ABSTRACT
Multidrug-resistant (MDR) Pseudomonas aeruginosa infections are a major clinical challenge. Many isolates are carbapenem resistant, which severely limits treatment options; thus, novel therapeutic combinations, such as imipenem-relebactam (IMI/REL), ceftazidime-avibactam (CAZ/AVI), ceftolozane-tazobactam (TOL/TAZO), and meropenem-vaborbactam (MEM/VAB) were developed. Here, we studied two extensively drug-resistant (XDR) P. aeruginosa isolates, collected in the United States and Mexico, that demonstrated resistance to IMI/REL. Whole-genome sequencing (WGS) showed that both isolates contained acquired GES β-lactamases, intrinsic PDC and OXA β-lactamases, and disruptions in the genes encoding the OprD porin, thereby inhibiting uptake of carbapenems. In one isolate (ST17), the entire C terminus of OprD deviated from the expected amino acid sequence after amino acid G388. In the other (ST309), the entire oprD gene was interrupted by an ISPa1328 insertion element after amino acid D43, rendering this porin nonfunctional. The poor inhibition by REL of the GES β-lactamases (GES-2, -19, and -20; apparent Ki of 19 ± 2 μM, 23 ± 2 μM, and 21 ± 2 μM, respectively) within the isolates also contributed to the observed IMI/REL-resistant phenotype. Modeling of REL binding to the active site of GES-20 suggested that the acylated REL is positioned in an unstable conformation as a result of a constrained Ω-loop.
KEYWORDS: Pseudomonas aeruginosa, relebactam, imipenem, GES β-lactamase, OXA β-lactamase, GES-19, GES-20, OprD deficient, relebactam resistance
INTRODUCTION
Pseudomonas aeruginosa is a health care-associated pathogen that causes severe infection in patients who are immunocompromised. Each year in the United States, more than 32,000 health care-associated P. aeruginosa infections occur, resulting in 2,700 deaths (https://www.cdc.gov/drugresistance/pdf/threats-report/pseudomonas-aeruginosa-508.pdf). Thus, the Centers for Disease Control and Prevention (CDC) designated P. aeruginosa a “serious threat” pathogen (https://www.cdc.gov/drugresistance/biggest-threats.html#pse). Overcoming antibiotic resistance, particularly carbapenem resistance, in multidrug-resistant (MDR) P. aeruginosa is a major clinical challenge, as diverse mechanisms are responsible for this phenotype. These mechanisms include drug inactivation by β-lactamases encoded by multiple bla genes, reduced drug uptake due to porin changes, increased drug removal by efflux pump overexpression, and reduced drug affinity by alterations in penicillin-binding proteins (PBPs) (1, 2).
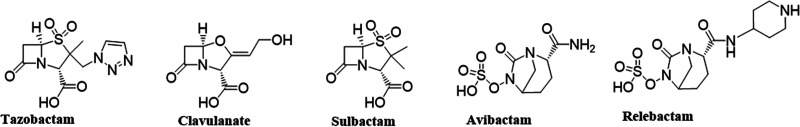
To combat the evolution of MDR in P. aeruginosa and other Gram-negative bacteria, novel therapeutics were developed. They include ceftazidime-avibactam (CAZ/AVI), ceftolozane-tazobactam (TOL/TAZO), meropenem-vaborbactam (MEM/VAB), and the diazabicyclooctane (DBO) β-lactamase inhibitor relebactam (REL), which is being used in combination with imipenem (IMI) and cilastatin. A comparison of β-lactam inhibitors is shown in Fig. 1. The latter β-lactam–β-lactamase inhibitor combination has microbiological and pharmacological properties that are designed to overcome common resistance mechanisms in P. aeruginosa and other MDR organisms. REL possesses a broad spectrum of activity that includes inhibition of class A, class C, and some class D β-lactamases (3–5). The U.S. Food and Drug Administration approved imipenem-cilastatin-relebactam (IMI/REL; Recarbrio) for the treatment of complicated urinary tract and intraabdominal infections and hospital-acquired and ventilator-associated bacterial pneumonia in adults (https://www.jwatch.org/fw116709/2020/06/07/fda-approves-imipenem-cilastatin-relebactam-recarbrio, https://www.jwatch.org/na49571/2019/08/02/fda-approves-imipenem-cilastatin-relebactam-recarbrio). The IMI/REL combination represents an alternative to CAZ/AVI, as the partner β-lactam is a member of the carbapenem class as opposed to an expanded-spectrum cephalosporin. It also has been shown that IMI/REL is not affected by drug efflux systems such as MexAB-OprM, MexCD-OprJ, and MexXY-OprM (6) or by substitutions in the Ω-loop of KPC-2 and KPC-3 β-lactamases that confer CAZ/AVI resistance (7–9).
FIG 1.
Structures of β-lactam-based β-lactamase inhibitors (tazobactam, clavulanate, and sulbactam) and diazabicyclooctane inhibitors (DBO; AVI and REL).
However, as is often the case with new antimicrobial agents, resistance to IMI/REL is observed in P. aeruginosa (10–15). A recent study of isolates collected in Spain demonstrated that susceptibility to IMI/REL was 97.3% (10). In contrast, multiple other surveillance studies report lower susceptibility to IMI/REL (∼90%, with ∼30% of all IMI-resistant P. aeruginosa isolates also being resistant to IMI/REL). In one of the largest studies to date, 14,813 P. aeruginosa isolates were collected by the SMART global surveillance program, spanning years 2009 to 2016 (6). Of those that tested IMI nonsusceptible, 3,747 isolates underwent further phenotypic and genotypic analysis. In that analysis, 1,200 (32%) remained nonsusceptible to IMI/REL; 42% of these encoded class B metallo-β-lactamases, and 11% encoded class A Guiana extended-spectrum (GES) enzymes, while class D carbapenemases were not detected. More specifically, another recent study demonstrated that REL did not restore susceptibility to GES-5-producing P. aeruginosa isolates (16).
Here, we present analysis that details the molecular and biochemical characterization of two IMI/REL-, CAZ/AVI-, TOL/TAZO-, and MEM/VAB-resistant P. aeruginosa isolates that were collected in the United States and Mexico as part of an international surveillance study. Our objective was to determine the mechanisms that contributed to this observed resistance phenotype. Both isolates contained relebactam-resistant class A GES, class C Pseudomonas-derived cephalosporinase (PDC; intrinsic β-lactamase of P. aeruginosa), and class D oxacillinase (intrinsic OXA) β-lactamases. Additionally, OprD alterations coupled with relebactam resistance of the GES β-lactamases in each isolate appear to play the defining role in resistance to IMI/REL. Of equal importance, we illustrate how the interplay of this constellation of resistance determinants also confers resistance to other contemporary new β-lactam–β-lactamase inhibitor combinations as well as other antibiotics.
RESULTS
AST.
Antimicrobial susceptibility testing (AST) results of the two highly resistant P. aeruginosa isolates are summarized in Tables 1 and 2. Both isolates (CLB 24388 and EM 2972704) were resistant to all antibiotics tested by broth microdilution, which include amikacin (AMK), aztreonam (ATM), cefepime (FEP), cefotaxime (TAX), CAZ, ciprofloxacin (CIP), gentamicin (GEN), IMI, levofloxacin (LVX), meropenem (MEM), piperacillin-tazobactam (TZP), and tobramycin (TOB). Broth macrodilution was next performed to accurately determine colistin (CST) susceptibility; both isolates displayed intermediate resistance (≤0.5 mg/liter for CLB 24388 and 1 mg/liter for EM 2972704). In addition, both isolates were resistant to MEM/VAB, TOL/TAZO, IMI/REL, and CAZ/AVI. The combination of CAZ/AVI/ATM in disk diffusion assays increased the zone diameters from 15 mm (CAZ/AVI alone) to 30 mm for CLB 24388 and from 6 mm (CAZ/AVI alone) to 30 mm for EM 2972704.
TABLE 1.
