Abstract
Background
The roles and clinical values of synaptojanin 2 (SYNJ2) in lung squamous cell carcinoma (LUSC) remain unclear.
Methods
A total of 2824 samples from multi-center were collected to identify the expression of SYNJ2 in LUSC by using Wilcoxon rank-sum test, t-test, and standardized mean difference (SMD), and 194 in-house samples were also included to validate SYNJ2 expression in LUSC. The clinical roles of SYNJ2 were investigated via receiver operating characteristic (ROC) curves, univariate Cox regression analysis, and Kaplan–Meier plots. The underlying mechanisms of SYNJ2 in LUSC were explored by gene set enrichment analysis and immune correlation analysis. Further, a pan-cancer analysis based on 10,238 sapiens was performed to promote the understating of the expression and clinical significance of SYNJ2 in multiple human cancers.
Results
SYNJ2 was found to be significantly upregulated in LUSC at both mRNA and protein levels (p < 0.05, SMD = 0.89 [95% CI 0.34–1.45]) via public and in-house samples. Overexpressed SYNJ2 predicted poor prognosis for LUSC patients (hazard ratio = 2.38 [95% CI 1.42–3.98]). The cancer-promoting effect of SYNJ2 may be related to protein digestion and absorption and extracellular matrix-receptor interaction. SYNJ2 expression was closely related to immune cell infiltration, indicating its role in the immune response. Moreover, the distinct expression levels and essential clinical relevance of SYNJ2 in a series of cancers were initially revealed in this study.
Conclusions
This study disclosed the clinical significance of SYNJ2 in LUSC and multiple cancers, demonstrating the novel and potential biomarker for predicting and treating cancers.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-022-01266-0.
Keywords: Gene expression, Prognosis, Prediction, Treatment, Biomarker, Immune
Introduction
Lung cancer is the most lethal malignant tumor in the world. As many as 1,796,144 people died of lung cancer worldwide in 2020, accounting for 18% of cancer-derived deaths [1, 2]. Lung squamous cell carcinoma (LUSC) is a common subtype of lung cancer characterized by a deficiency of known driver genes, late diagnosis, and high heterogeneity. Almost 50% of LUSC patients have developed metastases at diagnosis. The 5-year survival rates of LUSC patients with stages II, III, and IV disease are 32%, 13%, and 2%, respectively [3]. Molecular targeted therapy and immunotherapy are the main current treatment options to reduce LUSC mortality [4]. Progress in understanding the driver genes and drug resistance after systemic therapy is very slow.
A deficient endocytosis pathway is highly correlated with the development of tumor cells and drug tolerance [5]. Synaptojanin 2 (SYNJ2), a member of the polyphosphate 5-phosphatase family, inhibits clathrin-mediated endocytosis and functions in distinct ways compared to SYNJ1, despite the high degree of homology in their catalytic domains [6]. Genetic variation in SYNJ2 is widely found in human diseases, such as reduced cognitive ability [7], Alzheimer’s disease and depression [8], medulloblastoma [9], colorectal cancer [10], prostate cancer [11], hairy cell leukemia [12], glioma [13], and breast cancer [14]. Hereinto, SYNJ2 plays a critical role in glioma and breast cancer metastasis. In terms of lung cancer, only one study showed the relationship between SYNJ2 and lung cancer; after studying 1404 lung cancer patients, it revealed that people with high SYNJ2 transcript levels are prone to shorter survival times. The limitation of this study is that the authors only used the Kaplan–Meier Plotter [15], ignoring the heterogeneity among various subtypes of lung cancer, and based results on partial transcriptomic data [14]. Moreover, to our knowledge, the roles of SYNJ2 in LUSC have not been reported before. Therefore, the roles and biofunctions of SYNJ2 in LUSC remain largely unknown.
In this study, the mRNA and protein expression levels of SYNJ2 in LUSC were systematically investigated. Integrated analyses of 2824 samples from multiple databases, such as The Cancer Genome Atlas (TCGA), Gene Expression Omnibus, and ArrayExpress, were performed to understand the expression pattern of SYNJ2 in LUSC. SYNJ2 mRNA expression was assessed with in-house microarray, and its protein level in in-house LUSC tissue microarray samples was assessed using immunohistochemistry (IHC). Predictive and prognostic values were evaluated via receiver operator characteristic (ROC) curves and hazard ratio (HR), and the latent molecular mechanism was explored by functional enrichment with gene ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) [16–18]. Finally, a pan-cancer analysis based on 9781 samples was also performed in this study, contributing to the understanding of SYNJ2 in multiple cancers.
Materials and methods
This study was authorized by the Ethics Committee of The First Affiliated Hospital of Guangxi Medical University (No. 2015-KY-NSFC-019). In-house samples were obtained with the informed consent of the corresponding patients.
Datasets collection
For LUSC-related datasets, the data collection workflow is shown in Additional file 1. Using search keyword terms “lung AND (squamous OR NSCLC OR [non-small cell]) AND (mRNA OR gene),” publicly available high-throughput RNA sequencing and microarray data were obtained from the following databases: TCGA, Gene Expression Omnibus, ArrayExpress, Sequence Read Archive, Oncomine, PubMed, and Google Scholar. The inclusion criteria for datasets were as follows: (1) Homo sapiens-related study; (2) Microarrays or RNA-Seq datasets; and (3) Clear subtype of lung cancer. The exclusion criteria were as follows: (1) Repeated samples between datasets; (2) Unclear subtype of lung cancer samples; and (3) Unavailable raw data. As a result, 37 datasets (Additional file 2) containing 1435 LUSC and 1428 non-LUSC samples were included in this study. Moreover, six datasets involving 323 LUSC cases were obtained to explore whether the SYNJ2 expression was related to the overall survival (OS) of LUSC patients.
CCLE [19] database collects data from numerous cell lines of homo sapiens. The data (containing 457 samples) of the CCLE database was utilized to investigate the difference in SYNJ2 expression between 14 kinds of cancer lines. Datasets of 32 cancers from TCGA were downloaded from the Xena database on November 16, 2021, and 9054 cancer samples and corresponding 727 control samples (Additional file 3) were included for further analysis.
In-house mRNA microarray collection and experiment
As previously described [20], the total RNA of six in-house LUSC and control samples was extracted with TRIzol® Regent (Invitrogen, USA). The RNA purity and integrity were assessed using the OD260/OD280 ratio and the standard range of 1.8–2.1. The OD260/OD280 was greater than 1.8, and RNA was not degraded. Then the cDNA samples were synthesized and labeled. A Nanodrop ND-1000 (Agilent, California, USA) was used to detect the efficiency of fluorescent labeling. Then the chip (Aksomics Inc., Shanghai, China) was hybridized with the labeled probe under standard conditions. Finally, the results were converted into digital data for storage and analysis after scanning with an Agilent Microarray Scanner (Agilent p/n G2565BA).
In-house tissue microarrays collection and IHC experiment
The four tissue microarray sections (No. LUC1021, LUC1501, LUC1601, and LUC2281) used in this study were produced by Fanpu Biotech, Inc. (Guilin, China). They contained 167 LUSC and 21 non-cancerous tissues, collected from August 2017 to October 2017. All accepted samples had never been exposed to chemical therapy and radiotherapy and had been diagnosed as LUSC independently by two pathologists according to the World Health Organization (2015) lung tumor histological criteria.
