ABSTRACT
Posttranscriptional modifications to tRNA are critical elements for the folding and functionality of these adaptor molecules. Sulfur modifications in tRNA are installed by specialized enzymes that act on cognate tRNA substrates at specific locations. Most studied organisms contain a general cysteine desulfurase to mobilize sulfur for the synthesis of S-tRNA and other thio-cofactors. Bacillus subtilis and other Gram-positive bacteria encode multiple cysteine desulfurases that partner with specific sulfur acceptors in the biosynthesis of thio-cofactors. This metabolic layout suggests an alternate mode of regulation in these biosynthetic pathways. In this study, tRNA modifications were exploited as a readout for the functionality of pathways involving cysteine desulfurases. These analyses showed that the relative abundance of 2-thiouridine-modified tRNA (s2U) responds to sulfur availability in the growth medium in a dose-dependent manner. This study found that low sulfur concentrations lead to decreased levels of the s2U cysteine desulfurase YrvO and thiouridylase MnmA, without altering the levels of other cysteine desulfurases, SufS, NifS, and NifZ. Analysis of pathway metabolites that depend on the activity of cysteine desulfurases indicates that sulfur nutrient availability specifically impacts s2U accumulation while having no effect on the levels of other S-modified tRNA or activity levels of Fe-S enzymes. Collectively, these results support a model in which s2U tRNA serves as a marker for sulfur availability in B. subtilis.
IMPORTANCE The 2-thiouridine (s2U) tRNA modification is found ubiquitously across all domains of life. YrvO and MnmA, the enzymes involved in this modification, are essential in B. subtilis, confirming the well-established role of s2U in maintaining translational efficiency and, consequently, cellular viability. Herein, we show that in the model Gram-positive organism Bacillus subtilis, the levels of s2U are responsive to sulfur availability. Downregulation of the s2U biosynthetic components leads to lower s2U levels, which may serve as a signal for the slowing of the translational apparatus during cellular nutrient insufficiency. Our findings provide the basis for the identification of a potential bacterial mode of regulation during S-metabolite depletion that may use s2U as a marker of suboptimal metabolic status.
KEYWORDS: Bacillus subtilis, s2U, thiouridine, tRNA modification, YrvO, MnmA, thiouridylase, 2-thiouridine, cysteine desulfurase, thionucleoside, trmU
INTRODUCTION
tRNA provides the physical link for the decoding of genetic information during protein synthesis. The maturation of tRNAs involves a series of posttranscriptional modifications that alter the structure and function of these adaptor molecules, thereby guaranteeing the accuracy and efficiency of translation of the genetic code. More than 100 modifications have been described throughout the structure of tRNA (1). Chemical alterations to anticodon bases have long been defined as critical elements for recognizing degenerated codons within mRNA (2). It is also known that select tRNA modifications are essential for interactions with other components of the translational apparatus, including tRNA synthetases, elongation factor Tu (EF-Tu), and the ribosome (3–5). Thus, given the importance of these modifications, it is no surprise that defects in enzymes known to catalyze these modifications lead to either lethal phenotypes or metabolic defects resulting in compromised cellular viability (6–8).
The installment of tRNA modifications requires specialized enzymes that introduce expanded chemical functionalities within the bases and 2′-OH of tRNA at predictable locations. These enzymes recognize their cognate tRNA substrates and install chemical modifications through substitutions or addition reactions with complexities varying from simple methylations to the insertion of highly intricate structures like the queuosine (Q34) full base replacement and geranyl moieties (9, 10). Biosynthetic pathways responsible for these modifications are diverse in nature and number of enzymatic components, and in some cases, they diverge across species and even in organelles within the same species. These various pathways not only display distinct mechanistic strategies for modifying tRNA but also offer an opportunity for alternate regulatory mechanisms of these enzymes and, consequently, modulation of tRNA functionality.
In addition to their canonical role in translation, tRNAs in their fully and partially modified forms are also proposed to serve as regulatory entities and sensors of metabolic cellular status (11). These alternate roles are attributed to the fact that tRNA modifications vary in type and degree of modification in response to their growth phase, environmental signals, and nutritional conditions (12). These conditions are known to affect tRNA modifications either directly, by reacting with modified tRNA bases such as UV radiation and alkylating agents, or indirectly, by downregulating expression of or inactivating tRNA-modifying enzymes (13, 14). Thus, tRNA and their associated modified nucleosides can be exploited as markers of metabolic changes and potentially serve as intermediates in signaling events that allow cells to adjust translational capacity under suboptimal growth conditions.
Sulfur-containing modifications to tRNA are found in all organisms studied thus far (15, 16). In bacteria and eukaryotes, these modifications are dependent on the activity of cysteine desulfurases. These enzymes are known to utilize the free amino acid cysteine as the initial sulfur source for the biosynthesis of S-containing tRNA (17–19). They directly participate as sulfur donors for the biosynthesis of thio-modified tRNA, as observed for the 4-thiouridine (s4U8), 2-thiouridine (s2U34), and 2-methylthioadenosine (ms2A37) modifications (19). These enzymes are also involved in the synthesis of other thio-cofactors, including Fe-S clusters, thiamine, biotin, lipoic acid, and the molybdenum cofactor (20). Because of their role in Fe-S cluster formation, cysteine desulfurases also indirectly affect the synthesis of certain tRNA modifications that do not contain sulfur but rather are dependent on the activity of Fe-S cluster-containing tRNA-modifying enzymes, as in the case of the 2-methyladenosine (m2A37), epoxyqueuosine (oQ34), and Q modifications (19). Therefore, analysis of tRNA modifications across distinct cellular contexts provides an attractive metabolic probe to assess the direct and indirect participation of cysteine desulfurases and tRNA-modifying enzymes in sulfur trafficking pathways.
The involvement of cysteine desulfurases in pathways involving tRNA-modifying enzymes has been previously explored in bacteria. In Escherichia coli and Salmonella enterica, IscS is the master cysteine desulfurase that acts as the primary sulfur donor to all sulfur-containing modifications in tRNA, in addition to serving as the sulfur source for other thio-cofactors (17, 21, 22). The general involvement of IscS in several biochemical pathways, not limited to Fe-S clusters as initially described, stems from its ability to interact with a suite of sulfur acceptors with diverse folds and functions (23). Conversely, Bacillus subtilis and other Gram-positive organisms do not encode one master cysteine desulfurase like IscS. Instead, their genomes code for multiple cysteine desulfurases.
In organisms expressing multiple cysteine desulfurases, the genomic neighborhood of cysteine desulfurases gives insight into their distinct functions during the synthesis of thio-cofactors. B. subtilis, for example, expresses four functionally active cysteine desulfurases: SufS, NifZ, YrvO, and NifS. In previous work, our group investigated the selective reactivity of these enzymes and their corresponding sulfur acceptors. Our group established that NifZ and the thiouridylase ThiI are involved in the biosynthesis of s4U tRNA (24). Similarly, our group showed that YrvO can transfer sulfur directly to the MnmA thiouridylase during s2U tRNA formation, utilizing an abbreviated pathway that dispenses with the need for a sulfur relay system observed in other species (25). We and others determined that SufS partners with SufU, a zinc-dependent sulfurtransferase, in the general synthesis of Fe-S clusters (26, 27). Lastly, inactivation of the cysteine desulfurase NifS leads to defects in NAD biosynthesis (28), a phenotype that is justified by the proposed role of NifS in the activation of the Fe-S enzyme quinolinate synthase, NadA. Taken together, biochemical and genetic studies have demonstrated that these cysteine desulfurases partner with sulfur acceptors to perform dedicated roles. This metabolic layout suggests that the recruitment of multiple enzymes provides an alternate strategy for directing sulfur supply for thio-cofactor synthesis by uncoupling routes of sulfur trafficking.
