ABSTRACT
Herein, we report the in vivo evolution of imipenem/relebactam-resistance in KPC-producing K. pneumoniae (KPC-Kp) isolated from a critically-ill patient treated with multiple combination therapies based on ceftazidime-avibactam or meropenem-vaborbactam. Imipenem/relebactam-resistance was associated to meropenem-vaborbactam cross-resistance and was due to truncated OmpK35 and OmpK36 porins and increased copy of blaKPC copy number. Genome analysis demonstrated that imipenem/relebactam-resistant KPC-Kp harbored a second copy of blaKPC-carrying Tn4401 in a ColRNAI plasmid as a consequence of a transposition event.
KEYWORDS: cross-resistance, Tn4401, copy number, public health, transposition mechanisms
INTRODUCTION
Development of novel antimicrobial molecules represents a key to overcome clinical and microbiological challenge of the difficult-to-treat resistance (DTR) infections (1). The introduction of novel β-lactam/β-lactamase inhibitor combinations (BLICs) partially solved the pressing need to treat DTR (2). Imipenem-cilastatin-relebactam (IMI-REL) is a novel combination recently approved by the FDA in 2019 for patients with limited or no alternative therapeutic options (3). Although IMI-REL represents a valuable option for the treatment of infections due to DTR, emerging IMI-REL-resistant isolates have been recently reported (4). Herein, we characterized the evolution of the resistance to IMI-REL in a series of KPC-producing Klebsiella pneumoniae (KPC-Kp) following ceftazidime-avibactam (CAZ-AVI) antimicrobial exposure.
A 35-year-old woman suffering from systemic lupus erythematosus with end-stage renal disease and autoimmune liver cirrhosis was admitted for simultaneous liver-kidney transplantation. Surveillance rectal swabs on admission revealed that the patient was colonized by KPC-Kp (isolate KPC-Kp_TO1). After 25 days of hospitalization, patient presented with pneumonia, and subculture from broncho-alveolar lavage fluid yielded a CAZ-AVI-susceptible KPC-Kp strain (isolate KPC-Kp_TO2). According to the antimicrobial susceptibility pattern, ceftazidime-avibactam with fosfomycin was started for an overall therapy of 10 days with patient clinical improvement. On day 69, blood cultures collected yielded a KPC-Kp strain (isolate KPC-Kp_TO3) susceptible to imipenem, meropenem, and CAZ-AVI; thus, a treatment with meropenem in association with CAZ-AVI and tigecycline was started for an overall therapy of 21 days. On hospital day 92, patient underwent simultaneous liver-kidney transplantation, and meropenem-vaborbactam (MER-VAB) plus tigecycline were administered for 7 days as empirical therapy. After 16 days from transplantation, patient presented with fever and abdominal pain, and abdominal-pelvic computer tomography disclosed a peri-hepatic abscess that was promptly drained. Empirical therapy based on 3-h infusion of MER-VAB was started. Subculture from abscess drainage fluid yielded a CAZ-AVI-susceptible KPC-Kp strain (isolate TO4). According to antimicrobial susceptibility results, fosfomycin was added to MER-VAB for an overall therapy of 14 days with progressive clinical improvement. On day 134, patient presented with septic shock requiring fluid resuscitation and vasopressor support. Empirical therapy based on 3-h infusion of MER-VAB plus 4-h infusion of tigecycline and high-dose extended interval gentamicin was started. Blood cultures collected yielded a KPC-Kp strain (isolate TO5) resistant to MER-VAB and carbapenem and susceptible to CAZ-AVI. Based on clinical conditions and limited antimicrobial options, MER-VAB in association with gentamicin was continued, and despite initial clinical and analytic improvement, patient died 16 days later.
Antimicrobial susceptibility testing was performed using automated system MicroScan Walkaway-96 (Beckman Coulter, USA). The MICs for IMI-REL, MER-VAB, and CAZ-AVI were determined by using Microtiter Sensititre Plate MDRGN2F (Thermofisher, USA) and confirmed with MIC test strips (Liofilchem, Italy). MIC values were interpreted following European Committee for Antimicrobial Susceptibility Testing (EUCAST) breakpoints v11.0. Antimicrobial susceptibility patterns of KPC-Kp strains showed that all strains were susceptible to CAZ-AVI (MICs range 0.5–8 μg/L) and cefiderocol (MICs range 0.125–0.5 mg/L) (Table 1). At the same time, four out of five KPC-Kp strains were susceptible to MER-VAB (MIC range 1–2 mg/L), while KPC producer isolated from the last fosfomycin episode (named KPC-Kp_TO5) exhibited an increased MIC of 2–6-fold for IMI-REL, 7–8 -fold for MER-VAB, and 2–4 -fold for CAZ-AVI compared to parental strains.
