Abstract
Background
HIV-1 drug resistance genotyping is critical to the monitoring of antiretroviral treatment. Data on HIV-1 genotyping success rates of different laboratory specimen types from multiple sources is still scarce.
Methods
In this cross-sectional study, we determined the laboratory genotyping success rates (GSR) and assessed the correlates of genotyping failure of 6837 unpaired dried blood spot (DBS) and plasma specimens. Specimens from multiple studies in a resource-constrained setting were analysed in our laboratory between 2016 and 2019.
Results
We noted an overall GSR of 65.7% and specific overall GSR for DBS and plasma of 49.8% and 85.9% respectively. The correlates of genotyping failure were viral load (VL) < 10,000 copies/mL (aOR 0.3 95% CI: 0.24–0.38; p < 0.0001), lack of viral load testing prior to genotyping (OR 0.85 95% CI: 0.77–0.94; p = 0.002), use of DBS specimens (aOR 0.10 95% CI: 0.08–0.14; p < 0.0001) and specimens from routine clinical diagnosis (aOR 1.4 95% CI: 1.10–1.75; p = 0.005).
Conclusions
We report rapidly decreasing HIV-1 genotyping success rates between 2016 and 2019 with increased use of DBS specimens for genotyping and note decreasing median viral loads over the years. We recommend improvement in DBS handling, pre-genotyping viral load testing to screen samples to enhance genotyping success and the development of more sensitive assays with well-designed primers to genotype specimens with low or undetectable viral load, especially in this era where virological suppression rates are rising due to increased antiretroviral therapy roll-out.
Keywords: HIV-1, Genotypic resistance testing, Success rates, DBS, Plasma, Resource-limited settings
Importance
Genotypic resistance testing is crucial to HIV treatment as it guides on treatment decisions such as when to switch to which HIV regimens for individuals on HIV antiretroviral therapy. This is vital in achieving HIV virological suppression, a key outcome in minimizing the transmission of HIV. In resource-constrained settings, the use of DBS specimens has been popularised to circumvent logistical challenges associated with plasma, but genotyping success rates (GSR) of DBS are frustrating. We report lower GSR of DBS compared to plasma specimens and note that low GSR were associated with viral loads < 10,000 copies/mL, DBS specimens and specimens from routine clinical diagnosis. We highlight that as VLs continue to decrease due to suppression following ART rollout, we need more robust laboratory assays to genotype DBS specimens and emphasize the essence of proper handling of DBS specimens prior to genotyping. These findings are of fundamental public health importance as they may be used to improve laboratory performances in settings like ours and improve HIV treatment monitoring and outcomes.
Introduction
Sub-Saharan Africa remains the worst hit by the HIV/AIDS epidemic with 20.7 million of the global estimated 38 million HIV-infected individuals by 2020 [1]. A tremendous achievement has been realised in increasing access to antiretroviral therapy (ART) among individuals living with HIV [1]. This is a huge milestone in combating the epidemic since ART suppresses viral replication, reduces the morbidity associated with HIV/AIDS and curbs HIV transmission to uninfected individuals [2]. Increased access to ART comes with a challenge of increasing prevalence of HIV drug resistance (HIVDR) [3, 4] which compromises ART benefits [5]. Monitoring of HIVDR remains a priority in the WHO strategy for combating the HIV/AIDS epidemic. The new UNAIDS 2025 targets call for 95% of HIV-infected individuals becoming aware of their status, initiation of 95% of those with known HIV-positive status on treatment and having 95% of those on ART achieving virological suppression [6, 7]. To achieve these targets, virological monitoring and HIV genotypic resistance testing of individuals on ART are necessary as recommended by the WHO [7]. Viral load testing provides a basis for ART response monitoring and guides the need to switch drug regimen classes [2]. Guidelines in resource-rich settings recommend HIV-infected individuals to undergo viral load testing at ART initiation or modification. Due to transmitted HIVDR, genotypic resistance testing for mutations in the reverse transcriptase and protease genes is recommended prior to ART initiation or modification to guide selection of ART regimen [8]. In contrast, guidelines in resource-constrained settings like Uganda recommend viral load testing for adults 6 months post -ART initiation and subsequently every 12 months for those with viral load suppression. Also, genotypic resistance testing is only recommended for adult individuals failing on 2nd line and 3rd line regimes, and those failing on protease inhibitor-based or dolutegravir-containing regimens [2]. To circumvent economic barriers that impede ART monitoring in resource-constrained settings, the WHO has recommended a relatively cost-effective public health approach for surveillance of acquired [9] and pre-treatment [10] HIVDR in resource limited settings. However, this too, requires HIVDR genotyping.
HIV-1 genotypic testing is a nucleic acid amplification and sequencing test aimed at detecting the existence of HIVDR mutations in targeted regions of the HIV-genome such as protease, reverse transcriptase and integrase genes [11, 12]. It also examines HIV diversity and generates data used for molecular epidemiological and evolutionary studies [13, 14]. The interpretation of HIVDR genotyping data is done by a clinical expert or with the aid of an HIVDR database [15]. HIV genotyping consists of, extraction and purification of the viral nucleic acid, amplification of the target gene and sequencing of the amplicons.
Plasma specimens are preferred to dried plasma/serum and blood spot specimens for HIV genotyping in most research studies and clinical applications [16]. This is because plasma sequence data represents actively-replicating viruses which form a large proportion of viruses circulating in the body [17]. However, the need for cold-chain and associated transportation and storage costs have made plasma less appropriate in resource-constrained settings, especially rural areas. Consequently, dried blood spot (DBS) specimens offer a cheaper alternative to the conventional use of plasma for virological monitoring [18–20] and HIV genotypic resistance testing [21–25]. The preference of DBS to plasma specimens is because DBS specimens are easy to collect, process, transport and can be kept at ambient temperature for relatively longer time and thus do not require a costly and a not-readily available cold-chain in resource-constrained settings [18, 21, 22]. Furthermore, many studies report concordance between paired plasma and DBS specimens for HIV genotyping [21–23, 25, 26], although one study reported discordance based on next-generation sequencing [27]. Genotyping success rates of DBS specimens are, however, continually variable due to several factors.
Following the renewed global efforts to eliminate the public threat posed by HIV/AIDS [6], HIV genotypic drug resistance testing is crucial to the success of ART programmes. The clinical community is reliant on results of HIV genotypic tests in making important clinical decisions such as which drug regimen to offer and when to switch to another regimen class. It is therefore imperative for laboratories to ascertain the success rates of specimens they analyse. Currently in our setting, there is a paucity of data on the genotyping success rates of unpaired DBS and plasma specimens. In the current study, we retrieved the data of DBS and plasma specimens analysed in the MRC/UVRI and LSHTM virology laboratory between 2016 and 2019 and evaluated the genotyping success rates of both DBS and plasma specimens. In addition, we assessed the possible correlates of genotyping failure of both specimen types. This data is vital for quality assurance assessment and is informative in decisions for improving not just the performance of the laboratory assays but also the management of studies and clinical facilities from which specimens are obtained.
