Abstract
The chronological progression of brain atrophy over decades, from pre-symptomatic to dementia stages, has never been formally depicted in Alzheimer’s disease. This is mainly due to the lack of cohorts with long enough MRI follow-ups in cognitively unimpaired young participants at baseline. To describe a spatiotemporal atrophy staging of Alzheimer’s disease at the whole-brain level, we built extrapolated lifetime volumetric models of healthy and Alzheimer’s disease brain structures by combining multiple large-scale databases (n = 3512 quality controlled MRI from 9 cohorts of subjects covering the entire lifespan, including 415 MRI from ADNI1, ADNI2 and AIBL for Alzheimer’s disease patients). Then, we validated dynamic models based on cross-sectional data using external longitudinal data. Finally, we assessed the sequential divergence between normal aging and Alzheimer’s disease volumetric trajectories and described the following staging of brain atrophy progression in Alzheimer’s disease: (i) hippocampus and amygdala; (ii) middle temporal gyrus; (iii) entorhinal cortex, parahippocampal cortex and other temporal areas; (iv) striatum and thalamus and (v) middle frontal, cingular, parietal, insular cortices and pallidum. We concluded that this MRI scheme of atrophy progression in Alzheimer’s disease was close but did not entirely overlap with Braak staging of tauopathy, with a ‘reverse chronology’ between limbic and entorhinal stages. Alzheimer’s disease structural progression may be associated with local tau accumulation but may also be related to axonal degeneration in remote sites and other limbic-predominant associated proteinopathies.
Keywords: Alzheimer, MRI, atrophy, staging, lifespan
Using extrapolated lifetime volumetric models of healthy and Alzheimer’s disease brain structures, Planche et al. propose a staging of atrophy progression in Alzheimer’s disease including (i) hippocampus and amygdala; (ii) middle temporal gyrus; (iii) entorhinal cortex and other temporal areas; (iv) striatum and thalamus and (v) middle frontal, cingular, parietal, insular cortices and pallidum.
Graphical Abstract
Graphical Abstract.
Introduction
Alzheimer’s disease is a very slow progressing condition, likely to develop over three or four decades, from its preclinical phase to severe dementia and death.1,2 Unfortunately, to date, no longitudinal study has been long enough to describe such a slow progressing evolution through all these stages, and the model of Alzheimer’s disease progression over time remains partially hypothetical. However, providing insights into the spatiotemporal spreading of brain alterations, which will ultimately lead to the full spectrum of Alzheimer’s disease clinical symptoms, is crucial in the prospect of future therapeutic actions.
The current model of Alzheimer’s disease progression is mainly based on the Braak staging of Alzheimer’s disease tauopathy.3 This scheme was initially built on the concatenation of post-mortem pathological brain examinations from donors at all the stages of disease progression. Ongoing longitudinal tau-PET imaging studies support the Braak staging. However, the currently available follow-up is still relatively short, and only a few studies aimed at investigating the pre-symptomatic stages of the disease in cognitively unimpaired participants.4 Furthermore, first-generation tau-PET tracers do not allow the detailed investigation of medial temporal structures due to off-target binding in the choroid plexus, and PET-histological correlation studies showed that these tracers lack sensitivity and could only identify patients with Braak stages ≥4.5 Although the accumulation of Alzheimer’s disease tauopathy usually precedes measurable atrophy,6 the anatomical progression of brain atrophy in Alzheimer’s disease may not perfectly match with tau topography and atrophy can experimentally precede overt cell loss.7
Indeed, many other factors are likely to explain brain atrophy in Alzheimer’s disease: the vascular pathology and the neuroinflammation coupled with Alzheimer’s disease neuropathological changes,8 interactions with other proteinopathies,9 network disruption due to local pathology combined with axonal degeneration and distant atrophy,10 local replication rate of tau aggregates11 and regional differential vulnerabilities to tau pathology and hypometabolism.12 Thus, the chronological progression over decades of neurodegeneration and brain atrophy in Alzheimer’s disease has never been formally depicted at the whole-brain level, and it is unknown whether it strictly follows the Braak staging of Alzheimer’s disease tauopathy.
Because no single longitudinal data over decades is available, we proposed to take advantage of BigData sharing in neuroimaging by analysing MRI databases of both Alzheimer’s disease patients and healthy subjects at different ages covering the entire lifespan. From these cross-sectional data, we processed a massive number of MRI to generate extrapolated lifespan models of several brain structure volumes evolution. Then, we validated such an approach by comparing profiles extrapolated from these cross-sectional data with independent available ‘truly’ longitudinal data. Finally, we described an MRI staging of volume loss in Alzheimer’s disease by describing the timing (and severity) of significant divergence between healthy subjects and Alzheimer’s disease patients’ volumetric trajectories.
Methods
Datasets
Normal and Alzheimer’s disease trajectories of brain atrophy were estimated thanks to the aggregation of nine open-access databases. The number of subjects included in the present study is provided after QC:
– C-MIND: 236 images of control subjects from the C-MIND dataset (https://research.cchmc.org/c-mind/) are used in this study. All the 3D T1-weighted (T1w) MPRAGE high-resolution MRI were acquired at the same site on a 3 T scanner with spatial resolution of 1 mm3 acquired using a 32 channel SENSE head-coil.