In this investigation, we sought to gain insight into how sulfur source and concentration would impact the accumulation of these biosynthetic enzymes and their pathway metabolites. We analyzed the relative levels of tRNA modifications as a metabolic readout of the functionality of biosynthetic pathways involving three cysteine desulfurases in B. subtilis: YrvO, SufS, and NifZ. We show that sulfur availability provided in the media selectively impacts the levels of the s2U biosynthetic components, YrvO and MnmA, the accumulation of this thionucleoside, and select tRNAs targeted for this modification. Given the essentiality of these enzymes and the well-documented role of s2U in protein synthesis, we propose that downregulation of the s2U modification at various sulfur availabilities leads to changes in translational capacity in this organism.
RESULTS
Bacillus subtilis tRNA epitranscriptome is modulated by nutritional conditions.
In a recent study, 29 modifications were reported at various positions in B. subtilis tRNA (29). Analysis of genes encoding tRNA-modifying enzymes in the Colombos database showed variation in their expression across different growth conditions and phases. Most of these enzymes displayed a unique pattern of expression across different stimuli, suggesting that the landscape of tRNA modifications possibly adjusts its composition in response to cellular changes. Therefore, we first surveyed the B. subtilis tRNA epitranscriptome in an effort to expand upon these previous findings and determine the presence of tRNA modifications in B. subtilis cells cultured at different growth stages and media. This initial analysis ensured our ability to add to the evolving body of literature surrounding B. subtilis tRNA modifications. Using liquid chromatography-mass spectrometry (LC-MS) analysis of purified RNA nucleosides from B. subtilis cultures, we were able to confirm the prevalence of all base modifications recently reported by de Crécy-Lagard et al. (29). The various levels of these modifications were compared in rich and minimal medium cultures at early exponential phase, mid-exponential phase, and late exponential phase, corresponding to optical densities at 600 nm (OD600) of 0.5, 1, and 1.5, respectively (see Table S1 and S2 in the supplemental material). The relative levels of several modifications exhibited various patterns of accumulation (Table 1). These analyses also matched previous findings that show increased relative levels of 2-methylthio-N6-isopentenyladenosine (ms2i6A37) over its precursor form, N6-isopentenyladenosine (i6A37), as cells enter stationary phase (30). However, we attribute this result not to the apparent increase of the hypermodified base ms2i6A but instead to the decreased levels of i6A at the later growth stages, which decrease 2- and 10-fold in minimal and rich media, respectively. Another modification showing marked fluctuation is the Q hypermodification and its precursor nucleoside, oQ. Total RNA isolated from cells cultured in rich medium showed no detectable levels of oQ, and accumulation of this intermediate was observed only in minimal medium. Additionally, the relative levels of the Q modification were over 10-fold higher in rich medium. These variations are compatible with the requirement of cobalamin as a cofactor during the conversion of oQ to Q by the epoxyqueuosine reductase, QueG (31). The ratio of 5-methyaminomethyl-2-thiouridine (mnm5s2U34) to 5-carboxymethyaminomethyl-2-thiouridine (cmnm5s2U34) remains mostly balanced across growth stages in minimal medium, and was in agreement with previous reports (32), but drops at later growth stages in rich media. Overall, these initial analyses indicated that tRNA modifications fluctuate across growth stages and that the availability of nutrients, as assessed by rich and minimal media, also impacts the relative accumulation of these metabolites, some of which have been previously described. Therefore, when performing a comparative study on the effects of nutritional and environmental factors on the tRNA epitranscriptome, it is important to assess cultures grown under controlled conditions and analyzed at the same growth stage.
TABLE 1.
Relative levels of selected tRNA modifications in B. subtilis PS832 cells cultured in LB and MMa
| Modificationb | LB | MM |
|---|---|---|
| s4U | 0.08 (0.0) | 0.13 (0.05) |
| mnm5U* | 0.02 (0.02) | 0.06 (0.01) |
| mnm5s2U | 0.29 (0.07) | 0.24 (0.15) |
| i6A | 2.0 (0.48) | 1.81 (0.64) |
| ms2i6A* | 0.88 (0.23) | 0.50 (0.06) |
| Q* | 1.2 (0.38) | 0.09 (0.03) |
| oQ | ND | 0.6 (0.26) |
| cmnm5s2U | 0.3 (0.04) | 0.25 (0.02) |
| cmnm5U | 0.04 (0.02) | 0.07 (0.03) |
| mo5U* | 0.42 (0.09) | 0.19 (0.09) |
| m7G* | 0.02 (0.03) | 0.06 (0.01) |
tRNA isolated from cells cultured up to an OD600 of 0.5. LB, Luria-Bertani medium; MM, Spizizen’s medium. Asterisks indicate that there were statistically significant differences between modification levels from total RNA isolated from cells cultured in LB versus MM (P < 0.05). ND, not detected.
Relative levels of each modification were determined by normalizing the mass abundance associated with each modification to the mass abundance of dihydrouridine in the same sample. The reported averages and associated standard deviations were calculated based on data acquired from at least three independent experiments.
s2U-modified tRNA levels respond to sulfur availability.
Utilizing this workflow, we specifically analyzed tRNA modifications dependent on cysteine desulfurases using cultures grown in chemically defined media. Cells were harvested at mid-exponential phase (OD600 of 0.5), as this represents a growth stage in which overall nutrient levels have not yet been depleted. We hypothesized that the occurrence of multiple cysteine desulfurases provides a strategy for differential regulation of these enzymes and, consequently, differential accumulation of metabolites dependent on their activities. Therefore, we determined the relative abundance of tRNA modifications as a readout of cysteine desulfurase functionality and ultimately assessed their ability to either directly or indirectly promote chemical modifications within tRNA nucleosides. The relative degree of modification to tRNA was determined by analyzing individual digested tRNA species, in which the mass abundance of each modification was normalized by the mass abundance of dihydrouridine (D). This modification serves as an excellent internal normalizing standard due to its high abundance, stability, and virtual exclusivity in prokaryotic tRNA (33). We specifically interrogated whether varying the sulfur availability would impact the accumulation of tRNA modifications. Interestingly, under sulfur-replete conditions, the source of sulfur did not affect growth (Fig. S1) or the accumulation of sulfur-containing nucleosides (Table S3), indicating that B. subtilis can adjust its metabolism to multiple sulfur sources.