MICs of P. aeruginosa isolatesa
| Isolate no. | MIC (mg/liter) for: |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | ATM | FEP | TAX | CAZ | CIP | IMI | MEM | LVX | GEN | TZP | TOB | CAZ/AVI | MEM/VAB | TOL/TAZO | CST | IMI | IMI/REL | |
| CLB 24388 | >32 | >16 | >16 | >32 | >16 | >2 | >8 | >8 | >4 | >8 | >64/4 | >8 | 16/4 | 32/8 | 64/4 | ≤0.5 | 32 | 8/4 |
| EM 2972704 | >32 | >16 | >16 | >32 | >16 | >2 | >8 | >8 | >4 | >8 | >64/4 | >8 | >256/4 | >256/8 | >256/4 | 1 | 64 | 32/4 |
Antimicrobial susceptibility tests were interpreted according to 2021 CLSI criteria for Pseudomonas aeruginosa: for aztreonam (ATM), cefepime (FEP), and ceftazidime (CAZ), MIC of ≤8 mg/liter is susceptible (S), MIC of 16 mg/liter is intermediate (I), and MIC of ≥32 mg/liter is resistant (R); for meropenem (MEM) and imipenem (IMI), MIC of ≤2 mg/liter is S, MIC of 4 mg/liter is I, and MIC ≥ 8 mg/liter is R; for amikacin (AMK), MIC of ≤16 mg/liter is S, MIC of 32 mg/liter is I, and MIC of ≥64 mg/liter is R; for ciprofloxacin (CIP), MIC of ≤0.5 mg/liter is S, MIC of 1 mg/liter is I, and MIC of ≥2 mg/liter is R; for levofloxacin (LVX), MIC of ≤1 mg/liter is S, MIC of 2 mg/liter is I, and MIC of ≥4 mg/liter is R; for gentamicin (GEN) and tobramycin (TOB), MIC of ≤4 mg/liter is S, MIC of 8 mg/liter is I, and MIC of ≥16 mg/liter is R; for piperacillin-tazobactam (TZP), MIC of ≤16/4 mg/liter is S, MIC of 32/4 to 64/4 mg/liter is I, and MIC of ≥128/4 mg/liter is R. Colistin (CST) MICs were determined macrobroth dilution, I of ≤2 and R of ≥4. MICs done by MTS were ceftazidime-avibactam (CAZ/AVI), MIC of ≤8/4 mg/liter is S and MIC ≥ 16/4 mg/liter is R; meropenem-vaborbactam (MEM/VAB), MIC of ≤4/8 mg/liter is S, MIC of 8/8 mg/liter is I, and MIC ≥ 16/8 mg/liter is R; ceftolozane-tazobactam (TOL/TAZO), MIC of ≤4/4 mg/liter is S, MIC of 8/4 mg/liter is I, and MIC of ≥16/4 mg/liter is R. IMI/REL MIC of ≤2/4 mg/liter is S, MIC of 4/4 mg/liter is I, and MIC of ≥8/4 mg/liter is R. REL was tested at a fixed concentration of 4 mg/liter, while IMI was in doubling dilutions.
TABLE 2.
Zone diameters of P. aeruginosa isolatesa
| Isolate no. | Zone diameter (mm) for: |
|||
|---|---|---|---|---|
| CAZ | ATM | CAZ/AVI | CAZ/AVI/ATM | |
| CLB 24388 | 6 | 12 | 15 | 30 |
| EM 2972704 | 6 | 6 | 6 | 30 |
Disk diffusion assay was used for CAZ: S ≥ 18 mm, I = 15 to 17 mm, R ≤ 14 mm; ATM, S ≥ 22 mm, I = 16 to 21 mm, R ≤ 15 mm; CAZ/AVI, S ≥ 21 mm and R ≤ 20 mm; and CAZ/AVI/ATM testing, S ≥ 21 mm and R ≤ 20 mm.
Whole-genome sequencing (WGS).
Isolate CLB 24388 was determined to be sequence type 17 (ST17) according to the Pasteur multilocus sequence type (MLST) scheme. This strain contained the following β-lactamase (bla) genes: blaGES-2, blaOXA-5, blaOXA-50, and blaPDC-8. Isolate EM 2972704 belonged to ST309 and possessed multiple β-lactamase genes: blaGES-19, blaGES-20, blaOXA-2, blaOXA-50-like (OXA-50 A8T, T16A, K112E), and blaPDC-19a (Table 3).
TABLE 3.
Genes coding for β-lactam resistance determinants as detected by WGS of P. aeruginosa CLB 24388 and EM 2972704
| Isolate | MLST | β-lactamases |
AmpC regulators |
OprD | ||||
|---|---|---|---|---|---|---|---|---|
| PDC | GES | OXA | AmpD | AmpR | DacB (PBP4) | |||
| PAO1 | 549 | PDC-1 | NA | OXA-50 | WT | WT | WT | WT |
| CLB 24388 | 17 | PDC-8 | GES-2 | OXA-5, OXA-50 | A136V, G148A | WT | WT | Type T1-VIa-like, with 66 divergent aa after G388 |
| EM 2972704 | 309 | PDC-19a | GES-19, GES-20 | OXA-2 and OXA-50 A8T, T16A, K112E | Q44H, G148A | G283E, M288R | WT | Type T1-VII-like,a interrupted by ISPa1328 element containing a transposase gene after aa D43 |
OprD was interrupted after aa D43 by an ISPa1328 element containing a transposase (tnp) gene, followed by the continuation of OprD that is type T1-VII, differing by the following substitutions: starting the continuation of OprD there is a repeat insertion of 42L and 43D, P186G, 372(VDSSSS-YAGL-)383, and E437D (17). P. aeruginosa PAO1 genome sequence NC_002516.2 was used as the reference genome. Reference comparator amino acid sequences from PAO1 (WT, wild type) are the following: PDC-1, NP_252799.1; OXA-50, NP_254201.1; AmpD, NP_253212.1; AmpR, NP_252798.1; DacB, NP_251737; OprD, NP_249649.1. Reference sequences for other β-lactamases present are the following: PDC-8, WP_003118203.1; PDC-19, AEM44530.1; OXA-5, WP_032488483.1; OXA-2, WP_001007673.1; GES-2, WP_045912044.1; GES-19, WP_015059045.1; GES-20, WP_015059046.1.
We next examined other resistance determinants previously reported in P. aeruginosa (6, 11, 17–22). The AmpC regulator genes dacB, ampR, and ampD were analyzed (Table 3). Inactivation of dacB results in overproduction of AmpC (23). DacB (penicillin binding protein 4, a nonessential PBP) in both isolates was identical to the amino acid sequence of DacB in PAO1. However, AmpD in isolate CLB 24388 contained A136V and G148A substitutions while isolate EM 2972704 contained Q44H and G148A substitutions, and AmpR in isolate EM 2972704 contained G283E and M288R substitutions relative to the PAO1 sequence. The role of these substitutions in PDC expression is not currently known.
Changes relative to the WT PAO1 sequence were noted in the regulators for MexAB-OprM and MexXY-OprM (see Table S1 in the supplemental material), but their overall contribution to phenotype is unknown, as they have not been previously associated with resistance. Genetic determinants of fluoroquinolone and aminoglycoside resistance were also found, and the data are presented in Table S1.
The sequence of oprD in both isolates deviated from the wild-type (WT) P. aeruginosa PAO1 oprD gene extensively. In isolate CLB 24388, OprD was type T1-VIa (17); however, the 66 amino acids (aa) after G388 diverged from the WT OprD amino acid sequence. In isolate EM 2972704, the OprD porin was completely disrupted after amino acid D43 by an insertion sequence element, ISPa1328, containing a transposase (tnp) gene, after which came a duplication of amino acids 42 and 43 (LD), followed by the remaining sequence of OprD (24–27). PCR and sequence analysis of a larger than expected oprD amplicon (∼2,500 bp) in EM 2972704 established and confirmed this finding (see the supplemental material). This confirmation was required, as the oprD gene was split across 2 separate contigs. The first 129 bp (43 amino acids) of oprD was at the end of one contig (GenBank accession no. JAFFGW010000014.1, bp 221,915 to 222,044), while a repeat of amino acids 42 and 43 (LD) followed by the remaining oprD sequence was at the end of a different contig (GenBank accession no. JAFFGW010000034.1, bp 45,900 to 47,103) in the draft genome.
