ABSTRACT
Fungal infections are a major health concern because of limited antifungal drugs and development of drug resistance. Candida can develop azole drug resistance by overexpression of drug efflux pumps or mutating ERG11, the target of azoles. However, the role of epigenetic histone modifications in azole-induced gene expression and drug resistance is poorly understood in Candida glabrata. In this study, we show that Set1 mediates histone H3K4 methylation in C. glabrata. In addition, loss of SET1 and histone H3K4 methylation increases azole susceptibility in both C. glabrata and S. cerevisiae. This increase in azole susceptibility in S. cerevisiae and C. glabrata strains lacking SET1 is due to distinct mechanisms. For S. cerevisiae, loss of SET1 decreased the expression and function of the efflux pump Pdr5, but not ERG11 expression under azole treatment. In contrast, loss of SET1 in C. glabrata does not alter expression or function of efflux pumps. However, RNA sequencing revealed that C. glabrata Set1 is necessary for azole-induced expression of all 12 genes in the late ergosterol biosynthesis pathway, including ERG11 and ERG3. Furthermore, chromatin immunoprecipitation analysis shows histone H3K4 trimethylation increases upon azole-induced ERG gene expression. In addition, high performance liquid chromatography analysis indicated Set1 is necessary for maintaining proper ergosterol levels under azole treatment. Clinical isolates lacking SET1 were also hypersusceptible to azoles which is attributed to reduced ERG11 expression but not defects in drug efflux. Overall, Set1 contributes to azole susceptibility in a species-specific manner by altering the expression and consequently disrupting pathways known for mediating drug resistance.
KEYWORDS: azole, Candida glabrata, ERG11, epigenetics, H3K4 methylation, Set1, antifungal resistance, ergosterol, histone methylation, regulation of gene expression
INTRODUCTION
Candida infections are a major health concern due to the increased frequency of infections and the development of drug resistance (1, 2). Over the years, Candida glabrata has become the second most common cause of candidiasis (1–3). In some immunocompromised patients, such as diabetics, patients with hematologic cancer, organ transplant recipients, and the elderly, it is the most predominant Candida infection (2–6). The emergence of C. glabrata as a major pathogen is likely due to its intrinsic drug resistance to azole antifungal drugs and ability to quickly adapt and acquire clinical drug resistance during treatment (3, 7). The consequence of drug resistance leads to increases in health care costs, as well as lower success rates in treatment and an increase in mortality (8–10).
C. glabrata naturally has low susceptibility to azole drugs and consequently, echinocandins are the preferred drug choice for treating C. glabrata infections (11). C. glabrata can also acquire clinical resistance to azole drugs which is often due to overexpressing the ABC-transporter drug efflux pump Cdr1 or Pdh1 (Cdr2) caused by gain-of-function mutations in the transcription factor Pdr1 (7, 12–14). In other Candida species, acquired clinical azole resistance can also be due to overexpression of ERG11 due to gain-of-function mutations in the Upc2 transcription factor or mutations in ERG11 (15–17). However, for unknown reasons, ERG11 or UPC2 mutations are typically not found in clinically drug-resistant C. glabrata strains (7, 18–20).
Because pathogenic fungi can rapidly adapt to various cellular environments and xenobiotic drug exposures, epigenetic mechanisms are also likely contributing to altered gene expression profiles permissive for adaptation and drug resistance. Several studies in Candida albicans support this hypothesis and show that epigenetic factors such as histone acetyltransferases, CaGcn5 and CaRtt109, and histone deacetylases, CaRpd3 and CaHda1 are important for either fungal pathogenesis and/or drug resistance (21–25). In contrast, epigenetic factors that posttranslationally modify histones have not been extensively studied for their roles in drug resistance in C. glabrata. Nonetheless, Orta-Zavalz et al. have shown that deleting histone deacetylase, CgHST1, decreases susceptibility to fluconazole, which is likely attributed to an increase in transcript levels of CgPDR1 and CgCDR1 under untreated conditions (26). In addition, a recent publication by Filler et al., has indicated that C. glabrata strains with CgGCN5, CgRPD3, or CgSPP1 deleted have increased susceptibility to caspofungin when using high concentrations (27). However, no mechanistic understanding such as gene targets or changes in chromatin/histone modifications was provided for the caspofungin-hypersensitive phenotype.
Previous publications from our lab demonstrated that in Saccharomyces cerevisiae loss of Set1, a known histone H3K4 methyltransferase, has a hypersensitive growth defect in the presence of the antifungal metabolite, brefeldin A (BFA) and clinically used azole drugs (28, 29). We determined that hypersensitivity to BFA was due to a decrease in ergosterol levels in S. cerevisiae strains lacking histone H3K4 methylation. However, until this study, no mechanistic understanding has been provided why S. cerevisiae or C. glabrata strains lacking SET1 alter azole drug susceptibility. Furthermore, in C. albicans, loss of SET1 appears to alter virulence but not azole drug resistance (30). To determine whether an increase in azole susceptibility is conserved in a human fungal pathogen that is evolutionarily closer to S. cerevisiae than C. albicans, we investigated the role of Set1 and its mechanistic contribution to altering azole susceptibility in S. cerevisiae and C. glabrata.
Here, we show for the first time that Set1-mediates histone H3K4 mono-, di-, and trimethylation in C. glabrata and loss of Set1-mediated histone H3K4 methylation alters the azole drug susceptibility of C. glabrata similar to what is seen in S. cerevisiae. Interestingly, azole hypersusceptibility in S. cerevisiae lacking SET1 appears to be mediated by a decrease in the expression and function of ScPdr5, the CgCdr1 ortholog, and not a failure to induce ERG genes. However, hypersusceptibility to azole drugs in C. glabrata strains lacking Set1-mediated histone H3K4 methylation is not a consequence of altered expression levels of CgCDR1, CgPDR1 or their ability to efflux drugs. Interestingly, RNA sequencing (RNA-seq) and HPLC analysis revealed that CgSet1 is required for azole-induced expression of CgERG genes, including CgERG11 and CgERG3, so that proper ergosterol levels are maintained. Azole-induced gene expression was dependent on Set1 methyltransferase activity and associated with gene-specific increases in histone H3K4 trimethylation on CgERG11 and CgERG3 chromatin. In addition, clinical isolates lacking CgSET1 were hypersensitive to azoles and attributed to reduced CgERG11 expression but not reduced efflux pump function. Overall, we show that loss of Set1-mediated histone H3K4 methylation in S. cerevisiae and C. glabrata contributes to azole hypersusceptibility by distinct mechanisms where genes, known for drug resistance, are differentially altered. Identifying and understanding the epigenetic mechanisms contributing to altered drug efficacy in various fungal species will be important for the development of alternative drug targets for treating patients with fungal infections.
RESULTS
Loss of Set1-mediated histone H3K4 methylation in S. cerevisiae and C. glabrata increases azole drug susceptibility.
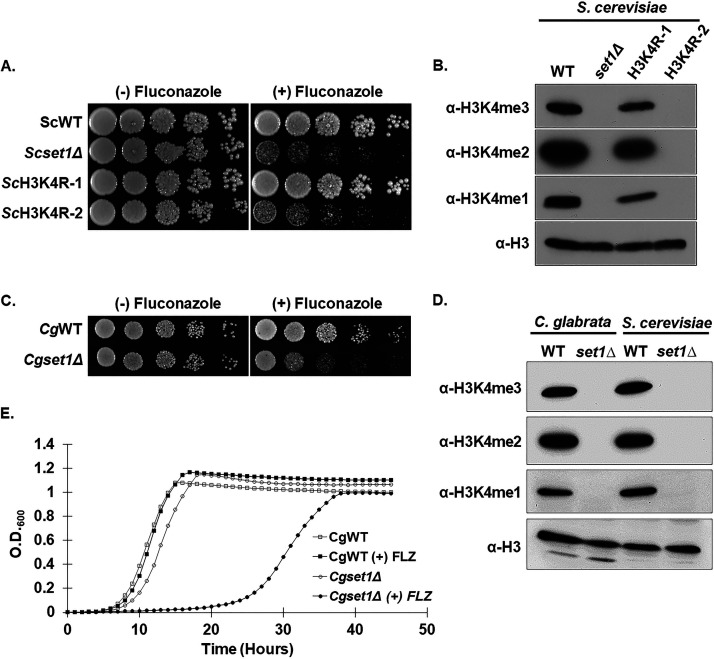
Set1 is a known SET domain-containing lysine histone methyltransferase that is conserved from yeast to humans and the enzymatic activity of the SET domain catalyzes mono-, di-, and trimethylation on histone H3 at lysine 4 (Lys4) (31, 32). Our previous work in S. cerevisiae has determined that loss of SET1 in the BY4741 background strain results in increased susceptibility to azole drugs, suggesting that H3K4 methylation is necessary for mediating wild-type azole drug resistance (29). To determine the role of histone H3K4 methylation in azole drug efficacy, we constructed histone H3K4R mutations in the BY4741 background strain. Because S. cerevisiae has two genes encoding histone H3, two yeast strains were constructed where a histone H3K4R mutation was integrated at one histone H3 gene keeping the other gene wild type (ScH3K4R-1), while the other strain contained H3K4R mutations integrated at both histone H3 genes (ScH3K4R-2; see Table S1 in the supplemental material). To determine whether a loss of histone H3K4 methylation altered azole drug sensitivity similar to a set1Δ (Scset1Δ) strain, a serial-dilution spot assay was performed. Both Scset1Δ and ScH3K4R mutant strains were grown in SC complete media and spotted onto SC agar plates with or without 8 μg/mL fluconazole (Fig. 1A). These data show that loss of histone H3K4 methylation by deleting ScSET1 or mutating histone H3 where both histone H3 genes are mutated at K4 (ScH3K4R-2), resulted in similar azole drug hypersensitivity compared to each other (Fig. 1A). To confirm that histone H3K4 methylation was abolished in these strains, Western blot analysis was performed using methyl-specific antibodies to detect histone H3K4 mono-, di-, and trimethylation (Fig. 1B). Histone H3 was used for a loading control (Fig. 1B). As expected, histone methylation was abolished in set1Δ and in H3K4R-2 mutation strains but not in the histone H3K4R-1 strain (Fig. 1B). Together, our data demonstrate that the presence of histone H3K4 methylation is critical for maintaining wild-type azole drug susceptibility.
FIG 1.
Loss of Set1-mediated mono-, di-, and trimethylation at histone H3K4 in S. cerevisiae and C. glabrata results in increased azole susceptibility and delayed growth in vitro. (A) Fivefold serial dilution spot assays of the indicated S. cerevisiae strains were grown on SC media with and without 8 μg/mL fluconazole and incubated at 30°C for 72 h. (B and D) Whole-cell extracts isolated from the indicated strains were immunoblotted using histone H3K4 methyl-specific mono-, di-, and trimethylation antibodies of whole-cell extracts isolated from the indicated strains. Histone H3 was used as a loading control. (C) Fivefold serial dilution spot assays of the indicated C. glabrata strains were grown on SC media with or without 32 μg/mL fluconazole and incubated at 30°C for 48 h. (E) Liquid growth curve assay of the indicated C. glabrata strains grown over 50 h with or without 32 μg/mL fluconazole.
