Abstract
Mutational processes in tumors create distinctive patterns of mutations, composed of neutral “passenger” mutations and oncogenic drivers that have quantifiable effects on the proliferation and survival of cancer cell lineages. Increases in proliferation and survival are mediated by natural selection, which can be quantified by comparing the frequency at which we detect substitutions to the frequency at which we expect to detect substitutions assuming neutrality. Most of the variants detectable with whole-exome sequencing in tumors are neutral or nearly neutral in effect, and thus the processes generating the majority of mutations may not be the primary sources of the tumorigenic mutations. Across 24 cancer types, we identify the contributions of mutational processes to each oncogenic variant and quantify the degree to which each process contributes to tumorigenesis. We demonstrate that the origination of variants driving melanomas and lung cancers is predominantly attributable to the preventable, exogenous mutational processes associated with ultraviolet light and tobacco exposure, respectively, whereas the origination of selected variants in gliomas and prostate adenocarcinomas is largely attributable to endogenous processes associated with aging. Preventable mutations associated with pathogen exposure and apolipoprotein B mRNA-editing enzyme activity account for a large proportion of the cancer effect within head-and-neck, bladder, cervical, and breast cancers. These attributions complement epidemiological approaches—revealing the burden of cancer driven by single-nucleotide variants caused by either endogenous or exogenous, nonpreventable, or preventable processes, and crucially inform public health strategies.
Keywords: cancer, tumor, single-nucleotide variants, mutational signatures, effect size, somatic mutation, selection, evolution, prevention, public health, molecular epidemiology
Introduction
In the past half-century, our understanding of the origins of cancers has progressed to a widespread acceptance that cancers are the outcome of an evolutionary process driven by mutation, consequent genetic variation, and natural selection for oncogenic variants (Nowell 1976; Merlo et al. 2006; Somarelli et al. 2020). Epidemiological studies have established both an association with age (Siegel et al. 2020) and causation by exposure to carcinogens (Smith et al. 2016), demonstrating that endogenous processes and exogenous mutagens can increase the rate of mutations (Barnes et al. 2018), create somatic genetic variation (Yates and Campbell 2012), and increase the incidence of cancer (Greaves 2015; Golemis et al. 2018). In recent years, large-scale analyses of whole-exome and whole-genome tumor sequencing have been able to recover characteristic tissue-specific signatures of these underlying mutagenic processes in the patterns of variants that have suffused cancer genomes (Alexandrov et al. 2020). However, the specific cancer-driver architecture within each kind of cancer tissue has also been demonstrated to be predictable (Hosseini et al. 2019) and, crucially, circumscribed (Venkatesan et al. 2017): cancer evolution is restricted to certain avenues, funneling specific variants at the rate that they are formed through a distinct process of fixation and then detection. Therefore, the causation of cancer by each mutational process is not determined solely by their effect on mutation rate nor upon the amount of somatic genetic variation they induce, but critically depends upon the degree to which the specific mutations they supply provide selective advantages to clonal lineages within tissues that give rise to cancer.
To evaluate selective advantages requires knowledge of mutation bias, for which characteristic patterns have long been attributed to specific tissues (Brash et al. 1991; Pfeifer et al. 2002; Poon et al. 2014; Pfeifer 2015). Over the last decade, the cancer genomics community has recognized that patterns of substitutions within genomic sequencing data are reflective of mutation bias and are associated with underlying mutational processes (Nik-Zainal et al. 2012; Koh et al. 2021). These patterns can be validated within model organisms under laboratory conditions and attributed to specific mutagenic sources (Segovia et al. 2015), and can be consistently deconvolved from the observed substitutions within exome and genome sequencing data, typically using a nonnegative matrix factorization mathematical framework (Grolleman et al. 2019). Application of these algorithms to whole-exome or whole-genome data recapitulates underlying mutation rates in their trinucleotide context without bias from natural selection because the vast majority of mutations are accumulating neutrally (Greenman et al. 2007; Cannataro and Townsend 2018). Nevertheless, Poulos et al (2018) demonstrated that known major driver mutations are statistically associated with specific mutational signatures. Therefore, specific mutagenic processes in different tissues are driving tumorigenesis via mutations in genes that confer a survival and proliferative advantage to somatic cells. Estimation of the effects of each mutagenic process on the development of cancer requires quantification of the effects of each somatic single-nucleotide variant (SNV) toward tumorigenesis.