Methods and materials
Study design and setting
In this cross-sectional study, we analysed the yield of genotyping success, herein referred to as genotyping success rates (GSR), with 6837 unpaired specimens of which 3836 were DBS and 3001 were plasma specimens that were brought to our laboratory for HIVDR genotyping between 2016 and 2019. All these specimens were coming from within Uganda except for cross-border DBS specimens that were shipped at ambient temperatures in sealed envelopes from Malawi. Our standard operation procedures for sample reception were used to accept the specimens (both plasma and DBS). Plasma specimens were accepted if they were correctly labelled, and the labels matched with the accompanying laboratory request forms. DBS specimens were accepted if they came with clear information that matched the paperwork, had at least three blood spots with a dark uniform colour and packed in glassine envelope, placed in a gas-impermeable, zip closure plastic bags [28, 29]. For studies that used plasma specimens, we accepted plasma with sufficient volumes in intact tubes (not leaking), stored at – 80 °C at collection points and transported in liquified nitrogen tanks within 3 days from the collection date [29]. The methods of transportation of laboratory specimens that we followed have been published elsewhere [30].
Data cleaning and processing
We retrieved sequencing data from our laboratory database and categorised specimen by year of sequencing and specimen type. We defined genotyping success of a specimen as completion of the entire process of specimen preparation, extraction, amplification, and generation of good quality HIV sequences (genotypes). Good quality sequences were those whose chromatograms were evenly-spaced peaks, each representing a single nucleotide and having no or very minimal baseline “noise,” if any [31]. The genotyping success rates refer to the percentage proportions of specimens whose specimens yield good quality nucleotide sequences, with the denominator being the total number of specimens presented for genotyping. Specimens were from different studies and clinical facilities in which individuals ranged from children to adults on ART. The dependent variable of interest was the genotyping result, either success or failure. The independent variables analysed were viral load, specimen type, source of the specimen (research study or routine clinical diagnosis). Research studies strictly adhere to clear standard operating procedures (SOPs) and possess proper facilities to ensure specimen integrity while specimens from routine clinical diagnosis came from health facilities, public clinics/laboratories which are not entirely research focused. As such, some facilities may have less stringent adherence to SOPs and insufficient infrastructure to ensure sample integrity. We excluded specimens whose genotyping results were missing in our database.
PCR amplification and HIV genotypic drug resistance testing
Viral load tests for both plasma and DBS specimens were done using Roche CoBAS TaqMan® platforms (Roche Molecular Systems, Branchburg, NJ USA) with methods described by Pollack et al. [32]. The modification made for DBS specimens was the incubation of two blood spots incubated for 10 min with 1 mL of sample pre-extraction (SPEX) buffer in a thermomixer set at 1000 rpm. The sample was then processed using the COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test v2.0 kit (Roche Molecular Systems, Branchburg, NJ) and the dried fluid spot procedure protocol (H12DFSP96) following the manufacturer’s instructions and as described earlier [32].
Both DBS and plasma specimens received in the laboratory were stored at -800C prior to HIVDR testing.
In processing the DBS specimens, we excised three spots using a DBS puncher, into a 2 mL NucliSENs lysis buffer (Biomereux, Germany), and lysis occurred on a roller mixer for 1 h at room temperature. For plasma specimens, 100 µL of specimen was added into a 2 mL NucliSENS lysis buffer and lysis done as already stated for DBS specimens. For both DBS and plasma specimens, we used a NucliSENS MiniMAG® (Biomerieux, Marcy l’Etoile, France) system to extract total nucleic, eluting 30 µL of RNA following the manufacturer’s instructions and our in-house validated assay. RNA from plasma specimens received in 2016 (before we introduced NucliSENS MiniMAG®) had been extracted using the Qiagen kit as previously described [33].
We used 10 µL of RNA for either type of specimens for reverse transcription and complementary DNA (cDNA) synthesis with superscript III high-fidelity one-step PCR kit (Invitrogen), followed by a nested PCR with 2 µL of the cDNA, using a validated inhouse assay [22, 29]. The HIV-1 polymerase sequences of the complete HIV-1 protease gene (1–99 amino acids) and the reverse transcriptase gene (1–320 amino acids) were realised from chromatogram data using either RECall [34] or Sequencher (Gene codes Corporation, Ann Arbor, MI, USA). For purposes of quality control, our laboratory subscribes to the virology quality assurance (VQA) programme: All sequences generated in the laboratory were examined for any possible cross-contamination by creating maximum-likelihood phylogenetic trees with 1000 bootstraps using RaxML [35]. The sequences that passed the HIVDR VQA test were analysed for HIV drug resistance mutations using the Stanford HIVdb [36].
Statistical analysis
We did statistical analyses using Stata v15 (StataCorp, USA). Medians and interquartile ranges (IQR) were used for continuous variables while frequencies and proportions were used for categorical variables. Univariate logistic regression analysis was done to examine the association of independent variables with the outcomes of interest. Only variables with a p < 0.2 in bivariate analysis were entered to adjusted multivariate logistic regression models. To be considered significantly associated with the outcome, only variables with a p < 0.05 were reported. We presented the results of adjusted odds ratios (aOR) with 95% confidence intervals (CI).
Results
Profile of DBS and plasma specimens analysed
Between 2016 and 2019, we tested 6837 specimens for HIV drug resistance, 3836 (56.1%) were DBS while 3001 specimens (43.9%) were plasma. Over this period, the number of specimens generally rapidly increased with a slight decline from 2018 to 2019. The number of DBS specimens exponentially increased over the years while the number of plasma specimens rapidly decreased (Table 1). Overall, research studies contributed 51.7% (3533/6837) while routine clinical diagnosis provided the rest of the specimens received in our laboratory. Most plasma specimens, 75.6% (2670/3533) were from research studies while most of the DBS specimens received, 90% (2973/3304), were from routine clinical diagnosis (Table 1). The increase in DBS was attributed to the roll out of DBS for the national viral load programme and the subsequent use of remnant DBS for HIVDR testing among patients failing ART in Uganda.
Table 1.
