Abstract
We examined the effectiveness of antisense RNA (as RNA) strategies for metabolic engineering of Clostridium acetobutylicum. Strain ATCC 824(pRD4) was developed to produce a 102-nucleotide asRNA with 87% complementarity to the butyrate kinase (BK) gene. Strain ATCC 824(pRD4) exhibited 85 to 90% lower BK and acetate kinase specific activities than the control strain. Strain ATCC 824(pRD4) also exhibited 45 to 50% lower phosphotransbutyrylase (PTB) and phosphotransacetylase specific activities than the control strain. This strain exhibited earlier induction of solventogenesis, which resulted in 50 and 35% higher final concentrations of acetone and butanol, respectively, than the concentrations in the control. Strain ATCC 824(pRD1) was developed to putatively produce a 698-nucleotide asRNA with 96% complementarity to the PTB gene. Strain ATCC 824(pRD1) exhibited 70 and 80% lower PTB and BK activities, respectively, than the control exhibited. It also exhibited 300% higher levels of a lactate dehydrogenase activity than the control exhibited. The growth yields of ATCC 824(pRD1) were 28% less than the growth yields of the control. While the levels of acids were not affected in ATCC 824(pRD1) fermentations, the acetone and butanol concentrations were 96 and 75% lower, respectively, than the concentrations in the control fermentations. The lower level of solvent production by ATCC 824(pRD1) was compensated for by ∼100-fold higher levels of lactate production. The lack of any significant impact on butyrate formation fluxes by the lower PTB and BK levels suggests that butyrate formation fluxes are not controlled by the levels of the butyrate formation enzymes.
Clostridium acetobutylicum is a gram-positive, spore-forming, obligate anaerobe that is capable of fermenting a wide variety of sugars to acids (acetate and butyrate) and solvents (ethanol, acetone, and butanol). This organism was used extensively for the production of acetone and butanol from the First World War to the late 1950s, when the petrochemical process became economically superior. Research into the fermentation process has since focused on genetic evaluation and metabolic engineering of solventogenic clostridia in order to develop strains that have enhanced solvent production capabilities, with the objective of reviving the industrial fermentation process.
In the past 10 years, considerable effort has been spent on the development of tools for metabolic engineering of C. acetobutylicum. A genetic transfer system has been developed (32, 33, 39, 52), and many genes have been cloned and sequenced (2, 3, 7, 8, 37, 42–44, 57, 60). While these tools have been used successfully for overexpression of homologous genes (2, 3, 33, 38, 59), a method for downregulation of enzyme production is an equally essential metabolic engineering tool. Recently, reductions in enzyme levels in C. acetobutylicum ATCC 824 have been achieved through gene inactivation with nonreplicative integrational plasmids (21, 22). This technique requires a DNA fragment from the host and a selection marker for each inactivated gene. While nonreplicative integrational plasmids are suitable for knocking out a single gene, they are not practical for more advanced metabolic engineering strategies. Here we report results of an investigation of antisense RNA (asRNA) strategies as alternative and possibly more flexible and empowering methods of enzyme level downregulation.
Gene expression in some naturally occurring systems is regulated through the action of small asRNA molecules. For example, a naturally occurring asRNA molecule has been implicated in the regulatory system of the glutamine synthetase gene (glnA) (16, 28) of a solventogenic clostridium strain, Clostridium sp. strain NCP262 (formerly C. acetobutylicum P262 [29]). It has been postulated that asRNA molecules function by hybridizing with complementary mRNA transcripts. It has been shown that the subsequent downregulation occurs by several possible mechanisms, including (i) inhibition of translation because the duplex RNA structure prevents access to the ribosome binding site, (ii) rapid degradation of the mRNA, possibly by duplex RNA-specific RNases, and (iii) inhibition of transcription of mRNA due to premature termination (11, 47, 48, 55). Although the mechanism of asRNA action is not completely understood, asRNA strategies have been used to downregulate levels of targeted gene products in prokaryotes (9, 14, 15, 31, 41). Most studies have focused on either determining the effectiveness of different asRNA strategies or elucidating the role of a structural gene targeted for downregulation. To our knowledge, only one study has examined the ability of asRNA strategies to alter primary metabolism. In this study, van den Berg et al. examined the role of hydrogenase in the metabolism of lactate by Desulfovibrio vulgaris (53), and the results suggested that artificial asRNA strategies are capable of affecting the primary metabolism of prokaryotes.
asRNA strategies may have a number of advantages over gene inactivation for metabolic engineering. In addition to rapid implementation, asRNA strategies can avoid the pitfalls of lethal mutations since complete inhibition of protein production is not likely. asRNA strategies may be used to inducibly repress protein production by using inducible promoters to transcribe asRNA. For example, Coleman et al. utilized the inducible lac promoter to decrease lipoprotein production 2- to 16-fold in the presence of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG) (9). In addition, the use of growth stage-specific promoters could result in enzyme downregulation during specific stages of a fermentation so that more advanced metabolic engineering objectives could be accomplished. Finally, asRNA strategies may be used to downregulate the products of multiple genes by expressing multiple asRNAs from a single plasmid.
In contrast to efforts directed toward determining the mechanism of asRNA action or the reduction in the amount of a single gene product, our goal was to determine the effectiveness of asRNA in redirecting the primary metabolism of C. acetobutylicum. Specifically, we attempted to reduce the levels of the enzymes responsible for butyrate formation in C. acetobutylicum. Butyrate is produced from the metabolic intermediate butyryl coenzyme A (butyryl-CoA) in two steps. In the first step, butyryl-CoA is converted to butyryl phosphate by phosphotransbutyrylase (PTB). In the second step, butyrate kinase (BK) converts butyryl phosphate to butyrate, with concomitant production of ATP. The genes ptb and buk, which code for PTB and BK, respectively, are organized in the order ptb-buk in a single operon (60). We developed two plasmid constructs designed to produce asRNA complementary to either the ptb gene or the buk gene. Since it is thought that butyrate levels may play a role in the induction of solventogenesis and, specifically, solvent formation by C. acetobutylicum (23, 27, 51), downregulation of PTB and BK was expected to have a significant impact on primary metabolism.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used are listed in Table 1. Plasmid pIMP1 (33) is an Escherichia coli-C. acetobutylicum shuttle vector and was used to develop a control C. acetobutylicum strain to account for the effects of host-plasmid interactions (58). Plasmid pSOS94 is an E. coli-C. acetobutylicum shuttle vector containing the promoter region of the ptb gene with a BamHI restriction site engineered 30 bp downstream of the transcriptional start site (49). Plasmid pRD4 was constructed to produce asRNA targeted against the buk gene (buk-asRNA) from C. acetobutylicum ATCC 824 (see below). Plasmid pRD1 was constructed to produce asRNA targeted against the ptb gene (ptb-asRNA) from C. acetobutylicum ATCC 824 (see below).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or referenceb |
|---|---|---|
| Strains | ||
| E. coli ER2275 | hsdR mcr recA1 endA1 | NEB |
| C. acetobutylicum ATCC 824 | Wild type | ATCC |
| Plasmids | ||
| pN5-1 | ptb buk | 59 |
| pIMP1 | ColE1 ORI, Apr, pIM13 ORI, MLSr | 33 |
| pSOS94 | ColE1 ORI, Apr, pIM13 ORI, MLSr, ptb promoter region | 49 |
| pRD1 | ColE1 ORI, Apr, pIM13 ORI, MLSr, ptb-asRNA | This study |
| pRD4 | ColE1 ORI, Apr, pIM13 ORI, MLSr, buk-asRNA | This study |
hsdR, host-specific restriction deficient; mcr, methylcytosine-specific restriction abolished; recA1, homologous recombination abolished; endA1, endonuclease abolished; ColE1 ORI, ColE1 origin of replication; Apr, ampicillin resistance; pIM13 ORI, gram-positive origin of replication; MLSr, macrolide-lincosamide-streptogramin resistance; ptb, PTB gene; buk, BK gene.
