ABSTRACT
The DNA-binding protein from starved cells, Dps, is a universally conserved prokaryotic ferritin that, in many species, also binds DNA. Dps homologs have been identified in the vast majority of bacterial species and several archaea. Dps also may play a role in the global regulation of gene expression, likely through chromatin reorganization. Dps has been shown to use both its ferritin and DNA-binding functions to respond to a variety of environmental pressures, including oxidative stress. One mechanism that allows Dps to achieve this is through a global nucleoid restructuring event during stationary phase, resulting in a compact, hexacrystalline nucleoprotein complex called the biocrystal that occludes damaging agents from DNA. Due to its small size, hollow spherical structure, and high stability, Dps is being developed for applications in biotechnology.
KEYWORDS: DNA-binding protein, Dps, ferritin, nucleoid-associated protein, stationary phase
INTRODUCTION
During typical bacterial growth and survival in the laboratory, a bacterial population that initially exists at low cell density transitions into a phase of rapid growth and cell division known as logarithmic (log) or exponential phase. As the population approaches high cell density, growth slows, and the population density levels out as cells enter stationary phase. The transition from log phase to stationary phase includes a series of environmental and cellular changes that must be managed, including lower nutrient availability, increased concentrations of metabolic waste products, nucleoid remodeling, and managing intracellular concentrations of important cofactors, including iron. To adapt to the changing environment and stresses of stationary phase, cells modify their gene expression patterns and protein levels. Curious about this phenomenon, Almirón and colleagues performed an SDS-PAGE experiment in which newly synthesized proteins in batch Escherichia coli cultures were labeled with radioactive methionine at several time points during log phase and stationary phase (1). One of the most highly labeled proteins as cells transitioned into stationary phase was Dps.
The DNA-binding protein from starved cells, Dps, is conserved across bacterial species (1–5) (Table 1). To date, a UniProt search of genes annotated as “dps” returns 93,962 prokaryotic proteins. Only one bacterial phylum, Thermomicrobia, is not represented in this list; of note, all gammaproteobacterial orders are represented. Most bacterial genomes contain one dps gene, but some contain as many as five (5–7). Additionally, some archaeal species have dodecameric, Dps-like ferritin proteins (8, 9).
TABLE 1.
Key Dps properties in several bacterial speciesa
| Species | Gene | Oligomer type (reference) | Presence of iron storage (reference) | Presence of ferroxidase (reference) | Preferred oxidant (reference) | No. of atoms in iron core (aerobic) (reference) | Presence of DNA binding (reference) | Presence of K-rich N terminus | Other ion bound (reference) | Other ion-binding site (reference) |
|---|---|---|---|---|---|---|---|---|---|---|
| Agrobacterium tumefaciens | dps | Dodecamer (36) | Y (36) | Y (36) | 500 (36) | N (36) | Y | |||
| Bacillus anthracis | dps1 | Dodecamer (182) | Y (182) | Y (39) | O2 (39) | 500 (39) | N (182) | N | ||
| dps2 | Dodecamer (182) | Y (182) | Y (39) | H2O2 (39) | 500 (39) | N (182) | N | |||
| Bacillus cereus | dps1 | Dodecamer (7) | Y (7) | N | ||||||
| dps2 | Dodecamer (7) | Y (7) | N | |||||||
| dps3 | Dodecamer (57) | Y (57) | Y (57) | N | ||||||
| Dimer (57) | ||||||||||
| Bacillus subtilis | mrgA | Dodecamer (145) | Y (145) | N | ||||||
| Borrelia burgdorferi | dps | Dodecamer (40) | Y (40) | Y (40) | 500 (40) | N (40) | Y | |||
| Campylobacter jejuni | dps | Dodecamer (37) | Y (37) | Y (37) | H2O2 (37) | 500 (37) | Y (91) | N | ||
| Deinococcus radiodurans | dps1 | Dodecamer (79) | Y (79) | Y (79) | H2O2 (183) | 250 (183) | Y (14) | Y | Ca(II) (14) | Ferroxidase center (14) |
| Co(II) (79) | C-term Co(II) site (79) | |||||||||
| N-term Co(II) site (79) | ||||||||||
| dps2 | Trimer (183) | N | Mn(II) (183) | |||||||
| Dimer (14) | Y (14) | Y (14) | Zn(II) (99) | N-term Zn(II) site (99) | ||||||
| Dodecamer (184) | Y (184) | Y (184) | H2O2 (183) | 400 (183) | Y (183) | Mn(II) (183) | ||||
| Escherichia coli | dps | Dodecamer (1) | Y (34) | Y (34) | H2O2 (34) | 500 (34) | Y (1) | Y | ||
| Helicobacter pylori | nap | Dodecamer (156) | Y (33) | Y (33) | 500 (33) | N (33) | N | |||
| Lactococcus lactis | dpsA | Dodecamer (53) | N (53) | Y (53) | Y | |||||
| Trimer (53) | N (53) | |||||||||
| Dimer (53) | N (53) | |||||||||
| dpsB | Dodecamer (53) | N (53) | Y (53) | Y | ||||||
| Listeria innocua | dps | Dodecamer (185) | Y (185) | Y (185) | H2O2 (38) | N (117) | N | |||
| Listeria monocytogenes | fri | Dodecamer (117) | Y (117) | Y (117) | N | |||||
| Microbacterium arborescens | aah | Dodecamer (116) | Y (116) | Y (116) | H2O2 (116) | N (116) | N | |||
| Mycobacterium smegmatis | dpsA | Dodecamer (186) | Y (2) | Y (2) | Y (186) | N | ||||
| Trimer (2) | N (2) | Y (2) | N (2) | |||||||
| dpsB | Dodecamer (27) | Y (187) | Y (187) | Y (27) | N | |||||
| Nostoc punctiforme | dps1 | Dodecamer (188) | Y (188) | Y (188) | H2O2 (188) | N (188) | N | |||
| Trimer (188) | ||||||||||
| dps2 | Dodecamer (188) | Y (188) | Y (188) | H2O2 (188) | Y (188) | Y | ||||
| Trimer (188) | ||||||||||
| dps3 | Dodecamer (188) | Y (188) | Y (188) | H2O2 (188) | Y (188) | Y | ||||
| dps4 | Dodecamer (89) | Y (89) | Y (89) | O2 (89) | Y (189) | N | Zn(II) (89) | Ferroxidase center (89) | ||
| dps5 | Dodecamer (189) | Y (189) | N | |||||||
| Porphyromonas gingivalis | dps | Dodecamer (77) | Y (40) | Y | ||||||
| Staphylococcus aureus | mrgA | Dodecamer (190) | Y (190) | Y (66) | N | |||||
| Staphylococcus suis | dpr | Dodecamer (26) | Y (191) | Y (26) | N | Co(II) (100) | Ferroxidase center (100) | |||
| Cu(II) (100) | Ferroxidase center (100) | |||||||||
| Mn(II) (100) | Ferroxidase center (100) | |||||||||
| Ni(II) (100) | Ferroxidase center (100) | |||||||||
| Zn(II) (97) | Ferroxidase center (97) | |||||||||
| Novel Zn(II) site (97) | ||||||||||
| Tb(II) (97) | Ferroxidase center (97) | |||||||||
| Streptomyces coelicolor | dpsA | Dodecamer (192) | Y (192) | Y (192) | H2O2 (192) | N (192) | N | |||
| dpsB | Dimer (192) | N | ||||||||
| dpsC | Dodecamer (192) | Y (192) | Y (192) | H2O2 (192) | N (192) | Y | ||||
| Sulfolobus solfataricus | dps | Dodecamer (42) | Y (42) | Y (42) | H2O2 (42) | 300 (42) | Y | Zn(II) (9) | ||
| Synechococcus sp. strain PCC 7942 | dpsA | Y (4) | Y (4) | H2O2 (4) | N (4) | N | ||||
| Thermosynechococcus elongatus | dpsA | Dodecamer (96) | Y (96) | Y (96) | 100 (96) | N | Zn(II) (87) | Ferroxidase center (87) | ||
| Trichodesmium erythraeum | dps | Dodecamer (95) | Y (95) | Y (95) | H2O2 (95) | 300 (95) | Y (95) | N | Phosphate (95) | |
C-term, C terminal; N-term, N terminal; Y, yes; N, no.
In most species, Dps functions as a ferritin, which is an iron-detoxifying and iron storage protein with ferroxidase activity. However, in some species, Dps is also a double-stranded DNA (dsDNA)-binding protein (Table 1). The DNA-binding and ferroxidase activities of Dps, in species with both functions, are biochemically discrete but function jointly to protect DNA and mediate stress tolerance (10–13).
Dps binding requires a minimum length of ∼90 bp of dsDNA in species in which it binds DNA, but otherwise, it has no well-defined DNA sequence-binding motifs or structural specificity (1, 14–18). In many organisms in which Dps functions as a dsDNA-binding protein, Dps becomes the major nucleoid-associated protein (NAP) during stationary phase (19–22). Like other NAPs, Dps modulates nucleoid shape and compaction (23).
During stationary phase, Dps and DNA form a tightly packed nucleoprotein complex called the biocrystal (1, 2, 24–27). Biocrystal formation is stationary-phase specific, requires Dps (11, 20, 22, 24, 25, 28–31), and, in E. coli, occurs gradually, beginning in early stationary phase and continuing until late stationary phase, at which point the nucleoid is organized into a hexacrystalline array (22, 25).
The ferritin properties of Dps are 3-fold. First, it is proposed that Dps stores iron and releases Fe(II) when needed (32–40). Second, Dps detoxifies excess iron in the cell using its ferroxidase activity to oxidize soluble Fe(II) to insoluble Fe(III), which is unavailable to participate in potentially damaging redox chemistry (see below). Third, as a ferroxidase, Dps uses H2O2 to oxidize ferrous iron to the ferric form, making it unique among ferritins (4, 8, 9, 26, 34–38, 41–44). Because using H2O2 as the oxidizing agent for iron results in its breakdown, Dps helps prevent the synthesis of reactive oxidative species (ROS), through the Fenton reaction, capable of damaging nucleic acids, proteins, and lipids (34).
Monomer structure.
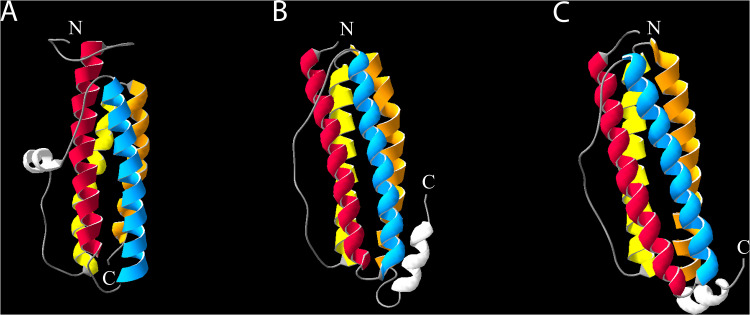
Dps was originally discovered in E. coli as a DNA-binding protein (1). When the X-ray crystal structure was determined, it became apparent that the Dps monomer, comprised primarily of a 4-helix bundle, shows striking similarity to the ferritin monomer despite a lack of sequence homology (Fig. 1) (18, 32). Ferritins are iron-sequestering proteins and are conserved throughout all three domains of life. Like ferritin, the E. coli Dps (Ec-Dps) monomer’s A and B helices are connected by a short loop; its C and D helices are similarly connected (18). The AB and CD helix pairs are connected by a longer loop, also akin to ferritins (18).
FIG 1.
The Dps monomer structure displays homology to ferritins. Shown are tertiary structures of Ec-Dps (PDB accession number 1DPS) (A), Ec-FtnA (PDB accession number 1EUM) (B), and Ec-Bfr (PDB accession number 3E1J) (C) (18, 180, 181). Homologous alpha helices are displayed in the same color: red, helix A; blue, helix B; yellow, helix C; orange, helix D. Nonhomologous helices are shown in white. N termini and C termini for each molecule are labeled with a white “N” and a white “C,” respectively.
There are some notable differences between Dps and ferritin monomers. The Ec-Dps monomer has an additional, smaller N-terminal helix (Fig. 1) (18). This helix is flexible, making it difficult to characterize using typical X-ray crystallography techniques. Ferritins lack this N-terminal helix, but they have an additional C-terminal helix (Fig. 1), which is postulated to contribute to the 24-mer organization in ferritins, compared to the Dps dodecamer (36).
Dodecamer structure.
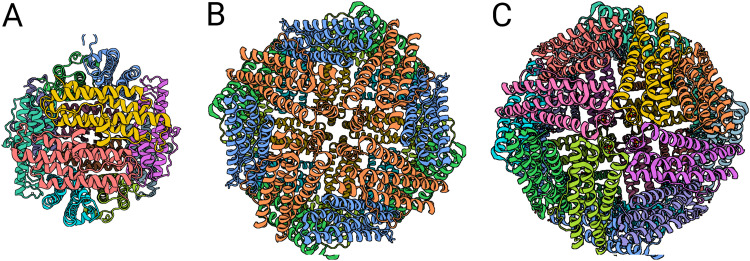
Dps monomers with a molecular weight of ∼19 kDa come together to form a dodecamer (1). This dodecamer, like the monomer, is structurally similar to ferritin oligomers (18). Both Dps and ferritins oligomerize into a hollow sphere, although ferritins contain 24 subunits, compared to 12 for Dps (2, 18, 27, 36, 45, 46). The Dps sphere, smaller than that formed by ferritins, is approximately 90 Å in diameter, with a 45-Å hollow core (9, 18, 27, 36, 45). Ferritins assemble into 120-Å-diameter spheres with 80-Å hollow cores (Fig. 2) (46). Symmetry also differs for the Dps dodecamer compared to the ferritin 24-mer: the Dps dodecamer has tetrahedral symmetry, in contrast to ferritin’s octahedral symmetry (18, 27, 36, 45–47).
FIG 2.
Quaternary structures of the E. coli Dps dodecamer and ferritin 24-mers. (A) Ec-Dps dodecamer (PDB accession number 1DPS); (B) Ec-FtnA 24-mer (PDB accession number 1EUM); (C) Ec-Bfr 24-mer (PDB accession number 3E1J) (18, 180, 181). Monomer subunits are shown in distinct colors. Ec-FtnA is a partial structure; mirrored subunits are shown in the same color. Images are scaled to approximate size differences between the 90-Å-diameter Dps dodecamer and the 180-Å-diameter ferritin 24-mers. (Created with BioRender.com.)