– NDAR: 382 of control subjects from the Database for Autism Research (NDAR) (https://ndar.nih.gov) are used in this study. The T1w 3D MRI were acquired on 1.5 T MRI and 3 T scanners. In our experiments, we used the NIHPD (http://www.bic.mni.mcgill.ca/nihpd/info/data_access.html) dataset and 197 images of control subjects from the Lab Study 19 of the National Database for Autism Research. For the NIHPD dataset, the 3D T1w spoiled gradient-recalled echo (SPGR) MRI were acquired at six different sites with 1.5 T systems by General Electric and Siemens Medical Systems with spatial resolution of 1 mm3. The 3D T1w MPRAGE MRI from the Lab Study 19 were scanned using a 3 T Siemens Tim Trio scanner at each site with spatial resolution of 1 mm3.
– ABIDE: 492 control subjects from the Autism Brain Imaging Data Exchange (ABIDE) dataset (http://fcon_1000.projects.nitrc.org/indi/abide/) are used in this study. The MRI are T1w MPRAGE acquired at 20 different sites on 3 T image and the details of the acquisition, informed consent and site-specific protocols are available on the website.
– ICBM: 294 normal subjects from the International Consortium for Brain Mapping (ICBM) dataset (http://www.loni.usc.edu/ICBM/) obtained through the LONI website are used in this study. The T1w MPRAGE MRI were acquired on a 1.5 T Philips GyroScan imaging system (Philips Medical Systems, Best, The Netherlands) with spatial resolution of 1 mm3.
– IXI: 549 normal control from the Information eXtraction from Images (IXI) database (http://brain-development.org/ixi-dataset/) are used in this study. The MRI are T1w images collected at three sites with 1.5 and 3 T scanners with spatial resolution close to 1 mm3.
– ADNI1&2: 404 control subjects and 332 Alzheimer’s disease patients from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu) Phases 1 and 2 are used in this study. These baseline MRI are T1w MPRAGE and SPGR acquired on 1.5 T scanners and 3 T at 60 different sites across the USA and Canada with reconstructed spatial resolution of 1 mm3. In ADNI 1&2, the diagnosis of Alzheimer’s disease was made according to NINCDS/ADRDA criteria for probable Alzheimer’s disease.13 Patients were included whatever their clinical presentation (typical or atypical Alzheimer’s disease), but all participants had abnormal memory functions at baseline, and Clinical Dementia Rating scale = 0.5 or 1. Alzheimer’s disease biomarkers analysis was optional. The complete list of inclusion/exclusion criteria is available here: http://adni.loni.usc.edu/methods/documents/.
– AIBL: 467 control subjects (857 images) and 83 Alzheimer’s disease patients (113 images) from the Australian Imaging, Biomarkers and Lifestyle (AIBL) database (http://www.aibl.csiro.au/) are used in this study. We used the longitudinal dataset as defined in Wen et al.14 and provided on the platform Clinica (http://www.clinica.run). These images are T1w acquired on 3 T MR scanners with the ADNI protocol (http://adni.loni.ucla.edu/research/protocols/mri-protocols) and with custom MPRAGE sequence on the 1.5 T scanners. In AIBL, the diagnosis of Alzheimer’s disease was made during clinical review panel meetings,15 according to NINCDS/ADRDA criteria for possible or probable Alzheimer’s disease .13
Image processing
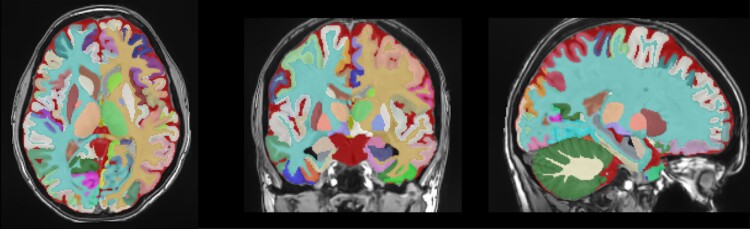
All the considered T1w MRI were processed with AssemblyNet.16 This software produces whole-brain segmentation of fine-grained structures using a large ensemble of deep neural networks. On the 132 structures produced by AssemblyNet following the Neuromorphometrics labels,17 we considered only 122 grey matter regions (61 left and 61 right). An illustration of the AssemblyNet segmentation is provided in Fig. 1. It included 9 subcortical structures, 17 frontal gyri/lobules, 8 temporal gyri/lobules, 6 parietal gyri/lobules, 8 occipital gyri/lobules, 6 gyri in the limbic cortex and 5 sub-regions of the insular cortex, ventricles and the cerebellum.
Figure 1.
Illustration of the AssemblyNet segmentation in the three axes. It included the bilateral segmentation of 9 subcortical structures, 17 frontal gyri/lobules, 8 temporal gyri/lobules, 6 parietal gyri/lobules, 8 occipital gyri/lobules, 6 gyri in the limbic cortex and 5 sub-regions of the insular cortex, the ventricles and the cerebellum.
The AssemblyNet pipeline included the following steps. After denoising,18 images were corrected for inhomogeneity,19 affine-registered into the Montreal Neurological Institute (MNI) space using ANTS20 and tissue-based intensity normalized.21 Then, the intracranial cavity was segmented using the NICE method.22 Afterwards, structure segmentation was achieved using 250 U-Nets through a multi-scale framework.
A multi-stage quality control procedure was performed by P.C., blinded of the subject’s group, as previously described.23,24 First, a visual assessment was done for all input images by checking screen shots of one sagittal, coronal and axial slice in the middle of the 3D volume. Images were rejected if partial head coverage, motion artefact, high distortion or abnormal noise level was detected. Then, a visual assessment of processing quality was carried out using the segmentation report which provides screenshots for each step of the pipeline. Images were rejected after this step in case of inaccurate registration in the MNI space, inaccurate intracranial cavity extraction, missing brain structures or over/under-segmentation of brain structures. The last control was performed by individually checking of all outliers (values higher/lower than 2 SD of the estimated model). For each outlier, the segmentation map was inspected using a 3D viewer. In case of segmentation failure, the subject was removed from the study.
