Abstract
Highly diversified astigmatic mites comprise many medically important human household pests such as house dust mites causing ∼1–2% of all allergic diseases globally; however, their evolutionary origin and diverse lifestyles including reversible parasitism have not been illustrated at the genomic level, which hampers allergy prevention and our exploration of these household pests. Using six high-quality assembled and annotated genomes, this study not only refuted the monophyly of mites and ticks, but also thoroughly explored the divergence of Acariformes and the diversification of astigmatic mites. In monophyletic Acariformes, Prostigmata known as notorious plant pests first evolved, and then rapidly evolving Astigmata diverged from soil oribatid mites. Within astigmatic mites, a wide range of gene families rapidly expanded via tandem gene duplications, including ionotropic glutamate receptors, triacylglycerol lipases, serine proteases and UDP glucuronosyltransferases. Gene diversification after tandem duplications provides many genetic resources for adaptation to sensing environmental signals, digestion, and detoxification in rapidly changing household environments. Many gene decay events only occurred in the skin-burrowing parasitic mite Sarcoptes scabiei. Throughout the evolution of Acariformes, massive horizontal gene transfer events occurred in gene families such as UDP glucuronosyltransferases and several important fungal cell wall lytic enzymes, which enable detoxification and digestive functions and provide perfect drug targets for pest control. This comparative study sheds light on the divergent evolution and quick adaptation to human household environments of astigmatic mites and provides insights into the genetic adaptations and even control of human household pests.
Keywords: astigmatic mites, comparative genomics, horizontal gene transfer, household pest adaptations, tandem gene duplication
Introduction
Astigmatic mites (suborder: Astigmata) are a group of rapidly evolving and successfully radiating small organisms including many medically important human household pests (Norton 1998; Sakata and Norton 2001; Domes et al. 2007; Dabert et al. 2010; Klimov and Oconnor 2013). Most astigmatic mites are commensals or parasites on hosts ranging from insects to birds and mammals; which many free-living species (especially house dust mites) are major human allergy-related mites and ∼1–2% of the world population (65–130 million people) suffers from allergic diseases caused by these mites (Arlian and Platts-Mills 2001; Colloff 2009; Thomas et al. 2010; Calderón, Kleine-Tebbe, et al. 2015; Miller 2019). Astigmata was considered to have evolved from oribatid mites (suborder: Oribatida) which are the numerically dominant microarthropods in soil environments, but the evolutionary dynamics underlying their emergence are still largely unclear. Moreover, their diversification into various lifestyles including reversal parasitism (Klimov and Oconnor 2013) remains to be thoroughly investigated at the genomic level. In this study, the high-quality genomes of six representative astigmatic mites, comprising two house dust mites, Dermatophagoides (D.) farinae and D. pteronyssinus, two parasitic mites, Psoroptes (P.) ovis and Sarcoptes (S.) scabiei, and two canonical storage mites, Blomia (B.) tropicalis and Tyrophagus (T.) putrescentiae (fig. 1A), were constructed for comparative genomics analysis.
Fig. 1.
Morphology of six astigmatic mites and phylogenetic tree of Acari (mites and ticks). (A) Taxonomy and diverse morphology of six astigmatic mites (manually painted). High-quality genomes of the six astigmatic mites with diverse morphologies were presented and summarized in table 1. Taxonomy assignment used the NCBI taxonomy version. S. scabiei is the var. canis. All the six mites are classified as human household pests in this study. (B) Phylogenetic tree of mites and ticks (subclass: Acari). Two major groups Acariformes and Parasitiformes in the subclass Acari were interrupted by the pseudoscorpion Cordylochernes scorpioides. The phylogenetic tree was constructed based on the alignment of 13,133 conserved amino-acid residues in 47 overlapped single and complete BUSCO protein sequences of 28 genomes by RAxML in a ML algorithm and 100 bootstrap replicates. The MSRs of the five lineages of mites and ticks are marked. *** P < 0.001.
Our phylogenomic analysis refuted the monophyly of mites and ticks (subclass: Acari) (Dabert et al. 2010; Pepato et al. 2010; Pepato and Klimov 2015; Lozano-Fernandez et al. 2019) and confirmed astigmatic mites as a rapidly evolving lineage under the monophyletic Acariformes. Massive horizontal gene transfer (HGT) events enabled gene novelties throughout the divergence of Acariformes with the involved genes (HGT genes) being perfect drug targets (Peixoto and Roos 2007; Manna and Harman 2016; Eme et al. 2017). After diverging from soil mites, astigmatic mites underwent rapid genome evolution and diversification through extensive gene family variations and frequent tandem gene duplications. These comprehensive findings should be useful for the future exploration of these astigmatic mites and the prevention of associated allergic diseases. Therefore, this comparative genomics study not only sheds light on the evolutionary origin and trajectory of allergic astigmatic mites but also paves the way for understanding the adaptation fundamentals and even designing control strategies for human household pests.
Results
Genome Assembly and Annotation
D. farinae and D. pteronyssinus are canonical house dust mites that cause human allergic diseases (Voorhorst et al. 1967; Peat et al. 1996; Thomas et al. 2002; Calderón, Linneberg, et al. 2015) (fig. 1A). P. ovis and S. scabiei are both parasitic mites of mammals; P. ovis is an ectoparasitic and non-burrowing mite mainly infesting in domesticated sheep (Sinclair and Kirkwood 1983; Hamilton et al. 2003), whereas S. scabiei can burrow into the skin of hosts such as humans and domestic dogs (Arlian 1989; Arlian and Morgan 2017) (fig. 1A). B. tropicalis was regarded as a canonical storage mite but is now considered a house dust mite mainly in tropical and subtropical areas (Guilleminault and Viala-Gastan 2017) (fig. 1A). T. putrescentiae is referred to as a mold mite or cheese mite and is considered a storage mite (Green and Woolcock 1978) (fig. 1A). In addition to D. farinae, D. pteronyssinus, B. tropicalis and T. putrescentiae, which are known as free-living allergic mites, two parasitic mites, P. ovis and S. scabiei, are also human allergy-related (Burgess et al. 2019; Shen et al. 2021).
Highly contiguous reference-quality genomes of four astigmatic mites (fig. 1A), D. farinae, D. pteronyssinus, B. tropicalis, and T. putrescentiae, were constructed using four next- and third-sequencing platforms (supplementary table S1, Supplementary Material online). The P. ovis genome deposited in the NCBI database (BioProject accession: PRJNA521406) was downloaded and reannotated, whereas raw sequencing data for S. scabiei (var. canis) genome were downloaded from the database (BioProject accession: PRJNA268368 and PRJNA304361), followed by reassembly and annotation. The completeness of these six genome assemblies ranged from 89.0% to 91.5%, and the N50 lengths ranged from 253,843 (S. scabiei) bp to 8,981,490 bp (D. farinae) (table 1). The genome sizes of the six astigmatic mites ranged from 56 Mb (S. scabiei) to 97 Mb (T. putrescentiae).
Table 1.
Overview of the Statistics of Genome Assemblies and Annotations of Six Astigmatic Mites.
| D. farinae | D. pteronyssinus | P. ovis | S. scabiei | B. tropicalis | T. putrescentiae | |
|---|---|---|---|---|---|---|
| Assembly features | ||||||
| Assembled genome size (bp) | 60,394,945 | 59,034,585 | 63,214,126 | 55,647,270 | 63,746,680 | 97,387,311 |
| BUSCOb completeness | 91.3% | 91.0% | 90.6% | 89.0% | 91.3% | 91.5% |
| BUSCOb duplication | 1.1% | 3.0% | 0.8% | 0.8% | 1.3% | 3.3% |
| Assembly type | scaffold | contig | contig | scaffold | scaffold | scaffold |
| Number of scaffolds or contigs | 10 | 131 | 93 | 574 | 116 | 176 |
| Maximum length | 16,911,843 | 3,357,599 | 5,538,194 | 1,295,475 | 11,105,103 | 6,764,852 |
| N50 length (bp) | 8,981,490 | 963,061 | 2,279,290 | 253,843 | 3,687,816 | 2,913,131 |
| N90 length (bp) | 4,381,017 | 219,916 | 605,092 | 58,941 | 311,152 | 467,811 |
| Total gap length (bp) | 274,781 | N. A.a | N. A.a | 19,550 | 318,601 | 747,038 |
| Annotation features | ||||||
| Number of genes | 15,457 | 14,999 | 14,688 | 14,763 | 16,869 | 23,793 |
| BUSCOb completeness | 92.0% | 91.9% | 90.9% | 91.1% | 92.9% | 90.7% |
| BUSCOb duplication | 2.7% | 4.5% | 1.4% | 0.9% | 2.4% | 4.6% |
| Repeat content | 15.79% | 14.24% | 18.87% | 9.03% | 9.54% | 13.03% |
N. A., not applicable.
BUSCO v3.1.0 with the database arthropoda_odb9.
A consistent MAKER annotation pipeline including both evidence-based annotation and ab initio prediction was performed on the six genomes of mites to avoid systematic biases. The completeness of the six proteomes ranged from 90.7% to 92.9% as assessed by BUSCO (Simão et al. 2015), showing the consistently high quality of annotations among the six astigmatic mite genomes (table 1). The protein-coding gene numbers of the six astigmatic mites ranged from 14,688 (P. ovis) to 23,793 (T. putrescentiae) (table 1). Notably, the canonical storage mite T. putrescentiae has approximately 55% more protein-coding genes than the other five mites.
Phylogenomic Analysis
It is controversial whether mites and ticks (subclass: Acari) comprising two major groups, Acariformes and Parasitiformes, are a monophyletic group (Dabert et al. 2010; Pepato et al. 2010; Pepato and Klimov 2015; Lozano-Fernandez et al. 2019). A phylogenomic analysis was performed with these six genome assemblies and other publicly available genomes (table 4, supplementary text S1, Supplementary Material online). In figure 1B, the phylogenetic tree of 26 mites and ticks, the pseudoscorpion Cordylochernes scorpioides and the fruit fly Drosophila melanogaster was constructed based on 13,133 conserved amino-acid residues in 47 overlapped single and complete BUSCO protein sequences and suggested that the phylogeny of Acariformes and Parasitiformes was interrupted by pseudoscorpion, which refuted the monophyly of mites and ticks. Meanwhile, the mean substitution rates (MSRs) of conserved amino-acid residues in the five lineages of mites and ticks were compared, and the significantly higher MSR (0.5632, P < 0.001, fig. 1B) of Astigmata confirmed astigmatic mites as a group of rapidly evolving species (Dabert et al. 2010).
