Abstract
Forskolin, a class of labdane-type diterpenoid, has significant medicinal value in anticancer, antiasthmatic, antihypertensive, and heart-strengthening treatments. The main source of natural forskolin is its extraction from the cork tissue of the root of Coleus forskohlii. However, conventional modes of extraction pose several challenges. In recent years, the construction of microbial cell factories to produce medicinal natural products via synthetic biological methods has effectively solved the current problems and is a research hotspot in this field. This review summarizes the recent progress in the heterologous synthesis of forskolin via synthetic biological technology, analyzes the current challenges, and proposes corresponding strategies.
Keywords: Forskolin, 13R-manoyl oxide, Cytochrome P450s, Synthetic biology
Introduction
Natural products synthesized by animals, plants, and microorganisms are a class of organic small-molecule compounds with structural and functional diversity, which are widely used in domains such as agriculture and medicine due to their important physiological functions and biological activities (Osbourn & Lanzotti, 2009; Newman & Cragg, 2016). Natural products can be primary or secondary metabolites. Natural products derived from plants are mostly alkaloids, polysaccharides, volatile oils, phenols, terpenoids, and lignans, whereas products derived from microorganisms and metabolites are mostly polysaccharides, enzymes, antibiotics, pigments, amino acids, organic acids, alcohols, and ketones. Furthermore, marine natural products include mostly sterols, unsaturated fatty acids, polysaccharides and glycosides, macrolides, and polyketides and alkaloids (Aharoni & Galili, 2011; Durazzo et al., 2018; Yan et al., 2018). Terpenoids with the molecular formula (C5H8)n, also known as isoprenoids, are the main plant secondary metabolites. Currently, more than 55 000 terpenoids are known (Harborne, 1995). Many plant-derived terpenoids have applications in medicine, such as the anticancer drug taxol (Shu et al., 2014) and the specific antimalarial drug artemisinin (Ansari et al., 2013). Some terpenoids are alternatives to aviation fuel, such as bisabolene and pinene (Peralta-Yahya & Keasling, 2010; Peralta-Yahya et al., 2011).
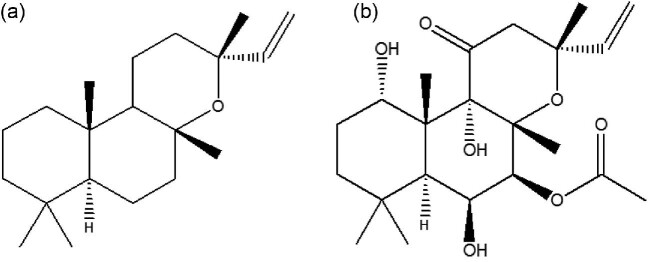
Forskolin is a labdane-type diterpenoid mainly found in the cork tissue of the root of Coleus forskohlii. Its molecular formula is C22H34O7 (Fig. 1) (Ammon & Müller, 1985). Its cyclic AMP (cAMP) booster effect makes it an effective treatment against glaucoma, tumors, HIV, obesity, hypertension, heart diseases, asthma, and cardiac complications (Yoneyama et al., 2002; Bodiwala et al., 2009; Virgona et al., 2010; Doseyici et al., 2014; Ponnam et al., 2014; Majeed et al., 2015; Sapio et al., 2017). An example of a forskolin derivative with good anticancer (Hylse et al., 2017) or anti-HIV (Bodiwala et al., 2009) effects is NKH477 (approved in japan as a heart failure treatment), which has been used to treat surgical complications, heart failure, and cerebral vasospasm (Kikura et al., 2004). 13R-manoyl oxide (13R-MO), the precursor of forskolin, was found in the roots of C. forskohlii (Pateraki et al., 2014). Fig. 1 shows the structures of 13R-MO and forskolin.
Fig. 1.
Schematic diagram of 13R-MO and forskolin. (A) Structure of 13R-MO. (B) Structure of forskolin.
Generally, the production methods of terpenoids include traditional plant extraction (Bajer et al., 2016), chemical synthesis (Du et al., 2015), and microbial heterologous synthesis (Liu et al., 2015). Forskolin is mainly derived from the plant C. forskohlii, but with a low extraction yield (0.013–0.728%) due to tedious separation steps and low purity (Asada et al., 2012; Harde & Singhal, 2012; Srivastava et al., 2017). The chemical synthesis of forskolin involved multistep reactions and large amounts of organic reagents, making the process more expensive and environmentally harmful than extraction (Corey & Jardine, 1989; Colombo et al., 1992). With the development of synthetic biology, the synthesis of terpenoids by microorganisms has emerged as an attractive alternative. Recently, 13R-MO and forskolin have been successfully expressed and synthesized in microorganisms. Current research focuses on how to efficiently obtain forskolin with high yield and high purity.
This paper introduces the research status of the heterologous synthesis of forskolin, its precursor 13R-MO, and 13R-MO derivatives in different microorganisms, describes the speed limiting steps of the efficient synthesis of forskolin, analyzes the existing problems in current research, and puts forward the corresponding strategies.
Biosynthesis of Forskolin
Key Enzymes Involved in Forskolin Biosynthesis
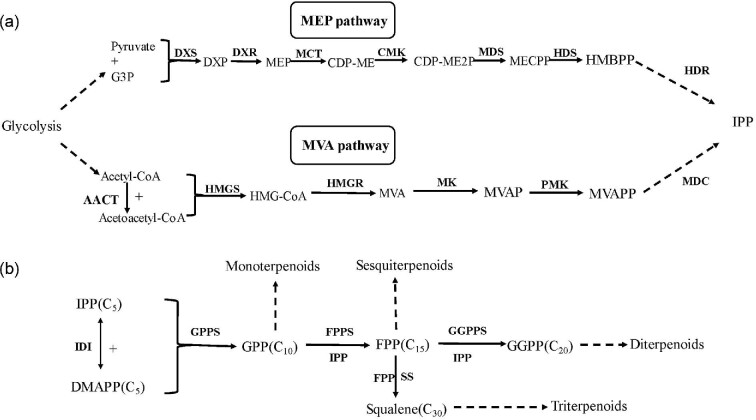
The general precursor for the synthesis of terpenoids is produced by the mevalonate pathway (MVA) (Bloch, 1992) and 2-methyl-4-phospho-d-erythroitol pathway (MEP) (Rohmer et al., 1993). Dimethylallyl pyrophosphate (DMAPP) and isopentenyl pyrophosphate (IPP) are the basic units for terpenoid synthesis (Katsuki & Bloch, 1967; Lynen, 1967). IPP and DMAPP are condensed head-to-tail into geranyl diphosphate (GPP). GPP then combines with another IPP to form 15-carbon farnesyl pyrophosphate (FPP), which condenses with yet another IPP to form geranylgeranyl pyrophosphate (GGPP). FPP can also form squalene via squalene synthase (ERG9). GPP, FPP, GGPP, and squalene are the precursors for the synthesis of monoterpenoids, sesquiterpenoids, diterpenoids, and triterpenoids, respectively. Exogenous terpene synthases cyclize and modify these precursors, producing the corresponding target terpenoids (Fig. 2).
Fig. 2.
