson O.; Nielsen P. E. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 2001, 19, 360–364. 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- Gruenheid S.; Moual H. Resistance to antimicrobial peptides in Gram-negative bacteria. FEMS Microbiol. Lett. 2012, 330, 81–89. 10.1111/j.1574-6968.2012.02528.x. [DOI] [PubMed] [Google Scholar]
- Zhao S.; Adamiak J. W.; Bonifay V.; Mehla J.; Zgurskaya H. I.; Tan D. S. Defining new chemical space for drug penetration into Gram-negative bacteria. Nat. Chem. Biol. 2020, 16, 1293–1302. 10.1038/s41589-020-00674-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati J. D.; Kleinekathöfer U.; Winterhalter M. How to Enter a Bacterium: Bacterial Porins and the Permeation of Antibiotics. Chem. Rev. 2021, 121, 5158–5192. 10.1021/acs.chemrev.0c01213. [DOI] [PubMed] [Google Scholar]
- Good L.; Sandberg R.; Larsson O.; Nielsen P. E.; Wahlestedt C. Antisense PNA effects in Escherichia coli are limited by the outer-membrane LPS layer. Microbiol-Sgm 2000, 146, 2665–2670. 10.1099/00221287-146-10-2665. [DOI] [PubMed] [Google Scholar]
- Ghosal A.; Nielsen P. E. Potent Antibacterial Antisense Peptide-Peptide Nucleic Acid Conjugates Against Pseudomonas aeruginosa. Nucleic Acid Nanotheranostics 2012, 22, 323–334. 10.1089/nat.2012.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejad A. J.; Shahrokhi N.; Nielsen P. E. Targeting of the Essential acpP, ftsZ, and rne Genes in Carbapenem-Resistant Acinetobacter baumannii by Antisense PNA Precision Antibacterials. Biomedicines 2021, 9, 429. 10.3390/biomedicines9040429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlecova Z.; Baldi L.; Hacker D.; Wurm F. M.; Klok H.-A. Comparative Study on the In Vitro Cytotoxicity of Linear, Dendritic, and Hyperbranched Polylysine Analogues. Biomacromolecules 2012, 13, 3127–3137. 10.1021/bm300930j. [DOI] [PubMed] [Google Scholar]
- Yavari N.; Goltermann L.; Nielsen P. E. Uptake, Stability, and Activity of Antisense Anti-acpP PNA-Peptide Conjugates in Escherichia coli and the Role of SbmA. ACS Chem. Biol. 2021, 16, 471–479. 10.1021/acschembio.0c00822. [DOI] [PubMed] [Google Scholar]
- Ghosal A.; Vitali A.; Stach J. E. M.; Nielsen P. E. Role of SbmA in the Uptake of Peptide Nucleic Acid (PNA)-Peptide Conjugates in E-coli. ACS Chem. Biol. 2013, 8, 360–367. 10.1021/cb300434e. [DOI] [PubMed] [Google Scholar]
- Puckett S. E.; Reese K. A.; Mitev G. M.; Mullen V.; Johnson R. C.; Pomraning K. R.; Mellbye B. L.; Tilley L. D.; Iversen P. L.; Freitag M.; Geller B. L. Bacterial Resistance to Antisense Peptide Phosphorodiamidate Morpholino Oligomers. Antimicrob. Agents Chemother. 2012, 56, 6147–6153. 10.1128/aac.00850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabas I. M.; Nielsen P. E. Effective Cellular Delivery of Antisense Peptide Nucleic Acid by Conjugation to Guanidinylated Diaminobutanoic Acid-Based Peptide Dendrons. Biomacromolecules 2020, 21, 472–483. 10.1021/acs.biomac.9b01227. [DOI] [PubMed] [Google Scholar]
- Mellbye B. L.; Puckett S. E.; Tilley L. D.; Iversen P. L.; Geller B. L. Variations in Amino Acid Composition of Antisense Peptide-Phosphorodiamidate Morpholino Oligomer Affect Potency against Escherichia coli In Vitro and In Vivo. Antimicrob. Agents Chemother. 2009, 53, 525–530. 10.1128/aac.00917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A. M.; Bonke G.; Larsen C. J.; Yavari N.; Nielsen P. E.; Franzyk H. Antibacterial Peptide Nucleic Acid-Antimicrobial Peptide (PNA-AMP) Conjugates: Antisense Targeting of Fatty Acid Biosynthesis. Bioconjugate Chem. 2016, 27, 863–867. 10.1021/acs.bioconjchem.6b00013. [DOI] [PubMed] [Google Scholar]