Figures 2 and 3 illustrate the genetic context of the GES β-lactamases in both isolates. From isolate CLB 24388, an approximately 10,000-bp segment of the genome sequence is shown in Fig. 2. This assembled segment details the positions of OXA-5 and GES-2 relative to the integrase genes. Figure 3 shows the genetic context of the GES β-lactamases in an approximately 10,000-bp segment of isolate EM 2972704. This assembled segment contains open reading frames for the integron integrase IntI1, GES-19, and GES-20 as well as OXA-2 and an IS6 family transposase. Other genes are associated with these segments and are illustrated in the figures.
FIG 2.
Detailed view of the integron-like element carrying GES and OXA β-lactamase genes in isolate CLB 24388. Open reading frames (ORFs) are represented by arrows. This segment (GenBank accession no. JAFFGY010000016.1, bp 400,688 to 410,007) contains the genes encoding a DDE-type integrase and a class 1 integron integrase (intI1), indicated by green arrows. Antimicrobial resistance genes are shown with red arrows. They include sul1, qacEΔ1, aac(3)-I family gene, aac(6′)-Ib4, blaOXA-5, an additional aac(6′)-Ib4, and blaGES-2. The deposited JAFFGY010000016.1 contig contains an assembly stop gap between blaGES-2 and blaOXA-5. PCR amplification, cloning, and sequencing of the stop gap sequence (see the supplemental material) identified an additional full-length aac(6′)-Ib4 gene with an alternative valine start codon.
FIG 3.
Detailed view of the integron-like element carrying the tandem GES genes in isolate EM 2972704. Open reading frames (ORFs) are represented by arrows. This segment (GenBank accession no. JAFFGW010000010.1, bp 72,401 to 81,619) contains genes encoding a class 1 integron integrase (intI1) and an IS6 family transposase, indicated by green arrows. Antimicrobial resistance genes are shown with red arrows. They include aac(6′)-33, blaGES-19, blaGES-20, aac(6′)-Ib4, blaOXA-2, aadA1, qacEΔ1, and sul1.
Immunoblot and ESI-MS analyses.
Immunoblotting of the purified fractions of different isoelectric points (pIs) from preparative isoelectric focusing (pIEF) gels demonstrated that the pI ∼6 fraction from both isolates produced a strong signal when probed with a newly prepared rabbit polyclonal anti-GES-2 antibody (Fig. 4a). Electrospray ionization-mass spectrometry (ESI-MS) analysis was also performed on the pI ∼6 fraction of EM 2972704 (Fig. 4b) to assess the relative quantities of the two GES variants. The expected molecular sizes of GES-19, 29,231 ± 3 Da, and GES-20, 29,247 ± 3 Da, were observed, and both β-lactamases were well expressed and present in approximately equal amounts.
FIG 4.
(a) Immunoblot of the purified fractions of different pIs from the two P. aeruginosa isolates probed with the polyclonal anti-GES-2 antibody. (b) A mass spectrometry analysis of the purified pI ∼6 fraction assessing the relative quantities of the two GES variants in EM 2972704. GES-19 and GES-20 were both well expressed and abundant. The expected molecular masses were GES-19, 29,231 ± 3 Da, and GES-20, 29,247 ± 3 Da.
β-Lactamase kinetics.
As the pI ∼6 fraction contained both GES-19 and GES-20, we cloned both blaGES genes into the pET-28(+) plasmid. Purified recombinant GES-2, GES-19, and GES-20 were then analyzed in steady-state kinetic experiments that are summarized in Table 4. To obtain the apparent Ki (Ki app) of REL, nitrocefin (NCF) was used as an indicator substrate, and the Km of NCF for each β-lactamase was determined. The Ki observed values were determined for each β-lactamase in a direct competition assay with NCF and corrected based on the Km value of NCF for each β-lactamase assayed to determine the Ki app. The Ki app values of purified GES-2, -19, and -20 β-lactamases were 19 ± 2 μM, 23 ± 2 μM, and 21 ± 2 μM, respectively (Table 4).
TABLE 4.
REL Ki app determinations from purified enzyme preparations
| Purified GES | NCF Km (μM) | REL Ki app (μM) |
|---|---|---|
| GES-2 | 3.6 ± 0.5 | 19 ± 2 |
| GES-19 | 29 ± 3 | 23 ± 2 |
| GES-20 | 37 ± 4 | 21 ± 2 |
DISCUSSION
WGS assisted in our understanding of the mechanisms of the XDR phenotype in both isolates, with particular interest in the IMI- and IMI/REL-resistant phenotypes. It is important to note the two P. aeruginosa isolates did not share the same lineage; isolate CLB 24388 was a member of ST17, and isolate EM 2972704 was a member of ST309.
Class D β-lactamases.
We identified the class D enzymes OXA-5 and OXA-50 (intrinsic oxacillinase of P. aeruginosa) in CLB 24388 and OXA-2 and a new variant of OXA-50 (A8T, T16A, K112E) in EM 2972704. The oxacillinases found in these isolates (OXA-2 and OXA-5, narrow-spectrum activity; OXA-50/50-like, additional weak imipenemase activity [28]) are resistant to inactivation by DBO β-lactamase inhibitors (AVI and REL) and VAB.
Class C β-lactamases.
The two isolates also possessed different variants of the intrinsic P. aeruginosa chromosomal class C β-lactamase, PDC-8 in CLB 24388 and PDC-19a in isolate EM 2972704. These PDC β-lactamases do not contain any of the reported substitutions that expand their normal spectrum of activity or lead to TOL/TAZO resistance (see Fig. S1 in the supplemental material) (29). However, in addition to their role in cephalosporin resistance, we propose that the PDC β-lactamases in these isolates serve to bind the carbapenems to various degrees and thereby contribute to the IMI-resistant phenotype (30, 31). We believe this is an underappreciated contributor of carbapenem resistance in many cases and could explain why isolates with inactivation of an outer membrane porin (OprD) and upregulation of an AmpC β-lactamase show resistance to IMI.
Normally PDC is expressed at low levels in P. aeruginosa unless there is an alteration in, or gene inactivation of, ampR, ampD, or dacB. However, we did not observe any of the reported mutations in these genes previously associated with increased PDC expression, such as inactivation of DacB (PBP4) or the D135N mutation in AmpR (23, 32–35). As it was hard to detect appreciable amounts of PDC β-lactamase by immunoblot assay in both isolates, quantitative reverse transcription-PCR (qRT-PCR) analysis was utilized to circumvent this issue. qRT-PCR demonstrated that mRNA levels of the PDC β-lactamases in CLB 24388 and EM 2972704 isolates were 22-fold and 27-fold lower, respectively, than those in a control P. aeruginosa isolate that contained a D135N substitution in AmpR and is known to overexpress PDC-16 (32) (Fig. S2), demonstrating the AmpCs in the two P. aeruginosa isolates were not overexpressed.
Class A β-lactamases.
CLB 24388 was found to contain blaGES-2, whereas EM 2972704 possessed two GES variants in tandem order, blaGES-19 followed by blaGES-20; the former encodes an extended-spectrum β-lactamase (ESBL) and the latter a carbapenemase. GES-2 has modest carbapenemase activity due to a G170N substitution relative to GES-1, as Poirel et al. were the first to demonstrate (36–38). GES-19 is similar to GES-1 but has one substitution in the mature protein, G243A (Ambler consensus numbering) (39). This G243A substitution was reported to increase both ATM and CAZ hydrolysis, thereby potentially challenging the efficacy of the CAZ/AVI/ATM combination (40). The importance of amino acid 243 with regard to increased ATM hydrolysis was first noted in GES-9, which contains a G243S substitution (41). However, GES-2 and GES-20 both retain the G at position 243. It is interesting that in two isolates that contained blaGES-19 and blaGES-26 in tandem, the triple combination exhibited potency (42), as was the case for both isolates reported here.
GES-20 is identical to GES-5 after the leader sequence has been removed. Notably, GES-20 contains a very important substitution in the mature protein, a serine at position 170, that confers carbapenem hydrolyzing ability (36). In addition, the S170 substitution present in GES-20 has also been linked to increased hydrolysis of TOL and decreased inhibition by tazobactam (43), which explains the very high MIC (>256/4 mg/liter) to TOL/TAZO in isolate EM 2972704. Lahiri et al. put forth that a blaGES-20 construct in an isogenic E. coli background remained susceptible to AVI inhibition. However, in this background, the E. coli blaGES-20 isolate demonstrated a CAZ MIC of 0.5 mg/liter and a CAZ/AVI MIC of 0.25 mg/liter (44). The high-level CAZ/AVI resistance that we observe in EM 2972704 may be a result of the tandem blaGES genes. Tandem carriage of blaGES has been described in detail before in similar transposons (42, 45). Khan et al. hypothesize that tandem expression influences the resistance phenotype, thereby producing a greater than additive effect (42). However, this is the first in-depth analysis of the tandem carriage of blaGES-19 and blaGES-20, especially as it relates to inhibitor resistance.
