Abstract
Streptomyces species are soil-dwelling bacteria that produce vast numbers of pharmaceutically valuable secondary metabolites (SMs), such as antibiotics, immunosuppressants, antiviral, and anticancer drugs. On the other hand, the biosynthesis of most SMs remains very low due to tightly controlled regulatory networks. Both global and pathway-specific regulators are involved in the regulation of a specific SM biosynthesis in various Streptomyces species. Over the past few decades, many of these regulators have been identified and new ones are still being discovered. Among them, a global regulator of SM biosynthesis named WblA was identified in several Streptomyces species. The identification and understanding of the WblAs have greatly contributed to increasing the productivity of several Streptomyces SMs. This review summarizes the characteristics and applications on WblAs reported to date, which were found in various Streptomyces species and other actinobacteria.
Keywords: Streptomyces, WhiB-like gene A (wblA), Secondary metabolite regulation
Introduction
Streptomyces are Gram-positive high G + C filamentous soil bacteria with superior characteristics in producing a variety of secondary metabolites (SMs), including many pharmaceutically valuable compounds, such as antibiotics, as well as anticancer, antiviral, and immunosuppressant agents (Bérdy, 2005; Kinghorn et al., 2009; Mann, 2001; Sivalingam et al., 2019) . Streptomyces SMs are synthesized by a group of enzymes encoded by their corresponding biosynthetic gene cluster (BGC). They are typically under tight and complicated regulation at the transcriptional level (Liu et al., 2013). The biosynthesis of Streptomyces SM is regulated through multiple regulatory pathways induced by both nutritional and environmental stimuli (Lee et al., 2005; Sun et al., 2017; van Wezel & McDowall, 2011). Although various global regulatory systems present in most Streptomyces species can control both morphological differentiation and SM production, the SM biosynthetic gene sets are also subject to pathway-specific regulation by linked regulatory genes (van der Heul et al., 2018). Most of these pathway-specific regulatory genes are transcriptionally regulated by a range of global regulatory networks in most Streptomyces species (Liu et al., 2013; Xia et al., 2020).
Among the global regulatory proteins, the WhiB-like (Wbl) family of proteins, which are only present in Actinobacteria, such as Streptomyces, Corynebacteria, and Mycobacteria, are a major class (Bush, 2018, refer to this reference for comprehensive review on Wbls). Following the initial characterization of WhiB in S. coelicolor (Chater, 1972; Davis & Chater, 1992), multiple paralogs were identified in many other Streptomyces species (Soliveri et al., 2000). Genome sequencing revealed the prevalence of Wbl paralogs throughout the phylum. Fourteen Wbl proteins were identified in S. coelicolor, with 11 encoded on the chromosome, and 3 encoded on the large linear plasmid, SCP1 (Bentley et al., 2002, 2004). The WblAsco is a WhiB4 ortholog, which is one of three Wbl-family members that regulate both differentiation and SM biosynthesis in S. coelicolor. In S. coeilcolor, a wblAsco mutant exhibits a defect in sporulation, with some aerial hyphae failing to sporulate and appearing thinner compared to the wild type (Aínsa et al., 2000; Fowler-Goldsworthy et al., 2011).
A Global Antibiotic Downregulator, WblAsco in S. coelicolor
The first biological function of WblA as an antibiotic downregulator was identified from somewhat unexpected experimental results. After the genome of S. coelicolor was sequenced nearly 20 years ago, Streptomyces interspecies DNA microarray analysis was applied to detect the global changes in mRNA abundance associated with the overproduction of the anticancer compound doxorubicin (DOX) in an S. peucetius overproducing industrial mutant (OIM) strain. S. coelicolor genome was the only available genome at the time to facilitate transcriptome analysis. The results showed that the wblAsco gene was a pleiotropic downregulator of antibiotic biosynthesis in S. coelicolor (Fowler-Goldsworthy et al., 2011). Comparative transcriptome analyses of cultures of the wild-type and OIM mutant strains of S. peucetius using S. coelicolor cDNA microarrays which was the only available genome at the time identified more than 100 S. coelicolor potential candidate genes that showed at least a twofold change in transcription between the wild-type and OIM mutant strains. After further analysis of the growth phase-dependent transcription profiles of these potential candidate genes, 20 genes exhibiting particularly large transcriptional changes between the two strains were selected and overexpressed individually in S. coelicolor (Kang et al., 2007).
Among them, SCO3579, which was previously proposed to be a whiB-like putative transcription factor gene, was identified and later named wblAsco in S. coelicolor (Soliveri et al., 2000). Although whiB is a developmental regulatory gene that was identified in S. coelicolor as being essential for the sporulation of aerial hyphae, the biological function of wblAsco in SM regulation was not determined (Soliveri et al., 2000). The overexpression of wblAsco inhibited the biosynthesis of actinorhodin (ACT), undecylprodigiosin (RED), and calcium-dependent antibiotic (CDA) in S. coelicolor. Moreover, transcripts encoded by pathway-specific activators of the three major S. coelicolor antibiotics (i.e. actIIORF4 for ACT, redD/Z for RED, and cdaR for CDA) were reduced in the wblAsco-overexpressing S. coelicolor, implying that wblAsco acts broadly to downregulate antibiotic biosynthesis in S. coelicolor (Kang et al., 2007).
During Streptomyces interspecies DNA microarray analysis, a previously-unidentified tetR family transcriptional regulatory gene (SCO1712), as well as a carbon flux regulating 6-phosphofructokinase gene (SCO5426), were also found to downregulate SM biosynthesis in the S. coelicolor wild-type strain as well as in a wblAsco deletion mutant (Kim et al., 2011; Lee et al., 2010). The S. coelicolor triple knockout mutant (ΔwblAsco, ΔSCO1712, and ΔSCO5426) showed the highest level of ACT production compared to any of the single and double knockout mutants in S. coelicolor, suggesting that wblAsco along with other wblAsco-independent regulatory and precursor pathway genes could be optimized synergistically for SM production in Streptomyces (Kim et al., 2011).