To determine whether an azole hypersensitive growth phenotype observed in S. cerevisiae is also conserved in the human fungal pathogen C. glabrata, the WT (CgWT) and a set1Δ (Cgset1Δ) strain were spotted on SC agar plates with and without 16 μg/mL fluconazole (Fig. 1C). Similar to what was observed in S. cerevisiae, deleting SET1 in C. glabrata 2001 (CBS138, ATCC 2001) showed an increase in azole susceptibility compared to a CgWT strain (Fig. 1C). In addition, the Cgset1Δ strain had a significant growth delay in liquid growth cultures compared to CgWT when treated with 32 μg/mL fluconazole (Fig. 1E). Western blot analysis showed that deleting CgSET1 abolished all histone H3K4 mono-, di-, and trimethylation confirming that CgSET1 is the sole histone H3K4 methyltransferase in C. glabrata (Fig. 1D). Altogether, our results show Set1-mediated histone H3K4 methylation in S. cerevisiae and C. glabrata is conserved and is necessary for maintaining a wild-type susceptibility to azole drugs.
Loss of C. glabrata Set1 complex members alters azole efficacy and histone H3K4 methylation.
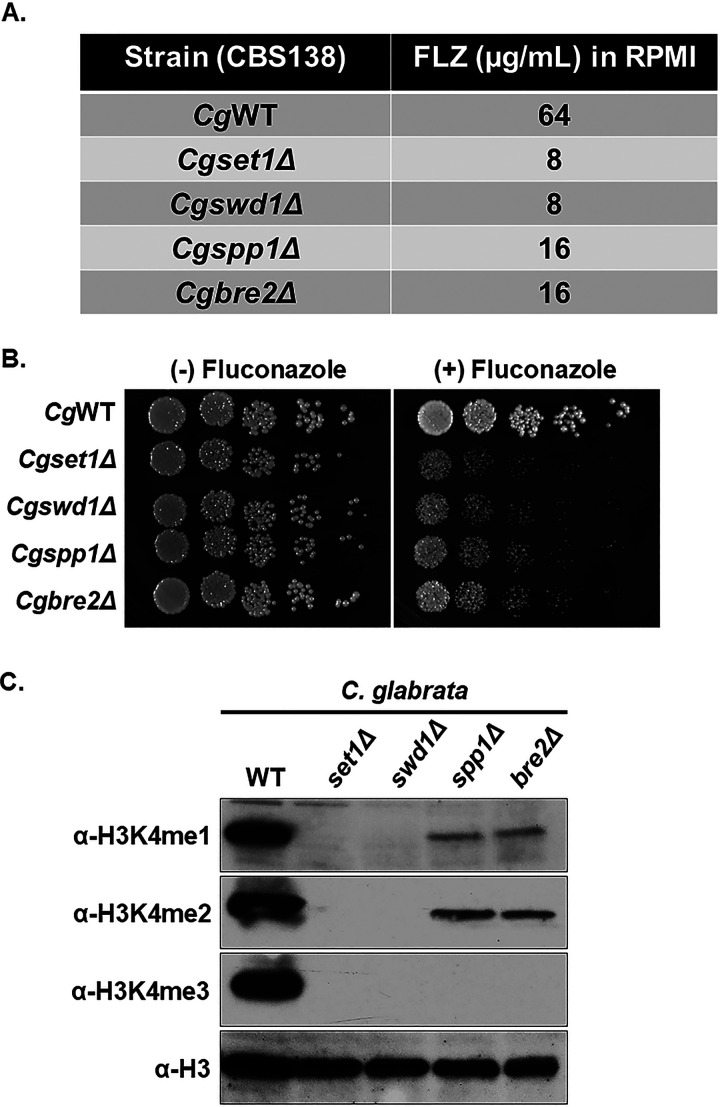
In S. cerevisiae, Set1 forms a complex referred to as the Complex Proteins Associated with Set1 or COMPASS. COMPASS forms a stable complex with eight proteins which includes the catalytic subunit Set1, Swd1, Swd2, Swd3, Spp1, Bre2, Sdc1, and Shg1 (33–35). Previous studies in S. cerevisiae have determined that Swd1, Swd2, Swd3, Spp1, Bre2, and Sdc1 are necessary for Set1 to properly catalyze the various states of histone H3K4 mono-, di-, and trimethylation (33–38). To determine whether COMPASS components are required for azole drug efficacy and Set1-mediated histone H3K4 methylation in C. glabrata, deletion strains lacking SET1, SPP1, BRE2, and SWD1 were generated, and the MIC (in RPMI media) of each strain was determined (Fig. 2A). Consistent with S. cerevisiae serial-dilution spot and liquid growth assays in Fig. 1, the Cgset1Δ strain showed increased susceptibility to fluconazole with an 8-fold difference in MIC compared to the CgWT strain (Fig. 2A). A Cgswd1Δ strain showed a MIC similar to that of the Cgset1Δ strain, while the MICs of Cgspp1Δ and Cgbre2Δ deletion strains were 4-fold different than the WT strain (Fig. 2A). All C. glabrata COMPASS deletions strains showed an increase in susceptibility to azole drugs on agar plates similar to S. cerevisiae COMPASS deletion strains except for the Scspp1Δ strain, which is likely due to differences in the histone H3K4 methylation status (Fig. 2B and C; see also Fig. S1A) (29, 33, 34, 36, 38).
FIG 2.
Deletion of Set1 complex members in C. glabrata results in increased azole susceptibility and loss of histone H3K4 methylation. (A) MIC assay of the indicated strains performed in RPMI 1640 media at 35°C, and results recorded after 48 h of incubation. (B) Fivefold serial dilution spot assays of the indicated C. glabrata strains were grown on SC plates with or without 32 μg/mL fluconazole. (C) Whole-cell extracts isolated from the indicated strains were immunoblotted using H3K4 methyl-specific mono-, di-, and trimethylation antibodies. Histone H3 was used as a loading control.
Western blot analysis determined that Cgswd1Δ strain lacked all forms of histone H3K4 methylation (Fig. 2C) which is also observed in Cgset1Δ and Scset1Δ strains (Fig. 2C and 1D). In contrast, deletion of CgSPP1 and CgBRE2 abolished all detectable levels of H3K4 trimethylation and significantly reduced the levels of histone H3K4 mono- and dimethylation. Taken together, our data show that when C. glabrata COMPASS subunits SET1 and SWD1 are deleted, global loss of histone H3K4 methylation is observed similar to what is seen when the subunits are deleted in S. cerevisiae (Fig. 2C) (33, 34, 36, 38). However, the Cgspp1Δ has a total loss of histone H3K4 trimethylation and significant loss of histone H3K4 mono- and dimethylation similar to the Cgbre2Δ and Scbre2Δ strains (Fig. 2C). For unknown reasons, the pattern of histone H3K4 methylation is different in the Scspp1Δ strain, which only has a reduction in histone H3K4 trimethylation but not mono- or dimethylation (33–39). Altogether, these results suggest that the COMPASS complex is needed to mediate proper histone H3K4 methylation and wild-type resistance to azole drugs.
Histone H3K4 methyltransferase activity of C. glabrata Set1 is necessary for wild-type growth on azole-containing media.
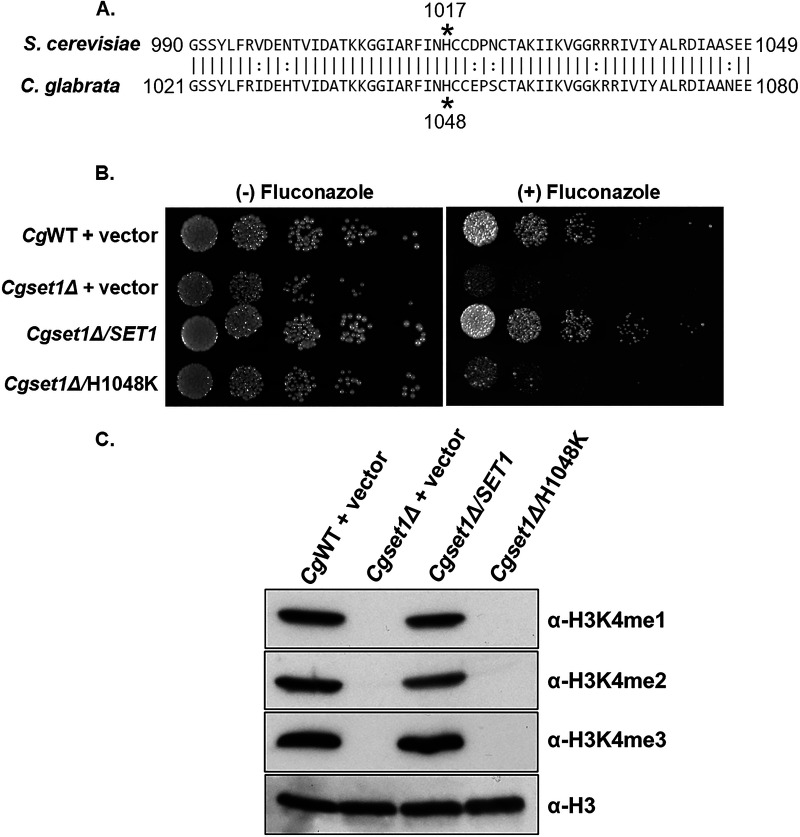
To confirm that altered azole efficacy in the Cgset1Δ strain was due to loss of SET1 and not a secondary mutation, a genomic fragment containing the CgSET1 promoter, 5′ untranslated region (UTR), open reading frame, and the 3′ UTR was amplified by PCR and cloned into the C. glabrata plasmid, pGRB2.0 (40). Because a H1017K mutation in the SET domain of S. cerevisiae Set1 is known to be catalytically inactive (28, 41, 42), we performed site-directed mutagenesis on pGRB2.0-CgSET1 and generated an analogous mutation in C. glabrata Set1 at H1048K determined using the sequence alignment in Fig. 3A. In addition, SET1 was deleted in C. glabrata 2001HTU (ATCC 200989) to utilize the ura3 auxotrophic marker (43). Importantly, Cg2001HTU lacking SET1 was hypersensitive to azole drugs similar to when SET1 was deleted in Cg2001 (Fig. 1C and Fig. 3B).
FIG 3.
The catalytic activity of the SET domain is necessary for Set1-mediated histone H3K4 methylation and alters azole susceptibility in C. glabrata. (A) Sequence alignment comparing the SET domain amino acid residues of S. cerevisiae and C. glabrata Set1. Asterisks indicate residues required for catalytic activity. (B) Fivefold serial dilution spot assays of the indicated C. glabrata strains were grown on SC plates with or without 32 μg/mL fluconazole. (C) Whole-cell extracts isolated from the indicated strains were immunoblotted using methyl-specific mono-, di-, and trimethylation antibodies. Histone H3 was used as a loading control.