Statistical analyses
To compensate for the variability introduced by head size difference, models were estimated on normalized volumes (% of total intracranial volume). Left and right volumes were added to obtain the final volume structure. Statistics were performed with MATLAB using default parameters.
Different strategies were considered to model the trajectories of each brain structure over time, as previously described.24 Briefly, the candidate models were tested from the simplest to the most complex: (i) a linear model, (ii) a quadratic model and (iii) a cubic model. A model was kept as a potential candidate only when simultaneously F-statistic based on ANOVA (i.e. model versus constant model) was significant (P < 0.05) and when all its coefficients were significant using t-statistic (P < 0.05). We finally used the Bayesian information criterion to compare the candidate models. This model selection procedure was applied to all the considered structures.
Afterwards, z-score and ‘distance’ between healthy and Alzheimer’s disease trajectories were computed on the estimated models. The prediction bounds were estimated with a confidence level of 95%. A brain structure was considered to be significantly smaller in Alzheimer’s disease compared with healthy ageing when the two structural trajectories diverged and when their 95% confidence intervals no longer overlapped.
To compare our lifespan cross-sectional models and longitudinal models over restricted periods, we followed a previously published strategy25 where cross-sectional and longitudinal models were compared using Spearman correlation over the age period defined by longitudinal models. We performed two experiments to analyse model similarities and atrophy pattern similarities. First, we estimated correlations between the models’ values for each structure. Second, we estimated the correlation between vectors containing cross-sectional volume shrinkage and longitudinal atrophy for all the structures simultaneously. These volume changes were estimated as the difference between the value at the starting age of longitudinal models and the final one.
Finally, the sequence of significant divergence of the different brain structures was listed in chronological order to obtain the MRI staging scheme.
Data availability
MRI raw data from the different cohorts are available online (see Acknowledgments). Other data are available from the corresponding author upon reasonable request.
Results
Dataset description
To study structures’ trajectories of healthy controls and Alzheimer’s disease across the entire lifespan, we compiled several open-access databases to construct two datasets. Their composition and characteristics are described in Table 1. After QC, 3512 MRI from healthy controls and 415 from Alzheimer’s disease patients remained for the analyses. MRI from the longitudinal AIBL database (n = 113) were used as an external testing dataset to validate our models. Consequently, we used 2665 MRI from healthy controls to estimate our healthy lifespan models. To infer Alzheimer’s disease models over the entire lifespan, we combined 1874 healthy controls younger than 55 years of age (all the subjects younger than 55 in the 2665 that were used for healthy ageing models) and 332 Alzheimer’s disease patients older than 55 to build our lifespan Alzheimer’s disease models.
Table 1.
Dataset description.
| Dataset | N | Gender | Age (years) |
|---|---|---|---|
| Healthy controls (total) | 3512 | − | − |
| C-MIND | 236 | F = 129; M = 107 | 8.44 [0.74–18.86] |
| NDAR | 382 | F = 174; M = 208 | 12.39 [1.08–49.92] |
| ABIDE | 492 | F = 84; M = 408 | 17.53 [6.50–52.20] |
| ICBM | 294 | F = 142; M = 152 | 33.75 [18–80] |
| IXI | 549 | F = 307; M = 242 | 48.76 [20.0–86.2] |
| OASIS | 298 | F = 187; M = 111 | 45.34 [18–94] |
| ADNI 1&2 | 404 | F = 203; M = 201 | 74.81 [56–90] |
| Longitudinal AIBL (467 subjects) | 857 | F = 485; M = 372 | 74.15 [60.5–92.4] |
| Alzheimer’s disease (total) | 415 | − | − |
| ADNI 1&2 | 332 | F = 151; M = 181 | 75.13 [55–91] |
| Longitudinal AIBL (83 subjects) | 113 | F = 66; M = 47 | 73.35 [55.5–93.4] |
This table provides the name of the dataset, the number (n) of considered images (after quality control), the gender proportion and the average ages and intervals in brackets.