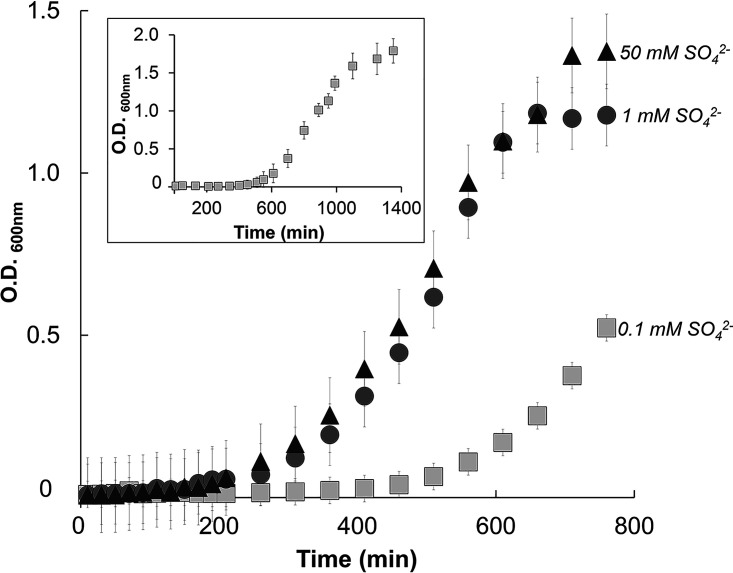
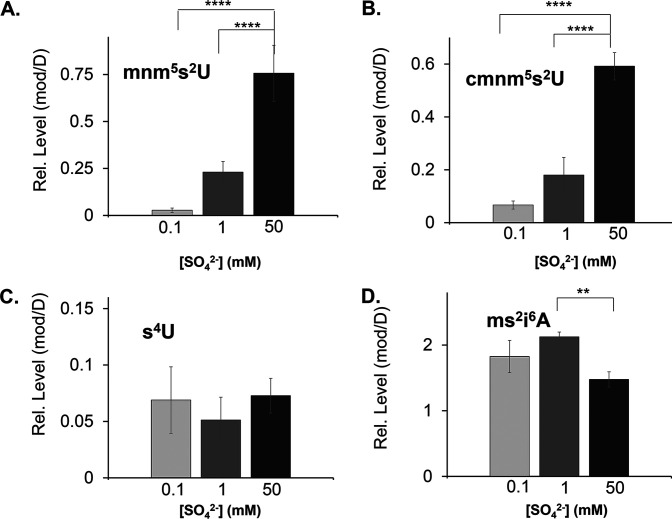
However, various concentrations of the sulfur source showed an impact on the rate of growth (Fig. 1). B. subtilis cells cultured under low sulfur concentrations (e.g., 0.1 mM sulfate) displayed prominent lag phase extension compared to the 1 mM and 50 mM “sulfur replete” growth rates. This growth profile suggests the presence of regulatory mechanisms under nutrient-deprived conditions. Relevant to the analysis of this study, we found that the relative levels of mmn5s2U and cmnm5s2U responded drastically to various sulfur concentrations (Fig. 2). In fact, the accumulation of hypermodified s2U derivatives showed a dose-dependent accumulation upon increasing concentrations of sulfur in the growth medium. Under these conditions in the same tRNA samples, the levels of sulfur-containing modifications whose synthesis depends on the activity of other cysteine desulfurases remained unchanged, suggesting a potential alternate mechanism for regulation of thio-cofactor synthesis (Fig. 2). It is worth mentioning that B. subtilis does not contain 2-thiocytidine (s2C32), and its genome does not encode the s2C-modifying biosynthetic enzyme, TtcA. Additionally, another S-modification reported for this organism, 2-methylthio-N6-threonylcarbamoyladenosine (ms2t6A37), does not accumulate to high enough levels to permit a semiquantitative assessment of its presence under the tested conditions. Collectively, these results show that the accumulation of s2U tRNA is responsive to nutritional growth conditions.
FIG 1.
Growth profile of B. subtilis PS832 cultured in the presence of various sulfur concentrations of (NH4)2SO4. Spizizen’s minimal media containing as the sole sulfur source either 0.1, 1, or 50 mM (NH4)2SO4. The full growth curve for 0.1 mM (NH4)2SO4 is represented in the inset. Growth was monitored through optical density (600 nm) with shaking at 300 rpm and 37°C. Values on the y axis have been adjusted for a path length of 1 cm. The reported averages and associated standard deviations were calculated based on data acquired from at least three independent experiments.
FIG 2.
Relative levels of S-tRNA modifications vary in B. subtilis cells cultured under various sulfur concentrations. B. subtilis PS832 was cultured to an OD of 0.5 in Spizizen’s minimal media containing different concentrations of (NH4)2SO4. Relative levels of modified tRNA nucleosides (mod) were analyzed by ultrahigh-pressure liquid chromatography-coupled mass spectrometry (UHPLC-MS) and normalized to the levels of dihydrouridine (D). tRNA modifications are shown for 5-methylaminomethyl-2-thiouridine (mnm5s2U) (A), 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U) (B), 4-thiouridine (s4U) (C), and 2-methylthio-N6-isopentenyl adenosine (ms2i6A) (D). A paired t test compared samples at 0.1, 1, and 50 mM (NH4)2SO4 (**, P ≤ 0.01; ****, P ≤ 0.0001). Values without an asterisk are considered statistically insignificant.
Levels of s2U biosynthetic enzymes respond to sulfur availability.
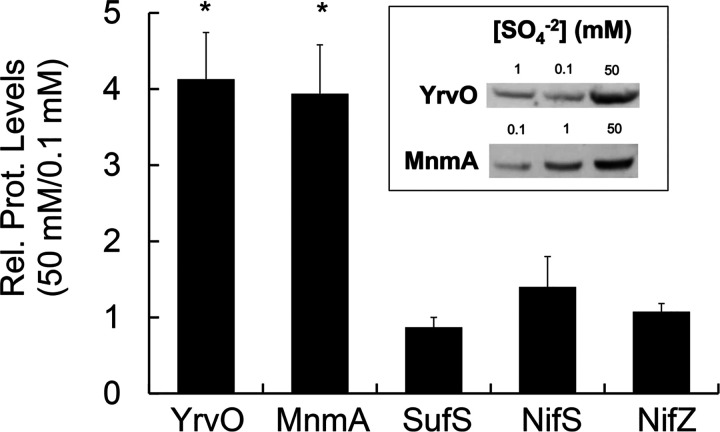
The decreased levels of s2U tRNA in response to sulfur limitation could be attributed to decreased levels of thiolation substrates (i.e., cysteine) and/or their biosynthetic enzymes. To investigate these possibilities, we determined intracellular levels of reduced cysteine, YrvO, and MnmA from cells cultured under distinct sulfur concentrations. Cell extracts from washed cell pellets were derivatized with monobromobimane, followed by high-performance liquid chromatography (HPLC) separation and quantification of the fluorescently labeled Cys-bimane adduct. These analyses showed steady levels of cysteine across various conditions (Table S4); that is, under controlled growth conditions, reduced cysteine varied from 0.35 to 0.39 mM, which is similar to previously reported intracellular reduced cysteine concentrations in B. subtilis (0.13 to 0.58 mM) (34, 35). Notably, changes in sulfur concentrations in the media did not dramatically impact intracellular levels of reduced cysteine, further iterating the tight regulatory network imposed on cysteine biosynthesis and degradation as previously described. While the levels of cysteine did not change under these various conditions, the levels of s2U biosynthetic enzymes, YrvO and MnmA, responded to sulfur availability in the growth medium in a dose-dependent manner. Further quantification of these enzymes in soluble cell extracts showed that cells cultured under low sulfur concentrations displayed reduced levels of both enzymes (Fig. 3). We then investigated if the decrease in accumulation of these enzymes was a result of downregulation at the transcriptional level. However, quantitative PCR (qPCR) analysis showed that the levels of yrvO and mnmA transcripts remain unaltered in cells cultured under both high and low sulfur conditions (Fig. S4). Interestingly, previous kinetic analysis reporting the reactivity of B. subtilis cysteine desulfurases toward their cysteine substrate revealed that YrvO has a lower Km for cysteine than the other desulfurases in this organism (24–26). Since the availability of intracellular cysteine is tightly controlled, regulation of the s2U pathway components’ expression or activity provides a rationale for the low levels of its final pathway metabolite, s2U tRNA.
FIG 3.
B. subtilis cells cultured under low sulfur concentrations show increased accumulation of YrvO and MnmA in soluble extracts. Western blots were used to determine the levels of B. subtilis YrvO, MnmA, SufS, NifS, and NifZ in 50 μg of soluble crude extracts prepared from B. subtilis. PS832 cells were cultured to an OD of 0.5 in Spizizen’s minimal media containing 0.1, 1, 17, and 50 mM (NH4)2SO4. Changes in relative protein levels were determined by the signal intensity from samples cultured in 50 mM (NH4)2SO4 over the signal intensity of samples cultured in 0.1 mM (NH4)2SO4 from the same gel. A paired t test compared samples at 0.1 and 50 mM from the same gel (*, P < 0.05); values without an asterisk are considered statistically insignificant. A representative blot is shown in the inset, and additional data sets are provided in Fig. S2.