Several assembly models for Dps have been proposed. In one model, the AB loop acts as a switch for the number of subunits in an oligomer: a rigid AB loop (like that found in Dps) directs the assembly of a dodecamer with 2- and 3-fold symmetry, while a flexible AB loop (like that found in bacterioferritin [Bfr] and ferritin [Ftn] proteins) directs the assembly of a ferritin 24-mer with 2-, 3-, and 4-fold symmetry (48). In another model, the N terminus of Dps may modulate Dps dodecamer formation and self-association during biocrystal formation (12, 49, 50). When examining the stepwise dynamics of dodecamer assembly, it has been suggested that some Dps species first form trimers and then form dodecamers, whereas others first form dimers and then form dodecamers; the difference roughly correlates with the length of the N-terminal helix (51). It is also notable that two arginine residues, R83 and R113 in Ec-Dps, have been found to be necessary for Ec-Dps dodecamer assembly (52).
Other quaternary structures.
While Dps proteins typically function as dodecamers, some Dps proteins can form smaller, semifunctional oligomers. Both Lactococcus lactis DpsA (Ll-DpsA) and Mycobacterium smegmatis Dps1 (Ms-Dps1) form stable dimers and trimers in addition to dodecamers (2, 53). The Ms-Dps1 trimer has ferroxidase activity, although it cannot store iron and does not bind DNA; the dodecamer performs all three functions (2, 50, 54). In some cases, the formation of nondodecameric oligomers is due to environmental conditions. The hexuronates d-glucuronate and d-galacturonate have been found to destabilize the Ec-Dps dodecamer (55). Notably, in E. coli, hexuronate concentrations are highest during log phase, when Dps is poorly expressed, and decrease significantly during stationary phase, when Dps is highly expressed (56). Additionally, Deinococcus radiodurans Dps1 (Dr-Dps1) forms DNA-binding dimers at low salt concentrations in vitro (12, 14), consistent with models of Dps-DNA-binding sensitivity to the cation concentration (see “Dps as a DNA-binding protein,” below). Bacillus cereus Dps3 (Bc-Dps3) is found primarily as a dimer but forms dodecamers upon the addition of Fe(II), suggesting an environmentally mediated mechanism for dodecamer assembly, akin to Dr-Dps1 (57).
Dps as a DNA-binding protein.
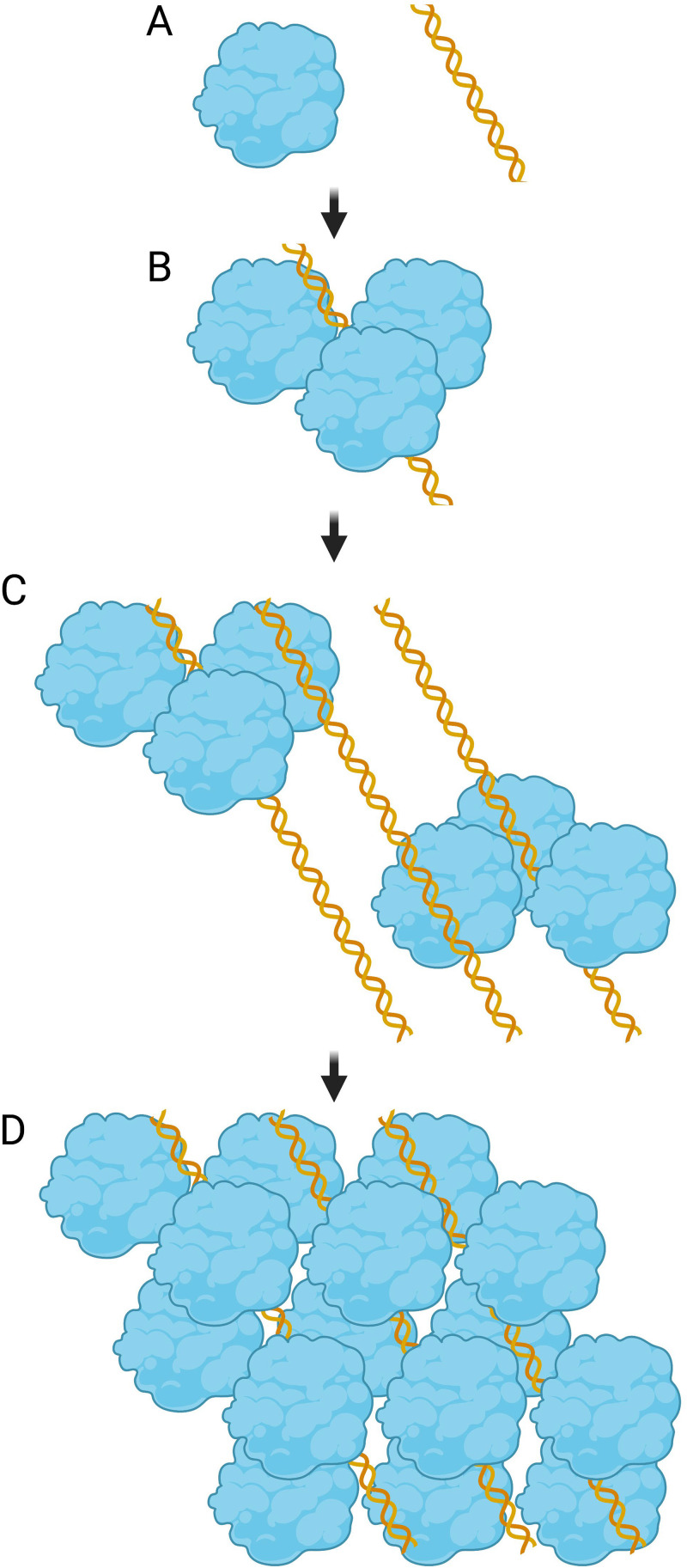
During stationary phase in organisms whose Dps is a DNA-binding protein, Dps and DNA assemble into a tight nucleoprotein complex called the biocrystal. The biocrystal forms a hexagonally packed assembly, with offset planar arrays stacked on top of one another, similar to oranges packed in a crate (Fig. 3) (1, 2, 24–27). While Ec-Dps can form hexacrystalline biocrystal-type structures with itself in vitro, the addition of DNA greatly accelerates the process (11, 49). However, the spacing of the Ec-Dps crystalline lattice is similar with and without DNA (11). Initially, during the formation of the biocrystal, Ec-Dps–DNA aggregates form toroidal (ring-shaped) structures (Fig. 4) (25). It has been hypothesized that Ec-Dps–DNA toroids are points of initial nucleation from which biocrystallization spreads until the nucleoid is largely restructured by Ec-Dps. The spacings of nucleoprotein complexes in early-stationary-phase toroids and late-stationary-phase biocrystals are similar, supporting this hypothesis (25).
FIG 3.
Model of E. coli Dps-DNA binding. Shown is a model of Ec-Dps DNA binding where blue shapes represent Dps dodecamers and yellow double helices represent dsDNA molecules. (A) A single Dps dodecamer is separate from dsDNA; (B) a triad of Dps dodecamers coalesces around a single dsDNA molecule; (C) multiple Dps dodecamer triads coalesce around dsDNA; (D) 3-dimensional Dps-DNA hexacrystalline array. The image is not to scale. (Model inspired by Grant and colleagues [18]; created with BioRender.com.)
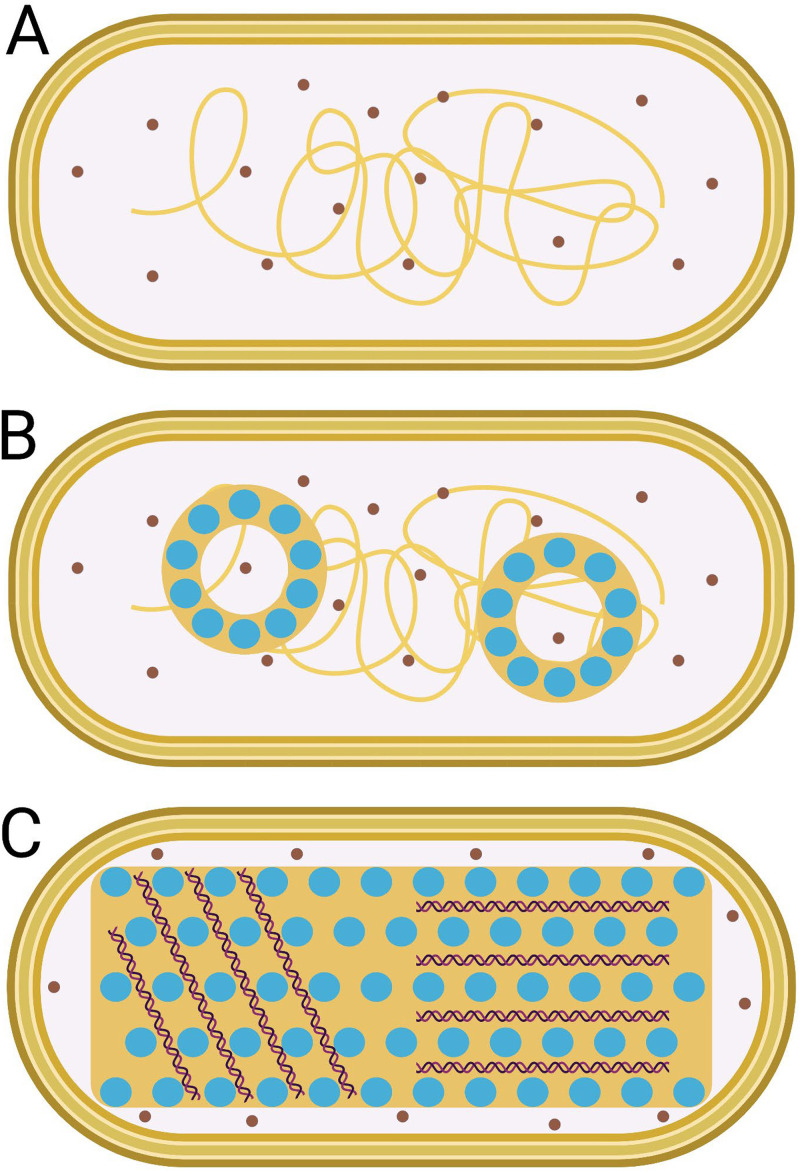
FIG 4.
E. coli Dps induces the formation of the stationary-phase-specific biocrystal. Shown is the E. coli nucleoid structure bounded within cell membranes, where brown circles represent ribosomes, yellow shapes represent DNA, and blue circles represent Dps dodecamers. (A) During log phase, chromatin is interspersed with the translation machinery. (B) During the transition between log phase and stationary phase, toroids composed of regularly spaced Dps-DNA nucleoprotein complexes form, which are segregated from ribosomes. (C) By late stationary phase, the nucleoid has been restructured to a regularly spaced “biocrystal” nucleoprotein complex, which is segregated from ribosomes. Double helices in panel C represent locally parallel DNA within the crystalline nucleoid. The image is not to scale. (Model inspired by Frenkiel-Krispin and colleagues [24, 25]; created with BioRender.com.)
Dps-DNA complexes are formed only during stationary phase in E. coli (Fig. 4) (11, 24, 28, 58), where one study found that Ec-Dps occupies over 50% of the nucleoid (22). This growth-phase-specific phenomenon is not due simply to increased Dps abundance during stationary phase because the overexpression of Ec-Dps during log phase does not result in biocrystal formation (24). Whether this stationary-phase specificity is due to unfavorable environmental conditions for Dps binding during log phase or reflects the concentration of other competing NAPs with a higher DNA affinity during log phase is not well understood.
Stationary-phase-specific nucleoid compaction is dependent on Dps (11, 20, 22, 24, 25, 28–31). To date, research suggests that Ec-Dps is unique among NAPs in that it is necessary for a proper nucleoid structure. The removal of any other NAP during log phase does not significantly change the log-phase nucleoid structure, but the removal of Ec-Dps significantly changes the structure of the stationary-phase nucleoid (59, 60). Without Dps, the late-stationary-phase nucleoid in E. coli is configured into a cholesteric (liquid crystalline) phase (24). While a cholesteric organization has been shown to reduce the accessibility of DNA molecules to a variety of damaging factors, suggesting an overarching necessity for DNA protection during late stationary phase, this nucleoid conformation also leads to a longer lag phase and other defects (61, 62). This is likely due to DNA being in a conformation that is more resistant to the remodeling to a log-phase chromatin structure that occurs upon inoculation into fresh medium.
Dps-dependent nucleoid compaction during the transition from log phase to stationary phase is gradual (22, 25). Nucleoid restructuring by Ec-Dps lags behind Ec-Dps binding (63, 64). This could explain the timing of the shift from toroids to a mature biocrystal between early and late stationary phases. Hysteresis, the phenomenon of a physical change lagging behind its inducing effect, appears to be a feature of the stationary-phase nucleoid, as nucleoid restructuring by integration host factor (IHF), the other major NAP during early stationary phase, also displays this property (64).
Not all Dps proteins with DNA-binding activity create a biocrystal. Some form noncrystalline aggregates, whereas others bind DNA without condensation (15, 53, 54). For example, the DNA-binding activity of Helicobacter pylori neutrophil-activating protein (HP-NAP), which is unique among Dps proteins for its positively charged exterior, is modulated by pH (15). Ms-Dps1, which does not induce DNA condensation and does not protect DNA from nuclease-induced cleavage, does protect DNA from hydroxyl radical-induced damage (54). However, the overexpression of either Ms-Dps protein in M. smegmatis results in nucleoprotein toroid formation (31).
The DNA-binding activity of several Dps species is modulated by environmental factors in vitro. While the DNA-binding and ferritin-like properties of Dps proteins are biochemically separable, incorporated Fe3+ enhances DNA-binding efficiency in some species (12, 65). In Staphylococcus aureus, stationary phase alone is not sufficient for nucleoid compaction, but oxidative stress or the overexpression of MrgA, the S. aureus Dps protein, results in a highly compacted nucleoid (30, 66). Additionally, treatment of the M. smegmatis stationary-phase nucleoid with RNase “loosens” the nucleoid structure (31). RNase-induced loosening may be due to decreased macromolecular crowding forces, with fewer macromolecules (RNA, DNA, and proteins) in the cytoplasm to promote nucleoid condensation (67).
Dps has no currently identified sequence or structural specificity for its DNA-binding activity. Ec-Dps binds DNA with a dissociation constant (Kd) of approximately 2 × 10−7 M, which is relatively low for a NAP (the Kd for specific binding of other major NAPs typically falls around 10−9 M [68–72]) and may explain the lack of observed sequence specificity (16, 18). It has been observed that Ec-Dps does not discriminate among linear dsDNA, circular dsDNA, and single-stranded RNA (ssRNA) in vitro (11, 73). Furthermore, Ec-Dps shows no preference between supercoiled and relaxed DNA (14). However, there appears to be a minimum size for high-affinity DNA binding: dsDNA fragments smaller than ∼90 bp do not bind Ec-Dps efficiently (18). Interestingly, 90 bp is about the length required to encircle a Dps dodecamer, a number that has been used to suggest a wrapping model for Dps-DNA binding (74).