Identification of brain structures diverging between healthy subjects and Alzheimer’s disease patients trajectories
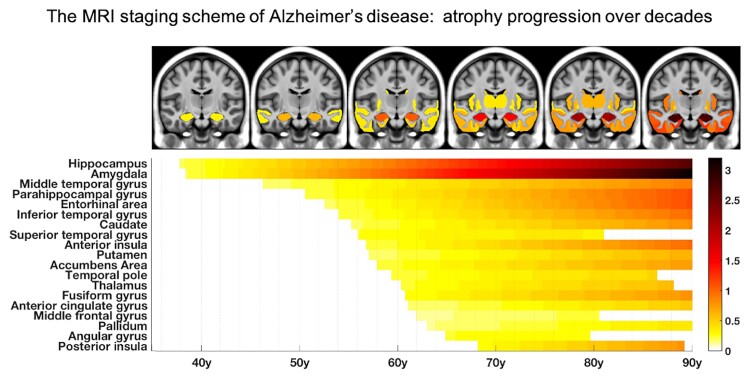
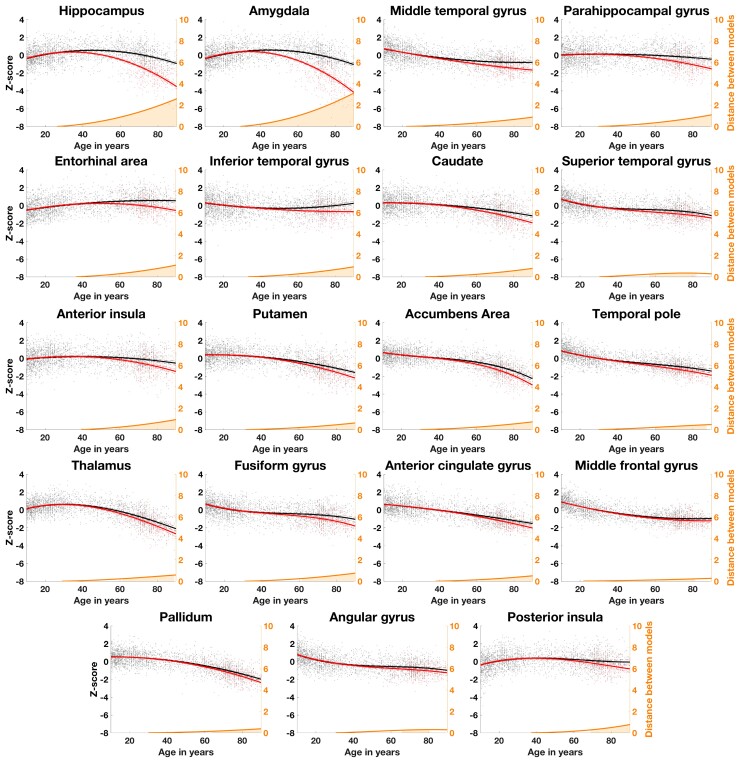
Figure 2 shows the 19 brain structures (out of the 61 grey matter structures we have tested using AssemblyNet) that significantly diverged during lifespan between Alzheimer’s disease and healthy ageing models. The theoretical starting age of statistically significant divergence between healthy and pathological models appears to be between 38 and 68 years old, depending on the structure. The most affected structures over time were the amygdala and the hippocampus (distance between healthy and ageing model at 90 years old = 3.20 and 2.65, respectively), followed by the entorhinal and parahippocampal cortices (distance = 1.18 and 1.13), the insula and the inferior temporal gyrus (distance = 0.96 and 0.96).
Figure 2.
Lifespan trajectories based on z-scores of normalized brain volumes for healthy ageing subjects (black line) and Alzheimer’s disease patients (red line). Black dots represent all healthy individuals and red dots Alzheimer’s disease patients. The prediction bounds of the models are estimated with a confidence level at 95%. The orange curve represents the distance between the healthy and pathological models. The orange area indicates the time period where the confidence intervals of both models do not overlap. Only models detected as significantly different between healthy ageing and Alzheimer’s disease are presented in this figure (19 brain structures out of 61 tested using AssemblyNet).
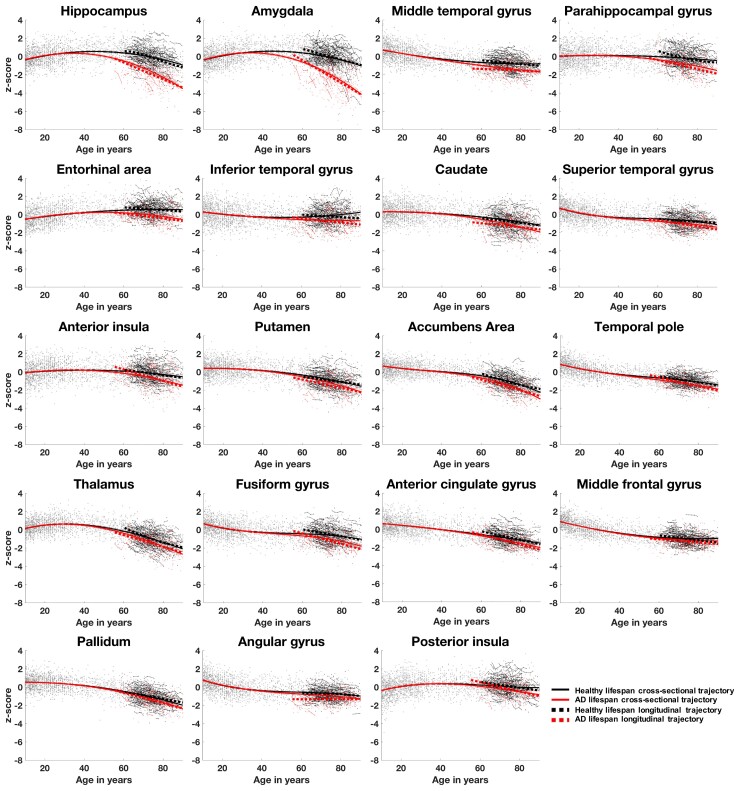
Comparison of cross-sectional and longitudinal lifespan models
For all the 19 structures detected as significantly different between normal ageing and Alzheimer’s disease, we noticed a significant correlation between cross-sectional and longitudinal models (P < 0.05) in the age range of [55.5–93.4] for Alzheimer’s disease patients and [60.5–92.4] for healthy subjects. The average correlation between the models’ values over these 19 structures was 0.91 for Alzheimer’s disease models and 0.82 for healthy ageing models. Moreover, we found significant correlations between cross-sectional volume shrinkage and longitudinal atrophy for both Alzheimer’s disease and healthy ageing trajectories (P < 0.05). The atrophy pattern correlation for these 19 structures was 0.90 for Alzheimer’s disease models and 0.85 for healthy ageing models. Figure 3 shows the graphical congruence of models based on cross-sectional and longitudinal data on all the 19 affected structures.
Figure 3.