Furthermore, transformation of pGRB2.0-CgSET1 into the Cg2001HTU/set1Δ strain was able to rescue azole hypersensitivity, while pGRB2.0-Cgset1H1048K did not rescue wild-type azole drug resistance, as shown by serial-dilution spot assays grown on SC agar plates with 32 μg/mL fluconazole (Fig. 3B). MIC assays under SC-ura conditions also show similar results (see Fig. S1B). Western blot analysis indicated that pGRB2.0-CgSET1 expression in Cg2001HTU/set1Δ strain restored histone H3K4 methylation to wild-type levels, while Cgset1H1048K did not rescue histone H3K4 methylation confirming that this mutation lacks catalytic activity similar to Scset1H1017K (Fig. 3C). Importantly, quantitative real-time PCR analysis (qRT-PCR) confirmed that the plasmids expressing CgSET1 and Cgset1H1048K were similar to the endogenously expressed SET1 (see Fig. S1C). To ensure Set1 protein is expressed, a 3×FLAG sequence was inserted between the endogenous promoter and ATG of pGRB2.0-cgSET1 WT and catalytically inactive plasmid. As expected, the strain expressing the 3×FLAG-Cgset1H1048K did not rescue H3K4 trimethylation and is hypersusceptible to fluconazole compared to a strain expressing 3×FLAG-CgSET1. Importantly, both wild-type and mutant SET1 constructs are expressed at a similar protein level (see Fig. S1D and E). This shows that loss of histone H3K4 methylation was not due to difference in expression levels but due to the catalytic inactivation of Cgset1H1048K. These data suggest that azole hypersusceptibility in Cgset1Δ strains are specifically due to the loss of SET1 and its catalytic activity.
Expression of drug efflux pumps are altered in a S. cerevisiae set1Δ strain but not in a C. glabrata set1Δ strain.
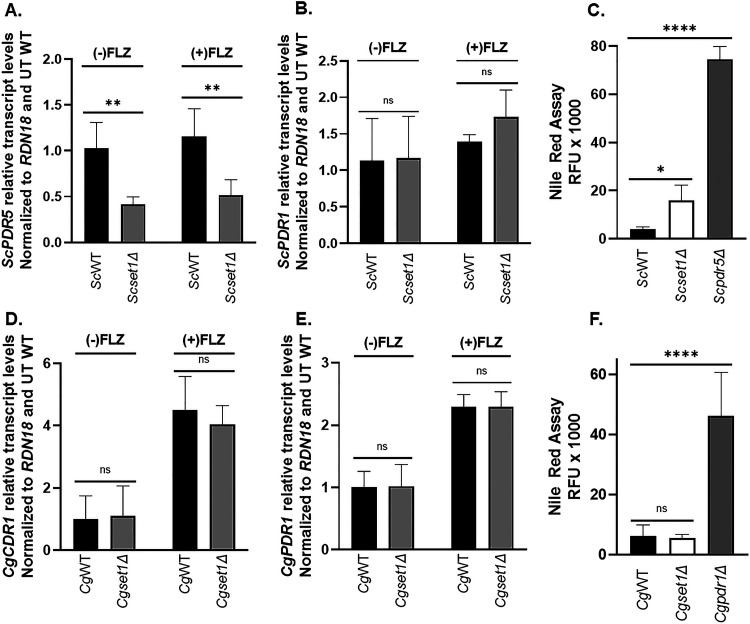
In Candida glabrata, the major mechanisms for changes in drug resistance are due to altered expression of CgCDR1, the main drug efflux pump, or gain-of-function mutations in CgPDR1, a gene that encodes the transcription factor for CgCDR1 (7, 12, 19, 20, 44). To determine whether altered drug resistance for Scset1Δ or Cgset1Δ strains are due to altered expression levels of the ATP-binding cassette (ABC) transporters ScPDR5 or CgCDR1, respectively, the transcript levels of ScPDR5 and CgCDR1 were analyzed by qRT-PCR (Fig. 4). Scset1Δ cells grown with or without azoles showed a significant decrease in ScPDR5 expression compared to WT cells (Fig. 4A). In addition, Scset1Δ cells did not have changes in ScPDR1 transcripts, a gene encoding the transcription factor for PDR5 (Fig. 4B). To determine whether drug efflux was reduced in Scset1Δ cells, a Nile red fluorescence-based assay was performed. Nile red, a fluorescent lipophilic stain, has been shown to be a substrate for the ABC-transporters and used as a proxy for drug efflux (45, 46). If Nile red is the substrate of an efflux pump (e.g., Pdr5), then cells expressing a functional efflux pump will not accumulate the dye and have low fluorescence. To show azole drugs induced efflux in S. cerevisiae, a Nile red fluorescence-based assay was performed on azole treated and untreated ScWT cells. As expected, azole treated ScWT cells had less fluorescence indicating an increase in Nile red efflux compared to ScWT untreated cells (see Fig. S2D). Consistent with decreased expression of PDR5 in Scset1Δ, azole-treated Scset1Δ cells had higher fluorescence than azole-treated ScWT cells, indicating reduced efflux (Fig. 4C). A Scpdr5Δ strain was used as a positive control to prevent drug efflux (Fig. 4C).
FIG 4.
Drug efflux pump expression and drug efflux is altered in a Scset1Δ but not Cgset1Δ. (A and B; D and E) Expression of the indicated genes was determined in ScWT, Scset1Δ, CgWT, and Cgset1Δ strain cells treated with or without 64 μg/mL fluconazole for 3 h by qRT-PCR analysis. Gene expression analysis was set relative to the untreated wild type, and expression was normalized to RDN18 mRNA levels. Data were analyzed from ≥3 biological replicates with three technical replicates each. Error bars represent the standard deviations (SD). (C and F) Red fluorescence units (RFU) were measured as output in a Nile red assay to determine the amount of drug efflux in the indicated strains with and without fluconazole. Scpdr5Δ and Cgpdr1Δ strains was used as controls. Data were analyzed from ≥3 biological replicates with three technical replicates each. Statistics were determined using the GraphPad Prism Student t test, version 9.2.0. ns, P > 0.05; *, P < 0.05; ****, P < 0.0001. Error bars represent SD.
In contrast, CgCDR1 or CgPDR1 expression was not altered in the Cgset1Δ cells compared to WT cells when grown with and without azoles (Fig. 4D and E). In addition, the transcript levels of other transporters CgSNQ2, CgYOR1, and CgPDH1 were also not altered, but a decrease in PDH1 transcripts was observed in Cgset1Δ cells upon azole treatment (see Fig. S2). However, previous studies have shown loss of CgPDH1 alone is not sufficient to lead to azole sensitivity (47). To determine whether drug efflux was functional in Cgset1Δ cells, a Nile red fluorescence-based assay was performed. As a control, a Cgpdr1Δ strain was used (Fig. 4F). A Cgpdr1Δ strain is known to disrupt the expression of CgCDR1 and subsequently prevent drug or Nile red efflux (15). To induce CgCDR1 expression levels, Cgset1Δ and wild-type cells were treated with fluconazole. Although azole treatment did reduce the amount of Nile red in CgWT cells compared to untreated cells (see Fig. S2E), there was no discernible difference in Nile red fluorescence between Cgset1Δ and CgWT cells upon azole treatment (Fig. 4F). These data show that cells lacking CgSET1 have similar efflux capabilities as CgWT cells in the presence or absence of azole treatment. Our results suggest that the increase in azole susceptibility in a Scset1Δ strain is a consequence of altered drug efflux expression, whereas azole hypersensitivity in a Cgset1Δ strain is not due to altered expression or function of drug efflux pumps but mediated by a different mechanism.
Loss of C. glabrata SET1 but not S. cerevisiae SET1 leads to decreased expression of genes involved in the late ergosterol biosynthesis pathway when treated with fluconazole.
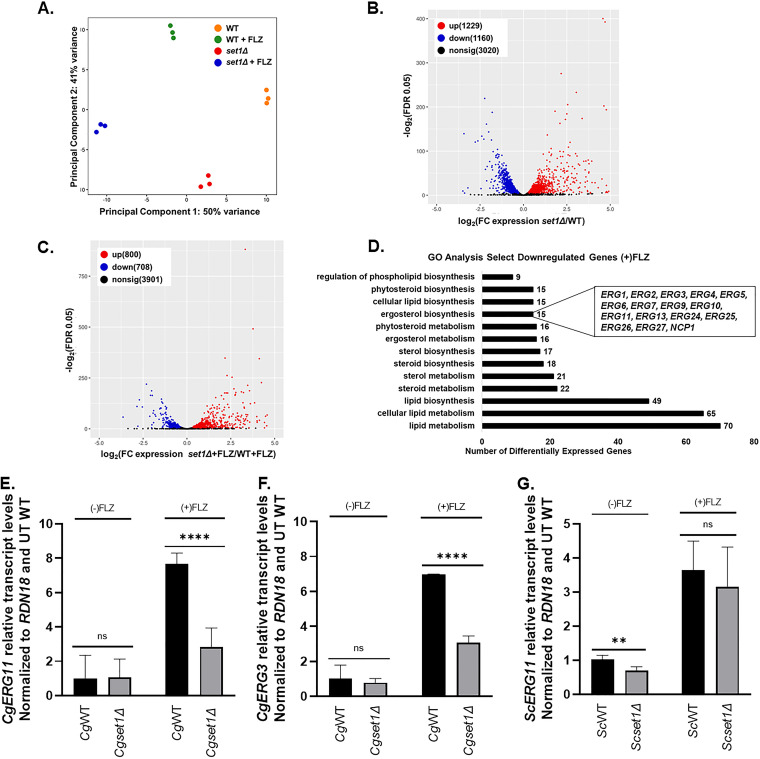
Because drug efflux function or transcript levels were not disrupted in a Cgset1Δ strain, RNA sequencing analysis was used to provide insight into what gene pathway might be disrupted in the Cgset1Δ strain and explain why a loss of SET1 alters azole drug efficacy. CgWT and Cgset1Δ strains were treated with 64 μg/mL fluconazole for 3 h in SC complete media, and RNA was extracted for RNA sequencing. Principal component analysis (PCA) and differentially expressed gene (DEG) analysis demonstrated by the volcano scatterplot (–log2 false discovery rate [FDR], y-axis) versus the fold change ([x-axis] of the DEGs) indicate that the untreated and treated CgWT strain is substantially and statistically different from the untreated and treated Cgset1Δ (Fig. 5A). DESeq2 analysis was used to identify the differentially expressed genes (DEGs) using a FDR of 0.05 (see Table S6). From this analysis, a total of 2389 genes were differentially expressed in Cgset1Δ versus CgWT strains under untreated conditions (Fig. 5B). Of 5,615 genes, 1,508 were differentially expressed under treated conditions, in which 800 (14.2%) genes were upregulated and 708 (12.6%) genes were downregulated in Cgset1Δ compared to CgWT strains (Fig. 5C). After applying a 1.4-fold cutoff to the data, we observed 1,619 genes differentially expressed in the untreated Cgset1Δ versus CgWT strains. In the treated strains, with a 1.4-fold cutoff, we observed 539 (9.6%) genes were downregulated in a Cgset1Δ versus CgWT strains, and 623 (11.1%) genes were upregulated. These data show that SET1 is important for maintaining proper gene expression in C. glabrata.
FIG 5.