Profile all laboratory specimens analysed between 2016 and 2019
| Overall total | For sample type | ||
|---|---|---|---|
| DBS | Plasma | ||
| Number analysed | 6837 | 3836 | 3001 |
| Proportion (%) | 100 | 56.1 | 43.9 |
| Successfully genotyped | 4489 | 1910 | 2579 |
| Genotyping success rate (%) | 4489/6837 (65.7) | 1910/3836 (49.8) | 2579/3001 (85.9) |
| Year of laboratory analysis N (%) | |||
| 2016 | 1990 | 89/1990 (4.5) | 1901/1990 (95.5) |
| 2017 | 1001 | 484/1001 (48.4) | 517/1001 (51.6) |
| 2018 | 1699 | 1302/1699 (76.6) | 397/1699 (23.4) |
| 2019 | 2147 | 1961/2147 (91.3) | 186/2147 (8.7) |
| Specimen source | |||
| Research studies | 3533 | 863/3533 (24.4) | 2670/3533 (75.6) |
| Routine clinical diagnosis | 3304 | 2973/3304 (90.0) | 331/3304(10.0) |
| Pre-genotyping viral load testing | |||
| Not done | 3893 | 1884/3893 (48.4) | 2009/3893 (51.6) |
| Done | 2944 | 1952/2944 (66.3) | 992/2944 (33.7) |
| Median viral load for known | 4.31log10 (3.57–4.93log10) | 4.12log10 (3.43–4.8log10) | 4.62 log (3.94–5.13log10) |
Viral load testing and viral load trends
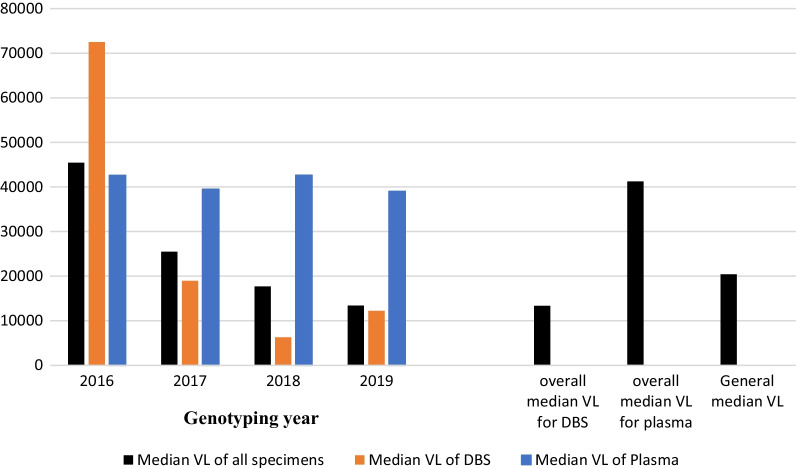
Of the 6837 specimens, pre-genotyping viral load testing had been done for 43.1% (2944/6837) of the specimens (Table 1). Out of 2944 specimens with pre-genotyping viral load testing results, 66.3% (1952/2944) were DBS specimens while the rest were plasma specimens. Each of the DBS specimens with viral load results was shipped with two DBS cards, one for viral load testing and another for genotypic resistance testing. The overall median viral load for all the specimens with prior VL testing results was 4.31 log10 [IQR: 3.57 log10-4.93 log10)]. Over the 4 years, the overall median VL of plasma specimens [4.62 log (IQR: 3.94–5.13 log10)] was higher than for DBS specimens [4.12 log10 (IQR: 3.43–4.8 log10)]. We noted median VLs of the specimens presented for genotyping rapidly decreasing over the years (Fig. 1). This trend corresponds to the increase in the proportion of the DBS specimens (with relatively lower median VLs) analysed.
Fig. 1.
Trends in the median VLs of the specimen genotyped over the years. Median VLs were determined for all specimens analysed in each year. Overall median VLs for DBS and plasma, and General median VLs represent values obtained for specimens over the 4 years
Genotyping success rates of HIV DBS and plasma specimens (2016–2019)
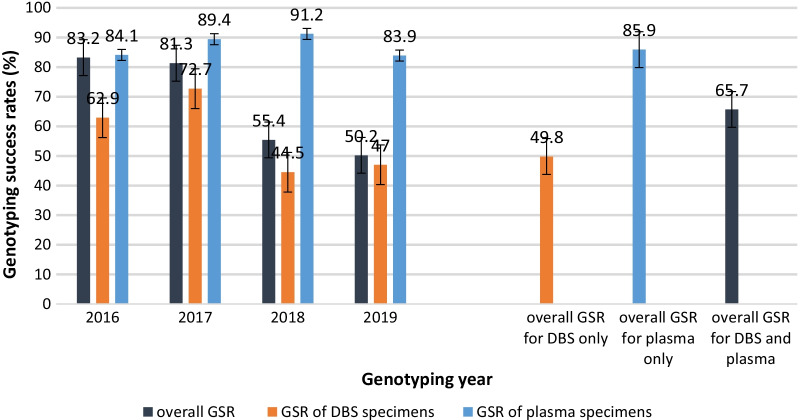
We successfully genotyped 4489 specimens between 2016 and 2019, thus an overall GSR of 65.7% for both specimen types over the 4-year period. From 2016 to 2019, the overall GSR (for both specimen types) generally decreased, with the highest overall GSR of 83.2% attained in 2016 and the lowest GSR of 50.2% recorded in 2019 (Fig. 2). We noted an overall DBS GSR of 49.6% which was considerably lower than 85.9% GSR of plasma over the analysis period (Fig. 2). The GSR of DBS specimens also decreased in the same period; from 62.9% in 2016 to 47% in 2019. There was however a gradual increase in GSR of DBS between 2016 and 2017, with highest GSR of 72.7% realised in 2017. Conversely, the GSR of plasma increased gradually from 2016 to 2018 and slightly decreased between 2018 and 2019 (Fig. 2).
Fig. 2.
Trends in the overall and genotyping success rates of DBS and plasma specimens from 2016 to 2019. Overall genotyping success rates (GSR) were obtained by analysing all specimens genotyped in the 4 years (2016–2019). The GSR was a proportion of specimens that yielded clean sequences (genotypes) with the denominator being the total number of specimens that we attempted to genotype in that year or in all the 4 years for the overall GSR (see Table 1 for denominator values). The analysis was also independently done for DBS and plasma specimens
Correlates of HIV-1 genotyping failure
In Table 2, we determined the independent correlates of genotyping failure as low viral loads, DBS specimens, lack of pre-genotyping viral load testing, and genotyping of specimens from routine clinical diagnosis.
Table 2.
