Abstract
The effect of three phenyl urea herbicides (diuron, linuron, and chlorotoluron) on soil microbial communities was studied by using soil samples with a 10-year history of treatment. Denaturing gradient gel electrophoresis (DGGE) was used for the analysis of 16S rRNA genes (16S rDNA). The degree of similarity between the 16S rDNA profiles of the communities was quantified by numerically analysing the DGGE band patterns. Similarity dendrograms showed that the microbial community structures of the herbicide-treated and nontreated soils were significantly different. Moreover, the bacterial diversity seemed to decrease in soils treated with urea herbicides, and sequence determination of several DGGE fragments showed that the most affected species in the soils treated with diuron and linuron belonged to an uncultivated bacterial group. As well as the 16S rDNA fingerprints, the substrate utilization patterns of the microbial communities were compared. Principal-component analysis performed on BIOLOG data showed that the functional abilities of the soil microbial communities were altered by the application of the herbicides. In addition, enrichment cultures of the different soils in medium with the urea herbicides as the sole carbon and nitrogen source showed that there was no difference between treated and nontreated soil in the rate of transformation of diuron and chlorotoluron but that there was a strong difference in the case of linuron. In the enrichment cultures with linuron-treated soil, linuron disappeared completely after 1 week whereas no significant transformation was observed in cultures inoculated with nontreated soil even after 4 weeks. In conclusion, this study showed that both the structure and metabolic potential of soil microbial communities were clearly affected by a long-term application of urea herbicides.
The environmental chemistry, fate, toxicology, and impact on soil fertility of phenylurea herbicides, used for selective control of annual weeds in fruit and field crops and noncrop areas, have already been studied in great detail (4, 5, 12). However, there is a serious lack of information on the effect of urea herbicides on soil microbial communities. In general, the effect of herbicides on soil microbial communities has often been studied by conventional methods based on cultivation of the microbial communities and on measurements of their metabolic activities (27, 34). Nowadays it is well known that more than 90% of the microorganisms existing in nature are refractory to selective enrichment cultures (33). To overcome the drawbacks of these culture-dependent methods, interest is currently focused on the use of molecular biological techniques, given their powerful capacity to allow the analysis of microorganisms in their natural habitats. In this context, analysis of the 16S rRNA molecule or its corresponding gene (16S rDNA) is by far the most widely used approach in the last decade (3). Of the 16S rDNA-based methods used for studying complex microbial populations, denaturing gradient gel electrophoresis (DGGE) has received the most attention and has been successfully applied to several natural habitats (15, 21, 29, 30). DGGE is based on the electrophoresis of PCR-amplified 16S rDNA fragments in polyacrylamide gels containing a linearly increasing gradient of denaturants. In DGGE gels, DNA fragments of the same length but with different base pair sequences can be separated (22).
In this paper, we describe the effect of three phenylurea herbicides (diuron, linuron, and chlorotoluron) on the soil microbial communities of a soil that has been treated regularly with these herbicides for more than 10 years. A culture-independent approach based on the PCR amplification of 16S rDNA genes followed by DGGE was used in combination with the determination of substrate utilization patterns of the microbial communities by using BIOLOG GN microplates.
MATERIALS AND METHODS
Soil.
The soil samples were taken from the Royal Research Station of Gorsem (Sint-Truiden, Belgium), an orchard that has been regularly treated with different urea herbicides since 1987. The soil plots investigated in this study were annually treated with diuron (3.75 kg/ha), a mixture of diuron (2 kg/ha), linuron (3 kg/ha), and simazin (2 liters/ha), or chlorotoluron (5 kg/ha). The soil treated with the mixture is referred to below as “soil treated with diuron+linuron” or “linuron-treated soil.” Soil from a nontreated plot at the same orchard was used as a control soil. All herbicides were applied without carrier compounds. The soil taken from each plot consists of 75% silt, 13% sand, and 12% clay. It has a cation exchange capacity CEC of 10.8 meq/100 g, 2.6% organic carbon, and a pH in water of 6.6. In March 1998, 20 soil samples (30 g each) were taken in different parts of each plot from 0 to 5 cm deep, using a soil auger. The samples from each plot were then mixed and stored overnight at 4°C. All data reported in this article were obtained from experiments done less than 24 h after sampling. The data in Fig. 4B were obtained with duplicate soil samples taken at different locations in the same plots in August 1998.
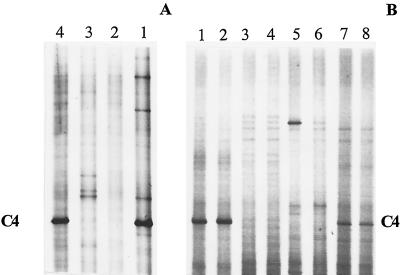
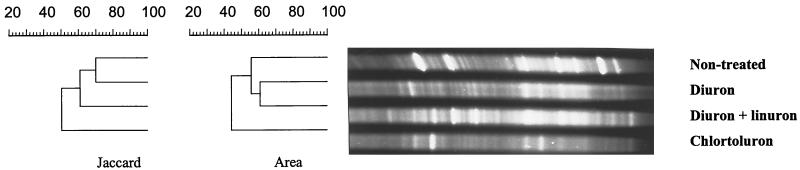
FIG. 4.