While there is no universally accepted model for Dps DNA-binding specificity, several studies have suggested DNA-binding motifs. One transcriptome sequencing (RNA-Seq) study suggested that Dps-binding regions are enriched for inverted repeats, overlap promoter islands significantly, tend to show increased structural flexibility, or overlap binding sites of other NAPs, particularly the factor for inversion stimulation (Fis), one of the major log-phase NAPs (17, 75). A SELEX-Seq (systematic evolution of ligands by exponential enrichment sequencing) experiment suggested a consensus sequence for linear DNA binding by Dps (76), but the universality of this sequence in vivo has yet to be confirmed.
There are 2 non-mutually exclusive models proposed for DNA-Dps interactions. The first involves interactions mediated through divalent cation bridges. This is supported by the observation that Dps will bind DNA only in a certain cationic range of ∼1.0 mM Mg2+ in vitro, which is abolished when EDTA is added to sequester cations (24, 53, 64). The second is that the short, lysine (K)-rich, N-terminal helices from three adjacent Dps dodecamers coalesce around a dsDNA molecule (Fig. 3) (10, 18, 49). Species with a K-rich N-terminal helix, including Ec-Dps, tend to also have DNA-binding activity, and species that lack a K-rich N-terminal helix tend to lack this activity (Table 1) (14, 26, 45, 65, 77, 78). The second model does have exceptions: Ms-Dps1 and Dr-Dps1 appear to require both the N-terminal and C-terminal regions to bind DNA, and HP-NAP is postulated to use its positively charged exterior to bind DNA (15, 50, 79, 80). Additionally, Agrobacterium tumefaciens Dps (At-Dps) has a positively charged N-terminal helix, although it is 11 amino acids shorter than the 20-amino-acid-long Ec-Dps N-terminal helix and does not bind DNA (36). The convergent evolution of different DNA-binding modes exhibited by different Dps species suggests a biological demand for DNA protection during stationary phase.
Interactions with other NAPs.
Log phase is characterized by rapid growth and cell division, which necessitates high levels of tightly regulated gene expression and, thus, ready access to the chromosome. This is reflected by the plurality of major NAPs during log phase and the resultant log-phase nucleoid structure. However, easy access to genetic material is not necessarily beneficial during stationary phase. Low nutrient availability and high population density shift the cell’s focus to maintenance and protection; this requires the Dps-dependent sequestration of DNA. The transition from a log-phase to a stationary-phase nucleoid structure requires a transition in NAP availability and perhaps particular interactions between log-phase and stationary-phase NAPs.
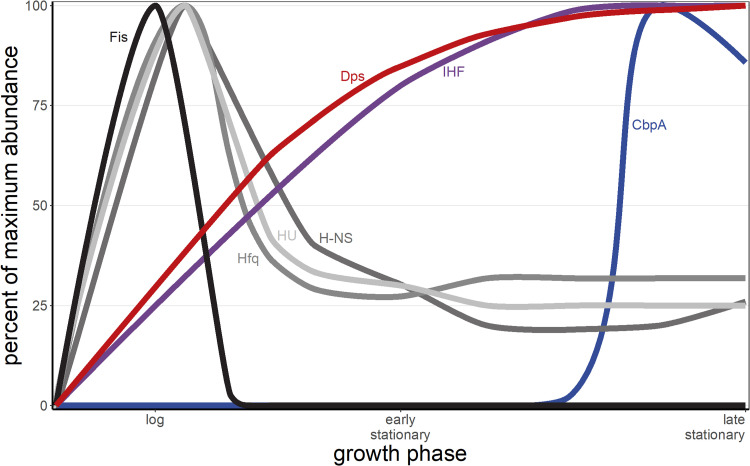
Dps interacts in various ways with other NAPs. Ec-Dps acts antagonistically to Fis as a nucleoid structural agent (58, 63, 75). Additionally, Dps and Fis expression has been found to invert between log and stationary phases in E. coli: Fis is highly expressed during log phase and below the limit of detection during stationary phase, while Dps is weakly expressed during log phase and highly expressed during stationary phase (Fig. 5) (19). Fis and H-NS each regulate Ec-Dps expression (81). Both Dps and curved DNA-binding protein A (CbpA), the two major NAPs present during late stationary phase in E. coli, self-aggregate and are postulated to cause nucleoid compaction by clustering distal DNA loci (82).
FIG 5.
Major E. coli NAP abundance by growth phase. NAPs are differentially expressed during different growth phases. Dps (red) and IHF (purple) protein abundances are low during log phase and high during stationary phase; Fis (black), H-NS (dark gray), HU (light gray), and Hfq (medium gray) are highly expressed during log phase and lowly expressed during stationary phase; and CbpA (blue) is lowly expressed until mid- to late stationary phase. The x axis is not linear with time. (Data inspired by Ali Azam et al. [19].)
The interactions between Dps and some of the major log-phase NAPs may suggest a mechanism for Dps accessing the chromosome throughout stationary phase. Perhaps Dps replaces other NAPs, including Fis, as they dissociate from the chromosome. If log-phase NAPs dissociate in a concentration-dependent manner, as previously reported (83, 84), this may provide the opportunity for Dps molecules to nucleate DNA locally before restructuring the entire chromosome into the biocrystal.
Dps as a ferritin.
Iron, a cofactor in many essential biological processes, can be critical in the generation of reactive oxidative species (ROS), which are capable of damaging a broad range of macromolecules (85). This is achieved primarily through the Fenton reaction:
Ferritins help the cell manage the dual nature of iron by converting it into its insoluble, less reactive form, storing this detoxified Fe(III), and releasing it when needed. Bacteria have two highly conserved ferritin proteins: ferritin (Ftn) and the heme-containing bacterioferritin (Bfr). Ftn and Bfr have little sequence homology, except at their ferroxidase sites, which are highly conserved (86). Dps, while ferritin like, has a number of notable differences compared to both canonical ferritins (86). Only a few Dps proteins, including the two Dps proteins of L. lactis, have been found not to possess ferritin activity; both of these proteins bind DNA (53). Dps uses H2O2 to oxidize Fe(II) to Fe(III), while traditional ferritins use O2 as the oxidant (4, 6, 34, 41). One H2O2 is metabolized for every two Fe(II) atoms oxidized, avoiding the production of hydroxyl radicals via Fenton chemistry (34). For some species with two or more Dps proteins, one may use O2 to oxidize Fe(II), and the other may use H2O2 (39, 87–89).
Dps dodecamers have iron entry pores that are unique among ferritins. These pores are negatively charged and located at the four 3-fold interfaces within the dodecamer (6, 18, 27). Interestingly, Dr-Dps1 has distinct iron exit channels that constantly release Fe(II) and contribute to DNA damage in vitro (79). When these iron exit channels are disrupted via mutation, Dr-Dps1 loses the ability to contribute to DNA damage, and instead, DNA is partially protected from iron-mediated cleavage (79).
The Dps dodecamer has 12 highly conserved ferroxidase centers, which can each oxidize two iron atoms simultaneously (34, 79, 88–91). Unlike ferritins, the Dps ferroxidase site is comprised of residues from two adjacent monomers: two histidine residues from one subunit and an aspartate and a glutamate from the other (6, 27, 41, 45, 51, 86, 92). This is distinct from ferritins, whose active sites are formed solely within each of the 24 monomers.
Once oxidized, iron is stored in the hollow core of the Dps dodecamer, at which point it organizes as microcrystals (92). While Dps can form a ferric core with O2 as an oxidant similar in size to that observed with H2O2, the O2 core formation reaction is less cooperative and leads to increased heterogeneity in ferric core size in a population of Dps dodecamers (34). A crystalline iron core is also observed in ferritin proteins (93). Two steps in Fe(III) reduction and release from Ec-Dps have been observed in biochemical experiments, which may indicate two populations of iron in the protein, perhaps one representing the bulk iron in the core and the other attached to the interior of the shell (92).
One Dps dodecamer typically contains up to 500 Fe(III) atoms in its internal cavity under aerobic culture conditions (32–40). When grown anaerobically, the ferric core contains ∼400 Fe(III) atoms; traditional ferritins can store up to ∼4,000 oxidized iron atoms per 24-mer (34, 46, 94). Several Dps species store fewer iron atoms: Thermosynechococcus elongatus Dps (Te-Dps) and Halobacterium salinarum DpsA (Hs-DpsA) each hold ∼100 Fe(III) atoms/dodecamer, and Trichodesmium erythraeum Dps (Te-Dps) and Sulfolobus solfataricus Dps (Ss-Dps) each store ∼300 Fe(III) atoms/dodecamer (42, 95, 96) (Table 1). The biological mechanism(s) for these discrepancies is unknown.
Some Dps proteins have been found to bind other metals, including zinc, calcium, cobalt, copper, nickel, manganese, and terbium, and one small charged molecule, phosphate (9, 14, 79, 87, 89, 95, 97–101). Each has been found to bind at the ferroxidase site and/or an allosteric (nonferroxidase) site, depending on the Dps species and noniron substrate (Table 1). Dr-Dps-1 contains 2 allosteric Co(II) sites, one near the C terminus and another near the N terminus of the protein, as well as an allosteric Zn(II) site in its longer-than-typical Dps N-terminal helix (79, 99). This can alter Dps action; for example, Zn(II) inhibits ferroxidase activity when bound to the iron site in Nostoc punctiforme Dps4 (Np-Dps4) (89). Additionally, it has been reported that phosphate can affect the crystallinity and chemical reactivity of ferritin cores, which may be due to interactions between the negatively charged phosphate and positively charged iron ions, suggesting that this molecule may serve to modulate these properties in the Dps core (102). Finally, Dps has been shown to protect Anabaena sp. strain PCC 7120 and E. coli from copper toxicity (Table 2), contributing further to a physiological role for Dps in binding noniron metals.
TABLE 2.
Dps as a stress response protein
| Type of stress | Species (reference[s]) | Growth phase (reference[s])a |
|---|---|---|
| Acid stress | Escherichia coli (152) | Log (193, 194) |
| Stationary (62, 193) | ||
| Streptococcus pyogenes | Log (195) | |
| Base stress | Escherichia coli | Log (62) |
| Streptococcus pyogenes | Log (195) | |
| Carbon limitation | Anabaena sp. PCC 7120 | Stationary (196) |
| Cold shock | Listeria monocytogenes (197) | |
| Streptococcus thermophilus (198, 199) | ||
| Copper stress | Anabaena sp. PCC 7120 | Stationary (135, 196) |
| Escherichia coli | Log (200) | |
| Stationary (62) | ||
| Endonucleases | Campylobacter jejuni (91) | |
| Helicobacter pylori (15) | ||
| Trichodesmium erythraeum (95) | ||
| Ethanol stress | Bacillus cereus (57) | |
| High NaCl | Anabaena sp. PCC 7120 | Stationary (135, 196) |
| Bacillus cereus (57) | ||
| Escherichia coli | Log (10) | |
| Legionella pneumophila | Stationary (115) | |
| High pressure | Escherichia coli | Stationary (201) |
| Heat stress | Anabaena sp. PCC 7120 | Stationary (135, 196) |
| Bacillus cereus | Log (7, 57) | |
| Escherichia coli | Log (10) | |
| Stationary (62) | ||
| Legionella pneumophila | Stationary (115) | |
| Iron excess | Escherichia coli | Log (10, 62) |
| Stationary (62) | ||
| Vibrio cholerae (125) | ||
| Iron limitation | Anabaena sp. PCC 7120 | Stationary (135, 196) |
| Escherichia coli | Stationary (K. Orban, unpublished results) | |
| Nitrogen limitation | Anabaena sp. PCC 7120 | Stationary (196) |
| Oxidative stress | Agrobacterium tumefaciens (36) | |
| Bacillus anthracis | Stationary (44) | |
| Bacillus cereus | Log (57) | |
| Bacillus subtilis | Log (104, 145) | |
| Stationary (145) | ||
| Campylobacter jejuni | Log (91) | |
| Escherichia coli | Log (1, 10, 194) | |
| Stationary (62, 146) | ||
| Helicobacter hepaticus (65) | ||
| Legionella pneumophila | Log (115) | |
| Listeria innocua (38) | ||
| Listeria monocytogenes | Log (159) | |
| Stationary (159) | ||
| Microbacterium arborescens | Log (116) | |
| Nostoc punctiforme (202) | ||
| Porphyromonas gingivalis (77) | ||
| Salmonella enterica serovar Typhimurium (108, 203) | Log (157) | |
| Staphylococcus aureus | Log (30) | |
| Stationary (13, 30) | ||
| Streptococcus mutans (204) | ||
| Streptococcus pyogenes | Log (195, 205) | |
| Streptococcus suis | Stationary (41, 206) | |
| Thermosynechococcus elongatus (43, 87) | ||
| Vibrio cholerae | Log (125) | |
| Stationary (125) | ||
| Phosphorus limitation | Anabaena sp. PCC 7120 | Stationary (135, 196) |
| UV and gamma irradiation | Anabaena sp. PCC 7120 | Stationary (135, 196) |
| Escherichia coli | Stationary (62) | |
| Staphylococcus aureus | Log (30) | |
| Visible-light stress | Nostoc punctiforme (127) | |
| Zinc excess | Escherichia coli | Transition (62) |
| Streptococcus pyogenes | Stationary (195) | |
For species that are not assigned a growth phase for the Dps-mediated stress response, the work was done either in vitro or on plates.
Dps expression.
The expression of Dps is primarily dependent on the growth phase. During log phase, dps expression is low; however, it is upregulated in response to oxidative stress (7, 66, 102–104). dps expression is upregulated during stationary phase relative to log phase (1, 19, 21, 104, 105).