General genomic features, including genome size, are summarized in (supplementary figure S1, Supplementary Material) online. Except for T. putrescentiae (97 Mb) and Euroglyphus (E.) maynei (with low completeness), the other astigmatic mites possess similar genome sizes of 60 ± 5 Mb, which are the smallest genome sizes among all sequenced genomes of mites and ticks (supplementary fig. S1, Supplementary Material online). In addition, repeat contents were compared (supplementary fig. S2, Supplementary Material online) and the genome sizes of five psoroptid mites (parvorder: Psoroptidia, D. farinae, D. pteronyssinus, P. ovis, S. scabiei var. suis and var. canis) were linearly correlated with total repeat contents or interspersed repeat contents (supplementary fig. S3, Supplementary Material online, R = 0.99 and 0.98, respectively). The publicly available genome of S. scabiei var. suis has a higher continuity but a lower completeness than that of S. scabiei var. canis (supplementary fig. S2, Supplementary Material online). Therefore, a subsequent study still used the re-assembled genome of S. scabiei var. canis for comparison. Based on conserved amino-acid alignment, an ultrametric time tree was constructed and revealed that these astigmatic mites evolved from oribatid mites ∼418 million years ago (MYA) and started diverging ∼278 MYA (supplementary fig. S4, Supplementary Material online).
Gene Family Evolution
To establish a well-resolved evolutionary analysis of the gene family, six proteomes generated from our annotated genomes, eight publicly available proteomes of oribatid, prostigmatic, and mesostigmatic mites (supplementary table S5, Supplementary Material online) and the proteome of D. melanogaster were assigned into orthogroups (or gene families) according to amino-acid sequence similarities.
A gene gain/loss analysis was performed, as shown in (figure 2A). We started our analysis from the monophyletic Acariformes, in which Prostigmata first evolved with four rapidly expanding gene families, including titin and sodium-dependent glucose transporter (supplementary table S6, Supplementary Material online), whereas only one uncharacterized protein family expanded in Oribatida and Astigmata (supplementary table S7, Supplementary Material online). Then, Oribatida further expanded 101 gene families, covering diverse detoxification and digestion gene families (supplementary table S8, Supplementary Material online). For the astigmatic mites, ten gene families rapidly expanded and enriched Gene Ontology (GO) terms related to ATP-binding cassette transporter (ABC), zinc finger, and ionotropic glutamate receptor (iGluR) (supplementary table S9, Supplementary Material online). Two orthogroups, cytochrome P450 clan 2 and glucosylceramidase contracted in astigmatic mites, with glucosylceramidase even decaying in S. scabiei (supplementary table S10 and supplementary text S2, Supplementary Material online).
Fig. 2.
Phylogenomic analysis revealed a wide range of genetic changes. (A) Analysis of gene gain or loss in the evolution of mites. The numbers in brackets on nodes indicate the number of orthogroups under expansion (+ and green) or contraction (- and red) and rapidly evolving orthogroups (* and blue). The ultrametric time tree was adapted from supplementary figure S4, Supplementary Material online, and the proteome of D. melanogaster (UniProt ID: UP000000803) was used as an outgroup. (B) Venn diagram of orthogroups of the six astigmatic mites. Proteomes were assigned into orthogroups using OrthoFinder2, and the overlapped orthogroups of the six astigmatic mites were then presented using a Venn diagram.
Within astigmatic mites, higher numbers of expanding and contracting gene families were found in T. putrescentiae and S. scabiei, respectively (fig. 2A). GO enrichment analysis of critical orthogroups involved in the evolution of the four psoroptid mites (D. farinae, D. pteronyssinus, P. ovis, S. scabiei) and the two canonical storage mites (B. tropicalis and T. putrescentiae) suggested divergent evolution of serine protease, iGluR, and various detoxification gene families including ABC transporter and cytochrome P450 (CYP) (supplementary tables S11–S21, supplementary text S2, Supplementary Material online). The gene loss of a phosphoenolpyruvate synthase (supplementary table S15, Supplementary Material online) in the psoroptid mites may be related to their ancestral parasitism-related metabolic changes (Klimov and Oconnor 2013). A wide range of rapidly expanding gene families were identified in T. putrescentiae, including several glycoside hydrolases, serine protease, heat shock protein 70 (HSP70), and detoxification gene families including UDP glucuronosyltransferase (UGT), CYP, and ABC (supplementary table 14, Supplementary Material online). In addition, other gene families with less apparent functions were also enriched and are discussed in (supplementary text S2, Supplementary Material online).
Species-specific orthogroups were identified in a Venn diagram (fig. 2B) and analyzed in GO enrichment (supplementary table S22–S28, Supplementary Material online). Notably, T. putrescentiae has a much larger number of species-specific orthogroups, including many proteases, glycoside hydrolases, CYP, and iGluRs, suggesting that this storage mite underwent more sophisticated adaptation during evolution. In addition, GO enrichment revealed species-specific serine proteases in B. tropicalis. Based on the analysis of gene family evolution, many gene families were further compared, especially the frequently enriched iGluR, digestive enzymes, and detoxification gene families.
Ionotropic Glutamate Receptors
The iGluR family is well known to sense a wide range of environmental changes or signals such as temperature, moisture, and taste (Rytz et al. 2013; van Giesen and Garrity 2017). All iGluRs of the six mites were collected for phylogenetic analysis (fig. 3A), in which P. ovis and S. scabiei displayed significantly fewer iGluR genes than the other four mites. The iGluRs in small Clusters a and b (fig. 3A) were identified as IR25a and IR93a for the phylogenetic analysis (fig. 3B), and their highly conserved regions showed homology to those of IR25a and IR93a of D. melanogaster (supplementary fig. S6, Supplementary Material online), respectively. Interestingly, IR25a and IR93a were adjacent in all six astigmatic mites (fig. 3B) but are located on two different chromosomes in the genome of D. melanogaster (chromosomes 2L and 3R, respectively).
Fig. 3.
Frequent tandem gene duplications of iGluRs. (A) Phylogenetic analysis of all iGluRs in the six astigmatic mites. All tandemly arrayed genes are connected using curved solid lines, whereas all proximally arrayed genes (separated by not more than 10 genes) are connected using dotted lines. Two large Clusters X and Y were highlighted, in which Cluster Y was further classified into Subclusters 1 and 2 (including the subcluster 2-1). Two small Clusters, a and b, were identified as IR25a and IR93a, respectively. (B) Phylogenetic analysis and similarity matrix of two adjacent iGluRs, IR25a and IR93a, of the six astigmatic mites. IR25a and IR93a of D. melanogaster were used as references. Tandem arrayed IR25a and IR93a were linked by solid curves in all six mites. The similarity matrix (BLOSUM62) was generated using the online tool SIAS with default parameters. (C) Gene synteny alignment of iGluRs in subcluster 2-1. Tandem gene duplications are highlighted; for example, such as five tandemly arrayed iGluRs in D. farinae are marked as iGluR (x5). The black turn arrow indicates reverse complement, and the red dotted lines indicate the genes of S. scabiei located on the opposite strand.
Frequent tandem gene duplications were observed in the iGluRs (fig. 3A), in which of a total of 285 iGluRs, 116 genes (40.7%) were tandemly arrayed and 37 genes (13.0%) were proximally arrayed (fig. 3A). Most tandem duplications occurred in Cluster Y, while gene clusters were evenly distributed among the six mites in Cluster X (fig. 3A). In Subcluster 1 under Cluster Y, more iGluR genes were identified in the two canonical storage mites than in the two house dust mites, whereas only one iGluR gene in P. ovis and none in S. scabiei were identified. In Subcluster 2, massive tandem duplications were observed, but only one iGluR gene was identified in T. putrescentiae. Since T. putrescentiae is the only storage mite and is far from humans or mammals, unlike the other five mites, we propose Cluster 2 iGluRs function in sensing signals related to humans or mammals. In Subcluster 2-1, gene synteny alignment identified tandemly arrayed iGluRs in D. farinae, D. pteronyssinus, S. scabiei and B. tropicalis, but only one iGluR each in P. ovis and T. putrescentiae (fig. 3C). Notably, nine divergent and tandemly arrayed iGluR genes were identified in B. tropicalis, while only one was identified in T. putrescentiae at the same gene locus (fig. 3C).
Among all identified iGluRs in this study, IR25a and IR93a (fig. 3B) are particularly important, because the essential function of these two genes in temperature and humidity sensing of the honeybee parasitic mite T. mercedesae has been demonstrated by rescuing temperature and humidity preference defects in fruit fly D. melanogaster IR25a and IR93a mutants (Lei et al. 2019). The gene family variations of HSP70s and aquaporins are shown in supplementary figure S7 and table S29, Supplementary Material online (supplementary text S3, Supplementary Material online).
Major Digestive Enzymes
The major digestive enzymes of the six astigmatic mites are summarized in (supplementary table S30, Supplementary Material online). Alpha amylase (EC 3.2.1.1) acts in the first step in the hydrolysis of starch to dextrin, and then alpha glucosidase (EC 3.2.1.20) catalyzes the hydrolysis of dextrin to glucose. No alpha amylase gene was identified in S. scabiei because of gene synteny, while the other four astigmatic mites except T. putrescentiae have duplicated alpha amylases (supplementary fig. S8A and B, Supplementary Material online). As a canonical storage mite, T. putrescentiae possessed more alpha glucosidases (21 genes) than the other five astigmatic mites, and gene expansion and diversification observed in the phylogenetic analysis (supplementary fig. S8C, Supplementary Material online) are consistent with its preference for a high-carbohydrate diet, as a canonical storage mite (Erban et al. 2009). Unexpectedly, alpha amylase was duplicated in D. farinae, D. pteronyssinus, P. ovis, and B. tropicalis but not the storage mite T. putrescentiae (supplementary fig. S8, Supplementary Material online). According to our metagenomic analysis, Neurospora crassa (a starch utilizer with highly stable amylase) (Tavano et al. 2008) was identified as the most abundant microorganism in T. putrescentiae (5.26% in abundance) but was not found in the other five astigmatic mites, implying that the alpha amylase of Neurospora crassa is an alternative to the alpha amylase gene duplication in T. putrescentiae.
