ABSTRACT
Pseudomonas aeruginosa and Staphylococcus aureus are two common pathogens causing chronic infections in the lungs of people with cystic fibrosis (CF) and in wounds, suggesting that these two organisms coexist in vivo. However, P. aeruginosa utilizes various mechanisms to antagonize S. aureus when these organisms are grown together in vitro. Here, we suggest a novel role for Psl in antagonizing S. aureus growth. Psl is an exopolysaccharide that exists in both cell-associated and cell-free forms and is important for biofilm formation in P. aeruginosa. When grown in planktonic coculture with a P. aeruginosa psl mutant, S. aureus had increased survival compared to when it was grown with wild-type P. aeruginosa. We found that cell-free Psl was critical for the killing, as purified cell-free Psl was sufficient to kill S. aureus. Transmission electron microscopy of S. aureus treated with Psl revealed disrupted cell envelopes, suggesting that Psl causes S. aureus cell lysis. This was independent of known mechanisms used by P. aeruginosa to antagonize S. aureus. Cell-free Psl could also promote S. aureus killing during growth in in vivo-like conditions. We also found that Psl production in P. aeruginosa CF clinical isolates positively correlated with the ability to kill S. aureus. This could be a result of P. aeruginosa coevolution with S. aureus in CF lungs. In conclusion, this study defines a novel role for P. aeruginosa Psl in killing S. aureus, potentially impacting the coexistence of these two opportunistic pathogens in vivo.
IMPORTANCE Pseudomonas aeruginosa and Staphylococcus aureus are two important opportunistic human pathogens commonly coisolated from clinical samples. However, P. aeruginosa can utilize various mechanisms to antagonize S. aureus in vitro. Here, we investigated the interactions between these two organisms and report a novel role for P. aeruginosa exopolysaccharide Psl in killing S. aureus. We found that cell-free Psl could kill S. aureus in vitro, possibly by inducing cell lysis. This was also observed in conditions reflective of in vivo scenarios. In accord with this, Psl production in P. aeruginosa clinical isolates positively correlated with their ability to kill S. aureus. Together, our data suggest a role for Psl in affecting the coexistence of P. aeruginosa and S. aureus in vivo.
KEYWORDS: Pseudomonas aeruginosa, Staphylococcus aureus, exopolysaccharide, Psl, polymicrobial, cystic fibrosis, cell lysis, wound
INTRODUCTION
Cystic fibrosis (CF) is a genetic disease that results in an abnormally thick mucus lining in the lung, which causes chronic pulmonary infections and reduced lung function, leading to high morbidity and mortality in this cohort (1, 2). Pseudomonas aeruginosa and Staphylococcus aureus are two dominant microorganisms colonizing CF airways (3, 4). These organisms are commonly coisolated from CF sputum samples, with one-third of people with CF being coinfected with both pathogens (5). P. aeruginosa and S. aureus are also coisolated from chronic wound environments (6). Coinfection with both pathogens correlates with increased disease severity (7–9).
Although they coexist in vivo, P. aeruginosa antagonizes S. aureus growth in vitro. The P. aeruginosa quorum-sensing system PQS (Pseudomonas quinolone signal) (10), protease LasA (11), iron sequestering pyoverdine (12), and rhamnolipid (13) all inhibit the planktonic growth of S. aureus in vitro.
Psl (polysaccharide synthesis locus) is a P. aeruginosa exopolysaccharide consisting of a pentasaccharide repeat including d-glucose1, d-mannose3, and l-rhamnose1 (14). There are two forms of Psl, a cell-associated form that is associated with the outer membrane and a cell-free form that is released into the extracellular environment. The molecular determinants for Psl localization are, however, unknown. The amount of Psl produced varies between P. aeruginosa strains and clinical isolates (15–17). Psl functions in early biofilm formation and maintaining biofilm structure (18). There have been several reports that Psl or other P. aeruginosa polysaccharides may be involved in interactions with different bacterial species, specifically, Staphylococcus spp. Staphylococcus protein A (SpA) can bind to P. aeruginosa clinical isolates via Psl and type IV pili (19). P. aeruginosa exopolysaccharides, including Psl and Pel, can disrupt established Staphylococcus epidermidis biofilms (20). It has also been reported that Psl does not affect the relative abundance of P. aeruginosa and S. aureus in a mature dual-species biofilm but does reduce S. aureus aggregate formation in the early stages of biofilm formation (21).
Given the above-cited reports that implicate Psl in P. aeruginosa and Staphylococcus sp. interactions, we decided to investigate the potential roles of Psl in mediating interactions with S. aureus. During the course of our study, we found that the cell-free form of Psl could induce S. aureus lysis. This is the first study to investigate the role of Psl in killing S. aureus.
RESULTS
P. aeruginosa Psl production antagonizes the growth of S. aureus.
To investigate the impact of Psl on S. aureus growth, we first performed a cross-streak assay (22). S. aureus strain USA300 was inoculated horizontally on solidified medium, and the P. aeruginosa strain was streaked vertically, perpendicularly intersecting the S. aureus streak (Fig. 1A). When cocultured with wild-type P. aeruginosa PAO1 in this assay, an inhibition zone of S. aureus growth was observed in the region neighboring the intersection. However, when cocultured with an isogenic psl promoter deletion (ΔPpsl) mutant that does not produce Psl, this inhibition zone was reduced, and S. aureus was able to grow almost to the point of intersection with P. aeruginosa (Fig. 1A). This suggests that S. aureus survival is negatively affected by P. aeruginosa Psl production.
FIG 1.
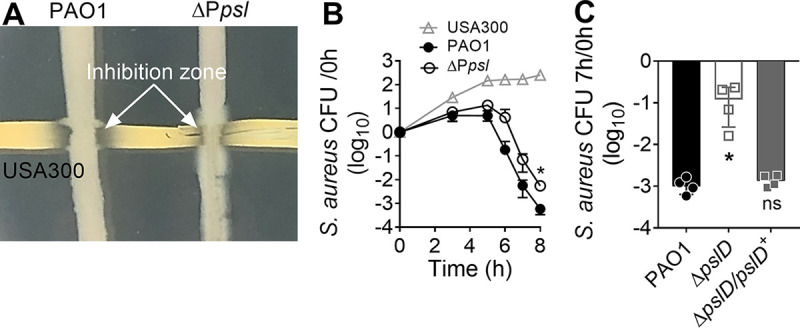
P. aeruginosa Psl production antagonizes growth of S. aureus. (A) S. aureus USA300 was grown in coculture with either P. aeruginosa PAO1 or ΔPpsl on solidified medium using a cross-streak assay. Arrows point to the inhibition zones of S. aureus growth that were observed after overnight incubation. (B) S. aureus survival over the course of 8 h when grown alone or cocultured with PAO1 or ΔPpsl. Data are means and standard deviations (SD); individual points indicate the means for biological replicates (N = 5; n = 3). Significance was determined using Student’s t test. *, P < 0.05 compared to PAO1. (C) S. aureus survival when cocultured for 7 h with PAO1, ΔpslD, or the complemented ΔpslD/pslD+ strain. N = 4; n = 3. Data are means and SD; individual points indicate biological replicates. S. aureus survival (B and C) is presented as CFU normalized to the starting CFU at 0 h. Significance was determined using a one-way ANOVA. *, P < 0.05 compared to PAO1.
We therefore hypothesized that Psl may have a role in antagonizing S. aureus. To further investigate this, S. aureus was grown in planktonic coculture with either P. aeruginosa PAO1 or the ΔPpsl mutant (23). S. aureus survival was quantified by enumerating CFU. After 5 h, S. aureus levels decreased when cocultured with both PAO1 and the ΔPpsl mutant, compared to when grown alone. However, when cocultured with the ΔPpsl mutant, S. aureus had increased survival (10- to 20-fold) compared to coculture with PAO1 (Fig. 1B). Similarly, S. aureus had increased survival when cocultured with a P. aeruginosa ΔpslD mutant, compared to the coculture with PAO1 (Fig. 1C). Introducing the wild-type pslD allele in trans (ΔpslD/pslD+) restored the activity of antagonizing S. aureus to that of PAO1 in coculture (Fig. 1C). Together, these data indicate that Psl production antagonizes S. aureus. The ΔPpsl mutant retained some antagonism toward S. aureus, likely due to other known killing P. aeruginosa mechanisms (10–13).
Cell-free Psl antagonizes the growth of multiple S. aureus isolates.