Graphical comparison of cross-sectional (whole lines) and longitudinal (dotted lines) trajectories based on z-scores of normalized brain volumes for healthy ageing (black lines and dots) and Alzheimer’s disease patients (red lines and dots). Only the 19 structures detected as significantly different between healthy and pathological conditions are represented here. The average correlation coefficients for these 19 structures were 0.91 for Alzheimer’s disease models and 0.82 for healthy ageing models.
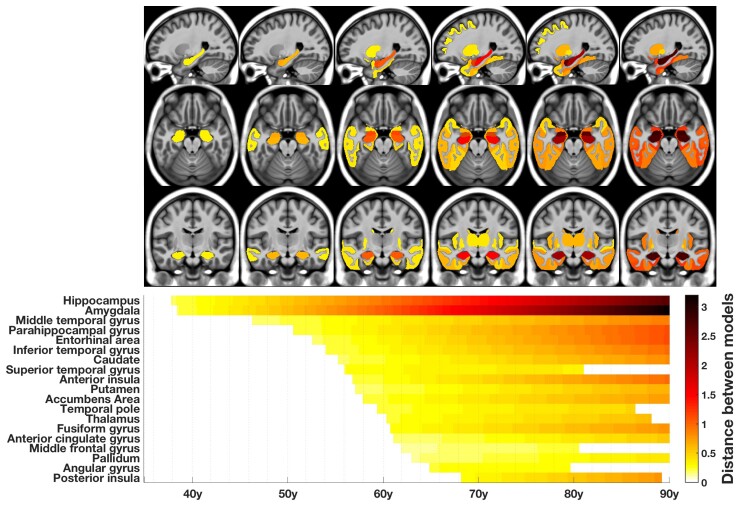
The MRI staging scheme
We then mapped the sequential divergence of healthy and Alzheimer’s disease trajectories in Fig. 4. Schematically, five major stages in Alzheimer’s disease structural progression appeared: (i) hippocampus and amygdala; (ii) middle temporal gyrus; (iii) entorhinal cortex, parahippocampal cortex and other temporal areas (inferior temporal gyrus, superior temporal gyrus, temporal pole and fusiform gyrus); (iv) thalamus and striatum (accumbens, caudate, putamen) and (v) middle frontal, anterior cingular, parietal (angular gyrus), insular cortices and pallidum.
Figure 4.
The MRI staging scheme of structural progression of Alzheimer’s disease. The figure represents the chronological progression of distances between healthy and Alzheimer’s disease trajectories. The severity of structural divergence is colour-coded according to the bar at the bottom right of the figure.
Discussion
In this study, we combined multiple large-scale MRI databases and whole-brain segmentation of fine-grained structures using a large ensemble of deep neural networks to describe the first exhaustive chronological structural progression of Alzheimer’s disease over decades and the differential severity of volumetric structure alterations. We finally proposed the following brain MRI staging scheme of structural progression in Alzheimer’s disease: (i) hippocampus and amygdala; (ii) middle temporal gyrus; (iii) entorhinal cortex, parahippocampal cortex and other temporal areas; (iv) striatum and thalamus and (v) middle frontal, cingular, parietal and insular cortices. The most severely affected structures during the entire course of Alzheimer’s disease were the amygdala and the hippocampus, followed by the entorhinal and parahippocampal cortices.
This MRI staging scheme of atrophy progression is close to but does not fully overlap with Braak staging of Alzheimer’s disease tauopathy.3 The main difference between these two staging models is the ‘reverse chronology’ between limbic and entorhinal stages (the earliest stages). Indeed, transentorhinal and entorhinal cortices are the first affected by tau neurofibrillary tangles (NFT) (Braak Stages I and II) before hippocampus [cornu ammonis 1 (CA1) pyramidal cells] and amygdala (Braak Stages III and IV). After these entorhinal and limbic stages, the sequential involvement of the thalamus and the striatum before the isocortical associative areas is concordant between MRI and Braak staging. However, Braak staging can be questioned. A recent 3D mapping of human tau pathology suggests, for instance, a more widespread distribution of NFT at the early stages of Alzheimer’s disease: similar levels of tau burden were found in entorhinal areas and the amygdala, the hippocampus, and the temporopolar cortex, supporting our MRI findings.26
Although tau distribution is usually correlated with focal atrophy in Alzheimer’s disease, recent tau-PET studies support our findings and the ‘early stage discrepancy hypothesis’ between atrophy and tau staging schemes. On the one hand, Mak et al.27 reported a disproportionate increase in tau accumulation in widespread regions compared to cortical atrophy that was restricted to the temporal areas during the same period. On the other hand, Harrison et al.28 reported that atrophy might exceed tau accumulation by including brain regions relatively unaffected by tauopathy. Furthermore, MRI-pathological studies failed to correlate the rate of hippocampal atrophy with early hippocampal tau NFT during ageing.29 In turn, the exceptional vulnerability of hippocampal CA1 pyramidal neurons to Alzheimer’s disease neuropathological changes30 may explain differential atrophy between the hippocampus and the entorhinal cortex over time.
Experimentally, when pathological tau is specifically expressed in the entorhinal cortex of transgenic mice, atrophy is described in the hippocampus, a remote unaffected brain region at the early stage of this model, through axon degeneration of the perforant pathway (major afference from the entorhinal cortex to the hippocampus).7 The pathological accumulation of tau oligomers is also known to impair axonal transport before NFT formation, inducing early axonal degeneration and the loss of synaptic transmission in the target structures.31,32 In the case of the amygdala, early atrophy may also be due to the presence of pathological tau in the perirhinal, postrhinal and transentorhinal cortices (Braak Stage I), which give rise to important afferent projections to the amygdala in primates.33 Finally, atrophy has to be considered as the result of complex cellular processes including cell death associated with tau accumulation, but also from axonal degeneration in remote sites. This diaschisis phenomenon may precede tau tangles in the limbic system, explaining the ‘early stage discrepancy’ between MRI and Braak staging.