Correlates of genotyping failure for all specimens analysed from 2016 to 2019
| Predictor Variable | Genotyping failure n/N (%) | Unadjusted OR | p-value | Adjusted OR | p-value |
|---|---|---|---|---|---|
| Specimen type | |||||
| DBS | 1926/3836 (50.2) | 1 | < 0.0001 | < 0.0001 | |
| Plasma | 422/3001 (14.1) | 0.16 (0.14–0.18) | 0.10 (0.08–0.14) | ||
| Pre-genotyping viral load testing | |||||
| Not done | 1398/3893 (35.9) | 1 | 0.002 | 0.023 | |
| Done | 950/2944 (32.3) | 0.85 (0.77–0.94) | 0.64 (0.13–0.56) | ||
| Viral load | |||||
| 1001–10,000 cps/mL | 634/1165 (54.4) | 1 | < 0.0001 | < 0.0001 | |
| > 10,000–50,000 cps/mL | 197/755 (25.4) | 0.28 (0.23–0.35) | 0.3 (0.24–0.38) | ||
| > 50,000–200,000 cps/mL | 81/609 (13.3) | 0.13 (0.10–0.17) | 0.14 (0.11–0.19) | ||
| > 200,000 cps/mL | 38/392 (9.7) | 0.09 (0.06–0.13) | 0.10 (0.07–0.14) | ||
| Specimen source | |||||
| Research studies | 745/3533 (21.1) | 1 | < 0.0001 | 0.005 | |
| Routine clinical diagnosis | 1603/3304 (48.5) | 3.5 (3.17–3.92) | 1.4 (1.10–1.75) | ||
Each denominator represents the number of specimens of that specific category
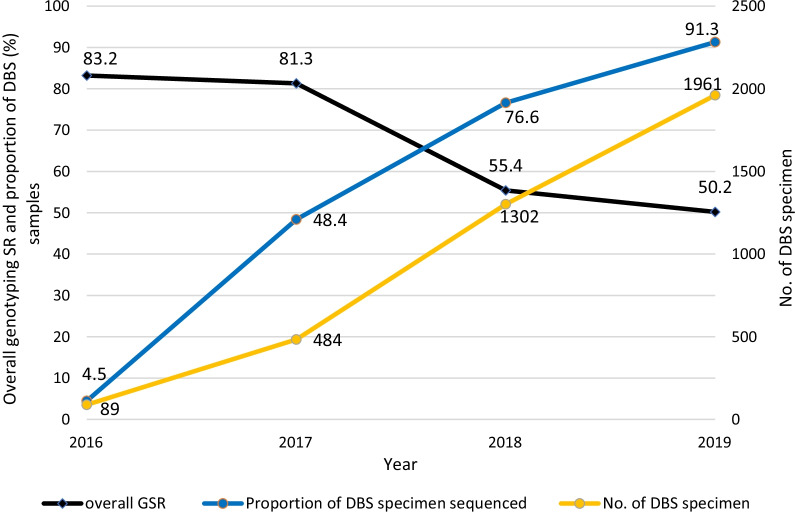
The specimen source was independently associated with genotyping failure. Specimens that originated from routine clinical diagnosis had significantly higher odds of failing the genotypic test compared to research studies (aOR 1.4 95% CI: 1.10–1.75, p = 0.005). In addition, we noted that the DBS specimens were 90% more likely to fail genotypic testing compared to plasma specimens (aOR 0.10 95% CI: 0.08–0.14; p = 0.0001). The overall genotyping success rates (GSR) decreased as both the number and proportion of DBS specimens we tested increased from 2016 to 2019 (Fig. 3).
Fig. 3.
Variations in genotyping success rates, number and proportion of DBS specimens genotyped with time. The proportion of DBS specimens represents the number of DBS specimens being the numerator and the denominator being the total number of specimens we attempted to genotype in a particular year (see Table 1). The overall genotyping success rates (GSR) decreased as both the number and proportion of DBS specimens we attempted to genotype increased from 2016 to 2019
Specimens analysed without pre-genotyping viral load testing failed genotypic resistance testing more frequently than those where pre-genotyping viral load testing had been done. Lack of pre-genotyping viral load testing significantly increased the odds of genotypic resistance testing by 36% (aOR 0.64 95% CI: 0.13–0.56; p = 0.001) (see Table 2).
Specimens with viral loads < 10,000 copies/mL were more likely to fail a genotyping test compared to those with viral loads > 10,000 copies/mL. Specimens with VL > 10,000–50,000 were 70% less likely to fail genotypic testing than specimens with VL < 10,000 copies/mL (aOR 0.3 (95% CI: 0.24–0.38, p < 0.0001). The odds of failing genotypic testing decreased with increased viral load. Specimens with VL > 200,000copies/mL had only 10% chances of failing a genotypic test (aOR 0.10 (95% CI:0.07–0.14), p < 0.0001) (Table 2).
Correlates of genotyping failure of DBS specimens
On realising the underwhelming genotypic success rates of DBS specimens, we assessed the possible predictors of genotyping failure and found that low viral loads, lack of pre-genotyping viral load testing and DBS specimens from routine clinical diagnosis correlated with genotyping failure (Table 3).
Table 3.
Correlates of DBS specimens genotyping failure
| Factor | Genotyping failure n (%) | Odds ratio (95% CI) | p-value | Adjusted OR [aOR (95% CI)] | p-value |
|---|---|---|---|---|---|
| Pre-genotyping viral load testing | |||||
| Not done | 1038/1885 (55.1) | 1 | < 0.0001 | 0.001 | |
| Done | 888/1952 (45.5) | 0.68 (0.6–0.77) | 0.71 (0.16–0.22) | ||
| Viral load | |||||
| 1000–10,000 cps/mL | 599/897 (66.8) | 1 | < 0.0001 | < 0.0001 | |
| > 10,000–50,000 cps/mL | 185/497 (37.2) | 0.29 (0.23–0.37) | 0.3 (0.24–0.38) | ||
| > 50,000–200,000 cps/mL | 73/344 (21.2) | 0.13 (0.11–0.18) | 0.14 (0.10–0.18) | ||
| > 200,000 cps/mL | 31/214 (14.5) | 0.08 (0.06–0.13) | 0.07 (0.06–0.13) | ||
| Specimen source | |||||
| Research studies | 48/137 (35.0) | 1 | < 0.0001 | 0.006 | |
| Routine clinical diagnosis | 1574/3279 (48.0) | 1.72 (1.36–2.18) | 1.4 (1.11–1.84) | ||
Each denominator represents the number of specimens of that specific category
DBS specimens that were analysed without pre-genotyping viral load testing had 29% more chances of failing genotyping testing than those DBS specimens with pre-genotyping viral load test results (aOR 0.71 95% CI: 0.16–0.22, p < 0.001).
Low viral loads < 10,000 copies/mL were significantly associated with DBS genotyping failure. We noted that the odds of genotyping failure of DBS specimens significantly decreased with increasing viral load. Specimens with VL > 200,000 copies/mL had the lowest odds of failing genotypic compared to specimens with lower VL (aOR 0.07 95% CI: 0.06–0.13, p < 0.0001).
Genotyping of DBS specimens from routine clinical diagnosis significantly increased the odds of failing the genotypic resistance testing (aOR 1.4 95% CI: 1.11–1.84, p = 0.006).
Discussion
HIV drug resistance genotyping has revolutionised the clinical management of individuals on ART because it informs decisions like when to implement regimen switch. Successful genotyping in the laboratory is critical to patient care. In this cross-sectional study, we examined the genotyping success rates of unpaired DBS and plasma specimens analysed in a WHO-designated HIVDR laboratory in a resource-constrained setting in Uganda and assessed the correlates of specimen genotyping failure.