The deletion of SET1 in C. glabrata alters global and local levels of gene expression under untreated and azole-treated conditions. The genomewide changes in gene expression under azoles were assessed using C. glabrata CBS138 (2001) WT and set1Δ strains. (A) PCA for WT and set1Δ azole-treated samples relative to WT untreated samples based on the counts per million. (B) Volcano plot showing the significance [−log2(FDR), y axis] versus the fold change (x axis) of the DEGs identified in the WT untreated samples relative to set1Δ untreated samples. (C) Volcano plot showing the significance [−log2(FDR), y axis] versus the fold change (x axis) of the DEGs identified in the set1Δ azole-treated samples relative to WT azole-treated samples. Genes with significant differential expression (FDR < 0.05) in panels B and C are highlighted in red and blue for up- and downregulated genes, respectively. Black highlighted genes are considered nonsignificant. (D) Genes from the RNA-seq data set that were statistically significantly enriched (FDR < 0.05) were used for GO term determination of Set1-dependent DEGs under azole conditions. Downregulated genes refer to the DEGs that are dependent on Set1 for activation either directly or indirectly. Significantly enriched groups of GO terms were identified as the DEGs from only set1Δ and WT azole-treated samples. (E to G) Expression of indicated genes was determined in WT and set1Δ strain cells in either C. glabrata or S. cerevisiae treated with 64 μg/mL fluconazole for 3 h by qRT-PCR analysis. Gene expression analysis was set relative to the untreated wild-type and expression was normalized to RDN18 mRNA levels. Data were analyzed from ≥3 biological replicates with three technical replicates each. Statistics were determined using the GraphPad Prism Student t test, version 9.2.0. ****, P < 0.0001; **, P < 0.01; ns, P > 0.05. Error bars represent the SD.
Because Set1-mediated histone H3K4 methylation is known to play a key role in gene activation, we focused our attention on genes downregulated in Cgset1Δ compared to CgWT strains. For azole treated strains, GO Term Finder of the gene sets that were downregulated found significant GO terms involved in lipid, steroid, and sterol/ergosterol metabolism or biosynthesis (Fig. 5D; see also Table S7). For untreated strains, GO Term Finder identified significant GO terms involved in lipid metabolism but not steroid and sterol/ergosterol metabolism or biosynthesis. Interestingly, our data showed that 12 of the 12 genes involved in the late ergosterol biosynthesis pathway are downregulated 1.4-fold or more in a Cgset1Δ compared to CgWT under azole-treated conditions (Fig. 5C; see also Fig. S4A and B and Table S6), whereas 5 of the 12 late pathway CgERG genes were downregulated in a Cgset1Δ compared to CgWT strains under untreated conditions using a 1.4-fold difference as a cutoff (Fig. 5D; see also Table S6). Two of these differentially expressed genes—ERG11, the gene that encodes the target of azoles, and ERG3, the gene that encodes the enzyme responsible for production of a toxic sterol when cells are treated with azoles—are known to play roles in azole drug resistance in various Candida species (17, 19, 48–50).
To validate results seen in RNA sequencing analysis, CgERG11 and CgERG3 transcript levels were analyzed by qRT-PCR. Our analysis showed that upon azole treatment, CgERG3 and CgERG11 transcript levels are induced in a CgWT strain (Fig. 5E and F), while loss of CgSET1 prevented CgWT induction of both CgERG11 and CgERG3 under azole treated conditions. Even though our untreated RNA sequencing data set did show minor changes in CgERG3 and CgERG11 transcript levels, we did not detect any significant changes between Cgset1Δ and CgWT cells when grown under untreated standard log-phase conditions using qRT-PCR analysis (Fig. 5E and F). We also performed gene expression analysis to determine whether CgERG gene transcript induction still depended on CgSet1 in saturated cultures. We show in both exponential and saturated cultures CgSet1 is necessary for CgERG3 and CgERG11 induction upon azole treatment (Fig. 5E and F; see also Fig. S3A and B). Because CgERG3 transcript levels were decreased, we do not anticipate azole sensitivity is due to an increase in toxic sterols but by the lack of induction of CgERG11 and other ERG genes resulting in lower total cellular ergosterol levels (51, 52). In addition, we examined transcript levels of ScERG11 in Scset1Δ cells. In contrast to Cgset1Δ, we observed a decrease in ScERG11 transcripts under untreated conditions, but no difference was detected upon azole treatment (Fig. 5G). The ScERG11 transcript data are consistent with what was previously published for Scset1Δ cells under untreated conditions (28). Altogether, our results show that the increase in azole susceptibility in a Scset1Δ strain is a consequence of decreased drug efflux expression, whereas azole hypersensitivity in a Cgset1Δ strain is not due to altered expression or function of drug efflux pumps but by decreased expression of genes needed for the late ergosterol pathway.
Set1-mediated histone H3K4 methylation is enriched on ERG gene chromatin and is required for azole induction of ERG genes.
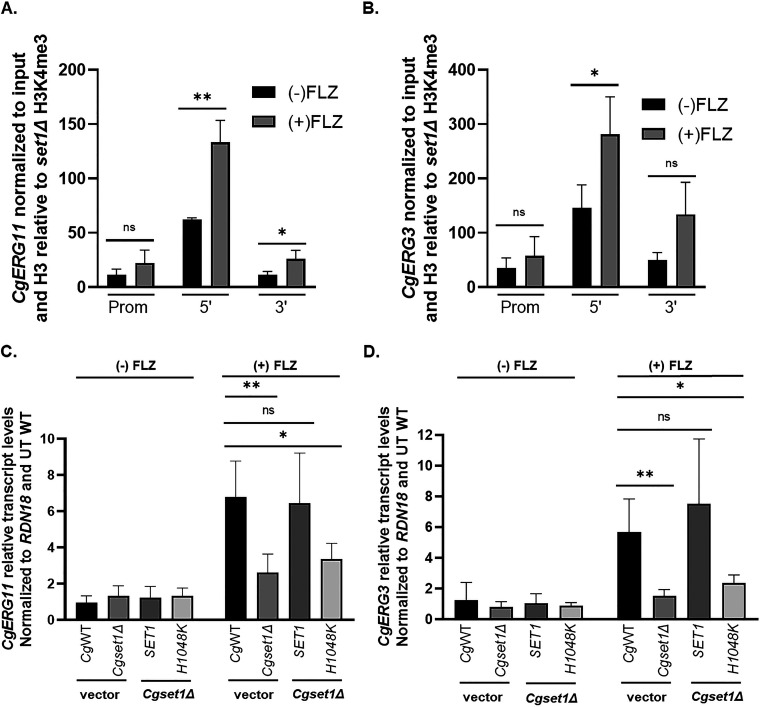
Because histone H3K4 trimethylation is associated with gene induction, we wanted to determine whether Set1 was directly catalyzing histone H3K4 methylation on chromatin at ERG loci. To determine whether histone H3K4 trimethylation was present at CgERG11 and CgERG3 chromatin, chromatin immunoprecipitation (ChIP) analysis was performed using histone H3K4 trimethyl-specific antibodies. As expected, histone H3K4 trimethylation is highly enriched at the 5′ ends of the open reading frame of CgERG11 and CgERG3 in untreated conditions and further enriched upon azole treatment corresponding to increased transcript levels of CgERG11 and CgERG3 in both exponential and saturated cell cultures (Fig. 6A and B; see also Fig. S3C and D).
FIG 6.
Set1-mediated histone H3K4 methylation is enriched on ERG gene chromatin and is required for azole induction of ERG genes. (A and B) ChIP analysis of histone H3K4 trimethylation levels at the promoter, 5′, and 3′ regions of ERG11 and ERG3 in a wild-type C. glabrata strain with or without 64 μg/mL fluconazole treatment. ChIP analysis was set relative to a set1Δ strain and normalized to histone H3 and DNA input levels. Data were analyzed from five biological replicates with three technical replicates each. *, P < 0.05. (C and D) Expression of indicated genes was determined in the indicated mutants treated with and without 64 μg/mL fluconazole for 3 h by qRT-PCR analysis. Gene expression analysis was set relative to the untreated wild-type containing an empty vector and expression was normalized to RDN18 mRNA levels. Data were analyzed from ≥3 biological replicates with three technical replicates each. Statistics were determined using the GraphPad Prism Student t test, version 9.2.0. ns, P > 0.05; *, P < 0.05; **, P < 0.01. Error bars represent the SD.
To confirm that this was due to the methyltransferase activity of Set1, we performed qRT-PCR transcript analysis using the Cg2001HTUset1Δ strain expressing pGRB2.0 only, pGRB2.0-CgSET1, and pGRB2.0-Cgset1H1048K. Cg2001HTU expressing pGRB2.0 only was used as our WT control. As shown in Fig. 6C and D, pGRB2.0-CgSET1 was able to induce ERG11 and ERG3 similar to WT cells under azole treatment, indicating that SET1 expression could rescue the ERG gene expression in the Cg2001HTUset1Δ strain. This rescue of ERG gene expression was dependent on the catalytic activity of Set1 since expression of pGRB2.0-Cgset1H1048K did not restore ERG gene expression under azole treatment. In addition, it looked similar to the Cg2001HTUset1Δ strain expressing pGRB2.0, indicating that the catalytic activity of Set1 is required for azole gene induction. Altogether, these data show that Set1-mediated histone H3K4 methylation directly targets the chromatin of ERG genes, and this epigenetic modification is required for azole induction of ERG genes.
Hypersensitivity to azole drugs is suppressed by exogenous ergosterol for C. glabrata set1Δ but not for a S. cerevisiae set1Δ.
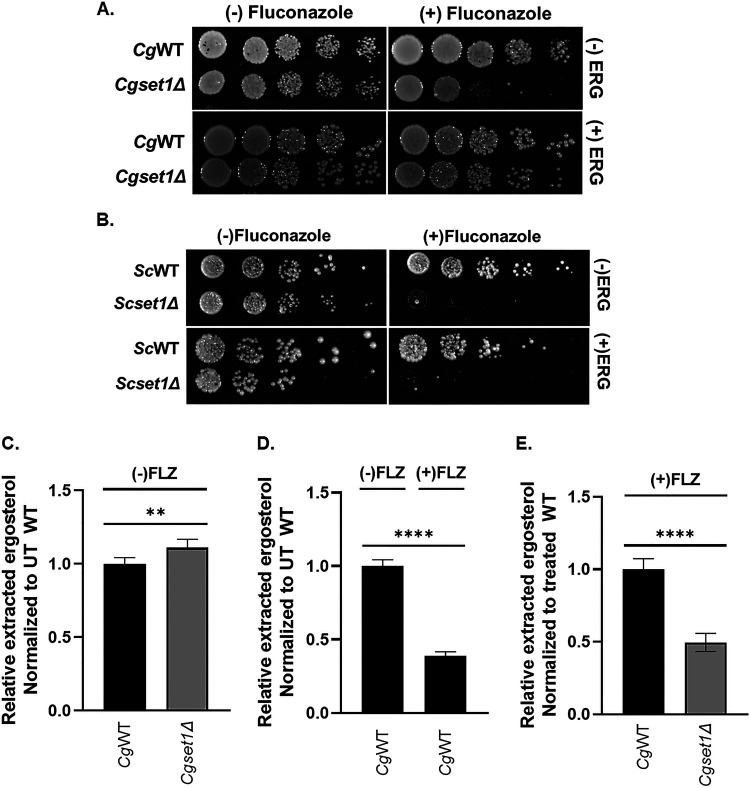
Based on our expression analysis and decreased expression of 12 genes involved in the late ergosterol biosynthesis, we predict further decreased endogenous ergosterol levels in C. glabrata strains lacking SET1, thus making the Cgset1Δ more susceptible to azole drugs. To determine whether endogenous ergosterol levels are reduced in Cgset1Δ strains upon azole treatment, CgWT and Cgset1Δ strains were plated on SC media supplemented with exogenous ergosterol with or without fluconazole. In support of our hypothesis and gene expression data, exogenous ergosterol suppressed azole hypersensitivity of a Cgset1Δ strain when grown in the presence of fluconazole while Cgset1Δ grown without ergosterol remained hypersensitive to azoles (Fig. 7A). Because azole hypersensitivity in a Scset1Δ is a consequence of altered ScPDR5 expression but not azole-induced gene expression, we hypothesized that exogenous sterols would not suppress azole hypersensitivity in the Scset1Δ. Based on serial-dilution spot assays, exogenous sterols did not suppress azole hypersensitivity in a Scset1Δ strain when grown in the presence of fluconazole and looked similar to Scset1Δ grown on SC media without ergosterol (Fig. 7B). A Scpdr5Δ was used as a control for azole hypersensitivity (see Fig. S2F).
