ABSTRACT
Clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas) systems are prokaryotic adaptive immune systems that have been well characterized biochemically, but in vivo spatiotemporal regulation and cell biology remain largely unaddressed. Here, we used fluorescent fusion proteins introduced at the chromosomal CRISPR-Cas locus to study the localization of the type I-F CRISPR-Cas system in Pseudomonas aeruginosa. When lacking a target in the cell, the Cascade complex is broadly nucleoid bound, while Cas3 is diffuse in the cytoplasm. When targeted to an integrated prophage, however, the CRISPR RNA (crRNA)-guided type I-F Cascade complex and a majority of Cas3 molecules in the cell are recruited to a single focus. Nucleoid association of the Csy proteins that form the Cascade complex is crRNA dependent and specifically inhibited by the expression of anti-CRISPR AcrIF2, which blocks protospacer adjacent motif (PAM) binding. The Cas9 nuclease is also nucleoid localized, only when single guide RNA (sgRNA) bound, which is abolished by the PAM-binding inhibitor AcrIIA4. Our findings reveal PAM-dependent nucleoid surveillance and spatiotemporal regulation in type I CRISPR-Cas that separates the nuclease-helicase Cas3 from the crRNA-guided surveillance complex.
IMPORTANCE CRISPR-Cas systems, the prokaryotic adaptive immune systems, are largely understood using structural biology, biochemistry, and genetics. How CRISPR-Cas effectors are organized within cells is currently not well understood. By investigating the cell biology of the type I-F CRISPR-Cas system, we show that the surveillance complex, which “patrols” the cell to find targets, is largely nucleoid bound, while Cas3 nuclease is cytoplasmic. Nucleoid localization is also conserved for class 2 CRISPR-Cas single protein effector Cas9. Our observation of differential localization of the surveillance complex and Cas3 reveals a new layer of posttranslational spatiotemporal regulation to prevent autoimmunity.
KEYWORDS: anti-CRISPR, bacterial cell biology, bacteriophages, CRISPR-Cas, spatial organization
INTRODUCTION
Bacteria have evolved a wide range of immune mechanisms, including the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) genes, to protect from bacteriophages and other mobile genetic elements (1). The adaptive CRISPR-Cas system is present in almost 85% of archaea and 40% of bacterial genomes sequenced. Currently, CRISPR-Cas systems are categorized into 2 broad classes, 6 types, and 33 subtypes (2). CRISPR-Cas systems acquire foreign DNA into the CRISPR array as new spacers and subsequently transcribe and process that array to generate CRISPR RNAs (crRNAs), which complex with Cas proteins. This crRNA-guided complex surveils the cell for a protospacer adjacent motif (PAM) and subsequent complementary base pairing with the crRNA, which triggers cleavage of the invading nucleic acid. In the case of type I CRISPR-Cas systems, which are the most abundant in bacteria (2, 3), a multisubunit crRNA-guided surveillance complex called Cascade recognizes the PAM and protospacer and then recruits a trans-acting helicase-nuclease (Cas3) for target degradation (4).
How CRISPR-Cas effectors are organized within cells is currently not well understood. To function efficiently as a defense system, CRISPR-Cas must rapidly recognize the incoming foreign DNA and destroy it before it becomes established. Previous in vitro studies using single-molecule imaging has shown that type I-E Cascade (which is the crRNA-guided complex of the I-E system) spends between 0.1 and 10 s scanning targets (4–6). A recent in vivo study using superresolution microscopy and single-molecule tracking of Cascade in live Escherichia coli suggested a timescale of 30 ms for target probing (7), and similar binding kinetics have also been suggested for Cas9 (8, 9). Cascade was suggested to spend approximately 50% of its search time on DNA and the rest distributed in the cytoplasm; however, the impacts of phage infection and Cas3 localization have not been determined. Additionally, chromatin immunoprecipitation sequencing (ChIP-seq) analysis of Cascade-DNA interactions in E. coli revealed that less than 5 bp of the crRNA-DNA interaction is sufficient to promote an association of Cascade at numerous sites in the genome (10). Genomic associations of CRISPR-Cas stand in contrast to another important class of defense systems, namely, restriction modification (R-M). Type I R-M complexes (HsdRMS) localize to the inner membrane in such a way that their activities are controlled spatially. Based on biochemical approaches, the methyltransferase is proposed to be on the cytoplasmic side of the inner membrane, while the restriction enzyme components are positioned in the periplasm (11, 12).
The type I-F CRISPR-Cas system of Pseudomonas aeruginosa has emerged as a powerful model for understanding various aspects of CRISPR-Cas biology, including a mechanistic understanding of type I systems (13–15), the discovery (16) and in vivo characterization (17, 18) of phage-encoded anti-CRISPR proteins (Acrs), and identification of regulatory pathways governing CRISPR-Cas (19–22). The I-F system of P. aeruginosa PA14 consists of two CRISPR loci and six Cas proteins, namely, Csy1 to Csy4, which form type I-F Cascade; Cas3 (a fusion of Cas2-3), a trans-acting nuclease/helicase protein; and Cas1, which drives spacer acquisition (13, 23). Structural and biochemical studies have shown that Csy1 to Csy4 assemble on a 60-nucleotide crRNA to form a 350-kDa seahorse-shaped crRNA-guided Cascade surveillance complex (14, 15, 24). Cascade recognizes DNA first via a Csy1-PAM interaction (G-G/C-C), leading to the destabilization of the DNA duplex, strand invasion, R-loop formation in the seed region, and downstream base pairing. Finally, DNA-bound Cascade complex triggers recruitment of Cas3 nuclease, which mediates processive degradation of the target DNA in a 3′ to 5′ direction (14, 15, 24).
Here, we address the cell biology of a naturally active P. aeruginosa type I-F CRISPR-Cas system by directly observing its subcellular localization using live cell microscopy. Using functional fluorescent fusions chromosomally integrated at the native locus, we show that the Csy1 and Csy4 proteins (Cas8 and Cas6 family members, respectively), which are part of the surveillance complex, are largely nucleoid bound, while the Cas3 nuclease is cytoplasmic. While phage infection did not induce changes in localization, both Cascade and Cas3 clearly localized to a stable intracellular target (i.e., a prophage). When Cascade is formed with a crRNA, it binds the nucleoid even in the absence of a target, but the individual Cas proteins do not. Nucleoid localization of Cascade, and Cas9, is mediated at the level of PAM recognition and is specifically disrupted by PAM-mimetic anti-CRISPR proteins. Taken together, our study suggests that the relatively promiscuous PAM-dependent (e.g., 5′-GG-3′) DNA search mechanism likely relegates these complexes to associate predominantly with the host genome, with no active mechanism for preventing host genome surveillance.
RESULTS
The majority of endogenous Cas3 molecules are recruited to CRISPR targets.
