ABSTRACT
The polymyxins display excellent in vitro antimicrobial activity against most Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii isolates, but their clinical utility has been limited because of class-specific toxicity problems. Therefore, new polymyxin analogs with improved safety properties are needed to combat serious infections caused by resistant Gram-negative pathogens. MRX-8 is a novel polymyxin B analog that displays reduced toxicity in in vitro and animal assays and is currently being evaluated in a phase 1 clinical trial. In this nonclinical study, the in vitro potency and spectrum of MRX-8 and comparators were evaluated against a large set of Gram-negative clinical isolates collected in the United States in 2017 to 2020. MRX-8, colistin, and polymyxin B exhibited nearly identical antimicrobial activities against the Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii isolate sets. MRX-8 MIC50 and MIC90 values were 0.12 and 0.25 mg/L, respectively, for the set of Enterobacterales isolates not intrinsically resistant to colistin and 0.5 and 1 mg/L, respectively, against both the A. baumannii and P. aeruginosa isolate sets. All three polymyxin-class compounds retained activity against meropenem-resistant and multidrug-resistant isolate subsets but were inactive against isolates displaying acquired or intrinsic resistance to polymyxins. These results support the continued development of MRX-8 to treat serious Gram-negative infections.
KEYWORDS: polymyxin, colistin, lipopeptide, Gram-negative, resistance, MRX-8
INTRODUCTION
Many recent reports and reviews have emphasized the need for the development of novel or improved antimicrobial agents to treat serious infections caused by Gram-negative species or groups like the Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii, which often display resistance to frontline drugs (1–4). Of particular concern are Gram-negative isolates that are multidrug resistant (MDR) or carbapenem resistant due to multifactorial mechanisms or the production of class B metallo-β-lactamases (3, 5).
The clinically relevant polymyxin antimicrobial class is composed of colistin (polymyxin E) and polymyxin B, which are structurally related polycationic molecules with a cyclic lipodecapeptide structure composed of a heptapeptide core, a tripeptide linear linker, and an N-terminal fatty acyl group. The structures of colistin and polymyxin B differ only at the R6 peptide position, which is d-phenylalanine in polymyxin B and d-leucine in colistin (6). Commercial preparations of polymyxin B and colistin also typically contain mixtures of components with related but distinct fatty acid groups (7). Three free amino groups are present within the heptapeptide core, and two free amino groups are present within the linker region. Both colistin and polymyxin B exhibit rapid in vitro bactericidal activity against many important Gram-negative pathogens. The mode of action is principally mediated by disruption of the bacterial outer membrane through binding of the lipopeptide cationic groups to lipopolysaccharide, followed by disruption of the inner membrane, which likely involves interaction with the fatty acid polymyxin tail (6, 8).
Gram-positive species and several Enterobacterales species (e.g., Providencia spp. and Proteus spp.) are intrinsically resistant to the polymyxins (9, 10). In contrast, Escherichia coli, Klebsiella spp., Citrobacter spp., Enterobacter spp., P. aeruginosa, and A. baumannii are intrinsically susceptible to the polymyxins but resistance can be acquired by various methods, including mutations in chromosomal genes or the acquisition of mobile genetic elements that contain mcr-1 or related genes encoding enzymes that modify the bacterial lipopolysaccharide structure (9, 11, 12).
Although the discovery of polymyxins was reported in 1947 (for a review, see reference 7), their clinical use was short-lived due to concerns about nephrotoxicity and neurotoxicity and the discovery of safer alternative antimicrobials (7, 13). Because of the recent rise in highly drug-resistant Gram-negative pathogens, however, there has been a resurgence in the clinical use of the polymyxins as drugs of last resort (14).
Several groups have launched research efforts to improve the clinical utility of the polymyxins with the goal of maintaining their excellent antimicrobial features while abrogating their toxicity (6, 14). Other groups have instead focused on designing truncated polymyxins, with decreased antimicrobial activity and decreased toxicity, that still maintain the ability to potentiate the entry of other antimicrobials through the Gram-negative outer membrane (8).
MRX-8 is a polymyxin-class antimicrobial under development that was designed using soft drug principles, in which the goal is to produce molecules that are “deactivated in a predictable and controllable way after achieving their therapeutic goal” (15). To that end, MRX-8 is an analog of polymyxin B in which the fatty acid tail is linked to the rest of the molecule by a polar ester group. MRX-8 maintains in vitro antimicrobial activity (reference 16 and see below). After exerting its antimicrobial effect in vivo, however, MRX-8 is intended to be converted by endogenous esterases into a nontoxic metabolite that displays minimized cell culture toxicity and animal nephrotoxicity properties, compared to polymyxin B (16).
Lepak et al. (17) recently investigated the pharmacodynamic (PD) properties of MRX-8, compared to polymyxin B, in mouse thigh and lung infection models involving Enterobacterales, P. aeruginosa, and A. baumannii strains. They found that MRX-8 exhibited efficacy in those animal models and that AUC/MIC and maximum drug concentration (Cmax)/MIC were the pharmacokinetic (PK)/PD indices that best correlated with the observed antimicrobial therapeutic effects.
The safety, tolerability, and human PK features of MRX-8 following intravenous dosing are currently being evaluated in a phase 1 clinical trial (ClinicalTrials registration no. NCT04649541), with results expected in 2022. In this study, we investigated the in vitro antimicrobial potency and spectrum of MRX-8, compared to colistin and polymyxin B, when tested against a large set of Gram-negative pathogens collected in the United States from 2017 to 2020.