The SYNJ2 protein was detected using the EnVision system (Mxim, Fuzhou, China), a sensitive two-step immunohistochemical technique. All procedures were performed according to the manufacturer’s instructions. Briefly, the antigen was prepared with ethylene diamine tetraacetic acid (pH 9.0), the primary antibody (anti-SYNJ2 rabbit anti-human polyclonal antibody, orb513930, Biorbyt, UK, 1:100 dilution) was incubated at 4℃ overnight. Horseradish peroxidase-labeled secondary antibody (ready-to-use, Long Island Antibody, Shanghai, China) was added and left to react at room temperature for 25 min. Protein was visualized using 3–3′-Diaminobenzidine (DAB, Maxin, Fuzhou, China). Two pathologists independently evaluated the slides following the scoring criteria described previously [21]. In detail, a score for staining degree equals 0, 1, 2, and 3 for no, weak, moderate, and strong staining, respectively; a score for the percentage of positive cells equals 0, 1, 2, 3, and 4 for < 26%, 26–50%, 51–75%, and > 75% positive staining cells, respectively. The ultimate SYNJ2 protein level was calculated by the product of the score for staining degree and the score for the percentage of positive cells.
Data preparation
Datasets collected from the public databases were normalized with quantile and log2(x + 1) using the “oligo” and “limma” packages [22, 23]. By removing batch effects using the “SVA” package [24], the 37 datasets included in this study were classified into 13 merged datasets based on the same platform (Additional file 2). Notably, the dataset “GSE6044” did not include SYNJ2 expression and was used only to explore the differential expression genes (DEGs) between LUSC and non-LUSC groups.
The SYNJ2 expression and its clinical relevance in LUSC
Wilcoxon rank-sum test, t-test, and standardized mean difference (SMD) were used to compare the difference in SYNJ2 expression between LUSC and non-LUSC groups. When the data of a merged dataset were normally distributed and with homogeneity of variance, the t-test should be applied; otherwise, the Wilcoxon rank-sum test would be utilized. SMD calculation was used for the integrative analysis of SYNJ2 expression between LUSC and non-LUSC groups. A random-effects model was established to calculate SMD, as the I2 value of the I2 test > 50%. The publication bias and robustness of SMD were evaluated by Begg’s test [25] and sensitivity analysis, respectively.
The area under the curve (AUC) of (ROC) curves and summary ROC curves was calculated for detecting the accuracy of SYNJ2 mRNA expression in distinguishing LUSC samples from non-LUSC samples. The univariate Cox regression analysis was used to explore the relevance of SYNJ2 expression to the prognosis of LUSC patients.
The biofunctions and mechanisms of SYNJ2 in LUSC
DEGs were defined with the absolute value of log2 (fold change) ≥ 1. The upregulated DEGs (Up-DEGs) of LUSC were DEGs with SMD > 0, while the downregulated DEGs (Down-DEGs) were DEGs with SMD < 0. All genes from all datasets above were analyzed for correlation with SYNJ2. Genes meeting the criteria of Spearman correlation coefficient ≥ 0.3 and p < 0.05 were identified as positively co-expressed genes of SYNJ2 (SYNJ2-PCEGs) in at least two raw datasets, while Spearman correlation coefficient ≤ –0.3 and p < 0.05 were identified as negatively co-expressed genes of SYNJ2 (SYNJ2-NCEGs). The upregulated positively co-expressed genes (Up-PCEGs) were obtained by the intersection of Up-DEGs and SYNJ2-PCEGs, and the downregulated negatively co-expressed genes (Down-NCEGs) were determined via the intersection of Down-DEGs and SYNJ2-NCEGs. The Up-PCEGs and Down-NCEGs were analyzed and visualized using the “clusterprofiler” package [26] that included items from the gene ontology analysis and KEGG [27] signal pathways (the enrichment items with adjusted p-value < 0.05 were regarded as significant items). CIBERSORT [28] was used to detect the correlation between SYNJ2 expression and infiltration levels of 22 types of immune cells.
The pan-cancer analysis of SYNJ2
The Wilcoxon rank-sum tests were utilized to evaluate the differential expression of SYNJ2 in multiple cancers. The AUC of ROC and summary ROC curves was also used to calculate the accuracy of SYNJ2 mRNA expression in distinguishing cancer samples from their control samples. Both the univariate Cox regression analysis and Kaplan–Meier curves were used to explore the relevance between SYNJ2 expression and the prognosis of cancer patients.
Using gene set enrichment analysis, KEGG signaling pathways of 32 cancers were explored, and the grouping criteria was the median levels of SYNJ2 expression. TIMER database [29] provided infiltration levels of several types of immune cells for patients included in TCGA, and the data were obtained for investigating the immune relevance of SYNJ2 in pan-cancer. Both tumor mutational burden (TMB) and microsatellite instability (MSI) data were downloaded from the research of Liu et al. [30], and the expression levels of 46 immune checkpoints were from the TCGA dataset included in this study.
Statistical analysis
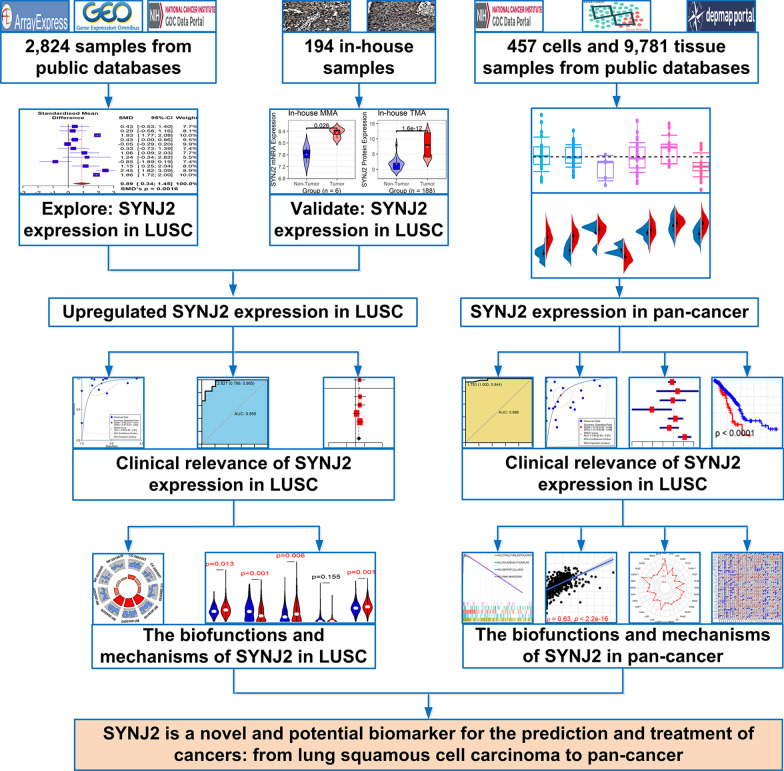
In addition to the statistical methods mentioned above, the Spearman correlation coefficient was utilized in all the correlation analyses for detecting the immune correlation of SYNJ2 in pan-cancer. An SMD value was considered significant if the corresponding 95% confidence interval (CI) did not exclude zero, while p < 0.1 indicated significant publication bias of SMD results. For HR, that 95% CI did not include 1 or p < 0.05 suggested statistical significance. All calculating processes and figures of this study were completed in R software (v4.1.0). The design of this study can be viewed in Fig. 1.
Fig. 1.