GES-20 is the most prevalent (84.4%) β-lactamase among IMI-resistant P. aeruginosa clinical isolates in Mexico (46) and is chromosomally encoded on embedded class 1 integrons, as are blaGES-19 and blaGES-20, contained in this study (Fig. 2 and 3). This is particularly concerning as widespread dissemination of blaGES-19 and blaGES-20 is much more likely given its genetic environment. This mechanism may not be the only cause of carbapenem resistance in the isolates from Mexico, as they did not assess the OprD porin function in these isolates.
A mass spectrometry analysis of the purified pI ∼6 fraction from EM 2972704 demonstrated that GES-19 and GES-20 were both well expressed and abundant (Fig. 4b). We next performed inhibitory enzyme kinetics on the purified GES-2, GES-19, and GES-20 β-lactamases to determine the effectiveness of REL inhibition (Table 4). We determined an elevated Ki app for all three enzymes (Ki app for GES-2, -19, and -20 β-lactamases of 19 ± 2 μM, 23 ± 2 μM, and 21 ± 2 μM, respectively).
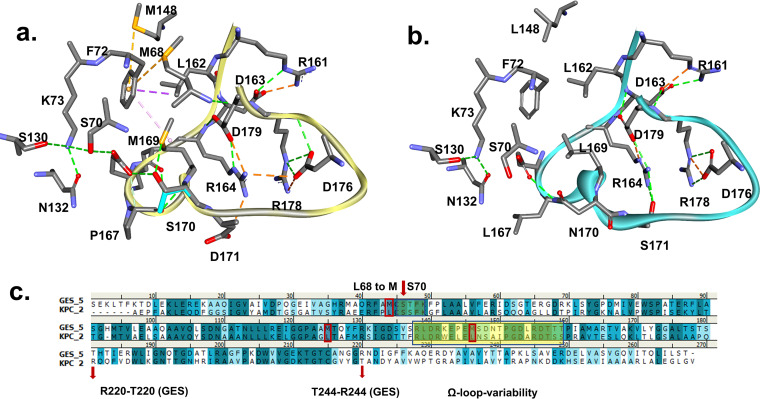
From previous studies, we know that REL is a potent inhibitor of KPC-2 and restores the susceptibility of IMI in clinical strains (47). The two β-lactamases (KPC and GES) share more than 50% similarity. However, we reasoned that amino acid variability in the Ω loop of GES-20 (GES-5) versus KPC-2 (Fig. 5) may explain the poor inhibition of GES-20 by REL. The most notable changes between the two are L167→P, L169→M, and N170→S in GES-20. In the GES-20 model, the hydrophobic interactions that are formed restrain the mobility of the Ω loop compared to KPC-2 (Fig. 5a and b). We note that the R164 interactions with D171 and D179 (GES-20) or S171 (KPC-2) are maintained. M169 in GES is in the same position as L169 in KPC. Similarly, M68 and M148 in GES are replaced by leucine residues in KPC (Fig. 5c). These changes allow the formation of bridging interactions. Furthermore, the hydrophobic interaction of L162 and F72 in the Ω loop of GES enzymes may stabilize these interactions even more, thereby restraining the Ω loop. This variability in the Ω loop sequence may also change the electrostatic properties of the surface of GES-20 (GES-5) (Fig. S3, top) as well as the active site S170 (Fig. S3, bottom), making the GES-20 active-site entrance more hydrophilic, smaller, and less shallow than that of the KPC-2 enzyme.
FIG 5.
Ω Loop variability between class A GES-20 (a) and KPC-2 (b) β-lactamases. The amino acid variability (c) in the Ω loop of KPC-2 versus GES-20 (GES-5) (L167→P, L169→M, and N170→S in GES-20) restrains the loop of GES-20. The R164 interactions with D179 and D171 or S171 are maintained. Methionine at position 169 replaces L in KPC-2. Similarly, L68 and L148 in KPC-2 are replaced by M in the GES enzyme. These changes facilitate a bridging interaction between the sulfur of M169, M68, M148, and the aromatic F72 in GES-20. Furthermore, the hydrophobic interaction of L162 and F72 may stabilize these interactions even more, restraining the Ω loop. The yellow rectangle highlights the Ω loop region, and the red arrows indicate areas of interest.
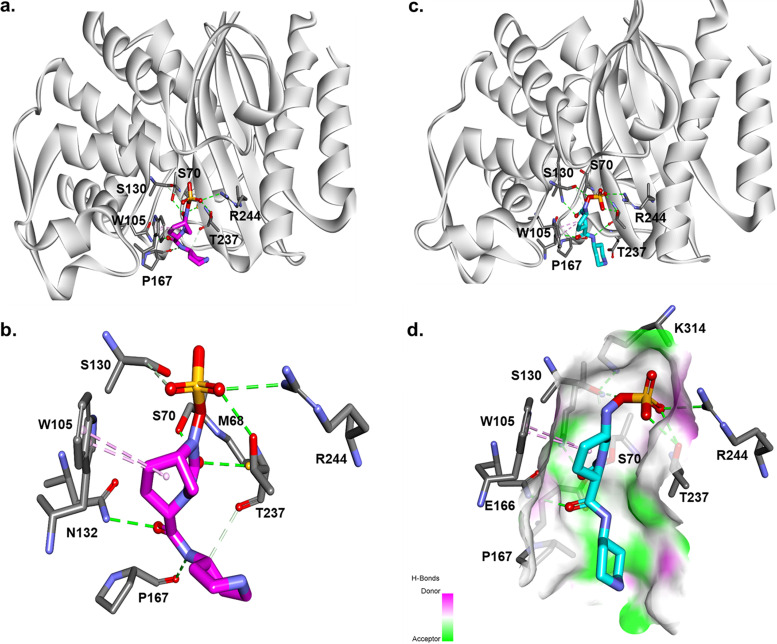
To explain why REL fails to sufficiently inactivate these members of the class A GES family, a model of REL docked into the active site of the GES-20 enzyme (Fig. 6) was generated. When the Michaelis-Menten complex of REL with GES-20 (Fig. 6a and b) is formed, the DBO establishes hydrophobic interactions with W105 and the piperidine ring with P167 (being positioned less than 4-Å distance from the pyrrolidine). The acyl-enzyme complex (Fig. 6c and d) suggests that REL is restricted between W105 and P167. The SO4−3 group creates interactions with R244 of GES-20 but with the carbonyl outside the oxyanion hole. Thus, REL forms an unstable complex. This unfavorable positioning may be responsible for the higher Ki app and inhibitor resistance.
FIG 6.
Molecular docking of REL as a Michaelis-Menten complex (a and b) and as an acyl enzyme (c and d) into the active site of GES-20 (GES-5). Hydrophobic interactions with W105 and the piperidine ring of REL with P167 (less than 4 Å distance) were made. The acyl-enzyme complex suggests that REL adopts an unfavorable position with the carbonyl outside the oxyanion hole, forming an unstable complex (c and d).
From the AST, we see that both isolates are extensively drug resistant (XDR). This troubling observation challenges us to propose potential therapeutic combinations to overcome this phenotype. We reasoned that as ATM is not hydrolyzed by most GES enzymes, if we inhibited the PDC β-lactamases with AVI, then the CAZ/AVI/ATM combination might produce a susceptible phenotype. For both isolates this was the case. The zone size of CAZ/AVI increased from 15 mm (CLB 24388) and 6 mm (EM 2972704) to 30 mm for both isolates with the addition of ATM, well into the susceptible range. The combination of both CAZ and ATM also allows for multiple PBP inactivations (PBP1a, PBP1b, and PBP3), augmenting susceptibility by completely affecting the “divisome” of P. aeruginosa.
OprD.