MEP and MVA pathways. (a) Related enzymes in the MEP pathway and MVA pathway. AACT, acetoacetyl CoA thiolase gene; HMGS, HMG-CoA synthase gene; HMGR, HMG-CoA reductase gene; MK, mevalonate kinase gene; PMK, phosphomevalonate kinase gene; MDC, 5-pyrophosphate methyl hydrovalerate decarboxylase gene; DXS, deoxyxylulose-5-phosphate synthase gene; DXR, deoxyxylulose-5-phosphate isomerase gene; MCT, 2-C-methyl-d-erythritol-4-cytidine monophosphate acyltransferase gene; CMK, 4-(5′-cytidine pyrophosphate)-2-C-methyl erythritol kinase gene; MDS, 2-C-methyl-erythritol-2,4-cyclic pyrophosphate synthetase gene; HDS, (E)-4-hydroxy-3-methylbutane-2-enyl diphosphate synthase gene; HDR, (E)-4-hydroxy-3-methylbutan-2-enyl diphosphate reductase gene. (b) Enzymes in the synthesis pathway of terpenoids. IDI, isoprene pyrophosphate isomerase; GPPS, geranyl diphosphate synthase gene; FPPS, farnesyl diphosphate synthase gene; GGPPS, geranylgeranyl diphosphate synthase gene; SS, squalene synthase.
In the early stages, researchers analyzed the extracts of the fibrous root culture of C. forskohlii to isolate and identify the possible precursors of forskolin biosynthesis (Bhat et al., 1977; Asada et al., 2012). In 2013, Zerbe et al. (Zerbe et al., 2013) assembled, classified, and analyzed the phylogenetic function of genes related to the forskolin synthesis pathway in the root of C. forskohlii via 454 and Illumina sequencing technology. Then in 2014, Pateraki et al. (Pateraki et al., 2014) analyzed the existing transcription group data and identified six candidate diterpene synthases (diTPS) CfTPS1 (KF444506), CfTPS2 (KF444507), CfTPS3 (KF444508), CfTPS4 (KF444509), CfTPS14 (AGN70881.1), and CfTPS15 (KF4710011). The six diTPS genes were heterologously expressed in Escherichia coli and Nicotiana benthamiana transient expression systems. GC–MS analysis showed that CfTPS1 and CfTPS2 are functionally different type II diTPS (the full-length sequence of CfTPS15 cannot be retrieved and has not been tested). An enzyme coupling assay revealed that CfTPS2 (which synthesizes the intermediate copal-8-ol diphosphate) combined with CfTPS3 resulted in the stereospecific formation of 13R-MO. Besides, the combination of CfTPS2 and CfTPS4 can also produce 13R-MO and its epimer13S-MO, while the other combinations cannot produce manoyl oxide. The discovery of CfTPS2 and CfTPS3 laid the foundation for subsequent research on other enzymes of the forskolin pathway.
The function of cytochrome P450 enzymes (P450s) and acetyltransferases were identified and studied via metabolomics, single-cell transcriptome research, and synthetic biological modular methods in 2017 (Pateraki et al., 2017). In-depth RNA sequencing and analysis of the root cork of C. forskohlii allowed the identification of 263 652 cDNA genes. A total of 29 cytochrome P450 candidate genes were screened and found based on the cytochrome P450 transcriptome relative expression levels in the root cork, and seven belonged to the CYP76AH subfamily, namely CfCYP76AH8 (KT382348), CfCYP76AH9 (KT382347), CfCYP76AH10 (KT382346), CfCYP76AH11 (KT382349), CfCYP76AH15 (KT382358), CfCYP76AH16 (KT382359), and CfCYP76AH17 (KT382360). Two acetyltransferase (ACT) candidate genes were also screened and found, CfACT1–6 (KT382361) and CfACT1–8 (KT382363). Among the candidates, five were full-length sequences (CfCYP76AH8, CfCYP76AH9, CfCYP76AH10, CfCYP76AH11, and CfCYP76AH17), whereas two (CfCYP76AH15 and CfCYP76AH16) were only partial cDNAs. Then, the cytochrome P450s and acetyltransferase candidates in C. forskohlii were functionally characterized via transient expression in N. benthamiana, and the intermediates were identified by GC–MS or HPLC–HRMS–SPE–NMR. CfCYP76AH15, CfCYP76AH8, and CfCYP76AH17 can catalyze 13R-MO to form the main product 11-oxo-manoyl oxide. CfCYP76AH15 is the most efficient and specific; however, CfCYP76AH8 and CfCYP76AH17 also effectively monooxidize the C-1 site. CfCYP76AH11 catalyzes the conversion of 13R-MO into 9-deoxy-7-deacetylforskolin. CfCYP76AH16 catalyzes the conversion of 13R-MO into 9-hydroxy-manoyl oxide. CfCYP76AH15 in combination with CfCYP76AH11 and CfCYP76AH16 can catalyze the conversion of 13R-MO into 7-deacetylforskolin. Thus, this combination of multifunctional cytochrome P450s appeared to constitute the optimal biosynthetic pathway for the specific formation of 7-deacetylforskolin from 13R-MO. The last enzyme catalyzes the acetylation of 7-deacylforskolin to form forskolin. Two ACT candidates, CfACT1–6 and CfACT1–8, catalyze the acetylation of 7-deacylforskolin. CfACT1–6 lacks specificity, thus its expression formed a wide range of acetylation products and only a small proportion of forskolin. In contrast, CfACT1–8 exhibited high activity and specificity, and effectively converted 7-deacylforskolin into forskolin without forming detectable byproducts.
Through joint efforts, the scientific researchers have identified the enzymes required for the biosynthesis pathway of forskolin.
Key Synthetic Pathway Involved in Forskolin Heterologous Biosynthesis
Transforming the metabolic network of chassis microorganisms to efficiently produce food, fuel, medicine, and health products is the current research hotspot in metabolic engineering and synthetic biology (Lee et al., 2018; Park et al., 2018). So far, E. coli, Cyanobacteria, and S. cerevisiae have been tried for forskolin biosynthesis.
The Synthetic Pathway of 13R-MO and its Optimization
As one of the model microorganisms, E. coli became the most widely used chassis strain and has been successfully used to synthesize a variety of diterpenes (Morrone et al., 2010). Nielsen et al. (Nielsen et al., 2014) engineered E. coli to produce 13R-MO by introducing the GGPP synthase AgGGPPS from Abies grandis, and CfTPS2 and CfTPS3 from C. forskohlii. They optimized the MEP pathway and achieved a production of 10 mg/l. This is also the first relevant report on the synthesis of 13R-MO in microorganisms.
Cyanobacteria have been successfully used to synthesize some simple compounds such as alcohols, sugars, and fatty acids (Savakis & Hellingwerf, 2015). Cyanobacteria have their own MEP pathway, which is very suitable for the production of terpenoids (Pattanaik & Lindberg, 2015). Synechocystis sp. PCC 6803 became a relatively mature model microorganism as it was the first cyanobacteria to have its whole genome sequenced (Ikeuchi & Tabata, 2001). Reconstructing the 13R-MO metabolic pathway in Synechocystis sp. PCC 6803, optimizing the MEP pathway and exploring the optimal induction conditions let to the production of 13R-MO with a yield of 0.45 mg/g DCW (Englund et al., 2015).