The design of this study. LUSC, lung squamous cell carcinoma
Results
Upregulated SYNJ2 expression was identified in LUSC
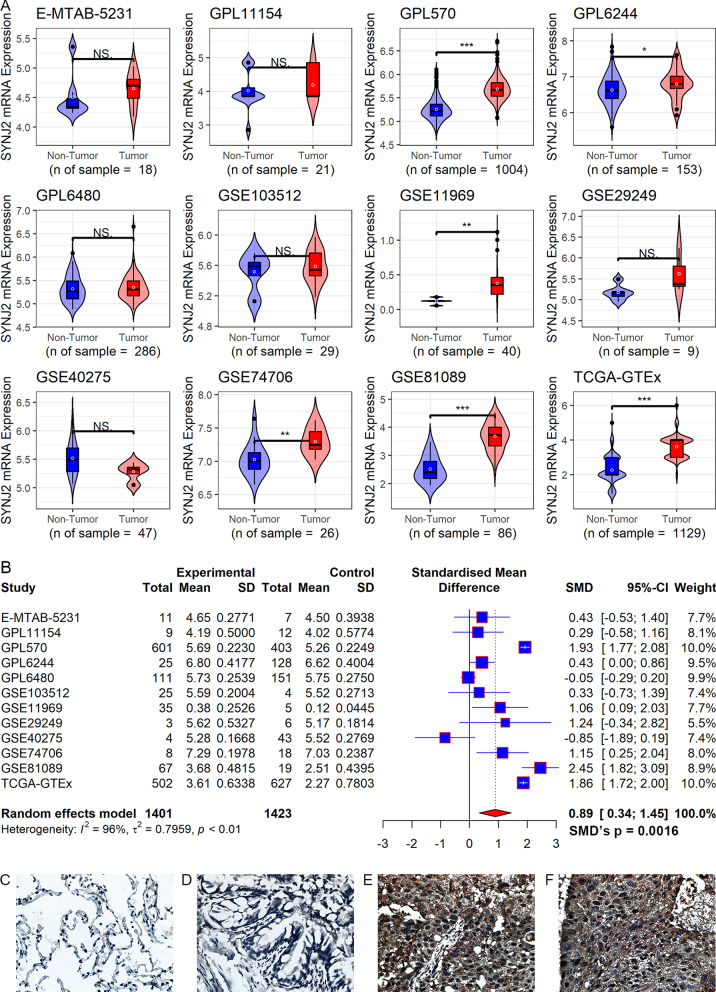
The statistical significance of the SYNJ2 expression difference between the LUSC and control groups was detected in 6 of 12 merged datasets included in this study, and 11 (except for “GSE40275”) of the 12 datasets indicated the SYNJ2 mRNA expression was higher in the LUSC group than that in the control group (Fig. 2A). Moreover, an integrative analysis including 2824 samples also suggested overexpression of SYNJ2 in LUSC (SMD = 0.89, 95% CI 0.34–1.45) (Fig. 2B), and there was no publication bias in the SMD results (Begg’s test, p = 0.784) (Additional file 4). The sensitivity analysis also determined the robustness of SMD results, as the results and heterogeneity of SMD did not change significantly after deleting any merged datasets (Additional file 5). At protein levels, it can be seen from Fig. 2C–F that SYNJ2 protein was significantly detected in not normal lung tissues (Fig. 2 C, D) but LUSC tissues (Fig. 2E, F). Further, by analyzing in-house samples, the upregulated SYNJ2 expression at both mRNA and protein levels in LUSC was validated (p < 0.05; Fig. 3A).
Fig. 2.
SYNJ2 expression between the LUSC group and control group. A The mRNA expression of SYNJ2 between the LUSC and control groups. *p < 0.05; **p < 0.01; ***p < 0.001; p values are based on the Wilcoxon rank-sum test or t-test. B The forest plot of mRNA expression of SYNJ2 between the LUSC and control groups. C The protein levels of SYNJ2 between the LUSC tissues (C, D) and normal lung tissues (E, F)
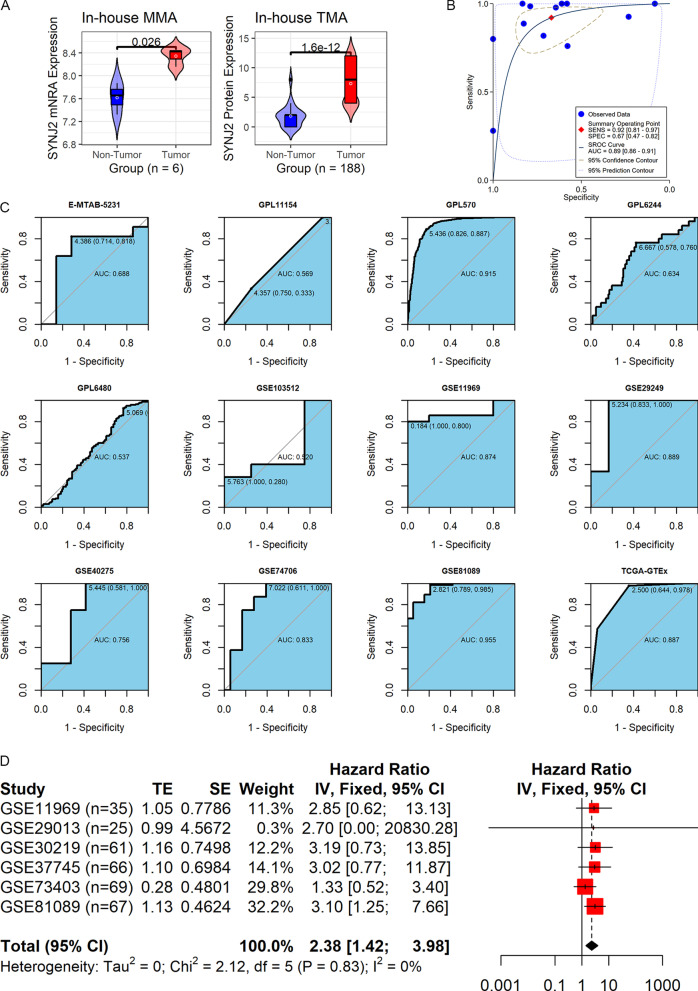
Fig. 3.
The expression, prediction ability, and prognosis relationship of SYNJ2 in LUSC. A The mRNA and protein levels of SYNJ2 expression in LUSC; The p values on the top of boxes are based on the Wilcoxon rank-sum test. B, C The prediction ability of SYNJ2 expression for LUSC. D SYNJ2 expression is related to the prognosis of patients with LUSC based on univariate Cox regression analysis results
Clinical relevance of SYNJ2 mRNA expression in LUSC
The sROC analysis revealed a high accuracy of SYNJ2 mRNA expression in distinguishing LUSC samples from non-LUSC samples (AUC = 0.89; Fig. 3B). At least moderate accuracy of SYNJ2 mRNA expression identifying LUSC was detected in seven datasets (AUC > 0.75; Fig. 3C).
A total of six datasets involving 323 cases were obtained for the prognostic analysis of SYNJ2 in LUSC. The results showed that patients with increased SYNJ2 expression tended to have poorer OS than those with decreased SYNJ2 expression (HR = 2.38, 95% CI 1.42–3.98) (Fig. 3D).