RESULTS
Cumulative distributions of MIC values for each isolate set tested against MRX-8, colistin, and polymyxin B are shown in Table 1, Table S1, and Table S2, respectively. Summary tables that display MIC50 and MIC90 values, MIC ranges, and percentages of susceptible, intermediate, or resistant isolates for various species are displayed in Table 2 and in Tables S3 to S6 in the supplemental material.
TABLE 1.
Cumulative MIC distributions for MRX-8 tested against the main species and organism groups
| Species/organism group (no. of isolates) | No. (cumulative %) of isolates inhibited with MIC of: |
MIC50 (mg/L) | MIC90 (mg/L) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.015 mg/L | 0.03 mg/L | 0.06 mg/L | 0.12 mg/L | 0.25 mg/L | 0.5 mg/L | 1 mg/L | 2 mg/L | 4 mg/L | 8 mg/L | 16 mg/L | 32 mg/L | >a | |||
| Enterobacterales (787) | |||||||||||||||
| Enterobacterales isolates not intrinsically resistant to colistin (685)b | 0 (0.0) | 2 (0.3) | 86 (12.8) | 450 (78.5) | 109 (94.5) | 20 (97.4) | 0 (97.4) | 1 (97.5) | 0 (97.5) | 3 (98.0) | 2 (98.2) | 1 (98.4) | 11 (100.0) | 0.12 | 0.25 |
| Citrobacter spp. (47) | 0 (0.0) | 10 (21.3) | 30 (85.1) | 6 (97.9) | 1 (100.0) | 0.12 | 0.25 | ||||||||
| Enterobacter cloacae species complex (52) | 0 (0.0) | 8 (15.4) | 27 (67.3) | 6 (78.8) | 1 (80.8) | 0 (80.8) | 0 (80.8) | 0 (80.8) | 1 (82.7) | 0 (82.7) | 0 (82.7) | 9 (100.0) | 0.12 | >32 | |
| Escherichia coli (261) | 0 (0.0) | 2 (0.8) | 46 (18.4) | 164 (81.2) | 40 (96.6) | 7 (99.2) | 0 (99.2) | 0 (99.2) | 0 (99.2) | 1 (99.6) | 1 (100.0) | 0.12 | 0.25 | ||
| Klebsiella aerogenes (23) | 0 (0.0) | 1 (4.3) | 15 (69.6) | 5 (91.3) | 2 (100.0) | 0.12 | 0.25 | ||||||||
| Klebsiella oxytoca (37) | 0 (0.0) | 6 (16.2) | 30 (97.3) | 1 (100.0) | 0.12 | 0.12 | |||||||||
| Klebsiella pneumoniae (265) | 0 (0.0) | 15 (5.7) | 184 (75.1) | 51 (94.3) | 9 (97.7) | 0 (97.7) | 1 (98.1) | 0 (98.1) | 1 (98.5) | 1 (98.9) | 1 (99.2) | 2 (100.0) | 0.12 | 0.25 | |
| Colistin-resistant (17) | 0 (0.0) | 3 (17.6) | 2 (29.4) | 1 (35.3) | 11 (100.0) | >32 | >32 | ||||||||
| Meropenem-resistant (10) | 0 (0.0) | 1 (10.0) | 3 (40.0) | 3 (70.0) | 0 (70.0) | 0 (70.0) | 0 (70.0) | 0 (70.0) | 1 (80.0) | 0 (80.0) | 0 (80.0) | 2 (100.0) | 0.25 | >32 | |
| MDR (54) | 0 (0.0) | 3 (5.6) | 34 (68.5) | 11 (88.9) | 3 (94.4) | 0 (94.4) | 0 (94.4) | 0 (94.4) | 1 (96.3) | 0 (96.3) | 0 (96.3) | 2 (100.0) | 0.12 | 0.5 | |
| Enterobacterales isolates intrinsically resistant to colistin (102)c | 0 (0.0) | 2 (2.0) | 100 (100.0) | >32 | >32 | ||||||||||
| Acinetobacter baumannii (264) | 0 (0.0) | 1 (0.4) | 3 (1.5) | 33 (14.0) | 58 (36.0) | 116 (79.9) | 35 (93.2) | 12 (97.7) | 1 (98.1) | 0 (98.1) | 2 (98.9) | 1 (99.2) | 2 (100.0) | 0.5 | 1 |
| Meropenem-resistant (74) | 0 (0.0) | 9 (12.2) | 20 (39.2) | 32 (82.4) | 9 (94.6) | 1 (95.9) | 0 (95.9) | 0 (95.9) | 1 (97.3) | 1 (98.6) | 1 (100.0) | 0.5 | 1 | ||
| Colistin-nonresistant (258) | 0 (0.0) | 1 (0.4) | 3 (1.6) | 33 (14.3) | 58 (36.8) | 116 (81.8) | 35 (95.3) | 12 (100.0) | 0.5 | 1 | |||||
| Colistin-resistant (6) | 0 (0.0) | 1 (16.7) | 0 (16.7) | 2 (50.0) | 1 (66.7) | 2 (100.0) | 16 | ||||||||
| MDR (104) | 0 (0.0) | 15 (14.4) | 23 (36.5) | 44 (78.8) | 15 (93.3) | 4 (97.1) | 0 (97.1) | 0 (97.1) | 1 (98.1) | 1 (99.0) | 1 (100.0) | 0.5 | 1 | ||
| Pseudomonas aeruginosa (263) | 0 (0.0) | 3 (1.1) | 9 (4.6) | 19 (11.8) | 166 (74.9) | 64 (99.2) | 2 (100.0) | 0.5 | 1 | ||||||
| Meropenem-resistant (31) | 0 (0.0) | 3 (9.7) | 3 (19.4) | 20 (83.9) | 5 (100.0) | 0.5 | 1 | ||||||||
| MDR (46) | 0 (0.0) | 1 (2.2) | 4 (10.9) | 6 (23.9) | 22 (71.7) | 12 (97.8) | 1 (100.0) | 0.5 | 1 | ||||||
Greater than the highest concentration tested.