Accumulation of other cysteine desulfurases and their metabolites does not respond to sulfur availability.
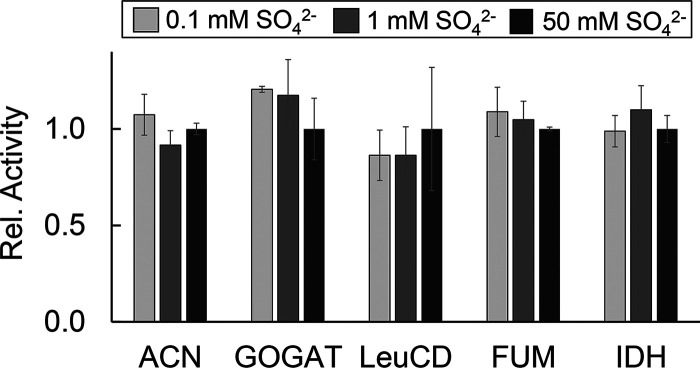
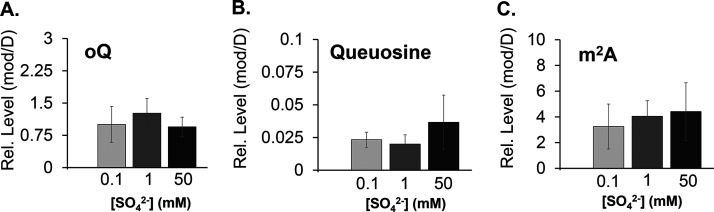
This regulatory response was not observed for the other three B. subtilis cysteine desulfurases. Analysis of the same crude extracts showed that the levels of NifS, NifZ, and SufS did not vary with sulfur concentration (Fig. 3). In fact, metabolites dependent on NifZ and SufS activity also remained unchanged under these conditions. Supporting this result, the levels of s4U, whose synthesis depends on NifZ and ThiI, did not vary with sulfur concentration (Fig. 2). Likewise, the metabolites presumably dependent on pathways involving SufS also remained unaltered under these various conditions. In B. subtilis, the general synthesis of Fe-S clusters is proposed to involve the Suf system (36). The functionality of this system can be assessed through the activity of Fe-S enzymes and through quantification of the levels of tRNA modifications that require Fe-S enzymes in their biosynthetic schemes (19). Under low sulfur concentrations, the activity of Fe-S enzymes aconitase (ACN), glutamine:2-oxoglutarate amidotransferase (GOGAT), and isopropyl malate dehydratase (LeuCD) remained unchanged. The same trend was also observed for enzymes that do not require Fe-S clusters or the activity of SufS, such as isocitrate dehydrogenase (IDH) and fumarase (FUM) (Fig. 4). Furthermore, relative levels of the tRNA modifications ms2i6A, oQ, Q, and m2A, which are synthesized by Fe-S enzymes proposed to rely on the Suf system, also remained unresponsive to these various sulfur concentrations (Fig. 2 and 5). Lastly, as expected, the levels of NifS did not change. The expression of this cysteine desulfurase has been shown to depend on the availability of nicotinic acid in a regulatory system dependent on the transcriptional regulator NadR, and its activity is known to impact the synthesis of NAD in B. subtilis (28).
FIG 4.
Varying the sulfur concentrations in the growth medium does not impact the activity of Fe-S enzymes. Soluble extracts from cells cultured in different sulfur concentrations were prepared anoxically and were subsequently tested for the activity of Fe-S enzymes aconitase (ACN), glutamine:2-oxoglutarate amidotransferase (GOGAT), and isopropylmalate isomerase (LeuCD) and non-Fe-S enzymes fumarase (FUM) and isocitrate dehydrogenase (IDH). The relative activities of ACN, GOGAT, LeuCD, FUM, and IDH were normalized to the activity values of cell extracts cultured in Spizizen’s media containing 17 mM (NH4)2SO4 (254, 32, 35, 584, 210 nmol min−1 mg−1, respectively). The reported averages and associated standard deviations were calculated based on data acquired from at least three independent experiments. A paired t test compared samples at 0.1, 1, and 50 mM (NH4)2SO4. None of the conditions showed statistically significant differences.
FIG 5.
Relative levels of epoxyqueuosine (oQ) (A), queuosine (Q) (B), and 2-methyl adenosine (m2A) (C) tRNA modifications from B. subtilis cells cultured under different sulfur concentrations. B. subtilis PS832 was cultured to an OD600 of 0.5 in Spizizen’s minimal media containing different concentrations of (NH4)2SO4. Total RNA was purified from these cultures, and the relative levels of individual tRNA nucleosides were analyzed by mass spectrometry and normalized to the levels of dihydrouridine. A paired t test compared samples at 0.1, 1, and 50 mM (NH4)2SO4. None of the conditions showed statistically significant differences.
Low levels of tRNA thiolation lead to low levels of tRNALys and tRNAGlu.
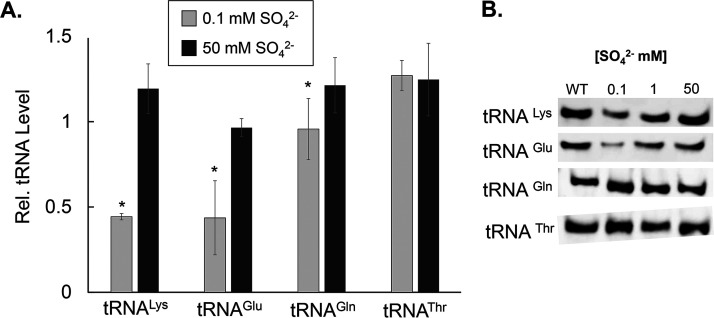
It has been previously demonstrated that hypomodified tRNA is targeted for degradation (2, 37, 38). Therefore, we sought to determine whether conditions that led to downregulation of the synthesis of s2U tRNA would ultimately lead to lower levels of its targets substrates, tRNAGlu,Gln,Lys. Northern blot analyses of total RNA samples isolated from cells cultured under low sulfur concentrations showed lower levels of accumulated tRNALys and tRNAGlu and a modest decrease of tRNAGln (Fig. 6). Importantly, under sulfur-limiting conditions, the extent to which these tRNA substrates are depleted was not nearly as dramatic as the effect observed in mnm5s2U under the same nutrient limitations. Control experiments included tRNAThr, which does not carry the s2U modification but is transcribed in the same operons as these selected tRNAs. While mechanisms triggering degradation of hypomodified tRNA have been described for s2U in E. coli (39), it is unknown if B. subtilis evokes a specific RNA nuclease under conditions of nutrient limitation or if this process fits under the umbrella of the B. subtilis general tRNA quality control system. Nevertheless, it is worth noting that the lower abundance of the s2U-modified tRNA under sulfur-depleted conditions is not accompanied by proportionally increased levels of the partially modified 5-methylaminomethyluridine (mnm5U34) and 5-carboxymethyaminomethyluridine (cmnm5U34) precursor tRNAs (Fig. S5). Taken together, these results support the presence of lower levels of these tRNA species under low-sulfur conditions and further highlight the importance of s2U for the stability of tRNAs carrying this modification.
FIG 6.