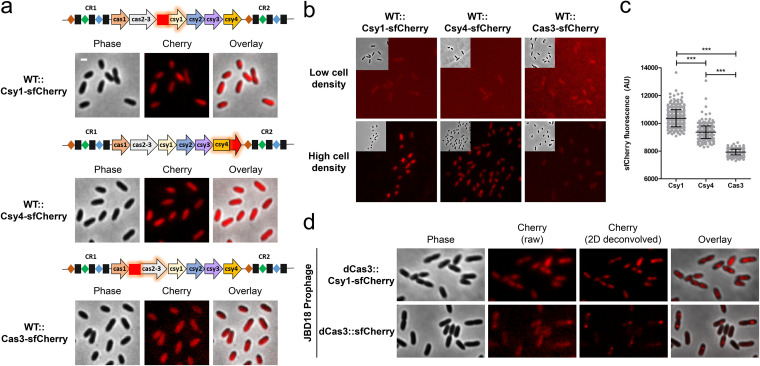
To investigate CRISPR-Cas subcellular localization, we focused on P. aeruginosa strain UCBPP-PA14 (denoted PA14), which has an active type I-F CRISPR-Cas system (25). We constructed PA14 strains in which Csy1 (Cas8), Csy4 (Cas6), and Cas3 are fused with sfCherry at their native locus. We assessed the functionality of the tagged strains using a panel of isogenic phages to read out CRISPR-Cas function, as follows: DMS3 (untargeted control), DMS3m (targeted by a natural spacer, CRISPR2 spacer 1), and DMS3m engineered phages that expresses Acr proteins AcrIF1, AcrIF2, AcrIF3, or AcrIF4 (17). All three fusions exhibited CRISPR-Cas activity similar to the wild type and were inhibited by Acr proteins (see Fig. S1 in the supplemental material). Western blot analysis, using α-mCherry antibodies, shows that the sfCherry fusions are expressed at the expected size (see Fig. S2 in the supplemental material). Live cell fluorescence microscopy enabled visualization of the distribution of the Cas proteins. All three proteins appeared diffuse in the cytoplasm under this condition (Fig. 1a). Of note, cells had to be grown to high cell density, which is known to be important for Cas protein expression in P. aeruginosa (Fig. 1b) (20, 26, 27). An analysis of fluorescent signal from single cells indicated that Csy1 and Csy4 protein levels are higher than Cas3 (Fig. 1c).
FIG 1.
Live-cell fluorescence imaging of P. aeruginosa PA14 expressing type I-F Cas protein reporters from endogenous locus. (a) Fluorescence microscopy of wild-type PA14 expressing Csy1-sfCherry, Csy4-sfCherry, or Cas3-sfCherry. (b) Comparison of fluorescence in cells expressing Csy1-sfCherry, Csy4-sfCherry, or Cas3-sfCherry at low cell density (optical density at 600 nm [OD600], 0.2 to 0.3) or high cell density (OD600, 1 to 1.5). (c) sfCherry fluorescence (arbitrary unit [AU]) in wild-type cells grown to high cell density expressing sfCherry-tagged Csy1, Csy4, or Cas3. More than 200 cells from each genotype were analyzed from two independent experiments. Means and standard deviations are shown. The statistical significance was calculated using an unpaired t test analysis (***, P < 0.0001). (d) Fluorescence microscopy of PA14 expressing Csy1-sfCherry (in dCas3 background) or dCas3-sfCherry with a JBD18 prophage. sfCherry fluorescence is shown in raw as well as deconvolved images. Scale bar, 1 μm.
To observe fluorescent Cas proteins localizing to target DNA in vivo, we generated lysogenic strains containing JBD18 as a prophage in PA14 strains expressing chromosomal Csy1-sfCherry (in catalytic dead Cas3 background) or dCas3-sfCherry. JBD18 naturally has five protospacer sequences with the correct PAM (25). Cells expressing dCas3 are incapable of antiphage activity (see Fig. S3 in the supplemental material) but can be recruited to the target sequence (19). A total of 23% of cells expressing Csy1-sfCherry (n = 492) and 27% of cells expressing dCas3-sfCherry (n = 348) formed fluorescent foci in the presence of the JBD18 prophage (Fig. 1d). Only one focus could be observed, presumably due to slow growth of cells at high density. However, cells undergoing division exhibited two foci. Notably, the foci formed by dCas3-sfCherry were quite discrete, suggesting that most of the cellular dCas3 protein is recruited to the targeted locus. Taken together, the data presented above show that endogenous levels of tagged I-F Cas proteins are functional, can be visualized, and can read out bona fide target DNA binding events in vivo, which apparently recruits multiple dCas3 proteins.
Cascade is nucleoid enriched while the Cas3 nuclease is cytoplasmic.
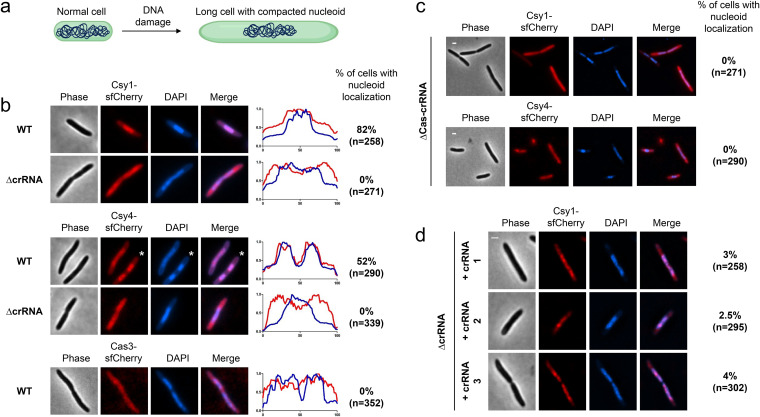
To determine the localization pattern of Cascade, we took an approach that can clearly differentiate membrane-associated, DNA-bound, and cytoplasmic proteins (28). Cells were treated with the DNA-damaging antibiotic nalidixic acid (NA). At the concentration used, P. aeruginosa continues to grow slowly and forms long cells with compacted nucleoids (schematic presented in Fig. 2a). In these nucleoid-compacted cells, DNA-localized proteins can be differentiated clearly from cytoplasmic and membrane proteins. We observed that Csy1-sfCherry (82% nucleoid localized, n = 258 cells) and Csy4-sfCherry (52% nucleoid localized, n = 290 cells) are enriched in the nucleoid, as evidenced by their colocalization with 4′,6-diamidino-2-phenylindole (DAPI) stain, while Cas3-sfCherry appeared diffuse in the cytoplasm (0% nucleoid localized, n = 352 cells) (Fig. 2b). To confirm that nucleoid localization of the Csy complex did not occur as a result of DNA damage caused by NA treatment, we treated cells with chloramphenicol (200 μg/mL), which causes nucleoid compaction as a result of translation inhibition, leading to transertion inhibition (29, 30), and monitored the localization of Csy1. Unlike NA-treated cells, chloramphenicol-treated cells are not elongated and thus the nucleoid compaction is only marginal. Still, in these nucleoid-compacted cells, Csy1 was found to be nucleoid enriched (see Fig. S4 in the supplemental material).
FIG 2.