Biofunctions and mechanisms prediction of SYNJ2 in LUSC
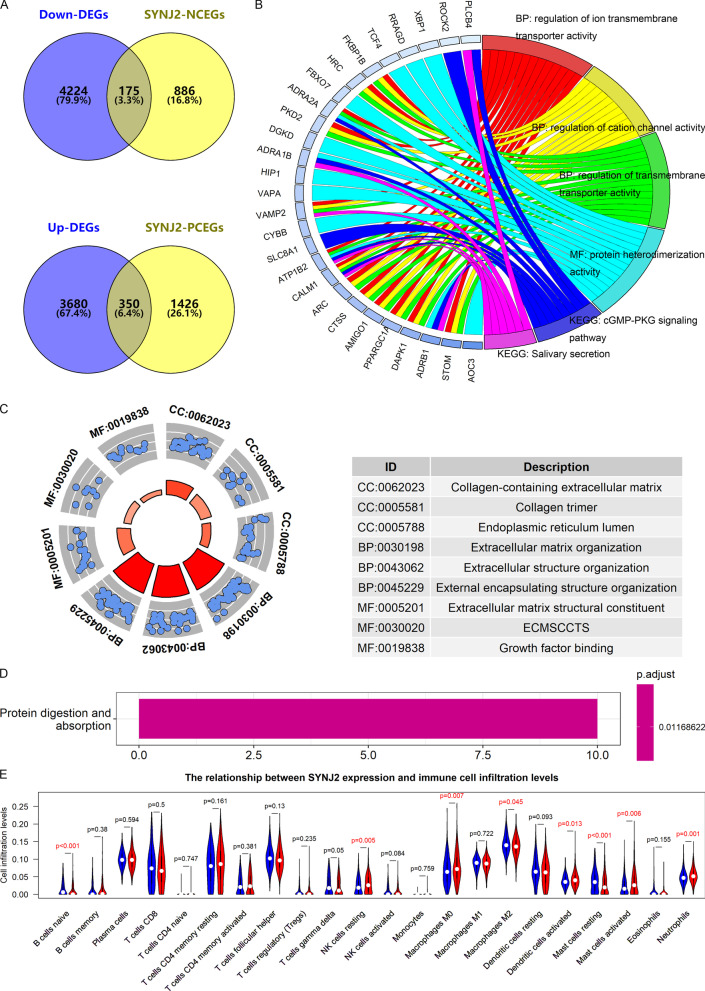
A total of 350 Up-PCEGs and 175 Down-NCEGs were identified (Fig. 4A). The analyses of gene ontology and KEGG showed that: (1) via Down-NCEGs, SYNJ2 may affect the regulation of ion transmembrane transporter activity (biological process), protein heterodimerization activity (molecular function), and cGMP-PKG signaling pathway (KEGG signaling pathway) (Fig. 4B). (2) based on Up-PCEGs, SYNJ2 was involved in cell components such as collagen, biological processes such as extracellular matrix organization; molecular functions such as extracellular matrix structural constituent (Fig. 4C); and signal pathways, such as protein digestion and absorption and extracellular matrix-receptor interaction (Fig. 4D).
Fig. 4.
Biofunctions and mechanisms prediction of SYNJ2 in lung squamous cell carcinoma. A Selection of downregulated negatively co-expressed genes (Down-NCEGs) and upregulated positively co-expressed genes (Up-PCEGs). B Gene ontology analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of Down-NCEGs. C Gene ontology analysis of Up-PCEGs. D KEGG analysis of Up-PCEGs. E The relationship between SYNJ2 expression and infiltration levels of immune cells; the blue “violin” refers to the low-SYNJ2 expression group, while the red “violin” refers to the high-SYNJ2 expression group
It can be seen from Fig. 4E that LUSC samples with SYNJ2 higher expression were detected with more resting NK cells, M0 macrophages, activated dendritic cells, activated mast cells, and neutrophils and less naive B cells, M2 macrophages, and resting mast cells.
Different SYNJ2 expression and its clinical relevance in various cancers
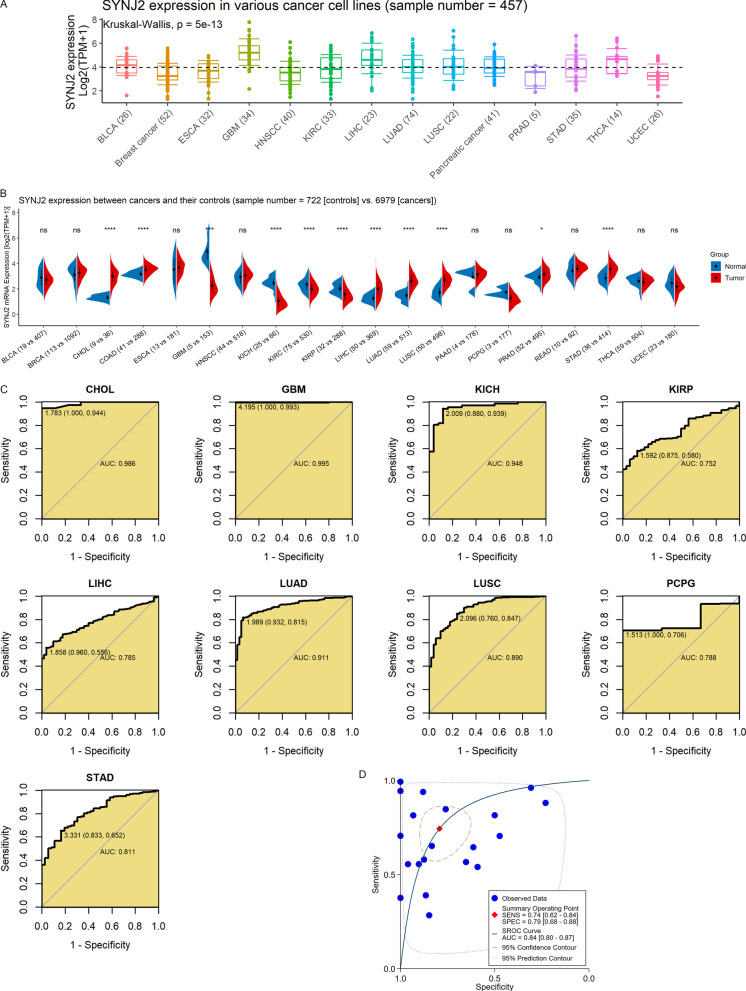
Differential expression of SYNJ2 in LUSC was identified, while its expression in multiple cancers and clinical relevance had not been discussed before. In this study, various SYNJ2 expression was identified between 14 cancer cell lines (Fig. 5A). By analyzing 9781 samples, compared to normal tissues, SYNJ2 was upregulated in cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), LUSC, prostate adenocarcinoma (PRAD), and stomach adenocarcinoma (STAD) and downregulated in glioblastoma multiforme (GBM), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), and kidney renal papillary cell carcinoma (KIRP) (p < 0.05; Fig. 5B). SYNJ2 expression made it feasible to differentiate cancer samples from corresponding normal samples with at least moderate accuracy (AUC > 0.75; Fig. 5C). The results of sROC analysis also demonstrate high accuracy of SYNJ2 expression in identifying 20 types of cancers from their controls (AUC = 0.87; Fig. 5D).
Fig. 5.