P. aeruginosa produces both cell-associated and cell-free Psl (24). The mechanism underlying Psl localization are, however, unknown. Prior studies with three classes of monoclonal antibodies directed against Psl suggested that one of the most functionally active anti-Psl antibodies (Cam-003) failed to bind to synthetic Psl oligosaccharides (25, 26). The authors suggested that Cam-003 bound to a Psl isoform that was found in the native polymer but not in the synthetic forms. Furthermore, mild alkaline treatment during carbohydrate purification eliminated Cam-003 binding, suggesting that Psl may contain lipid moieties that were not identified in the reported Psl structure (24, 25). To investigate the potential for lipid modification of Psl, we performed a limited genetic screen of transposon mutants (27) in nonessential genes annotated to be involved in lipid A biogenesis, acyl-transferases, or acyl carrier proteins. The goal was to identify mutants with altered Psl localization, aiming to uncover mechanisms involved in Psl association with the cell. Using an immunoblot assay, we found that wild-type PAO1 had an even distribution of Psl between the two forms, while many of the mutants screened had increased ratios of cell-free Psl to cell-associated Psl (Fig. 2A; also, see Table S2 in the supplemental material). Interestingly, several mutants that impacted Psl distribution were related to lipid A synthesis and modification, warranting future investigations of lipid A in mediating the association of Psl with the cell outer membrane.
FIG 2.
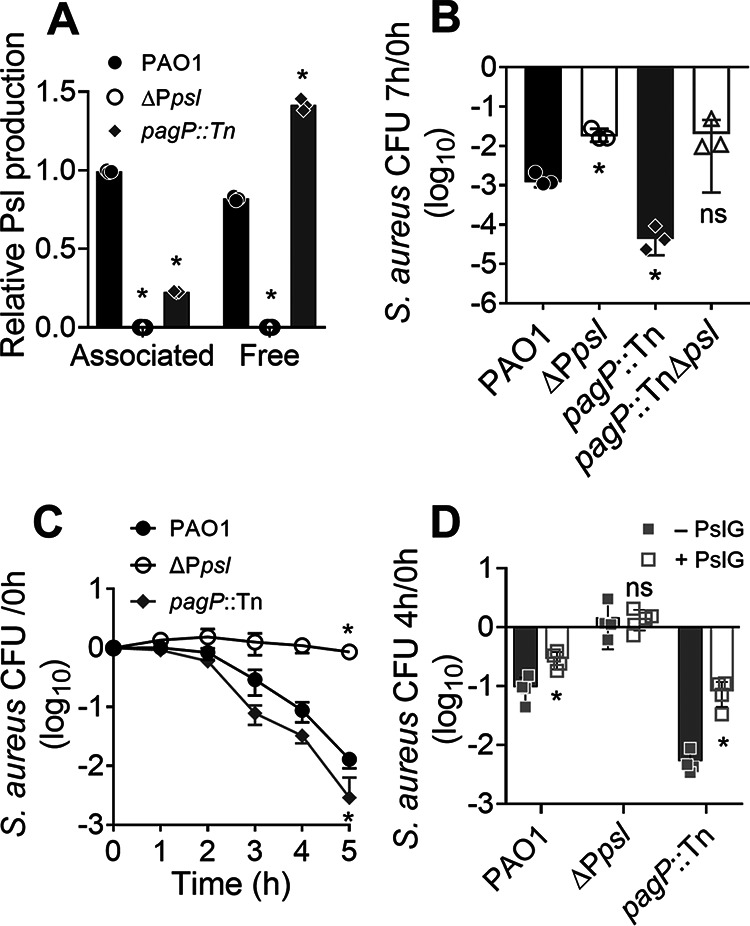
Cell-free Psl derived from P. aeruginosa spent medium antagonizes S. aureus growth. (A) Cell-associated (associated) and cell-free (free) Psl from P. aeruginosa was quantified by immunoblotting. The cell pellets and spent media of centrifuged overnight cultures were used as the source of cell-associated and cell-free Psl, respectively. Psl was quantified by densitometry of the blot and normalized to PAO1 cell-associated Psl. Data are means and SD; individual points indicate biological replicates (N = 3; n = 3). Significance was determined with one-way ANOVA. *, P < 0.05 compared to PAO1. (B) S. aureus survival when cocultured for 7 h with P. aeruginosa PAO1, ΔPpsl, pagP::Tn, or pagP::TnΔpsl strains. S. aureus survival is presented as CFU normalized to the starting CFU at 0 h. Data are means and SD; individual points indicate biological replicates (N = 3; n = 3). Significance was determined using Student’s t test compared to PAO1. *, P < 0.05; ns, not significant. (C) S. aureus survival when grown for 5 h in spent medium of PAO1 or the ΔPpsl or pagP::Tn strain that was diluted 1:1 in fresh medium. Data are means and SD; individual points indicate means for biological replicates (N = 4; n = 3). Significance was determined using Student’s t test. *, P < 0.05 compared to PAO1. (D) S. aureus was grown for 4 h in spent medium from PAO1, ΔPpsl, or pagP::Tn strains that had been pretreated with (+) or without (−) PslG. Data are means and SD; individual points indicate biological replicates (N = 4; n = 3). S. aureus survival (B and C) is presented as CFU normalized to the starting CFU at 0 h. Significance was determined using Student’s t test, compared to the no-PslG pretreated group. *, P < 0.05; ns, not significant.
To determine if a specific form of Psl was responsible for mediating the antagonism toward S. aureus, we used one of the mutants identified from the screen, a pagP transposon mutant (pagP::Tn PA1343::ISlacZ/hah), as it produces predominately cell-free Psl (Fig. 2A; Table S2). PagP is a palmitoyltransferase which transfers palmitate from outer membrane phospholipids to lipid A (28). We grew S. aureus in planktonic coculture with either P. aeruginosa PAO1 or the ΔPpsl or the pagP::Tn strain (Fig. 2B). As previously observed (Fig. 1B), coculture with the ΔPpsl mutant led to a significant increase in S. aureus survival, compared to coculture with PAO1. In contrast, when grown in coculture with the pagP::Tn mutant, S. aureus survival was significantly reduced (more than 10-fold) compared to that in coculture with PAO1 (Fig. 2B). To determine if this increased antagonism is dependent on Psl, a pslBCD deletion was created in the pagP::Tn background (pagP::TnΔpsl). We found that S. aureus survival when cocultured with the pagP::TnΔpsl strain was comparable to that seen with the ΔPpsl mutant (Fig. 2B). This suggests that the enhanced antagonism toward S. aureus from the pagP::Tn strain is dependent on Psl production. We also observed the same trends when other S. aureus strains, including LAC and a methicillin-sensitive S. aureus (MSSA) strain, were cocultured with either P. aeruginosa PAO1, ΔPpsl, or pagP::Tn (Fig. S1). The above data suggest an important role for cell-free Psl in antagonizing the growth of S. aureus during planktonic coculture.
Since cell-free Psl is secreted and appears to be the form that mediates S. aureus antagonism, we tested the ability of P. aeruginosa spent medium to antagonize S. aureus growth. Similar to planktonic coculture (Fig. 2B), S. aureus had reduced survival when grown in spent medium from PAO1 and the pagP::Tn strain, compared to growth in ΔPpsl spent medium, the latter of which did not show any changes in S. aureus survival when quantified by both CFU (Fig. 2C) and optical density at 600 nm (OD600) (Fig. S2). Furthermore, S. aureus survival was significantly reduced when grown in pagP::Tn spent medium, compared to PAO1 spent medium (Fig. 2C; Fig. S2).
PslG is a hydrolase that cleaves the polymeric chain of Psl into monomers (29). To further support the role of cell-free Psl in antagonizing S. aureus growth, P. aeruginosa spent medium was pretreated with PslG to degrade Psl (Fig. S3). S. aureus was supplemented with PslG treated spent media from PAO1 or the ΔPpsl or pagP::Tn strain and grown for a further 4 h. S. aureus survival significantly increased by 3- and 15-fold when grown with PAO1 and pagP::Tn PslG-treated spent medium, respectively, compared to spent medium without PslG (Fig. 2D). As expected, PslG treatment of ΔPpsl spent medium resulted in no change to S. aureus survival, indicating that PslG does not affect S. aureus growth (Fig. 2D).
Psl-mediated antagonism toward S. aureus is independent of other P. aeruginosa factors that impact S. aureus growth.