TABLE 1.
Antimicrobial susceptibility profiles of KPC-producing Klebsiella pneumoniae strains included in this studya
| Isolate | Sample | MIC (ug/mL) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOS | GEN | TGC | IPM | MEM | CAZ/AVI | MEM/VAB | IPM/REL | CFD | ||||
| KPC-KP_TO1 | Rectal swab | 64 | <2 | <=0.5 | >32 | >32 | 0.5 | 1 | 0.25 | 0.5 | ||
| KPC-KP_TO2 | BAL | ≤16 | <2 | <=0.5 | >32 | >32 | 2 | 2 | 0.5 | 0.5 | ||
| KPC-KP_TO3 | Blood | ≤16 | <2 | <=0.5 | 0.125 | 1 | 4 | 1 | 0.125 | 0.5 | ||
| KPC-KP_TO4 | ADF | >64 | <2 | ≤0.5 | >32 | >32 | 1 | 2 | 2 | 0.125 | ||
| KPC-KP_TO5 | Blood | >64 | <2 | ≤0.5 | >32 | >32 | 8 | ≥256 | 8 | 0.25 | ||
BAL, broncho-alveolar lavage fluid; ADF, abscess drainage fluid; FOS, fosfomycin; GEN, gentamicin; TGC, tigecycline; IPM, imipenem; MEM, meropenem; CAZ/AVI, ceftazidime-avibactam; MER/VAB, meropenem-vaborbactam; IPM/REL, imipenem/relebactam; CFD, cefiderocol. Resistance is shown in bold.
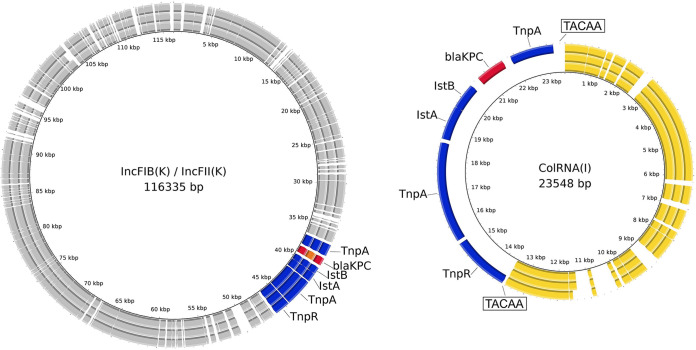
WGS was performed using Illumina iSeq100 (Illumina, USA) and Oxford Nanopore MinION (ONT, UK). Hybrid assemblies were performed using Unicycler v.0.4.7 and polished with Pilon v.1.23. Antimicrobial resistance genes, plasmid content, antimicrobial resistance genes, and MLST analysis were performed as previously described (5). Genomic analysis demonstrated that all KPC-Kp belonged to the Sequence Type ST512, harbored the same plasmid replicon types [i.e., ColRNAI, IncFIB (pQIL), IncFII(K)], and shared similar antimicrobial encoding genes (Table 2). In detail, analysis of β-lactamase genes showed that all strains harbored blaTEM1A, blaOXA-9, and blaSHV-11, while carbapenemase gene was present with blaKPC-3 in KPC-Kp_TO1 and KPC-Kp_TO5 strains, and blaKPC-66 in KPC-Kp_TO3. Analysis of porin genes showed that all strains harbored truncated ompK35 porin, while OmpK36 porin was present with GD134 insertion within IMI-REL-susceptible strains and resulted truncated in the IMI-REL-resistant KPC-Kp_TO5 strain. Sequence analysis of circular plasmids demonstrated that blaKPC-harboring Tn4401 transposon was present in single copy in IMI-REL-susceptible KPC-Kp, while it was present in double copies within IMI-REL-resistant KPC-Kp_TO5 (Fig. 1). In particular, Tn4401a transposon was located within IncFIB- IncFII(K) 115Kb plasmid in all sequentially-isolated KPC-Kp strain, while the second copy in the IMI-REL-resistant KPC-Kp_TO5 was located in a 25 Kb plasmid. Deep sequence analysis of 25 Kb plasmid demonstrated that the Tn4401 has been inserted into a ColRNAI plasmid common to parental KPC-Kp strains susceptible to IMI-REL and flanked by 5-bp direct repeats (DR) (Fig. 1).
TABLE 2.