From 2016 to 2019, we obtained an overall GSR of 65.7% for all specimens delivered for analysis in our laboratory, but the overall GSR for plasma was higher (85.9%) than that of DBS specimens (49.8%) over that 4-year period. The plasma genotyping success rate in this study was lower than the 100% genotyping success rate that was realised in our previous study [37]. Similarly, the genotyping success rate of DBS specimens reported here is also lower than that reported by Zhang et al. in a survey in Kenya and Tanzania [38]. The comparatively higher genotyping success rates in those studies could be attributed to them being well controlled research studies in which the study staff were well trained, monitored, and specimen collection, transportation and storage done according to well-designed protocols. In contrast, this study had specimens from both research studies and routine clinical diagnosis including cross-border specimens that required lengthy shipping procedures, with most specimens from routine clinical diagnosis contributing significantly to genotyping failure (p < 0.0001). Though not part of our analyses, we have in some cases received DBS specimens 3 months from the collection date. Aware of inadequate facilities in most of these facilities, it is possible that DBS specimens shipped to our laboratory for genotyping were probably not handled under optimum conditions that maintain sample integrity.
Studies have shown that better genotyping success rates are realised when DBS specimens are kept at ambient temperatures for 2 weeks and stored frozen at – 80 °C prior to shipping, and that prolonged storage for over 2 weeks at ambient temperature reduced genotyping success rates [22, 39]. Furthermore, most research studies prepared their DBS cards with venous blood while most routine clinical facilities preferred finger prick DBS. Parry et al. showed that venous DBS had better genotyping success rates compared to finger prick DBS [22]. This probably explains why specimens from research studies had better genotyping success rates. It is probable that the site of collection of blood for DBS specimen preparation could have impacted on the genotyping success rates of the DBS samples. It will be interesting for future studies to assess the various DBS preparation methods and their impact on GSR of DBS specimens.
Generally, the genotyping success rates of DBS specimens were substantially lower than those of plasma over the 4 years. Our analysis revealed that over the 4 years, the overall GSR rapidly decreased with increasing number of DBS specimens presented for genotyping. The observation that the overall GSRs were decreasing in a similar trend as genotyping success rates of DBS specimens, further asserts that DBS specimens, to a large extent, contributed to overall genotyping failure. Our logistic regression analysis showed that DBS were at 90% more odds of genotyping failure compared to plasma specimens (aOR 0.10; 95% CI: 0.08–0.14; p < 0.0001) correlating DBS specimens to genotyping failure. This probably arose from the large proportion of DBS specimens originating from routine clinical diagnosis that face significant challenges in terms of specimen collection, transportation, and storage. Moreover, DBS have considerably gained preference to plasma because DBS specimens are easy to collect, transport and do not require a cold chain [22, 40]. This explains why we received more DBS than plasma specimens between 2017 and 2019. The increase in DBS shipped to our laboratory was also attributed to the roll out of DBS for the national viral load programme and the subsequent use of remnant DBS for HIVDR testing among patients failing ART in Uganda. This popularity should be followed by precautions to ensure that specimen quality is not compromised on the way to the laboratory for genotyping. Evidence from other studies suggests that DBS specimen yield appreciable genotyping rates when specimens are properly handled to ensure nucleic acid integrity [38, 41].
The higher genotyping success rates of plasma compared to DBS specimens reported here mirrors with findings of other researchers [26, 42, 43]. The better GSR of plasma could probably be attributed to a higher rate of HIV-1 RNA degradation for DBS compared to plasma because of the extra processing time at ambient temperature. Temperature and humidity at which DBS specimens are stored affects the stability and integrity of nucleic acids embedded on filter papers [44]. Despite the lower GSR of DBS compared to plasma in our HIVDR genotyping laboratory, we cannot undermine the reliability of DBS specimens as a suitable alternative to plasma especially in resource-constrained settings. Studies by Rottinghaus et al. [45] and Diouara et al. [46] both corroborate the suitability of DBS for HIV drug resistance testing.
For both DBS and plasma specimens, we noted a significant association between VL levels and HIVDR genotyping rates: As expected, VLs < 10,000 copies/mL were associated with genotyping failure (p < 0.0001). A similar observation was also noted when DBS specimen were analysed alone. In studies that examined the use of DBS for genotyping, DBS with VL < 10,000 copies/mL had reduced genotyping success rates [22, 27]. Our findings also concur with studies that cited VL as the major determinant of concordance between plasma and DBS especially when viral load ≥ 5000 copies/mL [21, 25, 27, 47]. Studies analysing paired DBS and plasma specimens report significant correlation and strong concordance in RNA levels between plasma and DBS specimens [48, 49]. Similar to findings in this study, viral loads considerably influenced genotyping success rates in a survey carried out in Kenya and Tanzania where DBS specimens with VLs ≥ 5000 copies/mL achieved genotyping success rate of 90% [38]. Interestingly, in our study, a substantial proportion of specimens had VL < 10,000 copies/mL and could explain why the overall GSR was only 65.7%. This implies that in this era when virological suppression (VL < 1000 copies/mL) rates nearing or exceeding 90% have been observed [37, 41], and following recommendations to use more potent dolutegravir-containing regimens, genotyping success rates are likely to continue to decline. There is evidence in this study that shows median viral loads of specimens analysed in our laboratory rapidly decreasing from 2016 to 2019. This can be explained by rapid scale-up of ART following recommendations of the WHO and subsequent high VL suppression rates reported in our setting [41]. What is more intriguing is that even at VL < 1000 copies/mL, HIVDR variants can still be selected as reported in a Kenyan study in which plasma samples with VL ≤ 1000 copies/mL had a genotyping success rate of 32% [50]. Ultimately, robust techniques need to be adopted to ensure that genotyping assays are adapted to successfully genotype both plasma and DBS specimens at VL < 1000 copies/mL. Some studies have already demonstrated the possibility of successful genotyping at low and undetectable viremia [50–52].
The findings of our study also highlight the importance of virological monitoring. Our findings show that 53.9% of the specimens analysed were brought for genotyping without prior viral load testing. This is expected in our setting where viral load testing is not widely available for all HIV-infected individuals due to high costs. We noted significantly higher odds of genotyping failure in specimens in which prior viral load testing was not done, both in the general analysis of all samples (aOR 0.64 95% CI: 0.13–0.56; p = 0.001) and even when DBS specimens were analysed alone (OR 0.68 95% CI: 0.6–0.77; p < 0.0001). It is possible that the specimens that were not subjected to pre-genotyping VL testing had VL < 5000 copies/mL, the range at which genotyping failure is more likely [22, 38]. Pre-genotyping viral load testing could be essential in qualifying specimens for HIVDR genotyping. Without pre-genotyping VL testing, time and resources may be wasted on genotyping non-eligible specimens resulting in low GSR and increasing the turn-around time. A study in Malawi reported that a delayed turnaround time in their laboratories was associated with lack of viral load testing and the use of DBS compared to plasma [53]. In our unpublished data, we noted that paediatric plasma and DBS samples with substantial genotyping failure turned out to be virally suppressed when we did viral load testing.