Cascade is nucleoid-enriched in a crRNA-dependent manner while Cas3 is cytoplasmic. (a) A model for DNA damage-induced nucleoid compaction. (b) Fluorescence microscopy of nucleoid compacted wild-type or ΔcrRNA cells expressing Csy1-sfCherry or Csy4-sfCherry and nucleoid compacted wild-type cells expressing Cas3-sfCherry. Fluorescence intensity maps plotted against the long cell axis are shown. *, indicates which cell is plotted in the map. (c) Fluorescence microscopy of nucleoid-compacted ΔCas-crRNA cells expressing Csy1-sfCherry or Csy4-sfCherry from a plasmid. (d) Fluorescence microscopy of nucleoid-compacted ΔcrRNA cells expressing Csy1-sfCherry and synthetic crRNAs. Scale bars, 1 μm. For b and c, more than 125 nucleoid-compacted cells from each genotype were analyzed from two independent experiments.
We next asked whether Csy protein nucleoid localization is dependent on Cascade formation. To test this question, the same Csy1-sfCherry and Csy4-sfCherry labels were inserted into the chromosome of a ΔCRISPR strain that lacks all 35 spacers but still expresses all Cas proteins. Both Csy1-sfCherry (0% nucleoid localized, n = 271 cells) and Csy4-sfCherry (0% nucleoid localized, n = 339 cells) lost nucleoid localization in the ΔCRISPR mutant (Fig. 2b), suggesting that crRNA-mediated assembly of Cascade is essential for nucleoid localization. Additionally, ectopic expression of Csy1-sfCherry (0% nucleoid localized, n = 311 cells) or Csy4-sfCherry (0% nucleoid localized, n = 290 cells) from a plasmid in NA-treated cells that lack all components of the CRISPR-Cas system (ΔCRISPR-Cas) showed that they were not enriched in the nucleoid (Fig. 2c).
PA14 has a total of 35 distinct crRNAs produced from its 2 CRISPR loci. It is possible that one or more of these crRNAs with a partial match to the genomic DNA is responsible for the surveillance complex nucleoid localization. To determine the importance of crRNAs in nucleoid surveillance, we expressed designed crRNAs, which do not have a detectable sequence match in the PA14 genome in ΔCRISPR cells and asked if they can restore Csy1-sfCherry nucleoid localization. All three crRNAs partially restored Csy1-sfCherry nucleoid localization in ΔCRISPR cells. Restoration was observed only for a minority of cells (between 2% and 4%) in the three crRNA-expressing strains, possibly due to suboptimal crRNA expression. Still, partial restoration of Cascade nucleoid localization by ectopically expressed crRNA suggests that, once assembled, it is intrinsically capable of binding the nucleoid independent of the crRNA sequence (Fig. 2d). Together, these data indicate that nucleoid localization of Csy1 and Csy4 is not an intrinsic property of the proteins but occurs as a result of the formation of the Cascade complex, while the Cas3 nuclease is spread throughout the cell.
An anti-CRISPR that blocks PAM binding specifically prevents Cascade nucleoid localization.
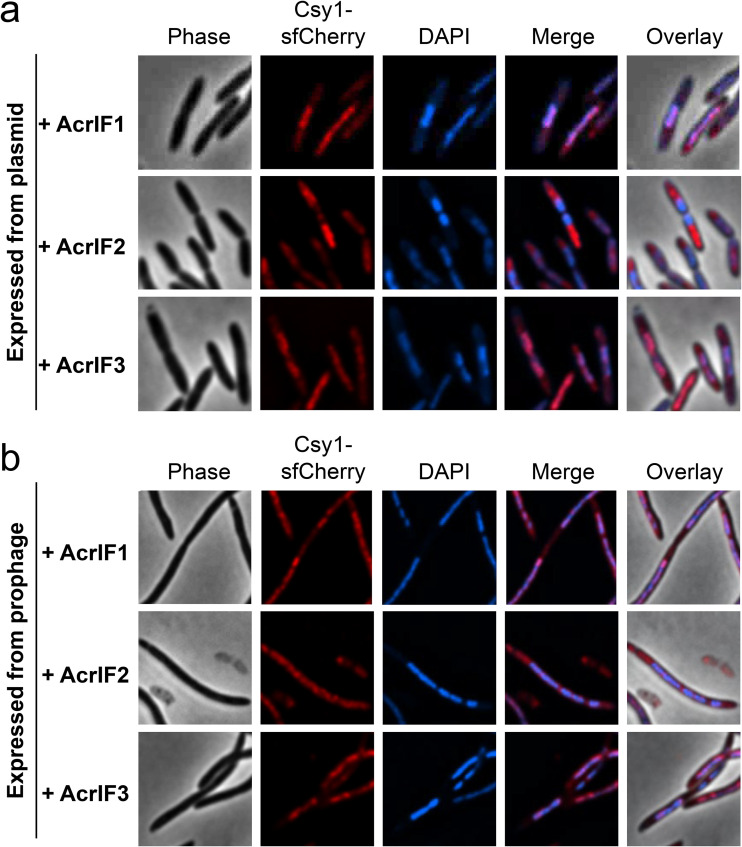
Cascade is nucleoid localized; however, whether this localization is through direct DNA binding or an interaction with host factors is unknown. The surveillance complex recognizes target DNA in two steps, as follows: (i) interaction with the “GG” PAM, which is mediated mostly by residues in Csy1, and (ii) base pairing between the crRNA with target DNA, which is mediated by the spacer region of the crRNA (14, 15, 24). Two type I-F Acr proteins have been identified that distinguish between these two binding mechanisms (14, 16, 24, 31, 32). AcrIF1 binds to Csy3 (Cas7), blocking crRNA-DNA target hybridization but does not occlude the PAM binding site, while AcrIF2 binds to the Csy1-Csy2 (Cas8-Cas5) heterodimer and specifically competes with PAM binding in target DNA (15, 24, 31). AcrIF3 was also utilized, which binds to Cas3 nuclease and prevents target cleavage but does not block stable DNA binding by Cascade (31, 33). Csy1-sfCherry localization was monitored in NA-treated nucleoid-compacted cells expressing one of these three anti-CRISPRs from a plasmid (Fig. 3a). Csy1-sfCherry localization to the nucleoid was largely unaffected when AcrIF1 (67% nucleoid localized, n = 277 cells) or AcrIF3 (71% nucleoid localized, n = 290 cells) were expressed. However, in cells expressing AcrIF2, nucleoid localization of Csy1-sfCherry is disrupted completely (0% nucleoid localized, n = 259 cells) (Fig. 3a and b). Similarly, when the three anti-CRISPRs were expressed from isogenic DMS3m prophages (17) integrated in the reporter strain, Csy1-sfCherry nucleoid localization was abrogated completely in the presence of AcrIF2 but not AcrIF1 or AcrIF3. Of note, prophage-containing cells were transformed with a plasmid expressing the C repressor in order to prevent possible excision during NA treatment. This process resulted in the formation of extremely long cells making it difficult to quantify the percentage of cells displaying nucleoid localization. To verify further that PAM mediates Cascade nucleoid localization, we expressed a recently characterized anti-CRISPR, AcrIF11 (34, 35), an enzyme that ADP-ribosylates Csy1 to prevent PAM binding, from a plasmid, and observed that it also disrupted nucleoid localization (0% nucleoid localized, n = 325 cells) (see Fig. S5 in the supplemental material). Together, these results suggest that PAM recognition is the dominant and direct factor that mediates Cascade localization to the nucleoid.