NEB, New England Biolabs, Beverly, Mass.; ATCC, American Type Culture Collection, Manassas, Va.
Growth conditions and maintenance.
E. coli ER2275 and plasmid-containing transformants were grown aerobically at 37°C in Luria-Bertani medium. C. acetobutylicum ATCC 824 and plasmid-containing transformants were grown anaerobically in clostridial growth medium (CGM) at 37°C (61). Colonies were obtained on agar-solidified reinforced clostridial medium (Difco Microbiological Media, VWR Scientific Products, Chicago, Ill.). Spores were obtained from >2-week-old cultures on solid media or in liquid media. Heat shocking at 70 to 80°C for 10 min was used to kill vegetative cells and to activate spores for growth in fresh media. C. acetobutylicum strains were stored as vegetative cells frozen at −85°C in CGM supplemented with 20% (vol/vol) glycerol or as spores in CGM at 4°C. Media were supplemented with the following antibiotics as required: for E. coli, ampicillin (50 μg/ml) and chloramphenicol (35 μg/ml); and for C. acetobutylicum, erythromycin (100 μg/ml). In the benchtop-scale fermentations, erythromycin was replaced by its pH-stable analog clarithromycin (100 μg/ml; Abbott Laboratories, North Chicago, Ill.) (36).
Analytical methods.
Cell growth was monitored by measuring the optical density at 600 nm (OD600) with a Beckman model DU 64 spectrophotometer as follows. Culture samples were diluted in fresh CGM to maintain the measured absorbance at 600 nm between 0.1 and 0.4. The OD600 was calculated by multiplying the absorbance at 600 nm by the dilution factor. The glucose and l-lactate contents of culture supernatants were determined with a model 2700 Biochemistry Analyzer (Yellow Springs Instruments, Yellow Springs, Ohio). d-Lactate contents were measured by using a previously described enzymatic procedure (18). Culture supernatants were also analyzed to determine their acid (acetate and butyrate) and solvent (acetone, butanol, ethanol, and acetoin) contents with a model Vista 6000 gas chromatograph (Varian Instruments, Palo Alto, Calif.) equipped with a column (2 mm by 6 ft) packed with Carbograph 1 AW 20 (Alltech Associates, Deerfield, Ill.) and a flame ionization detector.
Bioreactor experiments.
Large-scale pH-controlled batch fermentations were carried out in a Biostat M bioreactor (B. Braun, Allentown, Pa.) and a BioFlo II bioreactor (New Brunswick Scientific, Edison, N.J.) containing 1.5 and 4.0 liters (working volumes) of CGM, respectively, as previously described (37). The bioreactor inoculum was a preculture initiated with a colony from a freshly streaked plate or a heat-shocked colony from a plate that was 2 weeks old or older. The initial pH of the bioreactor was 6.2, and the pH was allowed to drop to 5.5 as the culture progressed. Subsequently, the pH was maintained at or above 5.5 by adding 6 M ammonium hydroxide. The reported time scale (tN) of the bioreactor experiments was normalized to account for differences in the lengths of lag phases (due to small variations in inoculum preparation and also due to strain differences) by setting tN = 0 h when the culture OD600 was 1.0. On this scale, values of tN less than 0 simply indicate that the OD600 was less than 1.0.
Crude lysates.
Cells were collected from 50-ml culture samples by centrifugation (10,000 × g, 10 min, 0°C) and then immediately frozen at −85°C. Frozen cells were thawed on ice for 1 h and resuspended to an OD600 of ∼20 in lysis buffer (10 mM potassium phosphate [pH 7.2], 1 mM dithiothreitol). Cells were then collected from 1 ml of the suspension by centrifugation and resuspended in 0.95 ml of lysis buffer. The cells were then lysed by sonication in an ice water bath by using a model W-225R cell disruptor (Heat Systems-Ultrasonics, Farmingdale, N.Y.) equipped with a microtip at power setting 4 and pulsed at 50% duty for 9 to 12 min. The disrupted cell suspension was centrifuged twice (15,000 × g, 20 min, 4°C) to remove the cell debris. Crude lysates were kept at 4°C until they were used in the enzyme assays described below.
Enzyme assays.
Enzyme assays were performed as soon as possible after crude lysates were prepared. The total protein content was determined by the method of Bradford (5) by using a protein assay kit (Bio-Rad, Hercules, Calif.). Bovine serum albumin was used as the standard. PTB activity was routinely assayed at room temperature in the butyryl phosphate-forming direction as previously described (62). Acetate kinase (AK) and BK activities were routinely assayed in the acetyl and butyryl phosphate-forming direction at 29°C by using a previously described method (45). Phosphotransacetylase (PTA) activity was determined at room temperature in the acetyl-CoA-forming direction by using the method of Brown et al. (6). Lactate dehydrogenase (LDH) activity was determined in the lactate-forming direction by using a modified version of a previously described method (10). Each LDH assay mixture contained 50 mM morpholinopropanesulfonic acid (MOPS) (pH 7.0), 3 mM fructose 1,6-diphosphate, 9 mM MgCl2, 0.1 mg of NADH per ml, and 10 mM pyruvate (pH 7). The reaction was initiated by adding pyruvate and was monitored by measuring the consumption of NADH at 340 nm (extinction coefficient, 6.22 mM−1 cm−1). The background activity was determined by replacing the crude extract with an equal volume of lysis buffer. Nonspecific activity was determined in the absence of pyruvate. One unit of PTB, BK, PTA, or AK activity was defined as the amount of enzyme that converted 1 μmol of substrate per min. One unit of LDH activity was defined as the amount of enzyme that converted 1 nmol of pyruvate per min. Enzyme specific activities are reported below as units of activity per milligram of protein in the crude lysate.
DNA isolation, manipulation, and transformation.
Plasmid DNA was prepared from E. coli by using standard procedures (46). Large-scale preparation of plasmid DNA from E. coli was performed by using a Qiagen Plasmid Kit (Qiagen Inc., Chatsworth, Calif.). Plasmid DNA was prepared from C. acetobutylicum by using an alkaline lysis protocol developed for C. acetobutylicum (24). Previously described methods were used for electrotransformation of E. coli (13) and C. acetobutylicum (32, 33). Transformation of C. acetobutylicum by pRD1 was carried out by using a modified protocol (90% of the glucose in the outgrowth medium was replaced with an equal amount of soluble starch). Restriction endonucleases, T4 DNA ligase, Vent DNA polymerase, and the DNA polymerase I-large (Klenow) fragment were obtained from New England Biolabs (Beverly, Mass.) and used according to the manufacturer’s specifications. A Geneclean II kit (Bio 101, Vista, Calif.) was used to purify DNA from solutions and agarose gels. Restriction sites were engineered into amplified DNA fragments by using a previously described PCR protocol (37).
Computer software.