(A) DGGE analysis of 16S rDNA fragments of pooled soil samples collected in March 1998, treated or not treated with urea herbicides. Lanes: 1, nontreated soil; 2, diuron; 3, diuron+linuron; 4, chlorotoluron. (B) DGGE analysis of individual soil samples collected in August 1998 from two different locations per herbicide-treated plot. Lanes: 1 and 2, nontreated; 3 and 4, diuron; 5 and 6, diuron+linuron; 7 and 8, chlorotoluron. The 16S rDNA genes were amplified with the primer set P63f plus P518r. Gels A and B were not run under exactly the same conditions, which explains the difference in distances between bands. C4 indicates the fragment that was excised and sequenced.
Viable counts and BIOLOG assays.
Suspensions of microorganisms were prepared from soil samples as reported previously (35). After appropriate dilution in sterile saline solution, the cell suspensions were used to determine the number of culturable heterotrophs and to inoculate BIOLOG microplates. The number of culturable heterotrophs, expressed as CFU, was determined by spreading 0.1 ml of the cell suspension onto R2A agar medium (Difco, Detroit, Mich.) amended with cycloheximide (200 μg/ml) to suppress fungal growth. Three replicates of each soil sample were spread on agar plates and incubated for 2 days at room temperature. The log-transformed CFU data were analyzed by one-way analysis of variance, and the means were compared by the Duncan multiple-range test.
To obtain a substrate utilization fingerprint of the microbial communities, three replicates of all the soil extracts were inoculated in BIOLOG GN microtiter plates (BIOLOG Inc., Hayward, Calif.) containing 95 different sole-carbon sources and a control without a carbon source. The BIOLOG GN plates were incubated at 28°C, and the optical density at 590 nm (OD590) in each well, produced from the reduction of tetrazolium violet, was recorded after several incubation times with a EL312e biokinetics reader and the KinetiCalc EIA application software release 2.03 (Bio-tek Instruments Inc., Winoosky, Vermont). Before the data were further processed, the OD590 at time zero was subtracted from the later readings, yielding the net OD590. The physiological versatility of the microbial community was evaluated by determining the number of carbon sources with a net OD590 of >0.4 (30). To reduce the dimensions of the highly multivariate data set and to reveal latent associations between C sources and the communities, principal-component analysis (PCA) was performed on the net OD590 by using the C sources as variables. In all analyses, three principal components (PCs) were retained and a varimax rotation of the PCs with Kaizer normalization was performed to facilitate the interpretation. Alternatively, relative similarity between the BIOLOG GN fingerprints of the soil communities was assessed by cluster analysis (CA) performed on the net OD590 by using the C sources as variables. The squared Euclidian distance was used as a dissimilarity index, and the average linkage between groups method was used to cluster the cases. The result of the CA is shown in a dendrogram. The statistical processing was performed with SPSS for Windows release 7.5.2.
Enrichment cultures.
Soil (5 g) was added to 95 ml of a mineral solution containing (milligrams per liter) MgSO4 · 6H2O, 98.50; CaCl2 · 2H2O, 5.88; ZnSO4 · 7H2O, 1.15; H3BO3, 1.16; MnSO4 · H2O, 1.69; CoCl2 · 6H2O, 0.24; MoO3, 0.10; FeSO4 · 7H2O, 2.78; and CuSO4 · 5H2O, 0.37. The pH was adjusted to around 7.0 with a phosphate buffer (KH2PO4, 10 mM; Na2HPO4, 10 mM). The urea herbicides, diluted in water and sterilized by filtration (0.25-μm-pore-size filters), were added as the sole source of carbon and nitrogen. The concentrations of the herbicides used were 15 mg/liter for diuron and 25 mg/liter for linuron and chlorotoluron. For each herbicide, the flasks were inoculated with the soil treated with the corresponding herbicide or with the nontreated soil. The disappearance of the herbicides was monitored over time by high-performance liquid chromatography analyses. The high-performance liquid chromatography system consisted of a Kontron liquid chromatograph with a DEGASYS DG-1310 system to degas the mobile phase, 3 Kontron 325 high-pressure pumps, a Kontron MSI 660 injector with a 20-μl loop, a Kontron DAD 495 (diode array detector), and a 450 MT2/DAD software system. A C18 reversed-phase column (Alltech, Deerfield, Ill.) was used. The mobile phase consisted of CH3OH, NH4H2PO4 (0.1 M, pH 3.8), and H2O (70:25:5); the flow rate was 0.75 ml/min; and the UV detector was set to 210 nm. Products were identified by comparison with authentic standards. After 10 days of incubation, DNA was extracted from 2 ml of each enrichment and DGGE analyses were carried out (see below). All enrichment culture experiments were done in triplicate.
DNA extraction and purification.