Quantification of the cancer effect of mutations requires estimation of their relative impact on cancer lineage survival and replication, an estimation that critically depends on an understanding of the baseline rate of mutation in the absence of natural selection (Cannataro and Townsend 2018). Ostrow et al. (2014) performed a comprehensive analysis of ratios of nonsynonymous change to synonymous change to quantify genome-wide natural selection in the somatic evolution of cancer. This decades-old approach has recently been adapted to the nuances of cancer evolution in several meaningful ways (Shpak and Lu 2016; Zhao et al. 2016), including taking tissue-specific trinucleotide mutational patterns into account (cf., Van den Eynden and Larsson 2017). Martincorena et al. (2017) performed an analysis using trinucleotide substitution rates and covariate-informed gene-level mutation rates to quantify gene-wide selection conferring enhanced proliferation and survival of cancer cell lineages. Temko et al. (2018) deconvolved the underlying mutational signatures in tumor sets, associated signatures and drivers, and quantified the relative intragenic selection of the somatic SNVs in a selection of high-burden driver genes. Cannataro, Gaffney, and Townsend (2018) quantified the site-specific selective effect of each somatic SNV during primary tumor development by determining the constituent mutational signatures driving mutation load in each tumor, coupling these rates with covariate-informed gene-level mutation rates, and quantifying their contribution to cancer cell lineage survival and reproduction in comparison to the convolved baseline mutation rate. These cancer drivers—and their relative effect—may be related back to the mechanisms driving genomic variation, that is, the processes behind the detected mutational signatures.
Mutagenic environmental exposures have been correlated to specific cancer incidences by epidemiological studies spanning the previous 70 years (Doll and Hill 1950; Loeb and Harris 2008). Recently, cancer incidence has also been correlated with tissue-specific stem-cell division numbers (Tomasetti and Vogelstein 2015; Tomasetti et al. 2017), which has been interpreted as evidence that cancers are mainly driven by endogenous, that is, aging or “bad luck”, effects. Other analyses dispute this conclusion, pointing out that it is confounded by the sensitivity of rapidly dividing tissues to exogenous mutational sources (Ashford et al. 2015; Wu et al. 2016), and by the exclusion of cancer types with known environmental causes (Wild et al. 2015). To determine the relative contributions of endogenous and exogenous processes on cancer phenotypes, tumor sequence data can be used to parameterize the magnitude of age-associated, exogenous, and preventable mutational processes that contribute to molecular variation and the consequent cancer effects of each mutation attributable to these processes on tumorigenesis. Such analyses of the evolutionary dynamics driving tumorigenesis back to the sources of the heterogeneity fueling cancer evolution are essential to the advancement of our understanding of oncogenesis and cancer prevention.
Here we analyze the signatures of mutational processes in diverse cancer types. We quantify the cancer effect size of consequent recurrent SNVs. We determine which cancer drivers in each tumor are attributable to processes that have been associated with preventable sources of mutagenesis. We quantify the contribution of each mutagenic process to cancer effect in individual patient tumors, and their relative contribution across tumors within sampled cancer types. We identify cancer types where the discrepancy between mutagenic input and cancer effect is largest, and smallest, and analyze which mutagenic processes are most proportionally discrepant with their cancer effect within each cancer type. This analysis enables comparison of the proportions of cancer effect attributable to age-associated processes to the proportions of cancer effect attributable to putatively preventable mutagenic processes such ultraviolet (UV) light exposure, tobacco smoking or chewing, and apolipoprotein B mRNA-editing enzyme (APOBEC) mutagenesis, addressing a longstanding controversy regarding the role of endogenous “bad luck” and exogenous exposure to tumorigenesis—and moreover, informing the benefits of prevention of mutation in the prevention of cancer.
New Approaches
In each tumor, the probability that a substitution of class j was produced via signature i is an entry of the probability source matrix
| (1) |
where is a vector of the proportional contributions to the tumor mutational burden of each biological signature (see Methods), and Ψ is a matrix defining the biologically relevant signatures, with each entry Ψij being the proportion of substitutions produced by signature i that are of trinucleotide context-specific substitution class j (e.g., A[C→T]T).
We define the cancer effect of each k substitution, γk, as in previous work (Cannataro, Gaffney, Stender, et al. 2018): we quantified mutation as occurring at an intrinsic rate μ per cell over the duration of somatic tissue evolution. Consequently, the expected number of substitutions for a given site in a tumor is the product of their origination rate N × μ times the probability that the mutated lineage spreads to fixation within the tumor cell population N. We assume that, at the time of sequencing, N is consistent across tumors within a given tumor type. We define this probability of fixation as u(s), where s is the population-genetic selection coefficient (Hartl and Clark 2007), leading to an enhanced flux λ = Nμ × u(s) of fixations of mutations. Because the probability of fixation of a neutral mutation is 1/N and the rate of neutral mutation within the population is N × μ, the rate of fixation of neutral mutations within a population is equal to μ.