P. aeruginosa utilizes various mechanisms to antagonize the growth of S. aureus, including PQS (10), pyoverdine (12), LasA (11), and rhamnolipid (13). Alginate, another exopolysaccharide produced by P. aeruginosa, has been found to promote P. aeruginosa and S. aureus coexistence by downregulating some of the above mechanisms in P. aeruginosa (23, 30). We therefore wanted to examine if Psl-mediated antagonism toward S. aureus was dependent on any of these mechanisms. There were no significant differences found in rhamnolipid and LasA activity in PAO1 and the ΔPpsl mutant (Fig. S4A to C). A deficiency in PQS or pyoverdine did not affect the ability of P. aeruginosa cell-free Psl to antagonize S. aureus growth during coculture (Fig. S4D). As controls, corresponding mutants for each mechanism showed expected loss of these antagonistic factors (see the supplemental methods and Fig. S4A to D). Collectively, these data indicate that cell-free Psl antagonizes the growth of S. aureus independently of rhamnolipid, LasA, PQS, and pyoverdine.
Cell-free Psl kills S. aureus by disrupting the cell envelope.
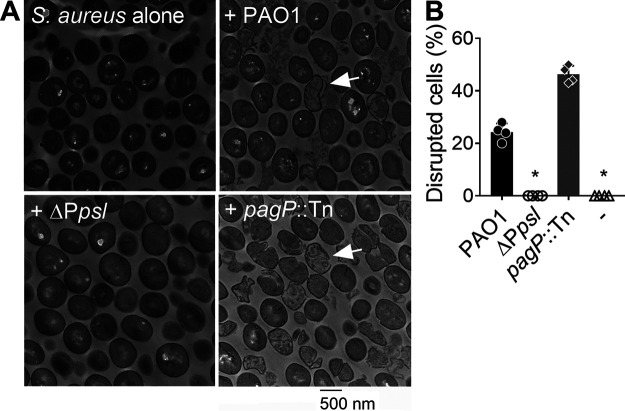
Exopolysaccharides isolated from bacteria and fungi can disrupt bacterial cell envelopes (31–33). We therefore hypothesized that Psl similarly caused S. aureus cell lysis. To test this, S. aureus was incubated with diluted spent medium from P. aeruginosa PAO1, ΔPpsl, or pagP::Tn for 2 h and processed for transmission electron microscopy (TEM). This time point was selected as S. aureus was still viable after 2 h of incubation with P. aeruginosa spent medium (Fig. 2C). TEM imaging revealed that S. aureus incubated with either PAO1 or pagP::Tn spent medium had disrupted cell membranes, while S. aureus incubated with ΔPpsl spent medium showed little evidence of cellular damage, similar to the no-treatment control (Fig. 3A; Fig. S5). Image analysis, which quantified the percentage of cells with membrane damage relative to the untreated control, revealed that approximately 25% of S. aureus cells incubated with PAO1 spent medium had disrupted cell envelopes (Fig. 3B). However, when incubated with spent medium from the pagP::Tn strain, the number of S. aureus cells with membrane damage increased to approximately 50% (Fig. 3B). This is consistent with our previous results which showed decreases in S. aureus cell numbers (Fig. 2C) and OD600 (Fig. S2) when the organism was grown in medium supplemented with P. aeruginosa spent medium, suggestive of cell lysis. The data indicate that cell-free Psl likely kills S. aureus by disrupting the cell envelope.
FIG 3.
Cell-free Psl kills S. aureus by disrupting the cell envelope. (A) S. aureus USA300 was incubated with spent medium from P. aeruginosa PAO1, ΔPpsl, or pagP::Tn for 2 h. Changes in cell morphology were visualized by TEM. Arrows indicate cells with disrupted cell envelopes. (B) Total and disrupted cell counts for each group were enumerated, and the percentage of disrupted cells within each group was calculated. Data are means and SD; individual points indicate biological replicates (N = 4; n = 3). Significance was determined using a one-way ANOVA compared to PAO1. *, P < 0.05.
Psl promotes S. aureus killing in conditions that mimic in vivo growth.
The above-described experiments were performed using planktonic cultures grown in a rich medium. We wanted to further determine if cell-free Psl could mediate S. aureus killing under in vivo-like conditions. Biofilm formation is one of the key factors contributing to P. aeruginosa and S. aureus chronic infections (34, 35). As such, we next examined if cell-free Psl could kill S. aureus grown in biofilms (Fig. 4A). Pre-established S. aureus biofilms were incubated for 5 h with spent medium from P. aeruginosa. Biofilm biomass was then quantified by crystal violet staining (36). Compared to the control group with no P. aeruginosa spent medium added, S. aureus biofilm biomass was significantly decreased when treated with both PAO1 and pagP::Tn spent media. No significant difference was found between the control and ΔPpsl spent media. This suggests that cell-free Psl also promotes killing of S. aureus in biofilms.
FIG 4.
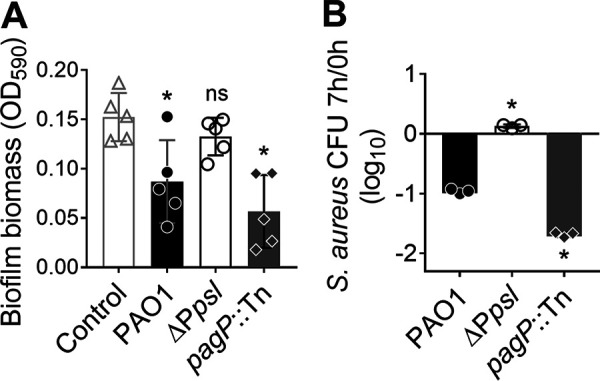
Cell-free Psl promotes S. aureus killing under in vivo-like conditions. (A) Twenty-four-hour S. aureus USA300 biofilms were incubated with or without P. aeruginosa PAO1, ΔPpsl, or pagP::Tn spent medium for 5 h. Biofilm biomass was quantified by crystal violet staining. Data are means and SD; individual points indicate biological replicates (N = 5; n = 3). Significance was determined using one-way ANOVA. *, P < 0.05, and ns, not significant, compared to the medium-only control. (B) S. aureus survival when cocultured with P. aeruginosa PAO1, ΔPpsl, or pagP::Tn in SCFM2 for 7 h. Data are means and SD; individual points indicate biological replicates (N = 3; n = 3). Significance was determined using Student’s t test. *, P < 0.05 compared to PAO1.
Synthetic CF sputum medium (SCFM2) mimics the CF sputum composition and has been used to culture both P. aeruginosa and S. aureus (23, 37). We quantified S. aureus survival when cocultured with P. aeruginosa PAO1, ΔPpsl, or pagP::Tn in SCFM2 for 8 h (Fig. 4B). Like our previous results in rich media (Fig. 2B), S. aureus survival significantly increased by 10-fold when cocultured with the ΔPpsl mutant, compared to coculture with PAO1 (Fig. 4B). Furthermore, S. aureus survival significantly decreased by 5-fold in coculture with the pagP::Tn mutant compared to coculture with PAO1 (Fig. 4B). This suggests that cell-free Psl may contribute to S. aureus killing in nutritional environments like a CF lung. Together, these data suggest that Psl-mediated killing of S. aureus occurs in conditions that are reflective of in vivo scenarios.
Increased cell-free Psl production in P. aeruginosa clinical isolates promotes S. aureus killing.
Some P. aeruginosa clinical isolates lack or have reduced Psl production (15–17). Given that cell-free Psl appears to kill S. aureus, it is possible that reduced Psl production in P. aeruginosa occurs during coevolution with S. aureus, facilitating polymicrobial infections. To test if there is a correlation between Psl production and S. aureus killing among P. aeruginosa clinical isolates, 16 P. aeruginosa isolates derived from CF sputum samples were collected from the Cure CF Columbus Translational Core at Nationwide Children’s Hospital (Fig. 5). Since mucoid P. aeruginosa can coexist with S. aureus, due to the overproduction of alginate (23), only nonmucoid P. aeruginosa isolates were evaluated. Psl production of each nonmucoid P. aeruginosa isolate was quantified by immunoblot assay. S. aureus USA300 was grown in planktonic coculture with each CF isolate, as well as PAO1 or the ΔPpsl or pagP::Tn mutant, and S. aureus survival was quantified. A correlation (R2 = 0.3578) was found between the cell-free Psl production of the P. aeruginosa strains and the cocultured S. aureus survival (Fig. 5A). Furthermore, 6 of the 16 P. aeruginosa CF isolates produced no Psl, and each had reduced S. aureus killing activity compared to PAO1 (Fig. 5B). This suggests that low Psl production in P. aeruginosa clinical isolates can result in enhanced survival when in coculture with S. aureus and that Psl production may be one factor involved in their coexistence during infections.