Species included Citrobacter amalonaticus/farmeri (1 isolate), Citrobacter freundii (1 isolate), C. freundii species complex (26 isolates), Citrobacter koseri (19 isolates), Enterobacter asburiae (1 isolate), Enterobacter cloacae (14 isolates), E. cloacae species complex (36 isolates), Enterobacter hormaechei (1 isolate), Escherichia coli (261 isolates), Klebsiella aerogenes (23 isolates), Klebsiella oxytoca (37 isolates), and Klebsiella pneumoniae (265 isolates).
Species included Morganella morganii (10 isolates), Proteus mirabilis (46 isolates), Proteus penneri (1 isolate), Proteus vulgaris group (5 isolates), Providencia rettgeri (7 isolates), Providencia stuartii (8 isolates), Serratia liquefaciens complex (3 isolates), and Serratia marcescens (22 isolates).
TABLE 2.
Antimicrobial activity of MRX-8 and comparator agents tested against Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa isolates
| Species and antimicrobial agent | No. of isolates | MIC findings (mg/L) |
Susceptibility results (%) using: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CLSI criteriaa |
EUCAST criteriaa |
|||||||||
| MIC50 | MIC90 | MIC range | S | I | R | S | I | R | ||
| Escherichia coli | ||||||||||
| MRX-8 | 261 | 0.12 | 0.25 | 0.03 to 16 | ||||||
| Colistin | 261 | 0.25 | 0.25 | 0.06 to 8 | 99.2 | 0.8 | 99.2 | 0.8 | ||
| Polymyxin B | 261 | 0.25 | 0.5 | 0.12 to 8 | 99.6 | 0.4 | ||||
| Amikacin | 261 | 2 | 8 | 0.5 to 16 | 100.0 | 0.0 | 0.0 | 98.9b | 1.1 | |
| Ceftazidime | 261 | 0.25 | 8 | 0.03 to >32 | 88.1 | 2.7 | 9.2 | 82.8 | 5.4 | 11.9 |
| Ceftazidime-avibactam (fixed at 4 mg/L) | 77 | 0.06 | 0.25 | ≤0.015 to 0.25 | 100.0 | 0.0 | 100.0 | 0.0 | ||
| Ceftriaxone | 261 | ≤0.06 | >8 | ≤0.06 to >8 | 83.5 | 0.0 | 16.5 | 83.5,c 83.5d | 0.0d | 16.5,c 16.5d |
| Gentamicin | 261 | 1 | >16 | ≤0.12 to >16 | 87.0 | 0.4 | 12.6 | 86.6b | 13.4 | |
| Levofloxacin | 260 | ≤0.03 | 16 | ≤0.03 to >16 | 68.8 | 0.8 | 30.4 | 68.8 | 0.8 | 30.4 |
| Meropenem | 261 | 0.015 | 0.03 | 0.008 to 8 | 99.6 | 0.0 | 0.4 | 99.6,c 99.6d | 0.4d | 0.4,c 0.0d |
| Piperacillin-tazobactam (fixed at 4 mg/L) | 261 | 2 | 8 | 1 to >128 | 96.6 | 1.5 | 1.9 | 94.3 | 5.7 | |
| Tigecycline | 261 | 0.12 | 0.25 | ≤0.06 to 1 | 100.0e | 0.0 | 0.0 | 98.9 | 1.1 | |
| Klebsiella pneumoniae | ||||||||||
| MRX-8 | 265 | 0.12 | 0.25 | 0.06 to >32 | ||||||
| Colistin | 265 | 0.12 | 0.25 | 0.12 to >32 | 98.1 | 1.9 | 98.1 | 1.9 | ||
| Polymyxin B | 265 | 0.25 | 0.5 | 0.12 to >32 | 98.1 | 1.9 | ||||
| Amikacin | 265 | 1 | 2 | 0.5 to >32 | 98.9 | 0.0 | 1.1 | 98.5b | 1.5 | |
| Ceftazidime | 265 | 0.25 | 16 | 0.03 to >32 | 88.3 | 1.1 | 10.6 | 87.5 | 0.8 | 11.7 |
| Ceftazidime-avibactam (fixed at 4 mg/L) | 102 | 0.12 | 0.25 | 0.03 to 1 | 100.0 | 0.0 | 100.0 | 0.0 | ||
| Ceftriaxone | 265 | ≤0.06 | >8 | ≤0.06 to >8 | 87.9 | 0.0 | 12.1 | 87.9,c 87.9d | 0.0d | 12.1,c 12.1d |
| Gentamicin | 265 | 0.25 | 0.5 | ≤0.12 to >16 | 94.0 | 0.4 | 5.7 | 94.0b | 6.0 | |
| Levofloxacin | 265 | 0.06 | 1 | ≤0.03 to >16 | 86.0 | 4.2 | 9.8 | 86.0 | 4.2 | 9.8 |
| Meropenem | 265 | 0.03 | 0.03 | 0.015 to >16 | 97.4 | 0.4 | 2.3 | 97.7,c 97.7d | 0.4d | 2.3,c 1.9d |
| Piperacillin-tazobactam (fixed at 4 mg/L) | 265 | 4 | 16 | 0.12 to >128 | 93.6 | 2.3 | 4.2 | 89.4 | 10.6 | |
| Tigecycline | 265 | 0.5 | 1 | ≤0.06 to 8 | 98.1e | 1.5 | 0.4 | |||
| Acinetobacter baumannii | ||||||||||
| MRX-8 | 264 | 0.5 | 1 | 0.03 to >32 | ||||||
| Colistin | 264 | 0.25 | 1 | 0.06 to >32 | 97.7 | 2.3 | 97.7 | 2.3 | ||
| Polymyxin B | 264 | 0.25 | 0.5 | 0.12 to 16 | 98.5 | 1.5 | ||||
| Amikacin | 264 | 4 | >32 | 0.5 to >32 | 83.3 | 1.9 | 14.8 | 79.2b | 20.8 | |
| Ceftazidime | 264 | 8 | >32 | 1 to >32 | 65.2 | 7.2 | 27.7 | |||