WblA Orthologs in Other Streptomyces Species
WblAspe in S. peucetius
Subsequently, a wblAsco ortholog named wblAspe from S. peucetius was identified through the construction of a total genomic DNA library of the above-mentioned S. peucetius OIM and the screening using a wblAsco gene probe. As expected, the production of both DXR and its precursor, daunorubicin (DNR) were improved through gene disruption of wblAspe from S. peucetius OIM (Table 1). Moreover, several putative wblAspe-dependent genes were also identified using interspecies DNA microarray analysis between the S. peucetius OIM and wblAspe-disrupted S. peucetius OIM. Among the putative wblAspe-dependent genes tested, a conserved hypothetical protein (SCO4967) further stimulated the production of DXR/DNR/aklavinone in the wblAspe-disrupted S. peucetius OIM. These results suggest that sequential genetic manipulation of the wblAspe and its dependent genes identified from comparative transcriptome analysis could provide an efficient and rational strategy for improving the titer of DXR/DNR in S. peucetius strain. This was the first example to improve the antibiotic-OIM strain using a microarray-driven reverse engineering approach in Streptomyces species (Noh et al., 2010).
Table 1.
WblA orthologs identified from various Streptomyces species
| Gene | Strain | Size | Identity (with WblAsco) | Compounds | SM production in DIS mutant (fold) | References |
|---|---|---|---|---|---|---|
| WblAsco | S. coelicolor M145 | 112 aa | 100% | Actinorhodin | Increased (5.6-fold) | Lee et al. (2010) |
| WblAspe | S. peucetius OIM-1 | 114 aa | 95% | Doxorubicin, daunorubicin | Increased (1.7-fold) | Kang et al. (2007), Noh et al. (2010) |
| WblAtmc | Streptomyces sp. CK4412 | 130 aa | 96% | Tautomycetin | Increased (3.2-fold) | Nah et al. (2012) |
| WblAgh | S. ghanaensis ATCC14672 | 128 aa | 96% | Moenomycin A | Increased (2.3-fold) | Rabyk et al. (2011) |
| Increased (2.5-fold) | Yan et al. (2020) | |||||
| WblAsro | S. roseosporus NRRL15998 | 116 aa | 90% | Daptomycin | Increased (1.5-fold) | Huang et al. (2017) |
| WblAsan | S. ansochromogenes 7100 | 112 aa | 96% | Nikkomycin (major) | Abolished | Lu et al. (2015) |
| Tylosin analogs (cryptic) | Activated | |||||
| WblAso | S. somaliensis SCSIO ZH66 | 124 aa | 85% | Violapyrone | Increased (NR) | Huang et al. (2016) |
| WblAsve | S. venezuelae ATCC15439 | 115 aa | 90% | Pikromycin | Increased (3.5-fold) | Yan et al. (2016) |
| WblAscb | Streptomyces sp. CB03234 | 131 aa | 93% | Tiancimycins | Increased (13.9-fold) | Nguyen et al. (2003) |
SM, secondary metabolite; DIS, disruption; NR, not reported.
WblAtmc in Streptomyces sp. CK4412
Tautomycetin (TMC) is a linear polyketide compound with a novel activated T cell-specific immunosuppressant and anticancer activities (Chae et al., 2004; Lee, Lee, et al., 2006; Niu et al., 2012). The whole-genome sequencing of the Streptomyces sp. CK4412 chromosome revealed the entire TMC (∼80-kb) BGC as well as the wblAsco ortholog gene named wblAtmc. The wblAtmc from Streptomyces sp. CK4412 showed 96% amino acid identity compared to a previously known wblAsco. (Figs. 1 & 2). The targeted gene disruption of wblAtmc in Streptomyces sp. CK4412 caused an approximately threefold higher TMC production titer than that in the wild-type strain. Moreover, transcription analyses of the two TMC pathway-specific positive regulatory genes, tmcN and tmcT, located within its BGC showed that the only tmcT expression was strongly downregulated by wblAtmc in Streptomyces sp. CK4412 (Nah et al., 2012). The tmcN expression was not affected by either deletion or overexpression of wblAtmc in Streptomyces sp. CK4412 (Nah et al., 2012). These results suggest that the TMC BGC regulatory network is controlled by two pathway-specific positive regulators, WblAtmc-dependent TmcT and WblAtmc-independent TmcN, in Streptomyces sp. CK4412 (Nah et al., 2012).
Fig. 1.
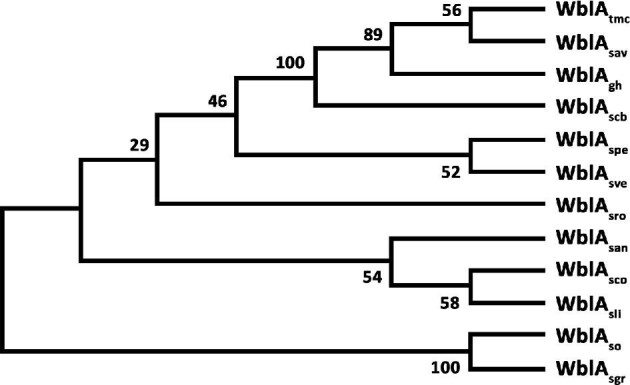
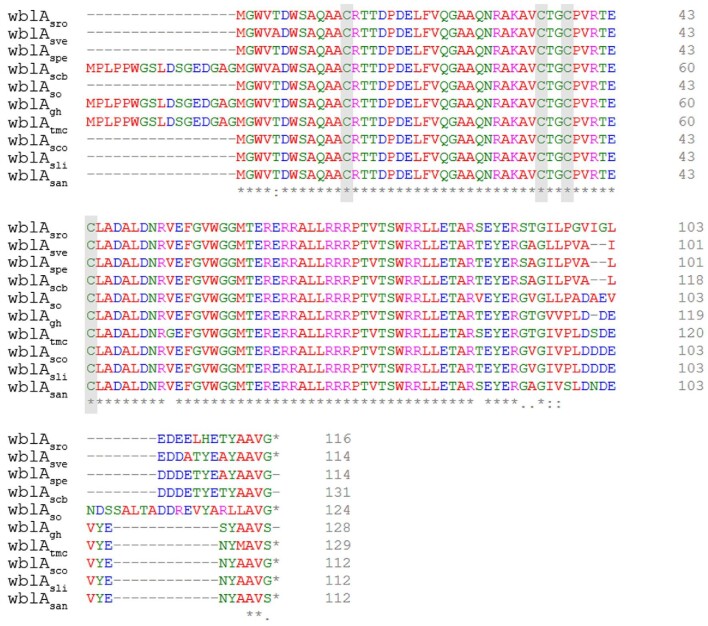
WblAs phylogenetic tree among Streptomyces species. Phylogenetic tree was built using the Mega X software by neighbor joining test by Bootstrap. WblAsco from S. coelicolor; wblAspe from S. peucetius; wblAtmc from Streptomyces sp. CK4412; wblAgh from S. ghanaensis; wblAsro from S. roseosporus; wblAsan from S. ansochromogenes; wblAso from S. somaliensis; wblAsve from S. venezuelae; wblAscb from Streptomyces sp. CB03234; wblAsav from S. avermitilis; and wblAsgr from S. griseus.