The Dps protein concentration is relatively low during log phase. Several studies have quantified the Ec-Dps concentration as <1,000 molecules per cell during log phase (19, 21, 105). Dps expression is controlled at the transcriptional, posttranscriptional, translational, and posttranslational levels during log phase. Transcription is repressed by the ferric uptake regulation protein (Fur) (106–109). During log phase, Ec-Dps is induced in response to a variety of stresses (3). This is not due to growth-phase-specific changes in the transcription machinery: E. coli σ70, the housekeeping sigma factor, and σs, the stationary-phase sigma factor, have similar affinities for the dps promoter in vitro (81). Instead, various stress-activated transcription factors modulate dps expression. OxyR, the oxidative stress response transcription factor, activates dps expression during log phase by binding upstream of the dps promoter and recruiting σ70 (102, 110, 111). This is modulated by the oxidative stress level encountered by the cell. Reduced OxyR has a significantly lower affinity for the dps promoter; i.e., oxidized OxyR induces dps expression (112). In addition to OxyR, dps expression is regulated by PerR, the peroxide regulon repressor (7, 66, 103, 113). dps is upregulated in response to iron depletion stress, iron excess stress, thermal stress, NaCl stress, ethanol stress, and gamma irradiation and in the presence of acetyl phosphate (42, 57, 96, 107, 114–119). The log-phase stressor concentration-dependent expression of Dps is similar to ferritin expression, which is low unless stressors are added (120).
Dps transcription is controlled by other NAPs during log phase. In E. coli, Fis inhibits dps promoter accessibility by RNA polymerase (RNAP) formed with σ70; Fis and σ70 are able to corepress transcription by σs (81). H-NS binds the −10 promoter region of dps, blocking σ70 from binding the promoter (81). Dps, which is an N-end rule degradation pathway substrate in E. coli, is rapidly degraded by ClpXP (121–124). During log phase, the Dps protein’s half-life is ∼10 min; this increases to ∼40 min with the addition of oxidative stress (121).
Dps is highly expressed during stationary phase, where several studies have quantified Dps levels in the range of hundreds of thousands of molecules per cell (19, 21, 105). As in log phase, Dps levels are controlled at the transcriptional, posttranscriptional, translational, and posttranslational levels during stationary phase. dps transcription is induced by σs, which directly activates dps expression by binding the −10 promoter region (1, 3, 110, 125). dps transcription is also controlled by other NAPs during stationary phase. IHF, the other major NAP during early stationary phase, has been found to cooperate with σs in the σs-mediated upregulation of Dps during early stationary phase (58, 110). At the posttranscriptional level, Dps degradation is not detected during stationary phase (121). In species with more than one dps gene, different dps loci are differentially regulated (7, 57, 126–130). Additionally, if one dps locus is knocked out in species with more than one dps gene, the expression of other dps loci may compensate for its absence (127).
Because Dps is a large family of proteins across many species, there are exceptions to these patterns. In Campylobacter jejuni, dps (Cj-dps) is constitutively expressed during log and stationary phases and is not upregulated in response to oxidative stress (37). Additionally, Borrelia burgdorferi Dps (Bb-Dps) is constitutively synthesized in both log and stationary phases with no change due to oxidative stress but is differentially expressed when incubated in mice (low expression) or ticks (high expression) (40). Finally, Porphyromonas gingivalis dps (Pg-dps) expression is not modulated by oxidative stress (131).
Dps as a regulator of gene expression.
One mechanism by which NAPs can affect gene expression is by altering nucleoid architecture (132). Like the other major nucleoid structural proteins, which affect gene expression through altering nucleoid structure, Ec-Dps is distributed throughout the nucleoid (1, 63, 133). Data also suggest that Dps may be a regulator of gene expression. When first identified, radiolabeled two-dimensional PAGE (2D-PAGE) showed many differences between newly synthesized proteins in E. coli wild-type and dps-null strains in late stationary phase, suggesting a role for Dps as a regulator of stationary-phase-specific gene expression (1). In addition, a series of promoterless lacZ fusions made in an arabinose-inducible dps background showed differential expression depending on the dps expression status (S. E. Finkel, unpublished results). Furthermore, a SELEX-Seq experiment identified 624 Dps-binding sites throughout the E. coli chromosome (76); from the locations of these sites, regulatory targets have been predicted (https://shigen.nig.ac.jp/ecoli/tec/). Dps in Salmonella enterica and Anabaena sp. PCC 7120 has been shown to affect global gene expression, although it is unknown whether the modulation effect is direct or indirect (134, 135). However, there are some conflicting data with respect to gene expression regulation. Antipov and colleagues (75) showed the differential regulation of genes by Dps between biological replicates in late-stationary-phase E. coli. These results may suggest regulatory plasticity modulated by Dps. However, an extensive regulatory study on Ec-Dps found no significant differences in expression via RNA-Seq or proteomics due to the presence or absence of Dps in log, stationary, or early long-term stationary phase (136).
There are several potential explanations for the differing Dps gene expression results, particularly across E. coli studies. First, the methodologies differ. Genetic experiments such as promoterless lacZ fusions, which examine larger-scale, population-level effects, may show different phenomena than sequencing experiments such as chromatin immunoprecipitation sequencing (ChIP-Seq) and mRNA-Seq, which examine changes at the molecular level. Similarly, biochemical experiments such as radiolabeled 2D-PAGE and mass spectrometry may show different phenomena than one another, as the former examines newly synthesized proteins and the latter probes the global protein distribution. Second, the strains being studied differ. The same mutation can produce a range of phenotypic changes, or no change at all, depending on the genetic background (137). This may be the case with Dps gene expression effects.
Macromolecular sequestration and phase separation.
The formation of the Dps-DNA biocrystal may be one example of a larger biological mechanism of protection. Ferritins also form crystalline assemblies, potentially indicating an evolutionary pressure to promote biocrystallization processes as a stress response (138). Ferritin crystals form when ferritin is overproduced in E. coli and cells are exposed to Fe(II), a potent source of oxidative stress that can damage macromolecules like DNA and proteins (61). This may indicate an evolutionary advantage in structural motifs that facilitate a transition of proteins into crystalline structures that protect cellular components through the rapid sequestration of valuable macromolecules from damaging agents. Additionally, biocrystallization has been suggested as a means to maintain homeostasis in stressful environments. For example, the nucleoids of dormant Bacillus spores are arranged into an SspC (small, acid-soluble spore protein C)-DNA crystalline lattice via toroid-mediated condensation, similar to the early-stationary-phase-specific nucleoid packaging mediated by Dps, although the packing of toroids and crystalline lattice formation in the spore are different than those mediated by Dps (139, 140). Because sporulation is induced in response to various stresses, the nucleoid repackaging occurring during this time supports the hypothesis of biocrystallization as a stress response mechanism.
The crystalline assembly of Dps-DNA complexes has been posited to create a distinct phase within the heterogeneous mixture of the nucleoid, a potentially important example of the role of phase separation in biological systems (141, 142). Early studies of Dps-DNA complexes in E. coli show a crystalline assembly (11, 24, 25), although the mechanism could be due to either liquid- or solid-phase separation. Additionally, Dps exhibits highly cooperative binding, which is emblematic of phase separation (142, 143). Finally, RNA polymerase can access DNA when bound by Dps (136), but nucleases and other DNA-damaging agents cannot as efficiently (1, 136). This observation is consistent with other phase-separated complexes, which can selectively concentrate enzymes and other factors (142). Just as Dps is highly expressed in stationary phase, phase separation is dependent on high concentrations of the proteins involved in the phase (142). Phase separation has been shown to play an important role in managing stress responses, including thermal and pH stress (144). This is notable when considering Dps-mediated phase separation, as Dps is a known contributor to stress responses (see below).
Stress response.
Dps confers resistance to several environmental stresses (Table 2), the most extensively studied of which is oxidative stress (1, 11, 30, 57, 62, 145, 146). It does so in three notable ways: (i) physical protection of DNA, (ii) sequestration of iron, and (iii) neutralization of H2O2. First, Dps specifically protects DNA from oxidative-stress-induced damage (10, 11, 57, 91, 146). This is akin to eukaryotic histone proteins, which physically protect DNA from oxidative stress (147). Moreover, Ec-Dps has been found to interact with DnaA to impede DNA replication initiation during log phase in periods of oxidative stress, suggesting a secondary regulatory role that is targeted at protecting DNA (148). Second, in its role as a ferritin, Dps protects the cell from oxidative stress by sequestering iron and, thus, preventing the formation of ROS. In addition to its physical sequestration of iron, Ec-Dps has also been found to interact with the iron-sulfur cluster protein YtfE to diminish YtfE-induced oxidative stress (149). This demonstrates a secondary regulatory role that is targeted toward reducing oxidative stress, which is a theme similar to that of how Dps protects DNA. The third way in which Dps protects the cell from oxidative stress is through the detoxification of H2O2. This is inherent in its ferritin function, as this protein’s preferred oxidant for iron is H2O2. Dps also confers resistance to other stressors, although the protective effects differ depending on the growth phase and species (Table 2).
In species with more than one dps gene, each gene may confer differential resistance to different stresses (7, 127, 128). In Bacillus cereus, two of its three Dps proteins (Bc-Dps1 and Bc-Dps2) act cooperatively to confer resistance to oxidative stress (7). In E. coli, both the ferritin and DNA-binding properties of Dps are required for full Dps-dependent DNA protection (10, 11). Ec-Dps significantly reduces the numbers of DNA strand breaks, abasic sites and ruptured/oxidized guanine, and GC→TA+TA→AT base mutations (146). This is due to Dps DNA protection, as the protein is not involved in the repair of oxidatively damaged DNA (146).
Other functions.
In addition to its ferritin and DNA-binding abilities, Dps has been identified in other important cellular functions. Dps has been identified in a screen for genes involved in biofilm formation, although no specific role has been classified, and one study of spontaneously occurring phage-tolerant E. coli identified Dps at the outer membrane, which may implicate Dps in an as-yet-undetermined role in each of these processes (3, 150–153). Dps has also been implicated in virulence (40, 77, 108, 125, 151, 154–162). This makes particular sense since iron acquisition can play a vital role in determining pathogenicity (163). Salt sensitivity, which is modulated by Dps (Table 2), is also highly correlated with virulence. The Dps protein from Helicobacter pylori, NAP, has been shown to impair human iron absorption and target iron to H. pylori during infection (164). Another Dps protein, Microbacterium arborescens amino acid hydrolase (AAH), catalyzes the cleavage and formation of amide bonds (78). Additionally, the overexpression of Ec-Dps has been found to impede colony growth on agar plates by 2- to 3-fold during log phase (148).
In some species, Dps has been found at the outer membrane, although the function of this localization is currently unclear (3, 150, 152, 153). In Synechococcus sp. strain PCC 7942, more DpsA is observed at the inner cell membrane during lag phase and log phase than during stationary phase (165). This makes sense in the context of stationary-phase-specific DNA binding, as more Dps should be observed in the nucleoid at that time, leaving fewer proteins available to participate in their outer membrane function(s). This, of course, assumes that Dps is able to move between the inner membrane and the nucleoid with relative freedom. The same study found DpsA localized at the cell membrane and the nucleoid (165). The authors of that study proposed that there are two “pools” of DpsA that function in Synechococcus: an insoluble, DNA-binding fraction at the nucleoid and a soluble, ferritin-active fraction at the membrane (165). Two pools of a Dps protein have been observed in D. radiodurans: Dr-Dps2, which can function either as the full-length gene product or as a truncated form (lacking the nonpolar portion of the N terminus that protrudes past the positively charged portion), is found in the full-length form at the membrane and in the truncated form in the nucleoid (80). Currently, the functional differences between the full-length and truncated forms of Dr-Dps2 are unclear; localization to different cellular components suggests broad functional differences.
Dps in nanotechnology.
Recently, Dps proteins, like ferritins, have been used for a range of nanotechnology applications. The “hollow-ball” structure of Dps and ferritin proteins makes them excellent candidates for nanotechnologies that require protein cages. It is advantageous to use Dps instead of canonical, 24-mer ferritins for several reasons. First, Dps is smaller, making it a better option when a smaller size is desirable (166, 167). Second, Dps is highly thermostable, making it easy to purify and often more durable (167).
Dps and ferritins have primarily been used in materials science and drug development and delivery. In materials science, horse spleen ferritin has been used as a nanoreactor (168), Ec-Dps has been used as a scaffold for nanodevice assembly (167), and Bacillus subtilis Dps (Bs-Dps) and Listeria innocua Dps (Li-Dps) have been used as catalysts for the formation of carbon nanotubes with a limited diameter distribution (169) and for platinum nanocluster formation for hydrogen production (170), respectively. Ec-Dps has also been used as a platform to experimentally reconstitute protein-protein interfaces (171), and Li-Dps has been used to synthesize CdSe nanoparticles with nanometric gaps (172) and to fabricate a “high-density, periodic silicon-nanodisc (Si-ND) array” for use in silicon quantum dot solar cells (173). In drug development and delivery, horse spleen and human ferritins have been used as platforms for antigen presentation (174), vaccine development (174), cancer immunotherapy development (175), and drug delivery (176) and as a magnetic resonance imaging (MRI) contrast agent (176). In recent years, there has been increasing interest in nanotechnologies for use in a range of applications such as tissue repair, drug delivery, and immunoassays (177). Combined with its smaller size and high thermostability, these demonstrated uses make Dps increasingly valuable in nanotechnology.
Future directions.
While much progress has been made in the understanding of the ferritin properties of Dps, there is much more to be learned. The dynamics of Dps ferric core organization are not well understood. If two iron subpopulations are present in the protein, it is important to understand the division between, dynamism within, and biological relevance of those populations. Furthermore, an investigation of the determining factors behind which Dps species bind other ions, metals, or small charged molecules awaits clarification.
In addition to ferritin activity, the dynamics of Dps dodecamer formation are still poorly understood. While data suggest that certain Dps species form dimers and/or trimers before the dodecamer forms, a more thorough inquiry is required to better understand these dynamics. The driving force behind the stable dimers/trimers and dodecamers formed by some Dps proteins may be the pH or salt concentration, as previously suggested (12, 14, 54). If that is the case, however, a comparative study of those Dps proteins that form stable dimers/trimers and those that do not is warranted to distinguish mechanisms of assembly.
The additional functions of Dps present interesting avenues of experimentation. Several studies have found Dps at the outer membrane. However, it is still unclear exactly why Dps is localized there. If Dps exists in a substantial quantity in the membrane, any movement or changes in the quantity or concentration may indicate its function there, whether ferritin, stress response, DNA-binding, or an additional, as-yet-undetermined activity. Examining membrane composition and permeability in dps-null strains, dps-overexpressing strains, as well as mutants for oligomerization, DNA-binding, and ferritin activities could shed further light on why Dps exists at the outer membrane.