Total DNA was isolated from soil samples by a method reported previously (6). This method was modified as follows. Soil (2 g) was added to a 14-ml polypropylene round-bottom tube (Falcon). To this, 3 g of beads (diameter, 0.10 to 0.11 mm) plus 6 ml of 10 mM Tris-HCl (pH 9) were added. The mixture was beaten three times for 90 s each with a bead beater (B. Braun Biotech International, Melsungen, Germany). After this, lysozyme was added at a final concentration of 5 mg/ml, and the samples were incubated for 15 min at 28°C with horizontal shaking. To achieve complete lysis, 1% sodium dodecyl sulfate was added and the samples were slowly mixed for 5 to 10 min. The supernatant was collected after centrifugation at 6,000 × g for 15 min at room temperature. Phenol extraction followed by chloroform-isoamyl alcohol purification was applied. The aqueous phase was transferred to a new tube containing isopropanol to 0.7 of its volume. Precipitation was performed for 1 h at room temperature. Alternatively, 2.5 volumes of ethanol (90%) was added for an overnight precipitation. The pellet (crude extract) was obtained by centrifugation at 12,000 × g for 30 min and resuspended in 250 μl of dionized water. A 100-μl aliquot of the crude extract was further purified with Wizard PCR preps (Promega, Madison, Wis.), and the purified DNA was finally recovered in 50 μl of deionized water.
PCR conditions.
A 2-μl volume of the extracted DNA was amplified by PCR with a 9600 thermal cycler (Perkin-Elmer, Norwalk, Conn.). The PCR mixture used contained 0.5 μM each primer, 100 μM each deoxynucleoside triphosphate, 10 μl of 10× Expand high-fidelity PCR buffer, 2 U of Expand high-fidelity DNA polymerase (Boehringer, Mannheim, Germany), 400 ng of bovine serum albumin (Boehringer) per μl, and sterile water to a final volume of 100 μl. The 16S rRNA genes from soil microbial communities were amplified by PCR with two different sets of primers. The first set consisted of primers P63f (5′CAGGCCTAACACATGCAAGTC3′, forward) and P518r, (5′ATTACCGCGGCTGCTGG3′, reverse). The specificity and efficacy of P63f were tested systematically with a variety of bacterial species and environmental samples, and it was found to be more useful for 16S rRNA gene amplification in ecological and systematic studies than are PCR amplimers that are currently more generally used (19). P518r was based on a universally conserved region (23). The length of the expected amplified fragment was 495 bp (the large fragment). The second set consisted of primer P338f (5′ACTCCTACGGGAGGCAGCAG3′, forward), which complements a region conserved among members of the domain Bacteria, and the reverse primer P518r described above (23). This set amplified a fragment of 210 bp (the small fragment). A GC clamp of 40 bases (23) was added to each forward primer. Samples were amplified as follows: 94°C for 5 min, followed by 30 cycles of 92°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min.
DGGE.
DGGE based on the method cited by Muyzer et al. (22) was performed with the D Gene System (Bio-Rad, Hercules, Calif.). PCR samples were loaded onto 6% (wt/vol) polyacrylamide gels in 1× TAE (20 mM Tris, 10 mM acetate, 0.5 mM EDTA [pH 7.4]) for the primer set P63f plus P518r or 8% polyacrylamide gels for the set P338f plus P518r. The polyacrylamide gels were made with a denaturing gradient ranging from 40 to 60% (where 100% denaturant contains 7 M urea and 40% formamide). The electrophoresis was run overnight at 60°C at 75 V for the primer set P63f plus P518r or at 35 V for the set P338f plus P518r. After the electrophoresis, the gels were soaked for 30 min in SYBR GreenI nucleic acid gel stain (1:10,000 dilution; FMC BioProducts, Rockland, Maine). The stained gel was immediately photographed on an UV transillumination table with a video camera module (Vilbert Lourmat, Marne la Vallée, France).
Statistical comparison of different DGGE patterns, run on the same gel, was done with GelCompar software 4.1 (Applied Maths, Kortrijk, Belgium). The similarity among the band patterns was calculated by using two similarity coefficients: (i) the coefficient of Jaccard (Sj), using band positions (for each pattern, Sj divides the number of corresponding bands by the total number of bands in both patterns); and (ii) the area-sensitive coefficient (SD), which is very similar to the coefficient of Jaccard but is modified to give more weight to matching bands.
Sequencing of DGGE fragments.
DNA fragments to be sequenced were excised from the gel, placed into sterile Eppendorf tubes containing 25 μl of sterilized water, and incubated overnight at 4°C. A 4-μl volume of the DNA diffused in water served as a template for PCR amplification. The amplified products were subjected to a new DGGE step to confirm their electrophoretic mobility. The PCR products were purified with a QIAquick PCR purification kit (QIAGEN GmbH, Hilden, Germany) before being sent for sequencing by Eurogentec (Liège, Belgium). The sequences were aligned to 16S rRNA sequences obtained from the National Centre for Biotechnology Information database by using the BLAST 2.0 search program (2).
Nucleotide sequence accession numbers.
The nucleotide sequences for fragments C4, C3, and L3 have been deposited in the GenBank database under accession no. AF103810, AF103811, and AF103812, respectively.
RESULTS
Analysis of soil microbial communities by using total counts and BIOLOG GN microplates.