Fig. 2.
Sequence alignments among the Streptomyces WblAs. Conserved (asterisk) and related (colon) amino acids were marked underneath. Gray box; cysteine conserved region.
WblAgh in S. ghanaensis
Another wblAsco ortholog named wblAgh was identified from the whole-genome sequencing of the moenomycin producer, S. ghanaensis ATCC14672. A deletion of wblAgh stimulated a more than twofold increase in moenomycins production along with inhibition of aerial mycelium sporulation. Moreover, the wblAgh overexpression in S. ghanaensis ATCC14672 decreased the moenomycin production by 50%, implying that WblAgh is a global antibiotic downregulator in S. ghanaensis ATCC14672. Since the moenomycin BGC in S. ghanaensis ATCC14672 did not contain any pathway-specific regulatory genes, the downstream target of WblAgh is not known. Although the regulation of putative Streptomyces subtilisin inhibitor (SSI) named SSFG_01 620 was proposed to be linked to the WblAgh deletion in S. ghanaensis ATCC14672, the detailed mechanism between WblAgh and SSFG_01 620 needs to be further pursued (Rabyk et al., 2011).
WblAsro in S. roseosporus
The wblAsco ortholog gene named wblAsro was also identified in the whole genome sequencing of the daptomycin producer S. roseosporus. Three types of strains, the wblAsro disruption strain, the complemented strain, and the overexpression strains, were generated to determine if it could affect the production of SMs as well as morphogenesis. The disruption mutant of wblAsro enhanced daptomycin production by more than 50% as well as blocked aerial mycelium sporulation. In contrast, overexpression of wblAsro resulted in a significant decrease in the daptomycin production titer. As expected, the transcription of the key daptomycin positive regulatory genes atrA, dptR2, and dptR3, and the structural gene, dptE, were increased remarkably in the wblAsro disruption mutant. These results suggest that wblAsro plays a key downregulatory role in controlling daptomycin biosynthesis (Huang et al., 2017).
WblAsan in S. ansochromogenes
Another wblAsco ortholog named wblAsan was found in S. ansochromogenes 7100 sequencing analysis. The wblAsan disruption mutant of S. ansochromogenes 7100 failed to sporulate as well as to produce nikkomycin, a major SM produced by S. ansochromogenes 7100 during fermentation. Moreover, two novel 16-membered tylosin-like macrolides were observed only in a fermentation broth of ΔwblAsan strain. These two compounds, which had different functional groups at the C6 position comparing with tylosin, exhibited similar antibacterial activities against several Gram-positive bacteria including Staphylococcus aureus and Bacillus cereus. Interestingly, however, these two compounds displayed much higher activity against S. pneumoniae than tylosin, suggesting that wblA ortholog disruption approach could activate cryptic compounds hidden in Streptomyces species, and the compounds identified by the ΔwblAsan in S. ansochromogenes might be used to broaden the application of tylosin (Lu et al., 2015).
WblAso in S. somaliensis
A wblAsco ortholog named wblAso was found in deep sea-derived S. somaliensis SCSIO ZH66 through the whole-genome sequencing analysis. To activate cryptic BGCs in S. somaliensis SCSIO ZH66, the wblAso from S. somaliensis SCSIO ZH66 was inactivated. Noticeable changes in SM production from the S. somaliensis ΔwblAso mutant were observed and the α-pyrone compound named violapyrone B (VLP B) was isolated. The VLP BGC consisted of a type III polyketide synthase (PKS) gene vioA and a pathway-specific positive regulatory gene vioB. The inactivation of vioB further led to the isolation of another four VLPs analogs, one novel SM and two improved anti-MRSA (methicillin-resistant S. aureus, MRSA) SMs compared to VLP B. Transcriptional analysis showed that wblAso seemed to regulate the expression levels of whi genes and wbl genes by different degrees, suggesting an intertwined regulatory mechanism of wblAso in SM biosynthesis as well as in morphological differentiation from S. somaliensis SCSIO ZH66. The wblAso inactivation-driven VLPs identification results imply that the wblAso orthologs would be effective targets for the activation of cryptic BGCs in marine-derived Streptomyces strains (Huang et al., 2016).
WblAsve in S. venezuelae
S. venezuelae ATCC15439 is a versatile producer for various macrolide antibiotics. The 12-membered and 14-membered ring macrolides are biosynthesized by pikromycin BGC (Lee, Park, et al., 2006; Oh & Kang, 2012; Xue et al., 1998; Zhang & Sherman, 2001). Because of low levels of pikromycin production, genetic engineering for titer improvement have been developed (Maharjan et al., 2008; Pyeon et al., 2017; Yi et al., 2018). The wblA ortholog gene named wblAsve was also found with a high degree of amino acid identity (90% with WblAsco) from S. venezuelae ATCC15439. Sporulation was blocked by a disruption of wblAsve in S. venezuelae ATCC15439. The production of pikromycin was increased by 3.5-fold in S. venezuelaeΔwblAsve and decreased by 2.5-fold in the wblAsve overexpressing strain (Woo et al., 2014), suggesting that the WblAsve controls both morphological differentiation and pikromycin production in S. venezuelae.