Compared to log phase, little is known about the stationary-phase nucleoid. This includes its structure, dynamics, and protein composition. More specifically, Dps-dependent nucleoid compaction during stationary phase may be due to cellular environmental conditions. Another model for this phenomenon is that Dps binds DNA when DNA is available to it, which is more likely after log phase when there are lower concentrations of other NAPs with which to compete. Furthermore, it is important to explore how the biocrystal forms. It is unknown what stimulates the formation of biocrystal precursor toroids: might this result from log-phase NAPs dissociating from DNA? It is also unknown whether the biocrystal forms at programmed chromosomal loci or if the process is more stochastic. Perhaps there is an intermediate mechanism by which preliminary, local nucleoid restructuring by Dps occurs when log-phase NAPs dissociate from the chromosome and secondary, global restructuring occurs in a programmed manner. If Dps truly has no discernible sequence or structural specificity, but the biocrystal forms reproducibly at certain loci, how is Dps directed to the sites to which it binds?
There is conflicting evidence with regard to Dps as a regulator of gene expression. Perhaps Dps does affect gene expression, as suggested by Almirón and colleagues’ 2D gel electrophoresis studies (1), but the stationary-phase intracellular environment is such that these changes cannot be detected by transcriptomic or proteomic techniques that focus on the global mRNA/protein population. If both gene expression as well as RNA and protein degradation slow during stationary phase, a larger relative shift in expression profiles may be necessary to outweigh the baseline from log phase and detect these phenomena during stationary phase. Alternatively, a posttranscriptional mechanism of gene expression regulation may be yet undiscovered; a direct interaction between Dps and mRNA may explain the biological relevance of the ssRNA-binding ability of Dps.
Until recently, the role of posttranslational modifications (PTMs) in NAPs had not been studied in bacteria (178). This is still largely the case for Dps proteins and stationary phase. While it has been shown that S. enterica Dps can be glycosylated (179) and that Dr-Dps2 can have its N terminus cleaved in vivo (80), it would be interesting to further study how and where Dps acquires PTMs and what the effects are, if any. The activity of Fis, to which Dps acts antagonistically, seems to be less subject to alterations by PTM than other log-phase NAPs. It has been hypothesized that this is due to Fis activity being more dependent on growth phase than the other NAPs, so PTMs would potentially be a redundant signal here (178). Because Dps activity is also highly regulated by growth phase, PTMs may not act as frequently on Dps as other NAPs. The results of a study of Dps PTMs may help bolster the Fis hypothesis or shed light on another factor in play.
Conclusion.
During stationary phase, the cell encounters an environment in which nutrient availability is more limited than during log phase. The cell has a biological imperative during this time to protect its genetic information from damaging agents, including ROS-inducing ferrous iron ions. Dps provides an elegant solution to this problem, both sequestering iron in its inner cavity and creating a phase-separated nucleoid that is less accessible to DNA-damaging agents. While stationary-phase-specific nucleoid compaction is surely impacted by the action of other NAPs, it requires Dps. This is likely because the cell needs a rapid switch to adapt its nucleoid to the pressures of stationary phase. In its role as a major NAP throughout stationary phase, Dps offers DNA protection against damaging agents and accessibility to “trusted” DNA-binding proteins such as RNA polymerase.
Dps is a highly conserved bacterial ferritin and NAP. It has unique ferritin and DNA-binding properties that make it not only interesting to study from a basic biological standpoint but also increasingly important in the development of nanotechnologies and drug delivery. Dps is involved in conferring resistance to a myriad of stresses. Whether Dps functions as a direct transcriptional regulator is not clearly understood; however, it is involved in regulating gene expression, even if indirectly through nucleoid restructuring. Additional studies will prove useful to understand the dynamics of the ferritin core of Dps; the process of Dps dodecamer formation; the structure, dynamics, and composition of the stationary-phase nucleoid; gene expression regulation by Dps; PTMs on Dps and other NAPs during stationary phase; and other functions of Dps, particularly as they pertain to virulence, phage resistance, biofilm formation, and presence in the cell membrane.
ACKNOWLEDGMENTS
We thank Christopher Corzett, Namita Shroff, Andrew Janiero, and Brandon Vong for helpful suggestions and comments on the manuscript.
This work was supported in part by U.S. Air Force Office of Scientific Research grant FA-9550-19-1-0249 to S.E.F. K.O. was supported in part by National Institutes of Health grant NIGMS T32-GM118289.
Contributor Information
Steven E. Finkel, Email: sfinkel@usc.edu.
Julie A. Maupin-Furlow, University of Florida
REFERENCES
- 1.Almirón M, Link AJ, Furlong D, Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 6:2646–2654. 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 2.Gupta S, Chatterji D. 2003. Bimodal protection of DNA by Mycobacterium smegmatis DNA-binding protein from stationary phase cells. J Biol Chem 278:5235–5241. 10.1074/jbc.M208825200. [DOI] [PubMed] [Google Scholar]
- 3.Lomovskaya OL, Kidwell JP, Matin A. 1994. Characterization of the σ38-dependent expression of a core Escherichia coli starvation gene, pexB. J Bacteriol 176:3928–3935. 10.1128/jb.176.13.3928-3935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peña MM, Bullerjahn GS. 1995. The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. Linkage of the Dps and bacterioferritin protein families. J Biol Chem 270:22478–22482. 10.1074/jbc.270.38.22478. [DOI] [PubMed] [Google Scholar]
- 5.Haikarainen T, Papageorgiou AC. 2010. Dps-like proteins: structural and functional insights into a versatile protein family. Cell Mol Life Sci 67:341–351. 10.1007/s00018-009-0168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiancone E, Ceci P. 2010. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: detoxification of iron and hydrogen peroxide and DNA binding. Biochim Biophys Acta 1800:798–805. 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Shu J-C, Soo P-C, Chen J-C, Hsu S-H, Chen L-C, Chen C-Y, Liang S-H, Buu L-M, Chen C-C. 2013. Differential regulation and activity against oxidative stress of Dps proteins in Bacillus cereus. Int J Med Microbiol 303:662–673. 10.1016/j.ijmm.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Ramsay B, Wiedenheft B, Allen M, Gauss GH, Lawrence CM, Young M, Douglas T. 2006. Dps-like protein from the hyperthermophilic archaeon Pyrococcus furiosus. J Inorg Biochem 100:1061–1068. 10.1016/j.jinorgbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Gauss GH, Benas P, Wiedenheft B, Young M, Douglas T, Lawrence CM. 2006. Structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like dimetal binding site within a DPS-like dodecameric assembly. Biochemistry 45:10815–10827. 10.1021/bi060782u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karas VO, Westerlaken I, Meyer AS. 2015. The DNA-binding protein from starved cells (Dps) utilizes dual functions to defend cells against multiple stresses. J Bacteriol 197:3206–3215. 10.1128/JB.00475-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minsky A, Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R. 1999. DNA protection by stress-induced biocrystallization. Nature 400:83–85. 10.1038/21918. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharyya G, Grove A. 2007. The N-terminal extensions of Deinococcus radiodurans Dps-1 mediate DNA major groove interactions as well as assembly of the dodecamer. J Biol Chem 282:11921–11930. 10.1074/jbc.M611255200. [DOI] [PubMed] [Google Scholar]
- 13.Ushijima Y, Yoshida O, Villanueva MJA, Ohniwa RL, Morikawa K. 2016. Nucleoid clumping is dispensable for the Dps-dependent hydrogen peroxide resistance in Staphylococcus aureus. Microbiology (Reading) 162:1822–1828. 10.1099/mic.0.000353. [DOI] [PubMed] [Google Scholar]
- 14.Grove A, Wilkinson SP. 2005. Differential DNA binding and protection by dimeric and dodecameric forms of the ferritin homolog Dps from Deinococcus radiodurans. J Mol Biol 347:495–508. 10.1016/j.jmb.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 15.Ceci P, Mangiarotti L, Rivetti C, Chiancone E. 2007. The neutrophil-activating Dps protein of Helicobacter pylori, HP-NAP, adopts a mechanism different from Escherichia coli Dps to bind and condense DNA. Nucleic Acids Res 35:2247–2256. 10.1093/nar/gkm077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer AS, Grainger DC. 2013. The Escherichia coli nucleoid in stationary phase. Adv Appl Microbiol 83:69–86. 10.1016/B978-0-12-407678-5.00002-7. [DOI] [PubMed] [Google Scholar]
- 17.Liu G, Ma Q, Xu Y. 2018. Physical properties of DNA may direct the binding of nucleoid-associated proteins along the E. coli genome. Math Biosci 301:50–58. 10.1016/j.mbs.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 18.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. 1998. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol 5:294–303. 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 19.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol 181:6361–6370. 10.1128/JB.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talukder AA, Ishihama A. 2014. Dps is a stationary phase-specific protein of Escherichia coli nucleoid. Adv Microbiol 04:1095–1104. 10.4236/aim.2014.415120. [DOI] [Google Scholar]
- 21.Ishihama A, Kori A, Koshio E, Yamada K, Maeda H, Shimada T, Makinoshima H, Iwata A, Fujita N. 2014. Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J Bacteriol 196:2718–2727. 10.1128/JB.01579-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talukder A, Ishihama A. 2015. Growth phase dependent changes in the structure and protein composition of nucleoid in Escherichia coli. Sci China Life Sci 58:902–911. 10.1007/s11427-015-4898-0. [DOI] [PubMed] [Google Scholar]
- 23.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8:185–195. 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 24.Frenkiel-Krispin D, Levin-Zaidman S, Shimoni E, Wolf SG, Wachtel EJ, Arad T, Finkel SE, Kolter R, Minsky A. 2001. Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J 20:1184–1191. 10.1093/emboj/20.5.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frenkiel-Krispin D, Ben-Avraham I, Englander J, Shimoni E, Wolf SG, Minsky A. 2004. Nucleoid restructuring in stationary-state bacteria. Mol Microbiol 51:395–405. 10.1046/j.1365-2958.2003.03855.x. [DOI] [PubMed] [Google Scholar]
- 26.Kauko A, Haataja S, Pulliainen AT, Finne J, Papageorgiou AC. 2004. Crystal structure of Streptococcus suis Dps-like peroxide resistance protein Dpr: implications for iron incorporation. J Mol Biol 338:547–558. 10.1016/j.jmb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Roy S, Saraswathi R, Chatterji D, Vijayan M. 2008. Structural studies on the second Mycobacterium smegmatis Dps: invariant and variable features of structure, assembly and function. J Mol Biol 375:948–959. 10.1016/j.jmb.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Yoshimura SH, Hizume K, Ohniwa RL, Ishihama A, Takeyasu K. 2004. Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res 32:1982–1992. 10.1093/nar/gkh512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeyasu K, Kim J, Ohniwa RL, Kobori T, Inose Y, Morikawa K, Ohta T, Ishihama A, Yoshimura SH. 2004. Genome architecture studied by nanoscale imaging: analyses among bacterial phyla and their implication to eukaryotic genome folding. Cytogenet Genome Res 107:38–48. 10.1159/000079570. [DOI] [PubMed] [Google Scholar]
- 30.Morikawa K, Ohniwa RL, Kim J, Takeshita SL, Maruyama A, Inose Y, Takeyasu K, Ohta T. 2007. Biochemical, molecular genetic, and structural analyses of the staphylococcal nucleoid. Microsc Microanal 13:30–35. 10.1017/S1431927607070080. [DOI] [PubMed] [Google Scholar]
- 31.Ghatak P, Karmakar K, Kasetty S, Chatterji D. 2011. Unveiling the role of dps in the organization of mycobacterial nucleoid. PLoS One 6:e16019. 10.1371/journal.pone.0016019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bozzi M, Mignogna G, Stefanini S, Barra D, Longhi C, Valenti P, Chiancone E. 1997. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J Biol Chem 272:3259–3265. 10.1074/jbc.272.6.3259. [DOI] [PubMed] [Google Scholar]
- 33.Tonello F, Dundon WG, Satin B, Molinari M, Tognon G, Grandi G, Del Giudice G, Rappuoli R, Montecucco C. 1999. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol Microbiol 34:238–246. 10.1046/j.1365-2958.1999.01584.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhao G, Ceci P, Ilari A, Giangiacomo L, Laue TM, Chiancone E, Chasteen ND. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells: a ferritin-like DNA-binding protein of Escherichia coli. J Biol Chem 277:27689–27696. 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto Y, Poole LB, Hantgan RR, Kamio Y. 2002. An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J Bacteriol 184:2931–2939. 10.1128/JB.184.11.2931-2939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ceci P, Ilari A, Falvo E, Chiancone E. 2003. The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage: X-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J Biol Chem 278:20319–20326. 10.1074/jbc.M302114200. [DOI] [PubMed] [Google Scholar]
