Abstract
Superoxide dismutases (SODs) are ancient enzymes of widespread importance present in all domains of life. Many insights have been gained into these important enzymes over the 50 years since their initial description, but recent studies in the context of microbial pathogenesis have resulted in findings that challenge long established dogmas. The repertoire of SODs that bacterial pathogens encode is diverse both in number and in metal dependencies, including copper, copper and zinc, manganese, iron, and cambialistic enzymes. Other bacteria also possess nickel dependent SODs. Compartmentalization of SODs only partially explains their diversity. The need for pathogens to maintain SOD activity across distinct hostile environments encountered during infection, including those limited for essential metals, is also a driver of repertoire diversity. SOD research using pathogenic microbes has also revealed the apparent biochemical ease with which metal specificity can change within the most common family of SODs. Collectively, these studies are revealing the dynamic nature of SOD evolution, both that of individual SOD enzymes that can change their metal specificity to adapt to fluctuating cellular metal availability, and of a cell’s repertoire of SOD isozymes that can be differentially expressed to adapt to fluctuating environmental metal availability in a niche.
Keywords: Superoxide dismutase (SOD), Evolution, Pathogenesis, Metalloenzymes, Metal specificity, Manganese, Iron
1. Introduction
As primordial environmental oxygen levels increased and life began to utilize the newly available oxygen, its potential to convert into toxic byproducts was unveiled. Simply the act of performing aerobic metabolism exposes organisms to internally generated reactive oxygen species (ROS) including superoxide (O2•−) [1,2]. Superoxide is toxic to the cell, targeting important molecules in the cell including iron-sulfur cluster containing proteins, metalloenzymes, and short-chain sugars [3–5]. While ROS generation in the context of respiration is accidental, the immune system actively harnesses the toxic properties of ROS to defend against microbial invaders. Immune cells including neutrophils and macrophages generate superoxide as part of the oxidative burst to kill microbes that breach the epithelial barrier [6–9]. Conversely, microbial pathogens need to adapt and evolve mechanisms to bypass the host’s oxidative immune response. The importance of the oxidative burst and the threat that ROS pose to potential invaders is revealed by chronic granulomatous disease (CGD). Individuals suffering from CGD have defects in the ability to generate an oxidative burst, preventing immune cells from clearing invaders and thus have enhanced susceptibility to infection [10–12].
Whether coping with endogenously or exogenously produced superoxide, both host and pathogen must protect themselves against this threat. Superoxide dismutases (SODs) are the primary means by which most organisms defend themselves against superoxide [6,13]. These redox active metalloenzymes are present in almost all forms of life, including humans, plants, pathogenic microbes, and non-pathogenic microbes [14,15]. The threat posed by superoxide and the importance of SODs is further revealed by their presence even in strict anaerobes [14]. Given their wide distribution and impact on human health, since their discovery in 1969 substantial insight has been gained into the function and distribution of SODs [16]. In particular, recent work studying these ubiquitous enzymes in the context of microbial pathogenesis has brought new insights but also revealed that there is still much to learn about these critical enzymes [17–19]. This review will focus on recent advances in our understanding of SODs, focusing primarily on the Fe/Mn family of SODs and how the evolution and distinct biological roles of these enzymes, has been revealed by studying them in the context of pathogenesis.
2. Biochemistry and metal specificity of SOD enzymes
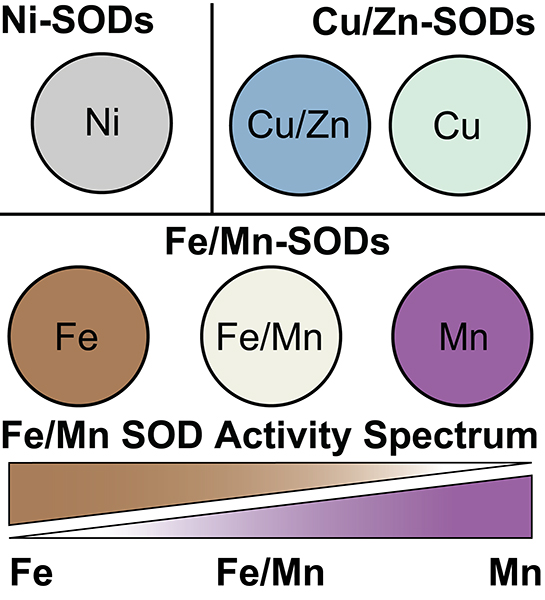
As enzymes, SODs catalyze the conversion of superoxide (O2•−) to oxygen (O2) and hydrogen peroxide (H2O2), which can be converted safely to water (H2O) by other enzymes within the cell [5,20]. The detailed molecular mechanism of SOD catalytic activity, and the key questions that remain to be resolved about how they function, have been previously reviewed [20]. Given the reaction they catalyze, it is unsurprising that nature has exploited redox active metal cofactors to perform this chemistry. What is surprising though is that three distinct catalytic centers utilizing different metal cofactors have evolved independently via functional convergence. Three structurally distinct SOD protein families exist, each that has activity with a specific set of metals. As a result, SODs are frequently discussed based on the metal that they have activity with, though enzymes active with different metals can belong to the same protein family. The SOD protein families include, those that are active with nickel (Ni), Ni-SODs, SODs that are active with copper (Cu) or Cu and zinc (Zn), Cu/Zn-SODs, and SODs that are active with either iron (Fe) or manganese (Mn), Fe/Mn-SODs (Figure 1) [20]. Ni-SODs were the most recently discovered and are the SODs about which least is known [21]. Found exclusively in bacteria, Ni-SOD distribution is more restricted than that of Cu/Zn- or Fe/Mn-SODs, being present primarily in marine cyanobacteria and actinomycetes [22,23]. While SODs differ in their metal utilization and fall into distinct protein families that evolved independently of one another, they overlap in their functional role within cells namely detoxifying superoxide. They also rely on the same chemical process; the sequential oxidation and reduction of substrate molecules in a ‘ping pong’ mechanism, whereby the redox-active metal cofactor undergoes sequential reduction and oxidation steps [20]. Given the disparate inherent reduction potentials of these catalytic metal ions (Eθ values ranging from 2.26 V for the Ni2+/Ni3+ redox couple to 0.15 V for the Cu2+/Cu+ couple), their different protein architectures must precisely and differentially regulate the reactivity of their respective metal ion to optimize SOD dismutation (optimal Eθ ~0.35 V) [20]. Indeed, this control of the metal’s reactivity by the protein is particularly important for Ni-SODs because the Ni redox couple lies well outside of the range that would enable it to react with the superoxide substrate [24]. The mechanism by which the reduction potential of the catalytic metal is optimized to facilitate dismutation remains enigmatic and is an area of active study.
Figure 1. The three superoxide dismutase (SOD) protein families and their metal-utilization.
SODs have arisen in three distinct protein families that each rely on a distinct set of metals for activity. These families are nickel SODs, copper and zinc SODs, and iron/manganese SODs. Recently, the Cu/Zn-SOD family was discovered to contain some enzymes that require only Cu for function. Similarly, the Fe/Mn-SOD family was realized to contain members that are specifically active with either Mn or Fe, but also enzymes that can use either metal for catalysis (cambialistic). Moreover, it has become apparent that metal-specificity in this family is best thought of as a spectrum rather than distinct bins. In the diagram spheres represent individual members of each SOD family and the metals they are active with.
SODs are usually cofactor specific, catalyzing superoxide dismutation using only their target metal cofactor(s) (Figure 1). Ni-SODs and Cu/Zn-SODs are absolutely dependent on their catalytic metal ions, Ni and Cu respectively, and are inactive when loaded with other metal ions in vitro [21,25]. Differing from the other SODs, the Fe/Mn-SOD family uniquely evolved the ability to use multiple different metals within a single protein framework [16,26–28]. Nonetheless, SODs from the Fe/Mn family were long regarded as being active exclusively with either Mn or Fe but not with both metals. This assumption was based on analyses of the cofactor specificity of a small number of early model systems, most notably the highly metal-specific Mn-SOD and Fe-SOD of Escherichia coli [29]. Similar observations were also made with the mitochondrial Fe/Mn SOD [30]. The discovery of Fe/Mn-SODs that are in fact cambialistic, i.e. active with either Mn or Fe, challenged this dogma of binary metal dependency [31]. Numerous cambialistic SODs have since been discovered in diverse organisms, and recent studies have shown that cambialism can be important physiologically in bacterial pathogens [19,28,31–35]. It is now recognized that the Fe/Mn-SOD family includes Fe-SODs, Mn-SODs, and cam-SODs [19,31,36–39]. Furthermore, the SOD enzymes characterized thus far show that these discrete definitions are not justified, with metal specificity instead lying on a spectrum, a continuum from Mn-specific to Fe-specific, with varying levels of cambialism in between these extremes (Figure 1) [28]. The simplistic initial model of binary metal specificity among Fe/Mn-SODs has likely restrained advances in our understanding of these enzymes, complicating both mechanistic biochemical studies and bioinformatic efforts to predict SODs metal utilization from sequence.