WblAscb in Streptomyces sp. CB03234
Streptomyces sp. CB03234 is a native producer of both ten-membered enediyne tiancimycins (TNMs) and diterpernoid tiancilactones (TNLs) (Dong et al., 2018; Yan et al., 2016). Among the TNMs discovered, both TNM-A and TNM-D showed superior cytotoxic activities against several cancer cell lines (Yan et al., 2016, 2018). Comparative transcription analysis between a wild-type and a streptomycin-induced ribosome engineered TNMs high-producer CB03234-S exhibited that the wblAscb transcription level was relatively higher than those of other wbl genes (Zhang et al., 2020). To overcome the low titer issue of these compounds, the wblAsco ortholog named wblAscb (tentatively named here) was disrupted via homologous recombination, resulting in 13.9- and 1.7-fold increases of TNM-A in CB3234S and CB3234-S, respectively (Zhang et al., 2020). The production of TNLs, a group of main fermentation metabolites in CB03234, was also affected by the deletion of wblAscb. Although the wblAscb in CB03234 deletion could improve the titers of TNMs significantly, the sporeless bald phenotypes could lead to the CB03234ΔwblAscb unsuitable for the scaled-up TNMs production. Since the existence of wblAscb is necessary for a spore formation but undesirable for TNMs overproduction, the NitR-ε-caprolactam (CPL) based inducible expression system for wblAscb was constructed in the CB03234ΔwblAscb. This cell factory system successfully maintained the overproduction of TNMs without any additional processes and recovered the normal development of sporulation upon induction (Zhang et al., 2020).
WblA Orthologs as SM Upregulators
Although most WblAs stated above negatively regulate SM biosynthesis in Streptomyces species, a couple of WblA orthologs such as WblAch in S. chattanoogensis L10 and WblAsxi in S. xiamenensis 318 have been reported to act as SM upregulators in their hosts (Bu et al., 2019; Yu et al., 2014). The wblAsxi overexpression under the Streptomyces constitutive promoter ermE*p stimulated the production of two tetramate macrolactams, ikarugamycin and capsimycin B in S. xiamenensis 318 (Bu et al., 2020). Similarly, the WblAch was proved to function as an activator of natamycin biosynthesis in S. chattanoogensis L10. The wblAch disruption downregulated natamycin production, while the wblAch overexpressed stimulated the natamycin yield by ∼30% (Yu et al., 2014). These findings suggest that some wblA orthologs are positively involved in the regulation of SM biosynthesis in some Streptomyces species.
Putative Regulatory Mechanism of WblA in Streptomyces
AdpA Binding to the wblA Promoter Region
Although many reports support that WblA is a general downregulator of SM biosynthesis in Streptomyces species, its regulatory target and mechanism are unclear. Although WblA has been proposed to be a transcriptional factor binding to a specific promoter of the target gene, there is no experimental evidence showing direct binding between WblA and the target promoter sequence (Bush, 2018). On the other hand, some studies on how the wblA gene expression is controlled have been reported. The upstream region of wblA in S. coelicolor was predicted to contain several putative AdpA binding motifs (Lee et al., 2013; Nguyen et al., 2003; Wolanski et al., 2011). AdpA is a key regulator controlling various processes involved in Streptomyces morphological differentiation and SM biosynthesis (Higo et al., 2012; Ohnishi et al., 2005). AdpAsc was shown to bind specifically the wblAsco upstream binding motifs through an electrophoretic mobility shift assay (EMSA), even though AdpAsc failed to bind to the mutated wblA upstream motif (Lee et al., 2013). Moreover, an AdpAsco disruption mutant showed increased wblAsc transcription in S. coelicolor, suggesting that AdpAsc negatively regulates wblAsco transcription in S. coelicolor (Fig. 3, Lee et al., 2013).
Fig. 3.
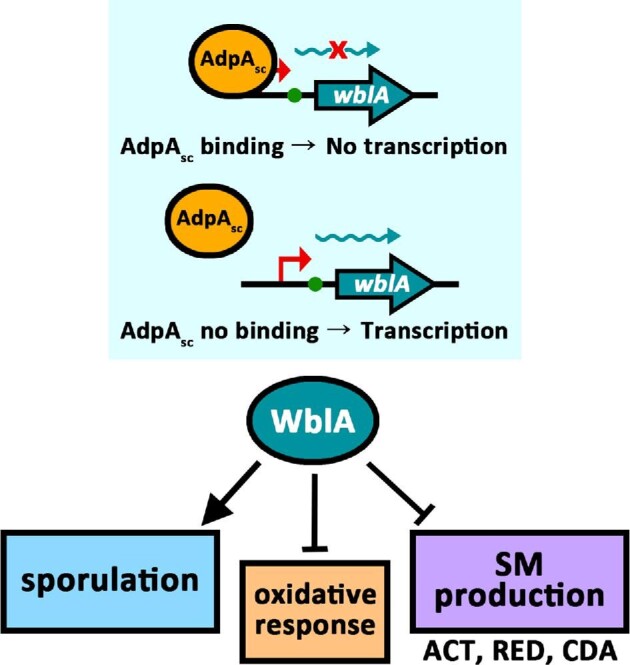
Regulatory network of WblA controlled by AdpAsc in S. coelicolor. ACT, actinorhodin; RED, undecylprodigiocin; CDA, calcium-dependent antibiotic.