SYNJ2 expression in pan-cancer and its prediction ability for multiple cancers. A SYNJ2 expression in various cancer cell lines. B Differential expression of SYNJ2 between normal cancer tissues; nsp > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; p-value is based on the Wilcoxon rank-sum test. C The prediction ability of SYNJ2 expression for multiple cancers. D The prediction ability of SYNJ2 expression for pan-cancers
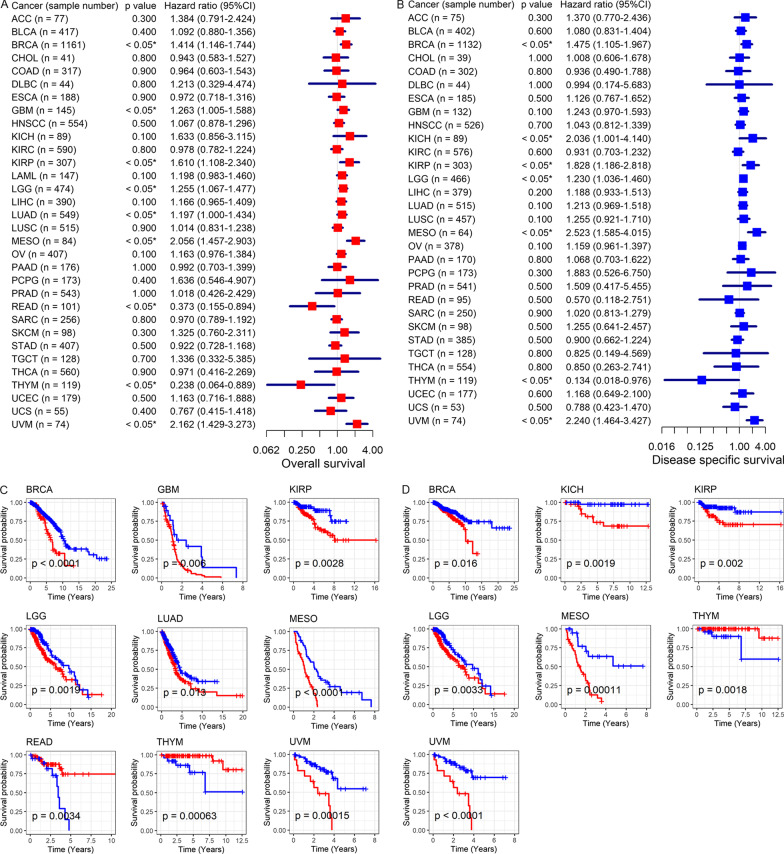
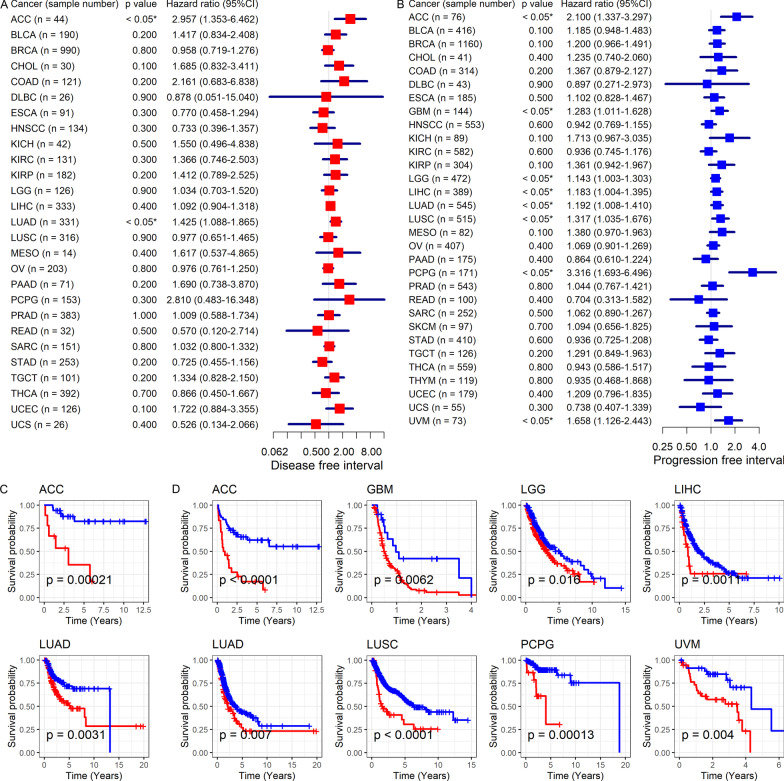
According to Cox analyses and Kaplan–Meier curves, SYNJ2 expression was associated with the prognosis of several cancers. In detail, high expression of SYNJ2 was related to the poor OS of patients with breast invasive carcinoma (BRCA), GBM, KIRP, LGG (brain lower grade glioma), LUAD, mesothelioma (MESO), and uveal melanoma (UVM) and poor disease specific survival (DSS) of patients with BRCA, KICH, KIRP, LGG, MESO, and UVM (hazard ratio [HR] > 1, p < 0.05; Fig. 6A–D). On the contrary, increased SYNJ2 expression was relevant to the good OS and DSS of THYM patients (HR < 1, p < 0.05; Fig. 6A–D). Moreover, upregulated expression of SYNJ2 represented unfavorable disease free interval (DFI) and progression free interval (PFI) for patients with adrenocortical carcinoma (ACC) and LUAD (HR > 1, p < 0.05; Fig. 7A–D); it also served as a risk factor for PFI of patients with GBM, LGG, LIHC, LUSC, pheochromocytoma and paraganglioma (PCPG), and UVM (HR > 1, p < 0.05; Fig. 7B, D). Notably, even though SYN2 expression was not found significantly correlated with the prognosis for LUSC in terms of OS, DSS, and DFI in the single TCGA dataset (Figs. 6, 7), the patients with elevated SYNJ2 expression had an unfavorable OS based on multiple datasets and 323 samples (Fig. 3D), and thus SYNJ2 was ultimately considered a risk factor for the prognosis of LUSC patients.
Fig. 6.
Relation of SYNJ2 expression with overall survival and disease-specific survival of cancers patients. A, B SYNJ2 expression is related to overall survival (A) and disease-specific survival (B) of cancers patients based on the results of univariate Cox regression analysis. C, D SYNJ2 expression is related to overall survival (C) and disease-specific survival (D) of cancers patients based on the results of Kaplan–Meier curves
Fig. 7.
Relation of SYNJ2 expression with disease-free interval and progression-free interval of cancers patients. A, B SYNJ2 expression is related to disease-free interval (A) and progression-free interval (B) of cancers patients based on the results of univariate Cox regression analysis. C, D SYNJ2 expression is related to disease-free interval (C) and progression-free interval (D) of cancers patients based on the results of Kaplan–Meier curves
Mechanisms prediction of SYNJ2 and the immune relevance of SYNJ2 in pan-cancer
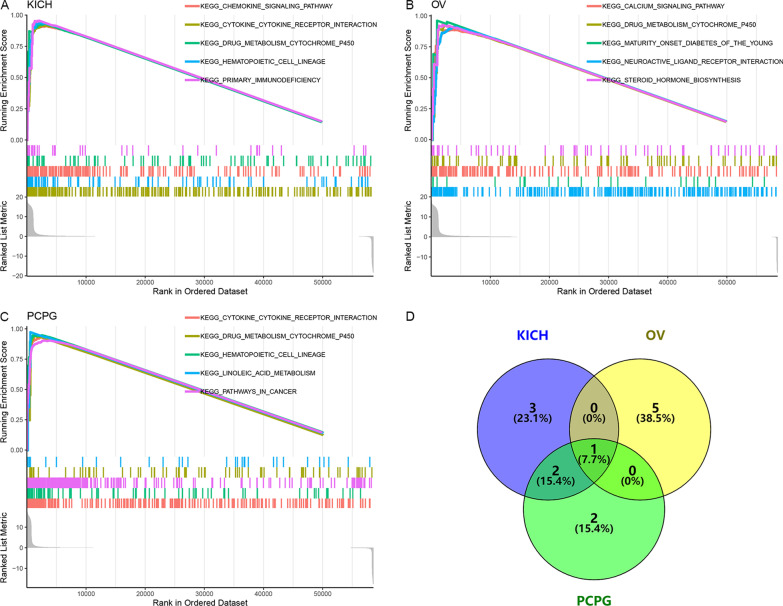
In three cancers—KICH, OV (ovarian serous cystadenocarcinoma), and PCPG, SYNJ2 were found related to at least five KEGG signaling pathways (Fig. 8A–C). Interestingly, “DRUG METABOLISM CYTOCHROME P450” was the overlap of three cancers (Fig. 8D), indicating that the important mechanism of SYNJ2 in various cancers may be related to this pathway.
Fig. 8.