Despite the challenges of using plasma specimens in our setting, they still yield better genotyping success rates than DBS. The use of DBS specimens is a plausible option to plasma and whole blood due to its advantages which include the ease of collection, transportation, and storage. The WHO recommends the use of DBS specimens for viral load monitoring in resource-constrained settings where infrastructural and logistic deficiencies hinder the use of plasma specimens. Interestingly, a viral load threshold of ≥ 1000 copies/mL used to define virological failure with plasma specimens is also applicable to DBS specimens [7, 16]. Studies collaborate this with findings of high sensitivity and specificity of DBS specimens at a VL ≥ 1000 copies/mL [32, 54]. Any form of specimen that is used in HIVDR genotyping requires maximum effort towards ensuring that specimen collection, processing, transportation, storage, and testing is done appropriately so that accurate and reliable results are obtained.
This cross-sectional study is limited by several factors: The specimens analysed came from multiple sources, some of which were routine clinical diagnosis laboratories, which are not research-focused, thus, strict adherence to SOPs for proper specimen collection and handling cannot be guaranteed. Therefore, the widespread genotyping failure in most of the specimens from routine clinical diagnosis could have led to over-estimation of genotyping failure rates. Also, our analyses did not involve paired plasma and DBS specimens and as such our comparisons could have been influenced by other demographic and clinical variations in individuals from whom the specimens were collected. Despite these shortcomings, the findings herein remain applicable to other laboratories in our setting and this study will set precedence for further research into strategies of improving genotyping rates. It is also worth noting that our study strength was the use of a large sample size of specimens analysed in our laboratory and the inclusion of both samples from research studies and non-research focused sources. To the best of our knowledge, this is the first study in our setting to determine the laboratory genotyping success rates and examine the correlates of genotyping failure of unpaired DBS and plasma specimens from multiple studies/sources.
Conclusions
We report decreasing overall HIVDR genotyping success rates with increased numbers of DBS specimens analysed between 2016 and 2019, for specimens collected in a resource-limited setting. While plasma specimens generally had higher GSR > 80% over this period, the GSR of DBS specimens decreased to as low as about 50%, compromising the genotyping performance of our laboratory. Genotyping failure correlated with the use of DBS specimens, low viral loads, lack of pre-genotyping viral load testing and the source of specimens being routine clinical diagnosis. The use of DBS specimens, in this resource-constrained setting, offers a viable alternative to plasma specimens for viral load testing and HIVDR monitoring. DBS is critical to the realisation of the ambitious UNAIDS 95-95-95 targets. We therefore recommend improvement in DBS handling, routine viral load testing prior to genotyping to screen samples with suppressed viral loads that are likely to fail genotyping tests. Highly sensitive assays should be developed to genotype specimens with low and undetectable viral load, especially in this era where VL suppression rates are rising following increased roll-out of ART.
Acknowledgements
We credit the Ministry of Health, Uganda for all the continuous technical and logistical support.
Author contributions
JO: Wrote the original manuscript text, and prepared figures and tables. NB, DL, JO and DS: Data curation and formal analysis. MN, SL, FN, SN, HS, JO, DL, NB and DS: Methodology. PK and DS: Funding acquisition; Project administration and Supervision. DS: Conceptualization and Investigation. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Bill and Melinda Gates Foundation, Investment ID INV-031335 to Deogratius Ssemwanga. Other funding was from the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) that is under the MRC/DFID Concordat Agreement and is also part of the EDCTP2 programme supported by the European Union.
Availability of data and materials
The datasets generated and analysed during the current study are not publicly available due to restriction policies and data protection policies of the MRC/UVRI & LSHTM but can be made available by the corresponding authors on reasonable request and on approval of the UVRI research ethics committee.
Declarations
Ethics approval and consent to participate
This study is under the HIV Molecular Epidemiology study that was approved by the Uganda Virus Research Institute Ethics committee [UVRI-REC Federal wide Assurance (FWA) No. 00001354] and the Uganda National Council for Science and Technology (UNCST FWA No. 00001293). Participants to all studies analysed were recruited voluntarily and submitted written informed consent. All studies used methods that were compliant with ethical research guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jonah Omooja, Email: Jonah.Omooja@mrcuganda.org.
Deogratius Ssemwanga, Email: Deogratius.Ssemwanga@mrcuganda.org.
References
- 1.UNAIDS. Fact sheet-Global HIV Statistics [Internet]. 2020 [cited 2020 Oct 11]. Available from: http://aidsinfo.unaids.org.
- 2.Ministry of Health. Consolidated Guidelines for the Prevention and Treatment of HIV and AIDS in Uganda [Internet]. MOH; 2020. Available from: https://elearning.idi.co.ug/pluginfile.php/5675/mod_page/content.
- 3.Gregson J, Tang M, Ndembi N, Hamers RL, Rhee S-Y, Marconi VC, et al. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. Lancet Infect Dis. 2016;16:565–575. doi: 10.1016/S1473-3099(15)00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ssemwanga D, Lihana RW, Ugoji C, Abimiku A, Nkengasong J, Dakum P, et al. Update on HIV-1 acquired and transmitted drug resistance in Africa. AIDS Rev. 2015;17:3–20. [PubMed] [Google Scholar]
- 5.Aves T, Tambe J, Siemieniuk RA, Mbuagbaw L. Antiretroviral resistance testing in HIV‐positive people. Cochrane Database Syst Rev [Internet]. 2018 [cited 2020 Oct 12];2018. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6517236/. [DOI] [PMC free article] [PubMed]
- 6.UNAIDS. Understanding FastTrack Accelerating Action to End the AIDS Epidemic by 2030. 2015.
- 7.World Health Organization. Consolidated guidelines on HIV prevention, testing, treatment, service delivery and monitoring: recommendations for a public health approach [Internet]. 2021 update. Geneva: World Health Organization; 2021 [cited 2021 Aug 23]. Available from: https://apps.who.int/iris/handle/10665/342899. [PubMed]
- 8.Department of Health and Human Services. Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV [Internet]. 2020 [cited 2020 Oct 11]. Available from: https://clinicalinfo.hiv.gov/sites/default/files/guidelines/documents/AdultandAdolescentGL.pdf.
- 9.WHO. HIV Drug Resistance: Surveillance of HIV drug resistance in Adults receiving ART (Acquired HIV drug resistance) [Internet]. 2014. Available from: www.who.int.