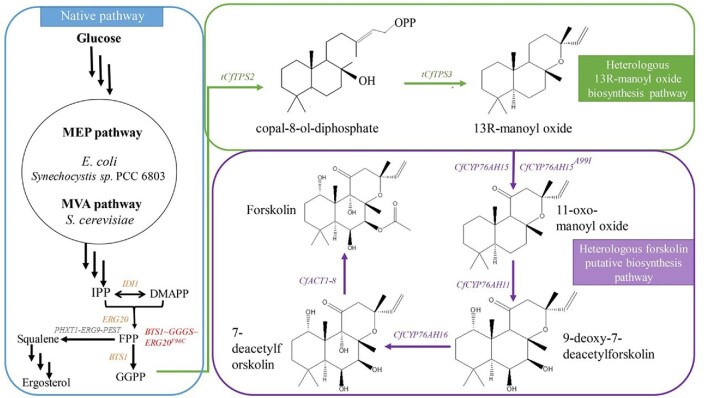
S. cerevisiae is also a model microorganism. As an industrial cell factory, it has a clear genetic background, and it has performed well in the industrial production of medicines, food additives, and bioenergy. Because of its mature eukaryotic expression system, it is more suitable for the expression of heterologous eukaryotic proteins. S. cerevisiae has an endogenous MVA pathway and can directly provide precursors for plant-derived terpenes (Vranová et al., 2013). Compared to E. coli, S. cerevisiae has the advantage of naturally expressing plant cytochrome P450s (Renault et al., 2014). Optimizing the precursor GGPP supply and knocking out the MCT1, WHI2, and GDH1 genes increased the yield of 13R-MO from 40 to 80 mg/l, but 13S-MO still existed as a by-product (Ignea, Athanasakoglou, et al., 2016a; Ignea, Ioannou, et al., 2016b). Our team implemented the initial synthesis of 13R-MO in S. cerevisiae by integrating the codon-optimized CfTPS2 and CfTPS3 and improved the 13R-MO yield to 167.1 mg/l by weakening ERG9 (MET3p-ERG9) and fed-batch fermenting in a shake flask (Guo et al., 2019). However, the performance of the chassis was limited and the biomass was too low even under the feed conditions, which hindered the synthesis of 13R-MO. Because of these drawbacks, our team applied a series of metabolic engineering strategies to produce 3 g/l 13R-MO via fed-batch fermentation in a 5 l bioreactor (Zhang et al., 2019). Our method included the overexpression of the MVA rate-limiting genes (tHMG1 and ERG20), the regulation of ERG9 expression at the transcription and protein levels (PHXT1-ERG9-PEST), the fusion of key genes in the MVA pathway (BTS1-GGGS-ERG20F96C), and the excision of the N-terminal plastid transit of CfTPS2 and CfTPS3 (Fig. 3).
Fig. 3.
Schematic overview of the 13R-MO and forskolin biosynthesis pathway. Native pathway genes are marked in orange, the Bts1p and Erg20F96Cp fusion protein is indicated in red, ERG9 was downregulated as shown in gray, heterologous 13R-MO biosynthesis genes are marked in green, and heterologous forskolin putative biosynthesis genes are marked in purple.
The Synthetic Pathway of 13R-MO Derivatives and its Optimization
Because of the substrate's promiscuity, structural modification of natural products is an effective way to obtain new natural products and improve their biological activity. Since enzymes involved in the heterologous synthesis of terpenoids sometimes show relaxed substrate specificity (Ignea, Athanasakoglou, et al., 2016a; Ignea, Ioannou, et al., 2016b), an option is to use alternative enzymes to produce intermediates in complex compound synthesis pathways. CYP76AH24 from Salvia pomifera oxidizes the C-12 of abietatriene to produce ferruginol and then continues to oxidize ferruginol at position C-11 to produce 11-hydroxy-ferruginol. Given that CYP76AH24 has more relaxed substrate specificity, it can hydroxylate the C-11β position of manoyl oxide to generate 11β-hydroxy-manoyl oxide (Ignea, Athanasakoglou, et al., 2016a; Ignea, Ioannou, et al., 2016b).
Exploring combinations of class I and class II diTPS from different sources, some diterpenoids that are more difficult to obtain from the environment were synthesized, such as 3β-hydroxy-manoyl oxide and 13-epi-manoyl oxide (Ignea et al., 2015). Reconstructing the synthetic pathway in yeast (using 8-hydroxy copalyl diphosphate synthase CcCLS from Cistus creticus combined with miltiradiene synthase SpMilS from S. pomifera and PtAO from Pinus taeda) yielded 1.1 mg/l of 3β-hydroxy-manoyl oxide.
The Synthetic Pathway of Forskolin and its Optimization
Besides the expression of cytochrome P450s, S. cerevisiae showed unique advantages for the conversion of 13R-MO into forskolin, such as the suitable intracellular environment and a complete membrane (Avalos et al., 2013).
Transferring four genes (CfCYP76AH15, CfCYP76AH11, CfCYP76AH16, and CfACT1–8), along with the cytochrome P450 reductase CfPOR (GenBank accession number KX151181) into S. cerevisiae EFSC4498 produced 350 mg/l of 13R-MO with fed-batch fermentation, resulting in 40 mg/l forskolin. This was the first total synthesis of forskolin in yeast (Pateraki et al., 2017). The limiting step in the forskolin production is obviously the expression of CfCYP76AHs. Recently, the expression of cytochrome P450s was optimized by protein engineering to improve the catalytic efficiency of plant-derived cytochrome P450s in microorganisms and increase the metabolic flow. In 2018, Forman et al. (Forman et al., 2018) identified the substrate recognition site of CfCYP76AH15 and developed an optimal mutant A99I via homology modeling and a semirational site-directed mutagenesis approach (Fig. 3). The activity of the mutant in engineered yeast increased by 5.6 times, and the yield of 11-oxo-manoyl oxide was twice that of the natural enzyme.
In summary, the MEP or MVA pathways produce the diterpeneoids universal precursor GGPP produced, the diterpeneoids synthase CfTPS2 and CfTPS3 convert it into the precursor 13R-MO, which undergoes six stereospecific oxidations to produce 7-deacetylforskolin via a series of cytochrome P450 monooxygenases (CfCYP76AH15, CfCYP76AH11, and CfCYP76AH16), cytochrome P450 reductase CfPOR (GenBank accession number KX151181), and finally the acetyltransferase CfACT1–8 acetylates the C-7 position of 7-deacetylforskolin to give forskolin. The product spectrum includes 13R-MO, 13R-MO derivatives, and forskolin (Table 1). There are two aspects of the current synthesis of forskolin in microbial hosts that can be improved: one is the amount of precursor 13R-MO produced. As mentioned above, our team has improved the yield of 13R-MO production. The second is the cytochrome P450s catalytic efficiency in microorganisms. As mentioned before, alternative enzymes such as CYP76AH24 have a more efficient catalytic effect on 13R-MO (Ignea, Athanasakoglou, et al., 2016a; Ignea, Ioannou, et al., 2016b). Besides, a semirational site-directed mutagenesis approach can improve the activity of cytochrome P450s, as exemplified by the site-directed mutagenesis of CfCYP76AH15 (Forman et al., 2018).
Table 1.