FIG 5.
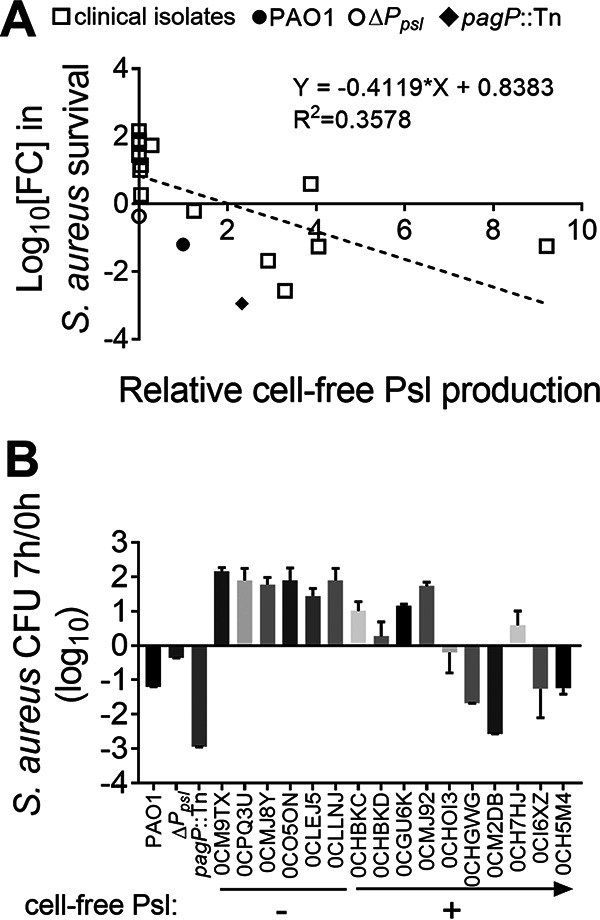
Psl production in P. aeruginosa clinical isolates positively correlates with the ability to kill S. aureus. S. aureus USA300 was cocultured with each of the 16 P. aeruginosa CF isolates, PAO1, and the ΔPpsl and pagP::Tn strains. The number of CFU at 7 h was divided by that at 0 h to quantify survival. (A) Linear regression analysis was performed to determine any correlation between the two parameters indicated. Cell-free Psl production by the designated strains was measured by immunoblot assay using Psl antibody and then normalized to PAO1. Individual points indicate biological replicates (N = 3; n = 3). (B) P. aeruginosa clinical isolates with low Psl production showed reduced S. aureus killing activity. Six of the CF isolates produced no cell-free Psl (−), while the other 10 produced variable amounts of cell-free Psl (+; the arrow indicates the increasing production of Psl), compared with that produced by PAO1. The number of CFU of S. aureus USA300 cocultured with each of the isolates, PAO1, and the ΔPpsl and pagP::Tn strains for 7 h was divided by that at 0 h to quantify S. aureus survival. Data are means and SD (N = 3; n = 3).
Purified cell-free Psl is sufficient to kill S. aureus.
Exopolysaccharides produced by some bacteria and fungi have antimicrobial activities (31–33). Our results demonstrate that cell-free Psl mediates the killing of S. aureus. However, this was determined from planktonic coculture of P. aeruginosa and S. aureus (Fig. 1 and 2A and B) or growth of S. aureus in P. aeruginosa spent medium (Fig. 2C and D), both conditions which could contain Psl-independent antagonists of S. aureus growth. We therefore tested if cell-free Psl purified from PAO1 and the pagP::Tn mutant could directly inhibit S. aureus growth. S. aureus survival was quantified by measuring the OD600 after 16 h of growth in medium supplemented with increasing concentrations of purified cell-free Psl (0 to 100 μg/mL). S. aureus grown alone, without the addition of cell-free Psl (untreated), was used as the control. S. aureus survival decreased as the concentration of purified Psl increased (Fig. 6A). The ΔPpsl strain also underwent the identical purification process, and the resulted extraction had no effect on S. aureus survival. In addition, we found that purified Psl originating from ΔpqsA and ΔpvdA mutants showed activity similar to that of Psl from the parent strains in killing S. aureus (Fig. S4E). This demonstrates that purified cell-free Psl, in a concentration-dependent manner, is sufficient to mediate the killing of S. aureus.
FIG 6.
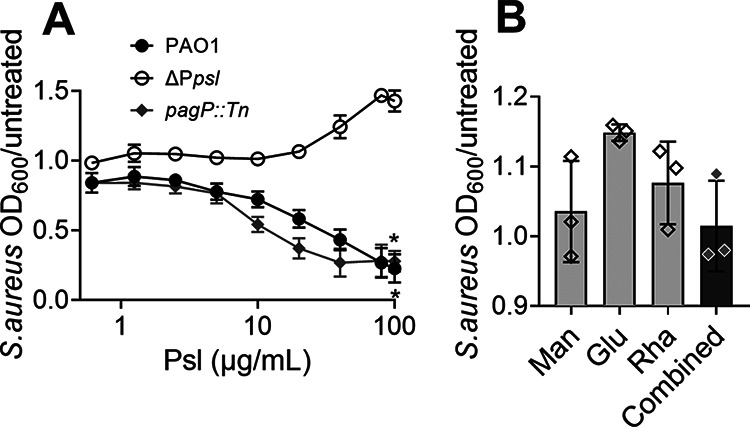
Purified cell-free Psl is sufficient to kill S. aureus. Survival of S. aureus grown in media supplemented with (A) increasing concentrations of purified cell-free Psl from PAO1 or the pagP::Tn mutant or cell-free products from the ΔPpsl mutant or (B) 60 μg/mL of d-mannose (Man), 20 μg/mL of d-glucose (Glu), 20 μg/mL of l-rhamnose (Rha), or a combination of the three monosaccharides (Combined). S. aureus grown without cell-free Psl or monosaccharides (untreated) was used as a control. OD600 was measured after 16 h. S. aureus survival is presented as OD600 normalized to that of the control. (A) The x axis is a log scale. Data are means and SD; individual points indicate the mean of biological replicates (N = 3; n = 3). Significance was determined using Student’s t test compared to the ΔPpsl mutant. *, P < 0.05.
Cell-free Psl consists of d-mannose, d-glucose, and l-rhamnose, at a ratio of 3:1:1, respectively (24). The influence of these individual monosaccharides on S. aureus survival was also evaluated. There was no change in survival of S. aureus grown in medium supplemented with either each monosaccharide separately or a combination (Fig. 6B). This indicates that an integral structure of Psl is critical for the S. aureus killing property.
DISCUSSION
P. aeruginosa and S. aureus often cause chronic coinfections that can lead to more severe disease outcomes (7–9). Understanding the dynamic between the two pathogens is key to successfully treating and eliminating such coinfections. Here, we demonstrate a novel role for P. aeruginosa exopolysaccharide Psl in killing S. aureus in vitro (Fig. 1). Specifically, we found that purified cell-free Psl mediated S. aureus killing (Fig. 6), likely by disrupting the cell envelope and causing cell lysis (Fig. 3). This antagonistic relationship would likely impact the coexistence of these two opportunistic pathogens in vivo.
Furthermore, we demonstrate that cell-free Psl can kill S. aureus in mature biofilms (Fig. 4A), suggesting that the antimicrobial properties of Psl are not limited to highly metabolically active, planktonic S. aureus. In support of this, P. aeruginosa extracellular Psl from spent medium can disrupt established S. epidermidis biofilms. Interestingly, this was independent of any bactericidal effect, as spent media of wild-type P. aeruginosa and a psl mutant were able to comparably inhibit the growth of planktonic S. epidermidis (20). This suggests that cell-free Psl may have different killing mechanisms for S. aureus and S. epidermidis. It is also possible that Psl-mediated killing of planktonic S. aureus and disruption of biofilms occur through distinct mechanisms. Conversely, Staphylococcus protein A (SpA) secreted by S. aureus can inhibit P. aeruginosa biofilm formation, and this can be prevented by Psl binding to SpA (19). However, we found that a spa transposon mutant (spa::Tn) was as susceptible as wild-type S. aureus to killing by PAO1 and pagP::Tn when grown in planktonic coculture (Fig. S6). This suggests that SpA does not play a role in the cell-free Psl-mediated killing of S. aureus described here.