The discovery of cambialistic members of the Fe/Mn-SOD family also raises critical questions about how distinct Mn-specific, Fe-specific, and cambialistic isozymes have evolved within a single metalloenzyme family. This diversity of metal specificities is achieved despite all members of the Fe/Mn-SOD family being closely related in sequence and adopting highly similar structures with near-identical metal coordination environments utilizing three histidines and one aspartate sidechain (the universally conserved His26, His81, Asp167 and His171, using the numbering system from E. coli Mn-SOD) [20,40,41]. A “redox tuning” hypothesis has been proposed to explain this phenomenon, in which key amino acid residues in the metal’s secondary coordination sphere (i.e. residues located near to the metal cofactor, but which make no direct contacts with it) tune each metal’s reduction potential to optimize catalysis [42,43]. In vitro data from numerous groups support this model. Biochemical and biophysical studies of numerous mutated forms of the E. coli SODs demonstrate that these second sphere residues do regulate the metal cofactor’s reduction potential and that this in turn correlated with catalytic activity [20,43]. Furthermore, it was recently shown that reciprocal switching of just two secondary coordination sphere residues between a pair of SODs from Staphylococcus aureus substantially inverts their metal specificity [28]. These two mutations were sufficient to make its Mn-SOD variant highly cambialistic, and to make its cambialistic-SOD highly Mn-specific. Manipulation of redox tuning via altering these second sphere residues is thus a relatively simple and possibly tractable mechanism by which evolution can shape SOD metal specificity. Yet, there are few structural clues as to how the chemistry of these non-coordinating residues regulates the metal’s reduction potential, due in part to the technical challenges involved in experimentally quantifying SOD reduction potentials with sufficient resolution to correlate redox changes with structure and activity of diverse mutant SODs.
Notably, the Fe/Mn-SOD family is not the only family of SODs whose metal usage has been informed by investigations studying host-pathogen interactions. The Cu/Zn SOD family had long been thought to absolutely require both Cu for catalysis and Zn for structural integrity and catalytic efficiency. However, recent work in Mycobacterium tuberculosis and Candida albicans demonstrated that some SODs within this family do not require Zn, again challenging long established dogma within the field [44,45]. Such discoveries have been recently reviewed and revitalize the field, urging work to be done to determine how widespread these Cu-only and Fe/Mn cambialistic SODs are across the domains of life [9,18,46].
3. Eukaryotic superoxide dismutase utilization
While all SODs carry out the same chemical reaction, many species across all domains of life frequently possess more than one of these critical enzymes. Mammals possess three SODs, SOD1, SOD2, and SOD3 [17]. SOD1 and SOD3 are both members of the Cu/Zn SOD family while SOD2 is from the Fe/Mn-SOD family. Many other animals possess a similar set of SODs, but their nomenclature can differ. Each of these SODs localize predominantly to distinct cellular locations with SOD1 mainly in the cytoplasm and the mitochondrial inter membrane space, SOD2 the mitochondrial matrix, and SOD3 associated with the extracellular matrix/cell surface. Similar to animals, plants also possess more than one SOD, each compartmentalized in a distinct cellular location [47–51]. As a result of this compartmentalization, these SODs have largely non-overlapping roles in the defense of the cell. Examples of these non-redundant roles are observed across multiple organisms including flies, mice, and humans. In Drosophila melanogaster, homozygous loss of either sod1 or sod2 is lethal [52–57]. Similarly, mice lacking the mitochondrial SOD2 die shortly after birth, further demonstrating this eukaryotic SOD’s importance [58,59]. In humans, genetic disorders have also revealed the non-overlapping roles of SODs [60,61]. Mutations in SOD1 are associated with familial amyotrophic lateral sclerosis (fALS), abnormalities in SOD2 are associated with mitochondrial stress and cancer while mutations in SOD3 are associated with vascular dysfunction including hypertension, diabetes, and atherosclerosis [20,57,62–66].
SODs also play important roles during infection by numerous eukaryotic microbes. Four Fe/Mn-SODs were identified in the causative agent of African sleeping sickness, Trypanosoma brucei, and overexpression of one of these homologues, SodB, increased survival of the closely related Trypanosoma cruzi during macrophage infection [67,68]. Based on phylogenetic evidence, the four SODs of T. brucei were likely acquired via at least two independent lateral gene transfer events followed by gene duplications [69]. Deletion of one of the SodA alleles from another eukaryotic pathogen, Leishmania amazonensis, resulted in its inability to replicate in macrophages and an attenuated ability to generate cutaneous lesions in mice [70]. Similar to protozoan pathogens, pathogenic fungi possess an expanded repertoire of SODs relative to their host organisms. For example, C. albicans and other pathogenic fungi possess an additional Mn-dependent SOD in the cytoplasm and an extracellular Cu-only SOD [45,71,72]. Loss of the Cu- only SOD reduces the ability of C. albicans to cause infection and the Mn-dependent SOD is induced in response to host-imposed copper limitation [72–74].
4. Superoxide dismutase utilization by pathogenic bacteria
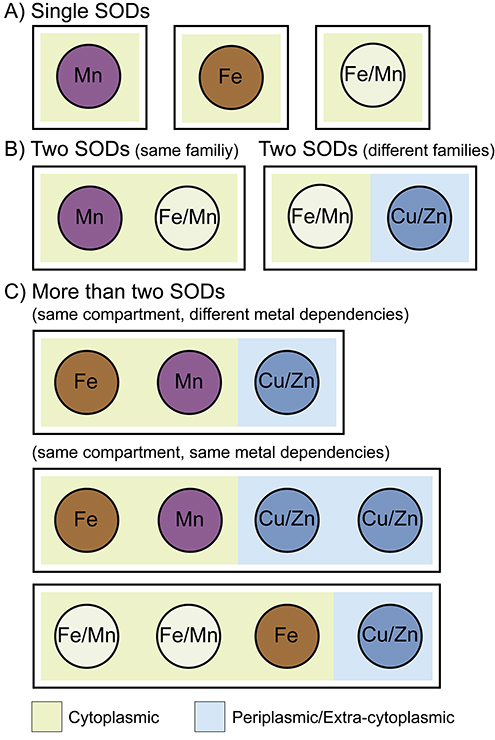
Compared with animals and plants, there is greater diversity in the repertoire of SODs possessed by bacteria. Most pathogenic bacteria possess one or more cytoplasmic SODs belonging to the Fe/Mn-SOD family [9,15]. These SODs are frequently referred to as SodA when Mn-dependent and SodB when Fe-dependent, with this nomenclature being derived from their initial discovery in E. coli. However, there is not always a one-to-one correlation between name and metal dependency and such annotations should be interpreted with care. The existence of cambialistic SODs further complicates the issue. In addition to Fe/Mn-SODs, some bacteria possess Cu/Zn SODs, which are generally referred to as SodC [46,75]. Differing from eukaryotes, studied bacterial Cu/Zn SODs are strictly extra-cytoplasmic [46]. In Gram-negative bacteria, Cu/Zn family members are found in the periplasm, while in Gram-positive bacteria they are frequently predicted to be attached to the cell surface via lipids [75,76]. For bacteria that encode multiple SODs, the diversity of number and overall repertoire varies significantly from genera to genera and can even differ between species of the same genera (Figure 2).
Figure 2. Superoxide dismutase (SOD) repertoires possessed by bacteria.
While the threat of superoxide and possession of SOD(s) is nearly universal among bacteria, the repertoire of enzymes they possess is extremely varied. (A) Bacteria can encode only a single SOD that can be either Mn-dependent (purple, e.g. Borrelia burgdorferi), Fe-dependent (brown, e.g. Campylobacter jejuni), or cambialistic (green, e.g. Streptococcus mutans). (B) When bacteria encode two SODs, they can either be from the same SOD family (Fe/Mn family, e.g. Staphylococcus aureus), or from different SOD families (Fe/Mn family, light grey, Cu/Zn family, blue e.g. Mycobacterium tuberculosis). (C) Finally, there are also many bacteria that encode more than two SODs. This can result in the presence of more than one SOD in a single cellular compartment and can result in enzymes from the same family, with either similar or distinct metal specificity, occupying the same compartment (Fe- and/or Mn-dependent in the cytoplasm, tan, Cu/Zn in the periplasm/extra-cytoplasmic space, light blue e.g. Escherichia coli, Salmonella enterica serovars and Bacilli). Despite carrying out the same chemical reactions, each SOD in these instances appears to uniquely contribute to the lifestyle of the harboring organisms.
While many pathogens encode multiple SODs, there are numerous examples of bacteria that possess only a single enzyme. As would be expected in these organisms, the single SOD is frequently important for virulence-associated phenotypes or infection [32,35,77–79]. Borrelia burgdorferi, the causative agent of Lyme Disease, encodes a single Mn-dependent SOD, SodA [80,81]. Loss of SodA leads to reduction of viability in B. burgdorferi after incubation with either neutrophils or macrophages as well as a complete loss of detectable bacteria in intradermally infected mice [77]. Similarly, Campylobacter spp., which are commonly associated with bacterial enteritis in humans and are found in the gastrointestinal tract of 60–80% of poultry broiler flocks worldwide, encode a single SOD also from the Fe/Mn-SOD family [82]. Deletion of the Fe-dependent SodB in Campylobacter coli leads to decreased bacterial survival on substrates essential to environmental transmission of this pathogen as well as decreased chick colonization [79]. In Campylobacter jejuni, Fe-dependent SodB mutants have decreased survival in human embryonic intestinal cells, indicating a role in intracellular survival [78,79]. Porphyromonas gingivalis and Streptococcus mutans, both oral pathogens, possess a single cambialistic SOD that contributes to aerotolerance, and thus survival, within the oral cavity [31–34]. Additionally, Streptococcus pneumoniae, the most common causative agent of pneumonia, as well as important gut microbes, the Bacteroidetes, each encode a single, cambialistic SOD [37,38,83]. Loss of SodA makes S. pneumoniae less invasive and less pathogenic as fewer bacteria are recovered from the blood and lungs of infected animals [35]. In pathogenic Bacteroidetes, it has been suggested that SOD is a virulence factor as the ability to withstand temporary tissue aeration would enhance the pathogenicity of these anaerobic microbes [84].