B. subtilis cells cultured under low sulfur concentrations show reduced levels of tRNALys and tRNAGlu. (A) B. subtilis PS832 was cultured to an OD of 0.5 in Spizizen’s minimal medium containing different concentrations of (NH4)2SO4. tRNA was purified from these cultures and the relative levels of tRNA known to contain s2U modifications (tRNALys, Glu, and Gln) were compared to the levels of a tRNA that is not modified at this position (tRNAThr). For each sample, 5 μg of total RNA was loaded in each gel lane, and representative samples of cells grown under various sulfur sources were quantified in the same gel. The relative accumulation of each cognate tRNA was determined by normalizing the signal intensity of each sample by the signal intensity of a tRNA sample extracted from cells cultured in Spizizen’s medium containing 17 mM (NH4)2SO4. Averages of ratios and associated standard deviations were calculated from blots generated from at least three experimental replicates. A paired t test compared samples at 0.1 and 50 mM from the same gel (*, P < 0.05). Values without an asterisk are considered statistically insignificant. (B) A representative blot is shown for each tRNA cognate and additional data sets are provided in Fig. S3.
DISCUSSION
The synthesis and reactivities of modified tRNA are known to be affected by nutritional and environmental changes (12). Here, we report that the levels of the essential s2U modification in tRNA vary with different sulfur concentrations provided in the growth medium. This phenomenon was specific to both s2U derivatives, namely, mnm5s2U and cmnm5s2U, and was not observed in other abundant thio-tRNA modifications. The near exclusivity of this dose-dependent response is relevant given the essential nature of this modification, and the broader implications of these findings highlight the complex nature of cellular physiological checkpoints. These findings provide a potential link between s2U’s long-understood and highly conserved role in directly influencing the speed and accuracy of translation (40–42) and its ability to respond to the available pool of cellular S-metabolites in a manner that is independent of other known B. subtilis sulfur trafficking systems.
Interestingly, the sulfur-mediated, dose-dependent effect impacting this modification did not show a proportionally inverse relationship to the partially modified forms of mnm5s2U and cmnm5s2U. In fact, the levels of these modifications only slightly increased at higher sulfur concentrations and not to the degree that their hypermodified counterparts did (Fig. S5). While modest increases were observed, a direct comparison between the levels of these hypomodified tRNAs and their thiolated forms is convoluted, as it has been shown that the E. coli Mnm machineries, MnmEG and MnmC, are able to use tRNALeu,Arg,Gly as substrates for their methylation and decarboxylation reactions in addition to the thiolation substrates, tRNALys,Glu,Gln (43). These results, however, support previous in vitro experiments promoting the synthesis of singly modified s2U and an absence of mnm5s2U formation when using bulk in vivo isolated tRNA containing both U34 and partially modified mnm5U34 tRNA (25). Furthermore, structural analysis of E. coli MnmA bound to the adenylated tRNA shows the presence of conserved, bulky residues that would hinder the binding of a partially modified tRNA substrate containing mnm5U or cmnm5U at the active site (44). Together with previously demonstrated structural and enzymatic data, the results presented herein support a model in which the s2U modification precedes installation of additional modifications at C5 of U34 in tRNALys,Glu,Gln. Notably, our tRNA analyses yielded no detectable levels of 5-aminomethyluridine (nm5U34) or 5-aminomethyl-2-thiouridine (nm5s2U34) intermediates. It is known that B. subtilis lacks an ortholog for MnmC, the enzyme responsible for the synthesis of these respective modified tRNAs. However, B. subtilis has been shown to efficiently catalyze the formation of mnm5s2U (32). Although the proposed pathway involves the transient formation of an nm5s2U intermediate, this species has not been detected in B. subtilis tRNA samples, suggesting the high efficiency of this proposed MnmC-like reaction.
Further analysis of tRNA also showed that low sulfur availability does not impact the levels of alternate tRNA modifications dependent on the activity of other cysteine desulfurases. The occurrence of distinct cysteine desulfurases in B. subtilis provides alternate points of regulation for the synthesis and use of thio-cofactors. The levels of SufS were unchanged under these conditions, indicating that its pathway components remain relatively stable. SufS is expressed with other Suf components and is proposed to be the primary, if not sole, Fe-S cluster biosynthetic system in this organism (36). The activities of Fe-S enzymes were unchanged; likewise, the levels of nucleosides whose syntheses are known to involve Fe-S enzymes were also unperturbed under identical conditions, suggesting that B. subtilis potentially prioritizes Fe-S metabolism to sustain critical biochemical reactions, even under sulfur-limiting conditions.
The occurrence of multiple cysteine desulfurases in B. subtilis and the involvement of a dedicated enzyme for the synthesis of s2U that is uncoupled from the general synthesis of Fe-S clusters suggest an alternate strategy to mobilize and regulate thio-cofactor biogenesis. The levels of these enzymes respond to sulfur availability and impact the accumulation of their pathway metabolites. Interestingly, the E. coli thiouridylase MnmA has been recently shown to coordinate an Fe-S cluster that provides the enzyme with enhanced reactivity (45). Whether this type of cofactor also occurs in B. subtilis MnmA merits investigation. Yet the utilization of an abbreviated s2U pathway in this organism provides an exciting model for elucidating the mechanistic steps during sulfur acquisition and transfer and potentially establishes additional points of regulation in s2U formation.
The low levels of s2U tRNA are attributed to the low availability of its biosynthetic enzymes. In this study, the quantification of cysteine desulfurases was determined in clear soluble cell extracts, allowing for accurate protein quantification of samples prior to Western blot analysis. The levels of other cysteine desulfurases in these samples did not vary across conditions, which unintentionally provided an internal standard for these analyses. Additionally, transcriptomic analyses revealed that levels of all B. subtilis cysteine desulfurases remained relatively consistent across all sulfur conditions, suggesting that regulation of YrvO and MnmA occurs at translational or posttranslational levels (Fig. S4). While the mode of regulation of s2U biosynthetic enzymes will be explored in subsequent investigations, it was outside the scope of the present study. However, it is known that genes encoding B. subtilis YrvO and MnmA are located immediately downstream of the master regulator of cysteine metabolism, CymR (46). The regulation of the promoter driving the expression of the cymR-yrvO-mnmA operon has been identified to be under the control of Spx and, consequently, sigA, which are known to respond to stressors affecting thiol-redox homeostasis, such as diamide and carbonyl electrophiles. Spx functions in sulfate-dependent control of organosulfur utilization operons through stimulation of cymR expression. Spx could affect the concentration of a metabolite, perhaps an intermediate of sulfur assimilation or organosulfur utilization, which serves as an effector of cymR expression control (47). Spx was also found to negatively regulate the expression of these operons in sulfate medium, in part by stimulating the expression of the cymR gene (48). In addition, trmU (mnmA) was the only tRNA-modifying gene affected by the RelA-dependent stringent response during norvaline-induced stress, suggesting that nutritional triggers other than sulfur availability may also lead to lower levels of s2U accumulation. Therefore, the genomic location of s2U biosynthetic genes and their coexpression with cymR provide us with a working model that links s2U to B. subtilis sulfur metabolism.
The dose-dependent accumulation of s2U with various sulfur concentrations supports a scenario in which s2U serves as a marker of sulfur availability. Previous studies demonstrated that both yrvO and mnmA are essential genes in B. subtilis, indicating that s2U tRNA is a critical modification enabling translational accuracy and cellular viability. In yeast, the degree of U34 thiolation reflects the availability of S-containing amino acids. This process ultimately utilizes phosphate homeostasis as a metabolic control point during amino acid insufficiency (8, 49). Notably, the yeast pathway involved in this modification is phylogenetically distinct from the ones described for bacteria (50). While the loss of U34 thiolation in yeast led to the characterization of broad metabolic defects and the downregulation of enzymes indispensable to central metabolism, this study explored the impact of sulfur nutrient limitation on sulfur trafficking pathways. Here, we showed that under conditions of sulfur limitation, B. subtilis cells display a growth delay phenotype accompanied by a remarkable decrease in the accumulation of YrvO and MnmA. This may be a result of a combination of not fully understood regulatory mechanisms in place during these nutrient conditions. It is also anticipated that other conditions and cellular processes may elicit the degradation of tRNA and regeneration of bases modified within tRNA molecules. For instance, B. subtilis contains a putative ortholog of the 4Fe-4S cluster thiouracil desulfidase TudS, which has been shown to catalyze the desulfurization of 4-thiouracil and 2-thiouracil (51).