| Ceftazidime-avibactam (fixed at 4 mg/L) | 91 | 8 | 32 | 1 to >32 | ||||||
| Ceftriaxone | 224 | >8 | >8 | 4 to >8 | 21.4 | 0.0 | 0.0 | |||
| Gentamicin | 264 | 1 | >16 | ≤0.12 to >16 | 75.4 | 5.7 | 18.9 | 75.4b | 24.6 | |
| Levofloxacin | 264 | 0.25 | >16 | ≤0.015 to >16 | 65.9 | 1.1 | 33.0 | 62.9 | 2.3 | 34.8 |
| Meropenem | 264 | 0.5 | >16 | 0.06 to >16 | 71.2 | 0.8 | 28.0 | 71.2,c 71.2d | 1.1d | 28.8,c 27.7d |
| Piperacillin-tazobactam (fixed at 4 mg/L) | 258 | 8 | >128 | ≤0.06 to >128 | 58.5 | 7.0 | 34.5 | |||
| Tigecycline | 264 | 0.5 | 4 | ≤0.06 to 8 | ||||||
| Pseudomonas aeruginosa | ||||||||||
| MRX-8 | 263 | 0.5 | 1 | 0.06 to 2 | ||||||
| Colistin | 263 | 0.5 | 1 | 0.12 to 2 | 100.0 | 0.0 | 100.0 | 0.0 | ||
| Polymyxin B | 263 | 0.5 | 1 | 0.12 to 2 | 100.0 | 0.0 | ||||
| Amikacin | 263 | 4 | 8 | ≤0.25 to >32 | 96.2 | 1.1 | 2.7 | 96.2b | 3.8 | |
| Ceftazidime | 263 | 2 | 32 | 0.25 to >32 | 82.1 | 4.2 | 13.7 | f | 82.1 | 17.9 |
| Ceftazidime-avibactam (fixed at 4 mg/L) | 92 | 2 | 4 | 0.25 to >32 | 98.9 | 1.1 | 98.9 | 1.1 | ||
| Ceftriaxone | 211 | >8 | >8 | 0.5 to >8 | ||||||
| Gentamicin | 263 | 2 | 8 | ≤0.12 to >16 | 87.8 | 6.5 | 5.7 | |||
| Levofloxacin | 263 | 0.5 | 8 | ≤0.03 to >16 | 68.4 | 11.8 | 19.8 | f | 68.4 | 31.6 |
| Meropenem | 263 | 0.5 | 8 | 0.015 to >16 | 82.5 | 5.7 | 11.8 | 82.5,c 82.5d | 8.7d | 17.5,c 8.7d |
| Piperacillin-tazobactam (fixed at 4 mg/L) | 263 | 4 | 128 | ≤0.06 to >128 | 79.8 | 7.6 | 12.5 | f | 79.8 | 20.2 |
| Tigecycline | 263 | 8 | >8 | 0.25 to >8 | ||||||
For infections originating from the urinary tract. For systemic infections, aminoglycosides must be used in combination with other active therapy.
Using meningitis breakpoints.
Using nonmeningitis breakpoints.
Using FDA breakpoints.
An arbitrary susceptible breakpoint of ≤0.001 mg/L and/or >50 mm has been published by EUCAST, indicating that susceptible should not be reported for this organism-agent combination and intermediate should be interpreted as susceptible-increased exposure.
Activity of MRX-8 and comparators against Enterobacterales species.
The activity of MRX-8 and comparator lipopeptides was first measured against Enterobacterales species that are not intrinsically resistant to colistin, including E. coli (Table 2), Klebsiella pneumoniae (Table 2), Citrobacter spp. (see Table S3), Enterobacter cloacae species complex (see Table S4), Klebsiella aerogenes (see Table S5), and Klebsiella oxytoca (see Table S6). Within each species or group of isolates, the MIC50 and MIC90 values for MRX-8, colistin, and polymyxin B agreed within 2-fold. For example, against the E. coli subset (Table 2), the MIC50 and MIC90 values for MRX-8, colistin, and polymyxin B were 0.12 and 0.25 mg/L, 0.25 and 0.25 mg/L, and 0.25 and 0.5 mg/L, respectively. There was also little variation in the different polymyxin MIC50 and MIC90 values among the various Enterobacterales species and groups. Except for the E. cloacae species complex isolate subset, the MIC50 and MIC90 values were 0.12 and 0.12 to 0.25 mg/L, respectively, for MRX-8 (Table 1), 0.12 to 0.25 and 0.25 mg/L, respectively, for colistin (see Table S1), and 0.25 and 0.25 to 0.5 mg/L, respectively, for polymyxin B (see Table S2). In contrast, the E. cloacae species complex isolate subset MIC90 values for MRX-8 (Table 1), colistin (see Table S1), and polymyxin B (see Table S2) were much higher (>32 mg/L for each lipopeptide) than those for the other species groups of Enterobacterales that are not intrinsically resistant to colistin. This difference is due to the larger percentage of Enterobacter species isolates that displayed acquired colistin resistance (19.2%) (see Table S4), compared to the other species.