DNA sequences were manipulated and analyzed by using The Gene Construction Kit (Textco Inc., West Lebanon, N.H.) and the BESTFIT algorithm of the Wisconsin Package, version 9.1 (Genetics Computer Group, Madison, Wis.). Metabolic flux analysis was performed by using software developed specifically for analysis of C. acetobutylicum fermentations (12), and pathway fluxes are presented below as follows. The glycolysis flux indicates glucose uptake and conversion to pyruvate via the glycolytic pathway. The lactate formation flux indicates conversion of pyruvate to lactate. The acetate and butyrate formation fluxes indicate conversion of acetyl-CoA and butyryl-CoA to acetate and butyrate, respectively. The acetone formation flux indicates conversion of acetoacetyl-CoA to acetone. The butanol formation flux indicates conversion of butyryl-CoA to butanol. Biomass variations were accounted for by using the OD600 to calculate specific fluxes, which are reported below as millimolar per hour per unit of OD600. The time-averaged specific fluxes are reported below for the following two stages of fermentation: early stage for tN < 10 h (exponential phase and the transition to the stationary phase of culture) and late stage for tN ≥ 10 h (stationary phase of culture).
Construction of pRD4.
Figure 1 shows the construction of plasmid pRD4 to produce buk-asRNA by using the ptb promoter. The goal for this construct was the production of a short asRNA molecule similar to many naturally occurring asRNAs. The target of the asRNA was selected so that it included 10 codons from the buk gene and 17 bp upstream of the ATG start codon (positions 1058 to 1103), including the putative ribosome binding site. The rho-independent terminator region of the naturally occurring asRNA targeted against the glnA gene (glnA-asRNA) (positions 1893 to 1909) (16) of Clostridium sp. strain NCP262 (formerly C. acetobutylicum P262 [29]) was selected to follow the buk antisense region and terminate transcription of the asRNA. In summary, the chimeric insert was designed to include a BamHI site, a 47-bp fragment from the 5′ end of the buk gene, and the 17-bp terminator region of the glnA-asRNA. The individual strands of the insert were chemically synthesized by workers at the Northwestern University Biotechnology Laboratory (Chicago, Ill.). Purified oligonucleotides were annealed at a final concentration of 0.2 μg/μl in 1× Vent DNA polymerase buffer by heating the preparation at 94°C for 10 min in a water bath and then slowly cooling it to room temperature overnight. The insert was then ligated to the ∼5.25-kb BamHI-AvaI fragment of pSOS94 containing the ptb promoter region. Large (≥3-kb) linear DNA was purified from the ligation mixture with a Geneclean II kit and subsequently treated with the DNA polymerase I-Klenow fragment to fill in recessed 3′ ends. The blunt-ended linear DNA was recircularized and transformed into E. coli ER2275. Ampicillin-resistant transformant colonies were screened for plasmid pRD4 with BamHI and HpaI digests (data not shown).
FIG. 1.
Construction of plasmids pRD4 and pRD1. For each plasmid, the locations and directions of transcription of relevant genes are indicated (arrows). Relevant restriction sites are shown. Abbreviations: Apr, ampicillin resistance gene; MLSr, macrolide-lincosamide-streptogramin resistance gene; repL, gene required for replication in gram-positive organisms.
Construction of plasmid pRD1.
Plasmid pRD1 was constructed as shown in Fig. 1 to produce ptb-asRNA by using the ptb promoter region. A 567-bp fragment of the ptb gene and its putative ribosome binding site were amplified by PCR by using plasmid pN5-1 (59) as the template. The upstream primer, NARUPPTB (5′-cctttggcgccTAATTAAATAGTAAAAGGG-3′), was designed by adding the first 11 nucleotides to a region upstream of the ptb ribosome binding site (positions 108 to 126) in order to introduce an EheI site (underlined region). The downstream primer, BAMDNPTB (5′-AACCTTTGGATccACAATTCCTATTGC-3′), was designed by substituting C for T at positions 651 and 652 in order to introduce a BamHI site (underlined region). The amplified fragment was sequentially digested with EheI and BamHI and ligated to the ∼5.0-kb BamHI-EheI fragment of pSOS94. The ligation mixture was used to transform E. coli ER2275. Ampicillin-resistant transformant colonies were screened for plasmid pRD1. The construction of plasmid pRD1 was verified by using BamHI and ScaI digests (data not shown).
RESULTS
Impact of putative buk-asRNA. (i) Development of strain ATCC 824(pRD4).
Plasmid pRD4 was designed to produce a 102-nucleotide buk-asRNA molecule with 87% complementarity in a 107-nucleotide region of the mRNA of the buk gene. The identities of ATCC 824(pRD4) transformants were verified by plasmid isolation and restriction mapping (data not shown). Strain ATCC 824(pRD4) sporulated both on solid media and in liquid media. Most colonies on solid media exhibited the star shape typical of sporulating clostridia. Furthermore, very little or no surface spreading of colonies indicated that a majority of the cells in each colony began the sporulation process. The representative bioreactor experiment performed with ATCC 824(pRD4) that is described below was carried out after multiple cycles of sporulation and activation by heat shock.
(ii) Enzyme levels in ATCC 824(pRD4).
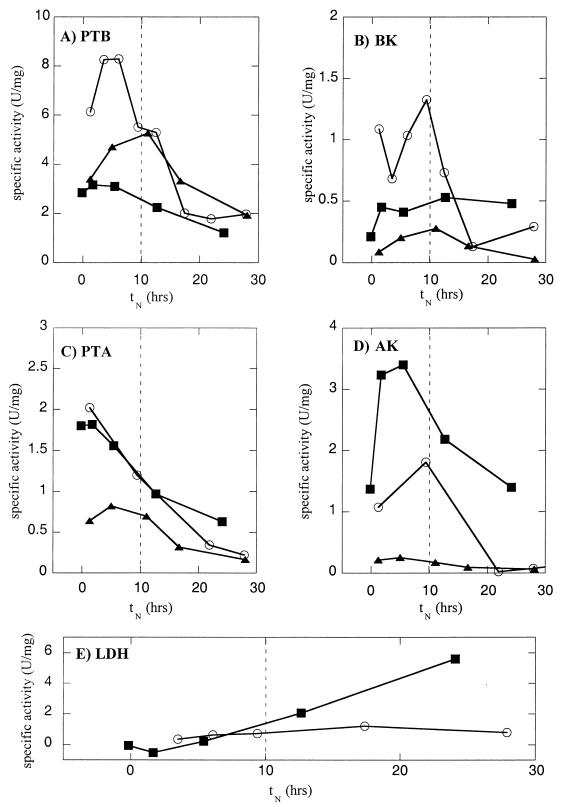
The impact of putative buk-asRNA on enzyme levels as determined by in vitro specific activities is summarized in Table 2. Since the butyrate formation enzymes have broad substrate specificities (25, 62), the levels of the acetate formation enzymes were also determined. Strain ATCC 824(pRD4) exhibited lower peak levels of acetate and butyrate formation enzymes than strain ATCC 824(pIMP1) exhibited. The ATCC 824(pRD4) peak levels of PTB and BK were ∼45 and ∼85% lower, respectively, than the control levels. In addition, the peak levels of PTA and AK were ∼50 and ∼90% lower, respectively, than the control levels. The time course profiles for specific enzyme activities from one fermentation are shown in Fig. 2. The levels of BK in ATCC 824(pRD4) were ∼80 to 90% lower than the levels of BK in the control during the exponential phase. The levels of PTB in ATCC 824(pRD4) were ∼50% lower than the levels of PTB in the control during the exponential growth phase but were comparable to the levels of PTB in the control during the stationary phase. The levels of PTA and AK in ATCC 824(pRD4) were ∼50 and ∼80% lower, respectively, than the levels of PTA and AK in the control during the exponential phase. The exponential-phase-specific downregulation observed is consistent with the expected activity of the ptb promoter (used to generate asRNA), as assessed in previous enzyme activity studies (1, 26, 56).
TABLE 2.