Overview of 13R-MO, 13R-MO Derivatives, and Forskolin Studies in Biosynthesis
| Host | Key pathways | Key enzymes | Product | Titer | References |
|---|---|---|---|---|---|
| E. coli | MEP | AgGGPPS, CfTPS2, CfTPS3 | 13R-manoyl oxide | 10 mg/l | Nielsen et al. (2014) |
| Synechocystis | MEP | CfTPS2, CfTPS3 | 13R-manoyl oxide | 0.45 mg/g DCW | Englund et al. (2015) |
| S. cerevisiae | MVA | CcCLS, SpMiLS, PtAO | 3β-OH-manoyl oxide | 4.87 mg/l | Ignea et al. (2015) |
| S. cerevisiae | MVA | CcCLS, SpMiLS | 13R-manoyl oxide | 80 mg/l | Ignea, Athanasakoglou, et al. (2016a), Ignea, Ioannou, et al. (2016b) |
| S. cerevisiae | MVA | CcCLS, SpMiLS, SpCYP76AH24 | 11β-OH-manoyl oxide | 21.9 mg/l | Ignea, Athanasakoglou, et al. (2016a), Ignea, Ioannou, et al. (2016b) |
| S. cerevisiae | MVA | CfTPS2, CfTPS3 | 13R-manoyl oxide | 350 mg/l | Andersen-Ranberg et al. (2016) |
| S. cerevisiae | MVA | CfTPS2, CfTPS3, CfCYP76AH15, CfCYP76AH11, CfCYP76AH16, CfPOR, CfACT1–8 | Forskolin | 40 mg/l | Pateraki et al. (2017) |
| S. cerevisiae | MVA | CfTPS2, CfTPS3, CfCYP76AH15 (A99I) , CfCYP76AH11, CfCYP76AH16 , CfPOR, CfACT1–8 | Forskolin | — | Forman et al. (2018) |
| S. cerevisiae | MVA | CfTPS2, CfTPS3, MET3p-ERG9 | 13R-manoyl oxide | 167.1 mg/l | Guo et al. (2019) |
| S. cerevisiae | MVA | tCfTPS2, tCfTPS3, PHXT1-ERG9-PEST, BTS1∼GGGS∼ERG20 (F96C) | 13R-manoyl oxide | 3 g/l | Zhang et al. (2019) |
Problems and Solutions in Forskolin Heterologous Biosynthesis
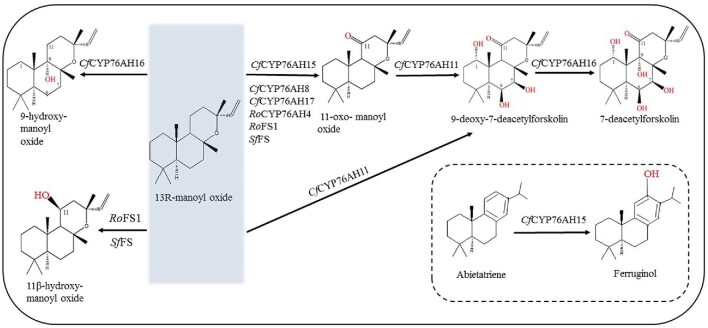
The engineering of S. cerevisiae allowed the biosynthesis of forskolin, in a synthetic pathway that involves the synergistic catalysis of multiple P450s. However, some issues remain. First, CfCYP76AH11, CfCYP76AH15, and CfCYP76AH16 can catalyze the oxidation of 13R-MO and the exact sequence of the catalytic oxidation of 13R-MO into forskolin is still unknown. Second, in the recent studies of the total putative synthesis pathway of forskolin, a large amount of the precursor 13R-MO and the intermediate 9-hydroxy-manoyl oxide accumulated, which hindered the subsequent catalytic oxidation of cytochrome P450 monooxygenase (Pateraki et al., 2017; Forman et al., 2018). Third, research on forskolin biosynthesis may need to identify and screen more cytochrome P450 monooxygenases. By analyzing the phylogeny of known functional P450s, cloning and expressing related P450 monooxygenase genes, and verifying their functions, we could identify P450 genes with higher catalytic specificity or promiscuity (Fig. 4). For instance, RoCYP76AH4 (Zi & Peters, 2013) from Rosemary officinalis can efficiently convert 13R-MO to 11-oxo-manoyl oxide, whereas RoFS1 and SfFS (Bozic et al., 2015) from Salvia fruticosa can produce 11β-hydroxy-manoyl oxide and 11-oxo-manoyl oxide. Fourth, the coordination of three cytochrome P450 monooxygenases in host cells is still a challenge. It is difficult to control the redox electron distribution environment, so it is necessary to continue to screen cytochrome P450s with higher catalytic efficiency and substrate specificity from C. forskohlii to shorten the total biosynthesis pathway of forskolin. Fifth, balancing the expression levels of cytochrome P450 reductase (CPR) and multiple cytochrome P450s is also challenging. Only one NADPH-dependent cytochrome P450 oxidoreductase was discovered in the forskolin biosynthesis pathway. More cytochrome P450 oxidoreductases with specificity and efficiency need to be found in C. forskohlii.
Fig. 4.
The promiscuity of cytochrome P450s in the forskolin biosynthesis pathway. The solid box indicates the related cytochrome P450s catalytic substrates and products; the dotted box indicates that CfCYP76AH15 can also catalyze the oxidation of abietatriene in addition to that of 13R-MO.
Obviously, the synergistic and efficient expression of multiple cytochrome P450s becomes the rate-limiting step in forskoline biosynthesis. When plant-derived cytochrome P450s are expressed in microorganisms, CPR is needed to provide electrons to complete the oxidation reaction (Gavira et al., 2013). The catalytic activity of cytochrome P450s is often affected by the following four aspects: (1) the catalytic properties of cytochrome P450s; (2) the matching of cytochrome P450s and CPR; (3) the efficiency of electron transfer from CPR to cytochrome P450s; and (4) the expression environment of cytochrome P450s and CPR. Therefore, this paper will summarize some expression strategies of plant cytochrome P450 enzymes in microorganisms, which may be beneficial to study the cytochromes P450 monooxygenases involved in the forskolin biosynthesis pathway (Fig. 5).
Fig. 5.
Cytochrome P450s expression strategies diagram.
The Choice of CPR and Balanced Expression Levels of P450s and CPR
CPR can influence the activity of cytochrome P450s by affecting the process of electron transfer (Pryor, 1996). Interaction with cytochrome P450s can also change the conformation of cytochrome P450s and affect the reaction process, thereby affecting the catalytic performance of cytochrome P450s (Sagadin et al., 2003). For example, some researchers tried to adapt to CPR from different sources (Zhu et al., 2018). To identify the best KO–KAH–CPR combination, two types of kaurene oxidase (KO), two types of kaurenoic acid hydroxylase (KAH), and five types of CPR were selected. Adjusting the copy number of KO–KAH–CPR led to an optimal combination that accumulated the final product and reduced the byproducts (Gold et al., 2018). Some researchers also screened mammalian cytochrome P450s and expressed new and interesting molecules in S. cerevisiae, providing guidance for the production of other terpenoids (Sarrade-Loucheur et al., 2020). By introducing highly effective cytochrome P450s and pairing them with various plant-derived CPR, the production of 11-oxo-β-amyrin and glycyrrhetinic acid GA was multiplied by nearly 1422 and 946.5, respectively (Wang et al., 2019). Screening 25 combinations of cytochrome P450s and CPR from five different sources greatly increased the yield of betulinic acid production (Jin et al., 2019).
Balancing the expression levels of cytochrome P450s and CPR is also important (Brown et al., 2015). The P450:CPR, P450:cytochrome b5 (CYB5), and P450:CPR:CYB5 ratios have an impact on the catalytic activity of cytochrome P450s (Zhang et al., 2007). In 2019, Lan et al. (Lan et al., 2019) constructed a dual-controllable system to balance the expression of cytochrome P450s and CPR and remarkably optimized the expression of cytochrome P450s and CPR, ultimately multiplying the yield of ganoderic acid production by 10.7. Thus, balancing the expression levels of CPR and cytochrome P450s with the least damage and the least loss is a key direction for future research.