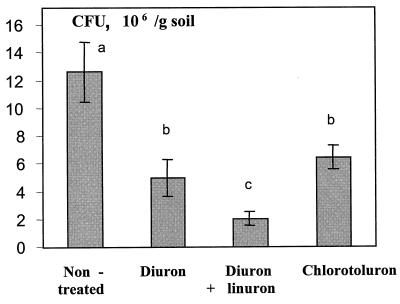
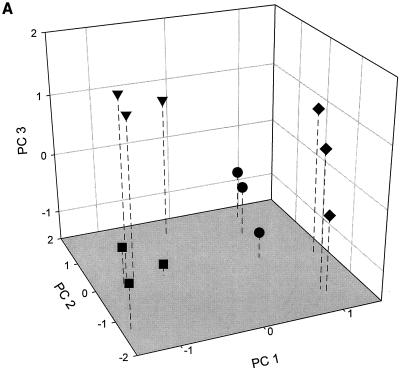
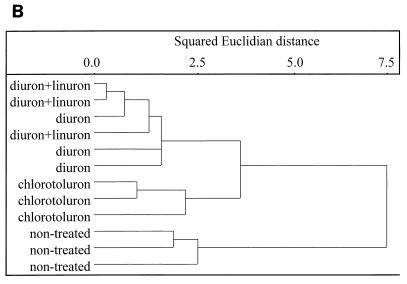
The effect of urea herbicides on soil microbial communities was first estimated by culture-dependent approaches. The numbers of total heterotrophic CFU in all three herbicide-treated soils were significantly smaller (P < 0.05) than those in the nontreated soil, but the differences were small (<1 log unit) (Fig. 1). The results of the PCA performed on the BIOLOG GN fingerprints of the different soil microbial communities, obtained after 53 h of incubation, are shown in Fig. 2A. It is clear that the functional abilities of the soil microbial communities were altered by the application of the herbicides. The explained variance amounted to 28.2% for PC 1, 17.4% for PC 2, and 10.8% for PC 3. The control soil and the soil treated with chlorotoluron were separated along PC 1 from the soils treated with diuron and diuron+linuron. Further separation of the BIOLOG fingerprints of the four soils occurred along PC 2 and PC 3. CA revealed a clear dissimilarity between the control soil and the soils treated with the herbicides (Fig. 2B). Furthermore, the soils treated with chlorotoluron were clearly distinguished from the other two treated soils whereas a clear distinction could not be made by CA between the soils treated with diuron or with diuron+linuron.
FIG. 1.
Viable heterotrophic bacterial counts from different soil samples treated or not treated with urea herbicides. Error bars represent standard deviations. Values with a different letter are significantly different from each other (P < 0.05).
FIG. 2.
PCA (A) and CA (B) performed on the BIOLOG GN fingerprints of the extracts of the soils treated with diuron (■), chlorotoluron (●), diuron+linuron (▾), and the control soil (⧫).
Analysis of soil microbial communities by DGGE.
In parallel with the culture-dependent approaches, DGGE analysis of 16S rDNA fragments was used to investigate the effect of the urea herbicides on the soil microbial communities. This was done after PCR amplification of the 16S rDNA genes from total soil DNA with the two sets of primers, described above. Figure 3 shows a DGGE analysis of the small 16S rDNA fragment (primers P338f and P518r) amplified from the three herbicide-treated soils and the one nontreated soil. Numerical analysis of the DGGE patterns with two similarity coefficients showed that the microbial communities of the treated and nontreated soils were different (Fig. 3). The DGGE patterns of the large 16S rDNA fragment (primers P63f and P518r) amplified from the same four soil samples are shown in Fig. 4A. There are fewer bands in these DGGE profiles than in the DGGE patterns of the small fragment. However, the discrimination between different soil samples was much clearer with this primer set. The preponderant bands seen on the gels can be considered the most abundant bacterial species in each soil sample. The fragment named C4 can, based on its intensity, be considered to represent the most dominant species in the nontreated and in the chlorotoluron-treated soil. Its sequence matched closely (96%) that of an uncultivated-soil bacterium named clone S111 (16). Since the corresponding 16S rDNA fragment was completely absent in the DGGE patterns of the soils treated with diuron and diuron+linuron, these treatments appear to cause the demise of this microbial species (Fig. 4A). To confirm the reproducibility of such meaningful information concerning the effect of herbicides on this abundant microbial species, duplicate soil samples were taken 5 months later (August 1998) from different locations within the same plots. As illustrated in Fig. 4B, the bacterial species corresponding to fragment C4 was again not observed among the predominant populations in the soils treated with diuron and with diuron+linuron.
FIG. 3.
DGGE analysis of 16S rDNA fragments of different soil samples treated or not treated with urea herbicides. The 16S rDNA genes were amplified with the primer set P338f plus P518r. For the dendrograms of community relatedness, the percent similarity was calculated on the basis of two band-based coefficients, the Jaccard and area-sensitive coefficients.
Biodegradation of urea herbicides by enrichment cultures.