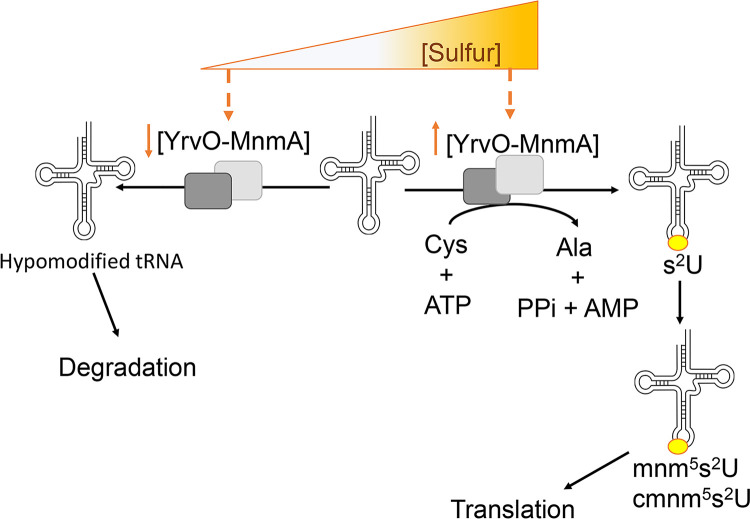
The results obtained in this study suggest that when cell growth and adequate translational efficiency are compromised through the combined loss of s2U biosynthetic components, unmodified substrate tRNAs, and thiolated U34 tRNA, metabolic output wanes and mirrors a manufactured “starvation state.” Under these conditions, lower levels of s2U biosynthetic enzymes are proposed to result in lower levels of s2U tRNA, leaving substrate tRNALys, Glu, Gln hypomodified (Fig. 7). These tRNAs are then likely targeted for degradation by a yet-unknown cellular quality control system. It is possible that an s2U-dependent response triggers a metabolic rewiring that allows for direct and indirect salvage of essential metabolites. Future studies aim to explore these interconnected regulatory pathways responsible for nutrient sensing in B. subtilis.
FIG 7.
Working model for regulation of s2U tRNA synthesis in B. subtilis. YrvO and MnmA catalyze the synthesis of s2U at position 34 of tRNALys, Glu, Gln. When cells are cultured under sulfur replete conditions, YrvO and MnmA promote the formation of s2U tRNA on U34, which is subsequently modified to cmnm5s2U and mnm5s2U as fully functional tRNAs that are indispensable to translation. Under sulfur-deplete conditions, the levels of YrvO and MnmA decrease significantly in a dose-dependent manner, leaving their thiolation substrate tRNAs hypomodified. These translationally inefficient surplus tRNAs are then proposed to be targeted for degradation, thereby impacting the overall translational apparatus.
MATERIALS AND METHODS
Media, medium additions, and chemicals.
All B. subtilis PS832 growths were cultured in Spizizen’s minimal media (52) consisting of 15 mM NH4Cl, 80 mM K2HPO4, 44 mM KH2PO4, 3.4 mM trisodium citrate dehydrate, 0.5% sucrose, various concentrations of (NH4)2SO4 as the sulfur source, and trace elements (60 μM MgCl2, 5 μM CaCl2, 5 μM FeCl2, 0.5 μM MnCl2, 1.25 μM ZnCl2, 0.25 μM CuCl2, 0.4 μM CoCl2, 0.3 μM Na2MoO4, final concentrations). Unless otherwise stated, all chemicals were purchased from Fisher Scientific and Sigma-Aldrich Inc.
B. subtilis growth and tRNA isolation.
B. subtilis PS832 was grown in LB medium or Spizizen’s medium at 37°C and 300 rpm in 0.5-L cultures. Growth was monitored by optical density at 600 nm (OD600). Unless otherwise indicated, cells were harvested at early log phase (OD600 = 0.5), and cell pellets were obtained through centrifugation at 8,200 × g for 10 min and frozen at −20°C until further use. Total RNA was extracted from cell pellets as reported previously (53). Briefly, RNA was extracted from resuspended cells using sodium acetate-saturated phenol (pH 4.3), followed by an additional extraction using a 24:1 chloroform/isoamyl alcohol mixture, and precipitated overnight with 100% ethanol at 4°C. RNA pellets were washed with 70% ethanol, air dried, and resuspended in Optima LC-MS-grade water for further analyses.
Northern blotting.
An aliquot of 5 μg of total purified RNA was separated on a 10% Tris-borate-EDTA (TBE) urea denaturing gel (10% acrylamide/Bis 19:1 wt/vol; 7 M urea). Electrophoresed RNA was transferred to a 0.45-μm Biodyne B precut modified nylon membrane at 250 mA for 2 h and then at 350 mA for 2 h, all performed at 4°C. Membranes were twice cross-linked for 60 s with 1,200 μJ of UV light, followed by prehybridization for 2 h at 42°C with ULTRAhyb-Oligo hybridization buffer. Membranes were subjected to 18 h of hybridization at 42°C with 10 pmol of the appropriate Cy5-labeled tRNA-specific oligonucleotide probe (Table S5). Membranes were washed twice with 2× SSC/SDS (0.3 M NaCl plus 0.03 M sodium citrate dihydrate and 0.1% sodium dodecyl sulfate; pH 7.0) low-stringency buffer for 5 min at 42°C, followed by two additional 0.1× SSC/0.1% SDS high-stringency buffer washes for 15 min at 37°C. Hybridized probe was detected using an Amersham AI600 imager (Amersham Biosciences) using the default Cy5 settings at 630 nm. All fluorescence intensities were quantified after digital capture using ImageJ software and normalized to B. subtilis PS832 grown in Spizizen’s minimal medium supplemented with 17 mM (NH4)2SO4.
Western blotting.
Protein crude extracts were prepared using a sonicator (Fisherbrand model 120 sonic dismembrator 4X) for 10 s at 50% amplitude, followed by centrifugation at 16,873 × g for 20 min. Protein concentrations of soluble crude extracts were determined using the Bio-Rad protein assay and a bovine serum albumin (BSA) standard curve. Aliquots of 50 μg of protein were separated using reducing 10% SDS-PAGE. Electrophoresed protein was transferred to a 0.2-μm nitrocellulose membrane for 1 h at 4°C and then blocked with 4% milk in 1× Tris-buffered saline with Tween 20 (TBST) for 1 h at room temperature. Blocked membranes were incubated with agitation at 4°C with rabbit custom antibodies against B. subtilis YrvO, MnmA, NifZ, NifS, and SufS (Fisher Scientific) used at a 1:1,000 or 1:20,000 dilution. Membranes were washed 6 times with 0.1% TBST at room temperature and then incubated at 4°C with horseradish peroxidase-conjugated goat anti-rabbit (Bio-Rad) secondary antibody for 90 min, followed by an additional set of 6 washes using 0.1% TBST at room temperature. Protein bands were detected by chemiluminescence using the automatic detection setting on an Amersham AI600 imager (Amersham Biosciences). Samples from cell cultures under various sulfate concentrations were loaded in the same gel, along with a purified protein standard. Signal intensity was quantified using ImageJ software and normalized to the levels detected for each protein in soluble extracts of B. subtilis PS832 grown in Spizizen’s minimal media supplemented with 17 mM (NH4)2SO4.
Digestion of total RNA and LC-MS sample preparation.