Manual Design
Constructing an artificial cytochrome P450–CPR fusion protein can improve electron transfer efficiency (Li et al., 2007; Schueckel et al., 2012; Scheps et al., 2013). In our previous study, we constructed a cytochrome P450s–CPR fusion protein by simulating natural cytochrome P450 fusion protein to synthesize protopanaxadiol. This strategy improved the catalytic activity of cytochrome P450s by four- to fivefold (Zhao et al., 2016). The strain was restricted by electron transfer during the production of oleanolic acid in Yarrowia lipolytica, so the cytochrome P450 CYP716A12 was fused with the NADPH–P450 reductase ATR1 and the yield of oleanolic acid increased from 92.1 to 129.9 mg/l (Li et al., 2020). In Y. lipolytica, the fusion of CYP706M1 with t46AtCPR1 (a 46-amino acid N-terminal truncation of AtCPR1) improved the conversion efficiency of the precursor and multiplied the yield of (+)-nootkatone production by nearly 6 times (Guo et al., 2018).
Rational Design of Proteins
By analyzing the structure of these enzymes and the amino acid residues in their active site using molecular modeling and protein engineering, we may obtain a highly efficient and substrate-specific cytochrome P450 monooxygenase. Chun Li et al. (Sun et al., 2020) predicted the key residues of CYP72A63 via homology modeling and molecular docking and speculated on the amino acid residues that affect the interaction between CYP72A63 and its substrate. Then, they mutated the amino acid residues of the predicted active center and improved the catalytic activity of the CYP72A63 mutant by improving proton transfer. As mentioned before, homology modeling and site-directed mutagenesis of CfCYP76AH15 improve the yield of 11-oxo-manoyl oxide production by twofold (Forman et al., 2018).
CPR Replacement
According to research reports, CYB5 can also provide electrons for cytochrome P450 oxidation, and the expression of CYB5 instead of CPR reduces ROS toxicity (Gilep et al., 2001; Zhang et al., 2007). CYB5 from Glycyrrhiza uralensis can increase the efficiency of glycyrrhetinic acid production by eightfold (Wang et al., 2019). IA low CPR level can improve cell health, while the interaction between cytochrome P450s and CYB5 can improve the reaction rate of cytochrome P450s and reduce the release of reactive oxygen. Expressing the new CYB5 identified from Artemisia annua improved the synthesis rate of artemisinin (Paddon et al., 2013).
Adapting to Different Chassis Cells
Plant-derived cytochrome P450s have been expressed in different chassis cells. For example, cytochrome P450-mediated catalysis was designed to optimize the synthesis of taxol precursors in E. coli. Regulating the expression of CYP725A4 and CPR lead to the biosynthesis of taxol. E. coli is also a suitable host to study P450-mediated chemistry (Biggs et al., 2016). However, the expression of cytochrome P450s in E. coli has certain limitations. For instance, E. coli lacks the complicated membrane structure compartments of eukaryotic cells, does not have a good cytochrome P450 enzyme and reductase expression system. Furthermore, it lacks good glycosylation modification function. Besides, cytochrome P450 enzyme binds to the E. coli cell membrane to form inclusion bodies and loses activity. These features increase the difficulty of expression of this type of enzyme in the E. coli system, resulting in low or inexistent expression levels (Zelasko et al., 2013; Ichinose et al., 2015). Our team also tried to express cytochrome P450s in E. coli, but it was unsuccessful. Therefore, the expression of cytochrome P450s in a prokaryotic system is still inadequate (Renault et al., 2014). Y. lipolytica, as a GRAS (generally regarded as safe) unconventional yeast, has become a potential platform for the production of highly hydrophobic compounds (Xu et al., 2016). When plant cytochrome P450s were coexpressed with endogenous CPR, no product accumulated in Y. lipolytica. Therefore, it may be necessary to coexpress heterologous CPR to produce exogenous terpenoids, which is different from its expression in S. cerevisiae. Studies have shown that plant cytochrome P450s and yeast endogenous CPRs can be well matched in S. cerevisiae (Li & Zhang, 2015). Generally, plant-derived cytochrome P450 oxidase and reductase have transmembrane domains, and the complete and complex membrane structure compartment of S. cerevisiae makes it suitable to synthesize terpenoids via cytochrome P450s. The most famous case is the final yield of artemisinic acid, which was as high as 25 g/l by balancing the expression levels of CYP71AV1 and CPR in S. cerevisiae (Sun et al., 2020).
Concluding Remarks and Perspectives
The significant medicinal value of forskolin is obvious and producing forskolin via heterologous expression is the current research hotspot. The total biosynthesis pathway of forskolin has been resolved, but there are still many challenges to achieve a high yield. The catalytic properties of cytochrome P450s themselves limit their expression in microorganisms. Besides, the synergy of multiple cytochrome P450s and the efficiency of electron transfer between cytochrome P450s and CPR also affect the efficiency of forskolin synthesis. In response to the cytochrome P450s expression problems involved in the forskolin pathway, this paper summarizes some common strategies to express plant cytochrome P450s in microorganisms. First, it would be best to determine the definite sequence of catalyzing 13R-MO to synthesize forskolin via molecular modeling and protein engineering or find a cytochrome P450 gene with higher catalytic specificity or promiscuity. Second, to improve the substrate specificity of the cytochrome P450s, reduce the accumulation of intermediate 9-hydroxy-manoyl oxide, and direct the metabolic flow to forskolin as much as possible, we can use molecular modeling to analyze the amino acid residues of the cytochrome P450s active sites, and protein engineering to make mutations and other modifications. Third, we can match different sources of CPR with cytochrome P450s and balance the expression levels of cytochrome P450s and CPRs from different sources, construct artificial cytochrome P450–CPR fusion proteins to improve the electron transfer rate, or continue to screen other CPRs from C. forskohlii. Finally, carrying out rational protein design is a popular choice, and looking for CPR replacements such as CYB5, which can promote electron transfer, has demonstrated its validity.
In short, based on the structural complexity, therapeutic value, and imperfect biosynthetic pathway of forskolin, the production of forskolin in microorganisms still needs to overcome great difficulties to achieve efficiency. The multiple cytochrome P450 CfCYP76AHs involved in the forskolin biosynthesis pathway should be well designed via synthetic biology, metabolic engineering, and protein engineering techniques to promote the effective conversion of 13R-MO to forskolin. Establishing a microbial cell factory with high production of forskolin is necessary to achieve its industrialization. Obviously, the subtle adaptation of the metabolic pathway with plant-derived cytochrome P450s to the microorganisms will be an important step in improving the synthesis of forskolin and other natural products.
Abbreviations
13R-MO, 13R-manoyl oxide; cAMP, cell adenosine monophosphate; MVA, mevalonate; MEP, 2-321 methyl-4-phospho-D-erythroitol pathway; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; 322 GPP, geranyl diphosphate; FPP, farnesyl pyrophosphate; GGPP, geranylgeranyl diphosphate.
Contributor Information
Haiyan Ju, School of Chemical Engineering and Technology, Tianjin University, Tianjin 300350, P. R. China.
Chuanbo Zhang, School of Chemical Engineering and Technology, Tianjin University, Tianjin 300350, P. R. China.