- 37.Ishikawa T, Mizunoe Y, Kawabata SI, Takade A, Harada M, Wai SN, Yoshida SI. 2003. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J Bacteriol 185:1010–1017. 10.1128/JB.185.3.1010-1017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su M, Cavallo S, Stefanini S, Chiancone E, Chasteen ND. 2005. The so-called Listeria innocua ferritin is a Dps protein. Iron incorporation, detoxification, and DNA protection properties. Biochemistry 44:5572–5578. 10.1021/bi0472705. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Kim K, Leighton T, Theil EC. 2006. Paired Bacillus anthracis Dps (mini-ferritin) have different reactivities with peroxide. J Biol Chem 281:27827–27835. 10.1074/jbc.M601398200. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, Anderson JF, Radolf JD, Fikrig E. 2007. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol Microbiol 63:694–710. 10.1111/j.1365-2958.2006.05550.x. [DOI] [PubMed] [Google Scholar]
- 41.Pulliainen AT, Haataja S, Kähkönen S, Finne J. 2003. Molecular basis of H2O2 resistance mediated by streptococcal Dpr. J Biol Chem 278:7996–8005. 10.1074/jbc.M210174200. [DOI] [PubMed] [Google Scholar]
- 42.Wiedenheft B, Mosolf J, Willits D, Yeager M, Dryden KA, Young M, Douglas T. 2005. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc Natl Acad Sci USA 102:10551–10556. 10.1073/pnas.0501497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franceschini S, Ceci P, Alaleona F, Chiancone E, Ilari A. 2006. Antioxidant Dps protein from the thermophilic cyanobacterium Thermosynechococcus elongatus. FEBS J 273:4913–4928. 10.1111/j.1742-4658.2006.05490.x. [DOI] [PubMed] [Google Scholar]
- 44.Tu WY, Pohl S, Gizynski K, Harwood CR. 2012. The iron-binding protein Dps2 confers peroxide stress resistance on Bacillus anthracis. J Bacteriol 194:925–931. 10.1128/JB.06005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haikarainen T, Tsou C-C, Wu J-J, Papageorgiou AC. 2010. Crystal structures of Streptococcus pyogenes Dpr reveal a dodecameric iron-binding protein with a ferroxidase site. J Biol Inorg Chem 15:183–194. 10.1007/s00775-009-0582-9. [DOI] [PubMed] [Google Scholar]
- 46.Harrison PM, Arosio P. 1996. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta 1275:161–203. 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 47.Roy S, Gupta S, Das S, Sekar K, Chatterji D, Vijayan M. 2003. Crystallization and preliminary X-ray diffraction analysis of Mycobacterium smegmatis Dps. Acta Crystallogr D Biol Crystallogr 59:2254–2256. 10.1107/s0907444903018742. [DOI] [PubMed] [Google Scholar]
- 48.Williams SM, Chandran AV, Prakash S, Vijayan M, Chatterji D. 2017. A mutation directs the structural switch of DNA binding proteins under starvation to a ferritin-like protein cage. Structure 25:1449–1454. 10.1016/j.str.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chiancone E. 2004. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res 32:5935–5944. 10.1093/nar/gkh915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy S, Saraswathi R, Gupta S, Sekar K, Chatterji D, Vijayan M. 2007. Role of N and C-terminal tails in DNA binding and assembly in Dps: structural studies of Mycobacterium smegmatis Dps deletion mutants. J Mol Biol 370:752–767. 10.1016/j.jmb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Roy S, Gupta S, Das S, Sekar K, Chatterji D, Vijayan M. 2004. X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules. J Mol Biol 339:1103–1113. 10.1016/j.jmb.2004.04.042. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Fu J, Chee SY, Ang EXW, Orner BP. 2011. Rational disruption of the oligomerization of the mini-ferritin E. coli DPS through protein-protein interface mutation. Protein Sci 20:1907–1917. 10.1002/pro.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stillman TJ, Upadhyay M, Norte VA, Sedelnikova SE, Carradus M, Tzokov S, Bullough PA, Shearman CA, Gasson MJ, Williams CH, Artymiuk PJ, Green J. 2005. The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding. Mol Microbiol 57:1101–1112. 10.1111/j.1365-2958.2005.04757.x. [DOI] [PubMed] [Google Scholar]
- 54.Ceci P, Ilari A, Falvo E, Giangiacomo L, Chiancone E. 2005. Reassessment of protein stability, DNA binding, and protection of Mycobacterium smegmatis Dps. J Biol Chem 280:34776–34785. 10.1074/jbc.M502343200. [DOI] [PubMed] [Google Scholar]
- 55.Bessonova TA, Shumeiko SA, Purtov YA, Antipov SS, Preobrazhenskaya EV, Tutukina MN, Ozoline ON. 2016. Hexuronates influence the oligomeric form of the Dps structural protein of bacterial nucleoid and its ability to bind to linear DNA fragments. Biophysics 61:825–832. 10.1134/S0006350916060075. [DOI] [Google Scholar]
- 56.Rothe M, Alpert C, Loh G, Blaut M. 2013. Novel insights into E. coli’s hexuronate metabolism: KduI facilitates the conversion of galacturonate and glucuronate under osmotic stress conditions. PLoS One 8:e56906. 10.1371/journal.pone.0056906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S-W, Chen C-Y, Tseng J-T, Liang S-H, Chen S-C, Hsieh C, Chen Y-H, Chen C-C. 2009. orf4 of the Bacillus cereus sigB gene cluster encodes a general stress-inducible Dps-like bacterioferritin. J Bacteriol 191:4522–4533. 10.1128/JB.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohniwa RL, Morikawa K, Kim J, Ohta T, Ishihama A, Wada C, Takeyasu K. 2006. Dynamic state of DNA topology is essential for genome condensation in bacteria. EMBO J 25:5591–5602. 10.1038/sj.emboj.7601414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohniwa RL, Muchaku H, Saito S, Wada C, Morikawa K. 2013. Atomic force microscopy analysis of the role of major DNA-binding proteins in organization of the nucleoid in Escherichia coli. PLoS One 8:e72954. 10.1371/journal.pone.0072954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zimmerman SB. 2006. Cooperative transitions of isolated Escherichia coli nucleoids: implications for the nucleoid as a cellular phase. J Struct Biol 153:160–175. 10.1016/j.jsb.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 61.Minsky A, Shimoni E, Frenkiel-Krispin D. 2002. Stress, order and survival. Nat Rev Mol Cell Biol 3:50–60. 10.1038/nrm700. [DOI] [PubMed] [Google Scholar]
- 62.Nair S, Finkel SE. 2004. Dps protects cells against multiple stresses during stationary phase. J Bacteriol 186:4192–4198. 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato YT, Watanabe S, Kenmotsu T, Ichikawa M, Yoshikawa Y, Teramoto J, Imanaka T, Ishihama A, Yoshikawa K. 2013. Structural change of DNA induced by nucleoid proteins: growth phase-specific fis and stationary phase-specific dps. Biophys J 105:1037–1044. 10.1016/j.bpj.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SY, Lim CJ, Dröge P, Yan J. 2015. Regulation of bacterial DNA packaging in early stationary phase by competitive DNA binding of Dps and IHF. Sci Rep 5:18146. 10.1038/srep18146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hong Y, Wang G, Maier RJ. 2006. Helicobacter hepaticus Dps protein plays an important role in protecting DNA from oxidative damage. Free Radic Res 40:597–605. 10.1080/10715760600618882. [DOI] [PubMed] [Google Scholar]
- 66.Morikawa K, Ohniwa RL, Kim J, Maruyama A, Ohta T, Takeyasu K. 2006. Bacterial nucleoid dynamics: oxidative stress response in Staphylococcus aureus. Genes Cells 11:409–423. 10.1111/j.1365-2443.2006.00949.x. [DOI] [PubMed] [Google Scholar]
- 67.Foley PL, Wilson DB, Shuler ML. 2010. Macromolecular crowding can account for RNase-sensitive constraint of bacterial nucleoid structure. Biochem Biophys Res Commun 395:42–47. 10.1016/j.bbrc.2010.03.128. [DOI] [PubMed] [Google Scholar]
- 68.Pinson V, Takahashi M, Rouviere-Yaniv J. 1999. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J Mol Biol 287:485–497. 10.1006/jmbi.1999.2631. [DOI] [PubMed] [Google Scholar]
- 69.Gulvady R, Gao Y, Kenney LJ, Yan J. 2018. A single molecule analysis of H-NS uncouples DNA binding affinity from DNA specificity. Nucleic Acids Res 46:10216–10224. 10.1093/nar/gky826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stella S, Cascio D, Johnson RC. 2010. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev 24:814–826. 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shao Y, Feldman-Cohen LS, Osuna R. 2008. Functional characterization of the Escherichia coli Fis-DNA binding sequence. J Mol Biol 376:771–785. 10.1016/j.jmb.2007.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S, Cosstick R, Gardner JF, Gumport RI. 1995. The specific binding of Escherichia coli integration host factor involves both major and minor grooves of DNA. Biochemistry 34:13082–13090. 10.1021/bi00040a020. [DOI] [PubMed] [Google Scholar]
- 73.Windbichler N, von Pelchrzim F, Mayer O, Csaszar E, Schroeder R. 2008. Isolation of small RNA-binding proteins from E. coli: evidence for frequent interaction of RNAs with RNA polymerase. RNA Biol 5:30–40. 10.4161/rna.5.1.5694. [DOI] [PubMed] [Google Scholar]
- 74.Joyeux M. 2015. Compaction of bacterial genomic DNA: clarifying the concepts. J Phys Condens Matter 27:383001. 10.1088/0953-8984/27/38/383001. [DOI] [PubMed] [Google Scholar]
- 75.Antipov SS, Tutukina MN, Preobrazhenskaya EV, Kondrashov FA, Patrushev MV, Toshchakov SV, Dominova I, Shvyreva US, Vrublevskaya VV, Morenkov OS, Sukharicheva NA, Panyukov VV, Ozoline ON. 2017. The nucleoid protein Dps binds genomic DNA of Escherichia coli in a non-random manner. PLoS One 12:e0182800. 10.1371/journal.pone.0182800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishihama A, Shimada T, Yamazaki Y. 2016. Transcription profile of Escherichia coli: genomic SELEX search for regulatory targets of transcription factors. Nucleic Acids Res 44:2058–2074. 10.1093/nar/gkw051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ueshima J, Shoji M, Ratnayake DB, Abe K, Yoshida S, Yamamoto K, Nakayama K. 2003. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect Immun 71:1170–1178. 10.1128/IAI.71.3.1170-1178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ping L, Büchler R, Mithöfer A, Svatoš A, Spiteller D, Dettner K, Gmeiner S, Piel J, Schlott B, Boland W. 2007. A novel Dps-type protein from insect gut bacteria catalyses hydrolysis and synthesis of N-acyl amino acids. Environ Microbiol 9:1572–1583. 10.1111/j.1462-2920.2007.01279.x. [DOI] [PubMed] [Google Scholar]
- 79.Kim SG, Bhattacharyya G, Grove A, Lee YH. 2006. Crystal structure of Dps-1, a functionally distinct Dps protein from Deinococcus radiodurans. J Mol Biol 361:105–114. 10.1016/j.jmb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 80.Santos SP, Cuypers MG, Round A, Finet S, Narayanan T, Mitchell EP, Romão CV. 2017. SAXS structural studies of Dps from Deinococcus radiodurans highlights the conformation of the mobile N-terminal extensions. J Mol Biol 429:667–687. 10.1016/j.jmb.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 81.Grainger DC, Goldberg MD, Lee DJ, Busby SJW. 2008. Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol Microbiol 68:1366–1377. 10.1111/j.1365-2958.2008.06253.x. [DOI] [PubMed] [Google Scholar]
- 82.Browning DF, Grainger DC, Busby SJ. 2010. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol 13:773–780. 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 83.Graham JS, Johnson RC, Marko JF. 2011. Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Res 39:2249–2259. 10.1093/nar/gkq1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hadizadeh N, Johnson RC, Marko JF. 2016. Facilitated dissociation of a nucleoid protein from the bacterial chromosome. J Bacteriol 198:1735–1742. 10.1128/JB.00225-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andrews SC. 2010. The ferritin-like superfamily: evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim Biophys Acta 1800:691–705. 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 87.Alaleona F, Franceschini S, Ceci P, Ilari A, Chiancone E. 2010. Thermosynechoccus [sic] elongatus DpsA binds Zn(II) at a unique three histidine-containing ferroxidase center and utilizes O2 as iron oxidant with very high efficiency, unlike the typical Dps proteins. FEBS J 277:903–917. 10.1111/j.1742-4658.2009.07532.x. [DOI] [PubMed] [Google Scholar]
- 88.Schwartz JK, Liu XS, Tosha T, Diebold A, Theil EC, Solomon EI. 2010. CD and MCD spectroscopic studies of the two Dps miniferritin proteins from Bacillus anthracis: role of O2 and H2O2 substrates in reactivity of the diiron catalytic centers. Biochemistry 49:10516–10525. 10.1021/bi101346c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Howe C, Moparthi VK, Ho FM, Persson K, Stensjö K. 2019. The Dps4 from Nostoc punctiforme ATCC 29133 is a member of His-type FOC containing Dps protein class that can be broadly found among cyanobacteria. PLoS One 14:e0218300. 10.1371/journal.pone.0218300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kauko A, Pulliainen AT, Haataja S, Meyer-Klaucke W, Finne J, Papageorgiou AC. 2006. Iron incorporation in Streptococcus suis Dps-like peroxide resistance protein Dpr requires mobility in the ferroxidase center and leads to the formation of a ferrihydrite-like core. J Mol Biol 364:97–109. 10.1016/j.jmb.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 91.Huergo LF, Rahman H, Ibrahimovic A, Day CJ, Korolik V. 2013. Campylobacter jejuni Dps protein binds DNA in the presence of iron or hydrogen peroxide. J Bacteriol 195:1970–1978. 10.1128/JB.00059-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ilari A, Ceci P, Ferrari D, Rossi GL, Chiancone E. 2002. Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J Biol Chem 277:37619–37623. 10.1074/jbc.M206186200. [DOI] [PubMed] [Google Scholar]
- 93.Haggis G. 1965. The iron oxide core of the ferritin molecule. J Mol Biol 14:598–602. 10.1016/s0022-2836(65)80210-8. [DOI] [PubMed] [Google Scholar]
- 94.Bevers LE, Theil EC. 2011. Maxi- and mini-ferritins: minerals and protein nanocages. Prog Mol Subcell Biol 52:29–47. 10.1007/978-3-642-21230-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Castruita M, Saito M, Schottel PC, Elmegreen LA, Myneni S, Stiefel EI, Morel FMM. 2006. Overexpression and characterization of an iron storage and DNA-binding Dps protein from Trichodesmium erythraeum. Appl Environ Microbiol 72:2918–2924. 10.1128/AEM.72.4.2918-2924.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reindel S, Schmidt CL, Anemüller S, Matzanke BF. 2002. Characterization of a non-haem ferritin of the archaeon Halobacterium salinarum, homologous to Dps (starvation-induced DNA-binding protein). Biochem Soc Trans 30:713–715. 10.1042/bst0300713. [DOI] [PubMed] [Google Scholar]