FIG 3.
AcrIF2 but not AcrIF1 or AcrIF3 blocks nucleoid localization of type I-F Cascade. Fluorescence microscopy of nucleoid-compacted wild-type cells producing Csy1-sfCherry and expressing anti-CRISPR proteins AcrIF1, AcrIF2, or AcrIF3 from a plasmid (a) or a prophage (b). For a, more than 125 nucleoid-compacted cells from each genotype were analyzed from two independent experiments.
Phage infection does not modulate Cascade localization.
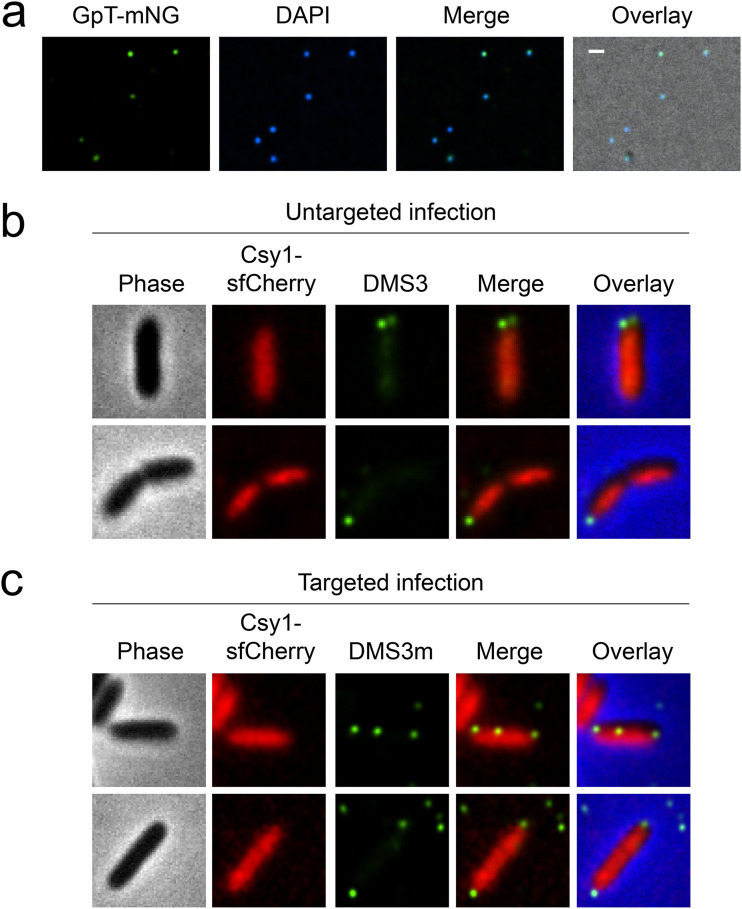
We next wanted to address the important question of whether CRISPR-Cas distribution responds to phage infection as a defense system. To enable visualization of infecting phage particles, we propagated DMS3 (0 protospacers) and DMS3m (1 protospacer) in a strain that expressed GpT, the major capsid protein, fused with the mNeonGreen fluorescent protein. This process allowed us to obtain fluorescent mosaic phage particles that contain labeled as well as unlabeled GpT proteins. We verified that 100% of fluorescent phages packed viral DNA, as evidenced by the colocalization of mNeonGreen and DAPI stain (Fig. 4a). Of note, the plaque-forming capacity of the labeled phages were comparable to that of unlabeled phages (see Fig. S6 in the supplemental material). The subcellular localization of Csy1-sfCherry and Cas3-sfCherry were assessed in the presence of labeled DMS3 (untargeted phage) or DMS3m (targeted phage) after 15 min of infection. Previous studies using lambda have shown that DNA ejection takes less than a minute (36). Thus, 15 min should be sufficient for DNA ejection. In both cases, the apparent diffuse localization of Csy1-sfCherry was not affected at a gross level, when comparing cells with fluorescent phage particles adjacent to the cell surface to those without (Fig. 4b and c). To check whether the presence of more protospacers would enable the recruitment of Cascade or Cas3, we infected Csy1-sfCherry (in dCas3 background) and dCas3-sfCherry-expressing cells with unlabeled JBD18 under the same conditions (see Fig. S7 in the supplemental material). Also, here, we did not observe any change in localization despite this genotype being the same where JBD18 prophage recognition was so striking (Fig. 1d). These data suggest that phage infection, irrespective of the presence or absence of a target sequence, does not significantly modulate the distribution of the Cascade or Cas3 as observed at this resolution.
FIG 4.
Phage infection does not affect Csy localization. (a) Fluorescence microscopy images of DMS3 phages with capsid labeled with GpT-mNeonGreen and DNA labeled with DAPI. Fluorescence microscopy of Csy1-sfCherry-producing cells (in dCas3 background) infected with labeled phages DMS3 (0 protospacers) (b) or DMS3m (1 protospacer) (c). Scale bar, 1 μm.
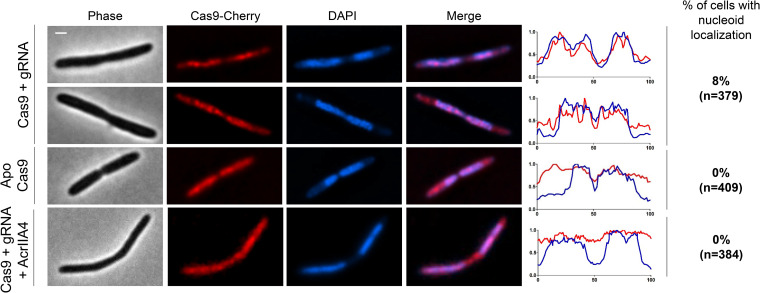
Nucleoid localization is conserved for Spy Cas9.
Having observed that the multisubunit type I-F Cascade is nucleoid enriched, we next wondered whether nucleoid localization is a conserved property of class 2 CRISPR-Cas single protein effectors. For this purpose, we chose Cas9 of Streptococcus pyogenes as a model protein, which coincidentally uses the same PAM as the I-F Cascade. To test the localization of SpyCas9, we used a plasmid that expressed a functional SpyCas9 fused with Cherry (37) in P. aeruginosa PAO1 and monitored its localization in the presence and absence of a crRNA in NA-treated nucleoid-compacted cells. While SpyCas9 was found to be diffuse in its Apo form (i.e., lacking a crRNA) (0% nucleoid localized, n = 409 cells), a minority of cells displayed nucleoid localization when SpyCas9 complexed with a crRNA, provided as a single guide RNA (4% nucleoid localized, n = 322 cells) (Fig. 5). When we expressed AcrIIA4, an anti-CRISPR that inhibits SpyCas9 by competing with the PAM-interacting domain (38, 39), nucleoid localization of guide RNA-bound SpyCas9 could not be observed in any cells (0% nucleoid localized, n = 384 cells) (Fig. 5). Since AcrIIA4 blocks PAM and seed interactions (40) and we lack an Acr protein that blocks only Cas9 single guide RNA (sgRNA):target hybridization (akin to AcrIF1), we cannot comment on whether PAM + seed binding is required here. These data, taken together with previous reports (7–9), suggest that genome surveillance via PAM for type I systems and PAM or PAM-seed interactions for type II systems drive the nucleoid localization of the CRISPR-Cas complex in their native environment.