In addition, regional atrophy in Alzheimer’s disease does not rely on a single biological process, but instead reflects the local impact of many different possible mechanisms leading to neurodegeneration. Mainly, limbic-predominant age-related TAR DNA-binding protein 43 encephalopathy (LATE) neuropathological changes are frequent during ageing and often associated with Alzheimer’s disease neuropathological changes. TAR DNA-binding protein 43 (TDP-43) inclusions are known to accelerate brain atrophy and mainly affect the amygdala (Stage 1) and the hippocampus (Stage 2). The frequent co-occurrence (up to 75% of cases) of these proteinopathies may explain the excess of limbic atrophy compared to entorhinal atrophy at the early stages of Alzheimer’s disease.34 Indeed, pathological-MRI correlation studies showed that the hippocampus and the amygdala were the most frequently affected regions in TDP-43 positive brains and that the magnitude of atrophy mediated by TDP-43 inclusions was similar to tau-related atrophy.35
From the conceptual point of view, it is essential to keep in mind that the present MRI staging scheme (such as the Braak staging scheme) only describes the most frequent ‘limbic-predominant’ and ‘typical’ progressions of Alzheimer’s disease.36,37 This model only describes the ‘average’ structural course of Alzheimer’s disease and does not apply to ‘atypical’ Alzheimer’s disease, such as logopenic primary progressive aphasia, posterior cortical atrophy or behavioural Alzheimer’s disease, which are driven by different patterns of atrophy and represent less common ‘extremes’ of the Alzheimer’s disease anatomical spectrum.38 Indeed, the methodology used in the present work does not take into account the recent description by Vogel et al.39 of distinct trajectories of tau deposition in Alzheimer’s disease. This work proposes an alternative to Braak’s model concerning tau spreading pattern, underlining the heterogeneity of Alzheimer’s disease development from one subject to another (not using pathological examination, but cross-sectional tau-PET with its limitations in terms of sensitivity). However, the most prevalent spatiotemporal trajectory of tau pathology was limbic in Vogel’s work, and spatial convergence was observed between Alzheimer’s disease trajectories. Furthermore, our MRI scheme of atrophy progression includes structures characterizing three of the four distinct trajectories of tau deposition, namely amygdala, hippocampus and entorhinal cortex (limbic trajectory), middle temporal gyrus, parahippocampal cortex and other temporal areas (lateral temporal trajectory) and parietal (angular gyrus) cortices (medial temporal lobe-sparing trajectory). Thus, our ‘average’ MRI staging scheme may not apply to all patients with Alzheimer’s disease, but to the greatest number, especially because cohorts such as ADNI 1&2 and AIBL included patients with ‘Alzheimer clinical syndrome’40 or ‘probable’ Alzheimer’s disease criteria13,41 (i.e. subjects with a single or multi-domain amnestic presentation, likely to have a limbic-predominant pattern of atrophy).
From the methodological point of view, we acknowledge that the estimated point of divergence between healthy and pathological trajectories remains hypothetical because of the lack of Alzheimer’s disease patients younger than 55 in the analysed cohorts. Another limitation of this study is that we cannot exclude that the anatomy of a structure of interest (the definition of boundaries on brain atlas, the precision of segmentation using MRI contrast, …) may impact the capability to depict volumetric modifications; this observation being particularly relevant for the entorhinal cortex.42 Thanks to an external validation using AIBL data, we have shown that our models based on cross-sectional ADNI data are reliable approximate of ‘true’ longitudinal trajectories, at least after 55 years old. Furthermore, the early atrophy in the amygdala and the hippocampus are consistent with a previous work from our group using a different method for structure segmentation.24 We also found substantial similarities with a previous article from another lab comparing the pattern of change of subcortical structures estimated on lifespan cross-sectional and longitudinal models in healthy ageing.25 Our study is, however, the first to assess the divergence between this healthy ageing trajectory and Alzheimer’s disease structural progression using inferred lifespan Alzheimer’s disease model, and at the whole-brain level.
To conclude, we have modelled the ‘average’ global structural progression of Alzheimer’s disease over the entire course of the disease. We found that the time course and the severity of structural alterations were close but did not completely overlap with Braak staging. We proposed a descriptive MRI staging scheme that will help better define the disease and interpret future studies looking at the differential vulnerabilities of brain structures to Alzheimer’s disease pathological processes.