Genomic characteristics of KPC-producing Klebsiella pneumoniae strainsa
| Isolate | ST | Antimicrobial resistance determinants |
Porins |
blaKPC copy no. | |||
|---|---|---|---|---|---|---|---|
| Beta-lactam | Aminoglycosides | ompK35 | ompK36 | Plasmid_replicons (InC) | |||
| KPC-KP_TO1 | 512 | blaKPC-3, blaTEM1A, blaOXA-9 | aadA2b, aac(6’)-Ib-cr | Truncated at aa 41 | GD insertion at aa 134-135 | ColRNAI, IncFIB (pQIL), IncFII(K) | 1b |
| KPC-KP_TO3 | 512 | blaKPC-66, blaTEM1A, blaOXA-9 | aadA2b, aac(6’)-Ib-cr | Truncated at aa 41 | GD insertion at aa 134-135 | ColRNAI, IncFIB (pQIL), IncFII(K) | 0,85 |
| KPC-KP_TO5 | 512 | blaKPC-3, blaTEM1A, blaOXA-9 | aadA2b, aac(6’)-Ib-cr | Truncated at aa 41 | Truncated at aa 310 | ColRNAI, IncFIB (pQIL), IncFII(K) | 4,59 |
ST, sequence type; ND, not determined; GD, glycine and aspartic acid.
Used as reference strain.
FIG 1.
Circular representation of the IncFIB/IncFII and ColRNAI plasmids in the KPC-producing Klebsiella pneumoniae included in this study. From outside to inside: KPC-Kp_TO5 strains, KPC-Kp_TO3, and KPC-Kp_TO1. The CDS are color-coded with the direction of transcription indicated by arrowheads. The Tn4401 transposons are displayed in blue, KPC-3 in red, KPC-66 in orange, ColRNAI backbone in yellow, and other CDS in gray. Boxes indicate direct repeats (DRs).
The blaKPC copy number was evaluated using the CT value of target gene (blaKPC) normalized to endogenous control gene (16S rDNA), as previously described (6). Relative quantification (RQ) analysis of blaKPC gene demonstrated that IMI-REL-resistant KPC-Kp_TO5 had a blaKPC copy number higher 4.59-fold than IMI-REL-susceptible KPC-Kp isolates (Table 1).
Comparative analysis based on Breseq software (7) demonstrated that Illumina reads of IMI-REL-susceptible strains aligned against IMI-REL-resistant KPC-Kp annotated genome identified respectively a total of 79 and 100 predicted mutations, most of them annotated as hypothetical proteins. Also, phylogenetic analysis generated based on the core genome SNPs analysis showed that KPC-Kp_TO3 and KPC-Kp_TO5 genomes differed by 175 and 167 SNPs with KPC-Kp_TO1, used as reference, respectively, thus demonstrating close genetic relationship.
In this study, we identified the mechanism at the basis of IMI-REL resistance in KPC-Kp isolated from a critically-ill patient treated with multiple antimicrobial combination therapies based on ceftazidime-avibactam or meropenem-vaborbactam. Our results are consistent with previous studies showing that increased level of expression of blaKPC gene in combination with reduced porins expression impacted the activity of IMI-REL, thus reducing its in vitro efficacy (4, 8, 9).
Clinical history showed that cross-resistance to meropenem-vaborbactam and IMI-REL emerged following multiple antimicrobial exposure to novel BLICs, thus suggesting the in vivo genomic adaption of K. pneumoniae. In line with this hypothesis, the first bacteremic episode was due to a KPC-Kp closely related to other KPC-Kp strains isolated from the same patient and that harbored a blaKPC-66 gene, a derivative of blaKPC-3 with an amino acid duplication at positions 167 and 168 (EL167-168). Although this variant did not confer resistance to IMI-REL, it should be noted that KPC-Kp carrying blaKPC-66 variant exhibited and increased MIC 3-fold for CAZ-AVI, reverted susceptibility to carbapenem in comparison to parental strain (i.e., KPC-Kp_TO1), and emerged after ceftazidime-avibactam-based therapy following pneumonia episode, thus suggesting a strength antimicrobial selection of mutated population after BLICs exposure in this anatomical site (10). These data are in accordance with previous studies showing that SNPs mutations within Ω –loop of the blaKPC gene conferring CAZ-AVI-resistance was associated to increased susceptibility to carbapenems (11–12).
Lastly, IMI-REL-resistance was associated with an increased MIC for meropenem-vaborbactam and ceftazidime-avibactam, thus suggesting that the mechanisms conferring this type of resistance could be related to a cross-resistance for novel BLICs. Further studies are necessary to elucidate the microbiological impact of different therapies based on novel BLICs to selected mutant-resistant strains and to clarify the impact of cross-resistance on the clinical outcome.
Data availability.