When bacteria encode two SODs, they can either be from different SOD families or both enzymes can belong to the same protein family. Legionella pneumophila, the causative agent in Legionnaires’ disease, and Mycobacterium tuberculosis, which is responsible for tuberculosis, both encode two SODs from different families, with one from the Fe/Mn family and one from the Cu/Zn family. While the Cu/Zn SOD in L. pneumophila is not essential for viability, it is critical for stationary phase survival and therefore perhaps also in extended dormancy between growth in hosts [85]. On the other hand, the Fe-dependent SodB in L. pneumophila is essential for viability, and presumptively virulence [86]. Similarly, early reports on SodA in M. tuberculosis concluded that it must be essential as attempts to delete it proved unsuccessful [87]. Reduced expression of SodA attenuated M. tuberculosis with ~100,000-fold fewer bacteria observed in the lungs and spleens of mice after infection [88]. M. tuberculosis SodC is important for survival of the oxidative burst during infection as sodC mutants are more susceptible to killing by activated macrophages [89]. Similar to eukaryotes, it is easy to hypothesize that the unique contribution of multiple SODs in L. pneumophila, M. tuberculosis, and other bacteria with similar arrangements is derived from compartmentalization. However, compartmentalization cannot fully explain why bacteria have multiple SODs, and varying metal specificities tend to be important in these cases. For example, S. aureus encodes two SODs, SodA and SodM, both of which belong to the Fe/Mn-SOD family and are present in the cytoplasm. SodA is strictly Mn-dependent and SodM is cambialistic displaying equal activity with Mn or Fe. Loss of SodA, SodM, or both SODs simultaneously results in reduced bacterial loads in a skin abscess model of infection [90,91]. In a systemic model of infection, loss of SodM but not of SodA results in reduced bacterial burdens in liver tissues [19]. Thus, despite localizing to the same cellular compartment, both of the staphylococcal SODs have unique non-overlapping roles during infection.
The diversity of SODs and unique contribution each SOD provides bacteria is further exemplified by the Enterobacteriaceae and Bacilli. The Enterobacteriaceae, including Escherichia coli and Shigella flexneri, have a common core set of SODs including one periplasmic Cu/Zn SOD, one cytoplasmic Mn-SOD, and one cytoplasmic Fe-SOD. For avian pathogenic E. coli O2 strain E058, SodA is important to virulence as mutants lacking SodA exhibit significantly increased ingestion and decreased survival in chicken macrophages [92]. In addition, mutants lacking SodA have a significant decrease in bacterial burdens in both single strain infections and competition experiments using chickens [92]. In S. flexneri, SodB mutants are more susceptible to killing by both mouse macrophages and human polymorphonuclear leukocytes [93]. Among Bacilli, both Bacillus cereus and Bacillus anthracis are predicted to encode four SODs, three that are from the Mn/Fe family (SodA1, SodA2, and SodS/Sod15) and one that is from the Cu/Zn family (SodC), with each of these enzymes differentially contributing to the organisms’ success [94–96]. In B. cereus 0–9, SodS is Fe-dependent and SodA1 and SodA2 are predicted to be Mn-dependent [94]. However, given sequence similarity to B. anthracis SODs, SodA2 could in fact be Fe-dependent [97]. Each of the four SODs in B. cereus 0–9 affect biofilm formation. However, the most significant reduction in the ability to form a biofilm is observed in a SOD null strain and a single sodA2 mutant, demonstrating SodA2’s importance [94]. Interestingly, deletion of Mn-dependent SodA1 leads to a delay in spore formation while deletion of Fe-dependent SodS leads to a complete loss of swarming abilities [94]. In B. anthracis, SodA1 and Sod15 are present in the exosporium [95,98,99]. Assuming similarity, these SODs might also be present in the exosporium of B. cereus, perhaps influencing these phenotypes. Finally, individual SodS and SodC mutants have drastically reduced lethality, even lower than that of a SOD-null strain, against the cotton bollworm host, Helicoverpa armigera, indicating the importance of these two SODs for pathogenicity in this host [94]. The fact that all four of these SODs are present in both of these Bacillus spp. indicates that having a diverse repertoire of SODs is particularly important for these organisms and increases their adaption and ability to handle ROS in diverse environments.
5. Understanding the Diversity of SODs
While the need to protect the cytoplasm and periplasm from ROS damage partially explains diversity associated with bacterial SODs, it does not explain all of it as there are many examples of bacteria expressing expanded repertoires of cytosolic and periplasmic SODs [9,93,100]. Again, we can glean insight into the roles of multiple SODs from the study of bacterial pathogens. When examining pathogenic and non-pathogenic bacteria from the same or related species, a common theme emerges, namely that more pathogenic strains/lineages/serovars in these genres tend to encode an extra SOD. This can be seen in the Bacilli, Staphylococci and Enterobacteriaceae. In Bacilli, the non-pathogenic Bacillus subtilis only encodes one SodA, while both B. anthracis and B. cereus encode two SodA, SodA1 and SodA2 [94,95,101,102]. Amongst the Staphylococci, all members encode SodA while only the more pathogenic S. aureus lineage encode the cambialistic sodM [103]. All Salmonellae encode sodC2 while only highly virulent strains in serovars such as Typhimurium, Enteritidis, Dublin, Choleraesuis, and Heidelberg possess the phage-encoded sodC1 [104–106]. This acquisition of a phage encoded SodC homolog is also seen in pathogenic E. coli including the highly virulent O157:H7 serotype [107–109].
Differing from cells in higher organisms, bacteria and other microbes do not fully control the nutrient content of the environment in which they reside. As all known SODs are dependent on metals for function, it stands to reason that the diversity of SODs possessed by bacteria is driven by the need to retain SOD activity in distinct environments. Beyond SODs, approximately 50% of all enzymes require a metal for function and thus metals are essential for life and for pathogens to cause infection [110–112]. Similar to leveraging the toxicity of ROS, the host restricts the availability of transition metals to prevent infection. This defense, known as nutritional immunity, was classically confined to the restriction of Fe, but has expanded to include Mn, Zn, and Cu [74,112,113]. Nutritional immunity can inactivate a myriad of metal-dependent bacterial enzymes, including SODs [91,114]. Despite this restriction within the host, pathogens must retain a defense against superoxide to survive the oxidative burst of immune cells. Recent work in S. aureus and the fungus C. albicans suggests that an enhanced repertoire of SODs enables pathogens to retain SOD activity when the host imposes metal starvation. In S. aureus, the Mn-dependent SodA is considered the primary staphylococcal SOD. However, host-imposed Mn starvation enacted in part by the immune effector calprotectin decreases SodA activity. Concurrent with this, S. aureus induces and derives most of its SOD activity from Fe-loaded cambialistic SodM [19]. As noted earlier in the liver of wild type mice, which sequester Mn away from S. aureus, SodM is necessary to achieve wild type bacterial burdens. However, in calprotectin-deficient mice, which fail to sequester Mn, SodA becomes critical to infect the liver, while SodM is dispensable [19]. C. albicans possesses two cytoplasmic SODs with differing metal specificity, namely a Cu/Zn SOD and a Mn-SOD [9]. Similar to S. aureus, it uses the differing metal specificity of these enzymes to retain a defense against superoxide in the host. During periods of Cu limitation, such as that which occurs in brain and kidney tissues, C. albicans switches from the Cu-requiring SOD to the Mn-dependent SOD [73]. Similar observations have also been made in laboratory strains of E. coli, with the importance of the Fe-dependent SodB and Mn-dependent SodA being dependent on the availability of their cognate metal, further suggesting that possessing multiple SODs with different metal specificities is a common survival strategy used by microbes [115–117]. While it remains to be tested, it seems reasonable to speculate that organisms with only a single, cambialistic SOD such as P. gingivalis, S. mutans, and S. pneumoniae may also be better poised than organisms with a single, metal specific SOD, to maintain a defense against oxidative stress in response to changing metal availability within the host [32–35].
While metal availability seems to be a driver of microbial SOD diversity, it does not appear to be the only pressure driving SOD expansion as some organisms have multiple SODs with the same metal dependency. In addition to the three core enterobacterial SODs, some Salmonella enterica serovars express four SODs, having gained an additional Cu/Zn SOD. Due to the order of discovery, the common Cu/Zn SOD is referred to as SodC2, while the additional enzyme is SodC1. While many studies have demonstrated the broad importance of SodC in this genus, it was less clear if encoding multiple SodC would provide unique superoxide protection derived from each enzyme. The SodC1 and SodC2 enzymes are both periplasmic SODs from the Cu/Zn family and were originally thought to provide overlapping protection from exogenous superoxide stress, however, now it is appreciated that they have non-overlapping, non-redundant roles. SodC2 mutants in Salmonella enterica serovar Choleraesuis have an intracellular survival defect relative to both wild type and SodC1 mutants in macrophages and human colon carcinoma cells [104]. Additionally, during in vivo competition experiments, double (sodC1/sodC2) mutants are not further attenuated from single (sodC1 or sodC2) mutants, supporting that these SODs provide complementary roles [104,118]. While these roles in S. enterica serovar Choleraesuis demonstrate that SodC2 is the primary SOD during macrophage and cell survival, many of the findings do not align with what has been seen in S. enterica serovar Typhimurium, suggesting that the contributions from each of the SodC may vary from strain to strain and/or serovar to serovar. In S. enterica serovar Typhimurium, deletion of SodC1, but not SodC2, results in a 7- to 10-fold reduction in virulence during intraperitoneal competitions [119–121]. The differences in effects on virulence between the two SodC in S. enterica serovar Typhimurium are likely due to two key differences in the proteins themselves, first, SodC1 forms a dimer like most other Cu/Zn SODs while SodC2 forms a monomer, and second, SodC1 tethers to the periplasm via peptidoglycan binding while SodC2 does not [119,122,123]. While there have been numerous advances in our understanding of the diversity of SODs, these studies also demonstrate that there is still much to be learned about the forces that drive expansion and diversification of the SOD repertoires possessed by microorganisms.