BldDsgh Binding to the wblAgh Promoter Region
Cyclic dimeric 3′–5′ guanosine monophosphate, c-di-GMP, is a ubiquitous second messenger controlling diverse cellular processes in bacteria (Römling et al., 2013). In Streptomyces, c-di-GMP plays a crucial role in a complex morphological differentiation by modulating the activity of the pleiotropic regulator BldD (Tschowri et al., 2014). Previous work on the transcriptional analysis of the BldD-targeted genes in non-Streptomyces actinobacterium Actinoplanes missouriensis showed that BldD repressed 12 genes, including a wblA ortholog (Mouri et al., 2017). The deletion of bldDgh led to an increase in wblAgh expression. While the bldDgh-deleted mutant of S. ghanaensis showed strongly reduced moenomycin A (MmA) production, the double mutant S. ghanaensis ΔbldDghΔwblAgh restored the capacity to produce MmA. This suggests that WblAgh plays a crucial role in the regulation of MmA biosynthesis and that BldDgh controls the expression of wblAgh. EMSA proved the direct control of wblAgh transcription by BldDgh. Increasing concentrations of BldDgh resulted in the formation of one nucleoprotein complex with the wblAgh promoter through EMSA. Moreover, binding of BldDgh to the wblAgh promoter was improved significantly in the presence of increasing concentrations of c-di-GMP. The specificity of this interaction is shown by the ability of the unlabeled wblAgh promoter to compete with the radiolabeled one for BldDgh (Makitrynskyy et al., 2020). This suggests that the high expression of wblA is mediated by the deletion of bldD, which leads to the repression of antibiotic synthesis (Yan et al., 2020).
Oxidative Stress Response by WblA
In addition to the Streptomyces SM regulation, WblA was also suggested to be involved in the oxidative stress response (Kim et al., 2012). Since WhcA, a WblA ortholog in C. glutamicum, was found to play a negative role in the oxidative stress response, wblAsco was speculated to have a similar role in S. coelicolor. A wblAsco-deletion mutant showed a less sensitive response to oxidative stress induced by the diamide present in the solid plate culture. Comparative real-time qRT-PCR analysis showed that the transcription levels of oxidative stress-related genes, including sodF, sodF2, sodN, trxB, and trxB2, were higher in the wblAsco-deletion mutant than the wild type, both in the absence and presence of oxidative stress. Moreover, expression of these target genes in the S. coelicolor wild type was stimulated only in the presence of oxidative stress, suggesting that WblAsco might play a negative role in the oxidative stress response of S. coelicolor, similar to that found in C. glutamicum WhcA (Kim et al., 2012).
Because WhcA was confirmed to interact with dioxygenase-encoding SpiA (stress protein interacting with WhcA) in C. glutamicum, a SpiA ortholog in S. coelicolor SCO2553 protein (named SpiAsco) was also proposed to interact with WblA in S. coelicolor. Using heterologous expression in Escherichia coli and in vitro pull-down assays, WblAsco was confirmed to bind to the SpiAsco, which was influenced by oxidants, such as diamide. These observations suggest that the interaction between WblAsco and SpiAsco is not only specific but also modulated by the redox status of the cell. Moreover, a spiAsco-disruption mutant exhibited a less sensitive response to the oxidative stress induced by the diamide present in solid plate culture. Real-time qRT-PCR analysis also showed that the transcription levels of oxidative stress response genes were higher in the spiAsco-deletion mutant than in wild-type S. coelicolor. These results show that SpiAsco negatively regulates WblA during the oxidative stress responses in S. coelicolor (Kim et al., 2013).
WblA Orthologs in Other Actinobacteria
The Gram-positive rare actinomycete Pseudonocardia autotrophica KCTC9441 was previously identified in the novel di-sugar-containing polyene compound producer, which was called NPP (Nystatin-like Pseudonocardia Polyene) (Kim et al., 2009). By whole-genome sequencing, a WblA ortholog was isolated and identified from P. autotrophica. WblApau showed 49% amino acid identity with various Streptomyces WblAs and 39% amino acid identity with a WblA ortholog, WhcA from C. glutamicum (Kim et al., 2014). Although no significant differences in NPP production were observed in the heterologous expression of wblAsco, a disruption of wblApau resulted in an approximately 80% increase in NPP production (Kim et al., 2014). These results suggest that the biological significance of wblApau might be similar to a previously known wblA from various Streptomyces strains, even though the amino acid identity was relatively low.
Corynebactirum rarely produces antibiotics and other SMs, there are no reports on WhcA-driven SM regulation. On the other hand, WhcA and other WhiB-like proteins appear to play key roles in the regulation of stress-related processes. WhcA appears to regulate negatively the genes involved in response to oxidative stress (Choi et al., 2009). WhcA was reported to interact directly with its partner, SpiA (Stress protein encoding a dioxygenase/oxidoreductase interacting with WhcA). This WhcA-SpiA interaction was confirmed experimentally to be disrupted in the presence of the oxidant diamide (Park et al., 2012). As stated above, this mechanism appears to be conserved in S. coelicolor, in which a SpiAsco was found to bind to WblAsco and downregulate the WblA-dependent oxidative stress response (Kim et al., 2013).
There is also a WblA ortholog named WhiB4mtb in Mycobacterium tuberculosis (Bush, 2018). The genome of M. smegmatis MC2 155, a model strain to study M. tuberculosis, has been sequenced (Mohan et al., 2015). The M. smegmatis MC2 155 and M. tuberculosis share significant similarities in their genome. The in silico analysis using antiSMASH 5.0 predicted presence of 18 SM BGCs in MC2 155. However, little is known on relation between SM regulation and WhiB4mtb. A major role of WhiB4mtb in Mycobacteria is believed to regulate redox-sensing and its homeostasis. A deletion of whiB4mtb leads to the hyper-induction of antioxidants, increased resistance to oxidative stress in vitro, and enhanced survival in macrophages (Alam et al., 2007; Chawla et al., 2012). WhiB4mtb also contains an O2- and NO-sensitive [4Fe–4S] cluster and the WhiB4mtb [4Fe–4S] cluster appears to be more sensitive to O2 than that other reported Wbls (Crack et al., 2009; Kudhair et al., 2017; Singh et al., 2007).