FIG 7.
Loss of SET1 in C. glabrata results in reduced endogenous ergosterol levels upon azole treatment. (A and B) Fivefold serial dilution spot assays of the indicated S. cerevisiae and C. glabrata strains were grown on SC plates with or without 8 or 32 μg/mL fluconazole, respectively, and 10 or 20 μg/mL ergosterol, respectively. (C to E) Total ergosterol was extracted from C. glabrata WT and set1Δ strains treated with or without 64 μg/mL fluconazole and analyzed by HPLC. The figure is depicted as a ratio of ergosterol to cholesterol and relative to untreated or treated WT. Data were generated from six biological replicates. Statistics were determined using the GraphPad Prism Student t test, version 9.2.0. ****, P < 0.0001; **, P < 0.01. Error bars represent the SD.
To determine whether total ergosterol levels were altered in Cgset1Δ compared to CgWT strains upon azole treatment, nonpolar lipids were extracted from Cgset1Δ and CgWT strains with or without fluconazole treatment. Total ergosterol levels were determined using high performance liquid chromatography (HPLC) analysis and cholesterol was used as an internal control. Consistent with our gene expression analysis, HPLC analysis showed no difference in total ergosterol levels between untreated Cgset1Δ and CgWT strains (Fig. 7C; see also Fig. S5), which is consistent with no changes in ERG gene expression under untreated conditions (Fig. 6). As expected, upon fluconazole treatment the ergosterol levels decreased for both Cgset1Δ and CgWT (Fig. 7D; see also Fig. S5). However, the fluconazole-treated Cgset1Δ strain had ~50% less ergosterol than the fluconazole-treated CgWT strain (Fig. 7E; see also Fig. S5), which is also consistent with our data showing that loss of SET1 alters ERG gene expression only under treated conditions (Fig. 6). Our results suggest that the increased azole susceptibility of a Cgset1Δ strain is a consequence of defects in ERG gene expression resulting in decreases in ergosterol levels, whereas azole hypersensitivity in a Scset1Δ strain is a result of decreased efflux pump expression and not changes in ergosterol levels.
Loss of SET1 in C. glabrata clinical isolates SM1 and resistant strain SM3 results in increased azole sensitivity and altered CgERG11 gene expression.
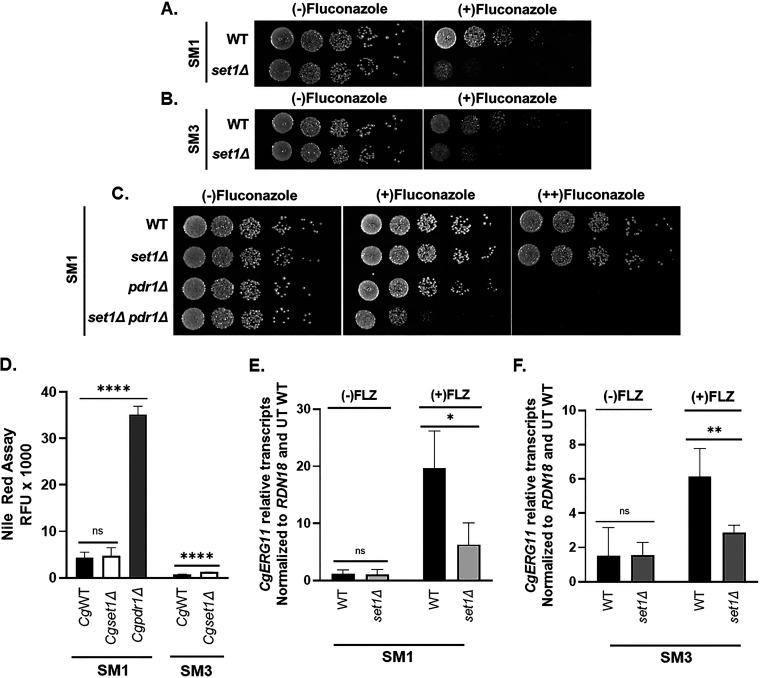
Because C. glabrata strains are genetically diverse in clinical isolates, we wanted to determine whether deleting SET1 in C. glabrata clinical isolates altered azole susceptibility. To determine the impact of deleting SET1 in C. glabrata clinical isolates, SET1 was deleted in the two characterized clinical isolates SM1 (fluconazole sensitive) and SM3 (fluconazole resistant) (53–55). Similar to the Cgset1Δ (Cg2001) strain, SM1 with CgSET1 deleted showed increased susceptibility to azole drugs as shown by serial-dilution spot assays (Fig. 8A). Furthermore, deleting CgSET1 from the clinically resistant SM3 strain, an isolate expressing a CgPDR1 gain-of-function mutation resulting in an increase in CgCDR1 expression, also resulted in increased susceptibility to azole treatment (Fig. 8B). Although deleting SET1 in the SM3 strain does not restore SM3 strains to SM1 azole susceptible levels, this is somewhat expected because Cg2001set1Δ strains do not alter PDR1 or CDR1 expression. Furthermore, a SM1set1Δpdr1Δ double deletion strain is more sensitive to fluconazole than either of the SM1pdr1Δ and SM1set1Δ when grown on plates with low concentrations of azole drugs (Fig. 8C). These genetic data are consistent with the idea that Set1 is altering another pathway and not drug efflux, which is consistent with our gene expression data. To determine whether SM1 and SM3 clinical isolates lacking SET1 had similar gene expression defects to Cg2001set1Δ strains, drug efflux function and CgERG11 expression were analyzed. The SM1set1Δ strain had similar Nile red fluorescence compared to SM1, indicating no defects in the function of drug efflux pumps (Fig. 8D). Although SM3set1Δ cells Nile red fluorescence was ~1.5-fold higher than SM3 cells, the overall drug efflux of both SM3 and SM3set1Δ strains were significantly more than SM1 and SM1set1Δ cells, a consequence of CDR1 overexpression (Fig. 8D). Similar to Cg2001set1Δ strains, the SM1set1Δ and SM3set1Δ strains failed to induce ERG11 expression under azole treatment (Fig. 8E and F).
FIG 8.
Loss of SET1 in C. glabrata clinical isolates SM1 and resistant strain SM3 results in increased azole sensitivity. (A to C) Fivefold serial dilution spot assays of the indicated C. glabrata clinical isolates were grown on SC plates with or without fluconazole and/or 20 μg/mL ergosterol. (D) RFU were measured as output in a Nile red assay to determine the amount of drug efflux in the indicated SM1 and SM3 clinical isolates treated with 64 or 256 μg/mL fluconazole. A Cgpdr1Δ strain was used as a control. Data were analyzed from ≥ 3 biological replicates with three technical replicates each. (E and F) Expression of ERG11 was determined from the indicated SM1 and SM3 clinical isolates treated with 64 and 256 μg/mL fluconazole, respectively, for 3 h by qRT-PCR analysis. Gene expression analysis was set relative to the untreated wild-type and expression was normalized to RDN18 mRNA levels. Data were analyzed from ≥3 biological replicates with three technical replicates each. Error bars represent the SD. Statistics were determined using the GraphPad Prism Student t test, version 9.2.0. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
DISCUSSION
In this study, we established that loss of Set1-mediated histone H3K4 methylation alters azole drug susceptibility in S. cerevisiae and C. glabrata by altering the expression of genes known to be involved in drug resistance. In S. cerevisiae, an azole-treated Scset1Δ strain shows decreased expression of ScPDR5 and consequently decreased drug efflux function, whereas the azole induction of ScERG11 was similar to WT strains. In contrast, decreased azole susceptibility in a Cgset1Δ strain was caused by a failure to completely induce CgERG genes under azole treatment and not a consequence of altered CgCDR1 and CgPDR1 expression levels or ability to efflux drugs. This azole-induced gene expression in C. glabrata was dependent on CgSet1 methyltransferase activity and correlated with gene-specific increases in histone H3K4 trimethylation on chromatin at ERG genes (see model, Fig. 9). Identifying and understanding how SET1 and likely other epigenetic factors contribute to species-specific drug susceptibility will be important for the development of alternative drug targets that could be used in combinatorial therapy for treating patients with drug-resistant fungal infections.
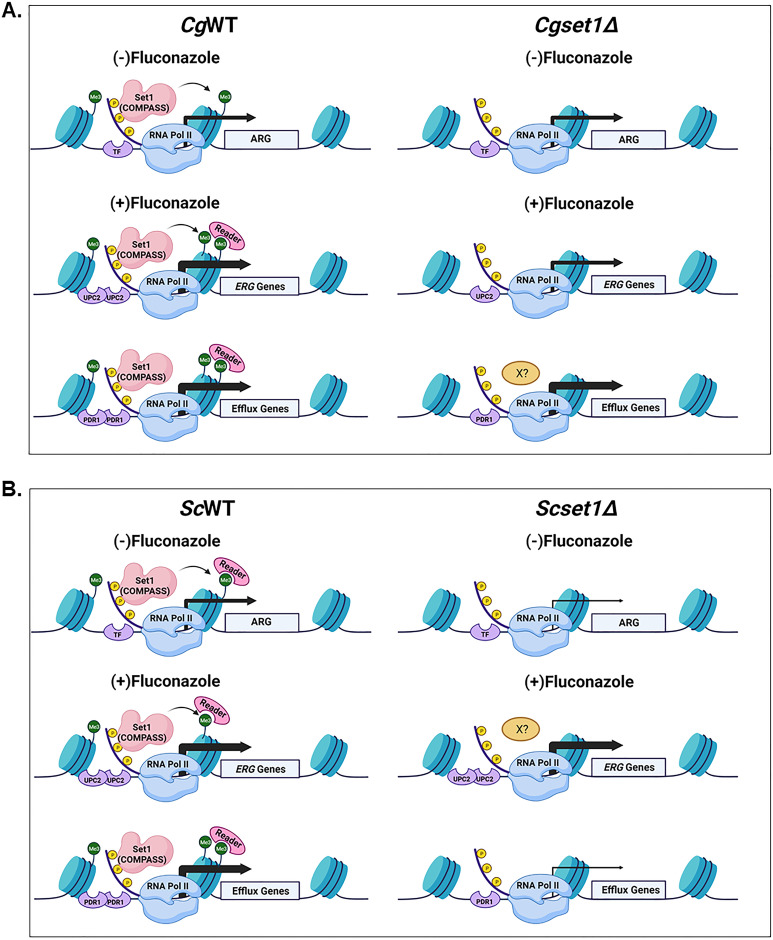
FIG 9.