FIG 5.
Nucleoid localization is conserved for SpyCas9. Fluorescence microscopy of nucleoid-compacted P. aeruginosa PAO1 cells producing Cas9-sgRNA, ApoCas9, or Cas9-sgRNA coexpressed with anti-CRISPR protein AcrIIA4. Fluorescence intensity maps plotted against the long cell axis are shown. Scale bar, 1 μm.
DISCUSSION
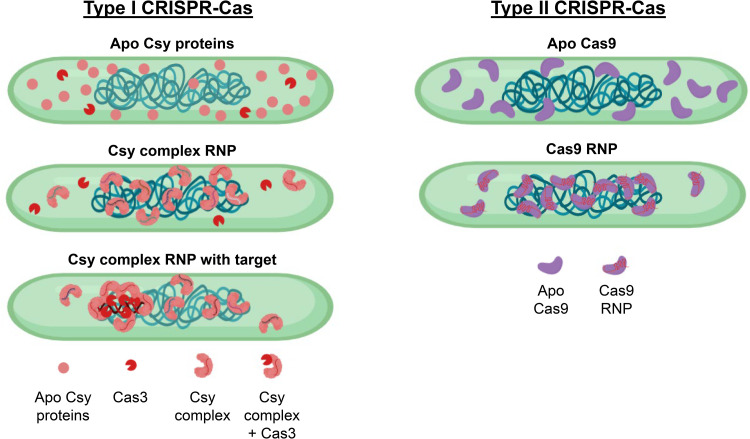
In this study, we report on the cell biology and subcellular organization of the P. aeruginosa type I-F system in its native host. We show that Csy1 and Csy4, which are used here as markers of type I-F Cascade, are enriched in the nucleoid, consistent with studies of type I-E Cascade (7), while the Cas3 nuclease-helicase is distributed in the cytoplasm (schematically illustrated in Fig. 6). Nucleoid localization is not an intrinsic property of these proteins when expressed alone or without crRNAs but is rather mediated by the assembly of Cascade. This finding is especially surprising regarding Csy1, as it and other Cas8 family members have been shown to bind DNA nonspecifically (5, 7, 41, 42). These observations indicate that Cascade spends most of its time scanning for a match, not in association with the Cas3 nuclease, perhaps avoiding a Cas3 “misfire.” A previous microscopy-based observation has shown that Cas3 is recruited to I-E Cascade only in the presence of a canonical protospacer (43). However, to the best of our knowledge, this study is the first one to examine the in vivo subcellular organization of Cas3, a universally conserved nuclease in type I systems, under physiologically relevant conditions. Our cell biology-based observations on posttranslational spatiotemporal regulation in CRISPR-Cas is consistent with structural and biophysical evidence suggesting Cas3 is recruited by Cascade only after target interaction (5, 43–45). At first glance, the subcellular organization of CRISPR-Cas seems like an inferior approach compared with type I R-M complexes which localize in the inner membrane (11, 12), likely minimizing host collateral damage and maximizing phage detection. Given this information, it remains to be seen whether Cascade/Cas9 surveillance of the genome is adaptive, as opposed to simply being a by-product of the basic PAM surveillance mechanisms of CRISPR-Cas systems, “forcing” these systems to localize in the genome.
FIG 6.
A model for subcellular organization of type I and type II CRISPR-Cas systems in bacteria. The subcellular localization of CRISPR-Cas systems are presented for type I (left) and type II (right) systems.
Our observation of differential localization of Cascade and Cas3 might present an additional regulatory strategy to prevent autoimmunity in type I systems, in a way that is not achievable for single effector class 2 systems (e.g., Cas9 and Cas12). Type II-A Cas9 of S. pyogenes, which mediates target recognition as well as cleavage as a single multidomain protein (46), is also enriched in the nucleoid, only when bound to a crRNA. In contrast to the Cas3 nuclease, localization of Cas9 in the nucleoid could pose autoimmunity risks. In fact, a recent study showed that uncontrolled induction Cas9 in S. pyogenes, which occurs in the absence of a naturally repressive single guide RNA, results in increased self-targeting and auto immunity (47). We have also observed that the activity of Listeria monocytogenes Cas9 is very low, at least under laboratory conditions (48, 49). It is possible that because of the single protein mediating DNA binding and cleavage, that the risks associated with Cas9-mediated toxicity could contribute to the observed bias toward type I systems in nature.
Our efforts to pin down the target recognition step that mediates nucleoid localization of Cascade as well as Cas9 were supported by experiments using Acr proteins, such as AcrIF2, AcrIF11, and AcrIIA4, which specifically block PAM binding, while AcrIF1 and AcrIF3 act downstream. Perhaps most notably, the inability of AcrIF1 to prevent nucleoid localization, together with observations that the crRNA sequence is not important, suggests that PAM binding, not base pairing, is driving nucleoid surveillance. Notably, previous studies using tiling electrophoretic mobility shift assay (EMSAs) have shown that Csy complex capable of binding to the seed region of a single-stranded target DNA were not blocked by AcrIF2 (31), and structural evidence shows that AcrIF1 does not occupy the PAM groove (24). Still, some base pairing in the seed region could contribute to binding strength (10), but we do not consider it necessary for nucleoid surveillance due to the abundance of PAM sequences in the genome (P. aeruginosa PA14 has 1.2 × 106 5′-GG-3′ PAM sites). In contrast to our observation, type I-E Cascade-DNA interactions were suggested to be mediated by PAM-dependent as well as PAM-independent interactions (7) because a mutant Cas8 (Csy1 in type I-F) that is incapable of recognizing PAM decreased, but did not completely abolish, its nucleoid localization (7). In our case, Csy1 nucleoid localization was not observed either in the presence of AcrIF2 or in the absence of crRNA or when Csy1 was expressed alone. Given the divergence of the Cas8 superfamily, it is possible that the mechanistic differences between type I-E and type I-F systems exist.
Lastly, while phage infection has been implicated in the upregulation of CRISPR-Cas enzymes in some bacteria and archaea (50–52), we did not observe significant upregulation or relocalization of Cascade or Cas3 during phage infection. Intriguingly, JBD18 could promote clustering of Cascade and Cas3 as a prophage but not as an infecting phage. Notably, the fluorescent foci formed by dCas3 were quite discrete, suggesting that most dCas3 molecules within the cell are recruited to the prophage, an observation we found surprising. Whether this truly reflects Cas3 recruitment events or is observed due to catalytic inactivation is worth further investigation. One possible reason for the absence of any Csy/Cas3 relocalization during lytic infection could be the transient nature of the interaction, with a relatively dynamic target like an injected phage genome (53). Additionally, Cascade complexes in PA14 are loaded with 35 distinct crRNAs, likely concealing the relocation of a minority of molecules, despite Csy and Cas3 relocalization being observable for a prophage. Dynamic relocation of molecules near the inner membrane during infection may require investigation using more sensitive imaging techniques, including single-molecule imaging and total internal reflection fluorescence (TIRF) microscopy (54).