Concluding remarks
WblAs are highly homologous global SM regulators present in most Streptomyces species as well as its closely related actinobacteria, of which the detailed mechanism needs to be further elucidated. Although WblAs typically regulate the SM BGC expression at the transcriptional level by controlling the target pathway-specific regulatory gene expression, there was no direct evidence showing direct binding between WblA and the target promoter sequence. The WblA is believed to be controlled by another global regulator such as AdpA and also involved in morphological differentiation and oxidative stress response through its iron-sulfur cluster. Considering the prevalence of WblAs and its conserved regulatory roles, the strategy for selective manipulation of WblAs should provide an efficient approach to improve the SM titer and discover cryptic SMs in actinobacteria.
Funding
This study was supported by the National Research Foundation of Korea (No. 2021R1A2C2012203). H.J. Nah was partially supported by the Basic Science Research Program through the National Research Foundation of Korea (No. 2018R1A6A3A01013335).
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Hee-Ju Nah, Department of Biological Engineering, Inha University, Incheon 22212, Republic of Korea.
Jihee Park, Department of Biological Engineering, Inha University, Incheon 22212, Republic of Korea.
Sisun Choi, Department of Biological Engineering, Inha University, Incheon 22212, Republic of Korea.
Eung-Soo Kim, Department of Biological Engineering, Inha University, Incheon 22212, Republic of Korea.
References
- Aínsa J. A., Ryding N. J., Hartley N., Findlay K. C., Bruton C. J., Chater K. F. (2000). WhiA, a protein of unknown function conserved among gram-positive bacteria, is essential for sporulation in Streptomyces coelicolor A3(2). Journal of Bacteriology, 182, 5470–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. S., Garg S. K., Agrawal P. (2007). Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: A [4Fe-4S] cluster co-ordinating protein disulphide reductase. Molecular Microbiology, 63, 1414–1431. [DOI] [PubMed] [Google Scholar]
- Bentley S. D., Brown S., Murphy L. D., Harris D. E., Quail M. A., Parkhill J., Barrell B. G., McCormick J. R., Santamaria R. I., Losick R., Yamasaki M., Kinashi H., Chen C. W., Chandra G., Jakimowicz D., Kieser H. M., Kieser T., Chater K. F. (2004). SCP1, a 356,023 bp linear plasmid adapted to the ecology and developmental biology of its host, Streptomyces coelicolor A3(2). Molecular Microbiology, 51, 1615–1628. [DOI] [PubMed] [Google Scholar]
- Bentley S. D., Chater K. F., Cerdeño-Tárraga A. M., Challis G. L., Thomson N. R., James K. D., Harris D. E., Quail M. A., Kieser H., Harper D., Bateman A., Brown S., Chandra G., Chen C. W., Collins M., Cronin A., Fraser A., Goble A., Hidalgo J., Hopwood, D. A. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature, 417, 141–147. [DOI] [PubMed] [Google Scholar]
- Bérdy J. (2005). Bioactive microbial metabolites. Journal of Antibiotics, 58, 1–26. [DOI] [PubMed] [Google Scholar]
- Bu X. L., Weng J. Y., He B. B., Xu M. J., Xu J. (2019). A novel adpA homologue negatively regulates morphological differentiation in Streptomyces xiamenensis 318. Applied and Environmental Microbiology, 85(7), e03107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu X. L., Weng J. Y., He-Lin Y., Xu M. J., Xu J. (2020). Three transcriptional regulators positively regulate the biosynthesis of polycyclic tetramate macrolactams in Streptomyces xiamenensis 318. Applied Microbiology and Biotechnology, 104, 701–711. [DOI] [PubMed] [Google Scholar]
- Bush M. J. (2018). The actinobacterial WhiB-like (Wbl) family of transcription factors. Molecular Microbiology, 110, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae W. J., Choi J. M., Yang J. J., Lee S. K. (2004). T Cell-specific immunosuppression using tautomycetin or PTD-conjugated protein drugs. Yonsei Medical Journal, 45, 978–990. [DOI] [PubMed] [Google Scholar]
- Chater K. F. (1972). A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. Journal of General Microbiology, 72, 9–28. [DOI] [PubMed] [Google Scholar]
- Chawla M., Parikh P., Saxena A., Munshi M., Mehta M., Mai D., Srivastava A. K., Narasimhulu K. V., Redding K. E., Vashi N., Kumar D., Steyn A. J., Singh A. (2012). Mycobacterium tuberculosis WhiB4 regulates oxidative stress response to modulate survival and dissemination in vivo. Molecular Microbiology, 85, 1148–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W. W., Park S. D., Lee S. M., Kim H. B., Kim Y. H., Lee H. S. (2009). The whcA gene plays a negative role in oxidative stress response of Corynebacterium glutamicum. FEMS Microbiology Letters, 290, 32–38. [DOI] [PubMed] [Google Scholar]
- Crack J. C., den Hengst C. D., Jakimowicz P., Subramanian S., Johnson M. K., Buttner M. J., Thomson A. J., Le Brun N. E. (2009). Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD. Biochemistry, 48, 12252–12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N. K., Chater K. F. (1992). The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Molecular and General Genetics, 232, 351–358. [DOI] [PubMed] [Google Scholar]
- Dong L. B., Rudolf J. D., Deng M. R., Yan X., Shen B. (2018). Discovery of the tiancilactone antibiotics by genome mining of atypical bacterial type II diterpene synthases. Chembiochem, 19(16), 1727–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler-Goldsworthy K., Gust B., Mouz S., Chandra G., Findlay K. C., Chater K. F. (2011). The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiology (Reading, England), 157, 1312–1328. [DOI] [PubMed] [Google Scholar]
- Higo A., Hara H., Horinouchi S., Ohnishi Y. (2012). Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Research, 19, 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Hou L., Li H., Qiu Y., Ju J., Li W. (2016). Activation of a plasmid-situated type III PKS gene cluster by deletion of a wbl gene in deepsea-derived Streptomyces somaliensis SCSIO ZH66. Microbial Cell Factories, 15, 116-016-0515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ma T., Tian J., Shen L., Zuo H., Hu C., Liao G. (2017). WblA, a pleiotropic regulatory gene modulating morphogenesis and daptomycin production in Streptomyces roseosporus. Journal of Applied Microbiology, 123, 669–677. [DOI] [PubMed] [Google Scholar]