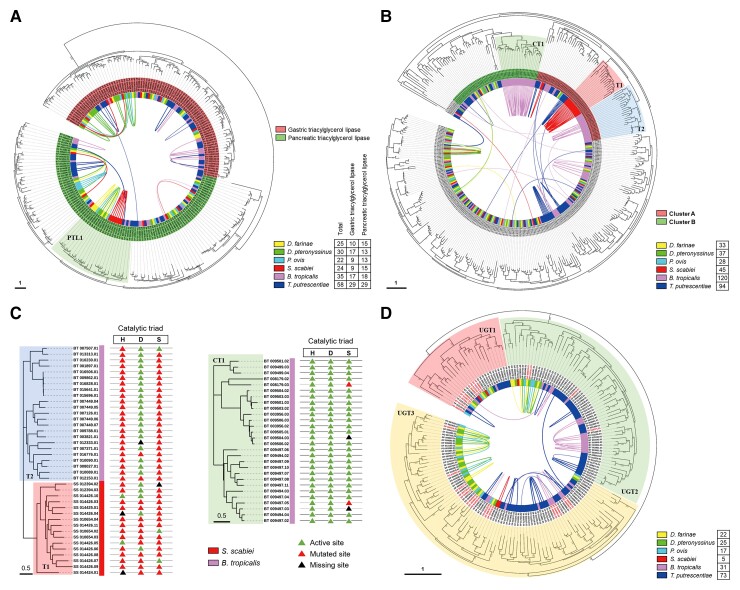
More genes of triacylglycerol lipase were identified in T. putrescentiae (58 genes), which was consistent with the fact that T. putrescentiae is referred to as a cheese mite and prefers food with high fat content (Erban et al. 2015). The phylogenetic tree of all triacylglycerol lipases of the six astigmatic mites confirmed the gene expansion in T. putrescentiae (fig. 4A). Among all 195 triacylglycerol lipase genes, 67 genes (34.4%) were tandemly arrayed and 11 genes (5.6%) were proximally arrayed (fig. 4A). In the marked Cluster PTL1, massive tandem gene duplications were identified in the four psoroptid mites, including 7 tandemly arrayed genes in S. scabiei. Of the 360 serine protease genes, 81 genes (22.5%) were tandemly arrayed and 33 genes (9.2%) were proximally arrayed (fig. 4B). In the phylogenetic tree (fig. 4B), two clusters of B. tropicalis-specific (CT1 and T2) and one cluster of S. scabiei-specific (T1) serine proteases were identified. None of the serine proteases in Clusters T1 and T2 possessed a complete and active catalytic triad, while 24 out of 28 serine proteases in Cluster CT1 possessed complete and active catalytic triads (Holt et al. 2003) (fig. 4C).
Fig. 4.
Remarkable tandem gene duplications in digestion and detoxification gene families. (A) Phylogenetic analysis and tandem gene duplications of triacylglycerol lipases in the six mites. Tandemly and proximally arrayed genes of glutamate receptors in six mites were connected using curved solid lines and dotted lines, respectively. A specific cluster with tandem gene duplications was marked as PTL1. (B) Phylogenetic analysis and tandem gene duplications of serine proteases. All serine proteases of the six astigmatic mites were identified and analyzed in a phylogenetic tree. Three species-specific clusters (T1, T2, and CT1) of serine proteases are highlighted. (C) Alignment of catalytic triads of serine proteases in three species-specific clusters. Three species-specific clusters of serine proteases in (figure 4B) were collected and aligned for their catalytic triads (H, histidine; D, aspartic acid; S, serine). Active, mutated, and missing sites are marked with colored triangles. (D) Phylogenetic analysis and tandem gene duplications of UGTs. UGTs of the six astigmatic mites were analyzed in a phylogenetic tree and divided into three large clusters, UGT1–3.
Detoxification Gene Families
Of the five major gene families responsible for detoxification, T. putrescentiae had the largest number of genes, while S. scabiei had the smallest (supplementary fig. S9, Supplementary Material online). The huge increase in the number of detoxification genes in the storage mite T. putrescentiae indicates its higher probability of encountering xenobiotics in living environments. Concurrent with the results of GO enrichment in T. putrescentiae, 85 CYP genes were identified, which was more than those in the other five mites. In the CYP family, the clans CYP3 and CYP4 are mainly responsible for the detoxification of xenobiotics (Zhang et al. 2018). Significant expansion in the CYP3 and CYP4 clans was observed in T. putrescentiae, while expansion only in the CYP3 clan was observed in B. tropicalis (supplementary fig. S10A, Supplementary Material online). S. scabiei had the fewest genes in both CYP3 and CYP4, with gene decay of the CYP4 gene (supplementary fig. S10B, Supplementary Material online). In the ABC family, D. farinae has more genes than D. pteronyssinus and P. ovis. The gene number variation mainly occurred in the G and C families, and both of the G and C families are related to drug resistance (Liu 2019). As visible in the highlighted Cluster 1 in supplementary figure S10C, Supplementary Material online, distinct and independent gene expansions of the ABCG family genes occurred among the six astigmatic mites. Similarly, an ABCB gene decayed in S. scabiei (supplementary fig. S10D, Supplementary Material online).
The variation in gene numbers was particularly apparent in the UGT family (supplementary fig. S9, Supplementary Material online). There were only 5 UGT genes identified in S. scabiei but 73 UGT genes in T. putrescentiae. To explore how the gene number varied among the six astigmatic mites, phylogenetic analysis was performed on all UGT genes of the six astigmatic mites (fig. 4D). Of the 172 UGTs, 73 genes (42.2%) were tandemly arrayed and 9 genes (5.2%) were proximally arrayed. Among them, 28 UGTs were single-exon genes (16.3% of the total 172 UGTs, highlighted with red labels). After we divided all the UGTs of the six astigmatic mites into three large clusters (UGT1–3), remarkable expansions in the two canonical storage mites B. tropicalis and T. putrescentiae were observed in UGT2 and UGT3, while significant gene expansions in the two house dust mites D. farinae and D. pteronyssinus were identified mainly in UGT3. However, the functional difference among these UGT clusters is still unknown.
Horizontal Gene Transfer
HGT is the movement of genetic information across mating barriers, such as DNA sharing between bacterial and animal genomes (Soucy et al. 2015; Husnik and McCutcheon 2018). A modified method based on Crisp (Crisp et al. 2015) was used to search HGT events, and 12 HGT genes of astigmatic mites were identified in five functional categories (table 2). All 12 HGT genes had the best match hits from unusual taxonomic groups, in which those of seven genes were from species mainly dwelling in soil environments including Actinobacteria, slime mold, and Bacillus (table 2). The conservation of these HGT genes was explored in Oribatida, Prostigmata, and springtails (supplementary table S32, Supplementary Material online). As many as 10 HGT genes were conserved in oribatid mites (supplementary table S32, Supplementary Material online), supporting that astigmatic mites evolved from soil oribatid mites, which is consistent with previous reports (Norton 1998; Sakata and Norton 2001; Domes et al. 2007).
Table 2.
HGT Events Identified in the Six Astigmatic Mites.
| Functional category | Description | Representativea | Length/aa | Best hit in NRb | Copy number | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E-value | Identity | Taxonomy | Df c | Dp c | Po c | Ss c | Bt c | Tp c | ||||
| Detoxification | UGT | DF_011775.01 | 430 | 2E−87 | 37.3% | Bacteria, Actinobacteria | 22 | 25 | 17 | 5 | 31 | 73 |
| Lysis of fungal cell wall | Chitinase 1 | DF_010318.01 | 459 | 5E−98 | 52.1% | Bacteria, Actinobacteria | 1 | 1 | 1 | 0 | 1 | 1 |
| Chitinase 2 | DF_000907.01 | 403 | 8E−102 | 55.4% | Bacteria, Actinobacteria | 1 | 1 | 1 | 1 | 1 | 1 | |
| Beta-1,3 glucanase | DF_001158.01 | 280 | 1E−65 | 43.1% | Eukaryota, Slime mold | 6 | 6 | 3 | 2 | 6 | 11 | |
| Chitosanase | DF_009713.01 | 259 | 1E−72 | 43.1% | Bacteria, Bacillus | 1 | 2 | 1 | 1 | 1 | 1 | |
| Lysis of bacterial cell wall | Peptidoglycan endopeptidase | DF_002569.01 | 150 | 2E−26 | 48.6% | Eukaryota, Fungi | 1 | 1 | 2 | 0 | 1 | 3 |
| Endolytic peptidoglycan transglycosylase | DF_000506.01 | 111 | 1E−26 | 50.1% | Bacteria, Sediminibacterium | 1 | 1 | 0 | 0 | 2 | 3 | |
| Bacterial resistance | Stress response protein | DF_013478.01 | 303 | 7E−28 | 49.2% | Bacteria, Bacillus | 1 | 1 | 1 | 1 | 1 | 1 |
| D-alanyl-D-alanine dipeptidase | BT_004312.01 | 352 | 2E−60 | 44.3% | Bacteria, Proteobacteria | 0 | 0 | 0 | 0 | 1 | 2 | |
| Others | Trans-aconitate 2-methyltransferase | DF_011983.01 | 310 | 1E−15 | 31.7% | Bacteria, Proteobacteria | 1 | 1 | 1 | 1 | 1 | 1 |
| Terpene synthase | DF_008370.02 | 415 | 9E−16 | 22.5% | Bacteria, Actinobacteria | 1 | 1 | 1 | 1 | 1 | 1 | |
| RNA 2′-phosphotransferase | DF_001392.01 | 190 | 2E−14 | 34.5% | Archaea, TACK group | 1 | 1 | 1 | 1 | 1 | 1 | |
One gene for each HGT event was selected as a representative gene to present the basic gene features. Three single-exon genes are marked with underlines. The protein sequences coded by these representative genes can be found in supplementary table S31, Supplementary Material online.
Best hit in the NCBI NR database (excluding Acariformes and springtails) with BLASTP. In additionally, rotifers were excluded for UGT and D-alanyl-D-alanine dipeptidase. More conservation information can be found in supplementary table S32, Supplementary Material online and best hits in other taxonomic groups were compared in supplementary table S33, Supplementary Material online. Updated on April 6th, 2022.
Df, D. farinae; Dp, D. pteronyssinus; Po, P. ovis; Ss, S. scabiei; Bt, B. tropicalis; Tp, T. putrescentiae.
Detoxification and UGTs
Phylogenetic analysis (fig. 5A) revealed that all three clusters of UGTs (fig. 4D) in the six astigmatic mites were clustered with those of two oribatid mites, three prostigmatic mites, rotifers, the springtail Folsomia and bacteria but not the closest UGTs of other arthropods (supplementary table S32, Supplementary Material online). The HGT of UGTs has been reported in bdelloid rotifers (Gladyshev et al. 2008). In the phylogenetic tree (fig. 5A), the UGT1 cluster was suggested as the outgroup to the other two clusters, UGT2 and UGT3. Therefore, we conclude that all UGTs of astigmatic mites were gained via HGT from some bacteria and suggest UGT1 as the most ancient cluster. Combined with the tandemly arrayed UGTs from B. tropicalis and T. putrescentiae among different clusters (fig. 4D), we speculate that the other two clusters of UGTs evolved from UGT1s through duplication and diversification. Since UGTs are important conjugative enzymes in detoxification, we consider that the HGT of UGTs in astigmatic mites contributes to their detoxification functions.
Fig. 5.
Phylogenetic analysis of HGT genes, UGTs and chitinases. (A) Phylogenetic analysis of UGTs from the six astigmatic mites. The closest UGTs from other taxonomic groups in the UniRef50 database, including those from bacteria, fungi, plants, other arthropods (excluding Acariformes) and other metazoa (excluding arthropods), were collected for the analysis with the UGTs of Acariformes. The rotifer sequences were collected from the NR database. (B) Phylogenetic analysis of chitinases in Cluster 1 (supplementary fig. S11A, Supplementary Material online) from the six astigmatic mites. The closest chitinases in the UniRef50 database from other taxonomic groups were collected for comparison of the chitinases of astigmatic and oribatid mites. All protein sequence accessions are listed in (supplementary tables S34 and S35, Supplementary Material online).