Unfractionated tRNA isolated from B. subtilis cells was digested into individual nucleosides for analysis. An aliquot of 60 μg of total RNA was heat denatured for 5 min at 95°C and digested for 4 h at 50°C using 5 μL of 0.25-U/μL nuclease P1 (Sigma-Aldrich) and 17 μL of 0.4 M NH4OAc (Ammonium acetate pH 5.3; Optima; LC-MS grade), followed by a 2-h dephosphorylation using recombinant shrimp alkaline phosphatase (New England BioLabs [NEB]) at 37°C. Digested nucleosides were spun at 16,873 × g and room temperature. The supernatant containing digested RNA was transferred to LC-MS glass vials and spikeds with 0.1% formic acid/2% methanol (MeOH; Optima; LC-MS grade) prior to further analysis.
LC-MS method run and tune parameters.
All LC-MS nucleoside analyses were performed in electrospray ionization (ESI)-positive mode using an Agilent Polaris C18-A HPLC column preequilibrated with 98% solvent A (water [Optima; LC-MS grade], 0.1% formic acid) and 2% solvent B (methanol [Optima; LC-MS grade], 0.1% formic acid) for 15 column volumes at a flow rate of 300 μL min−1. Tune file parameters used during runs were as follows: voltage, 4.01 kV; sheath gas flow rate arbitrary unit (arb), 47.00; auxiliary gas flow rate (arb), 30.00; sweep gas flow rate (arb), 0.00; capillary voltage, 2.00 V; capillary temperature, 350.00°C; and tube lens voltage, 49.89 V. Digested nucleosides were separated and detected over the course of a 39-min run using the gradient of solvent B as follows: 0 to 4 min, hold at 2% (vol/vol) solvent B; 4 to 25 min, increase solvent B from 2% to 100%; 25 to 33 min, hold at 100% solvent B; and 33 to 39 min, switch from 100% solvent B to 2% solvent B to allow column reequilibration at 2% solvent B.
HPLC detection of mBBr-labeled cellular l-cysteine.
The contents of reduced thiol l-cysteine were determined using monobromobimane (mBBr) as described by Sharma et al. (34). In short, cell pellets from a 0.5-L culture harvested at an OD600 of 0.5 were resuspended in 4.5 mL of 25 mM HEPES (pH 8). Aliquots of this resuspended pellet were used to determine the dry weight of the cell pellet, mBBr background, and cysteine bimane quantification. Controls were prepared by adding 5 mM N-ethylmaleimide (NEM) to block free thiol, followed by derivatization with 8 mM mBBr in warm acetonitrile. Reduced thiol samples were prepared by resuspension in 25 mM HEPES (pH 8), followed by direct incubation with 8 mM mBBr in warm acetonitrile. The mixture was incubated at 65°C for 20 min and quenched by the addition of 20 μL of 0.1 M HCl. The reactions were then centrifuged for 5 min, 100 μL of each supernatant was diluted 10 times with 2 mM HCl, and 10 μL was injected for high-performance liquid chromatography (HPLC) analysis. The HPLC method was conducted using a Waters Spherisorb C18 column (4.6 by 250 mm, 5 μΜ, 80 Å) using the gradient described in reference 34. The intracellular concentrations of l-cysteine derivatives were calculated based on the l-cysteine standard and normalized to cell dry weight.
ACKNOWLEDGMENTS
We thank Chris Tracy for his technical support with mass spectroscopy analysis and Nicole Kus for initial experimental growth optimization.
K.A.B. was a recipient of a graduate fellowship from the Wake Forest Center for Molecular Signaling. A.M.E. is a recipient of a T32 NIH predoctoral fellowship (GM 95440-9). This research was sponsored by the National Science Foundation (MCB-1716535).
Footnotes
Supplemental material is available online only.
Contributor Information
Patricia C. Dos Santos, Email: dossanpc@wfu.edu.
Elizabeth Anne Shank, University of Massachusetts Medical School.
REFERENCES
- 1.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46:D303–D307. 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motorin Y, Helm M. 2010. tRNA stabilization by modified nucleotides. Biochemistry 49:4934–4944. 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 3.Cusack S, Yaremchuk A, Tukalo M. 1996. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J 15:6321–6334. 10.1002/j.1460-2075.1996.tb01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voorhees RM, Mandal D, Neubauer C, Kohrer C, RajBhandary UL, Ramakrishnan V. 2013. The structural basis for specific decoding of AUA by isoleucine tRNA on the ribosome. Nat Struct Mol Biol 20:641–643. 10.1038/nsmb.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eargle J, Black AA, Sethi A, Trabuco LG, Luthey-Schulten Z. 2008. Dynamics of recognition between tRNA and elongation factor Tu. J Mol Biol 377:1382–1405. 10.1016/j.jmb.2008.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Tuck S, Bystrom AS. 2009. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 5:e1000561. 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker J, Kwon SY, Badenhorst P, East P, McNeill H, Svejstrup JQ. 2011. Role of elongator subunit Elp3 in Drosophila melanogaster larval development and immunity. Genetics 187:1067–1075. 10.1534/genetics.110.123893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. 2013. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154:416–429. 10.1016/j.cell.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillipson DW, Edmonds CG, Crain PF, Smith DL, Davis DR, McCloskey JA. 1987. Isolation and structure elucidation of an epoxide derivative of the hypermodified nucleoside queuosine from Escherichia coli transfer RNA. J Biol Chem 262:3462–3471. 10.1016/S0021-9258(18)61373-0. [DOI] [PubMed] [Google Scholar]
- 10.Dumelin CE, Chen Y, Leconte AM, Chen YG, Liu DR. 2012. Discovery and biological characterization of geranylated RNA in bacteria. Nat Chem Biol 8:913–919. 10.1038/nchembio.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persson BC. 1993. Modification of tRNA as a regulatory device. Mol Microbiol 8:1011–1016. 10.1111/j.1365-2958.1993.tb01645.x. [DOI] [PubMed] [Google Scholar]
- 12.Edwards AM, Addo MA, Dos Santos PC. 2020. Extracurricular functions of tRNA modifications in microorganisms. Genes (Basel) 11:907. 10.3390/genes11080907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Favre A, Yaniv M, Michelson AM. 1969. The photochemistry of 4-thiouridine in Escherichia coli t-RNA Val1. Biochem Biophys Res Commun 37:266–271. 10.1016/0006-291x(69)90729-3. [DOI] [PubMed] [Google Scholar]
- 14.Nair DP, Podgórski M, Chatani S, Gong T, Xi W, Fenoli CR, Bowman CN. 2014. The thiol-Michael addition click reaction: a powerful and widely used tool in materials chemistry. Chem Mater 26:724–744. 10.1021/cm402180t. [DOI] [Google Scholar]
- 15.Lauhon CT. 2006. Orchestrating sulfur incorporation into RNA. Nat Chem Biol 2:182–183. 10.1038/nchembio0406-182. [DOI] [PubMed] [Google Scholar]