Additionally, we must keep in mind that OprD is the main portal of entry for IMI and MEM into P. aeruginosa (6). Therefore, the isolates will manifest resistance to these two carbapenems. GES-20 could also significantly contribute to the observed phenotype by hydrolyzing any IMI that does find its way into the cell, as it is not effectively inhibited by REL. The presence of the GES-20 carbapenemase in isolate EM 2972704 is also the reason for higher IMI, IMI/REL, and MEM/VAB MICs relative to isolate CLB 24388.
Fluoroquinolone resistance and aminoglycoside-modifying enzymes.
Other mechanisms of resistance in both isolates were analyzed, and the data are presented in Table S1. Known mechanisms of resistance to fluoroquinolone and aminoglycoside antibiotics were found (19, 22, 48–50).
We provide evidence that two of the main drivers of carbapenem and carbapenem/β-lactam inhibitor resistance in these isolates is the presence of inhibitor-resistant GES carbapenemases and the loss of an important porin, OprD. In isolate EM 2972704, the ISPa1328 insertion element containing a transposase (tnp) gene, previously reported as being a causative agent in the inactivation of OprD (24–27), was coupled with GES-20, a robust carbapenemase. In isolate CLB 24388, the OprD is also disrupted and is accompanied by the presence of GES-2. Here, we report the limits of inhibition of certain class A GES carbapenemases by this novel inhibitor class that have not been well-characterized.
This work strongly supports the importance of establishing molecular surveillance programs that detect and define the basis of resistance, especially to novel agents as they are introduced into the clinic. This knowledge empowers clinicians who must choose empirical therapy, strengthens antimicrobial stewardship programs responsible for judicious drug utilization, and informs medicinal chemists as they devise strategies to combat MDR pathogens. This analysis also provides a snapshot of the complex mechanisms that result in an XDR phenotype.
MATERIALS AND METHODS
Selection of bacterial isolates.
P. aeruginosa isolates CLB 24388 and EM 2972704 were selected for characterization because of reduced susceptibility to IMI/REL in screening studies done by Merck and were sent to our reference lab for further characterization as metallo-β-lactamases, KPC, and class D carbapenemases were not found. Isolate CLB 24388 is a clinical isolate of P. aeruginosa from a urine specimen collected in 2004 (Texas), and isolate EM 2972704 (EM 100819) is a 2005 P. aeruginosa clinical isolate from Mexico; both were obtained from Eurofins (Lancaster, PA).
AST.
Susceptibility testing with standard antibiotics was performed by broth microdilution with MicroScan NM43 trays (Beckman Coulter Inc., Brea, CA). P. aeruginosa ATCC 27853 and E. coli ATCC 25922 were used as control strains. CST MICs were determined by broth macrodilution according to CLSI guidelines (51) and MICs for CAZ/AVI, TOL/TAZO, and MEM/VAB by MTS (MIC Test Strip; Liofilchem, Inc., Waltham, MA). IMI/REL MICs were determined by agar dilution using cation-adjusted Mueller-Hinton (MH) agar according to CLSI methods (51) utilizing a Steers replicator and a fixed concentration of 4 mg/liter REL. Disk diffusion assays for CAZ, ATM, CAZ/AVI, and CAZ/AVI/ATM were carried out as previously described (52, 53). MICs were interpreted according to the 2021 Clinical and Laboratory Standards Institute (CLSI) guidelines for P. aeruginosa (51).
Whole-genome sequencing.
Genomic DNA was extracted using the MasterPure Gram-positive DNA purification kit by following the manufacturer’s instructions (Epicentre, Madison, WI). The genomic libraries were prepared for sequencing using the Illumina Nextera XT kit (Illumina Inc., San Diego, CA) and the rapid barcoding kit (Oxford Nanopore) and then sequenced on MiSeq and MinION, respectively. For isolate EM 2972704, sequencing reads were used to generate a hybrid assembly using Unicycler (54). For isolate CLB 24388, only the MiSeq reads were used and assembled using SPAdes (55). The genomes were annotated using NCBI’s Prokaryotic Genome Annotation Pipeline (56). Multilocus sequence type (MLST) was determined using MLST 2.0 (available at the Center for Genomic Epidemiology, http://www.genomicepidemiology.org), and resistome analysis was performed using the analysis tools available online at the Pathosystems Resource Integration Center (PATRIC) (57–60). Resistance genes were manually confirmed by BLAST analysis if not a 100% match with 100% coverage of a known gene.
β-Lactamase purification from the P. aeruginosa isolates.
P. aeruginosa isolates CLB 24388 and EM 2972704 were each grown in 1 liter of super optimal broth (SOB) medium overnight at 37°C in a 225-rpm shaker. The bacterial cells were pelleted, frozen, and resuspended in 50 mL of 50 mM Tris-Cl (pH 7.4) containing 40 mg/liter lysozyme, 1 mM magnesium sulfate, and 1.0 U/mL Benzonase nuclease (Novagen). This suspension was incubated at room temperature for 30 min with stirring followed by 30 min of centrifugation at 12,000 rpm. The supernatant was then combined with 40 mL of sterile water, 5 mL of ampholines (pH range, 3 to 10) (Bio-Rad, Hercules, CA), and 4 g of Sephadex G-100 (GE Healthcare, Piscataway, NJ). A preparative isoelectric focusing (pIEF) gel was run at 8 W overnight. Areas with identified nitrocefinase activity were removed from the gel (pI ∼6 and pI ∼8 for each isolate), kept separate, and eluted with 2 mL of 10 mM phosphate-buffered saline (PBS; pH 7.4). pIEF elutions were further purified by size exclusion chromatography using a HiLoad 16/60 Superdex 75 gel filtration column (GE Healthcare, Piscataway, NJ). Eluted fractions were pooled, concentrated, and used for immunoblot analysis and electrospray ionization-mass spectrometry (ESI-MS) characterization.
Immunoblot analysis.
Samples of the purified β-lactamase fractions were analyzed from P. aeruginosa isolates CLB 24388 and EM 2972704. One microliter of the purified fraction was mixed with SDS-PAGE loading dye and loaded into each lane of an SDS-PAGE gel. Gels were run and transferred to a polyvinylidene difluoride (PVDF) membrane. PVDF membranes were probed with rabbit polyclonal anti-GES-2 antibodies at 0.1 μg/mL in 5% bovine serum albumin in Tris-buffered saline and processed as previously described (61).
ESI-MS.
The purified β-lactamase fraction pI ∼6 was also analyzed using a quadrupole time-of-flight (Q-TOF) Waters Synapt-G2-Si ESI-MS and Waters Acquity H class ultraperformance liquid chromatograph (UPLC) with a BEH C18 1.7-μm column (2.1 by 50 mm). The Synapt G2-Si spectrometer was calibrated with sodium iodide with a 50 to 2,000 m/z mass range. MassLynx V4.1 was used to deconvolute protein peaks. The tune settings for each sample were capillary voltage at 3 kV, sampling cone at 35 V, source offset at 35, source temperature of 100°C, desolvation temperature of 500°C, cone gas at 100 liters/h, desolvation gas at 800 liters/h, and nebulizer at 6.0. Mobile phase A was 0.1% formic acid (FA) in water. Mobile phase B was 0.1% FA in acetonitrile. The mass accuracy for this system is ±3 Da.
GES-19 and GES-20 purification.
GES-19 and GES-20 were individually purified based on a previously published method (62). Clones of the blaGES-19 and blaGES-20 genes lacking the first 18 residues were cloned in the pET-28(+) plasmid (GenScript, Piscataway, NJ) in which a tobacco etch virus (TEV) cleavage site preceded the blaGES coding sequence. These clones were transformed into Escherichia coli BL21(DE3), plated on lysogeny broth (LB) agar plates containing 50 μg/mL kanamycin (kan50), and grown overnight at 37°C. The following day, a single colony was inoculated into 5 mL LB (kan50) using a single isolated colony and grown overnight at 37°C in a shaker incubator. A volume of 2.5 mL of overnight culture was inoculated into 250 mL LB (kan50) and grown at 37°C until an optical density at 600 nm (OD600) of 0.6. Isopropyl-d-thiogalactopyranoside (IPTG) was then added to 0.25 mM and the culture incubated at 20°C overnight with shaking. The cells were harvested and stored at −20°C. The next day, the cells were resuspended in buffer A (50 mM Tris-Cl, pH 8.0, 200 mM NaCl) and sonicated at 40% amplitude (five cycles of 30 s with 1 min between cycles) followed by centrifugation for 60 min at 15,000 × g. The crude extracts were loaded onto a Ni-Sepharose column equilibrated with buffer A, the column was washed with five column volumes of buffer A (+20 mM imidazole), and His6-GES was eluted with buffer B (50 mM Tris-Cl, pH 8.0, 200 mM NaCl, 500 mM imidazole). Fractions containing His6-GES were combined and His6-TEV protease added (1:30 His6-TEV to His6-GES) and dialyzed for 16 h at 4°C against 100 volumes of buffer A. The protein was then loaded onto a Ni-Sepharose column to separate it from the His6 tag, the uncleaved fusion protein, and the His6-tagged TEV protease, allowing the GES β-lactamase to be collected in the flowthrough. The individual GES β-lactamases were then concentrated and dialyzed with 10 mM phosphate-buffered saline.