Model depicting the role of Set1 contributing to azole susceptibility in a species-specific manner (Biorender). (A and B) Transcriptional activation recruits TFs, RNA polymerase II, and the Set1 complex to increase histone H3K4 methylation and permit proper induction of gene expression. This increase in methylation could also recruitment other cofactors/epigenetic regulators (e.g., Set3 and/or SAGA complex) that contain “reader” domains that recognize and bind to the H3K4 methyl mark to help in expression. In addition, other factors (“X?”) could be utilized to bypass the requirement of Set1. (A) In C. glabrata, azole-resistant genes (ARG) such as efflux pumps and ERG genes are induced upon azole treatment. In Cgset1Δ strains azole induction of ERG gene expression consequently ergosterol levels are decreased, but expression or function efflux pumps are not altered. (B) Under azole-treated conditions, efflux pump expression and function are decreased, but ERG gene expression is induced similar to WT levels.
Set1 is the catalytic subunit of a multisubunit protein complex called COMPASS that mono-, di-, and trimethylates histone H3K4. In this study, we show that C. glabrata Set1 is the sole histone H3K4 methyltransferase under log-phase growth conditions since deletion of SET1 abolishes all forms of histone H3K4 methylation similar to what is seen in S. cerevisiae and C. albicans (30, 31). Additionally, deletion of the genes encoding C. glabrata COMPASS complex subunits Swd1 and Bre2 lead to a loss of histone H3K4 methylation similar to their S. cerevisiae counterparts (Fig. 2C) (33, 34, 36, 38). However, deleting SPP1 in C. glabrata abolishes all histone H3K4 trimethylation and significantly reduces the levels of histone H3K4 mono- and dimethylation, which is in contrast to what is found in a Scspp1Δ strain where only histone H3K4 trimethylation is disrupted but retains WT levels of mono- and dimethylation (33, 34, 36, 38). We assume this difference in histone H3K4 methylation pattern is due to how CgSpp1 assembles with the COMPASS complex allowing CgSpp1 to have a greater impact on the overall catalytic activity of COMPASS. Interestingly, this pattern of methylation appears to correlate with sensitivity to azole drugs (Fig. 2B; see also Fig. S1A), where the Scspp1Δ strain grows more similarly to a WT strain than to the Cgspp1Δ strain when grown on azole-containing plates.
Our published observation has determined that S. cerevisiae and C. glabrata show hypersensitivity to azole drugs when lacking SET1 (29). However, until this study, the mechanistic role of SET1 under azole treatment in S. cerevisiae or C. glabrata has not been investigated. Our previous publication showed loss of SET1 in S. cerevisiae altered expression of genes involved in early (e.g., HMG1) and late ergosterol biosynthesis (e.g., ERG11) under untreated log-phase growing conditions where decreased ergosterol levels were also observed (Fig. 5G) (28). In this study, we confirmed similar decreases in ERG11 expression in the Scset1Δ strain under untreated conditions. However, azole-induced ERG11 expression was induced in the Scset1Δ strain similar to that in the ScWT strain. In addition, adding back exogenous ergosterol did not suppress azole sensitivity, indicating no defects in ergosterol levels under azole treatment. This lack of suppression was not due to aerobic exclusion of sterols of the Scset1Δ strain because our previous publication showed that yeast lacking SET1 can take up exogenous sterols under aerobic conditions (28). Intriguingly, brefeldin A (BFA) hypersensitivity of a Scset1Δ strain is suppressed when treated with exogenous sterols indicating distinct differences between BFA treatment and azole treatment (28).
In contrast to the Scset1Δ strain, a Cgset1Δ strain showed changes in ERG gene expression under azole-treated conditions but not untreated conditions, suggesting that histone H3K4 methylation is needed for azole-induced gene induction and not basal level expression in C. glabrata (Fig. 5E and F). In addition, all 12 genes involved in the late ergosterol biosynthesis pathway were differentially expressed in the Cgset1Δ strain compared to the CgWT strain, as well as three genes in the early pathways ERG9, ERG10, and ERG13, but not HMG1, indicating the importance of Set1 for this azole-induced pathway in C. glabrata but not in S. cerevisiae. We suspect that under azole treatment another epigenetic factor is contributing this role in S. cerevisiae and thus bypassing the requirement of Set1.
Although regulation of ergosterol biosynthesis has been shown to be coupled to expression of ABC transport genes such as CgCDR1 and its transcription factor Pdr1 (56, 57), compensatory changes in CgCDR1 and CgPDR1 expression levels were not observed in a Cgset1Δ strain treated with azole drugs (Fig. 4D and E). Furthermore, when SET1 was deleted in the clinical isolates SM1 (azole sensitive) and SM3 (azole resistant), both clinical isolates showed increased hypersensitivity to azole drugs compared to their respective isolates. Again, hypersensitivity was likely due to decreased ERG gene expression and not because of altered efflux pump function. Even though the SM3set1Δ strain is considered less resistant than SM3, the SM3set1Δ strain was not altered to that of SM1’s sensitivity levels. We suspect this is because the loss of CgSET1 does not significantly alter the expression of CgPDR1 or CgCDR1 expression levels to impact efflux and thus altering SM3’s susceptibility but not resistance. More investigation will be needed to understand how CDR1 and/or PDR1 are epigenetically regulated in C. glabrata sensitive and clinically azole-resistant strains (26, 58).
Raman et al. reported that loss of SET1 in C. albicans did not alter azole sensitivity but did decrease virulence in mice which was attributed to decreased epithelial adherence (30). Interestingly, several genes encoding cell wall proteins and adhesion factors are also downregulated in a Cgset1Δ strain, as determined by RNA sequencing. In addition, C. albicans strains lacking ERG11 or ERG3 produce avirulent hyphae, decrease adherence to epithelial cells, and reduce virulence in oral mucosal infections and disseminated candidiasis (59–61). Based on these observations, we suspect that the loss of CgSET1 could alter the virulence of C. glabrata under azole treatment. Additional studies will be needed to determine the in vivo contribution of SET1 in C. glabrata and other Candida species.
Our data suggest that histone H3K4 methylation is an epigenetic mechanism to help induce ERG gene expression when C. glabrata strains are exposed to azole drugs. We propose histone H3K4 methylation and other epigenetic marks contribute to C. glabrata’s natural resistance to azole drugs. Interestingly, several histone deacetylases (HDACs), such as CaHda1, CaRpd3, and CaHos2, have been implemented in azole resistance in C. albicans (22, 23, 62–64). In addition, HDAC inhibitors have been shown to have a synergistic effect on cells when combined with azoles and echinocandins (62, 63, 65, 66). Interestingly, the treatment of C. albicans with trichostatin A (TSA) lacks the trailing effect observed in MIC assays when using azole drugs, and the lack of trailing effect was attributed to reduced CDR and ERG gene expression (63, 67). In a similar manner, the Cgset1Δ strain also lacked a trailing effect in our MIC assays (unpublished data), which we suspect is specifically due to the lack of azole-induced ERG gene expression since CDR1 expression was not altered (Fig. 4D and 5). Furthermore, treatment of drug-resistant fungal pathogens, including various isolates of C. glabrata, with a Hos2 inhibitor MGCD290 showed synergy with azole drugs, which converted the MICs of azole treatment from resistant to susceptible (65). Because Hos2 is known to be a key component of the Set3 complex and the Set3 complex is recruited to chromatin via Set1-mediated histone H3K4 methylation (68, 69), it is likely MGCD290 is mediating its effect with azoles through inhibiting azole-induced ERG gene expression.
The occurrence of multidrug-resistant strains is increasing across all Candida species. In addition, with the development and identification of multidrug-resistant fungal species such as C. auris, a pathogen of urgent concern for the Centers for Disease Control and Prevention, it is imperative to find alternative treatment options. Our study, along with others, provides compelling evidence that epigenetic modifiers are playing key roles in altering fungal pathogenesis and drug susceptibility. Understanding these epigenetic events and the pathways they impact is needed to develop new drug therapies so that current and newly emerging multidrug-resistant fungal pathogens can be effectively treated.
MATERIALS AND METHODS
Plasmids and yeast strains.
All plasmids and yeast strains are described in Tables S1 and S2. C. glabrata 2001 (CBS138, ATCC 2001) and C. glabrata 2001HTU (ATCC 200989) were purchased from the American Type Culture Collection (43). A genomic fragment containing the CgSET1 promoter, 5′ UTR, open reading frame, and 3′ UTR was amplified by PCR and cloned into the pGRB2.0 plasmid. The pGRB2.0 plasmid was purchased from Addgene. Standard, site-directed mutagenesis was used to generate Cgset1H1048K. A 3×FLAG sequence was inserted between the endogenous promoter and ATG of the pGRB2.0-CgSET1 and pGRB2.0-Cgset1H1048K using fusion-based PCR and restriction enzyme cloning. Candida glabrata SET1, BRE2, SWD1, SPP1, and PDR1 genes were deleted via standard homologous recombination. Briefly, drug-resistant selection markers were PCR amplified with Ultramer DNA Oligos (IDT) using pAG32-HPHMX6 (hygromycin) or pAG25-NATMX6 (nourseothricin).
Serial-dilution spot and liquid growth assays.
For serial-dilution spot assays, yeast strains were inoculated in SC media and grown to saturation overnight. Yeast strains were diluted to an optical density at 600 nm (OD600) of 0.1 and grown in SC media to log phase with shaking at 30°C. The indicated strains were spotted in 5-fold dilutions starting at an OD600 of 0.01 on untreated SC plates or plates containing 16, 32, or 64 μg/mL fluconazole (Sigma-Aldrich, St. Louis, MO). Plates were grown at 30° for 1 to 3 days. For growth assays, the indicated yeast strains were inoculated in SC media and grown to saturation overnight. Yeast strains were diluted to an OD600 of 0.1 and grown in SC media to log phase with shaking at 30°C. The indicated strains were diluted to an OD600 of 0.0001 in 100 μL SC media. Cells were left untreated or treated with 64 μg/mL fluconazole and grown for 50 h with shaking at 30°C. The cell density OD600 was determined every 1 h using the Bio-Tek Synergy 4 multimode plate reader.
Cell extract and Western blot analysis.
Whole-cell extraction and Western blot analysis to detect histone modifications were performed as previously described (36, 70). Histone H3K4 methylation-specific antibodies were used as previously described: H3K4me1 (Upstate, catalog no. 07-436; 1:2,500), H3K4me2 (Upstate, catalog no. 07-030; 1:10,000), and H3K4me3 (Active motif 39159, 1:100,000) (28, 71). Histone H3 antibodies were used as our loading control (Abcam, catalog no. ab1791; 1:10,000).
RNA sequencing analysis.