Peak acid formation enzyme levels
| Enzyme | Peak sp act (U/mg)a in:
|
||
|---|---|---|---|
| Strain 824(pIMP1) (control) | Strain 824(pRD4) | Strain 824(pRD1) | |
| PTB | 8.8 ± 0.5 | 4.7 ± 0.6 | 2.6 ± 0.6b |
| BK | 2.0 ± 0.6 | 0.31 ± 0.03 | 0.3 ± 0.1 |
| PTA | 1.6 ± 0.45 | 0.76 ± 0.06 | 1.1 ± 0.8 |
| AK | 3.1 ± 1.3 | 0.26 ± 0.01 | 2.1 ± 1.3 |
| LDH | 1.3 ± 0.06 | NDc | 4.1 ± 1.5 |
Mean ± standard error of the mean (n = 2) unless otherwise indicated.
Mean ± standard error of the mean (n = 3).
ND, not determined.
FIG. 2.
Evaluation of strains ATCC 824(pIMP1) (○), ATCC 824(pRD4) (▴), and ATCC 824(pRD1) (■): time course profiles of specific activities of PTB (A), BK (B), PTA (C), AK (D), and LDH (E). The vertical dashed lines indicate the transition from the early (exponential) stage to the late (stationary) stage.
(iii) Product formation by ATCC 824(pRD4).
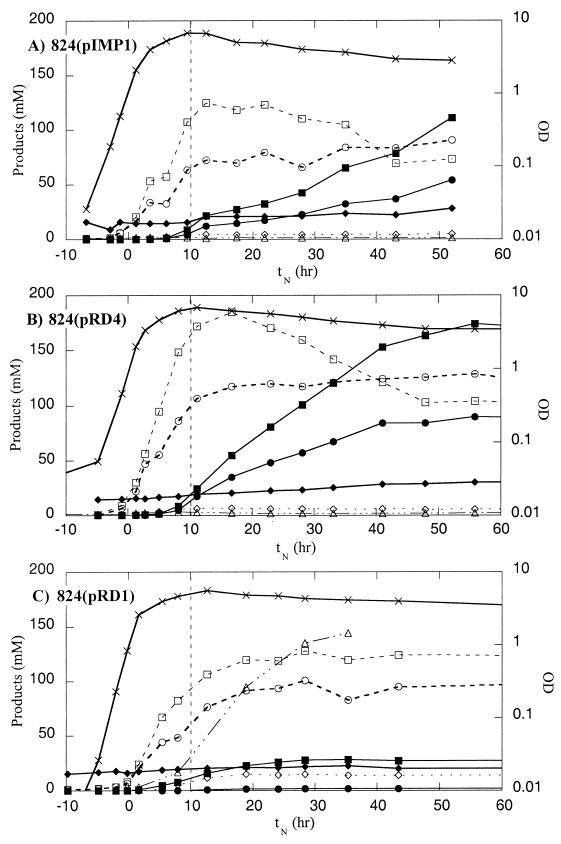
The impact of putatively produced buk-asRNA on growth and product formation is summarized in Table 3. Strains ATCC 824(pRD4) and ATCC 824(pIMP1) had similar growth rates and reached similar peak OD600. However, strain ATCC 824(pRD4) produced higher levels of all products than strain ATCC 824(pIMP1) produced. While the acid levels did vary significantly, the peak levels of acetate and butyrate produced by ATCC 824(pRD4) were 25% ± 19% and 34% ± 11% higher, respectively, than the ATCC 824(pIMP1) peak levels. Similarly, the final acetate and butyrate levels were higher in the ATCC 824(pRD4) fermentation than in the ATCC 824(pIMP1) fermentation. The solvent levels were also affected by the putative production of buk-asRNA. The final ethanol, acetone, and butanol concentrations were 24% ± 12%, 50% ± 32%, and 35% ± 18%, higher, respectively, in the ATCC 824(pRD4) fermentation than in the ATCC 824(pIMP1) fermentation. The time course product profiles of one fermentation (Fig. 3) also indicated that the kinetics of solvent formation were altered. Butanol was first detected ∼2 h earlier in the ATCC 824(pRD4) fermentation than in the control fermentation [tN = −1 h for ATCC 824(pRD4), compared to tN = 1.25 h for ATCC 824(pIMP1)]. Furthermore, acetone was first detected about ∼7 h earlier in the ATCC 824(pRD4) fermentation than in the control fermentation [tN = −1 h for ATCC 824(pRD4), compared to tN = 6 h for ATCC 824(pIMP1)].
TABLE 3.
Summary of product formation in bioreactor experiments
| Fermentation characteristic | Strain 824(pIMP1) (control)a | Strain 824(pRD4)a | Strain 824(pRD1)a |
|---|---|---|---|
| No. of expt | 2 | 2 | 4 |
| Growth rate (h−1) | 0.51 ± 0.01 | 0.53 ± 0.02 | 0.48 ± 0.04 |
| Maximum OD600 | 7.0 ± 0.18 | 7.1 ± 0.18 | 5.00 ± 0.3 |
| Ethanol concn (mM) | 11 ± 0.75 | 14 ± 1.0 | 5 ± 0.3 |
| Acetone concn (mM) | 50 ± 4.5 | 75 ± 14.5 | 2 ± 0.2 |
| Butanol concn (mM) | 114 ± 3 | 154 ± 20 | 28 ± 0.6 |
| Peak acetate concn (mM) | 106 ± 15.5 | 132 ± 4 | 98 ± 3 |
| Final acetate concn (mM) | 106 ± 15.5 | 126 ± 6 | 96 ± 3 |
| Peak butyrate concn (mM) | 135 ± 11 | 181 ± 4 | 123 ± 8 |
| Final butyrate concn (mM) | 81 ± 8 | 111 ± 10 | 117 ± 5 |
| Acetoin concn (mM) | 5 ± 0.65 | 5 ± 0.25 | 15 ± 1.0 |
| Lactate concn (mM) | <1 | <1 | 105 ± 16 |
Mean ± standard error of the mean.
FIG. 3.
Product formation patterns for strains ATCC 824(pIMP1) (A), ATCC 824(pRD4) (B), and ATCC 824(pRD1) (C): time course profiles of OD600 (×), acetate concentration (○), butyrate concentration (□), lactate concentration (▵), acetone concentration (●), butanol concentration (■), ethanol concentration (⧫), and acetoin concentration (◊). The vertical dashed lines indicate the transition from the early (exponential) stage to the late (stationary) stage.
(iv) Metabolic flux analysis of ATCC 824(pRD4).
In order to assess the impact of reduced levels of acid formation enzymes on carbon fluxes in ATCC 824(pRD4), we calculated specific pathway fluxes from the substrate and product concentration profiles. The fluxes are shown in Table 4 as the average specific fluxes during the early (tN < 10 h) and late (tN ≥ 10 h) stages of the fermentations. During the early stage of the fermentations, glucose was consumed primarily for growth and acid production. The carbon fluxes through the glycolysis pathway in strain ATCC 824(pRD4) were ∼20% higher than the carbon fluxes in strain ATCC 824(pIMP1). Similarly, the acetate and butyrate formation fluxes were ∼20% higher in strain ATCC 824(pRD4) than in strain ATCC 824(pIMP1). The acetone and butanol formation fluxes in ATCC 824(pRD4) were also ∼80 and ∼45% higher, respectively, than the acetone and butanol formation fluxes in ATCC 824(pIMP1). As growth ceased during the late stage of the fermentations, the glucose utilization patterns changed as follows. The specific fluxes through the glycolysis pathway in strain ATCC 824(pRD4) were ∼30% lower than the specific fluxes in strain ATCC 824(pIMP1), and the acetate and butyrate formation fluxes were ∼50 and ∼25% lower, respectively, in ATCC 824(pRD4) than in ATCC 824(pIMP1). While the acetone formation fluxes in ATCC 824(pRD4) were similar to those in ATCC 824(pIMP1), the butanol formation fluxes were ∼20% lower in ATCC 824(pRD4) than in ATCC 824(pIMP1).