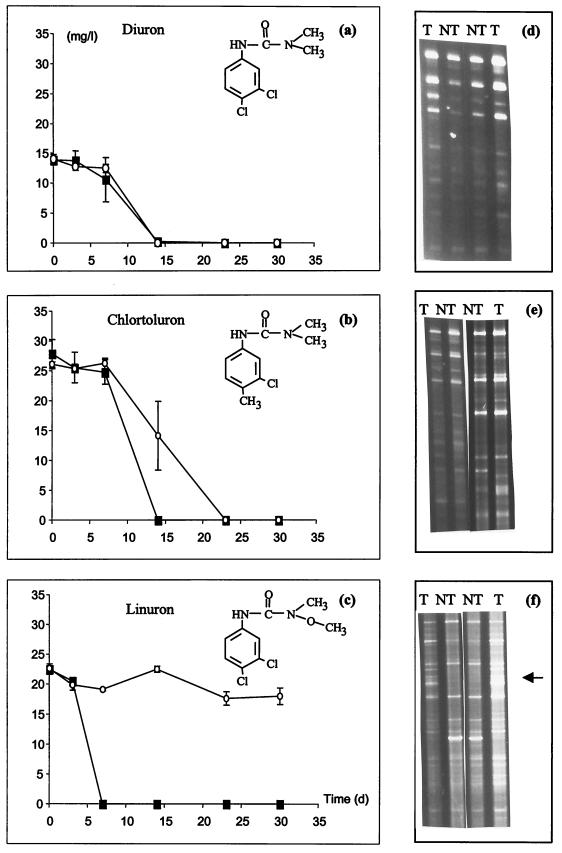
Enrichment studies with soil inocula taken from treated and nontreated soils were carried out to investigate whether there was a difference in the rate of biotransformation of the urea herbicides between soils with and without a history of herbicide treatment. In each case, the enrichment medium was inoculated with treated or nontreated soil in the presence of the corresponding herbicide, added as the sole source of carbon and nitrogen. Moreover, DGGE analyses were performed after 10 days of incubation to compare the 16S rDNA fingerprints of the enriched bacterial communities. Figure 5a through c depict the time curves corresponding to the transformation of diuron, chlorotoluron, and linuron, respectively. When the treated and nontreated soils were compared, there was no difference in the rate of transformation of diuron (Fig. 5a), a weak difference in the rate of transformation of chlorotoluron (Fig. 5b), and a strong difference in transformation of linuron (Fig. 5c). Indeed, whereas a total transformation of linuron was observed after 1 week of incubation in flasks inoculated with treated soil, no significant disappearance of linuron was observed in flasks inoculated with nontreated soil even after 4 weeks of incubation (Fig. 5c).
FIG. 5.
(a to c) Transformation of urea herbicides in enrichment cultures. For each herbicide, nontreated soil (○) and soil with a 10-year history of treatment (■) were used as the inoculum. (d to f) For each herbicide enrichment, the DGGE patterns from duplicate cultures were obtained after 10 days of incubation: T, enrichments inoculated with treated soil; NT, enrichments inoculated with nontreated soil. The 16S rDNA genes were amplified with primer set P63f and P518r. The arrow in panel f indicates a strong difference in the profile between treated and nontreated soil. a and d, diuron; b and e, chlorotoluron; c and f, linuron.
DGGE analyses of the enrichment cultures after 10 days were performed with primer set P63r and P518r (large fragment). Figure 5d through f show the DGGE patterns of duplicate enrichment cultures with diuron, chlorotoluron, and linuron, respectively, as the C and N source. The DGGE patterns of the enrichment cultures with diuron as the sole C and N source were closely related regardless of whether treated or nontreated soil was used as an inoculum (Fig. 5d). In the chlorotoluron enrichments, a few differences can be seen between the enrichments of treated and nontreated soil (Fig. 5e). However, for the linuron enrichments, there was a strong difference in the DGGE profiles between flasks inoculated with treated and nontreated soil (Fig. 5f). The results obtained with all herbicides showed that the degree of difference in herbicide transformation between the treated and nontreated soil was correlated with the degree of difference in their 16S rDNA fingerprints. The enrichment experiment was repeated with soil samples taken from the same plots 5 months later (August 1998), and showed very similar herbicide degradation results and DGGE patterns to those presented in Fig. 5 (data not shown).