Lysis of Fungal Cell Wall
HGTs involving a group of fungal cell wall lytic enzymes (table 2) were also identified in astigmatic mites, including two chitinases, three beta-1,3 glucanases and a chitosanase. It was reported in cotton that both chitinases and beta-1,3 glucanases participate in resistance to fungi (Xu et al. 2016). Chitosanase breaks down chitosan, one of the major components of the fungal cell wall (Fukamizo and Brzezinski 1997).
Two clusters of chitinases (supplementary fig. S11A, Supplementary Material online) in astigmatic mites were identified as HGT genes (table 2). The chitinases in Cluster 1 (chitinase 1) are close to those of Actinobacteria and oribatid mites but not other arthropods (fig. 5B and supplementary fig. S11B, Supplementary Material online), whereas the chitinases in Cluster 2 (chitinase 2) have protein homologs to those in other Acariformes species (oribatid and prostigmatic mites) and springtails (supplementary fig. S11B, table S32, Supplementary Material online). The phylogenetic analysis (supplementary fig. S11B, Supplementary Material online) supported that the two clusters of chitinases were two HGT genes independently originating from bacteria. The gene syntenies and splice sites (supplementary fig. S11C, Supplementary Material online) further supported the high conservation of the two chitinases among astigmatic mites. Gene synteny (supplementary fig. S11C, Supplementary Material online) also revealed a gene decay event of chitinase 1 in the genome of S. scabiei.
All beta-1,3 glucanases of mites could be divided into three clusters (supplementary fig. S12A, Supplementary Material online), of which Cluster 3 was expanded in D. farinae, D. pteronyssinus, B. tropicalis, and T. putrescentiae. Notably, all beta-1,3 glucanases in Cluster 3 are single-exon genes, except seven genes of T. putrescentiae (supplementary fig. S12A, Supplementary Material online). Initially, we considered three clusters of beta-1,3 glucanases to be independent HGT genes because they were all unusually matched to protein sequences from slime mold or bacteria; however, in further phylogenetic analysis of three clusters of beta-1,3 glucanases and their respective top matched protein sequences in the NCBI nr database (supplementary fig. S12B, Supplementary Material online), only Cluster 3 genes were clustered with those of unusual taxa, including slime mold and bacteria, whereas the Cluster 2 genes were clustered with those of oribatid mites, and the Cluster 1 genes were unique in astigmatic mites. This result suggested that the beta-1,3 glucanases in Cluster 3 are the original HGT genes, whereas those of the other two clusters are likely generated from Cluster 3 via duplication and mutation. In gene synteny alignment, we identified that an HGT-gained peptidoglycan endopeptidase was adjacent to the beta-1,3 glucanase in Cluster 1 (supplementary fig. S12C, Supplementary Material online). The absence of the S. scabiei gene in Cluster 3 was possibly caused by a gene decay event of Cluster 3-1 beta-1,3 glucanase and the triple tandemly arrayed beta-1,3 glucanases (Cluster 3-2) are specific in D. farinae and D. pteronyssinus (supplementary fig. S12D, Supplementary Material online). The triple tandemly arrayed beta-1,3 glucanases were considered nonexistent in D. farinae in a previous report (Waldron et al. 2019). The beta-1,3 glucanase of astigmatic mites has homologs from oribatid mites and springtails (supplementary table S32, Supplementary Material online).
Seven chitosanases of the six astigmatic mites (two genes in D. pteronyssinus but only one each in the other five species) were identified as HGT genes with 43.1% identity to that of Bacillus subtilis (UniProt accession: O07921) (table 2, supplementary fig. S13A, Supplementary Material online). To further confirm the HGT event, the seven chitosanases of astigmatic mites, two of oribatid mites and those reviewed in the UniProt database were phylogenetically analyzed (supplementary fig. S13A, Supplementary Material online). Chitosanases of the astigmatic and oribatid mites are clustered with those of bacteria (supplementary fig. S13A, Supplementary Material online). Gene synteny alignment and conserved splice sites (supplementary fig. S13B, Supplementary Material online) confirmed that the chitosanases of astigmatic mites were from a single HGT event. Three chitosanases, DF_Chitosanase, DP_Chitosanase-1, and PO_Chitosanase shared conserved gene synteny (supplementary fig. S13B, Supplementary Material online). For the other 4 chitosanases, conserved gene syntenies were observed among DP_Chitosanase-2, SS_Chitosanase, BT_Chitosanase, and TP_Chitosanase, but at the same gene locus, no chitosanase was identified in P. ovis and D. farinae (supplementary fig. S13B, Supplementary Material online). Therefore, we suggest that the chitosanase duplicated before the divergence of P. ovis, D. farinae, and D. pteronyssinus and then decayed in P. ovis and D. farinae but not D. pteronyssinus.
Lysis of Bacterial Cell Wall
Two HGT genes, peptidoglycan endopeptidase and endolytic peptidoglycan transglycosylase (table 2), are related to the lysis of peptidoglycan, the main component of the bacterial cell wall (Meroueh et al. 2006; Lee et al. 2018; Shin et al. 2020). Conserved gene synteny (supplementary fig. S12C, Supplementary Material online) revealed that the peptidoglycan endopeptidase and beta-1,3 glucanase in Cluster 1 (supplementary fig. S12A, Supplementary Material online) were tandemly arrayed in the genomes of the six astigmatic mites. The peptidoglycan endopeptidase was tandemly duplicated in both P. ovis and T. putrescentiae but decayed in S. scabiei (supplementary fig. S12C, Supplementary Material online). This peptidoglycan endopeptidase has been reported and explored in several astigmatic mites (Mathaba et al. 2002; Erban et al. 2013; Chan et al. 2015; Tang et al. 2015, 2017).
Discussion
Mites and ticks (subclass: Acari) comprise a wide range of pests of humans and other animals, and plants (Varma 1993; Xiong et al. 2020). Their monophyly was debated with more genetic resources available (Dabert et al. 2010; Pepato et al. 2010; Pepato and Klimov 2015; Lozano-Fernandez et al. 2019) and refuted by our phylogenomic analysis (fig. 1B). Our study suggests that Acariformes is a monophyletic group and further explored its divergence (fig. 6). Solifugae was suggested to be the sister group of Acariformes (Dabert et al. 2010), but the absence of genome assembly impedes our phylogenetic validation. Prior to the divergence of Acariformes and Parasitiformes, it was as short as only 1.93 million years after the root age with the outgroup D. melanogaster, which could not well support the monophyly like the situation in Deuterostomia (Kapli et al. 2021). Moreover, the three Parasitiformes mites shared more overlapping orthogroups with D. melanogaster than with Acariformes (supplementary fig. S5, Supplementary Material online), which revealed the large gene family difference between these two groups. In addition, no conserved HGT events with Parasitiformes species could support the monophyly.
Fig. 6.
Graphical illustration of the evolutionary history of astigmatic mites. In the monophyletic Acariformes, Prostigmata (known as notorious plant pests) first branched out, and then Astigmata evolved from Oribatida (known as soil mites). HGT-gained genes are shown in blue font, with UGT and fungal cell wall lytic enzymes bolded. In the phylogenetic tree of astigmatic mites, colored tables show the variation events of 12 gene families in three categories (corresponding to the table in the upper left corner). Red box, gene loss; green box, gene gain; star symbol *, tandem gene duplication; blank box, no change identified.
The evolutionary history of astigmatic mites was illustrated from the divergence of monophyletic Acariformes to their diversification, as shown in figure 6. Two HGT genes (UGT and chitinase 2) conserved among all three Acariformes lineages (Astigmata, Oribatida and Prostigmata) have homologs from springtail species known as abundant soil dwelling arthropods (supplementary table S32, Supplementary Material online). Because cariformes could not be the sister group of springtails for Arachnida monophyly (Howard et al. 2020) but shared common HGT genes, we deduced that the primitive Acariformes species live in the same soil environments as springtail species. In the monophyletic Acariformes, prostigmatic mites (including notorious plant pests) first branched out (fig. 6 and supplementary fig. S4, Supplementary Material online). Intriguingly, two gene families of titin and sodium-dependent glucose transporter rapidly expanded in Prostigmata (supplementary table S6, Supplementary Material online), which may contribute to their better locomotion for climbing on plants and absorption of glucose, respectively. Because the HGT gene beta-1,3 glucanase in Cluster 3 is also conserved in astigmatic and oribatid mites with springtails (supplementary fig. S12B, supplementary table S32, Supplementary Material online), we suggest that the common ancestor of astigmatic and oribatid mites dwelled in soil and that the rapidly evolving astigmatic mites (fig. 1B) diverged from Oribatida (soil mites).
HGT could contribute to a wide range of gene novelties in genomes (Nakabachi 2015; Simakov et al. 2015; Soucy et al. 2015; Husnik and McCutcheon 2018). Massive HGT events occurred throughout the divergence history of Acariformes and enabled many gene novelties (fig. 6, table 2). UGT, chitinase 2 (Cluster 2 in supplementary fig. S11A, Supplementary Material online) and terpene synthase (fig. 6) were gained through HGT in the ancestral Acariformes (before 463 MYA, supplementary fig. S4, Supplementary Material online). After the divergence of Prostigmata, the common ancestor of Oribatida and Astigmata (418–463 MYA, supplementary fig. S4, Supplementary Material online) acquired beta-1,3 glucanases, the other bacterial chitinase (chitinase 1), a chitosanase, a peptidoglycan endopeptidase and three other HGT genes (fig. 6). Chitinase, beta-1,3 glucanases and chitosanase play key roles in the digestion of fungal cell walls, while peptidoglycan endopeptidase participates in the lysis of the bacterial cell wall. These HGT-gained lytic enzymes for fungal and bacterial cell walls are considered important in the digestive functions of astigmatic mites after they settle in later established human household environments; in particular, house dust mites D. farinae and D. pteronyssinus are found feeding on human skin flakes colonized by microbes such as fungi and bacteria (Colloff et al. 1992; Colloff 2009; Portnoy et al. 2013). An RNA 2′-phosphotransferase is the only conserved HGT gene in astigmatic mites, and a D-alanyl-D-alanine dipeptidase is the specific HGT gene in two canonical storage mites B. tropicalis and T. putrescentiae (fig. 6, table 2 and supplementary Table S32, Supplementary Material online).