6. Evolution of the superoxide dismutases
SODs are very ancient enzymes. It seems likely that SODs evolved and diversified concomitant with the evolution of oxygenic photosynthesis. Prior to this, the highly reducing and sulfidic conditions in the early Earth’s atmosphere and oceans would have made widespread biological superoxide stress unlikely. The ancestors of cyanobacteria that first evolved oxygen-evolving metabolic pathways may have been the first organisms to experience such a persistent stress due to their creating elevated local dioxygen concentrations, making them likely candidates to have evolved the first SOD solution to this problem. This would date the origin of SODs to before the “Great Oxygenation Event”, the period in which atmospheric concentrations of dioxygen rose sharply as detected in the geological record around 2.4 billion years ago [124,125]. Atmospheric dioxygen would have made profound changes to the ecological availabilities of key biologically essential metals. While Fe would have undergone a dramatic decline in bioavailability during the transition from an anaerobic to an aerobic world due to its low aqueous solubility in the presence of oxygen, the opposite would be true for Cu and Zn [126]. Perhaps it is no coincidence that these metals were subsequently adopted for the disproportion of superoxide by an alternative SOD family [124].
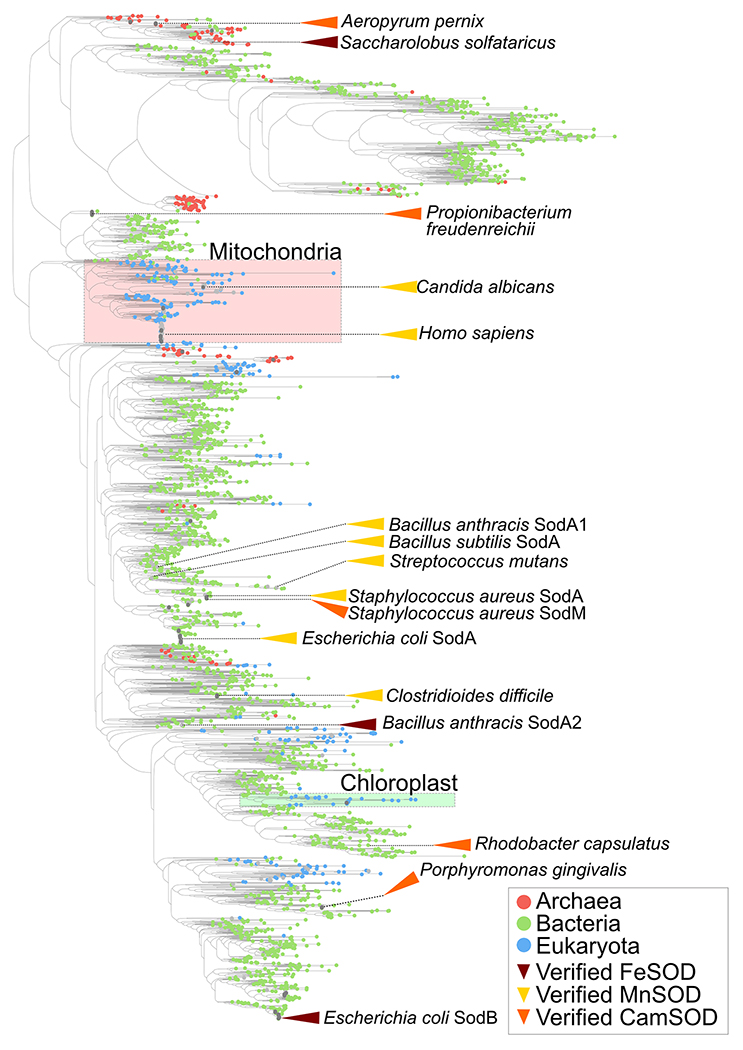
Due to the three SOD families being discrete (i.e., having distinct structural folds), their evolutionary history must be studied independently and thus varying amounts of bioinformatic analyses have been performed on each SOD type. The evolution of the Fe/Mn-SODs is by far the most extensively studied. This family is ubiquitous across biology, present in the genomes of Archaea, Bacteria and Eukarya (including endosymbiont-derived organellar homologs in the latter). They are even frequently observed in the genomes of obligate anaerobes. The preponderance of this SOD type supports the hypothesis that they are the most ancient lineage of SODs, which have spread through the tree of life through vertical transmission, and is consistent with their cofactor – whether Fe or Mn – being abundant on the early Earth prior to widespread oxygenation. Early bioinformatic analyses identified conserved sequence motifs that could differentiate two distinct sub-types of the Fe/Mn-SOD family, drew the conclusion that these sub-types correlated with the two metal specificities of SODs known at the time, Fe-SOD and Mn-SOD, and sought to use them to predict metal specificities of uncharacterized enzymes [47,124,127]. When linked with geological analyses, the common ancestor of these two groups was even proposed to be cambialistic [124]. However, more recent analyses of much larger Fe/Mn-SOD family sequence sets have demonstrated that their evolutionary history is more complex, especially regarding conservation and distribution of their metal specificities (Figure 3) [28]. Crucially, multiple different metal specificities – Mn-SOD, Fe-SOD and cam-SOD – are observed within both of the major phylogenetic groups [28]. This suggests that members of this enzyme family can switch metal specificities within relatively short evolutionary time scales. Indeed, mutagenesis studies of the S. aureus SODs, as described above, demonstrated the molecular mechanism by which such a switch in metal specificity has occurred during their differentiation from other Staphylococci through mutation of a limited number of amino acid residues that spatially localize to the metal’s secondary coordination sphere [28]. Given this capacity for altering metal specificity, we hypothesize that evolutionary switching of the preferred metal cofactor will have occurred regularly across the phylogenetic tree of Fe/Mn-SODs, and therefore, that metal specificity will not strictly correlate with the detected phylogenetic groups or group-specific amino acid motifs observed bioinformatically.
Figure 3. Phylogenetic relationships and metal specificities of the Fe/Mn-SOD family.
Phylogenetic tree illustrating the evolutionary relationship between the protein sequences of 2,691 members of the Fe/Mn-SOD family from across the tree of life [28], including isozymes from bacteria (green circles), archaea (red circles) and eukaryotes (blue circles). Groups of sequences that include the eukaryotic mitochondrial (red region) and chloroplast (green region) SODs, likely inherited from the ancestral organellar endosymbionts, are illustrated. Determined metal-specificities of characterized enzymes (MnSOD=yellow, FeSOD=brown; camSOD=orange) are annotated with triangles. Cofactor specificity does not correlate with SOD divergence suggesting that evolution can switch metal specificities over time within this SOD family.
If such switches in metal specificity are common, a key question is how such changes are selected through evolution. It has been hypothesized that changes in metal availability are likely a key selection pressure that drives such evolutionary changes [28]. Many SODs from eukaryotic pathogens are predicted to be Fe-specific enzymes rather than Mn-specific, a striking parallel with bacterial pathogens such as S. aureus (SodM) and B. anthracis (SodA2) that have gained SODs with Fe-dependent activity that are not present in their non-pathogenic relatives [69]. This suggest that acquisition of Fe-specific or cambialistic SODs may provide an important selective advantage during infection for both bacterial and eukaryotic pathogens. The mechanisms by which the Fe/Mn-SODs acquire their target cofactor inside cells has not been directly tested. No metallochaperones have been implicated in their metal delivery, and their metal loading is influenced by exogenous metal concentrations during both heterologous expression inside E. coli cells and, in some cases, in their own native host in vivo [28,31,117]. The latter observation is consistent with a model in which these SODs acquire their cofactor by competing against innate intracellular metal-buffers, rather than having their metal selectivity controlled by a third-party client metal-delivery system [128,129]. In such a model, it is tempting to speculate that changes in SOD metal specificity could be driven through changes in in vivo metal bioavailability, either during adaptation of a SOD to a new intracellular environment after duplication or horizontal gene transfer (HGT), or through a shift in ecological niche of its existing host. The dual SOD system of S. aureus may represent an example of the latter, in which acquisition and neofunctionalization of the cam-SOD coincided with the transition of S. aureus to an opportunistic pathogenic lifestyle and was concomitant with its acquisition of additional components of Fe acquisition and homeostasis [28]. This model is supported by the known immune effects on S. aureus metal acquisition during infection, in which it experiences Mn starvation through the metal sequestration activity of calprotectin released into its immediate environment by immune cells [91,113,130]. Nonetheless, the hypothesis that switches in SOD metal specificity can be driven by a switch in bacteria from an environmental lifestyle to becoming a pathogen awaits direct testing.
7. Conclusions
While SODs have been extensively studied, examining them through the lens of pathogenesis has revealed that, despite carrying out the same chemical reactions, individual SODs localized within the same compartment can and frequently do contribute differently to microbial success. This work has also revealed that there is still much to discover regarding these ubiquitous enzymes. The three SOD types (Fe/Mn, Cu/Zn and Ni) are clear examples of convergent evolution, demonstrating the crucial importance of superoxide detoxification in enabling life to survive and thrive in the presence of oxygen. As a result, the rigorous phylogenetic studies that are needed to dissect the evolutionary history of SODs must be performed independently for each SOD family. Nonetheless, prior analyses have demonstrated clear examples of family expansion via duplication and/or HGT often followed by neofunctionalization within all three protein families that have altered or expanded the repertoire of enzymes possessed by evolutionarily related organisms [22,28,131]. The extant distribution of each family has been driven by different selection pressures, including different accumulation and handling of essential metal ions by different organisms and the need to protect individual subcellular compartments. However, our limited understanding of the molecular features that dictate metal utilization and the uneven characterization of SODs from across the evolutionary landscape prevent a full understanding of SOD diversity, the underlying molecular rationale for their diversification, and any biological constraints there may be on neofunctionalization of metal specificity. The Fe/Mn-SOD family exhibits substantial flexibility in cofactor utilization, and evolution appears to have exploited this flexibility by switching the specificity of the Fe/Mn isozymes to ensure retention of SOD activity as metal availability changes within a given organism/niche. While this hypothesis is highly appealing based on the geological record and observations made in the context of pathogenesis, it is yet to be demonstrated that metal availability can select for altered SOD metal-specificity. Further work is necessary to test this hypothesis and determine the other evolutionary pressures that underly the extant diversification of SOD function and distribution.