- 16.Čavužić M, Liu Y. 2017. Biosynthesis of sulfur-containing tRNA modifications: a comparison of bacterial, archaeal, and eukaryotic pathways. Biomolecules 7:27. 10.3390/biom7010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauhon CT. 2002. Requirement for IscS in biosynthesis of all thionucleosides in Escherichia coli. J Bacteriol 184:6820–6829. 10.1128/JB.184.24.6820-6829.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson K, Lundgren HK, Hagervall TG, Bjork GR. 2002. The cysteine desulfurase IscS is required for synthesis of all five thiolated nucleosides present in tRNA from Salmonella enterica serovar typhimurium. J Bacteriol 184:6830–6835. 10.1128/JB.184.24.6830-6835.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng C, Black KA, Dos Santos PC. 2017. Diverse mechanisms of sulfur decoration in bacterial tRNA and their cellular functions. Biomolecules 7:33. 10.3390/biom7010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Black KA, Dos Santos PC. 2015. Shared-intermediates in the biosynthesis of thio-cofactors: mechanism and functions of cysteine desulfurases and sulfur acceptors. Biochim Biophys Acta 1853:1470–1480. 10.1016/j.bbamcr.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Lauhon CT, Kambampati R. 2000. The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J Biol Chem 275:20096–20103. 10.1074/jbc.M002680200. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz CJ, Djaman O, Imlay JA, Kiley PJ. 2000. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc Natl Acad Sci USA 97:9009–9014. 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi R, Proteau A, Villarroya M, Moukadiri I, Zhang L, Trempe JF, Matte A, Armengod ME, Cygler M. 2010. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol 8:e1000354. 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajakovich LJ, Tomlinson J, Dos Santos PC. 2012. Functional analysis of Bacillus subtilis genes involved in the biosynthesis of 4-thiouridine in tRNA. J Bacteriol 194:4933–4940. 10.1128/JB.00842-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black KA, Dos Santos PC. 2015. Abbreviated pathway for biosynthesis of 2-thiouridine in Bacillus subtilis. J Bacteriol 197:1952–1962. 10.1128/JB.02625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selbach B, Earles E, Dos Santos PC. 2010. Kinetic analysis of the bisubstrate cysteine desulfurase SufS from Bacillus subtilis. Biochemistry 49:8794–8802. 10.1021/bi101358k. [DOI] [PubMed] [Google Scholar]
- 27.Selbach BP, Chung AH, Scott AD, George SJ, Cramer SP, Dos Santos PC. 2014. Fe-S cluster biogenesis in Gram-positive bacteria: SufU is a zinc-dependent sulfur transfer protein. Biochemistry 53:152–160. 10.1021/bi4011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun D, Setlow P. 1993. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis nadB gene and a nifS-like gene, both of which are essential for NAD biosynthesis. J Bacteriol 175:1423–1432. 10.1128/jb.175.5.1423-1432.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Crécy-Lagard V, Ross RL, Jaroch M, Marchand V, Eisenhart C, Bregeon D, Motorin Y, Limbach PA. 2020. Survey and validation of tRNA modifications and their corresponding genes in Bacillus subtilis sp subtilis strain 168. Biomolecules 10:977. 10.3390/biom10070977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vold BS. 1978. Post-transcriptional modifications of the anticodon loop region: alterations in isoaccepting species of tRNA's during development in Bacillus subtilis. J Bacteriol 135:124–132. 10.1128/jb.135.1.124-132.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payne KA, Fisher K, Sjuts H, Dunstan MS, Bellina B, Johannissen L, Barran P, Hay S, Rigby SE, Leys D. 2015. Epoxyqueuosine reductase structure suggests a mechanism for cobalamin-dependent tRNA modification. J Biol Chem 290:27572–27581. 10.1074/jbc.M115.685693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moukadiri I, Villarroya M, Benitez-Paez A, Armengod ME. 2018. Bacillus subtilis exhibits MnmC-like tRNA modification activities. RNA Biol 15:1167–1173. 10.1080/15476286.2018.1517012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalluge JJ, Hashizume T, McCloskey JA. 1996. Quantitative measurement of dihydrouridine in RNA using isotope dilution liquid chromatography-mass spectrometry (LC/MS). Nucleic Acids Res 24:3242–3245. 10.1093/nar/24.16.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma SV, Arbach M, Roberts AA, Macdonald CJ, Groom M, Hamilton CJ. 2013. Biophysical features of bacillithiol, the glutathione surrogate of Bacillus subtilis and other Firmicutes. Chembiochem 14:2160–2168. 10.1002/cbic.201300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, Helmann JD. 2010. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci USA 107:6482–6486. 10.1073/pnas.1000928107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dos Santos PC. 2017. B. subtilis as a model for studying the assembly of Fe-S clusters in Gram-positive bacteria. Methods Enzymol 595:185–212. 10.1016/bs.mie.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Bjork GR, Huang B, Persson OP, Bystrom AS. 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13:1245–1255. 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21:87–96. 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 39.Klaiman D, Amitsur M, Blanga-Kanfi S, Chai M, Davis DR, Kaufmann G. 2007. Parallel dimerization of a PrrC-anticodon nuclease region implicated in tRNALys recognition. Nucleic Acids Res 35:4704–4714. 10.1093/nar/gkm494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. 2006. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol Cell 21:97–108. 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Ajitkumar P, Cherayil JD. 1988. Thionucleosides in transfer ribonucleic acid: diversity, structure, biosynthesis, and function. Microbiol Rev 52:103–113. 10.1128/mr.52.1.103-113.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J 20:4863–4873. 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moukadiri I, Garzon MJ, Bjork GR, Armengod ME. 2014. The output of the tRNA modification pathways controlled by the Escherichia coli MnmEG and MnmC enzymes depends on the growth conditions and the tRNA species. Nucleic Acids Res 42:2602–2623. 10.1093/nar/gkt1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Numata T, Ikeuchi Y, Fukai S, Suzuki T, Nureki O. 2006. Snapshots of tRNA sulphuration via an adenylated intermediate. Nature 442:419–424. 10.1038/nature04896. [DOI] [PubMed] [Google Scholar]
- 45.Zhou J, Lenon M, Ravanat JL, Touati N, Velours C, Podskoczyj K, Leszczynska G, Fontecave M, Barras F, Golinelli-Pimpaneau B. 2021. Iron-sulfur biology invades tRNA modification: the case of U34 sulfuration. Nucleic Acids Res 49:3997–4007. 10.1093/nar/gkab138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Even S, Burguiere P, Auger S, Soutourina O, Danchin A, Martin-Verstraete I. 2006. Global control of cysteine metabolism by CymR in Bacillus subtilis. J Bacteriol 188:2184–2197. 10.1128/JB.188.6.2184-2197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rochat T, Nicolas P, Delumeau O, Rabatinova A, Korelusova J, Leduc A, Bessieres P, Dervyn E, Krasny L, Noirot P. 2012. Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res 40:9571–9583. 10.1093/nar/gks755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi SY, Reyes D, Leelakriangsak M, Zuber P. 2006. The global regulator Spx functions in the control of organosulfur metabolism in Bacillus subtilis. J Bacteriol 188:5741–5751. 10.1128/JB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta R, Laxman S. 2020. tRNA wobble-uridine modifications as amino acid sensors and regulators of cellular metabolic state. Curr Genet 66:475–480. 10.1007/s00294-019-01045-y. [DOI] [PubMed] [Google Scholar]
- 50.Judes A, Bruch A, Klassen R, Helm M, Schaffrath R. 2016. Sulfur transfer and activation by ubiquitin-like modifier system Uba4*Urm1 link protein urmylation and tRNA thiolation in yeast. Microb Cell 3:554–564. 10.15698/mic2016.11.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou J, Pecqueur L, Aučynaitė A, Fuchs J, Rutkienė R, Vaitekūnas J, Meškys R, Boll M, Fontecave M, Urbonavičius J, Golinelli-Pimpaneau B. 2021. Structural evidence for a [4Fe-5S] intermediate in the non-redox desulfuration of thiouracil. Angew Chem Int Ed Engl 60:424–431. 10.1002/anie.202011211. [DOI] [PubMed] [Google Scholar]
- 52.Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA 44:1072–1078. 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edwards AM, Addo MA, Dos Santos PC. 2021. tRNA modifications as a readout of S and Fe-S metabolism. Methods Mol Biol 2353:137–154. 10.1007/978-1-0716-1605-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplemental material. Download jb.00009-22-s0001.pdf, PDF file, 0.5 MB (516KB, pdf)