The ratio of these two fluxes . Therefore,
| (2) |
quantifies the relative increase in cellular proliferation and survival conferred by observed somatic variants. We use the term cancer effect, or “scaled selection coefficient,” for the left hand side of equation (2), which can be estimated given knowledge of the flux of selected mutations and the intrinsic mutation rate, making a parallel to the classic derivation of scaled selection coefficient γ = 2Ns derived from population-genetic models like the Wright-Fisher and Moran models (Sawyer and Hartl 1992; Innan and Kim 2004; Bustamante 2005; Parsons and Quince 2007).
Considering a set V of variants that are present in the tumor, the proportion of total cancer effect in the tumor contributed by process i through variant k is
| (3) |
where the cancer effect of variant k, γk, that is attributable to a mutational process i is γkPi,j(k), the function j(·) maps any genomic substitution k or v to its trinucleotide-context class, and v indexes each variant in the tumor.
As in previous analyses (Cannataro, Gaffney, and Townsend 2018), we used a “soft” definition of fixation, which was that a variant was fixed as soon as it reached a high enough intratumor frequency to be detected during the typical whole-exome sequencing of data sets analyzed herein. Unlike previous analyses, we calculated the effect size for every variant recurrent in more than one tumor by maximizing the likelihood function
where μi, 1 ≤ i ≤ Z, is the rate of mutation to variant k for this tumor, and where M and Z are defined such that the variant is present in M tumors and there is an absence of any same-gene variants in tumors M + 1…Z (we exclude tumors with other variants in the same gene from the latter group due to the likelihood of reduced selection for subsequent same-gene mutations in these tumors). Each tumor-specific mutation rate was calculated by extracting the mutation rate in each trinucleotide context of each variant from the tumor-specific mutational signature weights (eq. 1) and convolving it with the gene-specific mutation rate as in Cannataro, Gaffney, and Townsend (2018).
To quantify the extent to which a mutational process i contributes to cancer effect through a given variant k across a cancer cohort, we calculated the mean value of αi,k across tumors, where the value of αi,k for a tumor without variant k is defined to be zero. To quantify the population-level proportion of cancer effect contributed by mutational process i, we calculated the total value of αi,k across all variants and tumors within a cancer cohort, and then calculated the proportion by which each process i contributes to this total attributed cancer effect.
Results
Proportional Contributions of Mutational Processes to Cancer Effect Can Be Calculated
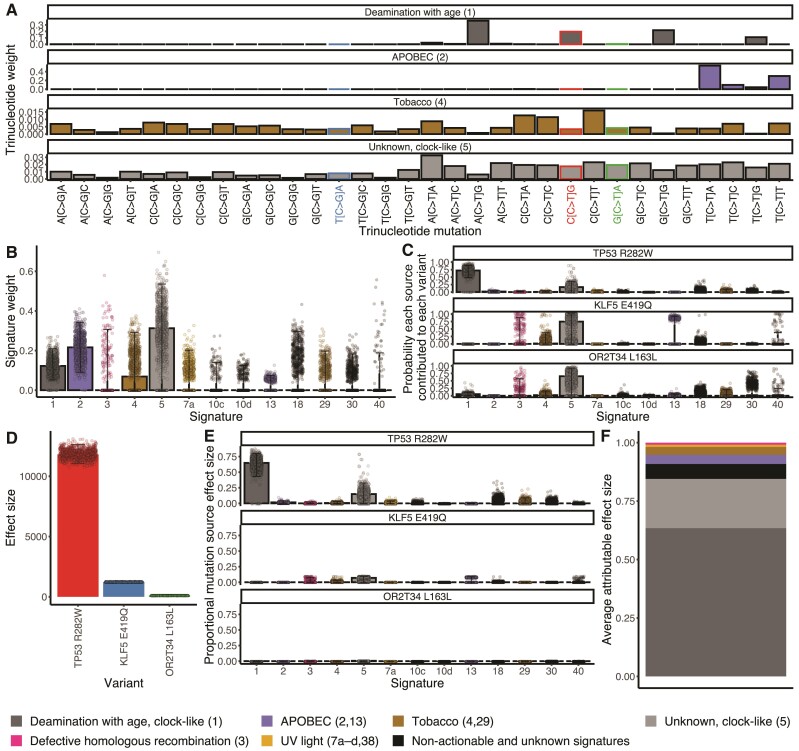
To determine the sources of mutagenesis occurring in tumor samples, we deconvolved the mutational burden of each tumor into the most likely distribution of attributed SNV mutational signatures (Petljak and Alexandrov 2016; Alexandrov and Zhivagui 2019). Applying signature deconvolution to 1,000 bootstrap resamples of a lung squamous-cell carcinoma (LUSC) tumor variant set from a single patient (TCGA-98-A53J-01A-11D-A26M-08) yielded four trinucleotide mutational signatures with median values >0 (age-associated clock-like Signatures 1 and 5, APOBEC-associated #2, and tobacco #4; fig. 1B), each contributing to the flux of SNVs in the tumor at a calculated weight (fig. 1A). The trinucleotide signature weight or combination of trinucleotide signature weights contributing to a specific variant (fig. 1A) times the proportion of mutational causation attributable to each