The study of SODs from pathogenic microbes has also revealed new insights into the molecular function of these enzymes, as well as their evolution. This work has revealed that the metal specificity of the Fe/Mn family of SODs is fluid and better thought of as a continuum rather than enzymes that fall strictly into an Fe-dependent or Mn-dependent bin. Studying SODs from pathogens has provided invaluable details regarding the secondary sphere residues that tune the metal’s catalytic power, and how this has been exploited by evolution to manipulate the SOD repertoire. Only with higher resolution and more dynamic information about the structures of diverse SODs of this type that cover the full range of the metal specificity spectrum, including of mutant forms with moderated specificity, will the field be able to understand both the molecular mechanism of catalysis and the mechanisms by which second sphere residues control metal specificity. A further goal for the field must be the development of improved methods for quantifying the reduction potential of the catalytic metal ions in vitro to enable direct testing of the redox tuning hypothesis. Such technical advances in the biochemical, biophysical, and structural characterization of SODs, combined with the power of bacterial genetics for cellular and infection studies and in silico evolutionary analysis of ever-expanding sequence databases, can glean new insights into the function of these globally crucial metalloenzymes. Despite over 50 years of work on SODs across all domains of life, these evolutionarily ancient enzymes still have new secrets to reveal.
Key Points.
Fe/Mn superoxide dismutases can use iron, manganese, or both metals for function.
Metal specificity in the Fe/Mn superoxide dismutase family is a spectrum.
Metal availability can shape an organism’s superoxide dismutase repertoire.
Distinct clades exist within the Fe/Mn superoxide dismutase family.
The metal specificity of an Fe/Mn superoxide dismutase can evolutionarily change.
8. Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 AI155611 and R21 AI149115), and a Vallee Scholar Award to T.E.K-F. This work was also supported by a grant (BB/S006818/1) from Biotechnology and Biological Sciences Research Council (BBSRC) to K.J.W. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH, Biotechnology and Biological Sciences Research Council, or Vallee Foundation.
9. References
- [1].Massey V, Strickland S, Mayhew SG, Howell LG, Engel PC, Matthews RG, Schuman M, Sullivan PA, The production of superoxide anion radicals in the reaction of reduced flavins and flavoproteins with molecular oxygen, Biochemical and Biophysical Research Communications. 36 (1969) 891–897. 10.1016/0006-291X(69)90287-3. [DOI] [PubMed] [Google Scholar]
- [2].Messner KR, Imlay JA, The Identification of Primary Sites of Superoxide and Hydrogen Peroxide Formation in the Aerobic Respiratory Chain and Sulfite Reductase Complex of Escherichia coli, Journal of Biological Chemistry. 274 (1999) 10119–10128. 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- [3].Imlay JA, Pathways of Oxidative Damage, Annual Review of Microbiology. 57 (2003) 395–418. 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- [4].Imlay JA, Cellular Defenses against Superoxide and Hydrogen Peroxide, Annual Review of Biochemistry. 77 (2008) 755–776. 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Beyer W, Imlay J, Fridovich I, Superoxide Dismutases, Progress in Nucleic Acid Research and Molecular Biology. 40 (1991) 221–253. 10.1016/S0079-6603(08)60843-0. [DOI] [PubMed] [Google Scholar]
- [6].Rigby KM, DeLeo FR, Neutrophils in innate host defense against Staphylococcus aureus infections, Seminars in Immunopathology. 34 (2012) 237–259. 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fenlon LA, Slauch JM, Phagocyte Roulette in Salmonella Killing, Cell Host & Microbe. 15 (2014) 7–8. 10.1016/j.chom.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Burton NA, Schürmann N, Casse O, Steeb AK, Claudi B, Zankl J, Schmidt A, Bumann D, Disparate Impact of Oxidative Host Defenses Determines the Fate of Salmonella during Systemic Infection in Mice, Cell Host & Microbe. 15 (2014) 72–83. 10.1016/j.chom.2013.12.006. [DOI] [PubMed] [Google Scholar]
- [9].Broxton CN, Culotta VC, SOD Enzymes and Microbial Pathogens: Surviving the Oxidative Storm of Infection, PLoS Pathog 12 (2016) e1005295. 10.1371/journal.ppat.1005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Quie PG, Kaplan EL, Page AR, Gruskay FL, Malawista SE, Defective Polymorphonuclear-Leukocyte Function and Chronic Granulomatous Disease in Two Female Children, N Engl J Med 278 (1968) 976–980. 10.1056/NEJM196805022781802. [DOI] [PubMed] [Google Scholar]
- [11].Buvelot H, Posfay-Barbe KM, Linder P, Schrenzel J, Krause K-H, Staphylococcus aureus, phagocyte NADPH oxidase and chronic granulomatous disease, FEMS Microbiology Reviews. (2016). 10.1093/femsre/fuw042. [DOI] [PubMed] [Google Scholar]
- [12].Roos D, Chronic granulomatous disease, Br Med Bull. 118 (2016) 50–63. 10.1093/bmb/ldw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hampton MB, Kettle AJ, Winterbourn CC, Involvement of superoxide and myeloperoxidase in oxygen-dependent killing of Staphylococcus aureus by neutrophils, Infect Immun. 64 (1996) 3512–3517. 10.1128/iai.64.9.3512-3517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Brioukhanov AL, Netrusov AI, Catalase and Superoxide Dismutase: Distribution, Properties, and Physiological Role in Cells of Strict Anaerobes, Biochemistry (Moscow). 69 (2004) 949–962. 10.1023/B:BIRY.0000043537.04115.d9. [DOI] [PubMed] [Google Scholar]
- [15].Lynch M, Kuramitsu H, Expression and role of superoxide dismutases (SOD) in pathogenic bacteria., Microbes and Infection / Institut Pasteur. 2 (2000) 1245–1255. https://doi.org/S1286-4579(00)01278-8 [pii]. [DOI] [PubMed] [Google Scholar]
- [16].McCord JM, Fridovich I, Superoxide Dismutase: An enzymic function for erythrocuprein (hemocuprein), Journal of Biological Chemistry. 244 (1969) 6049–6055. 10.1016/S0021-9258(18)63504-5. [DOI] [PubMed] [Google Scholar]
- [17].Miller AF, Superoxide dismutases: Ancient enzymes and new insights, FEBS Letters. 586 (2012) 585–595. 10.1016/j.febslet.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Robinett NG, Peterson RL, Culotta VC, Eukaryotic copper-only superoxide dismutases (SODs): A new class of SOD enzymes and SOD-like protein domains, Journal of Biological Chemistry. 293 (2018) 4636–4643. 10.1074/jbc.TM117.000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Garcia YM, Barwinska-Sendra A, Tarrant E, Skaar EP, Waldron KJ, Kehl-Fie TE, A Superoxide Dismutase Capable of Functioning with Iron or Manganese Promotes the Resistance of Staphylococcus aureus to Calprotectin and Nutritional Immunity, PLOS Pathogens. 13 (2017) e1006125. 10.1371/journal.ppat.1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sheng Y, Abreu IA, Cabelli DE, Maroney MJ, Miller AF, Teixeira M, Valentine JS, Superoxide dismutases and superoxide reductases, Chemical Reviews. 114 (2014) 3854–3918. 10.1021/cr4005296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Youn H-D, Kim E-J, Roe J-H, Hah YC, Kang S-O, A novel nickel-containing superoxide dismutase from Streptomyces spp, Biochemical Journal. 318 (1996) 889–896. 10.1042/bj3180889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sutherland KM, Ward LM, Colombero C-R, Johnston DT, Inter-domain horizontal gene transfer of nickel-binding superoxide dismutase, Geobiology. 19 (2021) 450–459. 10.1111/gbi.12448. [DOI] [PubMed] [Google Scholar]
- [23].Dupont CL, Neupane K, Shearer J, Palenik B, Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases, Environ Microbiol. 10 (2008) 1831–1843. 10.1111/j.1462-2920.2008.01604.x. [DOI] [PubMed] [Google Scholar]
- [24].Herbst RW, Guce A, Bryngelson PA, Higgins KA, Ryan KC, Cabelli DE, Garman SC, Maroney MJ, Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution, Biochemistry. 48 (2009) 3354–3369. 10.1021/bi802029t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ming LJ, Valentine JS, NMR studies of nickel(II)-substituted derivatives of bovine copper-zinc superoxide dismutase with nickel(II) bound in the copper site, J. Am. Chem. Soc. 112 (1990) 6374–6383. 10.1021/ja00173a028. [DOI] [Google Scholar]
- [26].Vance CK, Miller A-F, A Simple Proposal That Can Explain the Inactivity of Metal-Substituted Superoxide Dismutases, J. Am. Chem. Soc. 120 (1998) 461–467. 10.1021/ja972060j. [DOI] [Google Scholar]
- [27].Miller AF, Superoxide dismutases: active sites that save, but a protein that kills, Current Opinion in Chemical Biology. 8 (2004) 162–168. 10.1016/j.cbpa.2004.02.011. [DOI] [PubMed] [Google Scholar]
- [28].Barwinska-Sendra A, Garcia YM, Sendra KM, Baslé A, Mackenzie ES, Tarrant E, Card P, Tabares LC, Bicep C, Un S, Kehl-Fie TE, Waldron KJ, An evolutionary path to altered cofactor specificity in a metalloenzyme, Nat Commun. 11 (2020) 2738. 10.1038/s41467-020-16478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ose DE, Fridovich I, Superoxide dismutase. Reversible removal of manganese and its substitution by cobalt, nickel or zinc., Journal of Biological Chemistry. 251 (1976) 1217–1218. 10.1016/S0021-9258(17)33823-1. [DOI] [PubMed] [Google Scholar]