The sequences of KPC-Kp genomes included in this study were deposited at EMBL/EBI under the project study (EBI project PRJNA794363) with the following accession numbers: KPC-KP_TO1 (SAMN24648676), KPC-KP_TO3 (SAMN24648677), KPC-KP_TO5 (SAMN24648678).
ACKNOWLEDGMENT
This work was supported by Italian Ministry of Health (Ricerca Finalizzata, Giovani Ricercatori, GR-2018-12367572).
REFERENCES
- 1.Giannella M, Bussini L, Pascale R, Bartoletti M, Malagrinò M, Pancaldi L, Toschi A, Ferraro G, Marconi L, Ambretti S, Lewis R, Viale P. 2019. Prognostic utility of the new definition of difficult-to-treat resistance among patients with Gram-negative bloodstream infections. Open Forum Infect Dis 6:ofz505. 10.1093/ofid/ofz505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yahav D, Giske CG, Grāmatniece A, Abodakpi H, Tam VH, Leibovici L. 2020. New β-lactam-β-lactamase inhibitor combinations. Clin Microbiol Rev 34:e00115-20. 10.1128/CMR.00115-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouza E. 2021. The role of new carbapenem combinations in the treatment of multidrug-resistant Gram-negative infections. J Antimicrob Chemother 76:iv38–iv45. 10.1093/jac/dkab353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galani I, Souli M, Nafplioti K, Adamou P, Karaiskos I, Giamarellou H, Antoniadou A, Study Collaborators. 2019. In vitro activity of imipenem-relebactam against non-MBL carbapenemase-producing Klebsiella pneumoniae isolated in Greek hospitals in 2015–2016. Eur J Clin Microbiol Infect Dis 38:1143–1150. 10.1007/s10096-019-03553-8. [DOI] [PubMed] [Google Scholar]
- 5.Gaibani P, Lombardo D, Bussini L, Bovo F, Munari B, Giannella M, Bartoletti M, Viale P, Lazzarotto T, Ambretti S. 2021. Epidemiology of meropenem/vaborbactam resistance in KPC-producing Klebsiella pneumoniae causing bloodstream infections in Northern Italy, 2018. Antibiotics (Basel) 10:536. 10.3390/antibiotics10050536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombardo D, Ambretti S, Lazzarotto T, Gaibani P. 2022. In vitro activity of imipenem-relebactam activity against KPC-producing Klebsiella pneumoniae resistant to ceftazidime-avibactam and/or meropenem-vaborbactam. Clin Microbiol Infect. 10.1016/j.cmi.2022.01.025. [DOI] [PubMed] [Google Scholar]
- 7.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Landman D, Quale J. 2015. Activity of imipenem with relebactam against Gram-negative pathogens from New York City. Antimicrob Agents Chemother 59:5029–5031. 10.1128/AAC.00830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balabanian G, Rose M, Manning N, Landman D, Quale J. 2018. Effect of porins and blaKPC expression on activity of imipenem with relebactam in Klebsiella pneumoniae: can antibiotic combinations overcome resistance? Microb Drug Resist 24:877–881. 10.1089/mdr.2018.0065. [DOI] [PubMed] [Google Scholar]
- 10.Shields RK, Nguyen MH, Chen L, Press EG, Kreiswirth BN, Clancy CJ. 2018. Pneumonia and renal replacement therapy are risk factors for ceftazidime-avibactam treatment failures and resistance among patients with carbapenem-resistant Enterobacteriaceae infections. Antimicrob Agents Chemother 62:e02497-17. 10.1128/AAC.02497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carattoli A, Arcari G, Bibbolino G, Sacco F, Tomolillo D, Di Lella FM, Trancassini M, Faino L, Venditti M, Antonelli G, Raponi G. 2021. Evolutionary trajectories toward ceftazidime-avibactam resistance in Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother 65:e0057421. 10.1128/AAC.00574-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernández-García M, Castillo-Polo JA, Cordero DG, Pérez-Viso B, García- Castillo M, Saez de la Fuente J, Morosini MI, Cantón R, Ruiz-Garbajosa P. 2022. Impact of ceftazidime-avibactam treatment in the emergence of novel KPC variants in the ST307-Klebsiella pneumoniae high-risk clone and consequences for their routine detection. J Clin Microbiol 60:e0224521. 10.1128/jcm.02245-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences of KPC-Kp genomes included in this study were deposited at EMBL/EBI under the project study (EBI project PRJNA794363) with the following accession numbers: KPC-KP_TO1 (SAMN24648676), KPC-KP_TO3 (SAMN24648677), KPC-KP_TO5 (SAMN24648678).