- Kang S. H., Huang J., Lee H. N., Hur Y. A., Cohen S. N., Kim E. S. (2007). Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. Journal of Bacteriology, 189, 4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. G., Lee M. J., Seo J., Hwang Y. B., Lee M. Y., Han K., Sherman D. H., Kim E. S. (2009). Identification of functionally clustered nystatin-like biosynthetic genes in a rare actinomycetes, Pseudonocardia autotrophica. Journal of Industrial Microbiology & Biotechnology, 36, 1425–1434. [DOI] [PubMed] [Google Scholar]
- Kim H. J., Kim M. K., Jin Y. Y., Kim E. S. (2014). Effect of antibiotic down-regulatory gene wblA ortholog on antifungal polyene production in rare actinomycetes Pseudonocardia autotrophica. Journal of Microbiology and Biotechnology, 24, 1226–1231. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Lee H. N., Kim P., Lee H. S., Kim E. S. (2012). Negative role of wblA in response to oxidative stress in Streptomyces coelicolor. Journal of Microbiology and Biotechnology, 22, 736–741. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Lee H. N., Lee H. S., Kim P., Kim E. S. (2013). A WblA-binding protein, SpiA, involved in Streptomyces oxidative stress response. Journal of Microbiology and Biotechnology, 23, 1365–1371. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Lee H. N., Kim H. J., Kim E. S. (2011). Transcriptome analysis of an antibiotic downregulator mutant and synergistic actinorhodin stimulation via disruption of a precursor flux regulator in Streptomyces coelicolor. Applied and Environmental Microbiology, 77, 1872–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn A. D., Chin Y. W., Swanson S. M. (2009). Discovery of natural product anticancer agents from biodiverse organisms. Current Opinion in Drug Discovery and Development, 12, 189–196. [PMC free article] [PubMed] [Google Scholar]
- Kudhair B. K., Hounslow A. M., Rolfe M. D., Crack J. C., Hunt D. M., Buxton R. S., Smith L. J., Le Brun N. E., Williamson M. P., Green J. (2017). Structure of a Wbl protein and implications for NO sensing by M. tuberculosis. Nature Communications, 8, 2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. J., Karoonuthaisiri N., Kim H. S., Park J. H., Cha C. J., Kao C. M., Roe J. H. (2005). A master regulator sigmaB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Molecular Microbiology, 57, 1252–1264. [DOI] [PubMed] [Google Scholar]
- Lee H. N., Huang J., Im J. H., Kim S. H., Noh J. H., Cohen S. N., Kim E. S. (2010). Putative TetR family transcriptional regulator SCO1712 encodes an antibiotic downregulator in Streptomyces coelicolor. Applied and Environmental Microbiology, 76, 3039–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. N., Kim J. S., Kim P., Lee H. S., Kim E. S. (2013). Repression of antibiotic downregulator WblA by AdpA in Streptomyces coelicolor. Applied and Environmental Microbiology, 79, 4159–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Lee J. S., Kim S. E., Moon B. S., Kim Y. C., Lee S. K., Lee S. K., Choi K. Y. (2006). Tautomycetin inhibits growth of colorectal cancer cells through p21cip/WAF1 induction via the extracellular signal-regulated kinase pathway. Molecular Cancer Therapeutics, 5, 3222–3231. [DOI] [PubMed] [Google Scholar]
- Lee S. K., Park J. W., Kim J. W., Jung W. S., Park S. R., Choi C. Y., Kim E. S., Kim B. S., Ahn J. S., Sherman D. H., Yoon Y. J. (2006). Neopikromycin and novapikromycin from the pikromycin biosynthetic pathway of Streptomyces venezuelae. Journal of Natural Products, 69, 847–849. [DOI] [PubMed] [Google Scholar]
- Liu G., Chater K. F., Chandra G., Niu G., Tan H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiology and Molecular Biology Reviews, 77, 112–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Liao G., Zhang J., Tan H. (2015). Identification of novel tylosin analogues generated by a wblA disruption mutant of Streptomyces ansochromogenes. Microbial Cell Factories, 14, 173–015–0359–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan S., Oh T. J., Lee H. C., Sohng J. K. (2008). Heterologous expression of metK1-sp and afsR-sp in Streptomyces venezuelae for the production of pikromycin. Biotechnology Letters, 30, 1621–1626. [DOI] [PubMed] [Google Scholar]
- Makitrynskyy R., Tsypik O., Nuzzo D., Paululat T., Zechel D. L., Bechthold A. (2020). Secondary nucleotide messenger c-di-GMP exerts a global control on natural product biosynthesis in streptomycetes. Nucleic Acids Research, 48, 1583–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann J. (2001). Natural products as immunosuppressive agents. Natural Product Reports, 18, 417–430. [DOI] [PubMed] [Google Scholar]
- Mohan A., Padiadpu J., Baloni P., Chandra N. (2015). Complete genome sequences of a Mycobacterium smegmatis laboratory strain (MC2 155) and isoniazid-resistant (4XR1/R2) mutant strains. Genome Announcements, 3(1), e01520–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouri Y., Konishi K., Fujita A., Tezuka T., Ohnishi Y. (2017). Regulation of sporangium formation by BldD in the rare actinomycete Actinoplanes missouriensis. Journal of Bacteriology, 199(12), e00840–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nah J. H., Park S. H., Yoon H. M., Choi S. S., Lee C. H., Kim E. S. (2012). Identification and characterization of wblA-dependent tmcT regulation during tautomycetin biosynthesis in Streptomyces sp. CK4412. Biotechnology Advances, 30, 202–209. [DOI] [PubMed] [Google Scholar]
- Nguyen K. T., Tenor J., Stettler H., Nguyen L. T., Nguyen L. D., Thompson C. J. (2003). Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within theadpA gene. Journal of Bacteriology, 185, 7291–7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu M., Sun Y., Liu B., Tang L., Qiu R. (2012). Differential effects of tautomycetin and its derivatives on protein phosphatase inhibition, immunosuppressive function and antitumor activity. Korean Journal of Physiology and Pharmacology, 16, 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J. H., Kim S. H., Lee H. N., Lee S. Y., Kim E. S. (2010). Isolation and genetic manipulation of the antibiotic down-regulatory gene, wblA ortholog for doxorubicin-producing Streptomyces strain improvement. Applied Microbiology and Biotechnology, 86, 1145–1153. [DOI] [PubMed] [Google Scholar]