- [30].Yang M, Cobine PA, Molik S, Naranuntarat A, Lill R, Winge DR, Culotta VC, The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2, EMBO J. 25 (2006) 1775–1783. 10.1038/sj.emboj.7601064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Martin ME, Byers BR, Olson MOJ, Salin ML, Arceneaux JE, Tolbert C, A Streptococcus mutans superoxide dismutase that is active with either manganese or iron as a cofactor, Journal of Biological Chemistry. 261 (1986) 9361–9367. 10.1016/S0021-9258(18)67663-X. [DOI] [PubMed] [Google Scholar]
- [32].Nakayama K, Rapid viability loss on exposure to air in a superoxide dismutase-deficient mutant of Porphyromonas gingivalis, J Bacteriol. 176 (1994) 1939–1943. 10.1128/jb.176.7.1939-1943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lynch MC, Kuramitsu HK, Role of Superoxide Dismutase Activity in the Physiology of Porphyromonas gingivalis, Infect Immun. 67 (1999) 3367–3375. 10.1128/IAI.67.7.3367-3375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nakayama K, Nucleotide sequence of Streptococcus mutans superoxide dismutase gene and isolation of insertion mutants, J Bacteriol. 174 (1992) 4928–4934. 10.1128/jb.174.15.4928-4934.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yesilkaya H, Kadioglu A, Gingles N, Alexander JE, Mitchell TJ, Andrew PW, Role of Manganese-Containing Superoxide Dismutase in Oxidative Stress and Virulence of Streptococcus pneumoniae, Infect Immun. 68 (2000) 2819–2826. 10.1128/IAI.68.5.2819-2826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Meier B, Barra D, Bossa F, Calabrese L, Rotilio G, Synthesis of either Fe- or Mn-superoxide dismutase with an apparently identical protein moiety by an anaerobic bacterium dependent on the metal supplied, Journal of Biological Chemistry. 257 (1982) 13977–13980. 10.1016/S0021-9258(19)45329-5. [DOI] [PubMed] [Google Scholar]
- [37].Gregory EM, Characterization of the O2-induced manganese-containing superoxide dismutase from Bacteroides fragilis, Archives of Biochemistry and Biophysics. 238 (1985) 83–89. 10.1016/0003-9861(85)90143-2. [DOI] [PubMed] [Google Scholar]
- [38].Eijkelkamp BA, Morey JR, Ween MP, Ong CLY, McEwan AG, Paton JC, McDevitt CA, Extracellular zinc competitively inhibits manganese uptake and compromises oxidative stress management in Streptococcus pneumoniae, PLoS ONE. 9 (2014). 10.1371/journal.pone.0089427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Amano A, Shizukuishi S, Tamagawa H, Iwakura K, Tsunasawa S, Tsunemitsu A, Characterization of superoxide dismutases purified from either anaerobically maintained or aerated Bacteroides gingivalis, J Bacteriol. 172 (1990) 1457–1463. 10.1128/jb.172.3.1457-1463.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Edwards RA, Baker HM, Whittaker MM, Whittaker JW, Jameson GB, Baker EN, Crystal structure of Escherichia coli manganese superoxide dismutase at 2.1-Å resolution, J Biol Inorg Chem. 3 (1998) 161–171. 10.1007/s007750050217. [DOI] [Google Scholar]
- [41].Stallings WC, Powers TB, Pattridge KA, Fee JA, Ludwig ML, Iron superoxide dismutase from Escherichia coli at 3.1-A resolution: a structure unlike that of copper/zinc protein at both monomer and dimer levels., Proceedings of the National Academy of Sciences. 80 (1983) 3884–3888. 10.1073/pnas.80.13.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Grove LE, Xie J, Yikilmaz E, Miller A-F, Brunold TC, Spectroscopic and Computational Investigation of Second-Sphere Contributions to Redox Tuning in Escherichia coli Iron Superoxide Dismutase, Inorg. Chem. 47 (2008) 3978–3992. 10.1021/ic702412y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Vance CK, Miller A-F, Novel Insights into the Basis for Escherichia coli Superoxide Dismutase’s Metal Ion Specificity from Mn-Substituted FeSOD and Its Very High E m, Biochemistry. 40 (2001) 13079–13087. 10.1021/bi0113317. [DOI] [PubMed] [Google Scholar]
- [44].Spagnolo L, Törö I, D’Orazio M, O’Neill P, Pedersen JZ, Carugo O, Rotilio G, Battistoni A, Djinović-Carugo K, Unique Features of the sodC-encoded Superoxide Dismutase from Mycobacterium tuberculosis, a Fully Functional Copper-containing Enzyme Lacking Zinc in the Active Site, Journal of Biological Chemistry. 279 (2004) 33447–33455. 10.1074/jbc.M404699200. [DOI] [PubMed] [Google Scholar]
- [45].Gleason JE, Galaleldeen A, Peterson RL, Taylor AB, Holloway SP, Waninger-Saroni J, Cormack BP, Cabelli DE, Hart PJ, Culotta VC, Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense, Proceedings of the National Academy of Sciences. 111 (2014) 5866–5871. 10.1073/pnas.1400137111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schatzman SS, Culotta VC, Chemical Warfare at the Microorganismal Level: A Closer Look at the Superoxide Dismutase Enzymes of Pathogens, ACS Infectious Diseases. 4 (2018) 893–903. 10.1021/acsinfecdis.8b00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Grace SC, Phylogenetic distribution of superoxide dismutase supports an endosymbiotic origin for chloroplasts and mitochondria, Life Sciences. 47 (1990) 1875–1886. 10.1016/0024-3205(90)90399-C. [DOI] [PubMed] [Google Scholar]
- [48].Hayakawa T, Kanematsu S, Asada K, Purification and characterization of thylakoid-bound Mn-superoxide dismutase in spinach chloroplasts, Planta. 166 (1985) 111–116. 10.1007/BF00397393. [DOI] [PubMed] [Google Scholar]
- [49].del Río LA, Lyon DS, Olah I, Glick B, Salin ML, Immunocytochemical evidence for a peroxisomal localization of manganese superoxide dismutase in leaf protoplasts from a higher plant, Planta. 158 (1983) 216–224. 10.1007/BF01075257. [DOI] [PubMed] [Google Scholar]
- [50].Jackson C, Dench J, Moore AL, Halliwell B, Foyer CH, Hall DO, Subcellular Localisation and Identification of Superoxide Dismutase in the Leaves of Higher Plants, Eur J Biochem. 91 (1978) 339–344. 10.1111/j.1432-1033.1978.tb12685.x. [DOI] [PubMed] [Google Scholar]
- [51].Sevilla F, López-Gorgé J, del Río LA, Characterization of a Manganese Superoxide Dismutase from the Higher Plant Pisum sativum, Plant Physiol. 70 (1982) 1321–1326. 10.1104/pp.70.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Phillips JP, Tainer JA, Getzoff ED, Boulianne GL, Kirby K, Hilliker AJ, Subunit-destabilizing mutations in Drosophila copper/zinc superoxide dismutase: neuropathology and a model of dimer dysequilibrium., Proceedings of the National Academy of Sciences. 92 (1995) 8574–8578. 10.1073/pnas.92.19.8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Blackney MJ, Cox R, Shepherd D, Parker JD, Cloning and expression analysis of Drosophila extracellular Cu Zn superoxide dismutase, Bioscience Reports. 34 (2014) e00164. 10.1042/BSR20140133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Phillips JP, Campbell SD, Michaud D, Charbonneau M, Hilliker AJ, Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity., Proceedings of the National Academy of Sciences. 86 (1989) 2761–2765. 10.1073/pnas.86.8.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rogina B, Helfand SL, Cu, Zn superoxide dismutase deficiencyaccelerates the time course of an age-related markerin Drosophila melanogaster, Biogerontology. 1 (2000) 163–169. 10.1023/A:1010039813107. [DOI] [PubMed] [Google Scholar]
- [56].Kirby K, Hu J, Hilliker AJ, Phillips JP, RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress, Proceedings of the National Academy of Sciences. 99 (2002) 16162–16167. 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wang Y, Branicky R, Noë A, Hekimi S, Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling, The Journal of Cell Biology. (2018) jcb.201708007. 10.1083/jcb.201708007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li Y, Huang T-T, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ, Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase, Nat Genet. 11 (1995) 376–381. 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- [59].Lebovitz RM, Zhang H, Vogel H, Cartwright J, Dionne L, Lu N, Huang S, Matzuk MM, Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice., Proceedings of the National Academy of Sciences. 93 (1996) 9782–9787. 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fukai T, Ushio-Fukai M, Superoxide Dismutases: Role in Redox Signaling, Vascular Function, and Diseases, Antioxidants & Redox Signaling. 15 (2011) 1583–1606. 10.1089/ars.2011.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Miao L, St DK. Clair, Regulation of superoxide dismutase genes: Implications in disease, Free Radical Biology and Medicine. 47 (2009) 344–356. 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Van Remmen H, Williams MD, Guo Z, Estlack L, Yang H, Carlson EJ, Epstein CJ, Huang TT, Richardson A, Knockout mice heterozygous for sod2 show alterations in cardiac mitochondrial function and apoptosis, American Journal of Physiology-Heart and Circulatory Physiology. 281 (2001) H1422–H1432. 10.1152/ajpheart.2001.281.3.H1422. [DOI] [PubMed] [Google Scholar]