In addition to Psl, P. aeruginosa produces two other exopolysaccharides, alginate and Pel (38, 39). In contrast to the antimicrobial properties of Psl toward S. aureus observed here, alginate mediates P. aeruginosa and S. aureus coexistence (23, 30). Alginate overproduction, resulting in a mucoid phenotype, downregulates known antagonistic mechanisms in P. aeruginosa, including rhamnolipid, pyoverdine, and HQNO (2-heptyl-4-hydroxyquinoline N-oxide) (23). Interestingly, alginate and Psl share the same precursor, mannose-1-phosphate (40), leading to reduced Psl production when alginate is overproduced (41, 42). This is in accord with our finding that Psl plays the opposite role of alginate, by promoting S. aureus killing. This study focused exclusively on S. aureus antagonism mediated by nonmucoid P. aeruginosa. Prior studies showed that some nonmucoid clinical isolates harbor secondary mutations in algTU (43, 44), and we did not determine if isolates used in this study indeed harbored these mutations. Jones et al. (42), on the other hand, found that functional Psl, albeit at reduced levels, is still produced in some mucoid P. aeruginosa isolates, warranting further investigation of S. aureus killing by P. aeruginosa producing both Psl and alginate. Pel also contributes to P. aeruginosa and S. aureus polymicrobial interactions. It can reduce cross-linking within the biofilm matrix, enabling the expansion of P. aeruginosa in a dual-species biofilm with S. aureus (21). Secreted Pel can also remove established S. epidermidis biofilms (20). However, our data suggest that Pel is not directly involved in killing S. aureus (Fig. S7).
Our data also suggest that the structure of Psl may affect its antimicrobial activity. PslG-treated PAO1 and pagP::Tn spent media had no detectable Psl, similar to the ΔPpsl mutant (Fig. S3). However, while they both showed reduced S. aureus killing compared to untreated spent medium (Fig. 2D), S. aureus survival was still significantly reduced compared to that with ΔPpsl spent medium (Fig. 2D). This suggests that PslG-treated PAO1 and pagP::Tn spent media demonstrate intermediate S. aureus antimicrobial properties. It is possible that a higher-molecular-weight (MW) structure is needed for the antimicrobial property of cell-free Psl. One interpretation of these data is that PslG may partially disrupt Psl polymers such that it cannot be recognized by Psl antibodies, but these forms can have reduced, although some, S. aureus killing activity. Consistent with this, Maalej et al. found that a high-molecular-weight polysaccharide produced by Pseudomonas stutzeri showed antimicrobial activity toward S. aureus and other bacteria (31). This polysaccharide consists of a trisaccharide backbone chain containing glucose, mannose, and lactyl rhamnose, similar to P. aeruginosa Psl. It is likely that these two polysaccharides, both isolated from Pseudomonas species, share similar properties in killing S. aureus. Maalej et al. also compared antimicrobial abilities of the P. stutzeri polysaccharide in different MWs and found that higher-MW polysaccharides had increased Gram-positive bacterium-killing activity. In addition, we found that the monosaccharides making up cell-free Psl showed no killing of S. aureus (Fig. 6B). Together, these observations suggest that the structure of cell-free Psl may contribute to its antimicrobial activity.
From our TEM analysis, we propose that cell-free Psl damages S. aureus cell envelopes, causing cell lysis (Fig. 5). Our observations contribute to the growing evidence that exopolysaccharides produced by bacteria and fungi can have antimicrobial properties by disrupting cell envelopes (31–33). A polysaccharide isolated from Streptomyces virginiae, consisting of mannose, glucose, and galactose, also inhibits S. aureus growth (33). Interestingly, this polysaccharide has a sugar composition similar to that of Psl and a bactericidal concentration comparable to what we determined for cell-free Psl (Fig. 6). As mentioned above, the polysaccharide produced by P. stutzeri is antimicrobial toward S. aureus (31). It was speculated that the amphiphilicity of the polymer may provide structural affinity between the S. aureus cell wall and polysaccharide, presumably leading to lysis. Metal chelation was suggested as another potential mechanism of P. stutzeri polysaccharide permeabilization of the S. aureus cell envelope (31). This might also apply to the Psl-mediated killing of S. aureus, as Psl can sequester and store iron (45). Despite determining that cell-free Psl causes S. aureus cell lysis, we have yet to elucidate the mechanism. However, we observed no difference in PAO1 and ΔPpsl spent media in lysing heat-killed S. aureus (Fig. S4C). This suggests that killing requires active S. aureus growth/metabolism. Future investigations will focus on the mechanism(s) involved in Psl-mediated killing.
It is also possible that Psl can antagonize S. aureus by other mechanisms, in addition to cell lysis. The loss of cell-associated Psl leads to reduced aggregation (46). Cell aggregation promotes P. aeruginosa pyoverdine (47) and quorum sensing signal production, including PQS (48), both of which are involved in killing S. aureus. In addition, purified cell-associated Psl can act as a signal to stimulate cyclic di-GMP production in P. aeruginosa (49). It is therefore possible that Psl could affect the production of known S. aureus-killing factors by promoting cell aggregation and/or acting as a signal molecule in P. aeruginosa. However, this is not supported by our finding that purified Psl is sufficient to promote S. aureus killing and that pqs and pvdA mutants still showed Psl-dependent killing.
Psl is important for P. aeruginosa biofilm formation and contributes to chronic infections (18). However, several P. aeruginosa clinical isolates produce little or no Psl (15–17). Consistent with this, we found that 6 of the 16 examined CF isolates produced no Psl. A positive correlation was observed between Psl production and S. aureus killing activity among the isolates (Fig. 5), suggesting a role for Psl in affecting the coexistence of the two pathogens in CF lungs. Coexistence with S. aureus can be beneficial to P. aeruginosa. For example, S. aureus is predominant in early dual-species biofilms and can promote P. aeruginosa attachment and biofilm formation (50). We speculate that P. aeruginosa may undergo adaptation to reduce its antagonistic weapons, like Psl, to better coexist with S. aureus in vivo.
Taken together, our findings demonstrate that cell-free Psl produced by P. aeruginosa can disrupt the S. aureus cell envelope, leading to S. aureus cell death. Psl-mediated killing also occurs under in vivo-like conditions, including in SCFM2 and S. aureus biofilms. Consistent with this, the amount of Psl produced in P. aeruginosa clinical isolates positively correlates with their antistaphylococcal activity. This study demonstrates a novel role for exopolysaccharides in polymicrobial infections and highlights the need for future investigations in polymicrobial coevolution during chronic infections.
MATERIALS AND METHODS
Bacterial strains, primers, and growth conditions.