The CBS138 Cg2001 WT and set1Δ strains were inoculated in SC media and grown to saturation overnight. Cells were diluted to an OD600 of 0.1 and recovered to log phase for 3 h with shaking at 30°C. Prior to treatment, cells were collected for the untreated sample and zero time point. Cultures were treated at an OD600 of 0.2 with 64 μg/mL fluconazole (Sigma-Aldrich) dissolved in dimethyl sulfoxide as previously described (72). Cells were collected after 3 h. The total RNA of three biological replicates for each condition and sample was isolated by standard acid phenol purification and treated with DNase (Ambion), and the total RNA was purified using standard acid phenol purification. The quality of the RNA was tested using an Agilent Bioanalyzer 2100 using a High Sensitivity DNA Chip. The cDNA library was prepared by the Purdue Genomics Facility using the TruSeq stranded kit with poly(A) selection (Illumina) according to the manufacturer’s instructions. The software Trimmomatic v.0.39 was used to trim reads, removing adapters and low-quality bases (73). STAR v.2.5.4b was used to align reads to the C. glabrata CBS138 reference genome, version s02-m07-r23 (74). One mismatch was allowed per read. HTSeq v.0.6.1 was used to generate the gene count matrix on “intersection-nonempty” mode (75). R version 3.5.1 and Bioconductor release 3.6 were used to perform all statistical analyses on the RNA-seq data. The intersection of genes identified as statistically significantly differentially expressed with a Benjamini-Hochberg-corrected FDR of <5% by DESeq2 v.1.18.0 was used in downstream analyses (76, 77).
Quantitative real-time PCR analysis.
RNA was isolated from cells by standard acid phenol purification. Reverse transcription was performed using an All-in-One 5× RT Mastermix kit (ABM, Richmond, Canada). Primer Express 3.0 software was used for designing primers, and qRT-PCR was performed as previously described (28, 78, 79). At least three biological replicates, including three technical replicates, were performed for all samples. Data were analyzed using the comparative CT method (2–ΔΔCT), where RDN18 (18S rRNA) was used as an internal control. All samples were normalized to an untreated, untagged WT strain.
MIC assay.
MIC assays were performed based on a modified version of the CLSI method for testing yeast, 3rd edition (80). Briefly, yeast strains were inoculated in SC media and grown to saturation overnight. The indicated strains were diluted to an OD600 of 0.003 in in SC or RPMI media. Cells were mixed with fluconazole (Cayman Chemical) for a final volume of 100 μL per well in a 96-well polystyrene microplate, with concentrations of fluconazole ranging from 0 to 256 μg/mL. Plates were incubated at 35°C, and MICs were recorded at 24 h. MICs were determined visually where >90% of growth was inhibited.
Nile red assay.
Fluorescence-based Nile red assays were performed as previously described (45). Briefly, cells were grown overnight in SC media to saturation. Cells were back diluted to an OD600 of 0.1 and grown at 30°C for 6 h. Cells were collected and then washed with phosphate-buffered saline (PBS) twice and incubated in 1.5 mL of PBS plus 2% glucose for 1 h. Next, 2.87 μL of a 1-mg/mL stock of Nile red (Sigma) was added to each sample, followed by incubation at 30°C shaking for an additional 30 min. Samples were washed twice with PBS and placed in triplicate in a black 96-well flat-bottomed polystyrene microplate. Fluorescence was detected using a Bio-Tek Synergy 4 multimode plate reader using an excitation wavelength of 553 nm and an emission wavelength of 636 nm.
Chromatin immunoprecipitation.
ZipChIP was performed as previously described (71). Briefly, 50-mL cultures were grown to log phase (OD600 of 0.6) in SC complete media at 30°C shaking. Treated cells were dosed with 64 μg/mL fluconazole (Cayman Chemical) at an OD600 of 0.2 for 3 h. In addition, cultures were grown to saturation, back diluted to an OD600 of 0.6, treated with 64 μg/mL fluconazole for 3 h, and collected. Cells were formaldehyde cross-linked and harvested as previously described (71). Cell lysates were precleared with 5 μL of unbound protein G-magnetic beads for 30 min with rotation at 4°C. A total of 12.5 μL of precleared lysate was immunoprecipitated with 10 μL of protein G-magnetic beads (10004D; Life Technologies) conjugated to 1 μL of histone H3K4me3 antibody (Millipore, catalog no. 07-473) or histone H3 antibody (Abcam, catalog no. ab1791). Probe sets used in qRT-PCR are described in Table S5.
Ergosterol extraction.
Ergosterol extraction was performed previously described (28). Cultures were grown overnight in SC minimal media. Saturated cultures were backdiluted to an OD600 of 0.1 and were grown at 30°C to log phase (OD600 0.6) with or without 64 μg/mL fluconazole treatment. Sterols were extracted from yeast using 4 M potassium hydroxide in 70% (vol/vol) ethanol at 85°C for 1 h. After extraction, nonpolar lipids were separated using methanol two times. Nonpolar sterols were crystallized after evaporation of the N-hexane and dissolved in 100% methanol. Samples were then analyzed by HPLC using a C18 column with the flow rate set at 1 mL/min of 100% methanol. Ergosterol was detected at 280 nm. Cholesterol was added as an extraction internal control and was detected at 210 nm.
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health to S.D.B. (AI136995), the Purdue Department of Biochemistry Bird Stair Fellowship (to K.M.B.), Purdue Center for Cancer Research (grant P30CA023168: DNA Sequencing Shared Resource and Collaborative Core for Cancer Bioinformatics at Purdue), and The Walther Cancer Foundation and the IU Simon Cancer Center (grant P30CA082709). Additional funding support was provided by the NIFA 1007570 (S.D.B.).
We thank the Purdue Bioinformatics Core for their pipelines and bioinformatics software and P. David Rogers, St. Jude Children’s Research Hospital, for the C. glabrata SM1 and SM3 clinical isolates.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H, Ebbers J, Geurtz L, Stefanik D, Major Y, Edmond MB, Wenzel RP, Seifert H. 2014. Nosocomial bloodstream infections due to Candida spp. in the USA: species distribution, clinical features, and antifungal susceptibilities. Int J Antimicrob Agents 43:78–81. 10.1016/j.ijantimicag.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Slavin M, Nguyen Q, Marriott D, Playford EG, Ellis D, Sorrell T, Study AC, The Australian Candidemia Study . 2006. Active surveillance for candidemia. Emerg Infect Dis 12:1508–1516. 10.3201/eid1210.060389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chouhan S, Kallianpur S, Prabhu KT, Tijare M, Kasetty S, Gupta S. 2019. Candidal prevalence in diabetics and its species identification. Int J Appl Basic Med Res 9:49–54. 10.4103/ijabmr.IJABMR_259_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khatib R, Johnson LB, Fakih MG, Riederer K, Briski L. 2016. Current trends in candidemia and species distribution among adults: Candida glabrata surpasses C. albicans in diabetic patients and abdominal sources. Mycoses 59:781–786. 10.1111/myc.12531. [DOI] [PubMed] [Google Scholar]
- 6.Hachem R, Hanna H, Kontoyiannis D, Jiang Y, Raad I. 2008. The changing epidemiology of invasive candidiasis: Candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer 112:2493–2499. 10.1002/cncr.23466. [DOI] [PubMed] [Google Scholar]
- 7.Vermitsky JP, Edlind TD. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob Agents Chemother 48:3773–3781. 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelz RK, Lipsett PA, Swoboda SM, Diener-West M, Powe NR, Brower RG, Perl TM, Hammond JM, Hendrix CW. 2000. Candida infections: outcome and attributable ICU costs in critically ill patients. J Intensive Care Med 15:255–261. 10.1046/j.1525-1489.2000.00255.x. [DOI] [Google Scholar]
- 9.Rentz AM, Halpern MT, Bowden R. 1998. The impact of candidemia on length of hospital stay, outcome, and overall cost of illness. Clin Infect Dis 27:781–788. 10.1086/514955. [DOI] [PubMed] [Google Scholar]
- 10.Pfaller MA, Jones RN, Messer SA, Edmond MB, Wenzel RP, Grp SP. 1998. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE program. Diagn Microbiol Infect Dis 30:121–129. 10.1016/S0732-8893(97)00192-2. [DOI] [PubMed] [Google Scholar]
- 11.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1-50–50. 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whaley SG, Rogers PD. 2016. Azole resistance in Candida glabrata. Curr Infect Dis Rep 18:41. 10.1007/s11908-016-0554-5. [DOI] [PubMed] [Google Scholar]
- 13.Tscherner M, Schwarzmüller T, Kuchler K. 2011. Pathogenesis and antifungal drug resistance of the human fungal pathogen Candida glabrata. Pharmaceuticals (Basel) 4:169–186. 10.3390/ph4010169. [DOI] [Google Scholar]
- 14.Tsai HF, Krol AA, Sarti KE, Bennett JE. 2006. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob Agents Chemother 50:1384–1392. 10.1128/AAC.50.4.1384-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galkina KV, Okamoto M, Chibana H, Knorre DA, Kajiwara S. 2020. Deletion of CDR1 reveals redox regulation of pleiotropic drug resistance in Candida glabrata. Biochimie 170:49–56. 10.1016/j.biochi.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Warrilow AG, Mullins JG, Hull CM, Parker JE, Lamb DC, Kelly DE, Kelly SL. 2012. S279 point mutations in Candida albicans sterol 14-α demethylase (CYP51) reduce in vitro inhibition by fluconazole. Antimicrob Agents Chemother 56:2099–2107. 10.1128/AAC.05389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, Gygax SE, Morschhäuser J, Rogers PD. 2012. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell 11:1289–1299. 10.1128/EC.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanguinetti M, Posteraro B, Fiori B, Ranno S, Torelli R, Fadda G. 2005. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob Agents Chemother 49:668–679. 10.1128/AAC.49.2.668-679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD. 2016. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol 7:2173. 10.3389/fmicb.2016.02173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanglard D, Ischer F, Calabrese D, Majcherczyk PA, Bille J. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata are involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob Agents Chemother 43:2753–2765. 10.1128/AAC.43.11.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Kane CJ, Hyland EM. 2019. Yeast epigenetics: the inheritance of histone modification states. Biosci Rep 39:BSR20182006. 10.1042/BSR20182006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Cai Q, Mei H, Zhou X, Shen Y, Li D, Liu W. 2015. The Rpd3/Hda1 family of histone deacetylases regulates azole resistance in Candida albicans. J Antimicrob Chemother 70:1993–2003. 10.1093/jac/dkv070. [DOI] [PubMed] [Google Scholar]
- 23.Robbins N, Leach MD, Cowen LE. 2012. Lysine deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. Cell Rep 2:878–888. 10.1016/j.celrep.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shivarathri R, Tscherner M, Zwolanek F, Singh NK, Chauhan N, Kuchler K. 2019. The fungal histone acetyl transferase Gcn5 controls virulence of the human pathogen Candida albicans through multiple pathways. Sci Rep 9:9445. 10.1038/s41598-019-45817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopes da Rosa J, Boyartchuk VL, Zhu LJ, Kaufman PD. 2010. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc Natl Acad Sci USA 107:1594–1599. 10.1073/pnas.0912427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orta-Zavalza E, Guerrero-Serrano G, Gutierrez-Escobedo G, Canas-Villamar I, Juarez-Cepeda J, Castano I, De Las Penas A. 2013. Local silencing controls the oxidative stress response and the multidrug resistance in Candida glabrata. Mol Microbiol 88:1135–1148. 10.1111/mmi.12247. [DOI] [PubMed] [Google Scholar]