Wenyu Lu, School of Chemical Engineering and Technology, Tianjin University, Tianjin 300350, P. R. China; Key Laboratory of System Bioengineering (Tianjin University), Ministry of Education, Tianjin 300350, P. R. China; SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300350, P. R. China.
Author Contributions
This review was conceived, researched, and written by Haiyan Ju and Chuanbo Zhang. Wenyu Lu supervised the review and revised the manuscript strictly. All authors listed have read and approved the manuscript.
Funding
This work was financially supported by National Key Research and Development Project of China (2019YFA0905100) and the National Natural Science Foundation of China (21878220).
Conflicts of Interest
The authors declare no conflict of interest.
Additional information
Publisher's Note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aharoni A., Galili G. (2011). Metabolic engineering of the plant primary-secondary metabolism interface. Current Opinion in Biotechnology, 22, 239–244. [DOI] [PubMed] [Google Scholar]
- Ammon H. P., Müller A. B. (1985). Forskolin: From an ayurvedic remedy to a modern agent. Planta Medica, 51, 473–477. [DOI] [PubMed] [Google Scholar]
- Andersen-Ranberg J., Kongstad K. T., Nielsen M. T., Jensen N. B., Pateraki I., Bach S. S., Hamberger B., Zerbe P., Staerk D., Bohlmann J., Moller B. L., Hamberger B. (2016). Expanding the landscape of diterpene structural diversity through stereochemically controlled combinatorial biosynthesis. Angewandte Chemie, 128, 2182–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M. T., Saify Z. S., Sultana N., Ahmad I., Saeed-Ul-Hassan S., Tariq I., Khanum M. (2013). Malaria and artemisinin derivatives: An updated review. Mini-Reviews in Medicinal Chemistry, 13, 1879–1902. [DOI] [PubMed] [Google Scholar]
- Asada Y., Li W., Terada T., Kuang X. Z., Li Q., Yoshikawa T., Hamaguchi S., Namekata I., Tanaka H., Koike K. (2012). Labdane-type diterpenoids from hairy root cultures of C. forskohlii, possible intermediates in the biosynthesis of forskolin. Phytochemistry, 79, 141–146. [DOI] [PubMed] [Google Scholar]
- Avalos J. L., Fink G. R., Stephanopoulos G. (2013). Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nature Biotechnology, 31, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajer T., Ligor M., Ligor T., Buszewski B. (2016). Design of the extraction process for terpenes and other volatiles from allspice by solid-phase microextraction and hydrodistillation. Journal of Separation Science, 39, 769–775. [DOI] [PubMed] [Google Scholar]
- Bhat S. V., Bajqwa B. S., Dornauer H., Do Scusa N. J., Fehlhaber H.-W. (1977). Structures and stereochemistry of new labdane diterpiniods from Coleus forskohlii briq. Tetrahedron Letters, 18, 1669–1672. [Google Scholar]
- Biggs B. W., Lim C. G., Sagliani K., Shankar S., Stephanopoulos G., De Mey M., Ajikumar P. K. (2016). Overcoming heterologous protein interdependency to optimize P450-mediated taxol precursor synthesis in Escherichia coli. Proceedings of the National Academy of Sciences of the USA, 113, 3209–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch K. (1992). Sterol molecule: Structure, biosynthesis, and function. Steroids, 57, 378–383. [DOI] [PubMed] [Google Scholar]
- Bodiwala H. S., Sabde S., Mitra D., Bhutani K. K., Singh I. P. (2009). Anti-HIV diterpenes from C. forskohlii. Natural Products Communications, 4, 1173–1175. [PubMed] [Google Scholar]
- Bozic D., Papaefthimiou D., Brueckner K., de Vos R. C. H., Tsoleridis C. A., Katsarou D., Papanikolaou A., Pateraki I., Chatzopoulou F. M., Dimitriadou E., Kostas S., Manzano D., Scheler U., Ferrer A., Tissier A., Makris A. M., Kampranis S. C., Kanellis A. K. (2015). Towards elucidating carnosic acid biosynthesis in Lamiaceae: Functional characterization of the three first steps of the pathway in Salvia fruticosa and Rosmarinus officinalis. PLoS One, 10, e0124106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S., Clastre M., Courdavault V., O'Connor S. E. (2015). De novo production of the plant-derived alkaloid strictosidine in yeast. Proceedings of the National Academy of Sciences of the USA, 112, 3205–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M. I., Zinczuk J., Rúveda E. A. (1992). Synthetic routes to forskolin. Tetrahedron, 48, 963–1037. [Google Scholar]
- Corey E. J., Jardine P. D. S. (1989). A short and efficient enantioselective route to a key intermediate for the total synthesis of forskolin. Tetrahedron Letters, 30, 7297–7300. [Google Scholar]
- Doseyici S., Mehmetoglu I., Toker A., Yerlikaya F., Erbay E. (2014). The effects of forskolin and rolipram on cAMP, cGMP and free fatty acid levels in diet induced obesity. Biotechnic & Histochemistry, 89, 388–392. [DOI] [PubMed] [Google Scholar]
- Du K., Guo P., Chen Y., Cao Z., Wang Z., Tang W. J. (2015). Enantioselective palladium-catalyzed dearomative cyclization for the efficient synthesis of terpenes and steroids. Angewandte Chemie, 54, 3033–3037. [DOI] [PubMed] [Google Scholar]
- Durazzo A., D'Addezio L., Camilli E., Piccinelli R., Turrini A., Marletta L., Marconi S., Lucarini M., Lisciani S., Gabrielli P., Gambelli L., Aguzzi A., Sette S. (2018). From plant compounds to botanicals and back: A current snapshot. Molecules, 23, 1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund E., Andersenranberg J., Miao R., Hamberger B., Lindberg P. (2015). Metabolic engineering of Synechocystis sp. PCC 6803 for production of the plant diterpenoid manoyl oxide. ACS Synthetic Biology, 4, 1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman V., Bjerg-Jensen N., Dyekjær J. D., Moller B. L., Pateraki I. (2018). Engineering of CYP76AH15 can improve activity and specificity towards forskolin biosynthesis in yeast. Microbial Cell Factories, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavira C., Höfer R., Lesot A., Lambert F., Zucca J., Werck-Reichhart D. (2013). Challenges and pitfalls of P450-dependent (+)-valencene bioconversion by Saccharomyces cerevisiae. Metabolic Engineering, 18, 25–35. [DOI] [PubMed] [Google Scholar]
- Gilep A. A., Guryev O. L., Usanov S. A., Estabrook R. W. (2001). Reconstitution of the enzymatic activities of cytochrome P450s using recombinant flavocytochromes containing rat cytochrome b (5) fused to NADPH-cytochrome P450 reductase with various membrane-binding segments. Archives of Biochemistry and Biophysics, 390, 215–221. [DOI] [PubMed] [Google Scholar]
- Gold N. D., Fossati E., Hansen C. C., DiFalco M., Douchin V., Martin V. J. J. (2018). A combinatorial approach to study cytochrome P450 enzymes for de novo production of steviol glucosides in baker's yeast. ACS Synthetic Biology, 7, 2918–2929. [DOI] [PubMed] [Google Scholar]
- Guo X. Y., Liu J. J., Zhang C. B., Zhao F. L., Lu W. Y. (2019). Stepwise increase in the production of 13R-manoyl oxide through metabolic engineering of Saccharomyces cerevisiae. Biochemical Engineering Journal, 144, 73–80. [Google Scholar]