- Oh H. S., Kang H. Y. (2012). Total synthesis of pikromycin. Journal of Organic Chemistry, 77, 1125–1130. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y., Yamazaki H., Kato J. Y., Tomono A., Horinouchi S. (2005). AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Bioscience, Biotechnology, and Biochemistry, 69, 431–439. [DOI] [PubMed] [Google Scholar]
- Park J. S., Lee J. Y., Kim H. J., Kim E. S., Kim P., Kim Y., Lee H. S. (2012). The role of Corynebacterium glutamicum spiA gene in whcA-mediated oxidative stress gene regulation. FEMS Microbiology Letters, 331, 63–69. [DOI] [PubMed] [Google Scholar]
- Pyeon H. R., Nah H. J., Kang S. H., Choi S. S., Kim E. S. (2017). Heterologous expression of pikromycin biosynthetic gene cluster using Streptomyces artificial chromosome system. Microbial Cell Factories, 16, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabyk M., Ostash B., Rebets Y., Walker S., Fedorenko V. (2011). Streptomyces ghanaensis pleiotropic regulatory gene wblA(gh) influences morphogenesis and moenomycin production. Biotechnology Letters, 33, 2481–2486. [DOI] [PubMed] [Google Scholar]
- Römling U., Galperin M. Y., Gomelsky M. (2013). Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiology and Molecular Biology Reviews, 77, 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Guidry L., Narasimhulu K. V., Mai D., Trombley J., Redding K. E., Giles G. I., Lancaster J. R. Jr, Steyn, A. J. (2007). Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proceedings of the National Academy of Sciences of the United States of America, 104, 11562–11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivalingam P., Hong K., Pote J., Prabakar K. (2019). Extreme Environment Streptomyces: Potential Sources for New Antibacterial and Anticancer Drug Leads? International Journal of Microbiology, 2019, 5283948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliveri J. A., Gomez J., Bishai W. R., Chater K. F. (2000). Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology (Reading, England), 146 (Pt 2), 333–343. [DOI] [PubMed] [Google Scholar]
- Sun D., Liu C., Zhu J., Liu W. (2017). Connecting metabolic pathways: Sigma factors in Streptomyces spp. Front Microbiology, 8, 2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschowri N., Schumacher M. A., Schlimpert S., Chinnam N. B., Findlay K. C., Brennan R. G., Buttner M. J. (2014). Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell, 158, 1136–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heul H. U., Bilyk B. L., McDowall K. J., Seipke R. F., van Wezel G. P. (2018). Regulation of antibiotic production in Actinobacteria: New perspectives from the post-genomic era. Natural Product Reports, 35, 575–604. [DOI] [PubMed] [Google Scholar]
- van Wezel G. P., McDowall K. J. (2011). The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Natural Product Reports, 28, 1311–1333. [DOI] [PubMed] [Google Scholar]
- Wolanski M., Donczew R., Kois-Ostrowska A., Masiewicz P., Jakimowicz D., Zakrzewska-Czerwinska J. (2011). The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. Journal of Bacteriology, 193, 6358–6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M. W., Nah H. J., Choi S. S., Kim E. S. (2014). Pikromycin production stimulation through antibiotic down-regulatory gene disruption in Streptomyces venezuelae. Biotechnology and Bioprocess Engineering, 19, 973–977. [Google Scholar]
- Xia H., Li X., Li Z., Zhan X., Mao X., Li Y. (2020). The Application of regulatory cascades in Streptomyces: Yield enhancement and metabolite mining. Front Microbiology, 11, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Zhao L., Liu H. W., Sherman D. H. (1998). A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: Architecture of metabolic diversity. Proceedings of the National Academy of Sciences of the United States of America, 95, 12111–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Lu X., Sun D., Zhuang S., Chen Q., Chen Z., Li J., Wen Y. (2020). BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species. Molecular Microbiology, 113, 123–142. [DOI] [PubMed] [Google Scholar]
- Yan X., Chen J. J., Adhikari A., Teijaro C. N., Ge H., Crnovcic I., Chang C. Y., Annaval T., Yang D., Rader C., Shen B. (2018). Comparative studies of the biosynthetic gene clusters for anthraquinone-fused enediynes shedding light into the tailoring steps of tiancimycin biosynthesis. Organic Letters, 20, 5918–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Ge H., Huang T., Hindra, Yang, D., Teng Q., Crnovčić I., Li X., Rudolf J. D., Lohman J. R., Gansemans Y., Zhu X., Huang Y., Zhao L. X., Jiang Y., Van Nieuwerburgh F., Rader C., Duan Y., Shen B. (2016). Strain prioritization and genome mining for enediyne natural products. mBio, 7(6), e02104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J. S., Kim M., Kim E. J., Kim B. G. (2018). Production of pikromycin using branched chain amino acid catabolism in Streptomyces venezuelae ATCC 15439. Journal of Industrial Microbiology & Biotechnology, 45, 293–303. [DOI] [PubMed] [Google Scholar]
- Yu P., Liu S., Bu Q., Zhou Z., Zhu Z., Huang F., Li Y. (2014). WblAch, a pivotal activator of natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis L10, is positively regulated by AdpAch. Applied and Environmental Microbiology, 80, 6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gao D., Lin J., Zhu M., Zhuang Z., Duan Y., Zhu X. (2020). Construction of inducible genetic switch for the global regulator WblA to sustain both overproduction of tiancimycins and on-demand sporulation in Streptomyces sp. CB03234. ACS Synthetic Biology, 9, 1460–1467. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Sherman D. H. (2001). Isolation and structure determination of novamethymycin, a new bioactive metabolite of the methymycin biosynthetic pathway in Streptomyces venezuelae. Journal of Natural Products, 64, 1447–1450. [DOI] [PubMed] [Google Scholar]