- 27.Filler EE, Liu Y, Solis NV, Wang L, Diaz LF, Edwards JE, Filler SG, Yeaman MR. 2021. Identification of Candida glabrata transcriptional regulators that govern stress resistance and virulence. Infect Immun 89:e00146-20. 10.1128/IAI.00146-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.South PF, Harmeyer KM, Serratore ND, Briggs SD. 2013. H3K4 methyltransferase Set1 is involved in maintenance of ergosterol homeostasis and resistance to brefeldin A. Proc Natl Acad Sci USA 10:E1016–E1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serratore ND, Baker KM, Macadlo LA, Gress AR, Powers BL, Atallah N, Westerhouse KM, Hall MC, Weake VM, Briggs SD. 2018. A novel sterol-signaling pathway governs azole antifungal drug resistance and hypoxic gene repression in Saccharomyces cerevisiae. Genetics 208:1037–1055. 10.1534/genetics.117.300554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman S, Nguyen M, Zhang Z, Cheng S, Jia H, Weisner N, Iczkowski K, Clancy C. 2006. Candida albicans SET1 encodes a histone 3 lysine 4 methyltransferase that contributes to the pathogenesis of invasive candidiasis. Mol Microbiol 60:697–709. 10.1111/j.1365-2958.2006.05121.x. [DOI] [PubMed] [Google Scholar]
- 31.Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15:3286–3295. 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20:7137–7148. 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA 99:90–94. 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dehe PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodriguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, Shevchenko A, Rosaleny LE, Tordera V, Chavez S, Stewart AF, Geli V. 2006. Protein interactions within the Set1 complex and their roles in the regulation of histone 3-lysine 4-methylation. J Biol Chem 281:35404–35412. 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi YH, Westfield GH, Oleskie AN, Trievel RC, Shilatifard A, Skiniotis G. 2011. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci USA 108:20526–20531. 10.1073/pnas.1109360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. 2012. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. J Biol Chem 287:2652–2665. 10.1074/jbc.M111.280867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81:65–95. 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller JE, Canze M, Bryk M. 2006. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics 173:557–567. 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eissenberg JC, Shilatifard A. 2010. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol 339:240–249. 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zordan RE, Ren Y, Pan S-J, Rotondo G, De Las Peñas A, Iluore J, Cormack BP. 2013. Expression plasmids for use in Candida glabrata. G3 (Bethesda) 3:1675–1686. 10.1534/g3.113.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlichter A, Cairns BR. 2005. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J 24:1222–1231. 10.1038/sj.emboj.7600607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soares LM, Radman-Livaja M, Lin SG, Rando OJ, Buratowski S. 2014. Feedback control of Set1 protein levels is important for proper H3K4 methylation patterns. Cell Rep 6:961–972. 10.1016/j.celrep.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitada K, Yamaguchi E, Arisawa M. 1995. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene 165:203–206. 10.1016/0378-1119(95)00552-h. [DOI] [PubMed] [Google Scholar]
- 44.Vermitsky JP, Earhart KD, Smith WL, Homayouni R, Edlind TD, Rogers PD. 2006. Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol Microbiol 61:704–722. 10.1111/j.1365-2958.2006.05235.x. [DOI] [PubMed] [Google Scholar]
- 45.Tsao S, Weber S, Cameron C, Nehme D, Ahmadzadeh E, Raymond M. 2016. Positive regulation of the Candida albicans multidrug efflux pump Cdr1p function by phosphorylation of its N-terminal extension. J Antimicrob Chemother 71:3125–3134. 10.1093/jac/dkw252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivnitski-Steele I, Holmes A, Lamping E, Monk B, Cannon R, Sklar L. 2009. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal Biochem 394:87–91. 10.1016/j.ab.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whaley SG, Zhang Q, Caudle KE, Rogers PD. 2018. Relative contribution of the ABC transporters Cdr1, Pdh1, and Snq2 to azole resistance in Candida glabrata. Antimicrob Agents Chemother 62:e01070-18. 10.1128/AAC.01070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flowers SA, Colon B, Whaley SG, Schuler MA, Rogers PD. 2015. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother 59:450–460. 10.1128/AAC.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly SL, Lamb DC, Kelly DE. 1997. Sterol 22-desaturase, cytochrome P45061, possesses activity in xenobiotic metabolism. FEBS Lett 412:233–235. 10.1016/s0014-5793(97)00785-0. [DOI] [PubMed] [Google Scholar]
- 50.Morio F, Pagniez F, Lacroix C, Miegeville M, Le Pape P. 2012. Amino acid substitutions in the Candida albicans sterol Δ5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. J Antimicrob Chemother 67:2131–2138. 10.1093/jac/dks186. [DOI] [PubMed] [Google Scholar]
- 51.Kelly SL, Lamb DC, Kelly DE, Manning NJ, Loeffler J, Hebart H, Schumacher U, Einsele H. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett 400:80–82. 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 52.Watson PF, Rose ME, Ellis SW, England H, Kelly SL. 1989. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem Biophys Res Commun 164:1170–1175. 10.1016/0006-291x(89)91792-0. [DOI] [PubMed] [Google Scholar]
- 53.Caudle KE, Barker KS, Wiederhold NP, Xu L, Homayouni R, Rogers PD. 2011. Genomewide expression profile analysis of the Candida glabrata Pdr1 regulon. Eukaryot Cell 10:373–383. 10.1128/EC.00073-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magill SS, Shields C, Sears CL, Choti M, Merz WG. 2006. Triazole cross-resistance among Candida spp.: case report, occurrence among bloodstream isolates, and implications for antifungal therapy. J Clin Microbiol 44:529–535. 10.1128/JCM.44.2.529-535.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whaley SG, Caudle KE, Vermitsky JP, Chadwick SG, Toner G, Barker KS, Gygax SE, Rogers PD. 2014. UPC2A is required for high-level azole antifungal resistance in Candida glabrata. Antimicrob Agents Chemother 58:4543–4554. 10.1128/AAC.02217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vu B, Thomas G, Moye-Rowley W. 2019. Evidence that ergosterol biosynthesis modulates activity of the Pdr1 transcription factor in Candida glabrata. mBio 10:e00934-19. 10.1128/mBio.00934-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moye-Rowley WS. 2020. Linkage between genes involved in azole resistance and ergosterol biosynthesis. PLoS Pathog 16:e1008819. 10.1371/journal.ppat.1008819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu SJ, Chang YL, Chen YL. 2018. Deletion of ADA2 increases antifungal drug susceptibility and virulence in Candida glabrata. Antimicrob Agents Chemother 62:e01924-17. 10.1128/AAC.01924-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y, Liao M, Zhu C, Hu Y, Tong T, Peng X, Li M, Feng M, Cheng L, Ren B, Zhou X. 2018. ERG3 and ERG11 genes are critical for the pathogenesis of Candida albicans during the oral mucosal infection. Int J Oral Sci 10:9. 10.1038/s41368-018-0013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Becker JM, Kauffman SJ, Hauser M, Huang L, Lin M, Sillaots S, Jiang B, Xu D, Roemer T. 2010. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc Natl Acad Sci USA 107:22044–22049. 10.1073/pnas.1009845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyazaki T, Miyazaki Y, Izumikawa K, Kakeya H, Miyakoshi S, Bennett JE, Kohno S. 2006. Fluconazole treatment is effective against a Candida albicans erg3/erg3 mutant in vivo despite in vitro resistance. Antimicrob Agents Chemother 50:580–586. 10.1128/AAC.50.2.580-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L, Xu W. 2015. Histone deacetylase inhibitors for enhancing activity of antifungal agent: a patent evaluation of WO2014041424(A1). Expert Opin Ther Pat 25:237–240. 10.1517/13543776.2014.981256. [DOI] [PubMed] [Google Scholar]
- 63.Mai A, Rotili D, Massa S, Brosch G, Simonetti G, Passariello C, Palamara AT. 2007. Discovery of uracil-based histone deacetylase inhibitors able to reduce acquired antifungal resistance and trailing growth in Candida albicans. Bioorg Med Chem Lett 17:1221–1225. 10.1016/j.bmcl.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 64.Hnisz D, Majer O, Frohner IE, Komnenovic V, Kuchler K. 2010. The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog 6:e1000889. 10.1371/journal.ppat.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfaller MA, Messer SA, Georgopapadakou N, Martell LA, Besterman JM, Diekema DJ. 2009. Activity of MGCD290, a Hos2 histone deacetylase inhibitor, in combination with azole antifungals against opportunistic fungal pathogens. J Clin Microbiol 47:3797–3804. 10.1128/JCM.00618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfaller MA, Rhomberg PR, Messer SA, Castanheira M. 2015. In vitro activity of a Hos2 deacetylase inhibitor, MGCD290, in combination with echinocandins against echinocandin-resistant Candida species. Diagn Microbiol Infect Dis 81:259–263. 10.1016/j.diagmicrobio.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 67.Smith WL, Edlind TD. 2002. Histone deacetylase inhibitors enhance Candida albicans sensitivity to azoles and related antifungals: correlation with reduction in CDR and ERG upregulation. Antimicrob Agents Chemother 46:3532–3539. 10.1128/AAC.46.11.3532-3539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim T, Buratowski S. 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137:259–272. 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart AF. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev 15:2991–3004. 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fingerman IM, Wu CL, Wilson BD, Briggs SD. 2005. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J Biol Chem 280:28761–28765. 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harmeyer KM, South PF, Bishop B, Ogas J, Briggs SD. 2015. Immediate chromatin immunoprecipitation and on-bead quantitative PCR analysis: a versatile and rapid ChIP procedure. Nucleic Acids Res 43:e38. 10.1093/nar/gku1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agarwal AK, Rogers PD, Baerson SR, Jacob MR, Barker KS, Cleary JD, Walker LA, Nagle DG, Clark AM. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J Biol Chem 278:34998–35015. 10.1074/jbc.M306291200. [DOI] [PubMed] [Google Scholar]
- 73.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Givanna H Putri, Simon Anders, Paul Theodor Pyl, John E Pimanda, Fabio Zanini. 2022. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics, btac166. 10.1093/bioinformatics/btac166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Statist Methodol 57:289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 77.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, Serratore ND, Briggs SD. 2017. N-ICE plasmids for generating N-terminal 3×FLAG-tagged genes that allow inducible, constitutive or endogenous expression in Saccharomyces cerevisiae. Yeast 34:223–235. 10.1002/yea.3226. [DOI] [PubMed] [Google Scholar]
- 79.South PF, Fingerman IM, Mersman DP, Du HN, Briggs SD. 2010. A conserved interaction between the SDI domain of Bre2 and the Dpy-30 domain of Sdc1 is required for histone methylation and gene expression. J Biol Chem 285:595–607. 10.1074/jbc.M109.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard, 3rd ed. CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5 and Tables S1 to S5. Download aac.02250-21-s0001.pdf, PDF file, 1.1 MB (1.1MB, pdf)
Table S6. Download aac.02250-21-s0002.xlsx, XLSX file, 0.2 MB (204KB, xlsx)
Table S7. Download aac.02250-21-s0003.xlsx, XLSX file, 0.02 MB (25.2KB, xlsx)