TABLE 4.
Summary of metabolic flux analysis
| Pathway | Specific flux (mM h−1 unit of OD600−1)
|
|||||
|---|---|---|---|---|---|---|
| Strain 824(pIMP1)a
|
Strain 824(pRD4)a
|
Strain 824(pRD1)b
|
||||
| Early stage | Late stage | Early stage | Late stage | Early stage | Late stage | |
| Glycolysis | 5.6 ± 0.4 | 0.8 ± 0.03 | 6.6 ± 0.6 | 0.6 ± 0.1 | 5.1 ± 0.15 | 1.0 ± 0.03 |
| Acetate formation | 2.7 ± 0.07 | 0.5 ± 0.01 | 3.3 ± 0.3 | 0.24 ± 0.01 | 2.3 ± 0.12 | 0.35 ± 0.06 |
| Butyrate formation | 3.5 ± 0.05 | −0.2 ± 0.01 | 4.5 ± 0.4 | −0.15 ± 0.04 | 3.2 ± 0.2 | 0.30 ± 0.02 |
| Lactate formation | 0.11 ± 0.04 | 0.0 ± 0.0 | 0.15 ± 0.05 | −0.01 ± 0.01 | 0.5 ± 0.04 | 0.7 ± 0.09 |
| Acetone formation | 0.11 ± 0.05 | 0.30 ± 0.05 | 0.20 ± 0.01 | 0.28 ± 0.11 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| Butanol formation | 0.19 ± 0.06 | 0.64 ± 0.01 | 0.27 ± 0.01 | 0.52 ± 0.13 | 0.22 ± 0.01 | 0.13 ± 0.01 |
Mean ± standard error of the mean (n = 2).
Mean ± standard error of the mean (n = 3).
Impact of putative ptb-asRNA. (i) Development of strain ATCC 824(pRD1).
Plasmid pRD1 was developed to produce a 698-nucleotide ptb-asRNA molecule with 96% complementarity in a 656-nucleotide region of the mRNA of the ptb gene. Plasmid pRD1 was transformed into strain ATCC 824 by using a modified procedure, as described in Materials and Methods. When the standard procedure was used, no transformants were obtained. Glucose was replaced with soluble starch in the outgrowth medium in order to add a lag phase after electroporation. This lag phase was incorporated in order to prevent overgrowth by nontransformed cells and allow transformed cells extra time to recover from the electroporation procedure. The identities of erythromycin-resistant transformants as ATCC 824(pRD1) were verified by plasmid preparation and mapping of BamHI and ScaI sites (data not shown).
Strain ATCC 824(pRD1) exhibited characteristics that were not typical of wild-type C. acetobutylicum. ATCC 824(pRD1) cells did not appear to sporulate either on solid media or in liquid media. In addition, ATCC 824(pRD1) colonies did not appear to be star shaped, as is typical of sporulating clostridia. In fact, surface spreading on agar plates (which was indicative of vegetative growth) was observed for the majority of the colonies. Furthermore, colonies on solid media were short lived and could not be easily subcultured if the plate was more than 1 week old. All experiments with ATCC 824(pRD1) were performed by recovering frozen vegetative cells on solid media as individual colonies and then immediately subculturing them in liquid media.
(ii) Enzyme levels in ATCC 824(pRD1).
The impact of putative ptb-asRNA on enzyme levels as determined by in vitro specific activities is summarized in Table 2. Compared to strain ATCC 824(pIMP1), strain ATCC 824(pRD1) exhibited lower peak levels of the butyrate formation enzymes and higher peak levels of LDH. The ATCC 824(pRD1) peak levels of PTB and BK were ∼70 and ∼80% lower, respectively, than the ATCC 824(pIMP1) peak levels. The peak LDH specific activities of ATCC 824(pRD1) were ∼300% higher than the peak LDH specific activities of ATCC 824(pIMP1). The effects on PTA and AK were not conclusive since strain ATCC 824(pRD1) produced highly variable levels of both of these enzymes. The time course profiles of specific enzyme activities from one fermentation are shown in Fig. 2. The levels of PTB in ATCC 824(pRD1) were ∼50 to 80% lower than the levels of PTB in the control primarily during the exponential growth phase. The levels of BK in ATCC 824(pRD1) were also ∼30 to 60% lower than the levels of BK in the control but only early in the exponential phase. In contrast, the ATCC 824(pRD1) levels of LDH were up to ∼500% higher than the LDH levels in the control and peaked during the stationary phase of the fermentation. The exponential-phase-specific downregulation of PTB and BK observed is consistent with the expected activity of the ptb promoter, as assessed in previous enzyme activity studies (1, 26, 56).
(iii) Product formation by ATCC 824(pRD1).
The impact of putatively produced ptb-asRNA on growth and product formation is summarized in Table 3. While the growth rates of ATCC 824(pRD1) and ATCC 824(pIMP1) were essentially identical, the maximum OD600 of strain ATCC 824(pRD1) was ∼28% lower than the maximum OD600 of the control strain. During the growth-associated acidogenic phase, the peak levels of acetate and butyrate were similar in ATCC 824(pRD1) and ATCC(pIMP1). However, during the stationary phase no significant uptake of acids was observed in the ATCC 824(pRD1) fermentation. The effects of the putative ptb-asRNA on the levels of the solvents were greater. The final concentrations of ethanol, acetone, and butanol were 60% ± 4%, 96% ± 1%, and 75% ± 1% lower, respectively, in ATCC(pRD1) fermentations than in ATCC 824(pIMP1) fermentations. The reduction in solvent formation was compensated for by a ∼100-fold increase in lactate production by ATCC 824(pRD1), which accounted for nearly 20% of the glucose consumed in the fermentation. The time course product profiles from a representative fermentation demonstrated the effects on solvent formation kinetics (Fig. 3). Butanol was first detected at about the same time in both ATCC 824(pRD1) and ATCC 824(pIMP1) fermentations. However, acetone was first detected nearly 7 h later in the ATCC 824(pRD1) fermentation than in the ATCC 824(pIMP1) fermentation. [at tN = 12.7 h for ATCC 824(pRD1), compared to tN = 6 h for ATCC 824(pIMP1)]. Furthermore, production of lactic acid in the ATCC 824(pRD1) fermentation began at about the same time as butanol formation.
(iv) Metabolic flux analysis of ATCC 824(pRD1).