- 10.The WHO. Surveillance of HIV drug resistance in adukts initiating antiretroviral therapy (Pre-treatment HIV drug resistance) [Internet]. WHO; 2014 [cited 2020 Oct 11]. Available from: https://apps.who.int/iris/bitstream/handle/10665/112802/9789241507196_eng.pdf;jsessionid=AAC44A20E97B74CF7A52D52829125A02?sequence=1.
- 11.Hirsch MS, Conway B, D’Aquila RT, Johnson VA, Brun-Vézinet F, Clotet B, et al. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA Am Med Assoc. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch MS, Günthard HF, Schapiro JM, Brun-Vézinet F, Clotet B, Hammer SM, et al. Antiretroviral drug resistance testing in adult HIV-1 Infection: 2008 recommendations of an international AIDS Society–USA Panel. Clin Infect Dis. 2008;47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 13.Bbosa N, Ssemwanga D, Nsubuga RN, Salazar-Gonzalez JF, Salazar MG, Nanyonjo M, et al. Phylogeography of HIV-1 suggests that Ugandan fishing communities are a sink for, not a source of, virus from general populations. Sci Rep [Internet]. 2019;9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6355892/. [DOI] [PMC free article] [PubMed]
- 14.Abeler-Dörner L, Grabowski MK, Rambaut A, Pillay D, Fraser C. PANGEA-HIV 2: phylogenetics and networks for generalised epidemics in Africa. Curr Opin HIV AIDS. 2019;14:173–180. doi: 10.1097/COH.0000000000000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanford HIV drug resistance database. Major HIV-1 Drug Resistance Mutations [Internet]. 2015 [cited 2017 Aug 29]. Available from: https://hivdb.stanford.edu/pages/download/resistanceMutations_handout.pdf.
- 16.World Health Organization. WHO HIVResNet HIV drug resistance Laboratory Operational Framework Second Edition [Internet]. 2021 update. Geneva: World Health Organization; 2021 [cited 2021 Aug 23]. Available from: https://apps.who.int/iris/handle/10665/343175.
- 17.Escaich S, Ritter J, Rougier P, Lepot D, Lamelin JP, Sepetjan M, et al. Plasma viraemia as a marker of viral replication in HIV-infected individuals. AIDS. 1991;5:1189–1194. doi: 10.1097/00002030-199110000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Johannessen A, Trøseid M, Calmy A. Dried blood spots can expand access to virological monitoring of HIV treatment in resource-limited settings. J Antimicrob Chemother. 2009;64:1126–1129. doi: 10.1093/jac/dkp353. [DOI] [PubMed] [Google Scholar]
- 19.Lofgren SM, Morrissey AB, Chevallier CC, Malabeja AI, Edmonds S, Amos B, et al. Evaluation of a dried blood spot HIV-1 RNA program for early infant diagnosis and viral load monitoring at rural and remote health care facilities. AIDS. 2009;23:2459–2466. doi: 10.1097/QAD.0b013e328331f702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouma KN, Basavaraju SV, Okonji JA, Williamson J, Thomas TK, Mills LA, et al. Evaluation of quantification of HIV-1 RNA viral load in plasma and dried blood spots by use of the semiautomated cobas amplicor assay and the fully automated cobas ampliprep/TaqMan assay, version 2.0, in Kisumu, Kenya. J Clin Microbiol. 2013;51:1208–1218. doi: 10.1128/JCM.03048-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Youngpairoj AS, Masciotra S, Garrido C, Zahonero N, de Mendoza C, García-Lerma JG. HIV-1 drug resistance genotyping from dried blood spots stored for 1 year at 4°C. J Antimicrob Chemother. 2008;61:1217–1220. doi: 10.1093/jac/dkn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parry CM, Parkin N, Diallo K, Mwebaza S, Batamwita R, DeVos J, et al. Field study of dried blood spot specimens for HIV-1 drug resistance genotyping. J Clin Microbiol. 2014;52:2868–2875. doi: 10.1128/JCM.00544-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salimo AT, Ledwaba J, Coovadia A, Abrams EJ, Technau K-G, Kuhn L, et al. The use of dried blood spot specimens for HIV-1 drug resistance genotyping in young children initiating antiretroviral therapy. J Virol Methods. 2015;223:30–32. doi: 10.1016/j.jviromet.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenman J, Roberts T, Cohn J, Messac L. Dried blood spot in the genotyping, quantification and storage of HCV RNA: a systematic literature review. J Viral Hepatitis. 2015;22:353–361. doi: 10.1111/jvh.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziemniak C, Mengistu Y, Ruff A, Chen Y-H, Khaki L, Bedri A, et al. Use of dried-blood-spot samples and in-house assays to identify antiretroviral drug resistance in HIV-infected children in resource-constrained settings. J Clin Microbiol. 2011;49:4077–4082. doi: 10.1128/JCM.01004-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monleau M, Butel C, Delaporte E, Boillot F, Peeters M. Effect of storage conditions of dried plasma and blood spots on HIV-1 RNA quantification and PCR amplification for drug resistance genotyping. J Antimicrob Chemother. 2010;65:1562–1566. doi: 10.1093/jac/dkq205. [DOI] [PubMed] [Google Scholar]
- 27.Ji H, Li Y, Liang B, Pilon R, MacPherson P, Bergeron M, et al. Pyrosequencing dried blood spots reveals differences in HIV drug resistance between treatment naïve and experienced patients. PLoS ONE. 2013;8:e56170. doi: 10.1371/journal.pone.0056170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grüner N, Stambouli O, Ross RS. Dried blood spots—preparing and processing for use in immunoassays and in molecular techniques. J Vis Exp. 2015;52619. [DOI] [PMC free article] [PubMed]
- 29.Watera C, Ssemwanga D, Namayanja G, Asio J, Lutalo T, Namale A, et al. HIV drug resistance among adults initiating antiretroviral therapy in Uganda. J Antimicrob Chemother. 2021;76:2407–2414. doi: 10.1093/jac/dkab159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiyaga C, Sendagire H, Joseph E, McConnell I, Grosz J, Narayan V, et al. Uganda’s new national laboratory sample transport system: a successful model for improving access to diagnostic services for early infant HIV diagnosis and other programs. PLoS ONE. 2013;8:e78609. doi: 10.1371/journal.pone.0078609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.University of Michigan Medical School, Biomedical Research Core facilities. Interpretation of Sequencing Chromatograms | Sanger Sequencing/Fragment Analysis FAQs [Internet]. U-M Biomedical Research Core Facilities. 2020 [cited 2021 Aug 25]. Available from: https://brcf.medicine.umich.edu/cores/advanced-genomics/faqs/sanger-sequencing-faqs/interpretation-of-sequencing-chromatograms/.