corresponding cancer effect (fig. 1B) provides the probability each source contributed to each recurrent variant in this tumor (eq. 1, fig. 1C). In this instance, age-associated Signature 1 is the most likely contributor to tumor protein gene TP53 R282W, whereas age-associated Signature 5 is the most likely contributor to Kruppel-like factor gene KLF E419Q and the odorant receptor OR2T34 L163L. However, only a few of the mutations that occur in somatic tissue are thought to be selected for their effects on growth or survival, and therefore causative of cancer, and the level of causation is presumably quantitative—that is, driver mutations within a tumor are responsible to different degrees for the manifestation of a cancer phenotype (Cannataro, Gaffney, and Townsend 2018). In this case, TP53 R282W, a mutation in a well-known driver gene found in many cancer types (Wang and Sun 2017) has a higher cancer effect size than KLF E419Q, a variant within a gene shown to promote cancer cell proliferation and survival in bladder and breast cells (Chen et al. 2006; Zheng et al. 2009), and the olfactory receptor mutation OR2T34 L163L has negligible to no effect.
Fig. 1.
Calculating the cancer effects by mutational source for a LUSC, TCGA-98-A53J-01A-11D-A26M-08. The respective products of (A) the trinucleotide weights within each signature (Alexandrov et al. 2020), for example, a relatively frequent aging Signature 1 mutation, CCG → CTG, leads to TP53 R282W; a relatively infrequent aging Signature 1 mutation, TCA → TGA, leads to the KLF5 nonsynonymous mutation E419Q; and a relatively infrequent aging Signature 1 mutation, GCA → GTA, leads to the OR2T34 synonymous mutation L79L; and (B) the proportions of observed mutations in a tumor caused by each signature (aging [1], APOBEC [2], Tobacco [4], clock-like [5]) can be normalized to yield (C) the probability each source contributed to each variant in this tumor. Each of these probabilities serves as a weight to multiply by (D) the cancer effect size of each variant (Cannataro, Gaffney, and Townsend 2018), to yield (E) the probability-weighted portion of effect size for each variant attributable to each source of mutations, stacked to compose (F) the proportion of cancer causation attributable to each source of mutations averaged over bootstrapped resampling of mutational signature calls. Individual points on the plots represent single calculations from each bootstrap resampling. Bars represent median values. Whiskers represent 95% confidence intervals.
The product of the probability that each mutational source contributed to each variant in this tumor and the effect of the specific variant (fig. 1D) quantifies the probability-weighted cancer effect for each variant by each source (eq. 3, fig. 1E). Summing the probability-weighted cancer effect for each source across variants and averaging over bootstrap samples yields the proportion of cancer effect attributable to each source of mutations (fig. 1F). Age-associated mutational Signature 1 contributed the highest weight in TCGA-98-A53J-01A-11D-A26M-08, and led to the largest estimated effect through its high probability of being causative in the TP53 R282W mutation. Overall, the majority of the cancer effect within this tumor sample is attributable to endogenous processes that accumulate in a clock-like manner with age. Via deconvolution of the mutational signatures responsible for recurrent variants in cancer and calculation of the cancer effect sizes of the nucleotide substitutions driving cancer evolution, we have calculated which mutagenic sources fueling nucleotide variation can be attributed as proportionally causative of individual tumors in patients.