The metabolic flux analysis of ATCC 824(pRD1) fermentations revealed a major shift in carbon flow compared to ATCC 824(pIMP1) fermentations (Table 4). During the early stage, the fluxes through glycolysis and acetate and butyrate formation pathways in ATCC 824(pRD1) were 5 to 10% lower than those in ATCC 824(pIMP1). In contrast, the lactate formation fluxes were ca. fourfold higher in ATCC 824(pRD1) than in ATCC 824(pIMP1). While the acetone formation fluxes in ATCC 824(pRD1) were ∼90% lower than the acetone formation fluxes in ATCC 824(pIMP1), the butanol formation fluxes in ATCC 824(pRD1) were similar to the butanol formation fluxes in ATCC 824(pIMP1). As growth ceased and the fermentations entered the late stage, the glycolysis pathway fluxes in ATCC 824(pRD1) were ∼20% higher than the glycolysis pathway fluxes in ATCC 824(pIMP1), but the acetate formation fluxes were ∼20% lower than the control acetate formation fluxes. The most significant differences were in the lactate, butyrate, and solvent formation pathways. The lactate formation flux in the late stage of ATCC 824(pRD1) was significant in contrast to the zero flux of ATCC 824(pIMP1). The late-stage butyrate formation flux in ATCC 824(pRD1) was ∼50% higher and in the opposite direction compared to the ATCC 824(pIMP1) late-stage butyrate formation flux. This means that the butyrate formation enzymes continued to produce butyrate in ATCC 824(pRD1) instead of reutilizing butyrate for solvent production, as in ATCC 824(pIMP1). In addition, the acetone and butanol formation fluxes were ∼95 and ∼80% lower, respectively, in ATCC 824(pRD1) than in ATCC 824(pIMP1).
DISCUSSION
The goal of this investigation was to determine whether antisense strategies could be used to metabolically engineer strains of C. acetobutylicum. The putative production of asRNA resulted in strains that had downregulated enzyme levels. The asRNA strategies seem to be particularly effective considering that C. acetobutylicum ATCC 824 knockout mutants with mutations in pta (the PTA gene) and buk exhibited 80 to 90% reductions in enzyme activity compared to the control (22). While we demonstrated that it is possible to downregulate specific gene products, a more important feature of this work was the manipulation of cell metabolism. With regard to cell metabolism, although both strain ATCC 824(pRD4) and strain ATCC 824(pRD1) produced significant levels of butyrate, the two organisms exhibited completely different patterns of product formation.
Metabolic engineering of strain ATCC 824(pRD4).
Strain ATCC 824(pRD4) successfully downregulated BK production and exhibited 80 to 90% lower BK specific activities than the control exhibited. In addition, the AK specific activities of ATCC 824(pRD4) were also ∼80% lower than those of the control. The apparent nonspecific downregulation of AK levels can be explained by the homology between the buk and ack (AK) genes. The putative buk-asRNA exhibits 79% complementarity to the ack mRNA in a 105-nucleotide region. In addition to the levels of BK and AK, the levels of PTB and PTA were also affected in ATCC 824(pRD4). The effects on the PTB and PTA levels may be explained as follows. One mechanism of asRNA activity is enhancement of mRNA decay, perhaps due to RNA-RNA duplex-specific RNases (48, 55). Since PTB and BK are translated from a single mRNA transcript, any decrease in the buk mRNA levels could also have an impact on the production of PTB. In fact, reductions in the target mRNA level have been observed in some studies of artificial asRNA (9, 31, 41, 53). The same reasoning can explain the effects on PTA levels since the genes for PTA and AK are organized in a single operon with the same arrangement as ptb and buk (3).
The downregulation of the acid formation enzymes did not result in decreased production of acetate and butyrate throughout the fermentations. In fact, the acid formation pathway fluxes in ATCC 824(pRD4) were higher than the acid formation pathway fluxes in the control during the early stages. In contrast, during the late stages these fluxes were lower than the control fluxes. These patterns suggest that fluxes through the acetate and butyrate formation pathways are not controlled by enzyme levels. The metabolic flux and enzyme activity patterns observed in this study suggest that the metabolic branch points (acetyl-CoA and butyryl-CoA) that lead to acetate and butyrate formation may be rigidly (50) controlled in vivo. The possible rigidity of the acetyl-CoA and butyryl-CoA branch points is supported by the fact that even genetic knockouts of pta and buk continue to produce acetate and butyrate (22). Since the acetate and butyrate formation pathways represent crucial substrate level phosphorylation reactions, cells may have evolved to maintain the ability to generate ATP by regulating metabolic intermediate concentrations, as well as enzyme levels. The intracellular concentrations of acetyl-CoA and butyryl-CoA, which are substrates of the acetate and butyrate formation pathways, respectively, have been reported to vary significantly during different stages of a fermentation (4).
Although the asRNA strategy targeted against buk did not decrease butyrate production, solvent production was increased. The buk-asRNA cannot have a direct impact on the expression of solvent formation genes, which are induced just prior to the beginning of solvent formation (17, 19, 57). However, solvent formation genes were apparently induced earlier in ATCC 824(pRD4) than in the control, which resulted in the earlier appearance of acetone and butanol in ATCC 824(pRD4). The final solvent concentrations were also higher than the control concentrations. These data suggest that the putative production of buk-asRNA downregulated acid formation enzymes and indirectly induced solvent formation genes.
Since we examined antisense strategies as an alternative to gene inactivation, we also compared strain ATCC 824(pRD4) with the BK knockout mutant (strain PJC4BK) described by Green et al. (22) in order to assess the effectiveness of our antisense strategy. Strains PJC4BK and ATCC 824(pRD4) both exhibited ∼80% lower BK activity than their respective controls. However, PJC4BK exhibited elevated PTB, PTA, and AK activities, in contrast to the reduced activities exhibited by strain ATCC 824(pRD4). Strain PJC4BK also exhibited different patterns of product formation. PJC4BK produced lower levels of butyrate and acetone than strain ATCC 824(pRD4) produced. Surprisingly, PJC4BK and ATCC 824(pRD4) produced similar final levels of butanol (∼155 mM) and acetate (∼120 mM). Green et al. (22) also reported that 8% of the glucose in the preparation was converted to lactate, whereas strain ATCC 824(pRD4) produced negligible amounts of lactate. Furthermore, the product formation kinetics of PJC4BK and ATCC 824(pRD4) were also different. While both strains produced butanol earlier in the fermentations than their respective controls, strain PJC4BK produced acetone much later than ATCC 824(pRD4) produced acetone.
Metabolic engineering of ATCC 824(pRD1).
Strain ATCC 824(pRD1) successfully downregulated PTB production and exhibited 50 to 80% lower PTB specific activities than the control exhibited. In addition, strain ATCC 824(pRD1) also exhibited 30 to 60% lower BK specific activities than the control exhibited. This downregulation of downstream gene products has been observed in another antisense study (41). The simultaneous downregulation of BK can be explained by the fact that ptb and buk are transcribed in the same mRNA transcript. The interaction of the ptb-asRNA with the mRNA transcript could interfere with translation of both ptb and buk.
While strain ATCC 824(pRD1) exhibited downregulation of butyrate formation enzymes, the effects on product formation were more profound. As in strain ATCC 824(pRD4), reductions in PTB and BK levels did not lead to decreased butyrate production. In fact, the specific butyrate formation fluxes were indistinguishable from those of the control. We again reached the conclusions described above, namely, that the butyrate formation enzyme levels do not control butyrate production and that the in vivo carbon flux through the butyryl-CoA metabolic branch point is rigidly controlled.
However, in contrast to strain ATCC 824(pRD4), strain ATCC 824(pRD1) exhibited severely altered solvent production patterns. Induction of acetone formation was delayed compared to induction of butanol formation. During the stationary phase, while butanol production was significantly reduced, acetone production was virtually eliminated. Lactate production was also induced at the same time as butanol production. The threefold induction of LDH cannot completely explain the elevated lactate formation fluxes. In fact, the lactate formation fluxes in the early stage were elevated even though the LDH levels were similar to the control levels. Therefore, we hypothesized that elevated levels of the lactate precursors, pyruvate and NADH, must also play a role in controlling lactate formation fluxes. Actually, the product formation patterns exhibited by strain ATCC 824(pRD1) are very similar to the alcohologenic fermentations (characterized by butanol and lactate production but no acetone production) of wild-type strain ATCC 824. These alcohologenic fermentations have been observed in cultures (predominantly continuous cultures) with glycerol-glucose cofermentation (20, 54), carbon monoxide gassing (34, 35), and methyl viologen addition (40).