What is the physiological function of the nucleoid-enriched CRISPR-Cas enzymes? Several findings have suggested that CRISPR-Cas enzymes participate in cellular functions other than immunity. These functions include DNA repair, gene regulation, sporulation, genome evolution, and stress response (55–57); however, there is no strong evidence for these functions with I-F Cascade in P. aeruginosa. The molecular basis for most of these alternative functions of Cas proteins is not well understood. A recent study showing that SpyCas9 can repress its own promoter using a natural single guide opens the possibility that CRISPR-Cas systems can also function as intrinsic transcriptional regulators (47). It is therefore possible that crRNAs generated from degenerate self-targeting spacers might direct CRISPR-Cas enzymes in regulating host genes. However, binding of the E. coli cascade complex to hundreds of off-target sites does not affect gene expression globally (10). Alternatively, it could be concluded that the intrinsic PAM-sensing mechanism of DNA detection relegates Cascade and crRNA-loaded Cas9 to survey the genome, even if that function is neither adaptive directly for cellular functions or defense. Given that affirmative PAM recognition is important for function, as opposed to exclusion mechanisms (e.g., genome-wide methylation to prevent restriction enzyme binding), host genome surveillance likely reflects this limitation. Future studies examining these possibilities will provide important insights on the functions and evolutionary limitations of CRISPR-Cas systems in bacteria.
MATERIALS AND METHODS
Plasmids, phages, and growth media.
Plasmids and primer sequences used in this study are listed in Tables S1 and S2 in the supplemental material. P. aeruginosa UCBPP-PA14 (PA14) strains and Escherichia coli strains were grown on lysogeny broth (LB) agar or liquid at 37°C. To maintain the pHERD30T plasmid, the medium was supplemented with gentamicin (50 μg/mL for P. aeruginosa and 30 μg/mL for E. coli). Phage stocks were prepared as described previously (17). In brief, 3 mL SM buffer (100mM Sodium Chloride, 8mM Magnesium Sulfate, 0.01% Gelatin, 50mM Tris-HCl) was added to plate lysates of the desired purified phage and incubated at room temperature for 15 min. SM buffer containing phages was collected and 100 μL chloroform was added. This mixture was centrifuged at 10,000 × g for 5 min, and the supernatant containing phages was transferred to a storage tube with a screw cap and incubated at 4°C. Phages used in this study include DMS3, DMS3m, and engineered DMS3m phages encoding Acr proteins (17).
Construction of plasmids and strains.
Plasmids expressing sfCherry alone and sfCherry tagged with Cas3 or Cas9 were reported previously (37). Plasmids expressing Csy1-sfCherry and Csy4-sfCherry were constructed by Gibson assembly in a pHERD30T plasmid digested with SacI and PstI. These fusions have a ggaggcggtggagcc (G-G-G-G-A) linker sequence in between them. sCherry was amplified from SF-pSFFV-sfCherryFL1M3_TagBFP (kindly provide by Bo Huang lab, University of California San Francisco [UCSF]). csy1 and csy4 sequences were amplified from PA14. Csy1 and Cas3 are tagged at the N terminus, and Csy4 is tagged at the C terminus. A plasmid expressing AcrIIA4 was constructed by Gibson assembly in the MMBHE plasmid digested with HindIII and KpnI.
Endogenous Csy1-sfCherry and Cas3-sfCherry reporters were described previously (19). Csy4-sfCherry was constructed in a similar way. The sfCherry gene was inserted with csy4 of PA14 via allelic replacement. The recombination vector pMQ30, which contained sfCherry flanked by homology arms matching csy4, was introduced via conjugation. pMQ30-Csy4-sfCherry, which contains the sfCherry sequence flanked by 123 bp upstream and downstream of the csy4 stop codon, was cloned in the pMQ30 plasmid between HindIII and BamHI sites using Gibson assembly. pMQ30-Csy4-sfCherry contain the GGAGGCGGTGGAGCC sequence (encoding GGGGA) as a linker between sfCherry and csy4. The pMQ30-Csy4-sfCherry construct was introduced into PA14 strains of interest via allelic replacement to generate Csy4-sfCherry. Strains containing the appropriate insertion were verified via PCR.
crRNAs suitable for the type I-F system were expressed from I-F entry vector pAB04. Oligonucleotides with repeat-specific overhangs encoding the spacer sequences that do not have sequence homology with the PA14 genome are phosphorylated using T4 polynucleotide kinase (PNK) and cloned into the entry vectors using the BbsI sites. Sequences of the spacers are listed in Table S3 in the supplemental material. crRNAs are expressed without addition of the inducer arabinose.
Construction of PA14 lysogens.
Lysogens were obtained by first spotting phage onto a bacterial lawn and then streaking out surviving colonies from phage spots. These colonies were screened for phage resistance using a cross-streak method, and lysogeny was verified by prophage induction. For the maintenance of DMS3m-engineered lysogens that expresses Acr proteins AcrIF1, AcrIF2, and AcrIF3 during NA treatment, an arabinose-inducible pHERD30T plasmid expressing a C-repressor of DMS3 was present (17).
Live-cell imaging and image processing.
Fluorescence microscopy was carried out as described previously (37). Unless indicated, overnight cultures were diluted 1:10 in fresh LB medium and grown for 3 h. For compaction of the nucleoid, nalidixic acid (200 μg/mL) was added for 3 h or chloramphenicol (200 μg/mL) was added for the last 20 min. A total of 0.5 mL of cells was centrifuged, washed with 1:10 LB diluted with double-distilled water, and finally resuspended in 200 to 500 μL of 1:10 LB. Cell suspensions were placed onto 0.85% 1:10 LB agarose pads with uncoated coverslips. For DNA staining, DAPI (2 μg/mL) was added to the cell suspension for 10 min, washed twice with 1:10 LB, and finally resuspended. A Nikon Ti2-E inverted microscope equipped with the Perfect Focus System (PFS) and a Photometrics Prime 95B 25-mm camera were used for live-cell imaging. Time-lapse imaging was performed using a Nikon Eclipse Ti2-E instrument equipped with an Okolab cage incubator. Images were processed using NIS Elements advanced research (AR) software.
To measure single-cell fluorescence, regions of interest (ROIs) were drawn over the phase contrast images of endogenous sfCherry-tagged reporter strains using NIS Elements AR software. After the background was subtracted, the sfCherry ROI mean intensity values were obtained. The data were analyzed and presented as a scatterplot using GraphPad Prism.
For quantification of nucleoid localization, an intensity line was drawn over the middle long cell axis of the cells expressing sfCherry and stained with DAPI. Cells exhibiting overlapping intensity maps for sfCherry and DAPI were regarded as nucleoid localized. An analysis was performed for two independent experiments, and the percentage of nucleoid-localized cells was calculated as the mean value of two independent experiments.