- [63].Holley AK, Dhar SK, St DK. Clair, Curbing cancer’s sweet tooth: Is there a role for MnSOD in regulation of the Warburg effect?, Mitochondrion. 13 (2013) 170–188. 10.1016/j.mito.2012.07.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xu Y, Miriyala S, Fang F, Bakthavatchalu V, Noel T, Schell DM, Wang C, St Clair WH, St Clair DK, Manganese superoxide dismutase deficiency triggers mitochondrial uncoupling and the Warburg effect, Oncogene. 34 (2015) 4229–4237. 10.1038/onc.2014.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fukai T, Extracellular superoxide dismutase and cardiovascular disease, Cardiovascular Research. 55 (2002) 239–249. 10.1016/S0008-6363(02)00328-0. [DOI] [PubMed] [Google Scholar]
- [66].Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng H-X, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung W-Y, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak–Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH, Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis, Nature. 362 (1993) 59–62. 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- [67].Martínez A, Prolo C, Estrada D, Rios N, Alvarez MN, Piñeyro MD, Robello C, Radi R, Piacenza L, Cytosolic Fe-superoxide dismutase safeguards Trypanosoma cruzi from macrophage-derived superoxide radical, Proc Natl Acad Sci USA. 116 (2019) 8879–8888. 10.1073/pnas.1821487116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wilkinson SR, Prathalingam SR, Taylor MC, Ahmed A, Horn D, Kelly JM, Functional characterisation of the iron superoxide dismutase gene repertoire in Trypanosoma brucei, Free Radical Biology and Medicine. 40 (2006) 198–209. 10.1016/j.freeradbiomed.2005.06.022. [DOI] [PubMed] [Google Scholar]
- [69].Dufernez F, Yernaux C, Gerbod D, Noël C, Chauvenet M, Wintjens R, Edgcomb VP, Capron M, Opperdoes FR, Viscogliosi E, The presence of four iron-containing superoxide dismutase isozymes in Trypanosomatidae: Characterization, subcellular localization, and phylogenetic origin in Trypanosoma brucei, Free Radical Biology and Medicine. 40 (2006) 210–225. 10.1016/j.freeradbiomed.2005.06.021. [DOI] [PubMed] [Google Scholar]
- [70].Mittra B, Laranjeira-Silva MF, Miguel DC, Perrone J Bezerra de Menezes, N.W. Andrews, The iron-dependent mitochondrial superoxide dismutase SODA promotes Leishmania virulence, Journal of Biological Chemistry. 292 (2017) 12324–12338. 10.1074/jbc.M116.772624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Youseff BH, Holbrook ED, Smolnycki KA, Rappleye CA, Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress, PLoS Pathog 8 (2012) e1002713. 10.1371/journal.ppat.1002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Martchenko M, Alarco A-M, Harcus D, Whiteway M, Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene, Mol Biol Cell. 15 (2004) 456–467. 10.1091/mbc.e03-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Li CX, Gleason JE, Zhang SX, Bruno VM, Cormack BP, Culotta VC, Candida albicans adapts to host copper during infection by swapping metal cofactors for superoxide dismutase, Proc Natl Acad Sci USA. 112 (2015) E5336–E5342. 10.1073/pnas.1513447112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Besold AN, Gilston BA, Radin JN, Ramsoomair C, Culbertson EM, Li CX, Cormack BP, Chazin WJ, Kehl-Fie TE, Culotta VC, Role of Calprotectin in withholding zinc and copper from Candida albicans, Infect Immun. 86 (2017) e00779–17. 10.1128/IAI.00779-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].D’Orazio M, Folcarelli S, Mariani F, Colizzi V, Rotilio G, Battistoni A, Lipid modification of the Cu,Zn superoxide dismutase from Mycobacterium tuberculosis, Biochemical Journal. 359 (2001) 17–22. 10.1042/bj3590017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Benov L, Chang LY, Day B, Fridovich I, Copper, Zinc Superoxide Dismutase in Escherichia coli: Periplasmic Localization, Archives of Biochemistry and Biophysics. 319 (1995) 508–511. 10.1006/abbi.1995.1324. [DOI] [PubMed] [Google Scholar]
- [77].Esteve-Gassent MD, Elliott NL, Seshu J, sodA is essential for virulence of Borrelia burgdorferi in the murine model of Lyme disease, Molecular Microbiology. 71 (2009) 594–612. 10.1111/j.1365-2958.2008.06549.x. [DOI] [PubMed] [Google Scholar]
- [78].Pesci EC, Cottle DL, Pickett CL, Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni, Infect Immun. 62 (1994) 2687–2694. 10.1128/iai.62.7.2687-2694.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Purdy D, Cawthraw S, Dickinson JH, Newell DG, Park SF, Generation of a superoxide dismutase (SOD)-deficient mutant of Campylobacter coli : evidence for the significance of SOD in Campylobacter survival and colonization, Appl Environ Microbiol. 65 (1999) 2540–2546. 10.1128/AEM.65.6.2540-2546.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Troxell B, Xu H, Yang XF, Borrelia burgdorferi, a pathogen that lacks iron, encodes manganese-dependent superoxide dismutase essential for resistance to Streptonigrin, Journal of Biological Chemistry. 287 (2012) 19284–19293. 10.1074/jbc.M112.344903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Aguirre JD, Clark HM, McIlvin M, Vazquez C, Palmere SL, Grab DJ, Seshu J, Hart PJ, Saito M, Culotta VC, A manganese-rich environment supports superoxide dismutase activity in a Lyme Disease pathogen, Borrelia burgdorferi, Journal of Biological Chemistry. 288 (2013) 8468–8478. 10.1074/jbc.M112.433540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hermans D, Pasmans F, Messens W, Martel A, Van Immerseel F, Rasschaert G, Heyndrickx M, Van Deun K, Haesebrouck F, Poultry as a host for the zoonotic pathogen Campylobacter jejuni, Vector-Borne and Zoonotic Diseases. 12 (2012) 89–98. 10.1089/vbz.2011.0676. [DOI] [PubMed] [Google Scholar]
- [83].Pennington CD, Gregory EM, Isolation and reconstitution of iron- and manganese-containing superoxide dismutases from Bacteroides thetaiotaomicron, Journal of Bacteriology. 166 (1986) 528–532. 10.1128/jb.166.2.528-532.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Tally FP, Goldin BR, Jacobus NV, Gorbach SL, Superoxide dismutase in anaerobic bacteria of clinical significance, Infect Immun. 16 (1977) 20–25. 10.1128/iai.16.1.20-25.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].St John G, Steinman HM, Periplasmic copper-zinc superoxide dismutase of Legionella pneumophila: role in stationary-phase survival, J Bacteriol. 178 (1996) 1578–1584. 10.1128/jb.178.6.1578-1584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Sadosky AB, Wilson JW, Steinman HM, Shuman HA, The iron superoxide dismutase of Legionella pneumophila is essential for viability, J Bacteriol. 176 (1994) 3790–3799. 10.1128/jb.176.12.3790-3799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Dussurget O, Stewart G, Neyrolles O, Pescher P, Young D, Marchal G, Role of Mycobacterium tuberculosis Copper-Zinc Superoxide Dismutase, Infect Immun. 69 (2001) 529–533. 10.1128/IAI.69.1.529-533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Edwards KM, Cynamon MH, Voladri RKR, Hager CC, DeSTEFANO MS, Tham KT, Lakey DL, Bochan MR, Kernodle DS, Iron-cofactored Superoxide Dismutase Inhibits Host Responses to Mycobacterium tuberculosis, Am J Respir Crit Care Med. 164 (2001) 2213–2219. 10.1164/ajrccm.164.12.2106093. [DOI] [PubMed] [Google Scholar]
- [89].Piddington DL, Fang FC, Laessig T, Cooper AM, Orme IM, Buchmeier NA, Cu,Zn Superoxide Dismutase of Mycobacterium tuberculosis Contributes to Survival in Activated Macrophages That Are Generating an Oxidative Burst, Infect Immun. 69 (2001) 4980–4987. 10.1128/IAI.69.8.4980-4987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Karavolos MH, Horsburgh M, Ingham E, Foster S, Role and regulation of the superoxide dismutases of Staphylococcus aureus, Microbiology. 149 (2003) 2749–2758. 10.1099/mic.0.26353-0. [DOI] [PubMed] [Google Scholar]
- [91].Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP, Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus, Cell Host and Microbe. 10 (2011) 158–164. 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gao Q, Xia L, Wang X, Ye Z, Liu J, Gao S, SodA Contributes to the Virulence of Avian Pathogenic Escherichia coli O2 Strain E058 in Experimentally Infected Chickens, J Bacteriol. 201 (2019). 10.1128/JB.00625-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Franzon VL, Arondel J, Sansonetti PJ, Contribution of superoxide dismutase and catalase activities to Shigella flexneri pathogenesis, Infect Immun. 58 (1990) 529–535. 10.1128/iai.58.2.529-535.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zhang J, Wang H, Huang Q, Zhang Y, Zhao L, Liu F, Wang G, Four superoxide dismutases of Bacillus cereus 0–9 are non-redundant and perform different functions in diverse living conditions, World J Microbiol Biotechnol. 36 (2020) 12. 10.1007/s11274-019-2786-7. [DOI] [PubMed] [Google Scholar]