β-Lactamase kinetics and inhibition with REL.
The kinetic parameters of the purified proteins GES-2, GES-19, and GES-20 were determined by continuous assay at room temperature using an Agilent 8453 diode array spectrophotometer. Each determination was performed in 10 mM phosphate-buffered saline at pH 7.4. Measurements were obtained using nitrocefin (NCF) (Δε482 = 17,400 M−1 cm−1). The concentration of each protein was calculated by measuring the absorbance at 280 nm and using Beers’ law and the extinction coefficient calculated for each protein using the online program Expasy Protparam. The steady-state kinetic parameters Vmax and Km were obtained with nonlinear least-squared fit of the data (Michaelis-Menten equation) using Enzfitter (Biosoft Corporation, Ferguson, MO) and equation 1:
| (1) |
Ki app values were determined in a direct competition assay with NCF as previously described (63). In each case, initial velocities, v, were measured using a fixed amount of β-lactamase and increasing concentrations of REL against the indicator substrate (NCF = [S]). After the Ki (observed) values were obtained, the data were corrected based on equation 2 to account for the affinity of NCF for each β-lactamase to obtain the Ki app.
| (2) |
Molecular docking.
Structural representation of the β-lactamases was generated using the crystal coordinates of KPC-2 (PDB entry 2OV5) and GES-5 (GES-20) (PDB entry 4GNU) and the Build and Edit protocol of Discovery Studio 2020 (BIOVIA Dassault Systems, San Diego, CA) molecular modeling software. The crystallographic water molecules were not maintained during modeling. The KPC-2 and GES-5 (GES-20) β-lactamase structures were solvated and minimized to a root mean square of 0.01 Å using the conjugate gradient method. REL and acyl-REL were constructed using Fragment Builder tools and minimized using a Standard Dynamics Cascade protocol of D.S. 2020. The intact and acylated REL were automatically docked into the active site of GES-5 using CDOCKER module of D.S. 2020. To obtain acyl-enzyme complexes, the most favorable position of REL demonstrating the anticipated active-site contacts (e.g., a short distance [2 to 3 Å] between Ser70:O) was chosen. The complex between REL and the GES-5 β-lactamase was further minimized using a conjugate gradient method.
Data availability.
Raw sequence reads and genome assemblies are registered at NCBI under BioProject no. PRJNA342804 and are deposited in the SRA repository (SRR13607658 for CLB 24388 and SRR13610045 and SRR15291349 for EM 2972704) and in the GenBank WGS repository (WGS accession no. JAFFGY000000000 for CLB 24388 and JAFFGW000000000 for EM 2972704).
ACKNOWLEDGMENTS
The REL powder was provided by Merck & Co., Inc., Kenilworth, NJ. CL 24388 and EM 2972704 were purchased from Eurofins (Lancaster, PA). Purified GES-2 was a kind gift from Sergei Vakulenko.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) to R.A.B. under award number R01AI063517. This work was also funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services no. U19AI110819. In addition, a grant (MISP 58773) was provided by Merck Sharp & Dohme, Corp., Kenilworth, New Jersey, USA, to R.A.B. This study was also supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, award number 1I01BX001974, to R.A.B. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Department of Veterans Affairs.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Evans SR, Tran TTT, Hujer AM, Hill CB, Hujer KM, Mediavilla JR, Manca C, Domitrovic TN, Perez F, Farmer M, Pitzer KM, Wilson BM, Kreiswirth BN, Patel R, Jacobs MR, Chen L, Fowler VG, Chambers HF, Bonomo RA, Antibacterial Resistance Leadership Group . 2019. Rapid molecular diagnostics to inform empiric use of ceftazidime/avibactam and ceftolozane/tazobactam against Pseudomonas aeruginosa: PRIMERS IV. Clin Infect Dis 68:1823–1830. 10.1093/cid/ciy801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giske CG, Buaro L, Sundsfjord A, Wretlind B. 2008. Alterations of porin, pumps, and penicillin-binding proteins in carbapenem resistant clinical isolates of Pseudomonas aeruginosa. Microb Drug Resist 14:23–30. 10.1089/mdr.2008.0778. [DOI] [PubMed] [Google Scholar]
- 3.Drawz SM, Papp-Wallace KM, Bonomo RA. 2014. New β-lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother 58:1835–1846. 10.1128/AAC.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlowsky JA, Lob SH, Kazmierczak KM, Hawser SP, Magnet S, Young K, Motyl MR, Sahm DF. 2018. In vitro activity of imipenem/relebactam against Gram-negative ESKAPE pathogens isolated in 17 European countries: 2015 SMART surveillance programme. J Antimicrob Chemother 73:1872–1879. 10.1093/jac/dky107. [DOI] [PubMed] [Google Scholar]
- 5.Lob SH, Karlowsky JA, Young K, Motyl MR, Hawser S, Kothari ND, Gueny ME, Sahm DF. 2019. Activity of imipenem/relebactam against MDR Pseudomonas aeruginosa in Europe: SMART 2015–17. J Antimicrob Chemother 74:2284–2288. 10.1093/jac/dkz191. [DOI] [PubMed] [Google Scholar]
- 6.Young K, Painter RE, Raghoobar SL, Hairston NN, Racine F, Wisniewski D, Balibar CJ, Villafania A, Zhang R, Sahm DF, Blizzard T, Murgolo N, Hammond ML, Motyl MR. 2019. In vitro studies evaluating the activity of imipenem in combination with relebactam against Pseudomonas aeruginosa. BMC Microbiol 19:150. 10.1186/s12866-019-1522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes MD, Winkler ML, Taracila MA, Page MG, Desarbre E, Kreiswirth BN, Shields RK, Nguyen MH, Clancy C, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from beta-lactamase protein engineering. mBio 8:e00528-17. 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haidar G, Clancy CJ, Shields RK, Hao B, Cheng S, Nguyen MH. 2017. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum beta-lactamases. Antimicrob Agents Chemother 61:e02534-16. 10.1128/AAC.02534-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papp-Wallace KM, Mack AR, Taracila MA, Bonomo RA. 2020. Resistance to novel beta-lactam-beta-lactamase inhibitor combinations: the “price of progress.” Infect Dis Clin North Am 34:773–819. 10.1016/j.idc.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraile-Ribot PA, Zamorano L, Orellana R, del Barrio-Tofiño E, Sánchez-Diener I, Cortes-Lara S, López-Causapé C, Cabot G, Bou G, Martínez-Martínez L, Oliver A, Galán F, Gracia I, Rodríguez MA, Martín L, Sánchez JM, Viñuela L, García MV, Lepe JA, Aznar J, López-Hernández I, Seral C, Castillo-García FJ, López-Calleja AI, Aspiroz C, de la Iglesia P, Ramón S, Riera E, Pérez MC, Gallegos C, Calvo J, Quesada MD, Marco F, Hoyos Y, Horcajada JP, Larrosa N, José González J, Tubau F, Capilla S, Pérez-Moreno MO, Centelles MJ, Padilla E, Rivera A, Mirelis B, Rodríguez-Tarazona RE, Arenal-Andrés N, Ortega M.dP, Megías G, García I, Colmenarejo C, GEMARA-SEIMC/REIPI Pseudomonas Study Group , et al. 2020. Activity of imipenem-relebactam against a large collection of Pseudomonas aeruginosa clinical isolates and isogenic beta-lactam-resistant mutants. Antimicrob Agents Chemother 64:e02165-19. 10.1128/AAC.02165-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomis-Font MA, Cabot G, Sanchez-Diener I, Fraile-Ribot PA, Juan C, Moya B, Zamorano L, Oliver A. 2020. In vitro dynamics and mechanisms of resistance development to imipenem and imipenem/relebactam in Pseudomonas aeruginosa. J Antimicrob Chemother 75:2508–2515. 10.1093/jac/dkaa206. [DOI] [PubMed] [Google Scholar]