- Guo X. Y., Sun J., Li D. S., Lu W. Y. (2018). Heterologous biosynthesis of (+)-nootkatone in unconventional yeast Yarrowia lipolytica. Biochemical Engineering Journal, 137, 125–131. [Google Scholar]
- Harborne J. B. (1995). Dictionary of natural products. Phytochemistry, 38, 279–279. [Google Scholar]
- Harde S. M., Singhal R. S. (2012). Extraction of forskolin from C. forskohlii roots using three phase partitioning. Separation and Purification Technology, 96, 20–25. [Google Scholar]
- Hylse O., Maier L., Kucera R., Perecko T., Svobodova A., Kubala L., Paruch K., Svenda J. (2017). A concise synthesis of forskolin. Angewandte Chemie, 56, 12586–12589. [DOI] [PubMed] [Google Scholar]
- Ichinose H., Hatakeyama M., Yamauchi Y. (2015). Sequence modifications and heterologous expression of eukaryotic cytochromes P450 in Escherichia coli. Journal of Bioscience and Bioengineering, 120, 268–274. [DOI] [PubMed] [Google Scholar]
- Ignea C., Athanasakoglou A., Ioannou E., Georgantea P., Trikka F. A., Loupassaki S., Roussis V., Makris A. M., Kampranis S. C. (2016a). Carnosic acid biosynthesis elucidated by a synthetic biology platform. Proceedings of the National Academy of Sciences of the USA, 113, 3681–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignea C., Ioannou E., Georgantea P., Loupassaki S., Trikka F. A., Kanellis A. K., Makris A. M., Roussis V., Kampranis S. C. (2015). Reconstructing the chemical diversity of labdane-type diterpene biosynthesis in yeast. Metabolic Engineering, 28, 91–103. [DOI] [PubMed] [Google Scholar]
- Ignea C., Ioannou E., Georgantea P., Trikka F. A., Athanasakoglou A., Loupassaki S., Roussis V., Makris A. M., Kampranis S. C. (2016b). Production of the forskolin precursor 11 β -hydroxy-manoyl oxide in yeast using surrogate enzymatic activities. Microbial Cell Factories, 15, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Tabata S. (2001). Synechocystis sp. PCC 6803-a useful tool in the study of the genetics of cyanobacteria. Photosynthesis Research, 70, 73–83. [DOI] [PubMed] [Google Scholar]
- Jin C. C., Zhang J. L., Song H., Cao Y. X. (2019). Boosting the biosynthesis of betulinic acid and related triterpenoids in Yarrowia lipolytica via multimodular metabolic engineering. Microbial Cell Factories, 18, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuki H., Bloch K. (1967). Studies on the biosynthesis of ergosterol in yeast: Formation of methylated intermediates. Journal of Biological Chemistry, 242, 222. [PubMed] [Google Scholar]
- Kikura M., Morita K., Sato S. (2004). Pharmacokinetics and a simulation model of colforsin daropate, new forskolin derivative inotropic vasodilator, in patients undergoing coronary artery bypass grafting. Pharmacological Research, 49, 275–281. [DOI] [PubMed] [Google Scholar]
- Lan X. T., Yuan W., Wang M., Xiao H. (2019). Efficient biosynthesis of antitumor ganoderic acid HLDOA using a dual tunable system for optimizing the expression of CYP5150L8 and a ganoderma P450 reductase. Biotechnology and Bioengineering, 116, 3301–3311. [DOI] [PubMed] [Google Scholar]
- Lee H. M., Vo P. N. L., Na D. (2018). Advancement of metabolic engineering assisted by synthetic biology. Catalysts, 8, 619–619. [Google Scholar]
- Li D. S., Wu Y. F., Wei P. P., Gao X., Li M., Zhang C. B., Zhou Z. J., Lu W. Y. (2020). Metabolic engineering of Yarrowia lipolytica for heterologous oleanolic acid production. Chemical Engineering Science, 218, 115529. [Google Scholar]
- Li J., Zhang Y. S. (2015). Modulating betulinic acid production in Saccharomyces cerevisiae by managing the intracellular supplies of the co-factor NADPH and oxygen. Journal of Bioscience and Bioengineering, 119, 77–81. [DOI] [PubMed] [Google Scholar]
- Li S. Y., Podust L. M., Sherman D. H. (2007). Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain. Journal of the American Chemical Society, 129, 12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Guan N. Z., Li J. H., Shin H. D., Du G. C., Chen J. (2015). Development of GRAS strains for nutraceutical production using systems and synthetic biology approaches: Advances and prospects. Critical Reviews in Biotechnology, 37, 1–12. [DOI] [PubMed] [Google Scholar]
- Lynen F. (1967). Biosynthetic pathways from acetate to natural products. Pure and Applied Chemistry, 14, 137–167. [DOI] [PubMed] [Google Scholar]
- Majeed M., Nagabhushanam K., Natarajan S., Vaidyanathan P., Karri S. K., Jose J. A. (2015). Efficacy and safety of 1% forskolin eye drops in open angle glaucoma—an open label study. Saudi Journal of Ophthalmology, 29, 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D., Lowry L., Determan M. K., Hershey D. M., Xu M. M., Peters R. J. (2010). Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Applied Microbiology and Biotechnology, 85, 1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2016). Natural products as sources of new drugs from 1981 to 2014. Journal of Natural Products, 79, 629–661. [DOI] [PubMed] [Google Scholar]
- Nielsen M. T., Ranberg J. A., Christensen U., Christensen H. B., Harrison S. J., Olsen C. E., Hamberger B., Moller B. L., Norholm M. H. H. (2014). Microbial synthesis of the forskolin precursor manoyl oxide in an enantiomerically pure form. Applied and Environmental Microbiology, 80, 7258–7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. E., Lanzotti V. (2009). Plant-derived natural products: Synthesis, function, and application. Springer-Verlag. [Google Scholar]
- Paddon C. J., Westfall P. J., Pitera D. J., Pitera D. J., Benjamin K., Fisher K., McPhee D., Leavell M. D., Tai A., Main A., Eng D., Polichuk D. R., Teoh K. H., Reed D. W., Treynor T., Lenihan J., Fleck M., Bajad S., Dang G., Newman J. D. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature, 496, 528. [DOI] [PubMed] [Google Scholar]
- Park S. Y., Yang D., Ha S. H., Lee S. Y. (2018). Metabolic engineering of microorganisms for the production of natural compounds. Advanced Biosystems, 2, 1700190. [Google Scholar]
- Pateraki I., Andersen-Ranberg J., Hamberger B., Heskes A. M., Martens H. J., Zerbe P., Bach S. S., Moller B. L., Bohlmann J., Hamberger B. (2014). Manoyl oxide (13R), the biosynthetic precursor of forskolin, is synthesized in specialized root cork cells in C. forskohlii. Plant Physiology, 164, 1222–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateraki I., Andersen-Ranberg J., Jensen N. B., Wubshet S. G., Heskes A. M., Forman V., Hallstroom B., Hamberger B., Motawia M. S., Olsen C. E., Staerk D., Hansen J., Moller B. L., Hamberger B. (2017). Total biosynthesis of the cyclic AMP booster forskolin from C. forskohlii. eLife, 6, e23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanaik B., Lindberg P. (2015). Terpenoids and their biosynthesis in cyanobacteria. Life, 5, 269–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta-Yahya P. P., Keasling J. D. (2010). Advanced biofuel production in microbes. Biotechnology Journal, 5, 147–162. [DOI] [PubMed] [Google Scholar]