Gene set enrichment analyses of SYNJ2 in multiple cancers
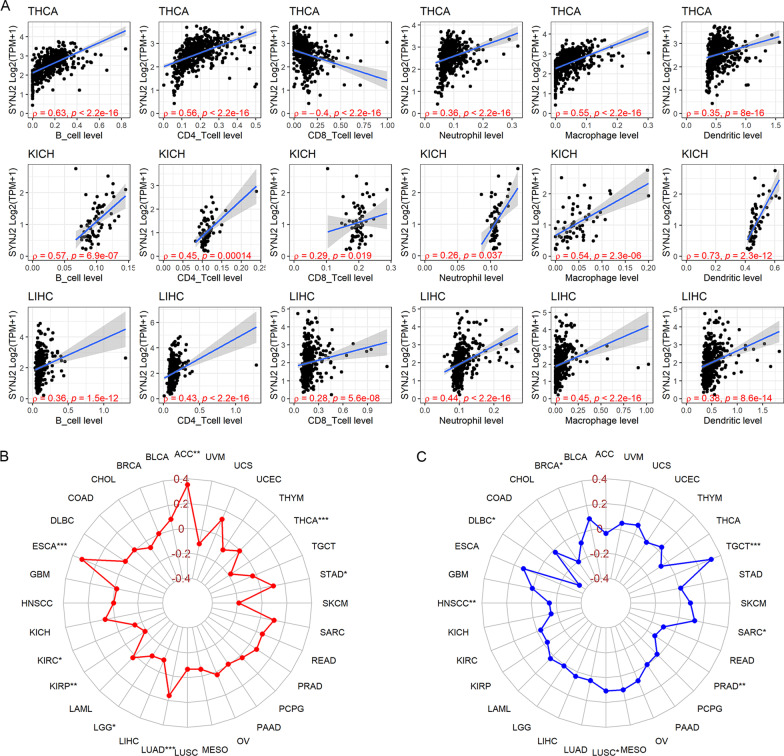
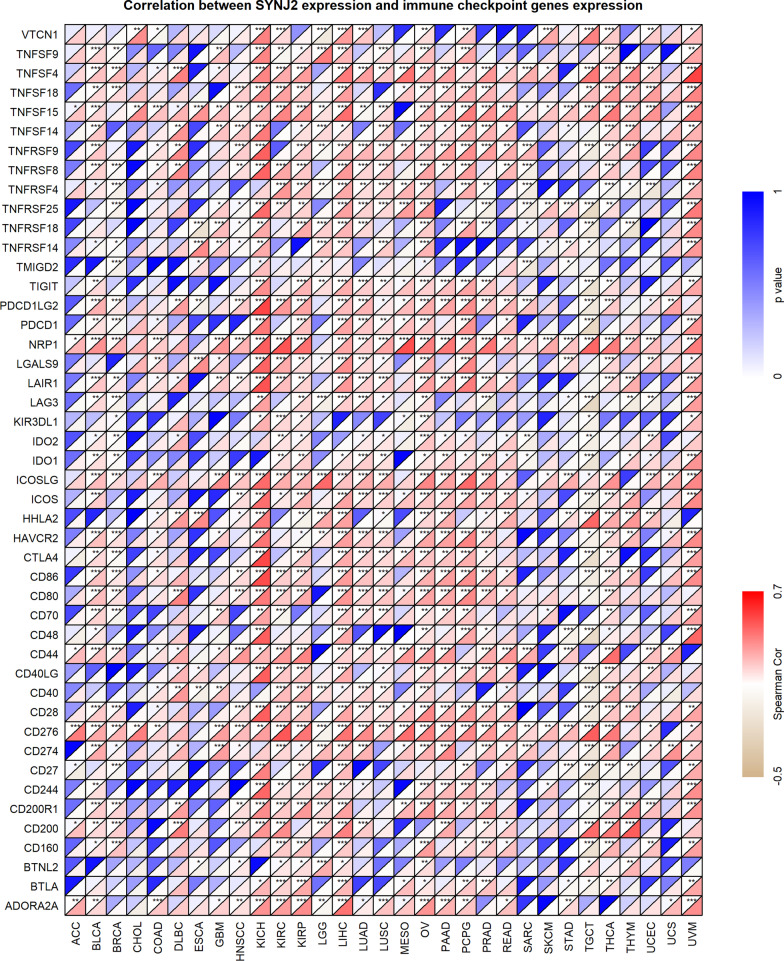
The immune response plays an essential role in anti-tumor, and thus the immune relevance of SYNJ2 in pan-cancer was investigated in this study. It can be viewed from Fig. 9A that SYNJ2 expression was positively (except for CD8 T cell in THCA) and closely related to infiltration levels of six immune cells in THCA, KICH, and LIHC. Moreover, the expression level of SYNJ2 was positively associated with TMB in ACC, esophageal carcinoma (ESCA), LUAD, and STAD; it was negatively related to TMB in KIRC, KIRP, LGG, and THCA (Fig. 9B). For MSI, SYNJ2 expression was positively relevant to LUSC, sarcoma (SARC), and testicular germ cell tumors (TGCT) and negatively associated with MSI in BRCA, DLBC (lymphoid neoplasm diffuse large B-cell lymphoma), HNSCC and prostate adenocarcinoma (PRAD) (Fig. 9C). Moreover, the significant relevance of SYNJ2 and immune checkpoints were defined in Fig. 10. For example, SYNJ2 demonstrated a positive association in KICH, LIHC, OV, PAAD, PCPG, and UVM and a negative association in TGCT with expression levels of not less than 15 immune checkpoints (Fig. 10).
Fig. 9.
Immune relevance of SYNJ2 in pan-cancer. A The relationship between SYNJ2 expression and infiltration levels of all six immune cells. B, C Spearman coefficient of SYNJ2 expression with tumor mutational burden (B) and microsatellite instability (C)
Fig. 10.
The expression relevance between SYNJ2 and immune checkpoints
Discussion
For the first time in this field, this study identified the expression and roles of SYNJ2 in LUSC. Based on the analysis of 3018 samples, SYNJ2 was significantly upregulated in LUSC at both mRNA and protein levels, and using multicenter samples, overexpressed SYNJ2 predicted poor prognosis for LUSC patients. The cancer-promoting effect of SYNJ2 may be related to protein digestion and absorption and extracellular matrix-receptor interaction. SYNJ2 expression was closely related to immune cell infiltration, indicating its role in the immune response. Moreover, this study initially investigated the expression levels and clinical relevance of SYNJ2 in pan-cancer, demonstrating the novel and potential biomarker for the prediction and treatment of cancers.
SYNJ2 is a synaptic binding protein with a role in endocytosis. Its involvement in human cancer has not been well-studied [6, 31]. Previous studies have reported that SYNJ2 is responsible for tumor initiation and progression. With genetic changes, SYNJ2 triggers colorectal cancer [10], medulloblastoma [9], prostate cancer [11], and hepatocellular carcinoma [32]. Overexpressed SYNJ2 boosts the invasion and migration of glioma [13] and breast cancer cells and prompts poor prognosis for cancer patients [14].
However, only one study by Ben-Chetrit et al. [14] has focused on the relationship between SYNJ2 and lung cancer. To verify the correlation between SYNJ2 and tumors, Ben-Chetrit et al. used an online informatics tool to show that high SYNJ2 transcript levels indicated a shorter lifespan for patients with lung cancer. Yet the study used only single-source data and limited samples, ignored genetic background, and simply evaluated the survival time. Moreover, distinct histological subtypes of lung cancer are characterized by specific morphological and molecular phenotypes. Thus SYNJ2 may play diverse roles in LUSC, lung adenocarcinoma, and small cell lung cancer, while the study of Ben-Chetrit et al. concentrated on all lung cancer rather than specific subtypes was not sufficient to understand the full spectrum of functions and mechanisms of action of SYNJ2 in LUSC. Therefore, the roles of SYNJ2 in LUSC remain largely unknown.