- 12.Karlowsky JA, Lob SH, Kazmierczak KM, Young K, Motyl MR, Sahm DF. 2020. In vitro activity of imipenem/relebactam against Enterobacteriaceae and Pseudomonas aeruginosa isolated from intraabdominal and urinary tract infection samples: SMART Surveillance United States 2015–2017. J Glob Antimicrob Resist 21:223–228. 10.1016/j.jgar.2019.10.028. [DOI] [PubMed] [Google Scholar]
- 13.Karlowsky JA, Lob SH, Raddatz J, DePestel DD, Young K, Motyl MR, Sahm DF. 2020. In vitro activity of imipenem/relebactam and ceftolozane/tazobactam against clinical isolates of gram-negative bacilli with difficult-to-treat resistance and multidrug-resistant phenotypes–SMART United States 2015–2017. Clin Infect Dis. 10.1093/cid/ciaa381. [DOI] [PubMed] [Google Scholar]
- 14.Karlowsky JA, Lob SH, Young K, Motyl MR, Sahm DF. 2021. In vitro activity of imipenem/relebactam against Gram-negative bacilli from pediatric patients-study for monitoring antimicrobial resistance trends (SMART) global surveillance program 2015–2017. J Pediatric Infect Dis Soc 10:274–281. 10.1093/jpids/piaa056. [DOI] [PubMed] [Google Scholar]
- 15.Lob SH, Karlowsky JA, Young K, Motyl MR, Hawser S, Kothari ND, Sahm DF. 2020. In vitro activity of imipenem-relebactam against resistant phenotypes of Enterobacteriaceae and Pseudomonas aeruginosa isolated from intraabdominal and urinary tract infection samples–SMART surveillance Europe 2015–2017. J Med Microbiol 69:207–217. 10.1099/jmm.0.001142. [DOI] [PubMed] [Google Scholar]
- 16.Mushtaq S, Meunier D, Vickers A, Woodford N, Livermore DM. 2021. Activity of imipenem/relebactam against Pseudomonas aeruginosa producing ESBLs and carbapenemases. J Antimicrob Chemother 76:434–442. 10.1093/jac/dkaa456. [DOI] [PubMed] [Google Scholar]
- 17.Ocampo-Sosa AA, Cabot G, Rodriguez C, Roman E, Tubau F, Macia MD, Moya B, Zamorano L, Suarez C, Pena C, Dominguez MA, Moncalian G, Oliver A, Martinez-Martinez L, Spanish Network for Research in Infectious Disease . 2012. Alterations of OprD in carbapenem-intermediate and -susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a Spanish multicenter study. Antimicrob Agents Chemother 56:1703–1713. 10.1128/AAC.05451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabot G, Ocampo-Sosa AA, Dominguez MA, Gago JF, Juan C, Tubau F, Rodriguez C, Moya B, Pena C, Martinez-Martinez L, Oliver A, Spanish Network for Research in Infectious Disease . 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Barrio-Tofino E, Lopez-Causape C, Cabot G, Rivera A, Benito N, Segura C, Montero MM, Sorli L, Tubau F, Gomez-Zorrilla S, Tormo N, Dura-Navarro R, Viedma E, Resino-Foz E, Fernandez-Martinez M, Gonzalez-Rico C, Alejo-Cancho I, Martinez JA, Labayru-Echverria C, Duenas C, Ayestaran I, Zamorano L, Martinez-Martinez L, Horcajada JP, Oliver A. 2017. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob Agents Chemother 61:e01589-17. 10.1128/AAC.01589-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horcajada JP, Montero M, Oliver A, Sorli L, Luque S, Gomez-Zorrilla S, Benito N, Grau S. 2019. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev 32:e00031-19. 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Causape C, Cabot G, Del Barrio-Tofino E, Oliver A. 2018. The versatile mutational resistome of Pseudomonas aeruginosa. Front Microbiol 9:685. 10.3389/fmicb.2018.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moya B, Dotsch A, Juan C, Blazquez J, Zamorano L, Haussler S, Oliver A. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog 5:e1000353. 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al Bayssari C, Diene SM, Loucif L, Gupta SK, Dabboussi F, Mallat H, Hamze M, Rolain JM. 2014. Emergence of VIM-2 and IMP-15 carbapenemases and inactivation of oprD gene in carbapenem-resistant Pseudomonas aeruginosa clinical isolates from Lebanon. Antimicrob Agents Chemother 58:4966–4970. 10.1128/AAC.02523-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Bayssari C, Valentini C, Gomez C, Reynaud-Gaubert M, Rolain JM. 2015. First detection of insertion sequence element ISPa1328 in the oprD porin gene of an imipenem-resistant Pseudomonas aeruginosa isolate from an idiopathic pulmonary fibrosis patient in Marseille, France. New Microbes New Infect 7:26–27. 10.1016/j.nmni.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Estepa V, Rojo-Bezares B, Azcona-Gutierrez JM, Olarte I, Torres C, Saenz Y. 2017. Characterisation of carbapenem-resistance mechanisms in clinical Pseudomonas aeruginosa isolates recovered in a Spanish hospital. Enferm Infecc Microbiol Clin 35:141–147. 10.1016/j.eimce.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Fournier D, Richardot C, Muller E, Robert-Nicoud M, Llanes C, Plesiat P, Jeannot K. 2013. Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J Antimicrob Chemother 68:1772–1780. 10.1093/jac/dkt098. [DOI] [PubMed] [Google Scholar]
- 28.Girlich D, Naas T, Nordmann P. 2004. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob Agents Chemother 48:2043–2048. 10.1128/AAC.48.6.2043-2048.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berrazeg M, Jeannot K, Ntsogo Enguene VY, Broutin I, Loeffert S, Fournier D, Plesiat P. 2015. Mutations in beta-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 59:6248–6255. 10.1128/AAC.00825-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beadle BM, McGovern SL, Patera A, Shoichet BK. 1999. Functional analyses of AmpC beta-lactamase through differential stability. Protein Sci 8:1816–1824. 10.1110/ps.8.9.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trehan I, Beadle BM, Shoichet BK. 2001. Inhibition of AmpC beta-lactamase through a destabilizing interaction in the active site. Biochemistry 40:7992–7999. 10.1021/bi010641m. [DOI] [PubMed] [Google Scholar]
- 32.Domitrovic TN, Hujer AM, Perez F, Marshall SH, Hujer KM, Woc-Colburn LE, Parta M, Bonomo RA. 2016. Multidrug resistant Pseudomonas aeruginosa causing prosthetic valve endocarditis: a genetic-based chronicle of evolving antibiotic resistance. Open Forum Infect Dis 3:ofw188. 10.1093/ofid/ofw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juan C, Macia MD, Gutierrez O, Vidal C, Perez JL, Oliver A. 2005. Molecular mechanisms of beta-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 49:4733–4738. 10.1128/AAC.49.11.4733-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuga A, Okamoto R, Inoue M. 2000. ampR gene mutations that greatly increase class C beta-lactamase activity in Enterobacter cloacae. Antimicrob Agents Chemother 44:561–567. 10.1128/AAC.44.3.561-567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moya B, Beceiro A, Cabot G, Juan C, Zamorano L, Alberti S, Oliver A. 2012. Pan-beta-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob Agents Chemother 56:4771–4778. 10.1128/AAC.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frase H, Toth M, Champion MM, Antunes NT, Vakulenko SB. 2011. Importance of position 170 in the inhibition of GES-type beta-lactamases by clavulanic acid. Antimicrob Agents Chemother 55:1556–1562. 10.1128/AAC.01292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. 2001. GES-2, a class A beta-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother 45:2598–2603. 10.1128/AAC.45.9.2598-2603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart NK, Smith CA, Frase H, Black DJ, Vakulenko SB. 2015. Kinetic and structural requirements for carbapenemase activity in GES-type beta-lactamases. Biochemistry 54:588–597. 10.1021/bi501052t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith CA, Caccamo M, Kantardjieff KA, Vakulenko S. 2007. Structure of GES-1 at atomic resolution: insights into the evolution of carbapenamase activity in the class A extended-spectrum beta-lactamases. Acta Crystallogr D Biol Crystallogr 63:982–992. 10.1107/S0907444907036955. [DOI] [PubMed] [Google Scholar]