In astigmatic mites, except for the HGT peptidoglycan endopeptidase, which was identified and explored (Mathaba et al. 2002; Erban et al. 2013; Chan et al. 2015; Tang et al. 2015, 2017), very few HGT events were reported previously. UGTs, chitinases, and other genes were reported as HGT genes in spider mites (Ahn et al. 2014; Wybouw et al. 2018; Snoeck et al. 2019). In this study, we identified many novel HGT events in Acariformes and provide perfect drug targets for the control of these mites (Peixoto and Roos 2007; Manna and Harman 2016; Eme et al. 2017). These HGT events were possibly mediated by some mite endosymbionts such as Wolbachia (Klasson et al. 2009; Husnik and McCutcheon 2018; de Miguel et al. 2019). Although 12 HGT events were identified in this study, only some of them were further analyzed.
Astigmatic mites are a group of rapidly evolving species with significantly higher MSRs (fig. 1B) than those of other lineages. Astigmatic mites evolved from Oribatida with rapid expansions of iGluRs and ABCs (supplementary table S9, Supplementary Material online), entered nests of birds and mammals and possibly became commensals of the host animals. We propose that astigmatic mites experienced two rounds of divergence. In the first round (approximately 278 MYA, supplementary fig. S4, Supplementary Material online), the ancestor of free-living storage mites, including Glycyphagoidea (e.g., B. tropicalis) and Acaroidea (e.g., T. putrescentiae), branched out with more gene family expansions especially in detoxification gene families (fig. 6) and split from the ancestor of psoroptid mites (parvorder: Psoroptidia) which are mostly parasitic mites. For the second round (approximately 212 MYA, supplementary fig. S4, Supplementary Material online), psoroptid mites diversified with tandemly duplicated triacylglycerol lipases and serine proteases (fig. 6). Afterward, with gene decay (fig. 6), the skin-burrowing mite S. scabiei developed a more obligate parasitic lifestyle than the sheep scab mite P. ovis, an ectoparasitic and non-burrowing mite mainly infesting on the fleece of sheep, while via gene family expansions (fig. 6, supplementary table S20, Supplementary Material online), the ancestor of house dust mites (e.g., D. farinae and D. pteronyssinus) diverged within Psoroptidia. Because psoroptid mites are closely related to the bodies of birds and mammals (Proctor 2003), the second round of divergence (approximately 212 MYA, supplementary fig. S4, Supplementary Material online) was considered a result of the emergence of feathers and hair, especially when the origin of feathers was estimated to be ∼165–250 MYA (Benton et al. 2019) and mammals appeared at least 178 MYA (Pickrell 2019). Because nonparasitic mites in the family Pyroglyphidae (including house dust mites) mainly live in the nests of birds and mammals (Arlian 2009), we suggest that massive gene duplications enabled their reversal (approximately 145 MYA, supplementary fig. S4, Supplementary Material online) from parasitic on animal bodies to free-living in surrounding environments (Klimov and Oconnor 2013).
In the diversification of astigmatic mites, a wide range of genetic variations in the genomes of six mites, including many gene family expansions via tandem gene duplications (Innan and Kondrashov 2010; Reams and Roth 2015), enabled their rapid genome evolution by acquiring new genes and quick adaptation to rather newly established and rapidly changing human household environments; for example, the expansions of five detoxification gene families in the storage mite T. putrescentiae could enable its tolerance to relatively high levels of toxins and contaminants. The gene family expansion of iGluRs frequently occurred in different mite lineages, which contributed to their more specific sensing of signals in living environments. In addition, digestion enzymes, especially triacylglycerol lipase and serine protease, were tandemly duplicated in astigmatic mites (fig. 4A and B). Extensive tandem gene duplications of triacylglycerol lipases in the six astigmatic mites facilitate the survival of these mites on a relatively high fat content in food (fig. 4A) (Klimov and Oconnor 2013). Regarding the species-specific inactive serine proteases identified in S. scabiei and B. tropicalis (fig. 4B and C), the role of those proteases in S. scabiei has been reported to be associated with immune evasion or inhibition of the host immune system (Holt et al. 2003; Bergström et al. 2009; Reynolds et al. 2014). It is possible that these proteases assist the canonical storage mite, but now the tropical house dust mite B. tropicalis better adapts to house dust environments than other storage mites, such as T. putrescentiae (Guilleminault and Viala-Gastan 2017; Santos da Silva et al. 2017). In the same line of thought, a new cluster of active serine proteases in B. tropicalis (fig. 4B) could be responsible for cleavage of tight junctions between epithelial cells in human skin, thereby contributing to its rising rates of allergy in the past decades (Wan et al. 2001; Santos da Silva et al. 2017). Meanwhile, many gene decay events occurred in both digestion and detoxification gene families of the skin-burrowing mite S. scabiei (fig. 6) because of its parasitic fate. There should be more diverse gene family variations participating in mite evolution, but this study could only cover these significantly enriched gene families.
Notably, in this study, even the most recent divergence between S. scabiei var. suis and var. canis occurred ∼23 MYA (supplementary fig. S4, Supplementary Material online), which is much earlier than the emergence of Homo sapiens (0.4–0.7 MYA) (Stringer 2016). Therefore, except for some species-specific events, such as the tandem duplications of serine proteases in B. tropicalis with uncertain occurrence times, all genetic events (fig. 6), including the HGTs and gene family variations in astigmatic mites, occurred prior to the establishment of human household environments. Therefore, we suggest that these genetic variations are not evolutionary responses to human household-related selection pressure, but possibly the genetic basis for the quick adaptation of these astigmatic mites to later established human household environments.
Conclusions
In this study, high-quality genomes of six allergic astigmatic mites contribute to a comparative genomics model for exploring their evolution and diversification. In the monophyletic Acariformes, prostigmatic mites known as plant pests first evolved from the primitive mites dwelling in soil environments; then, the rapidly evolving astigmatic mites diverged from soil oribatid mites and later diversified into storage mites, parasitic mites, and house dust mites. From Acariformes divergence to Astigmata diversification, many HGT events introduced functionally important genes into their genomes and enriched their adaptation capacities, especially in detoxification and digestion, and provided perfect drug targets for pest control. Within astigmatic mites, a wide range of gene family expansions via tandem duplications, especially in free-living mites, facilitated their rapid divergent evolution and quick adaptation to the rather new and rapidly changing human household environments, whereas many genes decayed in the parasitic mite S. scabiei as a result of parasitism. This comparative genomics study comprehensively illustrated the evolutionary dynamics of these allergic astigmatic mites and revealed the genetic fundamentals for how these mites diverge from soil mites, rapidly evolve, and quickly adapt to the human household environments, which vigorously expanded our knowledge to these medically important species and would ultimately facilitate the prevention of associated allergic diseases. Massive genomic insights into the adaptation of human household pests were provided by the divergent evolution of astigmatic mites and would be important for the further exploration and even design of effective pest control strategies.
Materials and Methods
Mite Culture and Purity Check
D. farinae, D. pteronyssinus, and T. putrescentiae were cultured in Shenzhen University (Shenzhen, China). The culture methods of D. farinae and D. pteronyssinus could be found in publications (Chan et al. 2015; Liu et al. 2018), and T. putrescentiae was cultured in the same way. Mite culture of B. tropicalis was performed in the Siriraj Dust Mite Center for Services and Research, Siriraj Hospital, Bangkok, Thailand. B. tropicalis mites were fed in the mixture of rat chow and wheat germ and cultured under 25 ± 3 °C and 75 ± 5% relative humidity. After the harvest of pure mite bodies of four species from the mite culture, monospecies of mites was confirmed during cultivation and at the final stage by observation of the morphology under a light microscope and verified with the pictorial keys (Colloff and Spieksma 1992). Furthermore, the species was confirmed by singleplex PCR with species-specific primers after genomic DNA extraction.
Sample Preparation of Genomic DNA and RNA
Genomic DNAs were extracted using the Qiagen Blood & Cell Culture DNA Maxi Kit (Qiagen, Germany). First, pure mite bodies were washed twice with Phosphate-buffered saline (PBS, pH 7.4) and homogenized into powder form by mortar and pestle with liquid nitrogen for keeping low temperature. The homogenized mite samples were then incubated at 50 °C for proteolysis following the manufacturer’s protocol. Genomic DNA was bound to the column, washed, and eluted by elution buffer, and precipitated in 70% ethanol cold in 4 °C. Finally, air-dry pellets of genomic DNA were dissolved overnight in UltraPure™ DNase/RNase-free distilled water (Thermo Fisher Scientific, USA) overnight at room temperature. The integrity of genomic DNA was determined by electrophoresis in 0.5% agarose gel and analyzed by Agilent 2100 Bioanalyzer (Agilent Technologies, USA), and the quantity was detected by Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) and Qubit Fluorometer (Thermo Fisher Scientific, USA).
For RNA extraction, the homogenized mite powder was transferred into tubes with TRIzol reagent (Invitrogen, USA), followed by phenol-chloroform extraction. The colorless upper aqueous phase was transferred to the column from the PureLink RNA Mini Kit (Thermo Fisher Scientific, USA) following the manufacturer’s protocol. After extraction, the concentration of RNA samples was quantified by Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) and Qubit Fluorometer (Thermo Fisher Scientific, USA). Then, with the RNA samples, SMARTer™ PCR cDNA Synthesis Kit (Takara Bio, Japan) was used to synthesize double-stranded cDNA. Both integrities of RNA and double-stranded cDNA were qualified by Agilent 2100 Bioanalyzer (Agilent Technologies, USA).
Genome and Transcriptome Sequencing
To assemble the genomes of D. farinae, D. pteronyssinus, T. putrescentiae and B. tropicalis, a variety of sequencing platforms were employed, and all new data of genomic DNA sequencing are listed in (supplementary table S1, Supplementary Material online). The genome sequencing data of S. scabiei (var. canis) were downloaded from NCBI database (BioProject accession: PRJNA268368). Besides, genome sequencing data of D. farinae and D. pteronyssinus (BioProject accession: PRJNA174061 and PRJNA388362, respectively) were also downloaded. For transcriptome sequencing of B. tropicalis and T. putrescentiae, both RNA sequencing and cDNA sequencing were performed on Illumina HiSeq 2500 by Groken Bioscience (Hong Kong) and paired-end 150-bp reads were generated. All data of newly sequenced transcriptome are listed in (supplementary table S2, Supplementary Material online), whereas transcriptome sequencing data of D. farinae, D. pteronyssinus, P. ovis, and S. scabiei were downloaded from the NCBI database (BioProject accession: PRJNA174061, PRJNA388362, PRJNA521406 and PRJNA304361, respectively). Quality control of both sequencing reads of genomic DNA and transcriptome was performed by FastQC v0.11.8.
Genome Assembly and Annotation
The genomes of D. farinae, D. pteronyssinus, B. tropicalis, and T. putrescentiae were de novo assembled and annotated. The genome assembly of P. ovis was directly downloaded from the NCBI database and then annotated, whereas the genome of S. scabiri (var. canis) was re-assembled and annotated. The detailed methods of genome assembly and annotation were described in (supplementary text S1, Supplementary Material online).