In this study, integrated analysis based on data covering more than 2824 samples revealed upregulated SYNJ2 mRNA expression in LUSC. Furthermore, in-house experiments verified a higher SYNJ2 expression in LUSC tissues than in non-tumorous lung tissues at mRNA and protein levels. In addition, this research not only demonstrated that SYNJ2 made it feasible to distinguish between LUSC and non-LUSC specimens but indicated that patients with high levels of SYNJ2 tend to live shorter lives. Thus, SYNJ2 was dramatically upregulated in LUSC and can predict both the cancer status and the poor prognosis for patients with LUSC, which has not been reported before, indicating the novelty of this study.
The expression patterns of SYNJ2 in different cancers were various, and its clinical relevance is conspicuous. Distinct SYNJ2 expression levels were identified in 14 types of cancer cell lines. Previous research showed differential expression levels of SYNJ2 between some cancers (e.g., BRCA [14] and LIHC [32]) and their control samples, and our study further comprehensively analyzed 20 cancers and revealed that increased and decreased SYNJ2 expression levels were defined in seven cancers (CHOL, COAD, LIHC, LUAD, LUSC, PRAD, and STAD) and four cancers (GBM, KICH, KIRC, and KIRP), respectively. SYNJ2 expression made it feasible to differentiate multiple types of cancers from their corresponding normal samples with at least moderate accuracy, suggesting its potential in identifying cancer status. In addition, elevated SYNJ2 expression was associated with the unfavorable prognosis (at least one of OS, DSS, DFI, and PFI) of cancers with ACC, BRCA, GBM, KICH, KIRP, LGG, LIHC, LUAD, LUSC, MESO, PCPG, and UVM, while it represented favorable OS and DSS of THYM patients. From these results, SYNJ2 generally served as a risk marker for the prognosis of cancer patients. Taken together, SYNJ2 may be a novel marker for predicting cancer status and prognosis of multiple cancers.
Few studies have focused on the molecular mechanistic understanding of SYNJ2 in human malignant tumors. According to previous studies, high expression of SYNJ2 was correlated with hepatocellular tumorigenesis via the CTCF/POLR2A-SYNJ2 axis [32] and with metastasis of glioma [13] and breast cancer through regulating the formation of lamellipodia and invadopodia [14]. In addition to these, SYNJ2 negatively regulated the PI3K/Akt pathway and was necessary for vesicle transport, focal adhesion, and lamellipodia as well as invadopodia formation, which led to cell survival, proliferation, invasion, and migration [14, 31]. Indeed, a similar finding can also be found in our study on LUSC (the proteins encoded by Up-PCEGs of SYNJ2 gathered in the extracellular matrix-receptor interaction). Our study also revealed that SYNJ2 might affect protein digestion and absorption (SYNJ2 protein is known to interact with the corresponding substrate, resulting in the translocation of the encoded protein to the plasma membrane and thereby inhibiting the clathrin-mediated endocytosis [32]). These findings implied that the roles SYNJ2 played in LUSC may be related to extracellular matrix-receptor interaction and protein metabolism; however, it requires further experiment exploration and verification. In addition, SYNJ2 was found related to the “DRUG METABOLISM CYTOCHROME P450” KEGG signaling pathway in several cancers (at least KICH, OV, and PCPG), indicating its druggable potential, which has also been demonstrated in BRCA before [14].
Little research has focused on the relevance between SYNJ2 and immunity before. However, based on the results of our study, SYNJ2 likely participated in the immune response by affecting filtration levels of immune cells, the mechanisms of which were complex with the fact that: (1) The infiltration levels of several antigen presenting cells (e.g., activated dendritic cells and neutrophils [33–37]), were significantly increased in LUSC samples with higher expression levels of SYNJ2. (2) SYNJ2 was positively associated with both infiltration levels of innate and acquired immune cells and expression levels of immune checkpoints in some cancers such as KICH and LIHC. (3) TMB and MSI were known to contribute to the production of new immune antigens and induction and promotion of immune responses [38, 39], and SYNJ2 showed its relationship with TMB and MSI in some cancers, implying its trigger role in this field. (4) Drugs targeting immune checkpoint block are promising in the treatment of cancers [40, 41]; SYNJ2 expression was significantly positively related to plenty of immune checkpoints in several cancers, suggesting that it may have similar clinical potential as immune checkpoints. Taken together, SYNJ2 was closely associated with the immune microenvironment and may serve as a potential marker of immune target treatment.
There are some limitations to this study. First, the number of in-house cohorts was small and must be expanded to include more clinical samples. Second, most of the in-house cases included in our study were at an early stage, did not have distant and lymph node metastases, and lacked clinical follow-up information, which limited our capacity to gain a comprehensive understanding of the clinical significance of SYNJ2 in LUSC. Last, a series of in vivo and in vitro experiments are needed to further explore the molecular mechanisms of action of SYNJ2 in LUSC. More scientific experiments in the future are required to address these limitations.
In conclusion, this study validated that SYNJ2 was significantly upregulated in LUSC tissues. Overexpressed SYNJ2 identities cancer status and predicts a poor prognosis for individuals with one type of multiple cancers, including LUSC. This research also disclosed the close association between SYNJ2 expression and the immune environment and the potential of this gene as a novel and potential biomarker for the prediction and treatment of multiple cancers, including LUSC.
Supplementary Information
Additional file 1. The collection workflow of datasets for calculating SYNJ2 expression.
Additional file 2. Datasets collected for LUSC.
Additional file 3. The details of the 32 cancers in this study.
Additional file 4. Funel plot with Begg’s test for publication bias in the SMD.
Additional file 5. The sensitivity analysis determined the robustness of SMD results.
Acknowledgements
We thank the Guangxi Key Laboratory of Medical Pathology for its technical support. We also thank the anonymous reviewers for their comments and suggestions, which greatly improved our article. The results shown in the study are in part based upon data generated by the ArrayExpress (https://www.ebi.ac.uk/arrayexpress/), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/), TCGA Research Network (www.cancer.gov/tcga), Genotype Tissue Expression (https://commonfund.nih.gov/GTEx), and DepMap [https://depmap.org/portal/]. The application of KEGG data in this study was approved by the Kanehisa Laboratories.
Abbreviations
- LUSC
Lung squamous cell carcinoma
- SYNJ2
Synaptojanin 2
- TCGA
The Cancer Genome Atlas
- IHC
Immunohistochemistry
- ROC
Receiver operator characteristic
- HR
Hazard ratio
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OS
Overall survival
- DEG
Differential expression gene
- SMD
Standardized mean difference
- AUC
Area under the curve
- Up-DEGs
Upregulated differential expression genes
- Down-DEG
Downregulated differential expression gene
- SYNJ2-PCEG
Positively co-expressed gene of SYNJ2
- SYNJ2-NCEG
Negatively co-expressed gene of SYNJ2
- Up-PCEG
Upregulated positively co-expressed gene
- Down-NCEG
Negatively co-expressed gene.
- TMB
Tumor mutational burden
- MSI
Microsatellite
- CI
Confidence interval
- DSS
Disease specific survival
- DFI
Disease free interval
- PFI
Progression free interval
Author contributions
All authors contributed to the study conception and design, and all of they commented, read and approved the submitted manuscript. Material preparation, data collection and analysis were performed by WH, GSL, LG, and HPL. The first draft of the manuscript was written by WH, GSL, LG, HPL, HFZ, JLK, and GC. All authors read and approved the final manuscript.
Funding
The study was supported by the funds of Project of Basic Capacity for Young and Middle-aged University Teachers in Guangxi (2019KY0102), Project of Basic Capacity for Young and Middle-aged University Teachers in Guangxi (2021KY0127), and Guangxi First-class Discipline Project for Pharmaceutical Sciences (GXFCDP-PS-2018).