All bacterial strains and plasmids are listed in Table 1. All primers used in this study are listed in Table S1. Gene deletion constructs were incorporated into the P. aeruginosa genome using homologous recombination as previously described (51). S. aureus was grown in lysogeny broth (LB; 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl). P. aeruginosa was grown in LB with no salt (LBNS). Overnight cultures were grown at 37°C with 200 rpm shaking for 16 h. Antibiotic concentrations were 10 μg/mL gentamicin and 100 μg/mL ampicillin for selection of Escherichia coli and 300 μg/mL carbenicillin for P. aeruginosa.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| NEB5α | New England Biolabs | |
| S17 | ||
| P. aeruginosa | ||
| PAO1 | WT P. aeruginosa | 55 |
| ΔPpsl mutant | psl production deficient; psl operon promoter deletion mutant | 55 |
| ΔpslD/V mutant | psl production deficient; ΔpslD with an empty vector pUCP18 | 24 |
| ΔpslD/pslD+ mutant | pUCP18::pslD was used to complement ΔpslD | 24 |
| PAO1/V | WT P. aeruginosa with an empty vector pUCP18 | 24 |
| pagP::Tn mutant | pagP transposon mutant (UWGC:PW3442, PA1343::ISlacZ/hah) | 27 |
| htrB1::Tn mutant | htrB1 transposon mutant (UWGC:PW1009, PA0011::ISlacZ/hah) | 27 |
| htrB2::Tn mutant | htrB2 transposon mutant (UWGC:PW6435, PA3242::ISlacZ/hah) | 27 |
| lpxO1::Tn mutant | lpxO1 transposon mutant (UWGC:PW8596, PA4512::ISphoA/hah) | 27 |
| lpxO2::Tn mutant | lpxO2 transposon mutant (UWGC:PW2702, PA0936::ISphoA/hah) | 27 |
| pagL::Tn mutant | pagL transposon mutant (UWGC:PW8859, PA4661::ISphoA/hah) | 27 |
| phoP::Tn mutant | phoP transposon mutant (UWGC:PW3128, PA1179::ISlacZ/hah) | 27 |
| phoQ::Tn mutant | phoQ transposon mutant (UWGC:PW3131, PA1180::ISlacZ/hah) | 27 |
| pmrB::Tn mutant | pmrB transposon mutant (UWGC:PW9023, PA4777::ISlacZ/hah) | 27 |
| pagP::TnΔpsl mutant | pagP transposon mutant with pslBCD deletion | This study |
| ΔpvdA mutant | pvdA deletion mutant | This study |
| ΔpqsA mutant | pqsA deletion mutant | This study |
| ΔpvdA ΔPpsl mutant | psl promoter and pvdA double-deletion strain | This study |
| ΔpqsA ΔPpsl mutant | psl promoter and pqsA double-deletion strain | This study |
| rhlA::Tn mutant | rhlA transposon mutant (UWGC:PW6886, PA3479::ISphoA/hah) | 27 |
| lasA::Tn mutant | lasA transposon mutant (UWGC:PW4282, PA1871::ISlacZ/hah) | 27 |
| ΔpelA mutant | pelA deletion mutant | 56 |
| 0CM2DB | CF clinical isolate | This study |
| 0CMJ92 | CF clinical isolate | This study |
| 0CPQ3U | CF clinical isolate | This study |
| 0CO5ON | CF clinical isolate | This study |
| 0CMJ8Y | CF clinical isolate | This study |
| 0CM9TX | CF clinical isolate | This study |
| 0CHBKC | CF clinical isolate | This study |
| 0CHBKD | CF clinical isolate | This study |
| 0CH5M4 | CF clinical isolate | This study |
| 0CLLNJ | CF clinical isolate, coisolated with S. aureus | This study |
| 0CLEJ5 | CF clinical isolate, coisolated with S. aureus | This study |
| 0CH7HJ | CF clinical isolate, coisolated with S. aureus | This study |
| 0CHGWG | CF clinical isolate, coisolated with S. aureus | This study |
| 0CGU6K | CF clinical isolate, coisolated with S. aureus | This study |
| 0CI6XZ | CF clinical isolate, coisolated with S. aureus | This study |
| 0CHOI3 | CF clinical isolate, coisolated with S. aureus | This study |
| S. aureus | ||
| USA300 | WT | 57 |
| MSSA | ATCC 29213, methicillin sensitive | ATCC |
| LAC | MRSA | 58 |
| spa::Tn mutant | spa transposon mutant (NE286, NARSA) | 57 |
| Plasmids | ||
| pEX18Gm | For allelic exchange in P. aeruginosa | |
| pΔpqsA | For pqsA deletion | This study |
| pΔpvdA | For pvdA deletion | This study |
| pMpsl-KO1 | For pslBCD deletion | 59 |
P. aeruginosa and S. aureus coculture.
Cross-streak and planktonic cocultures were performed as previously described with modifications (23). For the cross-streak coculture, overnight cultures of P. aeruginosa strains and S. aureus USA300 were normalized to an OD600 of 1.5. A loopful of S. aureus culture was streaked horizontally on lysogeny agar with no salt (LANS) followed by a vertical streak of P. aeruginosa. The cross-streaked plate was incubated overnight at 37°C. For the planktonic coculture, overnight cultures of P. aeruginosa and S. aureus were diluted to an OD600 of 0.05 and combined at a ratio of 1:1 in 2 mL of either fresh LBNS or SCFM2 (52). The coculture was incubated at 37°C with shaking at 200 rpm for up to 8 h. Aliquots were taken at 0, 3, 5, 6, 7, and 8 h, serially diluted to plate on Difco Pseudomonas isolation agar (PIA) and BBL mannitol salt agar (MSA) to enumerate CFU of P. aeruginosa and S. aureus, respectively. S. aureus survival at each time point was normalized to the number of CFU at 0 h.
Crude extraction of cell-free and cell-associated exopolysaccharide.
P. aeruginosa strains were grown overnight at 37°C and 200 rpm in 5 mL of LBNS and normalized to an OD600 of 2. Crude exopolysaccharide was isolated using a modified protocol (53). Briefly, the bacteria were centrifuged at 8,000 × g for 10 min at room temperature to harvest the spent medium and pellet. For cell-free exopolysaccharide, the spent medium after centrifugation was removed and passed through a 0.22-μm filter to remove any residual bacteria. For cell-associated exopolysaccharide, the bacterial pellet was suspended in 2 mL of 1.5 M NaCl, vortexed vigorously, and placed on a platform rocker at room temperature for 15 min. After extraction, the sample was centrifuged at 5,000 × g for 10 min at room temperature. The supernatant was removed and passed through a 0.22-μm filter to remove any residual bacteria.
Psl immunoblot assay.
Two microliters of the crude exopolysaccharide extracts and Psl standards (500 μg/mL, 250 μg/mL, 100 μg/mL, 50 μg/mL, and 10 μg/mL) were spotted onto a nitrocellulose membrane (0.2 μm; Bio-Rad) and incubated at room temperature with 5% skim milk for 1 h, Psl antibody (AstraZeneca; 1:3,000 dilution) for 1 h, and secondary antibody (goat anti-human IgG; 1:5,000 dilution; Abcam) for 1 h sequentially. Western blotting detection reagent (ECL) was added to the membrane for visualization via the ChemiDoc imaging system (Bio-Rad). Densitometry analysis was performed with ImageJ software, and results were compared to a standard curve of pure Psl.
S. aureus killing assay using P. aeruginosa spent medium.
Overnight cultures of P. aeruginosa and S. aureus were diluted to an OD600 of 0.05 and incubated at 37°C with shaking at 200 rpm for 7 h and 4 h, respectively. Psl-mediated antagonism was observed when S. aureus was grown in coculture with P. aeruginosa for 7 h, so we selected this time point to harvest P. aeruginosa spent medium. A P. aeruginosa culture was filter sterilized using a 0.22-μm filter (Fisherbrand) and diluted 1:1 in fresh LBNS. Four milliliters of stationary-phase S. aureus (OD600 = 1.5) was centrifuged, resuspended in 4 mL of diluted P. aeruginosa spent medium, and incubated at 37°C with shaking at 200 rpm for up to 4 h. Aliquots were taken every hour, and either absorbance (OD600) was measured using a spectrophotometer (Thermo Fisher), or the sample was serially diluted and CFUs were enumerated. For experiments with PslG-treated spent media, filter-sterilized P. aeruginosa spent medium was treated with 100 μM PslG for 1 h at 37°C to degrade cell-free Psl, prior to the addition of S. aureus. S. aureus survival at each time point was normalized to the CFU or OD600 at 0 h.
Cell-free Psl purification.
Psl was isolated by following an established protocol (24, 49) with modifications. Briefly, concentrated spent medium of P. aeruginosa was subjected to 3 rounds of ethanol precipitation, treatment with 0.1 mg/mL of DNase I, RNase A, and proteinase K, and 3 rounds of dialysis (Slide-A-Lyzer dialysis cassette; molecular weight cutoff [MWCO] of 3,500). Concentration and carbohydrate content of the resulting purified cell-free Psl were determined by immunoblotting and phenol-sulfuric acid assay (54), respectively.
S. aureus killing assay by purified Psl.
Overnight S. aureus culture was diluted to an OD600 of 0.05 and incubated at 37°C with shaking at 200 rpm for 4 h to reach an OD600 of 1.5. The culture was resuspended in fresh LBNS containing increasing concentrations (0, 2.5, 5, 10, 20, 40, 80, and 100 μg/mL) of purified cell-free Psl, and 200 μL was added into each well of a 96-well plate. To test if the monosaccharides of Psl contributed to S. aureus killing, 20 μg/mL l-rhamnose, 20 μg/mL d-glucose, 60 μg/mL d-mannose, or a combination of the three monosaccharides was added to S. aureus. Plates were incubated in a plate reader (SpectraMax i3x; Molecular Devices) at 37°C for 16 h. OD600 was measured every 30 min as an indication of survival and compared to that of the control (S. aureus grown alone). Relative S. aureus survival was measured by OD600 normalized to the control.