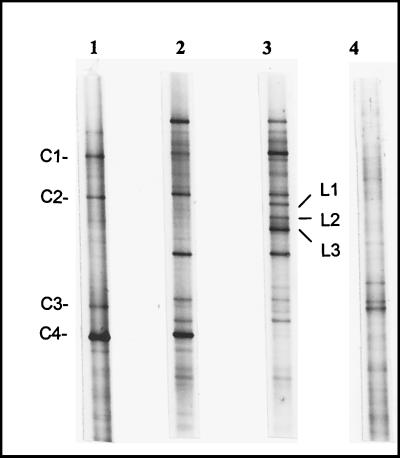
The striking results observed with linuron enrichments led us to compare the evolution of the microbial fingerprint before and after enrichments. Figure 6 shows the DGGE patterns of the soils treated or not treated with linuron and of their respective enrichment cultures after 10 days in the medium containing 25 mg of linuron per liter as the sole source of carbon and nitrogen. This experiment confirmed the fingerprints of the microbial communities of the linuron-treated and nontreated soil, as shown in Fig. 4A (lanes 3 and 1). Moreover, the same clear shifts in the communities during enrichment, which were different for the two different soils as shown in the previous experiment (Fig. 5f), were again demonstrated here. Fragment C4, found only in the nontreated soil, was maintained as a preponderant band in the corresponding linuron enrichment culture even after 10 days. This bacterial species was, however, completely absent in linuron enrichments inoculated with linuron-treated soil. Other fragments have also been sequenced; fragments C1 and C2 obtained from the nontreated soil showed high sequence similarity (98%) to Pseudomonas putida and Pseudomonas chlororaphis, respectively. These two fragments had appeared after enrichment in the linuron-treated soil (Fig. 6). Fragment C3, detected in the nontreated soil as well as in the soil treated with diuron+linuron, matched closely (96%) with an unidentified member of the β subclass of the Proteobacteria (19). In addition, fragments L1, L2, and L3 (Fig. 6), which seemed to be specific to the enrichments inoculated with the linuron-treated soil, were excised and sequenced. Fragments L1 and L2 showed high similarity (97%) to Pseudomonas mandelii and Pseudomonas jessenii, respectively. Fragment L3 matched closely (95% of similarity) with Variovorax sp.
FIG. 6.
Comparison of DGGE fingerprints between linuron treated and nontreated soil. Lanes: 1, nontreated soil before enrichment; 2 and 3, microbial communities enriched from nontreated and linuron-treated soil, respectively; 4, linuron-treated soil before enrichment. The enrichments were carried out in minimal medium containing 25 mg of linuron per liter. The 16S rDNA genes were amplified with primer set P63f plus P518r.
DISCUSSION
In this study we opted for the combination of culture-dependent methods (CFU, BIOLOG, and enrichments) and a culture-independent technique, DGGE, to gain a better understanding of the impact of urea herbicides on soil microbial communities. To achieve this goal, natural soil samples with a history of herbicide treatment (more than 10 years) were used. In the DGGE experiments, two sets of primers were used for the analysis of bacterial communities. Primer set P338f and P518r, first designed by Øvreas et al. (23), amplified a 210-bp fragment. This set seems to be very useful for surveys of bacterial diversity. Indeed, we have shown, by converting the DGGE banding patterns to a binary format for statistical comparisons, that the microbial communities in soils treated with different herbicides clustered differently. The numerical analysis of DGGE patterns with the primer set P338r and P518r provides an analytical tool to study the diversity of microbial communities of herbicide-treated soils. However, sequence information from such small rDNA fragments cannot readily be used to compare populations between environments, given that it is often not easy to place such short partial sequences accurately in phylogenetic trees, especially if the sequences lack close relatives in the database (17). Furthermore, it has been shown that the PCR amplification of 16S rDNA fragments can be biased by several parameters (24, 28). Most recently, it has been shown by Hansen et al. (11) that PCR amplification of 16S rDNA was highly biased, so that the rDNA from one species in four was preferentially amplified. To overcome such bias, those authors suggested the use of at least two different sets of primers (11). Because of these different reasons, it is highly recommended that at least two different primer sets be used to study the diversity of microbial communities by DGGE. We therefore used a second set of primers (P63r and P518r), and the corresponding DGGE profiles again showed that the genomic fingerprints of the bacterial communities of the treated and nontreated soils were different. Only this set of primers, which generated a larger, 495-bp fragment, was used to determine the sequence of the resulting DGGE bands. Of the sequenced fragments, C4 was the most intriguing. This most preponderant species in nontreated soil disappeared completely in soils treated with diuron or diuron+linuron and did not reappear after 10 days of incubation in enrichment media. The BLAST program identified a number of similar 16S rDNA fragments (percent similarity ranging from 90 to 96%), all of which have been identified as uncultivated soil bacteria. Two bacterial clones, clone S111 (16) and clone RB43 (18), showed the highest percent similarity to fragment C4, i.e., 96 and 94%, respectively. Both clones have been isolated from soil, and they both seem to belong to a new, widely distributed bacterial division named Acidobacterium (14), in spite of the large differences in characteristics and geographic locations of the different soils (17, 18). This new bacterial division has emerged as a result of the impact of culture-independent studies on the bacterial phylogeny (14). In this study, we report for the first time the effect of urea herbicides on such widely distributed nonculturable bacterial species. It is unclear whether these effects are caused by the herbicide molecules directly or indirectly by virtue of their impact on the ground cover. The disappearance of this dominant species was not due to local variability in the soils. First, the DGGE analyses of the soil samples collected in March 1998 were performed on pooled samples and thus represent average patterns per plot. Moreover, the DGGE patterns obtained from duplicate nonpooled soil samples collected 5 months later revealed that the same intense C4 band was present in the patterns of the control and chlorotoluron-treated soil but absent in the diuron- and linuron+diuron-treated soils (Fig. 4).