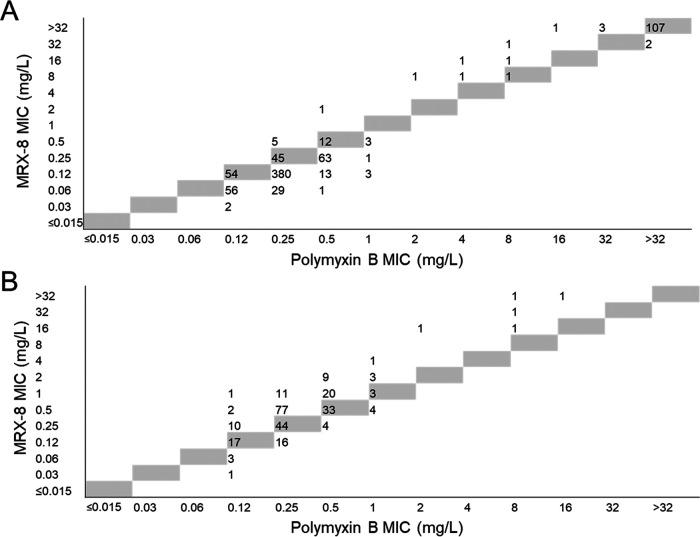
MRX-8 (Table 1) and colistin (see Table S1) were 2-fold more potent (MIC50 and MIC90 values for both antimicrobials of 0.12 and 0.25 mg/L, respectively) than polymyxin B (MIC50 and MIC90 of 0.25 and 0.5 mg/L, respectively) (see Table S2) against the combined set of Enterobacterales isolates not intrinsically resistant to colistin. This potency difference can be observed in the scatterplot of polymyxin B MIC values versus MRX-8 MIC values shown in Fig. 1A.
FIG 1.
Scatterplot of MRX-8 and polymyxin B MIC values when tested against Enterobacterales and Acinetobacter baumannii isolates. The figure displays a scatterplot of MIC values for MRX-8 and polymyxin B tested against Enterobacterales isolates (787 isolates) (A) and Acinetobacter baumannii isolates (264 isolates) (B). The gray cells represent identical MIC values for the two antimicrobials.
Against the meropenem-resistant subset of 10 Enterobacterales isolates from species that are not intrinsically resistant to colistin, the MIC50 and MIC90 values for MRX-8, colistin, and polymyxin B were 0.25 and >32 mg/L (Table 1), 0.12 and >32 mg/L (see Table S1), and 0.25 and 16 mg/L (see Table S2), respectively. Against the MDR subset of 54 Enterobacterales isolates from species that are not intrinsically resistant to colistin, the MIC50 and MIC90 values for MRX-8, colistin, and polymyxin B were 0.12 and 0.5 mg/L (Table 1), 0.12 and 0.5 mg/L (see Table S1), and 0.25 and 0.5 mg/L (see Table S2), respectively. MRX-8, colistin, and polymyxin B were all inactive (MIC50 values of ≥32 and MIC90 values of >32 mg/L) against the subset of 17 Enterobacterales isolates from species that are not intrinsically resistant to colistin but displayed acquired colistin resistance (Table 1; also see Tables S1 and S2 in the supplemental material).
Finally, we investigated the activity of MRX-8 and comparator lipopeptides against a subset of Enterobacterales species that are intrinsically resistant to colistin, such as Morganella morganii (10) (Table 1; also see Tables S1 and S2). As expected, MRX-8, colistin, and polymyxin B were all inactive against this isolate set (MIC90 values of >32 mg/L for all three antimicrobials).
Activity of MRX-8 and comparators against Acinetobacter baumannii.
The in vitro antimicrobial activities of MRX-8 and comparators against the A. baumannii isolate subset are displayed in Table 2. The MIC50 and MIC90 values for the three lipopeptides agreed within 2-fold (MIC50 and MIC90 ranges of 0.25 to 0.5 and 0.5 to 1 mg/L, respectively). Colistin and polymyxin B were 2-fold more potent than MRX-8 according to MIC50 values. This effect can be observed in the scatterplot of polymyxin B MIC values and MRX-8 MIC values shown in Fig. 1B. The three lipopeptides maintained their nearly equivalent potencies against the meropenem-resistant A. baumannii isolate subset. In vitro activity was lost for each antimicrobial, however, against the subset of colistin-resistant A. baumannii isolates, although 2 of the 6 colistin-resistant isolates remained intermediate to polymyxin B (Table 1; also see Tables S1 and S2). MDR status did not significantly affect the MIC50 and MIC90 values for any of the three polymyxins (Table 1; also see Tables S1 and S2).
Activity of MRX-8 and comparators against Pseudomonas aeruginosa.
The in vitro antimicrobial activities of MRX-8 and comparators against the P. aeruginosa isolate subset are displayed in Table 2. The MIC50 and MIC90 values for the three polymyxins were identical (MIC50 and MIC90 values of 0.5 and 1 mg/L, respectively), and the antimicrobial activities were unchanged against the subsets of meropenem-resistant and MDR P. aeruginosa isolates (Table 1; also see Tables S1 and S2).