For fluorescence intensity maps, an intensity line was drawn over the phase contrast images along the middle long cell axis. Fluorescence values of sfCherry and DAPI along the long axis were extracted. The data were analyzed and presented as a line plot using GraphPad Prism.
Western blotting.
Equal amounts of samples were collected and washed, and their proteins were separated on 10% SDS-polyacrylamide gels. Gels were subjected to Western blot analysis as described previously (49). α-mCherry (Abcam) was used for the detection of sfCherry proteins.
ACKNOWLEDGMENTS
We thank members of the Bondy-Denomy lab for helpful discussions. S.G. appreciates the helpful discussion with and support from Shweta Karambelkar and Bálint Csörgő.
This project in the Bondy-Denomy lab was supported by the UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation and an NIH Director’s Early Independence Award DP5-OD021344 and R01GM127489.
J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics and a scientific advisory board member and cofounder of Acrigen Biosciences.
Footnotes
Supplemental material is available online only.
Contributor Information
Joseph Bondy-Denomy, Email: joseph.bondy-denomy@ucsf.edu.
George O'Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526:55–61. 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 2.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P. 2019. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18:63–83. 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Der Oost J, Westra ER, Jackson RN, Wiedenheft B. 2014. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat Rev Microbiol 12:479–492. 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redding S, Sternberg SH, Marshall M, Gibb B, Bhat P, Guegler CK, Wiedenheft B, Doudna JA, Greene EC. 2015. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell 163:854–865. 10.1016/j.cell.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillard KE, Brown MW, Johnson NV, Xiao Y, Dolan A, Hernandez E, Dahlhauser SD, Kim Y, Myler LR, Anslyn EV, Ke A, Finkelstein IJ. 2018. Assembly and translocation of a CRISPR-Cas primed acquisition complex. Cell 175:934–946.e15. 10.1016/j.cell.2018.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue C, Zhu Y, Zhang X, Shin Y-K, Sashital DG. 2017. Real-time observation of target search by the CRISPR surveillance complex Cascade. Cell Rep 21:3717–3727. 10.1016/j.celrep.2017.11.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vink JNA, Martens KJA, Vlot M, McKenzie RE, Almendros C, Bonilla BE, Brocken DJW, Hohlbein J, Brouns SJJ. 2020. Direct visualization of native CRISPR target search in live bacteria reveals Cascade DNA surveillance mechanism. Mol Cell 77:39–50.e10. 10.1016/j.molcel.2019.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Martens KJA, van Beljouw SPB, van der Els S, Vink JNA, Baas S, Vogelaar GA, Brouns SJJ, van Baarlen P, Kleerebezem M, Hohlbein J. 2019. Visualisation of dCas9 target search in vivo using an open-microscopy framework. Nat Commun 10:3552. 10.1038/s41467-019-11514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones DL, Leroy P, Unoson C, Fange D, Ćurić V, Lawson MJ, Elf J. 2017. Kinetics of dCas9 target search in Escherichia coli. Science 357:1420–1424. 10.1126/science.aah7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper LA, Stringer AM, Wade JT. 2018. Determining the specificity of cascade binding, interference, and primed adaptation in vivo in the Escherichia coli type I-E CRISPR-Cas system. mBio 9:e02100-17. 10.1128/mBio.02100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holubová I, Vejsadová Š, Weiserová M, Firman K. 2000. Localization of the type I restriction–modification enzyme EcoKI in the bacterial cell. Biochem Biophys Res Commun 270:46–51. 10.1006/bbrc.2000.2375. [DOI] [PubMed] [Google Scholar]
- 12.Holubova I, Vejsadová Š, Firman K, Weiserova M. 2004. Cellular localization of type I restriction–modification enzymes is family dependent. Biochem Biophys Res Commun 319:375–380. 10.1016/j.bbrc.2004.04.184. [DOI] [PubMed] [Google Scholar]
- 13.Wiedenheft B, van Duijn E, Bultema JB, Bultema J, Waghmare SP, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck AJR, Heck A, Boekema EJ, Boekema E, Dickman MJ, Dickman M, Doudna JA. 2011. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA 108:10092–10097. 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo TW, Bartesaghi A, Yang H, Falconieri V, Rao P, Merk A, Eng ET, Raczkowski AM, Fox T, Earl LA, Patel DJ, Subramaniam S. 2017. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell 171:414–426.e12. 10.1016/j.cell.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rollins MCF, Chowdhury S, Carter J, Golden SM, Miettinen HM, Santiago-Frangos A, Faith D, Lawrence CM, Lander GC, Wiedenheft B. 2019. Structure reveals a mechanism of CRISPR-RNA-guided nuclease recruitment and anti-CRISPR viral mimicry. Mol Cell 74:132–142.e5. 10.1016/j.molcel.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–432. 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borges AL, Zhang JY, Rollins MF, Osuna BA, Wiedenheft B, Bondy-Denomy J. 2018. Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity. Cell 174:917–925.e10. 10.1016/j.cell.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landsberger M, Gandon S, Meaden S, Rollie C, Chevallereau A, Chabas H, Buckling A, Westra ER, van Houte S. 2018. Anti-CRISPR phages cooperate to overcome CRISPR-Cas immunity. Cell 174:908–916. 10.1016/j.cell.2018.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borges AL, Castro B, Govindarajan S, Solvik T, Escalante V, Bondy-Denomy J. 2020. Bacterial alginate regulators and phage homologs repress CRISPR–Cas immunity. Nat Microbiol 5:679–687. 10.1038/s41564-020-0691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J, Bassler BL. 2017. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci USA 114:131–135. 10.1073/pnas.1617415113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahator Dela S, Jianhe W, Zhang L-H. 2020. The ECF sigma factor PvdS regulates the type I-F CRISPR-Cas system in Pseudomonas aeruginosa. bioRxiv. 10.1101/2020.01.31.929752. [DOI]
- 22.Lin P, Pu Q, Wu Q, Zhou C, Wang B, Schettler J, Wang Z, Qin S, Gao P, Li R, Li G, Cheng Z, Lan L, Jiang J, Wu M. 2019. High-throughput screen reveals sRNAs regulating crRNA biogenesis by targeting CRISPR leader to repress Rho termination. Nat Commun 10:3728. 10.1038/s41467-019-11695-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rollins MF, Chowdhury S, Carter J, Golden SM, Wilkinson RA, Bondy-Denomy J, Lander GC, Wiedenheft B. 2017. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc Natl Acad Sci USA 114:E5113–E5121. 10.1073/pnas.1616395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chowdhury S, Carter J, Rollins MF, Golden SM, Jackson RN, Hoffmann C, Nosaka LAl, Bondy-Denomy J, Maxwell KL, Davidson AR, Fischer ER, Lander GC, Wiedenheft B. 2017. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell 169:47–57.e11. 10.1016/j.cell.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. 2012. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 194:5728–5738. 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson AG, Jackson SA, Taylor C, Evans GB, Salmond GPC, Przybilski R, Staals RHJ, Fineran PC. 2016. Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol Cell 64:1102–1108. 10.1016/j.molcel.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borges AL, Castro B, Govindarajan S, Solvik T, Escalante V, Bondy-Denomy J. 2019. CRISPR-Cas immunity repressed by a biofilm-activating pathway in Pseudomonas aeruginosa. bioRxiv. 10.1101/673095. [DOI]