- 40.Delbruck H, Bogaerts P, Kupper MB, Rezende de Castro R, Bennink S, Glupczynski Y, Galleni M, Hoffmann KM, Bebrone C. 2012. Kinetic and crystallographic studies of extended-spectrum GES-11, GES-12, and GES-14 beta-lactamases. Antimicrob Agents Chemother 56:5618–5625. 10.1128/AAC.01272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poirel L, Brinas L, Fortineau N, Nordmann P. 2005. Integron-encoded GES-type extended-spectrum beta-lactamase with increased activity toward aztreonam in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3593–3597. 10.1128/AAC.49.8.3593-3597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan A, Tran TT, Rios R, Hanson B, Shropshire WC, Sun Z, Diaz L, Dinh AQ, Wanger A, Ostrosky-Zeichner L, Palzkill T, Arias CA, Miller WR. 2019. Extensively drug-resistant Pseudomonas aeruginosa ST309 harboring tandem Guiana extended spectrum beta-lactamase enzymes: a newly emerging threat in the United States. Open Forum Infect Dis 6:ofz273. 10.1093/ofid/ofz273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poirel L, Ortiz De La Rosa JM, Kieffer N, Dubois V, Jayol A, Nordmann P. 2019. Acquisition of extended-spectrum beta-lactamase GES-6 leading to resistance to ceftolozane-tazobactam combination in Pseudomonas aeruginosa. Antimicrob Agents Chemother 63:e01809-18. 10.1128/AAC.01809-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lahiri SD, Bradford PA, Nichols WW, Alm RA. 2016. Structural and sequence analysis of class A beta-lactamases with respect to avibactam inhibition: impact of omega-loop variations. J Antimicrob Chemother 71:2848–2855. 10.1093/jac/dkw248. [DOI] [PubMed] [Google Scholar]
- 45.Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum beta-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother 53:4930–4933. 10.1128/AAC.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garza-Ramos U, Barrios H, Reyna-Flores F, Tamayo-Legorreta E, Catalan-Najera JC, Morfin-Otero R, Rodriguez-Noriega E, Volkow P, Cornejo-Juarez P, Gonzalez A, Gaytan-Martinez J, Del Rocio Gonzalez-Martinez M, Vazquez-Farias M, Silva-Sanchez J. 2015. Widespread of ESBL- and carbapenemase GES-type genes on carbapenem-resistant Pseudomonas aeruginosa clinical isolates: a multicenter study in Mexican hospitals. Diagn Microbiol Infect Dis 81:135–137. 10.1016/j.diagmicrobio.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 47.Papp-Wallace KM, Barnes MD, Alsop J, Taracila MA, Bethel CR, Becka SA, van Duin D, Kreiswirth BN, Kaye KS, Bonomo RA. 2018. Relebactam is a potent inhibitor of the KPC-2 beta-lactamase and restores imipenem susceptibility in KPC-producing Enterobacteriaceae. Antimicrob Agents Chemother 62:e00174-18. 10.1128/AAC.00174-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aghazadeh M, Rezaee MA, Nahaei MR, Mahdian R, Pajand O, Saffari F, Hassan M, Hojabri Z. 2013. Dissemination of aminoglycoside-modifying enzymes and 16S rRNA methylases among Acinetobacter baumannii and Pseudomonas aeruginosa isolates. Microb Drug Resist 19:282–288. 10.1089/mdr.2012.0223. [DOI] [PubMed] [Google Scholar]
- 49.Bruchmann S, Dotsch A, Nouri B, Chaberny IF, Haussler S. 2013. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob Agents Chemother 57:1361–1368. 10.1128/AAC.01581-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kos VN, Deraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clinical and Laboratory Standards Institute. 2021. M100-S31. Performance standards for antimicrobial susceptibility testing, 31st ed. Clinical and Laboratory Standards Institute, Malvern, PA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hujer AM, Long SW, Olsen RJ, Taracila MA, Rojas LJ, Musser JM, Bonomo RA. 2020. Predicting beta-lactam resistance using whole genome sequencing in Klebsiella pneumoniae: the challenge of beta-lactamase inhibitors. Diagn Microbiol Infect Dis 98:115149. 10.1016/j.diagmicrobio.2020.115149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marshall S, Hujer AM, Rojas LJ, Papp-Wallace KM, Humphries RM, Spellberg B, Hujer KM, Marshall EK, Rudin SD, Perez F, Wilson BM, Wasserman RB, Chikowski L, Paterson DL, Vila AJ, van Duin D, Kreiswirth BN, Chambers HF, Fowler VG, Jr, Jacobs MR, Pulse ME, Weiss WJ, Bonomo RA. 2017. Can ceftazidime-avibactam and aztreonam overcome beta-lactam resistance conferred by metallo-beta-lactamases in Enterobacteriaceae? Antimicrob Agents Chemother 61:e02243-16. 10.1128/AAC.02243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis JJ, Boisvert S, Brettin T, Kenyon RW, Mao C, Olson R, Overbeek R, Santerre J, Shukla M, Wattam AR, Will R, Xia F, Stevens R. 2016. Antimicrobial resistance prediction in PATRIC and RAST. Sci Rep 6:27930. 10.1038/srep27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, Gabbard JL, Gerdes S, Guard A, Kenyon RW, Machi D, Mao C, Murphy-Olson D, Nguyen M, Nordberg EK, Olsen GJ, Olson RD, Overbeek JC, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomas C, VanOeffelen M, Vonstein V, Warren AS, Xia F, Xie D, Yoo H, Stevens R. 2020. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res 48:D606–D612. 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wattam AR, Brettin T, Davis JJ, Gerdes S, Kenyon R, Machi D, Mao C, Olson R, Overbeek R, Pusch GD, Shukla MP, Stevens R, Vonstein V, Warren A, Xia F, Yoo H. 2018. Assembly, annotation, and comparative genomics in PATRIC, the all bacterial bioinformatics resource center. Methods Mol Biol 1704:79–101. 10.1007/978-1-4939-7463-4_4. [DOI] [PubMed] [Google Scholar]
- 60.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL. 2017. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542. 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winkler ML, Papp-Wallace KM, Hujer AM, Domitrovic TN, Hujer KM, Hurless KN, Tuohy M, Hall G, Bonomo RA. 2015. Unexpected challenges in treating multidrug-resistant Gram-negative bacteria: resistance to ceftazidime-avibactam in archived isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 59:1020–1029. 10.1128/AAC.04238-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gonzalez MM, Kosmopoulou M, Mojica MF, Castillo V, Hinchliffe P, Pettinati I, Brem J, Schofield CJ, Mahler G, Bonomo RA, Llarrull LI, Spencer J, Vila AJ. 2015. Bisthiazolidines: a substrate-mimicking scaffold as an inhibitor of the NDM-1 carbapenemase. ACS Infect Dis 1:544–554. 10.1021/acsinfecdis.5b00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bethel CR, Distler AM, Ruszczycky MW, Carey MP, Carey PR, Hujer AM, Taracila M, Helfand MS, Thomson JM, Kalp M, Anderson VE, Leonard DA, Hujer KM, Abe T, Venkatesan AM, Mansour TS, Bonomo RA. 2008. Inhibition of OXA-1 β-lactamase by penems. Antimicrob Agents Chemother 52:3135–3143. 10.1128/AAC.01677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Fig. S1 to S3, and Text S1. Download aac.01790-21-s0001.pdf, PDF file, 0.6 MB (576.3KB, pdf)
Data Availability Statement
Raw sequence reads and genome assemblies are registered at NCBI under BioProject no. PRJNA342804 and are deposited in the SRA repository (SRR13607658 for CLB 24388 and SRR13610045 and SRR15291349 for EM 2972704) and in the GenBank WGS repository (WGS accession no. JAFFGY000000000 for CLB 24388 and JAFFGW000000000 for EM 2972704).