Acknowledgements
The C-MIND data used in the preparation of this article were obtained from the C-MIND Data Repository created by the C-MIND study of Normal Brain Development. A listing of the participating sites and a complete listing of the study investigators can be found at https://research.cchmc.org/c-mind. The NDAR data used in the preparation of this manuscript were obtained from the NIH-supported National Database for Autism Research (NDAR). This is supported by the National Institute of Child Health and Human Development, the National Institute on Drug Abuse, the National Institute of Mental Health and the National Institute of Neurological Disorders and Stroke. A listing of the participating sites and a complete listing of the study investigators can be found at http://pediatricmri.nih.gov/nihpd/info/participating_centers.html. The ICBM data used in the preparation of this manuscript were supported by Human Brain Project grant PO1MHO52176-11 and Canadian Institutes of Health Research grant MOP-34996. The IXI data used in the preparation of this manuscript were supported by the UK. Engineering and Physical Sciences Research Council (EPSRC) GR/S21533/02—http://www.brain-development.org/. The ABIDE data used in the preparation of this manuscript were supported by ABIDE funding resources listed at http://fcon_1000.projects.nitrc.org/indi/abide/. The ADNI data used in the preparation of this manuscript were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). The ADNI is funded by the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics NV, Johnson & Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc., F. Hoffmann-La Roche, Schering-Plough, Synarc Inc., as well as non-profit partners, the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to the ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study was coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for NeuroImaging at the University of California, Los Angeles. This research was also supported by NIH grants P30AG010129, K01 AG030514 and the Dana Foundation. The OASIS data used in the preparation of this manuscript were obtained from the OASIS project funded by grants P50 AG05681, P01 AG03991, R01 AG021910, P50 MH071616, U24 RR021382 and R01 MH56584. See http://www.oasis-brains.org/for more details. The AIBL data used in the preparation of this manuscript were obtained from the AIBL study of ageing funded by the Commonwealth Scientific Industrial Research Organization (CSIRO; a publicly funded government research organization), Science Industry Endowment Fund, National Health and Medical Research Council of Australia (project grant 1011689), Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation and an anonymous foundation. See www.aibl.csiro.au for further details. We wish to thank all investigators of these projects who collected these datasets and made them freely accessible. This manuscript reflects the views of the authors and may not reflect the opinions or views of the database providers.
Glossary
Abbreviations
- ADNI =
Alzheimer’s Disease Neuroimaging Initiative
- AIBL =
Australian Imaging, Biomarkers and Lifestyle
- CA =
cornu ammonis
- LATE =
limbic-predominant age-related TDP-43 encephalopathy
- NFT =
neurofibrillary tangles
- TDP-43 =
TAR DNA-binding protein 43
Contributor Information
Vincent Planche, Univ. Bordeaux, CNRS, UMR 5293, Institut des Maladies Neurodégénératives, F-33000 Bordeaux, France; Centre Mémoire Ressources Recherches, Pôle de Neurosciences Cliniques, CHU de Bordeaux, F-33000 Bordeaux, France.
José V. Manjon, Instituto de Aplicaciones de las Tecnologías de la Información y de las Comunicaciones Avanzadas (ITACA), Universitat Politècnica de València, Camino de Vera s/n, 46022 Valencia, Spain
Boris Mansencal, Univ. Bordeaux, CNRS, UMR5800, Bordeaux INP, LABRI, F-33400 Talence, France.
Enrique Lanuza, Department of Cell Biology, University of Valencia, Burjassot 46100, Valencia, Spain.
Thomas Tourdias, Inserm U1215 - Neurocentre Magendie, Bordeaux F-33000, France; Service de Neuroimagerie diagnostique et thérapeutique, CHU de Bordeaux, F-33000 Bordeaux, France.
Gwenaëlle Catheline, Institut de Neurosciences Cognitives et Intégratives d’Aquitaine, CNRS, UMR 5287, University of Bordeaux, F-33000 Bordeaux, France.
Pierrick Coupé, Univ. Bordeaux, CNRS, UMR5800, Bordeaux INP, LABRI, F-33400 Talence, France.
Funding
This study was achieved within the context of the Human Brain Project, the French National Research Agency for the DeepvolBrain project (ANR-18-CE45-0013), the Laboratory of Excellence TRAIL ANR-10-LABX-57 for the BigDataBrain project and the CNRS/INSERM for the DeepMultiBrain project. V.P. received grants from Foundation Bettencourt Schueller (CCA-Inserm-Bettencourt). J.V.M. research was supported by the Spanish DPI2017-87743-R grant from the Ministerio de Economia, Industria y Competitividad of Spain. The sponsors did not participate in any aspect of the design or performance of the study, including data collection, management, analysis, and the interpretation or preparation, review and approval of the manuscript.
Competing interests
The authors declare no competing financial interests relative to the present study.
References
- 1. Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barthélemy NR, Li Y, Joseph-Mathurin N, et al. A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nat Med. 2020;26(3):398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. [DOI] [PubMed] [Google Scholar]
- 4. Jack CR, Wiste HJ, Weigand SD, et al. Predicting future rates of tau accumulation on PET. Brain. 2020;143(10):3136–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Soleimani-Meigooni DN, Iaccarino L, La Joie R, et al. 18F-flortaucipir PET to autopsy comparisons in Alzheimer’s disease and other neurodegenerative diseases. Brain. 2020;143(11):3477–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Joie RL, Visani AV, Baker SL, et al. Prospective longitudinal atrophy in Alzheimer’s disease correlates with the intensity and topography of baseline tau-PET. Sci Transl Med. 2020;12(524):eaau5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fung CW, Guo J, Fu H, Figueroa HY, Konofagou EE, Duff KE. Atrophy associated with tau pathology precedes overt cell death in a mouse model of progressive tauopathy. Sci Adv. 2020;6(42):eabc8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayley S, Hakim AM, Albert PR. Depression, dementia and immune dysregulation. Brain. 2021;144(3):746–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spires-Jones TL, Attems J, Thal DR. Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol. 2017;134(2):187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Villain N, Fouquet M, Baron JC, et al. Sequential relationships between grey matter and white matter atrophy and brain metabolic abnormalities in early Alzheimer’s disease. Brain. 2010;133(11):3301–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meisl G, Hidari E, Allinson K, et al. In vivo rate-determining steps of tau seed accumulation in Alzheimer’s disease. Sci Adv. 2021;7(44):eabh1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. La Joie R, Perrotin A, Barré L, et al. Region-specific hierarchy between atrophy, hypometabolism, and β-amyloid (Aβ) load in Alzheimer’s disease dementia. J Neurosci. 2012;32(46):16265–16273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–944. [DOI] [PubMed] [Google Scholar]