TEM imaging.
S. aureus cultures were treated with P. aeruginosa spent medium for 2 h, centrifuged, and washed with phosphate-buffered saline (PBS). Samples were fixed in 2.5% glutaraldehyde overnight and then embedded in 2% agarose for TEM processing. Samples were postfixed with 1% osmium tetroxide, stained en bloc with 1% aqueous uranyl acetate, dehydrated in a graded series of ethanol, and embedded in Eponate 12 epoxy resin (Ted Pella Inc.; 18012). Ultrathin sections were cut with a Leica EM UC6 ultramicrotome (Leica Microsystems; EM FC7) and collected on copper grids. Images were acquired with an FEI Technai G2 Spirit transmission electron microscope (FEI), a Macrofire (Optronics) digital camera, and AMT image capture software. Total and disrupted cell counts were manually enumerated. All intact and disrupted cells are individually labeled in Fig. S5.
Biofilm killing assay.
An overnight culture of S. aureus was diluted in fresh LB to an OD600 of 0.01. Two hundred microliters was aliquoted into each well of a 96-well plate and incubated at 37°C statically for 24 h. Biomass was washed with PBS 3 times, and the remaining attached biofilm was treated with diluted P. aeruginosa spent medium for 5 h. After the removal of planktonic cells and a PBS wash, the remaining biofilm was stained with 200 μL of 0.1% crystal violet for 15 min at room temperature. Biofilm was then washed with PBS 3 times, and crystal violet was extracted with 200 μL of 100% ethanol for 15 min at room temperature. OD590 was measured by a plate reader as an indication of biofilm mass.
Statistical analysis.
Statistical significance was determined using either a one-way analysis of variance (ANOVA) with Tukey’s post hoc test or Student’s t test. Analyses were performed using GraphPad Prism v.7 (GraphPad Software). Statistical significance was determined using a P value of <0.05.
ACKNOWLEDGMENTS
This study was supported by the National Institute of Allergy and Infectious Diseases (R01AI143916) to D.J.W., an American Heart Association Career Development Award (19CDA34630005) to E.S.G., and a Cure CF Columbus fellowship (C3LIU0219 and C3LIU0520) and a fellowship program for Advancing Research in Infection and Immunity to Y.L. This work was supported in part by the Cure CF Columbus Translational Core (C3TC). C3TC is supported by the Division of Pediatric Pulmonary Medicine, the Biopathology Center Core, and the Data Collaboration Team at Nationwide Children’s Hospital. Grant support was provided by The Ohio State University Center for Clinical and Translational Science (National Center for Advancing Translational Sciences, grant UL1TR002733) and by the Cystic Fibrosis Foundation (Research Development Program, grant MCCOY19RO).
We acknowledge resources from the Campus Microscopy and Imaging Facility (CMIF) and the OSU Comprehensive Cancer Center (OSUCCC) Microscopy Shared Resource (MSR) at The Ohio State University. We thank Ken Bayles and P. Lynne Howell for generously providing the S. aureus LAC strain and PslG hydrolase, respectively. We also acknowledge Antonio DiGiandomenico at AstraZeneca for providing us with Psl monoclonal antibodies.
Footnotes
Supplemental material is available online only.
Contributor Information
Daniel J. Wozniak, Email: Daniel.Wozniak@osumc.edu.
Mohamed Y. El-Naggar, University of Southern California
REFERENCES
- 1.Doring G, Flume P, Heijerman H, Elborn JS, Consensus Study Group. 2012. Treatment of lung infection in patients with cystic fibrosis: current and future strategies. J Cyst Fibros 11:461–479. 10.1016/j.jcf.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Nichols DP, Chmiel JF. 2015. Inflammation and its genesis in cystic fibrosis. Pediatr Pulmonol 50(Suppl 40):S39–S56. 10.1002/ppul.23242. [DOI] [PubMed] [Google Scholar]
- 3.Surette MG. 2014. The cystic fibrosis lung microbiome. Ann Am Thorac Soc 11(Suppl 1):S61–S65. 10.1513/AnnalsATS.201306-159MG. [DOI] [PubMed] [Google Scholar]
- 4.Wakeman CA, Moore JL, Noto MJ, Zhang Y, Singleton MD, Prentice BM, Gilston BA, Doster RS, Gaddy JA, Chazin WJ, Caprioli RM, Skaar EP. 2016. The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat Commun 7:11951. 10.1038/ncomms11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limoli DH, Hoffman LR. 2019. Help, hinder, hide and harm: what can we learn from the interactions between Pseudomonas aeruginosa and Staphylococcus aureus during respiratory infections? Thorax 74:684–692. 10.1136/thoraxjnl-2018-212616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, Stojadinovic O, Plano LR, Tomic-Canic M, Davis SC. 2013. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8:e56846. 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limoli DH, Yang J, Khansaheb MK, Helfman B, Peng L, Stecenko AA, Goldberg JB. 2016. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur J Clin Microbiol Infect Dis 35:947–953. 10.1007/s10096-016-2621-0. [DOI] [PubMed] [Google Scholar]
- 8.Serra R, Grande R, Butrico L, Rossi A, Settimio UF, Caroleo B, Amato B, Gallelli L, de Franciscis S. 2015. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev Anti Infect Ther 13:605–613. 10.1586/14787210.2015.1023291. [DOI] [PubMed] [Google Scholar]
- 9.Seth AK, Geringer MR, Galiano RD, Leung KP, Mustoe TA, Hong SJ. 2012. Quantitative comparison and analysis of species-specific wound biofilm virulence using an in vivo, rabbit-ear model. J Am Coll Surg 215:388–399. 10.1016/j.jamcollsurg.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 10.Reen FJ, Mooij MJ, Holcombe LJ, McSweeney CM, McGlacken GP, Morrissey JP, O’Gara F. 2011. The Pseudomonas quinolone signal (PQS), and its precursor HHQ, modulate interspecies and interkingdom behaviour. FEMS Microbiol Ecol 77:413–428. 10.1111/j.1574-6941.2011.01121.x. [DOI] [PubMed] [Google Scholar]
- 11.Kessler E, Safrin M, Olson JC, Ohman DE. 1993. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem 268:7503–7508. 10.1016/S0021-9258(18)53203-8. [DOI] [PubMed] [Google Scholar]
- 12.Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 187:554–566. 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bharali P, Saikia JP, Ray A, Konwar BK. 2013. Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: a novel chemotaxis and antibacterial agent. Colloids Surf B Biointerfaces 103:502–509. 10.1016/j.colsurfb.2012.10.064. [DOI] [PubMed] [Google Scholar]
- 14.Ryder C, Byrd M, Wozniak DJ. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10:644–648. 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zegans ME, DiGiandomenico A, Ray K, Naimie A, Keller AE, Stover CK, Lalitha P, Srinivasan M, Acharya NR, Lietman TM. 2016. Association of biofilm formation, Psl exopolysaccharide expression, and clinical outcomes in Pseudomonas aeruginosa keratitis: analysis of isolates in the Steroids for Corneal Ulcers Trial. JAMA Ophthalmol 134:383–389. 10.1001/jamaophthalmol.2015.5956. [DOI] [PubMed] [Google Scholar]
- 16.Thaden JT, Keller AE, Shire NJ, Camara MM, Otterson L, Huband M, Guenther CM, Zhao W, Warrener P, Stover CK, Fowler VG, Jr, DiGiandomenico A. 2016. Pseudomonas aeruginosa bacteremic patients exhibit nonprotective antibody titers against therapeutic antibody targets PcrV and Psl exopolysaccharide. J Infect Dis 213:640–648. 10.1093/infdis/jiv436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones CJ, Wozniak DJ. 2017. Psl produced by mucoid Pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. mBio 8:e00864-17. 10.1128/mBio.00864-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol 186:4466–4475. 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armbruster CR, Wolter DJ, Mishra M, Hayden HS, Radey MC, Merrihew G, MacCoss MJ, Burns J, Wozniak DJ, Parsek MR, Hoffman LR. 2016. Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. mBio 7:e00538-16. 10.1128/mBio.00538-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin Z, Yang L, Qu D, Molin S, Tolker-Nielsen T. 2009. Pseudomonas aeruginosa extracellular products inhibit staphylococcal growth, and disrupt established biofilms produced by Staphylococcus epidermidis. Microbiology (Reading) 155:2148–2156. 10.1099/mic.0.028001-0. [DOI] [PubMed] [Google Scholar]