- 32.Pollack TM, Duong HT, Truong PT, Pham TT, Do CD, Colby D. Sensitivity and specificity of two dried blood spot methods for HIV-1 viral load monitoring among patients in Hanoi, Vietnam. PLoS ONE. 2018;13:e0191411. doi: 10.1371/journal.pone.0191411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaleebu P, Kirungi W, Watera C, Asio J, Lyagoba F, Lutalo T, et al. Virological response and antiretroviral drug resistance emerging during antiretroviral therapy at three treatment centers in Uganda. PLoS ONE. 2015;10:e0145536. doi: 10.1371/journal.pone.0145536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woods CK, Brumme CJ, Liu TF, Chui CK, Chu AL, Wynhoven B, Hall TA, Trevino C, Shafer RW, et al. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol. 2012;50:1936–1942. doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42:1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Omooja J, Nannyonjo M, Sanyu G, Nabirye SE, Nassolo F, Lunkuse S, et al. Rates of HIV-1 virological suppression and patterns of acquired drug resistance among fisherfolk on first-line antiretroviral therapy in Uganda. J Antimicrob Chemother. 2019;74:3021–3029. doi: 10.1093/jac/dkz261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang G, DeVos J, Medina-Moreno S, Wagar N, Diallo K, Beard RS, et al. Utilization of dried blood spot specimens can expedite nationwide surveillance of HIV drug resistance in resource-limited settings. PLoS ONE. 2018;13:e0203296. doi: 10.1371/journal.pone.0203296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Phanuphak N, Henderson K, Nonenoy S, Srikaew S, Shattock AJ, et al. Scaling up of HIV treatment for men who have sex with men in Bangkok: a modelling and costing study. The Lancet HIV. 2015;2:e200–e207. doi: 10.1016/S2352-3018(15)00020-X. [DOI] [PubMed] [Google Scholar]
- 40.Bertagnolio S, Soto-Ramirez L, Pilon R, Rodriguez R, Viveros M, Fuentes L, et al. HIV-1 drug resistance surveillance using dried whole blood spots. Antivir Ther. 2007;12:107. doi: 10.1177/135965350701200114. [DOI] [PubMed] [Google Scholar]
- 41.Ssemwanga D, Asio J, Watera C, Nannyonjo M, Nassolo F, Lunkuse S, et al. Prevalence of viral load suppression, predictors of virological failure and patterns of HIV drug resistance after 12 and 48 months on first-line antiretroviral therapy: a national cross-sectional survey in Uganda. J Antimicrob Chemother. 2020;75:1280–1289. doi: 10.1093/jac/dkz561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hallack R, Doherty LE, Wethers JA, Parker MM. Evaluation of dried blood spot specimens for HIV-1 drug-resistance testing using the Trugene HIV-1 genotyping assay. J Clin Virol. 2008;41:283–287. doi: 10.1016/j.jcv.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 43.Johannessen A, Garrido C, Zahonero N, Naman E, de Mendoza C. HIV-1 drug resistance testing from dried blood spots collected in rural Tanzania using the ViroSeq HIV-1 genotyping system. J Antimicrob Chemother. 2011;66:260–264. doi: 10.1093/jac/dkq433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Auad JP, Rojas-Montes O, Maldonado-Rodriguez A, Alvarez-Muñoz MaT, Muñoz O, Torres-Ibarra R, et al. Use of dried plasma spots for HIV-1 viral load determination and drug resistance genotyping in Mexican patients. BioMed Res Int. 2015;2015:1–9. doi: 10.1155/2015/240407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rottinghaus EK, Ugbena R, Diallo K, Bassey O, Azeez A, DeVos J, et al. Dried blood spot specimens are a suitable alternative sample type for HIV-1 viral load measurement and drug resistance genotyping in patients receiving first-line antiretroviral therapy. Clin Infect Dis Oxford Academic. 2012;54:1187–1195. doi: 10.1093/cid/cis015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dried blood spots for HIV‐1 drug resistance genotyping in decentralized settings in Senegal—Diouara—2014—Journal of Medical Virology—Wiley Online Library [Internet]. [cited 2020 Oct 16]. 10.1002/jmv.23778. [DOI] [PubMed]
- 47.Masciotra S, Garrido C, Youngpairoj AS, McNulty A, Zahonero N, Corral A, et al. High concordance between HIV-1 drug resistance genotypes generated from plasma and dried blood spots in antiretroviral-experienced patients. AIDS. 2007;21:2503–2511. doi: 10.1097/QAD.0b013e3281c618db. [DOI] [PubMed] [Google Scholar]
- 48.David S, Sachithanandham J, Jerobin J, Parasuram S, Kannangai R. Comparison of HIV-1 RNA level estimated with plasma and DBS samples: a pilot study from India (South) Indian J Med Microbiol. 2012;30:403–406. doi: 10.4103/0255-0857.103759. [DOI] [PubMed] [Google Scholar]
- 49.Vidya M, Saravanan S, Rifkin S, Solomon SS, Waldrop G, Mayer KH, et al. Dried blood spots versus plasma for the quantitation of HIV-1 RNA using a real-Time PCR, m2000rt assay. J Virol Methods. 2012;181:177–181. doi: 10.1016/j.jviromet.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 50.Kantor R, Delong A, Schreier L, Reitsma M, Kemboi E, Orido M, et al. HIV second-line failure and drug resistance at high- and low-level viremia in Western Kenya: AIDS. 2018;1. [DOI] [PMC free article] [PubMed]
- 51.Gonzalez-Serna A, Min JE, Woods C, Chan D, Lima VD, Montaner JSG, et al. Performance of HIV-1 drug resistance testing at low-level viremia and its ability to predict future virologic outcomes and viral evolution in treatment-naive individuals. Clin Infect Dis. 2014;58:1165–1173. doi: 10.1093/cid/ciu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta S, Taylor T, Patterson A, Liang B, Bullard J, Sandstrom P, et al. A robust PCR protocol for HIV drug resistance testing on low-level viremia samples. Biomed Res Int. 2017;2017:1–6. doi: 10.1155/2017/4979252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minchella PA, Chipungu G, Kim AA, Sarr A, Ali H, Mwenda R, et al. Specimen origin, type and testing laboratory are linked to longer turnaround times for HIV viral load testing in Malawi. PLoS ONE. 2017;12:e0173009. doi: 10.1371/journal.pone.0173009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pannus P, Claus M, Gonzalez MMP, Ford N, Fransen K. Sensitivity and specificity of dried blood spots for HIV-1 viral load quantification: a laboratory assessment of 3 commercial assays. Medicine (Baltimore) 2016;95:e5475. doi: 10.1097/MD.0000000000005475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analysed during the current study are not publicly available due to restriction policies and data protection policies of the MRC/UVRI & LSHTM but can be made available by the corresponding authors on reasonable request and on approval of the UVRI research ethics committee.