Another striking result revealed by sequencing DGGE fragments concerned fragment L3 (95% similarity to Variovorax sp.), detected in enrichments inoculated with linuron-treated soil (Fig. 6). Given that this bacterial species was found only in enrichments where linuron was degraded, one can assume that it was involved in the transformation of linuron. This hypothesis is supported by several previous studies, which have shown that bacterial strains of the genus Variovorax are involved in the transformation of aromatic compounds such as the herbicide 2,4-dichlorophenoxyacetic acid (16) and homovanillate (1). The role in the degradation of linuron by this strain and the other strains that appeared (L1 and L2 in Fig. 6), closely related to P. mandelii and P. jessenii, will be investigated further.
The results obtained by DGGE were in accordance with those obtained by BIOLOG after 53 h of incubation. Indeed, both methods showed that the fingerprints of the microbial communities of the nontreated and herbicide-treated soils differed from each other. However, at later incubation times, the differences among the BIOLOG GN fingerprints tended to disappear. As an example, the physiological versatility of the microbial communities in the different soils was compared at 53 and 165 h of incubation. The important differences in physiological versatility between the different soils, observed after 53 h of incubation of the BIOLOG GN plates, had disappeared after 165 h (data not shown). These results are in accordance with the DGGE profiles obtained with enrichment cultures inoculated with treated or nontreated soils (Fig. 5). Although the DGGE patterns of the different soils before enrichment differed substantially, the DGGE patterns of the corresponding enrichment cultures were quite similar after 10 days, except for those inoculated with the linuron-treated soil.
The shifts in microbial communities after cultivation, detected by both DGGE and BIOLOG assays, can be explained by the fact that organisms which grow rapidly in the presence of high concentrations (like pseudomonads) respond well in BIOLOG assays (10) and enrichment cultures. A typical example observed in this study concerned the two Pseudomonas species (bands C1 and C2) detected in the nontreated soil but not in treated soil before enrichment culture. After 10 days of enrichment, these two species appeared with almost the same signal intensity in DGGE patterns of all linuron enrichments (Fig. 6). In this context, it is interesting that previous reports have concluded that the meaning of differences in community-level physiological profiles remains unclear, even if the fraction of the community that responds in the assay was accurately defined (10). Recently, the analysis of the microbial communities of the BIOLOG GN microplates by DGGE or temperature gradient gel electrophoresis showed that carbon source utilization profiles obtained with BIOLOG GN plates do not necessarily reflect the functional potential of the numerically dominant members of the microbial community used as the inoculum. Indeed, changes in the structure of the microbial community had occurred during the BIOLOG assay (26).
The study of biodegradation of the herbicides in enrichment cultures showed that there was no significant difference between treated and nontreated soil for the transformation of diuron and chlorotoluron. On the other hand, a striking difference was observed with linuron, since this herbicide persisted for more than 4 weeks in enrichment cultures inoculated with nontreated soil. This different behavior of the three herbicides in the enrichment cultures could be explained by the fact that different enzymes are involved in the biotransformation of urea herbicides. One can speculate that there are two different aryl acyl amidases. The first, specific for the transformation of linuron (N′-methoxy) (Fig. 5c), leads to the production of N,O-dimethylhydroxylamine, a compound identified by thin-layer chromatography during transformation of linuron by cell extracts of Bacillus sphaericus ATCC 12123 (7). The second aryl acylamidase leads to the formation of a dimethylamine from diuron and chlorotoluron (1,1-dimethyl) (Fig. 5a and b). This hypothesis is in accordance with the results obtained previously with B. sphaericus, which was able to degrade linuron and monolinuron but not diuron and several other nonmethoxy herbicides (32). The hypothesis that different enzymes are involved in the transformation of diuron and linuron is supported by the results obtained by Roberts et al. (25), who enriched a stable mixed bacterial culture capable of degrading the herbicide linuron. This culture was also able to degrade related herbicides such as monolinuron but was unable to degrade 1,1-dimethyl-substituted ureas, such as monuron, and diuron. However, it must be noted that other transformation pathways, involving the demethylation of the urea group as a first enzymatic reaction, cannot be excluded, given that the transformation of diuron can lead to the formation of N′-(3,4-dichlorophenyl)-N-methylurea and 3,4-dichlorophenylurea in ground water and surface water (9).
In conclusion, our data have shown that an effect of urea herbicides on the soil microbial community could be observed by using both PCA plus cluster analysis of BIOLOG fingerprints and cluster analysis of DGGE profiles. More specifically, it seemed that certain species were either eliminated or stimulated by the application of the herbicides, especially linuron. Moreover, sequencing of DGGE bands allowed us to observe specific, possibly important changes in the species composition of the dominant soil microbial populations, caused by human impact, which would not be observed by culture-dependent techniques.
ACKNOWLEDGMENTS
This work was supported by grant G.O.A. 1997-2002 of the “Ministerie van de Vlaamse Gemeenschap, Bestuur Wetenschappelijk Onderzoek” (Belgium) and by an F.W.O. project grant (1998-2001). E. M. Top is also indebted to the Fund for Scientific Research of Flanders (F.W.O-Vlaanderen) for a position as Research Associate (Onderzoeksleider), and for an equipment grant “Krediet aan Navorsers, 1995.”