Availability of data and materials
The datasets supporting the conclusions of this article are available in the Gene Expression Omnibus [https://www.ncbi.nlm.nih.gov/gds/], the ArrayExpress [https://www.ebi.ac.uk/arrayexpress/], TCGA Research Network [www.cancer.gov/tcga], Genotype Tissue Expression [https://commonfund.nih.gov/GTEx], and DepMap [https://depmap.org/portal/]. Direct persistent links for each public dataset were shown in the Sheet 1 of the Additional file 2. In-house data can be obtained from the corresponding author for reasonable reasons.
Declarations
Ethics approval and consent to participate
All methods were carried out in accordance with relevant guidelines and regulations (declarations of helsinki). This study was approved by the Ethics Committee of The First Affiliated Hospital of Guangxi Medical University (No. 2015-KY-NSFC-019). In-house samples were obtained with the written informed consent of the corresponding patients.
Consent for publication
Not applicable.
Competing interests
Wei Hou, Guo-Sheng Li, Li Gao, Hui-Ping Lu, Hua-Fu Zhou, Jin-Liang Kong, Gang Chen, Shuang Xia, and Hong-Yu Wei declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wei Hou and Guo-Sheng Li contributed as co-first authors
Contributor Information
Shuang Xia, Email: xiassxia@163.com.
Hong-Yu Wei, Email: weihongyu-789@163.com.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Jakobsen E, Olsen KE, Bliddal M, et al. Forecasting lung cancer incidence, mortality, and prevalence to year 2030. BMC Cancer. 2021;21:985. doi: 10.1186/s12885-021-08696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang BY, Huang JY, Chen HC, et al. The comparison between adenocarcinoma and squamous cell carcinoma in lung cancer patients. J Cancer Res Clin Oncol. 2020;146:43–52. doi: 10.1007/s00432-019-03079-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhang S, Sun K, Zheng R, Zeng H, Wang S, Chen R, Wei W, He J. Cancer incidence and mortality in China, 2015. JNCC. 2020;1:2–11. doi: 10.1016/j.jncc.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 6.Rusk N, Le PU, Mariggio S, et al. Synaptojanin 2 functions at an early step of clathrin-mediated endocytosis. Curr Biol. 2003;13:659–663. doi: 10.1016/S0960-9822(03)00241-0. [DOI] [PubMed] [Google Scholar]
- 7.Lopez LM, Harris SE, Luciano M, et al. Evolutionary conserved longevity genes and human cognitive abilities in elderly cohorts. Eur J Hum Genet. 2012;20:341–347. doi: 10.1038/ejhg.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasparoni G, Bultmann S, Lutsik P, et al. DNA methylation analysis on purified neurons and glia dissects age and Alzheimer's disease-specific changes in the human cortex. Epigenetics Chromatin. 2018;11:41. doi: 10.1186/s13072-018-0211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang P, Guo YD, Zhang HW. Identification of hub genes in pediatric medulloblastoma by multiple-microarray analysis. J Mol Neurosci. 2020;70:522–531. doi: 10.1007/s12031-019-01451-4. [DOI] [PubMed] [Google Scholar]
- 10.Du Q, Guo X, Zhang X, et al. SYNJ2 variant rs9365723 is associated with colorectal cancer risk in Chinese Han population. Int J Biol Mark. 2016;31:e138–e143. doi: 10.5301/jbm.5000182. [DOI] [PubMed] [Google Scholar]
- 11.Rossi MR, Hawthorn L, Platt J, et al. Identification of inactivating mutations in the JAK1, SYNJ2, and CLPTM1 genes in prostate cancer cells using inhibition of nonsense-mediated decay and microarray analysis. Cancer Genet Cytogenet. 2005;161:97–103. doi: 10.1016/j.cancergencyto.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Spaenij-Dekking EH, Van Delft J, Van Der Meijden E, et al. Synaptojanin 2 is recognized by HLA class II-restricted hairy cell leukemia-specific T cells. Leukemia. 2003;17:2467–2473. doi: 10.1038/sj.leu.2403174. [DOI] [PubMed] [Google Scholar]
- 13.Chuang YY, Tran NL, Rusk N, et al. Role of synaptojanin 2 in glioma cell migration and invasion. Cancer Res. 2004;64:8271–8275. doi: 10.1158/0008-5472.CAN-04-2097. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Chetrit N, Chetrit D, Russell R, et al. Synaptojanin 2 is a druggable mediator of metastasis and the gene is overexpressed and amplified in breast cancer. Sci Signal. 2015;8:ra7. doi: 10.1126/scisignal.2005537. [DOI] [PubMed] [Google Scholar]
- 15.Gyorffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Furumichi M, Sato Y, et al. KEGG: integrating viruses and cellular organisms. Nucl Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghandi M, Huang FW, Jane-Valbuena J, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao L, Xiong DD, He RQ, et al. Identifying TF-miRNA-mRNA regulatory modules in nitidine chloride treated HCC xenograft of nude mice. Am J Transl Res. 2019;11:7503–7522. [PMC free article] [PubMed] [Google Scholar]
- 21.Huang WT, He RQ, Li XJ, et al. miR146a5p targets TCSF and influences cell growth and apoptosis to repress NSCLC progression. Oncol Rep. 2019;41:2226–2240. doi: 10.3892/or.2019.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 26.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucl Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucl Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csolle MP, Ooms LM, Papa A, et al. PTEN and other PtdIns(3,4,5)P3 lipid phosphatases in breast cancer. Int J Mol Sci. 2020;21:9189. doi: 10.3390/ijms21239189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang R, Mo WJ, Huang LS, et al. Identifying the prognostic risk factors of synaptojanin 2 and its underlying perturbations pathways in hepatocellular carcinoma. Bioengineered. 2021;12:855–874. doi: 10.1080/21655979.2021.1890399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wculek SK, Cueto FJ, Mujal AM, et al. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. 2020;20:7–24. doi: 10.1038/s41577-019-0210-z. [DOI] [PubMed] [Google Scholar]
- 34.Mysore V, Cullere X, Mears J, et al. FcgammaR engagement reprograms neutrophils into antigen cross-presenting cells that elicit acquired anti-tumor immunity. Nat Commun. 2021;12:4791. doi: 10.1038/s41467-021-24591-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Leun AM, Thommen DS, Schumacher TN. CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer. 2020;20:218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borst J, Ahrends T, Babala N, et al. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18:635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 38.Galuppini F, Dal Pozzo CA, Deckert J, et al. Tumor mutation burden: from comprehensive mutational screening to the clinic. Cancer Cell Int. 2019;19:209. doi: 10.1186/s12935-019-0929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petitprez F, Meylan M, de Reynies A, et al. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front Immunol. 2020;11:784. doi: 10.3389/fimmu.2020.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The collection workflow of datasets for calculating SYNJ2 expression.
Additional file 2. Datasets collected for LUSC.
Additional file 3. The details of the 32 cancers in this study.
Additional file 4. Funel plot with Begg’s test for publication bias in the SMD.
Additional file 5. The sensitivity analysis determined the robustness of SMD results.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the Gene Expression Omnibus [https://www.ncbi.nlm.nih.gov/gds/], the ArrayExpress [https://www.ebi.ac.uk/arrayexpress/], TCGA Research Network [www.cancer.gov/tcga], Genotype Tissue Expression [https://commonfund.nih.gov/GTEx], and DepMap [https://depmap.org/portal/]. Direct persistent links for each public dataset were shown in the Sheet 1 of the Additional file 2. In-house data can be obtained from the corresponding author for reasonable reasons.