- 28.Hershko-Shalev T, Odenheimer-Bergman A, Elgrably-Weiss M, Ben-Zvi T, Govindarajan S, Seri H, Papenfort K, Vogel J, Altuvia S. 2016. Gifsy-1 prophage IsrK with dual function as small and messenger RNA modulates vital bacterial machineries. PLoS Genet 12:e1005975. 10.1371/journal.pgen.1005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Helvoort JM, Kool J, Woldringh CL. 1996. Chloramphenicol causes fusion of separated nucleoids in Escherichia coli K-12 cells and filaments. J Bacteriol 178:4289–4293. 10.1128/jb.178.14.4289-4293.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Govindarajan S, Elisha Y, Nevo-Dinur K, Amster-Choder O. 2013. The general phosphotransferase system proteins localize to sites of strong negative curvature in bacterial cells. mBio 4:e00443-13. 10.1128/mBio.00443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR. 2015. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature 526:136–139. 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxwell KL, Garcia B, Bondy-Denomy J, Bona D, Hidalgo-Reyes Y, Davidson AR. 2016. The solution structure of an anti-CRISPR protein. Nat Commun 7:13134. 10.1038/ncomms13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Yao D, Xu JG, Li AR, Xu J, Fu P, Zhou Y, Zhu Y. 2016. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat Struct Mol Biol 23:868–870. 10.1038/nsmb.3269. [DOI] [PubMed] [Google Scholar]
- 34.Niu Y, Yang L, Gao T, Dong C, Zhang B, Yin P, Hopp AK, Li D, Gan R, Wang H, Liu X, Cao X, Xie Y, Meng X, Deng H, Zhang X, Ren J, Hottiger MO, Chen Z, Zhang Y, Liu X, Feng Y. 2020. A type I-F anti-CRISPR protein inhibits the CRISPR-Cas surveillance complex by ADP-ribosylation. Mol Cell 80:512–524.e5. 10.1016/j.molcel.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Marino ND, Zhang JY, Borges AL, Sousa AA, Leon LM, Rauch BJ, Walton RT, Berry JD, Joung JK, Kleinstiver BP, Bondy-Denomy J. 2018. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 362:240–242. 10.1126/science.aau5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novick SL, Baldeschwieler JD. 1988. Fluorescence measurement of the kinetics of DNA injection by bacteriophage lambda into liposomes. Biochemistry 27:7919–7924. 10.1021/bi00420a050. [DOI] [PubMed] [Google Scholar]
- 37.Mendoza SD, Nieweglowska ES, Govindarajan S, Leon LM, Berry JD, Tiwari A, Chaikeeratisak V, Pogliano J, Agard DA, Bondy-Denomy J. 2020. A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature 577:244–248. 10.1038/s41586-019-1786-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong D, Guo M, Wang S, Zhu Y, Wang S, Xiong Z, Yang J, Xu Z, Huang Z. 2017. Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature 546:436–439. 10.1038/nature22377. [DOI] [PubMed] [Google Scholar]
- 39.Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J. 2017. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 168:150–158.e10. 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin J, Jiang F, Liu J-J, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, Doudna JA. 2017. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv 3:e1701620. 10.1126/sciadv.1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders APL, Dickman MJ, Doudna JA, Boekema EJ, Heck AJR, van der Oost J, Brouns SJJ. 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18:529–536. 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 42.Sashital DG, Wiedenheft B, Doudna JA. 2012. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell 46:606–615. 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westra ER, van Erp PBG, Künne T, Wong SP, Staals RHJ, Seegers CLC, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJJ, van der Oost J. 2012. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46:595–605. 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao Y, Luo M, Hayes RP, Kim J, Ng S, Ding F, Liao M, Ke A. 2017. Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas system. Cell 170:48–60.e11. 10.1016/j.cell.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y, Luo M, Dolan AE, Liao M, Ke A. 2018. Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science 361:eaat0839. 10.1126/science.aat0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang F, Doudna JA. 2017. CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529. 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 47.Workman RE, Pammi T, Nguyen BTK, Graeff LW, Smith E, Sebald SM, Stoltzfus MJ, Euler CW, Modell JW. 2021. A natural single-guide RNA repurposes Cas9 to autoregulate CRISPR-Cas expression. Cell 184:675–688.e19. 10.1016/j.cell.2020.12.017. [DOI] [PubMed] [Google Scholar]
- 48.Osuna BA, Karambelkar S, Mahendra C, Sarbach A, Johnson MC, Kilcher S, Bondy-Denomy J. 2020. Critical anti-CRISPR locus repression by a bi-functional Cas9 inhibitor. Cell Host Microbe 28:23–30.e5. 10.1016/j.chom.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osuna BA, Karambelkar S, Mahendra C, Christie KA, Garcia B, Davidson AR, Kleinstiver BP, Kilcher S, Bondy-Denomy J. 2020. Listeria phages induce Cas9 degradation to protect lysogenic genomes. Cell Host Microbe 28:31–40.e9. 10.1016/j.chom.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quax TEF, Voet M, Sismeiro O, Dillies M-A, Jagla B, Coppée J-Y, Sezonov G, Forterre P, van der Oost J, Lavigne R, Prangishvili D. 2013. Massive activation of archaeal defense genes during viral infection. J Virol 87:8419–8428. 10.1128/JVI.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young JC, Dill BD, Pan C, Hettich RL, Banfield JF, Shah M, Fremaux C, Horvath P, Barrangou R, VerBerkmoes NC. 2012. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS One 7:e38077. 10.1371/journal.pone.0038077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. 2010. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J Mol Biol 395:270–281. 10.1016/j.jmb.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 53.Shao Q, Hawkins A, Zeng L. 2015. Phage DNA dynamics in cells with different fates. Biophys J 108:2048–2060. 10.1016/j.bpj.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shashkova S, Leake MC. 2017. Single-molecule fluorescence microscopy review: shedding new light on old problems. Biosci Rep 37:BSR20170031. 10.1042/BSR20170031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss D, Sampson T. 2014. CRISPR-Cas systems: new players in gene regulation and bacterial physiology. Front Cell Infect Microbiol 4:37. 10.3389/fcimb.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hille F, Charpentier E. 2016. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc B 371:20150496. 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ratner HK, Sampson TR, Weiss DS. 2015. I can see CRISPR now, even when phage are gone: a view on alternative CRISPR-Cas functions from the prokaryotic envelope. Curr Opin Infect Dis 28:267–274. 10.1097/QCO.0000000000000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6; Tables S1 to S3. Download jb.00105-22-s0001.pdf, PDF file, 0.6 MB (643KB, pdf)