Conclusion.
During the development and study of strains ATCC 824(pRD4) and ATCC 824(pRD1), we established that antisense strategies can be used to downregulate specific protein production and alter the primary metabolism of C. acetobutylicum. Due to the elaborate nature of the metabolic regulatory systems, downregulation of a single pathway is unlikely to directly alter the yield of a specific product. However, our study of strains ATCC 824(pRD4) and ATCC 824(pRD1) revealed that secondary effects, such as induction of solvent formation genes, can result in even more substantial changes in metabolic activity. The success of the relatively simple strategies used during the development of strains ATCC 824(pRD4) and ATCC 824(pRD1) suggests that the antisense strategies to alter cell metabolism are very versatile. While the ability to target specific proteins for downregulation needs to be examined in more detail, antisense strategies are emerging as useful metabolic engineering tools.
ACKNOWLEDGMENTS
This work was supported in part by National Science Foundation grant BES-9632217 and by National Institutes of Health Pre-doctoral Biotechnology Training grant GM 08449.
We thank Philippe Soucaille for plasmid pSOS94.
REFERENCES
- 1.Andersch W, Bahl H, Gottschalk G. Level of enzymes involved in acetate, butyrate, acetone, and butanol formation by Clostridium acetobutylicum. Eur J Appl Microbiol Biotechnol. 1983;18:327–332. [Google Scholar]
- 2.Boynton Z L, Bennett G N, Rudolph F B. Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl–coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1996;178:3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boynton Z L, Bennett G N, Rudolph F B. Cloning, sequencing, and expression of genes encoding phosphotransacetylase and acetate kinase from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1996;62:2758–2766. doi: 10.1128/aem.62.8.2758-2766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boynton Z L, Bennett G N, Rudolph F B. Intracellular concentrations of coenzyme A and its derivatives from Clostridium acetobutylicum ATCC 824 and their roles in enzyme regulation. Appl Environ Microbiol. 1994;60:39–44. doi: 10.1128/aem.60.1.39-44.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Ann Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brown T D K, Jones-Mortimer M C, Kornberg H L. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol. 1977;102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 7.Cary J W, Petersen D J, Papoutsakis E T, Bennett G N. Cloning and expression of Clostridium acetobutylicum ATCC 824 acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A-transferase in Escherichia coli. Appl Environ Microbiol. 1990;56:1576–1583. doi: 10.1128/aem.56.6.1576-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cary J W, Petersen D J, Papoutsakis E T, Bennett G N. Cloning and expression of Clostridium acetobutylicum phosphotransbutyrylase and butyrate kinase genes in Escherichia coli. J Bacteriol. 1988;170:4613–4618. doi: 10.1128/jb.170.10.4613-4618.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman J, Green P J, Inouye M. The use of RNAs complementary to specific mRNAs to regulate the expression of individual bacterial genes. Cell. 1984;37:429–436. doi: 10.1016/0092-8674(84)90373-8. [DOI] [PubMed] [Google Scholar]
- 10.Contag P R, Williams M G, Rogers P. Cloning of a lactate dehydrogenase gene from Clostridium acetobutylicum B643 and expression in Escherichia coli. Appl Environ Microbiol. 1990;56:3760–3765. doi: 10.1128/aem.56.12.3760-3765.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delihas N. Regulation of gene expression by trans-encoded antisense RNAs. Mol Microbiol. 1994;15:411–414. doi: 10.1111/j.1365-2958.1995.tb02254.x. [DOI] [PubMed] [Google Scholar]
- 12.Desai, R. P., L. N. Nielsen, and E. T. Papoutsakis. Metabolic flux analysis of C. acetobutylicum fermentation using nonlinear constraints. J. Biotechnol., in press.
- 13.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellison M J, Kelleher III R J, Rick A. Thermal regulation of β-galactosidase synthesis using anti-sense RNA directed against the coding portion of the mRNA. J Biol Chem. 1985;260:9085–9087. [PubMed] [Google Scholar]
- 15.Engdahl H M, Hjalt T A, Wagner E G. A two unit antisense RNA cassette test system for silencing of target genes. Nucleic Acids Res. 1997;25:3218–3227. doi: 10.1093/nar/25.16.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fierro-Monti I P, Reid S J, Woods D R. Differential expression of a Clostridium acetobutylicum antisense RNA: implications for regulation of glutamine synthetase. J Bacteriol. 1992;174:7642–7647. doi: 10.1128/jb.174.23.7642-7647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer R J, Helms J, Dürre P. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J Bacteriol. 1993;175:6959–6969. doi: 10.1128/jb.175.21.6959-6969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gawehn K, Bergmeyer H U. d-Lactate. In: Bergmeyer H U, editor. Methods of enzymatic analysis. 2nd ed. Vol. 3. New York, N.Y: Academic Press, Inc.; 1974. pp. 1493–1495. [Google Scholar]
- 19.Gerischer U, Dürre P. mRNA analysis of the adc gene region of Clostridium acetobutylicum during the shift to solventogenesis. J Bacteriol. 1992;174:426–433. doi: 10.1128/jb.174.2.426-433.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girbal L, Soucaille P. Regulation of Clostridium acetobutylicum metabolism as revealed by mixed-substrate steady-state continuous cultures: role of NADH/NAD ratio and ATP pool. J Bacteriol. 1994;176:6433–6438. doi: 10.1128/jb.176.21.6433-6438.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green E M, Bennett G N. Inactivation of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. Appl Biochem Biotechnol. 1996;58:213–221. doi: 10.1007/BF02941702. [DOI] [PubMed] [Google Scholar]
- 22.Green E M, Boynton Z L, Harris L M, Rudolph F B, Papoutsakis E T, Bennett G N. Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology. 1996;142:2079–2086. doi: 10.1099/13500872-142-8-2079. [DOI] [PubMed] [Google Scholar]
- 23.Grupe H, Gottschalk G. Physiological events in Clostridium acetobutylicum during the shift from acidogenesis to solventogenesis in continuous culture and presentation of a model for shift induction. Appl Environ Microbiol. 1992;58:3896–3902. doi: 10.1128/aem.58.12.3896-3902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris L. M.S. thesis. Evanston, Ill: Northwestern University; 1997. [Google Scholar]
- 25.Hartmanis M G N. Butyrate kinase from Clostridium acetobutylicum. J Biol Chem. 1987;262:617–621. [PubMed] [Google Scholar]
- 26.Hartmanis M G N, Gatenbeck S. Intermediary metabolism in Clostridium acetobutylicum: levels of enzymes involved in the formation of acetate and butyrate. Appl Environ Microbiol. 1984;47:1277–1283. doi: 10.1128/aem.47.6.1277-1283.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hüsemann M H W, Papoutsakis E T. Solventogenesis in Clostridium acetobutylicum fermentations related to carboxylic-acid and proton concentrations. Biotechnol Bioeng. 1988;32:843–852. doi: 10.1002/bit.260320702. [DOI] [PubMed] [Google Scholar]