Collection of Genome Assemblies
Genome assemblies of other 20 species of mites and ticks, and the pseudoscorpion Cordylochernes scorpioides were directly downloaded from NCBI assembly database (supplementary table S4, Supplementary Material online). The genome assembly of Drosophila (D.) melanogaster (NCBI accession: GCF_000001215.4) was used as an outgroup, because other species are all belong to Arachnida which is considered as a monophyletic group (Howard et al. 2020). Completeness of genome assemblies was assessed by BUSCO v3.1.0 (Simão et al. 2015) with database arthropoda_odb9 (supplementary fig. S1, Supplementary Material online). Other assembly statistics was assessed by QUAST v5.0.2 (Gurevich et al. 2013) (supplementary fig. S1, Supplementary Material online). Repeat annotation was performed by de novo prediction with RepeatModeler v2.0.1 and masking with RepeatMasker v4.0.8 (Tarailo-Graovac and Chen 2009) (RepBase edition 20181026). In the de novo prediction step in RepeatModeler v2.0.1 (Smit and Hubley 2008), RECON v1.05 (Bao and Eddy 2002) and RepeatScout v1.0.6 (Price et al. 2005) were used for the prediction of repeat families in the genome assemblies.
Phylogenetic Analysis of Genome Assemblies
Based on the genome completeness assessment above, 47 common single and complete BUSCO proteins of 28 genomes were collected and aligned by the MAFFT (Katoh et al. 2002) and then edited in Gblocks (Castresana 2000) with the options “-b4 = 5 -b5 = h” to generate a sequence alignment of 13,133 conserved amino-acid residues.
Then the sequence alignment was used to construct the phylogenetic tree (supplementary fig. S1, Supplementary Material online) in maximum likelihood (ML) algorithm and 100 bootstrap replicates by RAxML v8.2.12 with the options “-m PROTCATWAG -f a -# 100”. The MSRs among five phylogenetic lineages (fig. 1B) were compared with the outgroup D. melanogaster by relative-rate tests using RRTree v1.1.11 (Robinson-Rechavi and Huchon 2000).
The ultrametric time tree (supplementary fig. S4, Supplementary Material online) was constructed by BEAST v2.6.6 (Bouckaert et al. 2014) using Yule Model and calibrated by the divergence times of Drosophila melanogaster-Tetranychus urticae (mean time: 605 MYA) and Tetranychus urticae-Platynothrus peltifer (mean time: 526 MYA) provided by TIMETREE (Kumar et al. 2017), and final edited by the online tool Interactive Tree of Life (iTOL) (Letunic and Bork 2021).
Collection of Proteomes
The genomes of D. farinae, D. pteronyssinus, P. ovis, S. scabiei (var. canis), B. tropicalis, and T. putrescentiae were all annotated by our group. Combined with the other eight proteomes of mites downloaded from NCBI GenBank or UniProt database, a total of 14 proteomes of mites were collected, and the quality was assessed by BUSCO v3.1.0 (Simão et al. 2015) with database arthropoda_odb9 (supplementary table S5, Supplementary Material online). The publicly available genome of S. scabiei var. suis has a higher continuity but a lower completeness than that of S. scabiei var. canis (supplementary fig. S2, Supplementary Material online). So, the proteome of the re-assembled genome of S. scabiei var. canis was selected for comparison. For D. tinctorium, a one-protein-per-gene proteome (UniProt ID: UP000285301) was downloaded from UniProt database. The high duplication rates of T. urticae (18.8%), V. destructor (52.2%), V. jacobsoni (48.3%) were caused by too many redundant isoforms included in the proteomes, then their protein-per-gene proteomes were extracted for analysis. Besides, the high duplication rate in D. tinctorium (9.8%) was probably caused by the high heterozygosity of the sample, similar as that of the genome.
Phylogenomic Analysis and GO Enrichment Analysis
Phylogenomic orthology and gene gain/loss analysis (Hahn et al. 2005) was performed among 14 proteomes of mites and D. melanogaster (UniProt ID: UP000000803) as an outgroup by OrthoFinder v2.5.4 (Emms and Kelly 2015, 2019) and CAFÉ v4.2 (De Bie et al. 2006). With the proteome of D. melanogaster as outgroup, all 14 proteomes of mites were assigned into orthogroups according to the protein similarities on sequence level (supplementary fig. S5, Supplementary Material online). These orthogroups were also considered as gene families.
GO enrichment analysis was performed by the online tool DAVID (david.ncifcrf.gov). Rapidly evolving gene families and species-specific orthogroups were collected, of which the genes were mapped to the proteome of D. melanogaster (UniProt ID: UP000000803) to acquire UniProt accession identifiers for GO enrichment using BLASTP v2.9.0 (McGinnis and Madden 2004) with E-value cutoff as 1E-6, to understand the evolutionary history of gene families.
Collection and Comparison of Gene Families
All proteins of the six astigmatic mites were searched by BLASTP v2.9.0 (McGinnis and Madden 2004) with reference proteins in Swiss-Prot database at E-value cutoff of 1E-6. After manual curation based on transcriptome data to confirm intron–exon split sites and filtering out annotated proteins with shorter than 50% average length of the gene family, all proteins of the six astigmatic mites identified in the target gene families were aligned and drawn into a phylogenetic tree. Sequence alignment was performed by CLUSTAL W (Thompson et al. 1994) and MUSCLE (Edgar 2004) in MEGA v10.2.2 (Kumar et al. 2018), and all phylogenetic trees were constructed by MEGA v10.2.2 (Kumar et al. 2018) with ML algorithm in the Jones–Taylor–Thornton (JTT) model, 80% site coverage and 100 bootstrap replicates, and then edited by online tool Interactive Tree of Life (iTOL) (Letunic and Bork 2021). The similarity matrix (BLOSUM62) of genes was analyzed and generated by the online tool Sequence Identity And Similarity (SIAS, http://imed.med.ucm.es/Tools/sias.html) using default parameters.
If two genes were located adjacently on genome, and no other gene was located between them, these two genes were both considered as tandemly arrayed genes generated via tandem duplication (Qiao et al. 2019). If two genes were proximally arrayed (separated by no more than 10 genes) but no transposable element was identified as adjacent to these proximally arrayed genes, we consider these genes as generated via anciently tandem duplication events (Qiao et al. 2019). Frequent tandemly arrayed genes were identified in gene families, like iGluRs of astigmatic mites, and connected with curve lines in phylogenetic trees edited by the online tool iTOL (Letunic and Bork 2021) with manually validated gene synteny information. If one gene is both tandemly arrayed with a gene and proximally arrayed with another gene, this gene was classified as tandemly arrayed gene.
Identification of HGT Events
A method based on the Crisp (Crisp et al. 2015) was performed to identify the HGT events in the six astigmatic mites. All proteins from bacteria and eukaryotes (excluding all metazoa) in UniRef50 database (UniProt) were collected and built as reference databases, while all proteins from other metazoa (excluding all arthropods) were collected and built as a comparison database.
The HGT index was calculated by dividing the bit score of the best bacteria or other eukaryotes (excluding all metazoa) match by that of the best other metazoa (excluding all arthropods) match. If one gene has a HGT index ≥ 2, or has match hits in bacteria or eukaryotes (excluding all metazoa) databases but not in other metazoa (excluding all arthropods) database with BLASTP v2.9.0 (McGinnis and Madden 2004) (E-value cutoff set as 1E-6), this gene was considered as a candidate HGT gene. All candidate HGT genes were further validated via BLASTP (McGinnis and Madden 2004) to non-redundant (NR) protein database (online version). If the top non-Acariformes matches of one candidate HGT gene to NCBI NR database are mainly from bacteria or eukaryotes (excluding all metazoa), but not other arthropods or other metazoa (excluding all arthropods), this candidate HGT gene was considered as acquired via HGT.
In this study, massive horizontal gene transferring events in astigmatic mites were identified and summarized in table 2. For better reproducibility, the protein sequences encoded by the representative HGT genes were listed in supplementary table S31, Supplementary Material online. When the conservation of HGT genes (supplementary table S32, Supplementary Material online) was explored, we found that all HGT genes were conserved in at least two independently cultured mites, which alleviated the concern of contamination. The best hits in different taxonomic groups in NCBI NR database were compared in supplementary table S33, Supplementary Material online, in which the unusual similarity differences were apparently incongruent with the tree of life and suggested those genes were acquired via HGT. Moreover, all HGT genes were searched in the genome assemblies and annotations of Parasitiformes in the NCBI database, but no homolog was found.
To further validate these HGT genes in the phylogenetic analysis, sequence alignment was performed by CLUSTAL W (Thompson et al. 1994) and MUSCLE (Edgar 2004), and all phylogenetic trees were constructed by MEGA v10.2.2 (Kumar et al. 2018) with ML algorithm in the JTT model, 80% site coverage and 100 bootstrap replicates, and then edited by the online tool, iTOL (Letunic and Bork 2021). IQ-TREE 2 (Minh et al. 2020) was used with options “-st AA -m TEST -bb 1000 -alrt 1000” to test and validate the phylogenetic results. Phylogenetic noise-like long branch attraction effect (Townsend et al. 2012) was carefully filtered using the classic SAW method (Siddall and Whiting 1999).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank the staff members in the Shenzhen Key Laboratory of Allergy and Immunology of the Shenzhen University and Siriraj Dust Mite Center for Services and Research of the Mahidol University for the mite authentication and culturing work. This work was made possible by grants: (i) General Research Fund from Research Grants Council of Hong Kong (Reference numbers: 475113, 14175617, 14119219, 14119420); (ii) Health and Medical Research Fund from Food and Health Bureau of Hong Kong (Reference numbers: 06171016, 07181266); (iii) Continuation project of Joint Research Fund for Overseas Chinese Scholars and Scholars in Hong Kong and Macao Young Scholars (Reference number: 31729002); (iv) National Natural Science Foundation of China (Reference numbers: 31729002, 81971514); (v) Shenzhen Science and Technology Plan Project (Reference number: KQTD20170331145453160, GJHZ20190822095605512, SGDX20201103095609027); (vi) Shenzhen Nanshan District Science and Technology Project (Reference number: LHTD20180007).
Contributor Information
Qing Xiong, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong; Hong Kong Bioinformatics Centre, The Chinese University of Hong Kong, Hong Kong.
Angel Tsz-Yau Wan, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong; Hong Kong Bioinformatics Centre, The Chinese University of Hong Kong, Hong Kong.
Xiaoyu Liu, Shenzhen Key Laboratory of Allergy and Immunology, School of Medicine, Shenzhen University, China.
Cathy Sin-Hang Fung, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong.
Xiaojun Xiao, Shenzhen Key Laboratory of Allergy and Immunology, School of Medicine, Shenzhen University, China.