- 97.Havukainen H, Haataja S, Kauko A, Pulliainen AT, Salminen A, Haikarainen T, Finne J, Papageorgiou AC. 2008. Structural basis of the zinc- and terbium-mediated inhibition of ferroxidase activity in Dps ferritin-like proteins. Protein Sci 17:1513–1521. 10.1110/ps.036236.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haikarainen T, Tsou C-C, Wu J-J, Papageorgiou AC. 2010. Structural characterization and biological implications of di-zinc binding in the ferroxidase center of Streptococcus pyogenes Dpr. Biochem Biophys Res Commun 398:361–365. 10.1016/j.bbrc.2010.06.071. [DOI] [PubMed] [Google Scholar]
- 99.Romão CV, Mitchell EP, McSweeney S. 2006. The crystal structure of Deinococcus radiodurans Dps protein (DR2263) reveals the presence of a novel metal centre in the N terminus. J Biol Inorg Chem 11:891–902. 10.1007/s00775-006-0142-5. [DOI] [PubMed] [Google Scholar]
- 100.Haikarainen T, Thanassoulas A, Stavros P, Nounesis G, Haataja S, Papageorgiou AC. 2011. Structural and thermodynamic characterization of metal ion binding in Streptococcus suis Dpr. J Mol Biol 405:448–460. 10.1016/j.jmb.2010.10.058. [DOI] [PubMed] [Google Scholar]
- 101.Ardini M, Fiorillo A, Fittipaldi M, Stefanini S, Gatteschi D, Ilari A, Chiancone E. 2013. Kineococcus radiotolerans Dps forms a heteronuclear Mn-Fe ferroxidase center that may explain the Mn-dependent protection against oxidative stress. Biochim Biophys Acta 1830:3745–3755. 10.1016/j.bbagen.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 102.Wei Q, Minh PNL, Dötsch A, Hildebrand F, Panmanee W, Elfarash A, Schulz S, Plaisance S, Charlier D, Hassett D, Häussler S, Cornelis P. 2012. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res 40:4320–4333. 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun 69:3744–3754. 10.1128/IAI.69.6.3744-3754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dowds BC, Murphy P, McConnell DJ, Devine KM. 1987. Relationship among oxidative stress, growth cycle, and sporulation in Bacillus subtilis. J Bacteriol 169:5771–5775. 10.1128/jb.169.12.5771-5775.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ohniwa RL, Ushijima Y, Saito S, Morikawa K. 2011. Proteomic analyses of nucleoid-associated proteins in Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus. PLoS One 6:e19172. 10.1371/journal.pone.0019172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bsat N, Chen L, Helmann JD. 1996. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J Bacteriol 178:6579–6586. 10.1128/jb.178.22.6579-6586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reindel S, Schmidt CL, Anemüller S, Matzanke BF. 2005. Expression and regulation pattern of ferritin-like DpsA in the archaeon Halobacterium salinarum. Biometals 18:387–397. 10.1007/s10534-005-3713-y. [DOI] [PubMed] [Google Scholar]
- 108.Velayudhan J, Castor M, Richardson A, Main-Hester KL, Fang FC. 2007. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol Microbiol 63:1495–1507. 10.1111/j.1365-2958.2007.05600.x. [DOI] [PubMed] [Google Scholar]
- 109.Shrivastava AK, Pandey S, Dietz KJ, Singh PK, Singh S, Rai R, Rai LC. 2016. Overexpression of AhpC enhances stress tolerance and N 2-fixation in Anabaena by upregulating stress responsive genes. Biochim Biophys Acta 1860:2576–2588. 10.1016/j.bbagen.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 110.Altuvia S, Almirón M, Huisman G, Kolter R, Storz G. 1994. The dps promoter is activated by OxyR during growth and by IHF and σS in stationary phase. Mol Microbiol 13:265–272. 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 111.Rothe M, Alpert C, Engst W, Musiol S, Loh G, Blaut M. 2012. Impact of nutritional factors on the proteome of intestinal Escherichia coli: induction of OxyR-dependent proteins AhpF and Dps by a lactose-rich diet. Appl Environ Microbiol 78:3580–3591. 10.1128/AEM.00244-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Toledano MB, Kullik I, Trinh F, Baird PT, Schneider TD, Storz G. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897–909. 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 113.Bsat N, Herbig A, Casillas‐Martinez L, Setlow P, Helmann JD. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol 29:189–198. 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 114.de Castro Ferreira IG, Rodrigues MM, da Silva Neto JF, Mazzon RR, do Valle Marques M. 2016. Role and regulation of ferritin-like proteins in iron homeostasis and oxidative stress survival of Caulobacter crescentus. Biometals 29:851–862. 10.1007/s10534-016-9956-y. [DOI] [PubMed] [Google Scholar]
- 115.Yu M, Ren J, Zeng Y, Zhou S, Lu Y. 2009. The Legionella pneumophila Dps homolog is regulated by iron and involved in multiple stress tolerance. J Basic Microbiol 49:S79–S86. 10.1002/jobm.200800357. [DOI] [PubMed] [Google Scholar]
- 116.Pesek J, Büchler R, Albrecht R, Boland W, Zeth K. 2011. Structure and mechanism of iron translocation by a Dps protein from Microbacterium arborescens. J Biol Chem 286:34872–34882. 10.1074/jbc.M111.246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Polidoro M, De Biase D, Montagnini B, Guarrera L, Cavallo S, Valenti P, Stefanini S, Chiancone E. 2002. The expression of the dodecameric ferritin in Listeria spp. is induced by iron limitation and stationary growth phase. Gene 296:121–128. 10.1016/s0378-1119(02)00839-9. [DOI] [PubMed] [Google Scholar]
- 118.Wolfe AJ, Chang D-E, Walker JD, Seitz-Partridge JE, Vidaurri MD, Lange CF, Prüss BM, Henk MC, Larkin JC, Conway T. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol Microbiol 48:977–988. 10.1046/j.1365-2958.2003.03457.x. [DOI] [PubMed] [Google Scholar]
- 119.Williams E, Lowe TM, Savas J, DiRuggiero J. 2007. Microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus exposed to gamma irradiation. Extremophiles 11:19–29. 10.1007/s00792-006-0002-9. [DOI] [PubMed] [Google Scholar]
- 120.Nandal A, Huggins CCO, Woodhall MR, McHugh J, Rodríguez-Quiñones F, Quail MA, Guest JR, Andrews SC. 2010. Induction of the ferritin gene ftnA of Escherichia coli by Fe2+-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol Microbiol 75:637–657. 10.1111/j.1365-2958.2009.06977.x. [DOI] [PubMed] [Google Scholar]
- 121.Stephani K, Weichart D, Hengge R. 2003. Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli. Mol Microbiol 49:1605–1614. 10.1046/j.1365-2958.2003.03644.x. [DOI] [PubMed] [Google Scholar]
- 122.Flynn JM, Neher SB, Kim Y-I, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11:671–683. 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 123.Ninnis RL, Spall SK, Talbo GH, Truscott KN, Dougan DA. 2009. Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli. EMBO J 28:1732–1744. 10.1038/emboj.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schmidt R, Zahn R, Bukau B, Mogk A. 2009. ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol Microbiol 72:506–517. 10.1111/j.1365-2958.2009.06666.x. [DOI] [PubMed] [Google Scholar]
- 125.Xia X, Larios-Valencia J, Liu Z, Xiang F, Kan B, Wang H, Zhu J. 2017. OxyR-activated expression of Dps is important for Vibrio cholerae oxidative stress resistance and pathogenesis. PLoS One 12:e0171201. 10.1371/journal.pone.0171201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li X, Sandh G, Nenninger A, Muro-Pastor AM, Stensjö K. 2015. Differential transcriptional regulation of orthologous dps genes from two closely related heterocyst-forming cyanobacteria. FEMS Microbiol Lett 362:fnv017. 10.1093/femsle/fnv017. [DOI] [PubMed] [Google Scholar]
- 127.Moparthi VK, Li X, Vavitsas K, Dzhygyr I, Sandh G, Magnuson A, Stensjö K. 2016. The two Dps proteins, NpDps2 and NpDps5, are involved in light-induced oxidative stress tolerance in the N2-fixing cyanobacterium Nostoc punctiforme. Biochim Biophys Acta 1857:1766–1776. 10.1016/j.bbabio.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 128.Facey PD, Hitchings MD, Saavedra-Garcia P, Fernandez-Martinez L, Dyson PJ, Del Sol R. 2009. Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol Microbiol 73:1186–1202. 10.1111/j.1365-2958.2009.06848.x. [DOI] [PubMed] [Google Scholar]
- 129.Chowdhury RP, Gupta S, Chatterji D. 2007. Identification and characterization of the dps promoter of Mycobacterium smegmatis: promoter recognition by stress-specific extracytoplasmic function sigma factors σH and σF. J Bacteriol 189:8973–8981. 10.1128/JB.01222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ekman M, Sandh G, Nenninger A, Oliveira P, Stensjö K. 2014. Cellular and functional specificity among ferritin-like proteins in the multicellular cyanobacterium Nostoc punctiforme. Environ Microbiol 16:829–844. 10.1111/1462-2920.12233. [DOI] [PubMed] [Google Scholar]
- 131.Diaz PI, Slakeski N, Reynolds EC, Morona R, Rogers AH, Kolenbrander PE. 2006. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J Bacteriol 188:2454–2462. 10.1128/JB.188.7.2454-2462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McLeod SM, Johnson RC. 2001. Control of transcription by nucleoid proteins. Curr Opin Microbiol 4:152–159. 10.1016/s1369-5274(00)00181-8. [DOI] [PubMed] [Google Scholar]
- 133.Azam TA, Hiraga S, Ishihama A. 2000. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells 5:613–626. 10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 134.Calhoun LN, Kim JN, Ren Y, Song JJ, Kwon M, Department of Poultry Science. 2011. The DNA-binding protein Dps functions as a global regulator in starved Salmonella enterica serovar Enteritidis during starvation. Int J Microbiol Res 3:136–147. 10.9735/0975-5276.3.3.136-147. [DOI] [Google Scholar]
- 135.Narayan OP, Kumari N, Bhargava P, Rajaram H, Rai LC. 2016. A single gene all3940 (Dps) overexpression in Anabaena sp. PCC 7120 confers multiple abiotic stress tolerance via proteomic alterations. Funct Integr Genomics 16:67–78. 10.1007/s10142-015-0467-7. [DOI] [PubMed] [Google Scholar]
- 136.Janissen R, Arens MMA, Vtyurina NN, Rivai Z, Sunday ND, Eslami-Mossallam B, Gritsenko AA, Laan L, de Ridder D, Artsimovitch I, Dekker NH, Abbondanzieri EA, Meyer AS. 2018. Global DNA compaction in stationary-phase bacteria does not affect transcription. Cell 174:1188–1199. 10.1016/j.cell.2018.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mullis MN, Matsui T, Schell R, Foree R, Ehrenreich IM. 2018. The complex underpinnings of genetic background effects. Nat Commun 9:3548. 10.1038/s41467-018-06023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Harrison P, Fischbach F, Hoy T, Haggis G. 1967. Ferric oxyhydroxide core of ferritin. Nature 216:1188–1190. 10.1038/2161188a0. [DOI] [PubMed] [Google Scholar]
- 139.Dittmann C, Han HM, Grabenbauer M, Laue M. 2015. Dormant Bacillus spores protect their DNA in crystalline nucleoids against environmental stress. J Struct Biol 191:156–164. 10.1016/j.jsb.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 140.Frenkiel-Krispin D, Sack R, Englander J, Shimoni E, Eisenstein M, Bullitt E, Horowitz-Scherer R, Hayes CS, Setlow P, Minsky A, Wolf SG. 2004. Structure of the DNA-SspC complex: implications for DNA packaging, protection, and repair in bacterial spores. J Bacteriol 186:3525–3530. 10.1128/JB.186.11.3525-3530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Abbondanzieri EA, Meyer AS. 2019. More than just a phase: the search for membraneless organelles in the bacterial cytoplasm. Curr Genet 65:691–694. 10.1007/s00294-018-00927-x. [DOI] [PubMed] [Google Scholar]
- 142.Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, Fuxreiter M. 2018. Protein phase separation: a new phase in cell biology. Trends Cell Biol 28:420–435. 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vtyurina NN, Dulin D, Docter MW, Meyer AS, Dekker NH, Abbondanzieri EA. 2016. Hysteresis in DNA compaction by Dps is described by an Ising model. Proc Natl Acad Sci USA 113:4982–4987. 10.1073/pnas.1521241113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA. 2017. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168:1028–1040.e19. 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen L, Helmann JD. 1995. Bacillus subtilis MrgA is a Dps (PexB) homologue: evidence for metalloregulation of an oxidative-stress gene. Mol Microbiol 18:295–300. 10.1111/j.1365-2958.1995.mmi_18020295.x. [DOI] [PubMed] [Google Scholar]
- 146.Martinez A, Kolter R. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol 179:5188–5194. 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ljungman M, Hanawalt PC. 1992. Efficient protection against oxidative DNA damage in chromatin. Mol Carcinog 5:264–269. 10.1002/mc.2940050406. [DOI] [PubMed] [Google Scholar]
- 148.Chodavarapu S, Gomez R, Vicente M, Kaguni JM. 2008. Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol Microbiol 67:1331–1346. 10.1111/j.1365-2958.2008.06127.x. [DOI] [PubMed] [Google Scholar]
- 149.Silva LSO, Baptista JM, Batley C, Andrews SC, Saraiva LM. 2018. The di-iron RIC protein (YtfE) of Escherichia coli interacts with the DNA-binding protein from starved cells (Dps) to diminish RIC protein-mediated redox stress. J Bacteriol 200:e00527-18. 10.1128/JB.00527-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lacqua A, Wanner O, Colangelo T, Martinotti MG, Landini P. 2006. Emergence of biofilm-forming subpopulations upon exposure of Escherichia coli to environmental bacteriophages. Appl Environ Microbiol 72:956–959. 10.1128/AEM.72.1.956-959.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Theoret JR, Cooper KK, Zekarias B, Roland KL, Law BF, Curtiss R, Joens LA. 2012. The Campylobacter jejuni dps homologue is important for in vitro biofilm formation and cecal colonization of poultry and may serve as a protective antigen for vaccination. Clin Vaccine Immunol 19:1426–1431. 10.1128/CVI.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu L, Lin X, Peng X. 2009. From proteome to genome for functional characterization of pH-dependent outer membrane proteins in Escherichia coli. J Proteome Res 8:1059–1070. 10.1021/pr800818r. [DOI] [PubMed] [Google Scholar]