- 21.Chew SC, Yam JKH, Matysik A, Seng ZJ, Klebensberger J, Givskov M, Doyle P, Rice SA, Yang L, Kjelleberg S. 2018. Matrix polysaccharides and SiaD diguanylate cyclase alter community structure and competitiveness of Pseudomonas aeruginosa during dual-species biofilm development with Staphylococcus aureus. mBio 9:e00585-18. 10.1128/mBio.00585-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machan ZA, Pitt TL, White W, Watson D, Taylor GW, Cole PJ, Wilson R. 1991. Interaction between Pseudomonas aeruginosa and Staphylococcus aureus: description of an anti-staphylococcal substance. J Med Microbiol 34:213–217. 10.1099/00222615-34-4-213. [DOI] [PubMed] [Google Scholar]
- 23.Limoli DH, Whitfield GB, Kitao T, Ivey ML, Davis MR, Grahl N, Hogan DA, Rahme LG, Howell PL, O’Toole GA, Goldberg JB. 2017. Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio 8:e00186-17. 10.1128/mBio.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol 73:622–638. 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Mo KF, Wang Q, Stover CK, DiGiandomenico A, Boons GJ. 2013. Epitope mapping of monoclonal antibodies using synthetic oligosaccharides uncovers novel aspects of immune recognition of the Psl exopolysaccharide of Pseudomonas aeruginosa. Chemistry 19:17425–17431. 10.1002/chem.201302916. [DOI] [PubMed] [Google Scholar]
- 26.DiGiandomenico A, Warrener P, Hamilton M, Guillard S, Ravn P, Minter R, Camara MM, Venkatraman V, Macgill RS, Lin J, Wang Q, Keller AE, Bonnell JC, Tomich M, Jermutus L, McCarthy MP, Melnick DA, Suzich JA, Stover CK. 2012. Identification of broadly protective human antibodies to Pseudomonas aeruginosa exopolysaccharide Psl by phenotypic screening. J Exp Med 209:1273–1287. 10.1084/jem.20120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 100:14339–14344. 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thaipisuttikul I, Hittle LE, Chandra R, Zangari D, Dixon CL, Garrett TA, Rasko DA, Dasgupta N, Moskowitz SM, Malmstrom L, Goodlett DR, Miller SI, Bishop RE, Ernst RK. 2014. A divergent Pseudomonas aeruginosa palmitoyltransferase essential for cystic fibrosis-specific lipid A. Mol Microbiol 91:158–174. 10.1111/mmi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu S, Su T, Wu H, Liu S, Wang D, Zhao T, Jin Z, Du W, Zhu MJ, Chua SL, Yang L, Zhu D, Gu L, Ma LZ. 2015. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res 25:1352–1367. 10.1038/cr.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price CE, Brown DG, Limoli DH, Phelan VV, O’Toole GA. 2020. Exogenous alginate protects Staphylococcus aureus from killing by Pseudomonas aeruginosa. J Bacteriol 202:e00559-19. 10.1128/JB.00559-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maalej H, Boisset C, Hmidet N, Colin-Morel P, Buon L, Nasri M. 2017. Depolymerization of Pseudomonas stutzeri exopolysaccharide upon fermentation as a promising production process of antibacterial compounds. Food Chem 227:22–32. 10.1016/j.foodchem.2017.01.079. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Xu Z, Guo Z, Zhao Z, Zhao Y, Wang X. 2018. Structural investigation of a polysaccharide from the mycelium of Enterobacter cloacae and its antibacterial activity against extensively drug-resistant E. cloacae producing SHV-12 extended-spectrum beta-lactamase. Carbohydr Polym 195:444–452. 10.1016/j.carbpol.2018.04.114. [DOI] [PubMed] [Google Scholar]
- 33.He F, Yang Y, Yang G, Yu L. 2010. Structural investigation of an antibacterial polysaccharide from Streptomyces virginia H03. Z Naturforsch C J Biosci 65:317–321. 10.1515/znc-2010-5-602. [DOI] [PubMed] [Google Scholar]
- 34.Agarwal A, Singh KP, Jain A. 2010. Medical significance and management of staphylococcal biofilm. FEMS Immunol Med Microbiol 58:147–160. 10.1111/j.1574-695X.2009.00601.x. [DOI] [PubMed] [Google Scholar]
- 35.Hoiby N, Ciofu O, Bjarnsholt T. 2010. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 5:1663–1674. 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 36.O’Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp 2011:2437. 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barraza JP, Whiteley M. 2021. A Pseudomonas aeruginosa antimicrobial affects the biogeography but not fitness of Staphylococcus aureus during coculture. mBio 12:e00047-21. 10.1128/mBio.00047-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol 51:675–690. 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 39.Fyfe JA, Govan JR. 1980. Alginate synthesis in mucoid Pseudomonas aeruginosa: a chromosomal locus involved in control. J Gen Microbiol 119:443–450. 10.1099/00221287-119-2-443. [DOI] [PubMed] [Google Scholar]
- 40.Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol 2:167. 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma L, Wang J, Wang S, Anderson EM, Lam JS, Parsek MR, Wozniak DJ. 2012. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ Microbiol 14:1995–2005. 10.1111/j.1462-2920.2012.02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones CJ, Ryder CR, Mann EE, Wozniak DJ. 2013. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J Bacteriol 195:1637–1644. 10.1128/JB.02190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sautter R, Ramos D, Schneper L, Ciofu O, Wassermann T, Koh CL, Heydorn A, Hentzer M, Hoiby N, Kharazmi A, Molin S, Devries CA, Ohman DE, Mathee K. 2012. A complex multilevel attack on Pseudomonas aeruginosa algT/U expression and algT/U activity results in the loss of alginate production. Gene 498:242–253. 10.1016/j.gene.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciofu O, Lee B, Johannesson M, Hermansen NO, Meyer P, Hoiby N. 2008. Investigation of the algT operon sequence in mucoid and non-mucoid Pseudomonas aeruginosa isolates from 115 Scandinavian patients with cystic fibrosis and in 88 in vitro non-mucoid revertants. Microbiology (Reading) 154:103–113. 10.1099/mic.0.2007/010421-0. [DOI] [PubMed] [Google Scholar]
- 45.Yu S, Wei Q, Zhao T, Guo Y, Ma LZ. 2016. A survival strategy for Pseudomonas aeruginosa that uses exopolysaccharides to sequester and store iron to stimulate Psl-dependent biofilm formation. Appl Environ Microbiol 82:6403–6413. 10.1128/AEM.01307-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, Christensen LD, Jensen PO, Nielsen AK, Parsek M, Wozniak D, Molin S, Tolker-Nielsen T, Hoiby N, Givskov M, Bjarnsholt T. 2011. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One 6:e27943. 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visaggio D, Pasqua M, Bonchi C, Kaever V, Visca P, Imperi F. 2015. Cell aggregation promotes pyoverdine-dependent iron uptake and virulence in Pseudomonas aeruginosa. Front Microbiol 6:902. 10.3389/fmicb.2015.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darch SE, Simoska O, Fitzpatrick M, Barraza JP, Stevenson KJ, Bonnecaze RT, Shear JB, Whiteley M. 2018. Spatial determinants of quorum signaling in a Pseudomonas aeruginosa infection model. Proc Natl Acad Sci USA 115:4779–4784. 10.1073/pnas.1719317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 109:20632–20636. 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alves PM, Al-Badi E, Withycombe C, Jones PM, Purdy KJ, Maddocks SE. 2018. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog Dis 76:fty003. 10.1093/femspd/fty003. [DOI] [PubMed] [Google Scholar]
- 51.Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5:30. 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci USA 112:4110–4115. 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiba A, Sugimoto S, Sato F, Hori S, Mizunoe Y. 2015. A refined technique for extraction of extracellular matrices from bacterial biofilms and its applicability. Microb Biotechnol 8:392–403. 10.1111/1751-7915.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masuko T, Minami A, Iwasaki N, Majima T, Nishimura S, Lee YC. 2005. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem 339:69–72. 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. 2006. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol 188:8213–8221. 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bose JL, Lehman MK, Fey PD, Bayles KW. 2012. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One 7:e42244. 10.1371/journal.pone.0042244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirisits MJ, Prost L, Starkey M, Parsek MR. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 71:4809–4821. 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, Table S1, and Fig. S1-S7. Download jb.00076-22-s0001.pdf, PDF file, 0.5 MB (517.5KB, pdf)