We thank K. Smalla for sharing practical tips concerning the DGGE analysis, S. Maertens for technical assistance, T. Tanghe for his advice and help with the HPLC analyses, and T. Deckers, Royal Research Station of Gorsem (Sint Truiden, Belgium), for allowing and helping us to collect the soil samples.
REFERENCES
- 1.Allison N, Turner J E, Wait R. Degradation of homovanillate by a strain of Variovorax paradoxus via ring hydroxylation. FEMS Microbiol Lett. 1995;134:213–219. doi: 10.1111/j.1574-6968.1995.tb07940.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bromilow R H, Evans A A, Nicholls P H, Todd A D, Briggs G G. The effect on soil fertility of repeated applications of pesticides over 20 years. Pestic Sci. 1996;48:63–72. [Google Scholar]
- 5.Caux P Y, Kent R A, Fan G T, Grande C. Canadian water quality guidelines for linuron. Environ Toxicol Water Qual. 1998;13:1–41. [Google Scholar]
- 6.El Fantroussi S, Mahillon J, Naveau H, Agathos S N. Introduction and PCR detection of Desulfomonile tiedjei in soil microcosms. Biodegradation. 1997;8:125–133. doi: 10.1023/a:1008262426800. [DOI] [PubMed] [Google Scholar]
- 7.Engelhardt G, Wallnöffer P R, Plapp R. Identification of N,O-diethylhydroxylamine as a microbial degradation product of the herbicide linuron. Appl Microbiol. 1972;23:664–666. doi: 10.1128/am.23.3.664-666.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Field J A, Reed R L, Sawyer T E, Martinez M. Diuron and its metabolites in surface water and ground water by solid phase extraction and in-vial elution. J Agric Food Chem. 1997;45:3897–3902. [Google Scholar]
- 10.Garland J L. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol Ecol. 1997;24:289–300. [Google Scholar]
- 11.Hansen M C, Nielsen T T, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 12.Hill G D, McGahen J W, Baker H, Finnerty D W, Bingerman C W. The fate of substituted urea herbicides in agricultural soils. Agron J. 1955;47:93–104. [Google Scholar]
- 13.Hoagland R E, Zablotowicz R M. Rhizobacteria with exceptionally high aryl acylamidase activity. Pestic Biochem Physiol. 1995;52:190–200. [Google Scholar]
- 14.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-dependent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen S, Øvreas L, Daae F L, Torsvik V. Diversity in methane enrichments from an agricultural soil revealed by DGGE separation of PCR amplified 16S rDNA fragments. FEMS Microbiol Ecol. 1998;26:17–26. [Google Scholar]
- 16.Kamagata Y, Fulthorpe R R, Tamura K, Takami H, Forney L J, Tiedje J M. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol. 1997;63:2266–2272. doi: 10.1128/aem.63.6.2266-2272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann T, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 19.Marchesi J R, Sato T, Weightman A J, Martin T A, Fry J C, Hiom S J, Wade W G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitsui H, Gorlach K, Lee H, Hattori R, Hattori T. Phylogenetic analysis of the soil bacterial community using a collection. J Microbiol Methods. 1997;30:103–110. [Google Scholar]
- 21.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two Californian estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer G, de Waal E C, Uitterlinden A. Profiling of complex microbial populations using denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Øvreas L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic lake Saelevannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reysenbach A L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts S J, Walker A, Parekh N R, Welch S J, Waddington M J. Studies on a mixed bacterial culture from soil which degrades the herbicide linuron. Pestic Sci. 1993;39:71–78. [Google Scholar]
- 26.Smalla K, Wachtendorf U, Heuer H, Liu W, Forney L. Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl Environ Microbiol. 1998;64:1220–1225. doi: 10.1128/aem.64.4.1220-1225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somerville L, Greaves M P. Pesticide effects on soil microflora. New York, N.Y: Taylor and Francis; 1987. [Google Scholar]
- 28.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallaeys T, Topp E, Muyzer G, Macheret V, Laguerre G, Rigaud A, Soulas G. Evaluation of denaturing gradient gel electrophoresis in the detection of 16S rDNA sequence variation in rhizobia and methanotrophs. FEMS Microbiol Ecol. 1997;24:279–285. [Google Scholar]
- 31.Verschuere L, Fievez V, Van Vooren L, Verstraete W. The contribution of individual populations to the BIOLOG pattern of model microbial communities. FEMS Microbiol Ecol. 1997;24:353–362. [Google Scholar]
- 32.Wallnöfer P. The decomposition of urea herbicides by Bacillus sphaericus, isolated from soil. Weed Res. 1969;9:333–339. [Google Scholar]
- 33.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;344:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 34.Wardle D A, Parkinson D. Effects of three herbicides on soil microbial biomass and activity. Plant Soil. 1990;122:21–28. [Google Scholar]
- 35.Wünsche L, Bruggemann L, Babel W. Determination of substrate utilization patterns of soil microbial communities: an approach to assess population changes after hydrocarbon pollution. FEMS Microbiol Ecol. 1995;17:295–305. [Google Scholar]