No colistin-resistant P. aeruginosa isolates were present within the randomly selected set tested in this study (Table 2). However, we measured MIC values for the three polymyxins against a JMI Laboratories colistin-resistant P. aeruginosa isolate (collection no. 991784). All three polymyxins were inactive against this isolate, which displayed modal MIC values for MRX-8, colistin, and polymyxin B of >32 mg/L, 32 mg/L, and 8 mg/L, respectively (data not shown).
DISCUSSION
In this study, the in vitro antimicrobial activity of MRX-8 and polymyxin comparators was measured against a large set of clinically relevant Gram-negative pathogens, including various Enterobacterales species, A. baumannii, and P. aeruginosa. MRX-8, colistin, and polymyxin B exhibited identical or nearly identical in vitro antimicrobial activities against almost all of the Gram-negative species and groups tested, including MDR and meropenem-resistant subsets. Against the Enterobacterales isolates not intrinsically resistant to colistin, MRX-8 was 2-fold more potent than polymyxin B according to MIC50 and MIC90 values. In contrast, MRX-8 was about 2-fold less potent than polymyxin B against the A. baumannii set and equipotent against the P. aeruginosa isolate set according to MIC50 and MIC90 values. In general, all three polymyxins exhibited potent in vitro activity against the pathogen groups, e.g., the MRX-8 MIC50 and MIC90 values were 0.12 and 0.25 mg/L, respectively, against the set of Enterobacterales isolates not intrinsically resistant to colistin and 0.5 and 1 mg/L, respectively, against the A. baumannii and P. aeruginosa isolate sets. MRX-8, colistin, and polymyxin B were all inactive against isolate subsets that displayed acquired or intrinsic resistance to colistin.
MRX-8 merits additional study for the potential treatment of serious infections caused by Gram-negative pathogens. This conclusion is based on the following considerations. First, the in vitro potency and spectrum of MRX-8 are similar to those of colistin and polymyxin B. Second, the unique soft drug structure of MRX-8, featuring an ester bond that is cleaved by enzymes in vivo, is designed to decrease the potential for nephrotoxicity, which is a significant treatment-limiting side effect of colistin and polymyxin B use. Although further work is needed, in vitro and in vivo toxicology studies have generated favorable data in support of this hypothesis (16). Third, recent animal data suggest that MRX-8 exhibits PK/PD properties equivalent or superior to those of colistin and polymyxin B (17).
Finally, when evaluating the potential clinical utility of MRX-8, it is important to consider the existing and potential future colistin resistance rates within target pathogen groups, because MRX-8, like other polymyxins, is inactive against such resistant isolates. Global longitudinal rates of colistin resistance have recently been reviewed for the Enterobacterales (18), A. baumannii (19), and P. aeruginosa (20). In general, European Committee on Antimicrobial Susceptibility Testing (EUCAST) colistin susceptibility rates remain high for these target pathogens, but significant global variation exists. Against the Enterobacterales, colistin susceptibility rates were generally >90%, but increased resistance has been noted for Klebsiella spp. and Enterobacter spp. (9, 18). Against A. baumannii isolates, colistin susceptibility was >93% overall in each global region examined from 2005 to 2016, although resistance rates did increase over this period, particularly in Europe (9, 19). Importantly, colistin resistance rates are also typically higher against carbapenem-resistant A. baumannii isolate subsets than against randomly selected isolate sets. In contrast to the Enterobacterales and A. baumannii findings, P. aeruginosa susceptibility rates for colistin have remained >99% from 2005 to the present (9, 20).
In summary, we have shown that MRX-8, which is a lipopeptide designed to abrogate the cytotoxic properties common to other polymyxins, displayed in vitro antimicrobial activity against Enterobacterales, A. baumannii, and P. aeruginosa clinical isolates that was identical or nearly identical to the activities of colistin and polymyxin B.
MATERIALS AND METHODS
Bacterial isolates.
A total of 1,314 nonduplicate, Gram-negative clinical isolates from the SENTRY Antimicrobial Surveillance Program (21) were randomly selected from 77 medical centers located in all 9 U.S. Census Bureau divisions in 2017 to 2020 (see Table S7 in the supplemental material). The isolate sets were composed of various Enterobacterales species (including some species intrinsically resistant to polymyxins), Acinetobacter baumannii-calcoaceticus species complex (referred to as A. baumannii), and Pseudomonas aeruginosa. All organisms were isolated from various documented infection types (see Table S8), and only 1 isolate per patient infection episode was included in the surveillance collection. The number of isolates tested per species was arbitrary and should not be interpreted as representing the clinical prevalence of the species for these infection types. Species identification was performed at the participating medical centers and confirmed at the monitoring laboratory (JMI Laboratories, North Liberty, IA, USA) using standard microbiological methods and matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker, Billerica, MA, USA).