- 28.Janssen P J, Jones W A, Jones D T, Woods D R. Molecular analysis and regulation of the glnA gene of the gram-positive anaerobe Clostridium acetobutylicum. J Bacteriol. 1988;170:400–408. doi: 10.1128/jb.170.1.400-408.1988. . (Errata, 170:3319, 1988; 171:6394, 1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson J L, Toth J, Santiwatanakul S, Chen J S. Cultures of “Clostridium acetobutylicum” from various collections comprise Clostridium acetobutylicum, Clostridium beijerinckii, and two other distinct types based on DNA-DNA reassociation. Int J Syst Bacteriol. 1997;47:420–424. doi: 10.1099/00207713-47-2-420. [DOI] [PubMed] [Google Scholar]
- 30.Jones D T, Woods D R. Acetone-butanol fermentation revisited. Microbiol Rev. 1986;50:484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kernodle D S, Voladri R K, Menzies B E, Hager C C, Edwards K M. Expression of an antisense hla fragment in Staphylococcus aureus reduces alpha-toxin production in vitro and attenuates lethal activity in a murine model. Infect Immun. 1997;65:179–184. doi: 10.1128/iai.65.1.179-184.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mermelstein L D, Papoutsakis E T. In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1993;59:1077–1081. doi: 10.1128/aem.59.4.1077-1081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mermelstein L D, Welker N E, Bennett G N, Papoutsakis E T. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Bio/Technology. 1992;10:190–195. doi: 10.1038/nbt0292-190. [DOI] [PubMed] [Google Scholar]
- 34.Meyer C L, McLaughlin J K, Papoutsakis E T. The effect of CO on growth and product formation in batch cultures of Clostridium acetobutylicum. Biotechnol Lett. 1985;7:37–42. [Google Scholar]
- 35.Meyer C L, Roos J W, Papoutsakis E T. Carbon monoxide gassing leads to alcohol production and butyrate uptake without acetone formation in continuous cultures of Clostridium acetobutylicum. Appl Microbiol Biotechnol. 1986;24:159–167. [Google Scholar]
- 36.Morimoto S, Takahashi Y, Watanabe Y, Omura S. Chemical modification of erythromycins. I. Synthesis and antibacterial activity of 6-O-methylerythromycin. Am J Antibiot. 1984;37:187–189. doi: 10.7164/antibiotics.37.187. [DOI] [PubMed] [Google Scholar]
- 37.Nair R V, Bennett G N, Papoutsakis E T. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1994;176:871–885. doi: 10.1128/jb.176.3.871-885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nair R V, Papoutsakis E T. Expression of plasmid-encoded aad in Clostridium acetobutylicum M5 restores vigorous butanol production. J Bacteriol. 1994;176:5843–5846. doi: 10.1128/jb.176.18.5843-5846.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oultram J D, Minton N P, Brehm J K, Thompson D E, Swinfield T J, Loughlin M. Introduction of plasmids into whole cells of Clostridium acetobutylicum by electroporation. FEMS Microbiol Lett. 1988;56:83–88. [Google Scholar]
- 40.Peguin S, Soucaille P. Modulation of carbon and electron flow in Clostridium acetobutylicum by iron limitation and methyl viologen addition. Appl Environ Microbiol. 1995;61:403–405. doi: 10.1128/aem.61.1.403-405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestka S, Daugherty B L, Jung V, Hotta K, Pestka R K. Anti-mRNA: specific inhibition of translation of single mRNA molecules. Proc Nat Acad Sci USA. 1984;81:7525–7528. doi: 10.1073/pnas.81.23.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen D J, Bennett G N. Cloning of the Clostridium acetobutylicum ATCC 824 acetyl coenzyme A acetyltransferase (thiolase; EC 2.3.1.9) gene. Appl Environ Microbiol. 1991;57:2735–2741. doi: 10.1128/aem.57.9.2735-2741.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen D J, Cary J W, Vanderleyden J, Bennett G N. Sequence and arrangement of genes encoding enzymes of the acetone-production pathway of Clostridium acetobutylicum ATCC 824. Gene. 1993;123:93–97. doi: 10.1016/0378-1119(93)90545-e. [DOI] [PubMed] [Google Scholar]
- 44.Petersen D J, Welch R W, Walter K A, Mermelstein L D, Papoutsakis E T, Rudolph F B, Bennett G N. Cloning of an NADH-dependent butanol dehydrogenase gene from Clostridium acetobutylicum. Ann N Y Acad Sci. 1991;646:94–98. doi: 10.1111/j.1749-6632.1991.tb18567.x. [DOI] [PubMed] [Google Scholar]
- 45.Rose I A. Acetate kinase of bacteria (acetokinase) Methods Enzymol. 1955;1:591–595. [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Simons R W. Naturally occurring antisense RNA control—a brief review. Gene. 1988;72:35–44. doi: 10.1016/0378-1119(88)90125-4. [DOI] [PubMed] [Google Scholar]
- 48.Simons R W, Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- 49.Soucaille, P. Unpublished data.
- 50.Stephanopoulos G, Vallino J J. Network rigidity and metabolic engineering in metabolite overproduction. Science. 1991;252:1675–1681. doi: 10.1126/science.1904627. [DOI] [PubMed] [Google Scholar]
- 51.Terracciano J S, Kashket E R. Intracellular conditions required for the initiation of solvent production by Clostridium acetobutylicum. Appl Environ Microbiol. 1986;52:86–91. doi: 10.1128/aem.52.1.86-91.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Truffaut N, Hubert J, Reysset G. Construction of shuttle vectors useful for transforming Clostridium acetobutylicum. FEMS Microbiol Lett. 1989;49:15–20. doi: 10.1016/0378-1097(89)90334-0. [DOI] [PubMed] [Google Scholar]
- 53.van den Berg W A, van Dongen W M, Veeger C. Reduction of the amount of periplasmic hydrogenase in Desulfovibrio vulgaris (Hildenborough) with antisense RNA: direct evidence for an important role of this hydrogenase in lactate metabolism. J Bacteriol. 1991;173:3688–3694. doi: 10.1128/jb.173.12.3688-3694.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasconcelos I, Girbal L, Soucaille P. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol. 1994;176:1443–1450. doi: 10.1128/jb.176.5.1443-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner E G, Simons R W. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 56.Walter K A. Ph. D. thesis. Evanston, Ill: Northwestern University; 1994. [Google Scholar]
- 57.Walter K A, Bennett G N, Papoutsakis E T. Molecular characterization of two Clostridium acetobutylicum ATCC 824 butanol dehydrogenase isozyme genes. J Bacteriol. 1992;174:7149–7158. doi: 10.1128/jb.174.22.7149-7158.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walter K A, Mermelstein L D, Papoutsakis E T. Host-plasmid interactions in recombinant strains of Clostridium acetobutylicum ATCC 824. FEMS Microbiol Lett. 1994;123:335–342. [Google Scholar]
- 59.Walter K A, Mermelstein L D, Papoutsakis E T. Studies of recombinant Clostridium acetobutylicum with increased dosages of butyrate formation genes. Ann N Y Acad Sci. 1994;721:69–72. doi: 10.1111/j.1749-6632.1994.tb47377.x. [DOI] [PubMed] [Google Scholar]
- 60.Walter K A, Nair R V, Cary J W, Bennett G N, Papoutsakis E T. Sequence and arrangement of two genes of the butyrate-synthesis pathway of Clostridium acetobutylicum ATCC 824. Gene. 1993;134:107–111. doi: 10.1016/0378-1119(93)90182-3. [DOI] [PubMed] [Google Scholar]
- 61.Wiesenborn D P, Papoutsakis E T, Rudolph F B. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl Environ Microbiol. 1988;54:2717–2722. doi: 10.1128/aem.54.11.2717-2722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiesenborn D P, Rudolph F B, Papoutsakis E T. Phosphotransbutyrylase from Clostridium acetobutylicum ATCC 824 and its role in acidogenesis. Appl Environ Microbiol. 1989;55:317–322. doi: 10.1128/aem.55.2.317-322.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]