- Goltermann L.; Yavari N.; Zhang M.; Ghosal A.; Nielsen P. E. PNA Length Restriction of Antibacterial Activity of Peptide-PNA Conjugates in Escherichia coli Through Effects of the Inner Membrane. Front. Microbiol. 2019, 10, 1032. 10.3389/fmicb.2019.01032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbalán N. S.; Adler C.; De Cristóbal R. E.; Pomares M. F.; Delgado M. A.; Vincent P. A. The tolC locus affects the expression of sbmA through Sigma E activity increase. FEMS Microbiol. Lett. 2010, 311, 185–192. 10.1111/j.1574-6968.2010.02090.x. [DOI] [PubMed] [Google Scholar]
- Mattiuzzo M.; Bandiera A.; Gennaro R.; Benincasa M.; Pacor S.; Antcheva N.; Scocchi M. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 2007, 66, 151–163. 10.1111/j.1365-2958.2007.05903.x. [DOI] [PubMed] [Google Scholar]
- Frimodt-Moller J.; Campion C.; Nielsen P. E.; Lobner-Olesen A. Translocation of non-lytic antimicrobial peptides and bacteria penetrating peptides across the inner membrane of the bacterial envelope. Curr. Genet. 2021, 68, 83. 10.1007/s00294-021-01217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison S.; Wallace J.; Harris F.; Phoenix D. Amphiphilic alpha-helical antimicrobial peptides and their structure/function relationships. Protein Pept. Lett. 2005, 12, 31–39. 10.2174/0929866053406084. [DOI] [PubMed] [Google Scholar]
- Maloy W. L.; Kari U. P. Structure-Activity Studies on Magainins and Other Host-Defense Peptides. Biopolymers 1995, 37, 105–122. 10.1002/bip.360370206. [DOI] [PubMed] [Google Scholar]
- Yeaman M. R.; Yount N. Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Frimodt-Moller J.; Koulouktsis A.; Charbon G.; Otterlei M.; Nielsen P. E.; Lobner-Olesen A. Activating the Cpx response induces tolerance to antisense PNA delivered by an arginine-rich peptide in Escherichia coli. Mol Ther-Nucl Acids 2021, 25, 444–454. 10.1016/j.omtn.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdish D.; Davidson D.; Hancock R. A re-evaluation of the role of host defence peptides in mammalian immunity. Curr. Protein Pept. Sci. 2005, 6, 35–51. 10.2174/1389203053027494. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. W.; Sahl H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Hansen F.; Olsen S. S.; Heltberg O.; Justesen U. S.; Fuglsang-Damgaard D.; Knudsen J. D.; Hammerum A. M. Characterization of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. Microb. Drug Resist. 2014, 20, 316–324. 10.1089/mdr.2013.0157. [DOI] [PubMed] [Google Scholar]
- Brolin C.; Lim E. W. K.; Grizot S.; Olsen C. H.; Yavari N.; Krag T. O.; Nielsen P. E. Approaches for Systemic Delivery of Dystrophin Antisense Peptide Nucleic Acid in the mdx Mouse Model. Nucleic Acid Nanotheranostics 2021, 31, 208–219. 10.1089/nat.2020.0856. [DOI] [PubMed] [Google Scholar]
- McMahon B. M.; Mays D.; Lipsky J.; Stewart J. A.; Fauq A.; Richelson E. Pharmacokinetics and tissue distribution of a peptide nucleic acid after intravenous administration. Antisense Nucleic Acid Drug Dev. 2002, 12, 65–70. 10.1089/108729002760070803. [DOI] [PubMed] [Google Scholar]
- Ganguly S.; Chaubey B.; Tripathi S.; Upadhyay A.; Neti P. V. S. V.; Howell R. W.; Pandey V. N. Pharmacokinetic analysis of polyamide nucleic-acid-cell penetrating peptide conjugates targeted against HIV-1 transactivation response element. Oligonucleotides 2008, 18, 277–286. 10.1089/oli.2008.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelesidis T.; Falagas M. E. The safety of polymyxin antibiotics. Expert Opin. Drug Saf. 2015, 14, 1687–1701. 10.1517/14740338.2015.1088520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand I.; Hilpert K.; Hancock R. E. W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.