Isolates were categorized as MDR if they were nonsusceptible to at least one antimicrobial from ≥3 drug classes using Clinical and Laboratory Standards Institute (CLSI) breakpoints (10). For the A. baumannii isolate set, the antimicrobial classes were extended-spectrum cephalosporins (ceftriaxone, ceftazidime, or cefepime), carbapenems (meropenem, doripenem, or imipenem), piperacillin-tazobactam (fixed at 4 mg/L), quinolones (ciprofloxacin or levofloxacin), aminoglycosides (gentamicin, tobramycin, or amikacin), polymyxins (colistin or polymyxin B), tetracyclines (tetracycline, doxycycline, or minocycline), and ampicillin-sulbactam (2:1). For the P. aeruginosa isolate set, the antimicrobial classes were cephalosporins (ceftazidime or cefepime), carbapenems (meropenem, doripenem, or imipenem), piperacillin-tazobactam (fixed at 4 mg/L), quinolones (ciprofloxacin or levofloxacin), aminoglycosides (gentamicin, tobramycin, or amikacin), and polymyxins (colistin or polymyxin B). For the set of Enterobacterales isolates that are not intrinsically resistant to colistin (E. coli, Enterobacter spp., Citrobacter spp., and Klebsiella spp.), the antimicrobial classes were extended-spectrum cephalosporins (cefotaxime, ceftriaxone, ceftazidime, or cefepime), carbapenems (meropenem, doripenem, or imipenem), piperacillin-tazobactam (fixed at 4 mg/L), quinolones (ciprofloxacin or levofloxacin), aminoglycosides (gentamicin, tobramycin, or amikacin), polymyxins (colistin or polymyxin B), and tigecycline. The MIC data for MDR categorizations were obtained from the SENTRY Antimicrobial Surveillance Program (21). A subset of MIC data for comparator antimicrobials is displayed in Table 2 and Tables S3 to S6. We also tested the activity of MRX-8 and comparators against a rare colistin-resistant P. aeruginosa isolate (JMI 991784) collected in 2017.
Susceptibility testing methods.
MRX-8 powder was supplied by MicuRx Pharmaceuticals. Colistin (catalog no. C4461) and polymyxin B (catalog no. 1547007) were obtained from Sigma-Aldrich and United States Pharmacopeia, respectively. MRX-8, colistin, and polymyxin stocks were made in water.
Susceptibility to MRX-8 and comparator agents was measured using current CLSI methods (10, 22). The test medium was cation-adjusted Mueller-Hinton broth. Non-tissue-culture-treated polystyrene plates were used. CLSI and EUCAST interpretive criteria were applied according to current guidelines (10, 23). U.S. FDA product package insert interpretive criteria were used for tigecycline (24).
The current EUCAST interpretive criteria for colistin against Enterobacterales, P. aeruginosa, and A. baumannii isolates are ≤2 mg/L for susceptible and ≥4 mg/L for resistant (23). In contrast, CLSI categorizes an Enterobacterales, P. aeruginosa, or A. baumannii isolate as resistant if it exhibits a colistin or polymyxin MIC value of ≥4 mg/L (10). CLSI does not currently recognize a susceptible category for colistin or polymyxin B against any of these organism groups or species. Rather, isolates from these groups and species that display a colistin or polymyxin B MIC value of ≤2 mg/L are categorized as intermediate (10).
JMI Laboratories followed current CLSI quality assurance practices when performing the susceptibility tests. MIC values were validated by concurrently testing CLSI- and EUCAST-recommended (10, 25) ATCC or National Collection of Type Cultures (NCTC) quality control (QC) reference strains. The QC strains included E. coli ATCC 25922, P. aeruginosa ATCC 27853, K. pneumoniae ATCC BAA 1705, K. pneumoniae ATCC 700603, and E. coli NCTC 13846. QC ranges for the tested reference strains were the criteria published by CLSI or EUCAST (10, 25). The inoculum density during susceptibility testing was monitored by bacterial colony counts.
Data availability.
Data will be made available upon reasonable request.
ACKNOWLEDGMENTS
We thank Amy Chen and Judy Oberholser for their help during the preparation of the manuscript.
This study was performed by JMI Laboratories and supported by MicuRx Pharmaceuticals Inc., which included funding for services related to preparing the manuscript. This study was partially funded by CARB-X.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the Department of Health and Human Services Office of the Assistant Secretary for Preparedness and Response.
JMI Laboratories contracted to perform services in 2021 for AbbVie Inc., Affinity Biosensors, AimMax Therapeutics, Inc., Alterity Therapeutics, Amicrobe, Inc., Arietis Pharma, Armata Pharmaceuticals, Inc., Astrellas Pharma Inc., Basilea Pharmaceutica AG, Becton, Dickinson and Company, bioMérieux, Inc., Boost Biomes, Brass Dome Ventures Ltd., Bravos Biosciences, Bugworks Research Inc., Centers for Disease Control and Prevention, Cerba Research, Cidara Therapeutics, Cipla Ltd., ContraFect Corp., CXC7, Diamond V, Enveda Biosciences, Fedora Pharmaceuticals, Inc., Fimbrion Therapeutics, First Light Diagnostics, Forge Therapeutics, Inc., Fox Chase Cancer Center, GlaxoSmithKline plc, Harvard University, Institute for Clinical Pharmacodynamics, International Health Management Associates, Inc., Iterum Therapeutics plc, Janssen Research & Development, Johnson & Johnson, Kaleido Biosciences, Inc., Laboratory Specialists, Inc., Meiji Seika Pharma Co., Ltd., Melinta Therapeutics, Menarini Group, Merck & Co., Inc., MicuRx Pharmaceuticals Inc., Mutabilis, Nabriva Therapeutics, National Institutes of Health, Novome Biotechnologies, Omnix Medical Ltd., Paratek Pharma, Pattern Bioscience, Pfizer Inc., Prokaryotics Inc., Pulmocide Ltd., QPEX Biopharma, Inc., Roche Holding AG, Roivant Sciences, SeLux Diagnostics, Inc., Shionogi Inc., Sinovent Pharmaceuticals, Inc., SNIPR Biome ApS, Spero Therapeutics, Summit Therapeutics, Inc., T2 Biosystems, TenNor Therapeutics, Thermo Fisher Scientific, University of Southern California, University of Wisconsin, USCAST, U.S. Food and Drug Administration, Venatorx Pharmaceutics, Inc., Weill Cornell Medicine, and Wockhardt Ltd.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. [Google Scholar]