- Peralta-Yahya P. P., Ouellet M., Chan R., Mukhopadhyay A., Keasling J. D., Lee T. S. (2011). Identification and microbial production of a terpene-based advanced biofuel. Nature communications, 2, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnam D., Shilpi S., Srinivas K., Suiab L., Alam S., Amtul Z., Arigari N. K., Jonnala K. K., Siddiqui L., Dubey V., Tiwari A. K., Balasubramanian S., Khan F. (2015). Synthesis of cyclic 1,9-acetal derivatives of forskolin and their bioactivity evaluation. European Journal of Medicinal Chemistry, 87, 735–744. [DOI] [PubMed] [Google Scholar]
- Pryor W. A. (1996). Cytochrome P450: Structure, mechanism, and biochemistry. Free Radical Biology and Medicine, 21, 251. [Google Scholar]
- Renault H., Bassard J. E., Hamberger B., Werck-Reichhart D. (2014). Cytochrome P450-mediated metabolic engineering: Current progress and future challenges. Current Opinion in Plant Biology, 19, 27–34. [DOI] [PubMed] [Google Scholar]
- Rohmer M., Knani M., Simonin P., Sutter B., Sahm H. (1993). Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochemical Journal, 295, 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagadin T., Riehm J. L., Milhim M., Hutter M. C., Bernhardt R. (2003). Binding modes of CYP106A2 redox partners determine differences in progesterone hydroxylation product patterns. Communications Biology, (2018) 1, 1414–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapio L., Gallo M., Illiano M., Chiosi E., Naviglio D., Spina A., Naviglio S. (2017). The natural cAMP elevating compound forskolin in cancer therapy: Is it time? Journal of Cellular Physiology, 232, 922–927. [DOI] [PubMed] [Google Scholar]
- Sarrade-Loucheur A., Ro D. K., Faure R., Remaud-Simeon M., Truan G. (2020). Synthetic derivatives of (+)-epi-α-bisabolol are formed by mammalian cytochromes P450 expressed in a yeast reconstituted pathway. ACS Synthetic Biology, 9, 368–380. [DOI] [PubMed] [Google Scholar]
- Savakis P., Hellingwerf K. J. (2015). Engineering cyanobacteria for direct biofuel production from CO2. Current Opinion in Biotechnology, 33, 8–14. [DOI] [PubMed] [Google Scholar]
- Scheps D., Malca S. H., Richter S. M., Marisch K., Nestl B. M., Hauer B. (2013). Synthesis of omega-hydroxy dodecanoic acid based on an engineered CYP153A fusion construct. Microbial Biotechnology, 6, 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueckel J., Rylott E. L., Grogan G., Bruce N. C. (2012). A gene-fusion approach to enabling plant cytochromes P450 for biocatalysis. Chembiochem, 13, 2758–2763. [DOI] [PubMed] [Google Scholar]
- Shu C., Li R. X., Yin Y. J., Yin D. Y., Gu Y. Q., Ding L., Zhong W. Y. (2014). Synergistic dual-targeting hydrogel improves targeting and anticancer effect of taxol in vitro and in vivo. Chemical Communications, 50, 15423–15426. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Misra A., Mishra P., Shukla P., Kumar M., Sundaresan V., Negi K. S., Agrawal P. K., Rawat A. K. S. (2017). Molecular and chemotypic variability of forskolin in C. forskohlii briq., a high value industrial crop collected from western himalayas (India). RSC Advances, 7, 8843–8851. [Google Scholar]
- Sun W. T., Xue H. J., Liu H., Lv B., Yu Y., Wang Y., Huang M., Li C. (2020). Controlling chemo- and regioselectivity of a plant P450 in yeast cell towards rare licorice triterpenoid biosynthesis. ACS Catalysis, 10, 4253–4260. [Google Scholar]
- Virgona N., Taki Y. K., Umegaki K. (2010). A rapid HPLC with evaporative light scattering method for quantification of forskolin in multi-herbal weight-loss solid oral dosage forms. Die Pharmazie, 65, 322–326. [PubMed] [Google Scholar]
- Vranová E., Coman D., Gruissem W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annual Review of Plant Biology, 64, 665–700. [DOI] [PubMed] [Google Scholar]
- Wang C. X., Su X. Y., Sun M. C., Zhang M. T., Wu J. J., Xing J. M., Wang Y., Xue J. P., Liu X., Sun W., Chen S. L. (2019). Efficient production of glycyrrhetinic acid in metabolically engineered Saccharomyces cerevisiae via an integrated strategy. Microbial Cell Factories, 18, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Qiao K. J., Ahn W. S., Stephanopoulos G. (2016). Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proceedings of the National Academy of Sciences of the USA, 113, 10848–10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Liu Q. K., Jacobsen S. E., Tang Y. (2018). The impact and prospect of natural product discovery in agriculture new technologies to explore the diversity of secondary metabolites in plants and microorganisms for applications in agriculture. EMBO Reports, 19, e46824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Sugiyama A., Satoh Y., Takahara A., Nakamura Y., Hashimoto K. (2002). Cardiovascular and adenylate cyclase stimulating effects of colforsin daropate, a water-soluble forskolin derivative, compared with those of isoproterenol, dopamine and dobutamine. Circulation Journal, 66, 1150–1154. [DOI] [PubMed] [Google Scholar]
- Zelasko S., Palaria A., Das A. (2013). Optimizations to achieve high-level expression of cytochrome P450 proteins using Escherichia coli expression systems. Protein Expression and Purification, 92, 77–87. [DOI] [PubMed] [Google Scholar]
- Zerbe P., Hamberger B., Yuen M. M. S., Chiang A., Sandhu H. K., Madilao L. L., Nguyen A., Hamberger B., Bach S. S., Bohlmann J. (2013). Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiology, 162, 1073–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. B., Ju H. Y., Lu C. Z., Zhao F. L., Liu J. J., Guo X. Y., Wu Y. F., Zhao G. R., Lu W. Y. (2019). High-titer production of 13R-manoyl oxide in metabolically engineered Saccharomyces cerevisiae. Microbial Cell Factories, 18, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. M., Im S. C., Waskell L. (2007). Cytochrome b (5) increases the rate of product formation by cytochrome p450 2B4 and competes with cytochrome p450 reductase for a binding site on cytochrome p450 2B4. Journal of Biological Chemistry, 282, 29766–29776. [DOI] [PubMed] [Google Scholar]
- Zhao F. L., Bai P., Liu T., Li D. S., Zhang X. M., Lu W. Y., Yuan Y. J. (2016). Optimization of a cytochrome P450 oxidation system for enhancing protopanaxadiol production in Saccharomyces cerevisiae. Biotechnology and Bioengineering, 113, 1787–1795. [DOI] [PubMed] [Google Scholar]
- Zhu M., Wang C. X., Sun W. T., Zhou A. Q., Wang Y., Zhang G. L., Zhou X. H., Huo Y. X., Li C. (2018). Boosting 11-oxo-ss-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants. Metabolic Engineering, 45, 43–50. [DOI] [PubMed] [Google Scholar]
- Zi J. C., Peters R. J. (2013). Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the lamiaceae. Organic & Biomolecular Chemistry, 11, 7650–7652. [DOI] [PMC free article] [PubMed] [Google Scholar]