Nat Malainual, Department of Parasitology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand.
Jinpao Hou, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong; Centre for Microbial Genomics and Proteomics, The Chinese University of Hong Kong, Hong Kong.
Lingyi Wang, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong.
Mingqiang Wang, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong; Hong Kong Bioinformatics Centre, The Chinese University of Hong Kong, Hong Kong.
Kevin Yi Yang, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong; Hong Kong Bioinformatics Centre, The Chinese University of Hong Kong, Hong Kong.
Yubao Cui, Department of Clinical Laboratory, Wuxi People’s Hospital Affiliated to Nanjing Medical University, Wuxi, China.
Elaine Lai-Han Leung, Macau Institute for Applied Research in Medicine and Health, Macau University of Science and Technology, Macau.
Wenyan Nong, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong.
Soo-Kyung Shin, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong.
Shannon Wing-Ngor Au, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong.
Kyoung Yong Jeong, Institute of Allergy, Department of Internal Medicine, College of Medicine, Yonsei University, Seoul, Korea.
Fook-Tim Chew, Department of Biological Sciences, National University of Singapore, Singapore.
Jerome Ho-Lam Hui, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong.
Ting-Fan Leung, Department of Paediatrics, The Chinese University of Hong Kong, Hong Kong.
Anchalee Tungtrongchitr, Department of Parasitology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand.
Nanshan Zhong, State Key Laboratory of Respiratory Disease, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China.
Zhigang Liu, Shenzhen Key Laboratory of Allergy and Immunology, School of Medicine, Shenzhen University, China.
Stephen Kwok-Wing Tsui, School of Biomedical Sciences, The Chinese University of Hong Kong, Hong Kong; Hong Kong Bioinformatics Centre, The Chinese University of Hong Kong, Hong Kong; Centre for Microbial Genomics and Proteomics, The Chinese University of Hong Kong, Hong Kong.
Data Availability
All data are deposited in NCBI database under BioProject numbers (PRJNA174061 for D. farinae, PRJNA388362 for D. pteronyssinus, PRJNA702011 for B. tropicalis, PRJNA706095 for T. putrescentiae).
References
- Ahn SJ, Dermauw W, Wybouw N, Heckel DG, Van Leeuwen T. 2014. Bacterial origin of a diverse family of UDP-glycosyltransferase genes in the Tetranychus urticae genome. Insect Biochem Mol Biol. 50:43–57. [DOI] [PubMed] [Google Scholar]
- Arlian LG. 1989. Biology, host relations, and epidemiology of Sarcoptes scabiei. Annu Rev Entomol. 34:139–159. [DOI] [PubMed] [Google Scholar]
- Arlian LG. 2009. Chapter 42 – Chiggers and other disease-causing mites. In: Resh VH, Cardé RT, editors. Encyclopedia of insects. 2nd ed. San Diego: Academic Press. p. 152–156. [Google Scholar]
- Arlian LG, Morgan MS. 2017. A review of Sarcoptes scabiei: past, present and future. Parasites & Vectors 10:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlian LG, Platts-Mills TA. 2001. The biology of dust mites and the remediation of mite allergens in allergic disease. J Allergy Clin Immunol. 107:S406–413. [DOI] [PubMed] [Google Scholar]
- Bao Z, Eddy SR. 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12:1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MJ, Dhouailly D, Jiang B, McNamara M. 2019. The early origin of feathers. Trends Ecol Evol. 34:856–869. [DOI] [PubMed] [Google Scholar]
- Bergström FC, Reynolds S, Johnstone M, Pike RN, Buckle AM, Kemp DJ, Fischer K, Blom AM. 2009. Scabies mite inactivated serine protease paralogs inhibit the human complement system. J Immunol. 182:7809–7817. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 10:e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess STG, Marr EJ, Bartley K, Nunn FG, Down RE, Weaver RJ, Prickett JC, Dunn J, Rombauts S, Van Leeuwen T, et al. 2019. A genomic analysis and transcriptomic atlas of gene expression in Psoroptes ovis reveals feeding- and stage-specific patterns of allergen expression. BMC Genomics 20:756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón MA, Kleine-Tebbe J, Linneberg A, De Blay F, Hernandez Fernandez de Rojas D, Virchow JC, Demoly P. 2015. House dust mite respiratory allergy: an overview of current therapeutic strategies. J Allergy Clin Immunol Pract. 3:843–855. [DOI] [PubMed] [Google Scholar]
- Calderón MA, Linneberg A, Kleine-Tebbe J, De Blay F, Hernandez Fernandez de Rojas D, Virchow JC, Demoly P. 2015. Respiratory allergy caused by house dust mites: what do we really know? J Allergy Clin Immunol. 136:38–48. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chan TF, Ji KM, Yim AK, Liu XY, Zhou JW, Li RQ, Yang KY, Li J, Li M, Law PT, et al. 2015. The draft genome, transcriptome, and microbiome of Dermatophagoides farinae reveal a broad spectrum of dust mite allergens. J Allergy Clin Immunol. 135:539–548. [DOI] [PubMed] [Google Scholar]
- Colloff MJ. 2009. Dust Mites. Springer. [Google Scholar]
- Colloff MJ, Ayres J, Carswell F, Howarth PH, Merrett TG, Mitchell EB, Walshaw MJ, Warner JO, Warner JA, Woodcock AA. 1992. The control of allergens of dust mites and domestic pets: a position paper. Clin Exp Allergy 22(Suppl 2):1–28. [DOI] [PubMed] [Google Scholar]
- Colloff MJ, Spieksma FT. 1992. Pictorial keys for the identification of domestic mites. Clin Exp Allergy 22:823–830. [DOI] [PubMed] [Google Scholar]
- Crisp A, Boschetti C, Perry M, Tunnacliffe A, Micklem G. 2015. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 16:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabert M, Witalinski W, Kazmierski A, Olszanowski Z, Dabert J. 2010. Molecular phylogeny of acariform mites (Acari, Arachnida): strong conflict between phylogenetic signal and long-branch attraction artifacts. Mol Phylogenet Evol. 56:222–241. [DOI] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22:1269–1271. [DOI] [PubMed] [Google Scholar]
- de Miguel T, Zhu O, Villa TG. 2019. Horizontal gene transfer between Wolbachia and animals. In: Horizontal gene transfer. Springer. p. 227–234. [Google Scholar]
- Domes K, Althammer M, Norton RA, Scheu S, Maraun M. 2007. The phylogenetic relationship between Astigmata and Oribatida (Acari) as indicated by molecular markers. Exp Appl Acarol. 42:159–171. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eme L, Gentekaki E, Curtis B, Archibald JM, Roger AJ. 2017. Lateral gene transfer in the adaptation of the anaerobic parasite blastocystis to the gut. Curr Biol. 27:807–820. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erban T, Di Presa CA, Kopecky J, Poltronieri P, Hubert J. 2013. PCR detection of the 14.5 antibacterial NlpC/P60-like Dermatophagoides pteronyssinus protein in Dermatophagoides farinae (Acari: Pyroglyphidae). J Med Entomol. 50:931–933. [DOI] [PubMed] [Google Scholar]
- Erban T, Erbanova M, Nesvorna M, Hubert J. 2009. The importance of starch and sucrose digestion in nutritive biology of synanthropic acaridid mites: alpha-amylases and alpha-glucosidases are suitable targets for inhibitor-based strategies of mite control. Arch Insect Biochem Physiol. 71:139–158. [DOI] [PubMed] [Google Scholar]
- Erban T, Rybanska D, Hubert J. 2015. Population growth of the generalist mite Tyrophagus putrescentiae (Acari: Acaridida) following adaptation to high- or low-fat and high- or low-protein diets and the effect of dietary switch. Environ Entomol. 44:1599–1604. [DOI] [PubMed] [Google Scholar]
- Fukamizo T, Brzezinski R. 1997. Chitosanase from Streptomyces sp. strain N174: a comparative review of its structure and function. Biochem Cell Biol. 75:687–696. [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Meselson M, Arkhipova IR. 2008. Massive horizontal gene transfer in Bdelloid Rotifers. Science 320:1210–1213. [DOI] [PubMed] [Google Scholar]
- Green WF, Woolcock AJ. 1978. Tyrophagus putrescentiae: an allergenically important mite. Clin Allergy 8:135–144. [DOI] [PubMed] [Google Scholar]
- Guilleminault L, Viala-Gastan C. 2017. Blomia tropicalis: a house dust mite in the tropics. Rev Mal Respir. 34:791–801. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, De Bie T, Stajich JE, Nguyen C, Cristianini N. 2005. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 15:1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton K, Nisbet A, Lehane M, Taylor M, Billingsley PF. 2003. A physiological and biochemical model for digestion in the ectoparasitic mite, Psoroptes ovis (Acari: Psoroptidae). Int J Parasitol. 33:773–785. [DOI] [PubMed] [Google Scholar]
- Holt DC, Fischer K, Allen GE, Wilson D, Wilson P, Slade R, Currie BJ, Walton SF, Kemp DJ. 2003. Mechanisms for a novel immune evasion strategy in the scabies mite Sarcoptes scabiei: a multigene family of inactivated serine proteases. J Invest Dermatol. 121:1419–1424. [DOI] [PubMed] [Google Scholar]
- Howard RJ, Puttick MN, Edgecombe GD, Lozano-Fernandez J. 2020. Arachnid monophyly: morphological, palaeontological and molecular support for a single terrestrialization within Chelicerata. Arthropod Struct Dev. 59:100997. [DOI] [PubMed] [Google Scholar]
- Husnik F, McCutcheon JP. 2018. Functional horizontal gene transfer from bacteria to eukaryotes. Nat Rev Microbiol. 16:67–79. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11:97–108. [DOI] [PubMed] [Google Scholar]
- Kapli P, Natsidis P, Leite DJ, Fursman M, Jeffrie N, Rahman IA, Philippe H, Copley RR, Telford MJ. 2021. Lack of support for Deuterostomia prompts reinterpretation of the first Bilateria. Sci Adv 7:eabe2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Ki K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP. 2009. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics 10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimov PB, Oconnor B. 2013. Is permanent parasitism reversible?—critical evidence from early evolution of house dust mites. Syst Biol. 62:411–423. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34:1812–1819. [DOI] [PubMed] [Google Scholar]