- 153.Li H, Wang B-C, Xu W-J, Lin X-M, Peng X-X. 2008. Identification and network of outer membrane proteins regulating streptomysin [sic] resistance in Escherichia coli. J Proteome Res 7:4040–4049. 10.1021/pr800310y. [DOI] [PubMed] [Google Scholar]
- 154.Evans DJ, Evans DG, Takemura T, Nakano H, Lampert HC, Graham DY, Granger DN, Kvietys PR. 1995. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect Immun 63:2213–2220. 10.1128/iai.63.6.2213-2220.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Montemurro P, Nishioka H, Dundon WG, de Bernard M, Del Giudice G, Rappuoli R, Montecucco C. 2002. The neutrophil‐activating protein (HP‐NAP) of Helicobacter pylori is a potent stimulant of mast cells. Eur J Immunol 32:671–676. . [DOI] [PubMed] [Google Scholar]
- 156.Zanotti G, Papinutto E, Dundon WG, Battistutta R, Seveso M, Del Giudice G, Rappuoli R, Montecucco C. 2002. Structure of the neutrophil-activating protein from Helicobacter pylori. J Mol Biol 323:125–130. 10.1016/S0022-2836(02)00879-3. [DOI] [PubMed] [Google Scholar]
- 157.Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. 2004. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect Immun 72:1155–1158. 10.1128/IAI.72.2.1155-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Thumiger A, Polenghi A, Papinutto E, Battistutta R, Montecucco C, Zanotti G. 2006. Crystal structure of antigen TpF1 from Treponema pallidum. Proteins 62:827–830. 10.1002/prot.20828. [DOI] [PubMed] [Google Scholar]
- 159.Olsen KN, Larsen MH, Gahan CGM, Kallipolitis B, Wolf XA, Rea R, Hill C, Ingmer H. 2005. The Dps-like protein Fri of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiology (Reading) 151:925–933. 10.1099/mic.0.27552-0. [DOI] [PubMed] [Google Scholar]
- 160.Amedei A, Cappon A, Codolo G, Cabrelle A, Polenghi A, Benagiano M, Tasca E, Azzurri A, D’Elios MM, Del Prete G, de Bernard M. 2006. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J Clin Invest 116:1092–1101. 10.1172/JCI27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Polenghi A, Bossi F, Fischetti F, Durigutto P, Cabrelle A, Tamassia N, Cassatella M, Montecucco C, Tedesco F, de Bernard M. 2007. The neutrophil-activating protein of Helicobacter pylori crosses endothelia to promote neutrophil adhesion in vivo. J Immunol 178:1312–1320. 10.4049/jimmunol.178.3.1312. [DOI] [PubMed] [Google Scholar]
- 162.Kottakis F, Befani C, Asiminas A, Kontou M, Koliakos G, Choli-Papadopoulou T. 2009. The C-terminal region of HPNAP activates neutrophils and promotes their adhesion to endothelial cells. Helicobacter 14:177–179. 10.1111/j.1523-5378.2009.00678.x. [DOI] [PubMed] [Google Scholar]
- 163.Zughaier SM, Cornelis P. 2018. Editorial: role of iron in bacterial pathogenesis. Front Cell Infect Microbiol 8:344. 10.3389/fcimb.2018.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ciacci C, Sabbatini F, Cavallaro R, Castiglione F, Di Bella S, Iovino P, Palumbo A, Tortora R, Amoruso D, Mazzacca G. 2004. Helicobacter pylori impairs iron absorption in infected individuals. Dig Liver Dis 36:455–460. 10.1016/j.dld.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 165.Durham KA, Bullerjahn GS. 2002. Immunocytochemical localization of the stress-induced DpsA protein in the cyanobacterium Synechococcus sp. strain PCC 7942. J Basic Microbiol 42:367–372. . [DOI] [PubMed] [Google Scholar]
- 166.Yamashita I, Iwahori K, Kumagai S. 2010. Ferritin in the field of nanodevices. Biochim Biophys Acta 1800:846–857. 10.1016/j.bbagen.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 167.He D, Marles-Wright J. 2015. Ferritin family proteins and their use in bionanotechnology. N Biotechnol 32:651–657. 10.1016/j.nbt.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gálvez N, Sánchez P, Domínguez-Vera JM. 2005. Preparation of Cu and CuFe Prussian blue derivative nanoparticles using the apoferritin cavity as nanoreactor. Dalton Trans 2005:2492–2494. 10.1039/b506290j. [DOI] [PubMed] [Google Scholar]
- 169.Kramer RM, Sowards LA, Pender MJ, Stone MO, Naik RR. 2005. Constrained iron catalysts for single-walled carbon nanotube growth. Langmuir 21:8466–8470. 10.1021/la0506729. [DOI] [PubMed] [Google Scholar]
- 170.Kang S, Lucon J, Varpness ZB, Liepold L, Uchida M, Willits D, Young M, Douglas T. 2008. Monitoring biomimetic platinum nanocluster formation using mass spectrometry and cluster-dependent H2 production. Angew Chem Int Ed Engl 47:7845–7848. 10.1002/anie.200802481. [DOI] [PubMed] [Google Scholar]
- 171.Cornell TA, Ardejani MS, Fu J, Newland SH, Zhang Y, Orner BP. 2018. A structure-based assembly screen of protein cage libraries in living cells: experimentally repacking a protein-protein interface to recover cage formation in an assembly-frustrated mutant. Biochemistry 57:604–613. 10.1021/acs.biochem.7b01000. [DOI] [PubMed] [Google Scholar]
- 172.Okuda M, Suzumoto Y, Iwahori K, Kang S, Uchida M, Douglas T, Yamashita I. 2010. Bio-templated CdSe nanoparticle synthesis in a cage shaped protein, Listeria-Dps, and their two dimensional ordered array self-assembly. Chem Commun (Camb) 46:8797–8799. 10.1039/c0cc03298k. [DOI] [PubMed] [Google Scholar]
- 173.Budiman MF, Hu W, Igarashi M, Tsukamoto R, Isoda T, Itoh KM, Yamashita I, Murayama A, Okada Y, Samukawa S. 2012. Control of optical bandgap energy and optical absorption coefficient by geometric parameters in sub-10 nm silicon-nanodisc array structure. Nanotechnology 23:e065302. 10.1088/0957-4484/23/6/065302. [DOI] [PubMed] [Google Scholar]
- 174.Li CQ, Soistman E, Carter DC. 2006. Ferritin nanoparticle technology. A new platform for antigen presentation and vaccine development. Ind Biotechnol 2:143–147. 10.1089/ind.2006.2.143. [DOI] [Google Scholar]
- 175.Wu H, Wang J, Wang Z, Fisher DR, Lin Y. 2008. Apoferritin-templated yttrium phosphate nanoparticle conjugates for radioimmunotherapy of cancers. J Nanosci Nanotechnol 8:2316–2322. 10.1166/jnn.2008.177. [DOI] [PubMed] [Google Scholar]
- 176.Uchida M, Terashima M, Cunningham CH, Suzuki Y, Willits DA, Willis AF, Yang PC, Tsao PS, McConnell MV, Young MJ, Douglas T. 2008. A human ferritin iron oxide nano-composite magnetic resonance contrast agent. Magn Reson Med 60:1073–1081. 10.1002/mrm.21761. [DOI] [PubMed] [Google Scholar]
- 177.Gupta AK, Gupta M. 2005. Synthesis and surface engineering of iron oxide nanoparticles for biomedical applications. Biomaterials 26:3995–4021. 10.1016/j.biomaterials.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 178.Dilweg IW, Dame RT. 2018. Post-translational modification of nucleoid-associated proteins: an extra layer of functional modulation in bacteria? Biochem Soc Trans 46:1381–1392. 10.1042/BST20180488. [DOI] [PubMed] [Google Scholar]
- 179.Hanna ES, Roque-Barreira M-C, Bernardes ES, Panunto-Castelo A, Sousa MV, Almeida IC, Brocchi M. 2007. Evidence for glycosylation on a DNA-binding protein of Salmonella enterica. Microb Cell Fact 6:11. 10.1186/1475-2859-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Stillman TJ, Hempstead PD, Artymiuk PJ, Andrews SC, Hudson AJ, Treffry A, Guest JR, Harrison PM. 2001. The high-resolution X-ray crystallographic structure of the ferritin (EcFtnA) of Escherichia coli; comparison with human H ferritin (HuHF) and the structures of the Fe3+ and Zn2+ derivatives. J Mol Biol 307:587–603. 10.1006/jmbi.2001.4475. [DOI] [PubMed] [Google Scholar]
- 181.Crow A, Lawson TL, Lewin A, Moore GR, Le Brun NE. 2009. Structural basis for iron mineralization by bacterioferritin. J Am Chem Soc 131:6808–6813. 10.1021/ja8093444. [DOI] [PubMed] [Google Scholar]
- 182.Papinutto E, Dundon WG, Pitulis N, Battistutta R, Montecucco C, Zanotti G. 2002. Structure of two iron-binding proteins from Bacillus anthracis. J Biol Chem 277:15093–15098. 10.1074/jbc.M112378200. [DOI] [PubMed] [Google Scholar]
- 183.Santos SP, Mitchell EP, Franquelim HG, Castanho MARB, Abreu IA, Romão CV. 2015. Dps from Deinococcus radiodurans: oligomeric forms of Dps1 with distinct cellular functions and Dps2 involved in metal storage. FEBS J 282:4307–4327. 10.1111/febs.13420. [DOI] [PubMed] [Google Scholar]
- 184.Cuypers MG, Mitchell EP, Romão CV, McSweeney SM. 2007. The crystal structure of the Dps2 from Deinococcus radiodurans reveals an unusual pore profile with a non-specific metal binding site. J Mol Biol 371:787–799. 10.1016/j.jmb.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 185.Chiancone E, Ilari A, Stefanini S, Tsernoglou D. 2000. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat Struct Biol 7:38–43. 10.1038/71236. [DOI] [PubMed] [Google Scholar]
- 186.Gupta S, Pandit SB, Srinivasan N, Chatterji D. 2002. Proteomics analysis of carbon-starved Mycobacterium smegmatis: induction of Dps-like protein. Protein Eng 15:503–511. 10.1093/protein/15.6.503. [DOI] [PubMed] [Google Scholar]
- 187.Williams SM, Chatterji D. 2017. Flexible aspartates propel iron to the ferroxidation sites along pathways stabilized by a conserved arginine in Dps proteins from Mycobacterium smegmatis. Metallomics 9:685–698. 10.1039/c7mt00008a. [DOI] [PubMed] [Google Scholar]
- 188.Howe C, Ho F, Nenninger A, Raleiras P, Stensjö K. 2018. Differential biochemical properties of three canonical Dps proteins from the cyanobacterium Nostoc punctiforme suggest distinct cellular functions. J Biol Chem 293:16635–16646. 10.1074/jbc.RA118.002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Moparthi VK, Moparthi SB, Howe C, Raleiras P, Wenger J, Stensjö K. 2019. Structural diffusion properties of two atypical Dps from the cyanobacterium Nostoc punctiforme disclose interactions with ferredoxins and DNA. Biochim Biophys Acta 1860:148063. 10.1016/j.bbabio.2019.148063. [DOI] [PubMed] [Google Scholar]
- 190.Ushijima Y, Ohniwa RL, Maruyama A, Saito S, Tanaka Y, Morikawa K. 2014. Nucleoid compaction by MrgAAsp56Ala/Glu60Ala does not contribute to staphylococcal cell survival against oxidative stress and phagocytic killing by macrophages. FEMS Microbiol Lett 360:144–151. 10.1111/1574-6968.12598. [DOI] [PubMed] [Google Scholar]
- 191.Niven DF, Ekins A. 2001. Iron content of Streptococcus suis and evidence for a dpr homologue. Can J Microbiol 47:412–416. 10.1139/w01-027. [DOI] [PubMed] [Google Scholar]
- 192.Hitchings MD, Townsend P, Pohl E, Facey PD, Jones DH, Dyson PJ, Del Sol R. 2014. A tale of tails: deciphering the contribution of terminal tails to the biochemical properties of two Dps proteins from Streptomyces coelicolor. Cell Mol Life Sci 71:4911–4926. 10.1007/s00018-014-1658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Choi SH, Baumler DJ, Kaspar CW. 2000. Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157:H7. Appl Environ Microbiol 66:3911–3916. 10.1128/AEM.66.9.3911-3916.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Barth E, Gora KV, Gebendorfer KM, Settele F, Jakob U, Winter J. 2009. Interplay of cellular cAMP levels, σS activity and oxidative stress resistance in Escherichia coli. Microbiology (Reading) 155:1680–1689. 10.1099/mic.0.026021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tsou CC, Chiang-Ni C, Lin YS, Chuang WJ, Lin MT, Liu CC, Wu JJ. 2008. An iron-binding protein, Dpr, decreases hydrogen peroxide stress and protects Streptococcus pyogenes against multiple stresses. Infect Immun 76:4038–4045. 10.1128/IAI.00477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Narayan OP, Kumari N, Rai LC. 2010. Heterologous expression of Anabaena PCC 7120 all3940 (a Dps family gene) protects Escherichia coli from nutrient limitation and abiotic stresses. Biochem Biophys Res Commun 394:163–169. 10.1016/j.bbrc.2010.02.135. [DOI] [PubMed] [Google Scholar]
- 197.Hébraud M, Guzzo J. 2000. The main cold shock protein of Listeria monocytogenes belongs to the family of ferritin-like proteins. FEMS Microbiol Lett 190:29–34. 10.1016/S0378-1097(00)00310-4. [DOI] [PubMed] [Google Scholar]
- 198.Nicodeme M, Perrin C, Hols P, Bracquart P, Gaillard J-L. 2004. Identification of an iron-binding protein of the Dps family expressed by Streptococcus thermophilus. Curr Microbiol 48:51–56. 10.1007/s00284-003-4116-3. [DOI] [PubMed] [Google Scholar]
- 199.Perrin C, Guimont C, Bracquart P, Gaillard JL. 1999. Expression of a new cold shock protein of 21.5 kDa and of the major cold shock protein by Streptococcus thermophilus after cold shock. Curr Microbiol 39:342–347. 10.1007/s002849900469. [DOI] [PubMed] [Google Scholar]
- 200.Thieme D, Grass G. 2010. The Dps protein of Escherichia coli is involved in copper homeostasis. Microbiol Res 165:108–115. 10.1016/j.micres.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 201.Malone AS, Chung Y-K, Yousef AE. 2006. Genes of Escherichia coli O157:H7 that are involved in high-pressure resistance. Appl Environ Microbiol 72:2661–2671. 10.1128/AEM.72.4.2661-2671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Li X, Mustila H, Magnuson A, Stensjö K. 2018. Homologous overexpression of NpDps2 and NpDps5 increases the tolerance for oxidative stress in the multicellular cyanobacterium Nostoc punctiforme. FEMS Microbiol Lett 365:fny198. 10.1093/femsle/fny198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pacello F, Ceci P, Ammendola S, Pasquali P, Chiancone E, Battistoni A. 2008. Periplasmic Cu,Zn superoxide dismutase and cytoplasmic Dps concur in protecting Salmonella enterica serovar Typhimurium from extracellular reactive oxygen species. Biochim Biophys Acta 1780:226–232. 10.1016/j.bbagen.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 204.Yamamoto Y, Higuchi M, Poole LB, Kamio Y. 2000. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J Bacteriol 182:3740–3747. 10.1128/JB.182.13.3740-3747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Brenot A, King KY, Caparon MG. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol Microbiol 55:221–234. 10.1111/j.1365-2958.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- 206.Pulliainen AT, Kauko A, Haataja S, Papageorgiou AC, Finne J. 2005. Dps/Dpr ferritin-like protein: insights into the mechanism of iron incorporation and evidence for a central role in cellular iron homeostasis in Streptococcus suis. Mol Microbiol 57:1086–1100. 10.1111/j.1365-2958.2005.04756.x. [DOI] [PubMed] [Google Scholar]