- 2.World Health Organization. 2021. Global Antimicrobial Resistance and Use Surveillance System (GLASS) report: 2021. World Health Organization, Geneva, Switzerland. https://www.who.int/publications/i/item/9789240027336. [Google Scholar]
- 3.Theuretzbacher U. 2017. Global antimicrobial resistance in Gram-negative pathogens and clinical need. Curr Opin Microbiol 39:106–112. doi: 10.1016/j.mib.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 4.Paterson DL, Bonomo RA. 2019. Multidrug-resistant Gram-negative pathogens: the urgent need for 'old' polymyxins. Adv Exp Med Biol 1145:9–13. doi: 10.1007/978-3-030-16373-0_2. [DOI] [PubMed] [Google Scholar]
- 5.Bassetti M, Di Pilato V, Giani T, Vena A, Rossolini GM, Marchese A, Giacobbe DR. 2020. Treatment of severe infections due to metallo-β-lactamases-producing Gram-negative bacteria. Future Microbiol 15:1489–1505. doi: 10.2217/fmb-2020-0210. [DOI] [PubMed] [Google Scholar]
- 6.Vaara M. 2019. Polymyxins and their potential next generation as therapeutic antibiotics. Front Microbiol 10:1689. doi: 10.3389/fmicb.2019.01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nang SC, Azad MAK, Velkov T, Zhou QT, Li J. 2021. Rescuing the last-line polymyxins: achievements and challenges. Pharmacol Rev 73:679–728. doi: 10.1124/pharmrev.120.000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaara M. 2019. Polymyxin derivatives that sensitize Gram-negative bacteria to other antibiotics. Molecules 24:249. doi: 10.3390/molecules24020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivas P, Rivard K. 2017. Polymyxin resistance in Gram-negative pathogens. Curr Infect Dis Rep 19:38. doi: 10.1007/s11908-017-0596-3. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2021. Performance standards for antimicrobial susceptibility testing; 31st ed. M100Ed31. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Bialvaei AZ, Samadi Kafil H. 2015. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin 31:707–721. doi: 10.1185/03007995.2015.1018989. [DOI] [PubMed] [Google Scholar]
- 12.Ling Z, Yin W, Shen Z, Wang Y, Shen J, Walsh TR. 2020. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother 75:3087–3095. doi: 10.1093/jac/dkaa205. [DOI] [PubMed] [Google Scholar]
- 13.Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin Microbiol Rev 21:449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenhard JR, Bulman ZP, Tsuji BT, Kaye KS. 2019. Shifting gears: the future of polymyxin antibiotics. Antibiotics (Basel) 8:42. doi: 10.3390/antibiotics8020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodor N, Buchwald P. 2000. Soft drug design: general principles and recent applications. Med Res Rev 20:58–101. doi:. [DOI] [PubMed] [Google Scholar]
- 16.Gordeev MF. 2018. Polymyxin soft drug MRX-8 with potential to address the class nephrotoxicity, abstr 6. In 3rd Int Conf Polymyxins, Madrid, Spain. [Google Scholar]
- 17.Lepak AJ, Wang W, Andes DR. 2020. Pharmacodynamic evaluation of MRX-8, a novel polymyxin, in the neutropenic mouse thigh and lung infection models against Gram-negative pathogens. Antimicrob Agents Chemother 64:e01517. doi: 10.1128/AAC.01517-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castanheira M, Deshpande LM, Mendes RE, Canton R, Sader HS, Jones RN. 2019. Variations in the occurrence of resistance phenotypes and carbapenemase genes among Enterobacteriaceae isolates in 20 years of the SENTRY Antimicrobial Surveillance Program. Open Forum Infect Dis 6(Suppl 1):S23–S33. doi: 10.1093/ofid/ofy347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gales AC, Seifert H, Gur D, Castanheira M, Jones RN, Sader HS. 2019. Antimicrobial susceptibility of Acinetobacter calcoaceticus-Acinetobacter baumannii complex and Stenotrophomonas maltophilia clinical isolates: results from the SENTRY Antimicrobial Surveillance Program (1997–2016). Open Forum Infect Dis 6(Suppl 1):S34–S46. doi: 10.1093/ofid/ofy293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shortridge D, Gales AC, Streit JM, Huband MD, Tsakris A, Jones RN. 2019. Geographic and temporal patterns of antimicrobial resistance in Pseudomonas aeruginosa over 20 years from the SENTRY Antimicrobial Surveillance Program, 1997–2016. Open Forum Infect Dis 6(Suppl 1):S63–S68. doi: 10.1093/ofid/ofy343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuhrmeister AS, Jones RN. 2019. The importance of antimicrobial resistance monitoring worldwide and the origins of SENTRY Antimicrobial Surveillance Program. Open Forum Infect Dis 6(Suppl 1):S1–S4. doi: 10.1093/ofid/ofy346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed. M07Ed11. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.EUCAST. 2021. Breakpoint tables for interpretation of MICs and zone diameters, version 11.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf.
- 24.Pfizer. 2018. Tygacil package insert. Pfizer, New York, NY. https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/211158s000lbl.pdf. [Google Scholar]
- 25.EUCAST. 2021. Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST, version 11.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/QC/v_11.0_EUCAST_QC_tables_routine_and_extended_QC_pdf.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S8. Download aac.00139-22-s0001.pdf, PDF file, 0.2 MB (201.5KB, pdf)
Data Availability Statement
Data will be made available upon reasonable request.