- Lee M, Batuecas MT, Tomoshige S, Domínguez-Gil T, Mahasenan KV, Dik DA, Hesek D, Millán C, Usón I, Lastochkin E, et al. 2018. Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 115:4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Liu Q, Kadowaki T. 2019. Honey bee parasitic mite contains the Sensilla-rich sensory organ on the foreleg tarsus expressing ionotropic receptors with conserved functions. Front Physiol. 10:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucl Acids Res. 49:W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. 2019. ABC family transporters. In: Liu X, Pan G, editors. Drug transporters in drug disposition, effects and toxicity. Singapore: Springer Singapore. p. 13–100. [Google Scholar]
- Liu XY, Yang KY, Wang MQ, Kwok JSL, Zeng X, Yang ZY, Xiao XJ, Lau CPY, Li Y, Huang ZM, et al. 2018. High-quality assembly of Dermatophagoides pteronyssinus genome and transcriptome reveals a wide range of novel allergens. J Allergy Clin Immunol. 141:2268–2271. [DOI] [PubMed] [Google Scholar]
- Lozano-Fernandez J, Tanner AR, Giacomelli M, Carton R, Vinther J, Edgecombe GD, Pisani D. 2019. Increasing species sampling in chelicerate genomic-scale datasets provides support for monophyly of Acari and Arachnida. Nat Commun. 10:2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S, Harman A. 2016. Horizontal gene transfer of a Chlamydial tRNA-guanine transglycosylase gene to eukaryotic microbes. Mol Phylogenet Evol. 94:392–396. [DOI] [PubMed] [Google Scholar]
- Mathaba LT, Pope CH, Lenzo J, Hartofillis M, Peake H, Moritz RL, Simpson RJ, Bubert A, Thompson PJ, Stewart GA. 2002. Isolation and characterisation of a 13.8-kDa bacteriolytic enzyme from house dust mite extracts: homology with prokaryotic proteins suggests that the enzyme could be bacterially derived. FEMS Immunol Med Microbiol. 33:77–88. [DOI] [PubMed] [Google Scholar]
- McGinnis S, Madden TL. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucl Acids Res. 32:W20–W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. 2006. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci U S A 103:4404–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD. 2019. The role of dust mites in allergy. Clin Rev Allergy Immunol. 57:312–329. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A. 2015. Horizontal gene transfers in insects. Curr Opin Insect Sci. 7:24–29. [DOI] [PubMed] [Google Scholar]
- Norton RA. 1998. Morphological evidence for the evolutionary origin of Astigmata (Acari: Acariformes). Exp Appl Acarol. 22:559–594. [Google Scholar]
- Peat JK, Tovey E, Toelle BG, Haby MM, Gray EJ, Mahmic A, Woolcock AJ. 1996. House dust mite allergens. A major risk factor for childhood asthma in Australia. Am J Respiratory Critical Care Med. 153:141–146. [DOI] [PubMed] [Google Scholar]
- Peixoto L, Roos DS. 2007. Genomic scale analysis of lateral gene transfer in Apicomplexan parasites: insights into early eukaryotic evolution, host-pathogen interaction and drug target development. BMC Bioinformatics 8:S5. [Google Scholar]
- Pepato AR, da Rocha CEF, Dunlop JA. 2010. Phylogenetic position of the acariform mites: sensitivity to homology assessment under total evidence. BMC Evol Biol. 10:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepato AR, Klimov PB. 2015. Origin and higher-level diversification of acariform mites – evidence from nuclear ribosomal genes, extensive taxon sampling, and secondary structure alignment. BMC Evol Biol. 15:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. 2019. How the earliest mammals thrived alongside dinosaurs. Nature 574:468–473. [DOI] [PubMed] [Google Scholar]
- Portnoy J, Miller JD, Williams PB, Chew GL, Miller JD, Zaitoun F, Phipatanakul W, Kennedy K, Barnes C, Grimes C, et al. 2013. Environmental assessment and exposure control of dust mites: a practice parameter. Ann Allergy, Asthma Immunol.: Off Publ Amn College Allergy, Asthma, & Immunol. 111:465–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21:i351–i358. [DOI] [PubMed] [Google Scholar]
- Proctor HC. 2003. Feather mites (Acari: Astigmata): ecology, behavior, and evolution. Annu Rev Entomol. 48:185–209. [DOI] [PubMed] [Google Scholar]
- Qiao X, Li Q, Yin H, Qi K, Li L, Wang R, Zhang S, Paterson AH. 2019. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reams AB, Roth JR. 2015. Mechanisms of gene duplication and amplification. Cold Spring Harbor Perspect Biol. 7:a016592–a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds SL, Pike RN, Mika A, Blom AM, Hofmann A, Wijeyewickrema LC, Kemp D, Fischer K. 2014. Scabies mite inactive serine proteases are potent inhibitors of the human complement lectin pathway. PLoS Negl Trop Dis 8:e2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Huchon D. 2000. RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics 16:296–297. [DOI] [PubMed] [Google Scholar]
- Rytz R, Croset V, Benton R. 2013. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol. 43:888–897. [DOI] [PubMed] [Google Scholar]
- Sakata T, Norton RA. 2001. Opisthonotal gland chemistry of early-derivative oribatid mites (Acari) and its relevance to systematic relationships of Astigmata. Int J Acarol. 27:281–292. [Google Scholar]
- Santos da Silva E, Asam C, Lackner P, Hofer H, Wallner M, Silva Pinheiro C, Alcântara-Neves NM, Ferreira F. 2017. Allergens of Blomia tropicalis: an overview of recombinant molecules. Int Arch Allergy Immunol. 172:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Chen Y, Wei W, Xiong L, Tao Y, Xiao J, Liu S, He X, Du X, Gu X, et al. 2021. Comparative analysis of the allergenic characteristics and serodiagnostic potential of recombinant chitinase-like protein-5 and -12 from Sarcoptes scabiei. Parasites Vectors 14:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J-H, Sulpizio AG, Kelley A, Alvarez L, Murphy SG, Fan L, Cava F, Mao Y, Saper MA, Dörr T. 2020. Structural basis of peptidoglycan endopeptidase regulation. Proc Natl Acad Sci U S A 117:11692–11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddall ME, Whiting MF. 1999. Long-branch abstractions. Cladistics 15:9–24. [Google Scholar]
- Simakov O, Kawashima T, Marlétaz F, Jenkins J, Koyanagi R, Mitros T, Hisata K, Bredeson J, Shoguchi E, Gyoja F, et al. 2015. Hemichordate genomes and deuterostome origins. Nature 527:459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Sinclair A, Kirkwood A. 1983. Feeding behaviour of Psoroptes ovis. Vet Rec. 112:65. [DOI] [PubMed] [Google Scholar]
- Smit AF, Hubley R. 2008. RepeatModeler Open-1.0. Available from: http://www.repeatmasker.org.
- Snoeck S, Pavlidi N, Pipini D, Vontas J, Dermauw W, Van Leeuwen T. 2019. Substrate specificity and promiscuity of horizontally transferred UDP-glycosyltransferases in the generalist herbivore Tetranychus urticae. Insect Biochem Mol Biol. 109:116–127. [DOI] [PubMed] [Google Scholar]
- Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nat Rev Genet. 16:472–482. [DOI] [PubMed] [Google Scholar]
- Stringer C. 2016. The origin and evolution of Homo sapiens. Philos Trans R Soc Lond B Biol Sci. 371:20150237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang VH, Stewart GA, Chang BJ. 2017. Dermatophagoides pteronyssinus lytFM encoding an NlpC/P60 endopeptidase is also present in mite-associated bacteria that express LytFM variants. FEBS Open Bio 7:1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang VH, Stewart GA, Chang BJ. 2015. House dust mites possess a polymorphic, single domain putative peptidoglycan d, l endopeptidase belonging to the NlpC/P60 Superfamily. FEBS Open Bio 5:813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 25:4.10.11–14.10.14. [DOI] [PubMed] [Google Scholar]
- Tavano O, Pessela B, Goulart A, Fernández-Lafuente R, Guisan J, Monti R. 2008. Stabilization of an Amylase from Neurospora crassa by immobilization on highly activated supports. Food Biotechnol. 22:262–275. [Google Scholar]
- Thomas WR, Hales BJ, Smith WA. 2010. House dust mite allergens in asthma and allergy. Trends Mol Med. 16:321–328. [DOI] [PubMed] [Google Scholar]
- Thomas WR, Smith W-A, Hales BJ, Mills KL, O’Brien RM. 2002. Characterization and immunobiology of house dust mite allergens. Int Arch Allergy Immunol. 129:1–18. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JP, Su Z, Tekle YI. 2012. Phylogenetic signal and noise: predicting the power of a data set to resolve phylogeny. Systc Biol. 61:835–835. [DOI] [PubMed] [Google Scholar]
- van Giesen L, Garrity PA. 2017. More than meets the IR: the expanding roles of variant Ionotropic Glutamate Receptors in sensing odor, taste, temperature and moisture. F1000Res 6:1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma MRG. 1993. Ticks and mites (Acari). In: Lane RP, Crosskey RW, editors. Medical insects and arachnids. Dordrecht: Springer Netherlands. p. 597–658. [Google Scholar]
- Voorhorst R, Spieksma FTM, Varekamp H, Leupen M, Lyklema A. 1967. The house-dust mite (Dermatophagoides pteronyssinus) and the allergens it produces. Identity with the house-dust allergen. J Allergy 39:325–339. [Google Scholar]
- Waldron R, McGowan J, Gordon N, Mitchell EB, Fitzpatrick DA, Doyle S. 2019. Characterisation of three novel β-1,3 glucanases from the medically important house dust mite Dermatophagoides pteronyssinus (airmid). Insect Biochem Mol Biol. 115:103242. [DOI] [PubMed] [Google Scholar]
- Wan H, Winton HL, Soeller C, Taylor GW, Gruenert DC, Thompson PJ, Cannell MB, Stewart GA, Garrod DR, Robinson C. 2001. The transmembrane protein occludin of epithelial tight junctions is a functional target for serine peptidases from faecal pellets of Dermatophagoides pteronyssinus. Clin Exp Allergy 31:279–294. [DOI] [PubMed] [Google Scholar]
- Wybouw N, Van Leeuwen T, Dermauw W. 2018. A massive incorporation of microbial genes into the genome of Tetranychus urticae, a polyphagous arthropod herbivore. Insect Mol Biol. 27:333–351. [DOI] [PubMed] [Google Scholar]
- Xiong Q, Wan ATY, Tsui SK. 2020. A mini-review of the genomes and allergens of ites and ticks. Curr Protein Pept Sci. 21:114–123. [DOI] [PubMed] [Google Scholar]
- Xu X, Feng Y, Fang S, Xu J, Wang X, Guo W. 2016. Genome-wide characterization of the β-1,3-glucanase gene family in Gossypium by comparative analysis. Sci Rep. 6:29044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chen W, Li Z, Ma L, Yu J, Wang H, Liu Z, Xu B. 2018. Identification and characterization of three new cytochrome P450 genes and the use of RNA isnterference to evaluate their roles in antioxidant defense in Apis cerana cerana Fabricius. Front Physiol. 9:1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are deposited in NCBI database under BioProject numbers (PRJNA174061 for D. farinae, PRJNA388362 for D. pteronyssinus, PRJNA702011 for B. tropicalis, PRJNA706095 for T. putrescentiae).