- [95].Cybulski RJ, Sanz P, Alem F, Stibitz S, Bull RL, O’Brien AD, Four Superoxide Dismutases Contribute to Bacillus anthracis Virulence and Provide Spores with Redundant Protection from Oxidative Stress, Infect Immun. 77 (2009) 274–285. 10.1128/IAI.00515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wang Y, Mo X, Zhang L, Wang Q, Four superoxide dismutase (isozymes) genes of Bacillus cereus, Ann Microbiol. 61 (2011) 355–360. 10.1007/s13213-010-0149-6. [DOI] [Google Scholar]
- [97].Tu WY, Pohl S, Gray J, Robinson NJ, Harwood CR, Waldron KJ, Cellular iron distribution in Bacillus anthracis, Journal of Bacteriology. 194 (2012) 932–940. 10.1128/JB.06195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Liu H, Bergman NH, Thomason B, Shallom S, Hazen A, Crossno J, Rasko DA, Ravel J, Read TD, Peterson SN, Yates J, Hanna PC, Formation and Composition of the Bacillus anthracis Endospore, J Bacteriol. 186 (2004) 164–178. 10.1128/JB.186.1.164-178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Steichen C, Chen P, Kearney JF, Turnbough CL, Identification of the Immunodominant Protein and Other Proteins of the Bacillus anthracis Exosporium, J Bacteriol. 185 (2003) 1903–1910. 10.1128/JB.185.6.1903-1910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Valderas MW, Hart ME, Identification and Characterization of a Second Superoxide Dismutase Gene (sodM) from Staphylococcus aureus, Journal of Bacteriology. 183 (2001) 3399–3407. 10.1128/JB.183.11.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Casillas-Martinez L, Setlow P, Alkyl hydroperoxide reductase, catalase, MrgA, and superoxide dismutase are not involved in resistance of Bacillus subtilis spores to heat or oxidizing agents, J Bacteriol. 179 (1997) 7420–7425. 10.1128/jb.179.23.7420-7425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Inaoka T, Matsumura Y, Tsuchido T, Molecular Cloning and Nucleotide Sequence of the Superoxide Dismutase Gene and Characterization of Its Product from Bacillus subtilis, J Bacteriol. 180 (1998) 3697–3703. 10.1128/JB.180.14.3697-3703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wright Valderas M, Gatson JW, Wreyford N, Hart ME, The superoxide dismutase gene sodM is unique to Staphylococcus aureus: Absence of sodM in coagulase-negative staphylococci, Journal of Bacteriology. 184 (2002) 2465–2472. 10.1128/JB.184.9.2465-2472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Ammendola S, Ajello M, Pasquali P, Kroll JS, Langford PR, Rotilio G, Valenti P, Battistoni A, Differential contribution of sodC1 and sodC2 to intracellular survival and pathogenicity of Salmonella enterica serovar Choleraesuis, Microbes and Infection. 7 (2005) 698–707. 10.1016/j.micinf.2005.01.005. [DOI] [PubMed] [Google Scholar]
- [105].Fang FC, DeGroote MA, Foster JW, Baumler AJ, Ochsner U, Testerman T, Bearson S, Giard J-C, Xu Y, Campbell G, Laessig T, Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases, Proceedings of the National Academy of Sciences. 96 (1999) 7502–7507. 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Figueroa-Bossi N, Bossi L, Inducible prophages contribute to Salmonella virulence in mice, Mol Microbiol. 33 (1999) 167–176. 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- [107].Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR, Genome sequence of enterohaemorrhagic Escherichia coli O157:H7, Nature. 409 (2001) 529–533. 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- [108].Hayashi T, Complete Genome Sequence of Enterohemorrhagic Eschelichia coli O157:H7 and Genomic Comparison with a Laboratory Strain K-12, DNA Research. 8 (2001) 11–22. 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- [109].D’Orazio M, Scotti R, Nicolini L, Cervoni L, Rotilio G, Battistoni A, Gabbianelli R, Regulatory and structural properties differentiating the chromosomal and the bacteriophage-associated Escherichia coli O157:H7 Cu, Zn Superoxide Dismutases, BMC Microbiol. 8 (2008) 166. 10.1186/1471-2180-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Waldron KJ, Rutherford JC, Ford D, Robinson NJ, Metalloproteins and metal sensing, Nature. 460 (2009) 823–830. 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- [111].Waldron KJ, Robinson NJ, How do bacterial cells ensure that metalloproteins get the correct metal?, Nature Reviews Microbiology. 7 (2009) 25–35. 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- [112].Hood MI, Skaar EP, Nutritional immunity: Transition metals at the pathogen-host interface, Nature Reviews Microbiology. 10 (2012) 525–537. 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kehl-Fie TE, Skaar EP, Nutritional immunity beyond iron: a role for manganese and zinc, Current Opinion in Chemical Biology. 14 (2010) 218–224. 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Hammer ND, Skaar EP, The impact of metal sequestration on Staphylococcus aureus metabolism, Current Opinion in Microbiology. 15 (2012) 10–14. 10.1016/j.mib.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Anjem A, Varghese S, Imlay JA, Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli, Molecular Microbiology. 72 (2009) 844–858. 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Pugh SY, DiGuiseppi JL, Fridovich I, Induction of superoxide dismutases in Escherichia coli by manganese and iron, J Bacteriol. 160 (1984) 137–142. 10.1128/jb.160.1.137-142.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Beyer WF, Fridovich I, In vivo competition between iron and manganese for occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli, Journal of Biological Chemistry. 266 (1991) 303–308. 10.1016/S0021-9258(18)52435-2. [DOI] [PubMed] [Google Scholar]
- [118].Sansone A, Watson PR, Wallis TS, Langford PR, Kroll JS, The role of two periplasmic copper- and zinc-cofactored superoxide dismutases in the virulence of Salmonella choleraesuis, Microbiology. 148 (2002) 719–726. 10.1099/00221287-148-3-719. [DOI] [PubMed] [Google Scholar]
- [119].Krishnakumar R, Craig M, Imlay JA, Slauch JM, Differences in Enzymatic Properties Allow SodCI but Not SodCII To Contribute to Virulence in Salmonella enterica Serovar Typhimurium Strain 14028, J Bacteriol. 186 (2004) 5230–5238. 10.1128/JB.186.16.5230-5238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Ho TD, Figueroa-Bossi N, Wang M, Uzzau S, Bossi L, Slauch JM, Identification of GtgE, a Novel Virulence Factor Encoded on the Gifsy-2 Bacteriophage of Salmonella enterica Serovar Typhimurium, J Bacteriol. 184 (2002) 5234–5239. 10.1128/JB.184.19.5234-5239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ho TD, Slauch JM, Characterization of grvA, an Antivirulence Gene on the Gifsy-2 Phage in Salmonella enterica Serovar Typhimurium, J Bacteriol. 183 (2001) 611–620. 10.1128/JB.183.2.611-620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Krishnakumar R, Kim B, Mollo EA, Imlay JA, Slauch JM, Structural Properties of Periplasmic SodCI That Correlate with Virulence in Salmonella enterica Serovar Typhimurium, J Bacteriol. 189 (2007) 4343–4352. 10.1128/JB.00010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Tidhar A, Rushing MD, Kim B, Slauch JM, Periplasmic superoxide dismutase SodCI of S almonella binds peptidoglycan to remain tethered within the periplasm: SodCI binds peptidoglycan, Molecular Microbiology. 97 (2015) 832–843. 10.1111/mmi.13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Kirschvink JL, Gaidos EJ, Bertani LE, Beukes NJ, Gutzmer J, Maepa LN, Steinberger RE, Paleoproterozoic snowball Earth: Extreme climatic and geochemical global change and its biological consequences, Proceedings of the National Academy of Sciences. 97 (2000) 1400–1405. 10.1073/pnas.97.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Boden JS, Konhauser KO, Robbins LJ, Sánchez-Baracaldo P, Timing the evolution of antioxidant enzymes in cyanobacteria, Nat Commun. 12 (2021) 4742. 10.1038/s41467-021-24396-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Moore EK, Jelen BI, Giovannelli D, Raanan H, Falkowski PG, Metal availability and the expanding network of microbial metabolisms in the Archaean eon, Nature Geosci. 10 (2017) 629–636. 10.1038/ngeo3006. [DOI] [Google Scholar]
- [127].Wintjens R, Gilis D, Rooman M, Mn/Fe superoxide dismutase interaction fingerprints and prediction of oligomerization and metal cofactor from sequence, Proteins. 70 (2007) 1564–1577. 10.1002/prot.21650. [DOI] [PubMed] [Google Scholar]
- [128].Capdevila DA, Edmonds KA, Giedroc DP, Metallochaperones and metalloregulation in bacteria, Essays In Biochemistry. 61 (2017) 177–200. 10.1042/EBC20160076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Foster AW, Osman D, Robinson NJ, Metal Preferences and Metallation, Journal of Biological Chemistry. 289 (2014) 28095–28103. 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Corbin WJ, D. B, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Anderson KL, Dunman PM, Gerads R, Caprioli R, Nacken W, Chazin EP Skaar, Metal Chelation and Inhibition of Bacterial Growth in Tissue Abscesses, Science. 319 (2008) 962–966. 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- [131].Wright GSA, Bacterial Evolutionary Precursors of Eukaryotic Copper–Zinc Superoxide Dismutases, Molecular Biology and Evolution. 38 (2021) 3789–3803. 10.1093/molbev/msab157. [DOI] [PMC free article] [PubMed] [Google Scholar]