- 14. Wen J, Thibeau-Sutre E, Diaz-Melo M, et al. Convolutional neural networks for classification of Alzheimer's disease: overview and reproducible evaluation. Med Image Anal. 2020;63:101694. [DOI] [PubMed] [Google Scholar]
- 15. Ellis KA, Bush AI, Darby D, et al. The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: Methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int Psychogeriatr. 2009;21(4):672–687. [DOI] [PubMed] [Google Scholar]
- 16. Coupé P, Mansencal B, Clément M, et al. AssemblyNet: A large ensemble of CNNs for 3D whole brain MRI segmentation. Neuroimage. 2020;219:117026. [DOI] [PubMed] [Google Scholar]
- 17. Klein A, Tourville J. 101 labeled brain images and a consistent human cortical labeling protocol. Front Neurosci. 2012;6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manjón JV, Tohka J, Robles M. Improved estimates of partial volume coefficients from noisy brain MRI using spatial context. Neuroimage. 2010;53(2):480–490. [DOI] [PubMed] [Google Scholar]
- 19. Tustison NJ, Avants BB, Cook PA, et al. N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manjón JV, Tohka J, García-Martí G, et al. Robust MRI brain tissue parameter estimation by multistage outlier rejection. Magn Reson Med. 2008;59(4):866–873. [DOI] [PubMed] [Google Scholar]
- 22. Manjón JV, Eskildsen SF, Coupé P, Romero JE, Collins DL, Robles M. Nonlocal intracranial cavity extraction. Int J Biomed Imaging. 2014;2014:820205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coupé P, Catheline G, Lanuza E, Manjón JV. Alzheimer’s disease neuroimaging initiative. Towards a unified analysis of brain maturation and aging across the entire lifespan: A MRI analysis. Hum Brain Mapp. 2017;38(11):5501–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coupé P, Manjón JV, Lanuza E, Catheline G. Lifespan changes of the human brain in Alzheimer’s disease. Sci Rep. 2019;9(1):3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fjell AM, Westlye LT, Grydeland H, et al. Critical ages in the life course of the adult brain: Nonlinear subcortical aging. Neurobiol Aging. 2013;34(10):2239–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yushkevich PA, López MM, Iñiguez de Onzoño Martin MM, et al. Three-dimensional mapping of neurofibrillary tangle burden in the human medial temporal lobe. Brain. 2021;144(9):2784–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mak E, Bethlehem RAI, Romero-Garcia R, et al. In vivo coupling of tau pathology and cortical thinning in Alzheimer’s disease. Alzheimers Dement (Amst). 2018;10:678–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harrison TM, La Joie R, Maass A, et al. Longitudinal tau accumulation and atrophy in aging and Alzheimer disease. Ann Neurol. 2019;85(2):229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erten-Lyons D, Dodge HH, Woltjer R, et al. Neuropathologic basis of age-associated brain atrophy. JAMA Neurol. 2013;70(5):616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. West MJ, Coleman PD, Flood DG, Troncoso JC. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer’s disease. Lancet. 1994;344(8925):769–772. [DOI] [PubMed] [Google Scholar]
- 31. Gyparaki MT, Arab A, Sorokina EM, et al. Tau forms oligomeric complexes on microtubules that are distinct from tau aggregates. Proc Natl Acad Sci USA. 2021;118(19):e2021461118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Combs B, Mueller RL, Morfini G, Brady ST, Kanaan NM. Tau and axonal transport misregulation in tauopathies. Adv Exp Med Biol. 2019;1184:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDonald AJ. Cortical pathways to the mammalian amygdala. Prog Neurobiol. 1998;55(3):257–332. [DOI] [PubMed] [Google Scholar]
- 34. Josephs KA, Martin PR, Weigand SD, et al. Protein contributions to brain atrophy acceleration in Alzheimer’s disease and primary age-related tauopathy. Brain. 2020;143(11):3463–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bejanin A, Murray ME, Martin P, et al. Antemortem volume loss mirrors TDP-43 staging in older adults with non-frontotemporal lobar degeneration. Brain. 2019;142(11):3621–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Planche V, Bouteloup V, Mangin JF, et al. Clinical relevance of brain atrophy subtypes categorization in memory clinics. Alzheimers Dement. 2021;17(4):641–652. [DOI] [PubMed] [Google Scholar]
- 37. Planche V, Coupé P, Helmer C, et al. Evolution of brain atrophy subtypes during aging predicts long-term cognitive decline and future Alzheimer’s clinical syndrome. Neurobiol Aging. 2019;79:22–29. [DOI] [PubMed] [Google Scholar]
- 38. Vogel JW, Hansson O. Subtypes of Alzheimer’s disease: Questions, controversy, and meaning. Trends Neurosci. 2022;45:342–345, S0166-2236(22)00033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vogel JW, Young AL, Oxtoby NP, et al. Four distinct trajectories of tau deposition identified in Alzheimer’s disease. Nat Med. 2021;27(5):871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jack CR, Bennett DA, Blennow K, et al. NIA-AA research framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14(4):535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fung YL, Ng KET, Vogrin SJ, et al. Comparative utility of manual versus automated segmentation of hippocampus and entorhinal cortex volumes in a memory clinic sample. J Alzheimers Dis. 2019;68(1):159–171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
MRI raw data from the different cohorts are available online (see Acknowledgments). Other data are available from the corresponding author upon reasonable request.