Mutagenic Input and Cancer Effect From Each Source Can Differ Substantially within Tumors
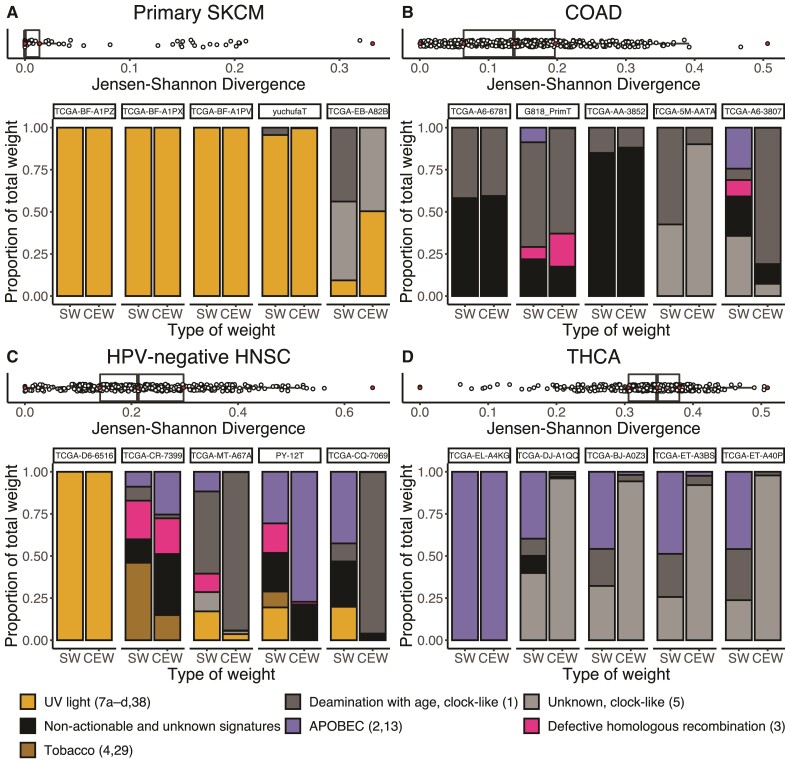
The match between the proportional input to total mutations by each mutagenic source (fig. 1B) and the proportional cancer effect arising from each mutagenic source (fig. 1F) varies in each patient’s tumor (fig. 2). Quantifying the degree of match between proportional mutational input and proportion of cancer causation by the Jensen–Shannon Divergence (JSD), we found the tumor type with the lowest median mismatch to be primary skin cutaneous melanoma (primary SKCM, fig. 2A). The mutational input to the majority of the primary SKCM tumors could be entirely attributed to UV radiation (7a–d, 38). Accordingly, all of the cancer effect from the somatic SNVs was attributable to these signatures as well. This match resulted in JSD values of zero for the majority of SKCM tumors. Similarly, the lowest-JSD tumor for human papillomavirus negative head-and-neck squamous-cell carcinoma (HPV-negative HNSC) exhibited all mutational weight and effect size attributable to UV signatures. Indeed, the tissue of origin of this particular head-and-neck cancer was the sun-exposed lip (Grossman et al. 2016).
Fig. 2.
Box plots of the median JSD between the proportional input of each mutagenic source and the proportion that each signature contributed to the total cancer effect in each tumor among 1,000 bootstrap samples, accompanied by the proportions of observed mutations in a tumor caused by each signature (signature weight, SW) and the proportions of cancer causation attributable to each source of mutations (cancer effect weight, CEW), for the median JSD resampling of five tumors (filled dots) bounding the quartiles of the JSD among tumors, for four cancer types that bound the tertiles of median JSD among cancer types: (A) the cancer type with the least median divergence between the proportional input of each mutagenic source and the proportion that each signature contributed to the total cancer effect, primary SKCM, (B) Colon adenocarcinoma (COAD), (C) HPV-negative HNSC, and (D) the cancer type with the greatest median divergence between the proportional input of each mutagenic source and the proportion that each signature contributed to the total cancer effect, Thyroid carcinoma (THCA).
Other tumors, such as TCGA-EB-A82B in primary SKCM, exhibited a greater mismatch between processes driving mutation accumulation and the processes contributing high-effect variants. In the bootstrap sample with the median JSD value for this tumor, 44% of somatic mutations to TCGA-EB-A82B were attributable to age-associated Signature #1; in contrast, over 99% of the cancer effect is attributable to signatures associated with UV light and clock-like Signature #5. These mismatches are even more frequent in 24 other tumor types analyzed (colon adenocarcinoma, fig. 2B; and human papillomavirus negative head-and-neck squamous-cell carcinoma, fig. 2C—both tumor types with intermediate median JSD values—and thyroid cancer, exhibiting the greatest median mismatch across tumor types, fig. 2D; supplemental fig. S1, Supplementary Material online).
Mutagenic Input and Cancer Effect From Each Source Can Differ Enormously Among Oncogenic Variants Within Each Cancer Type
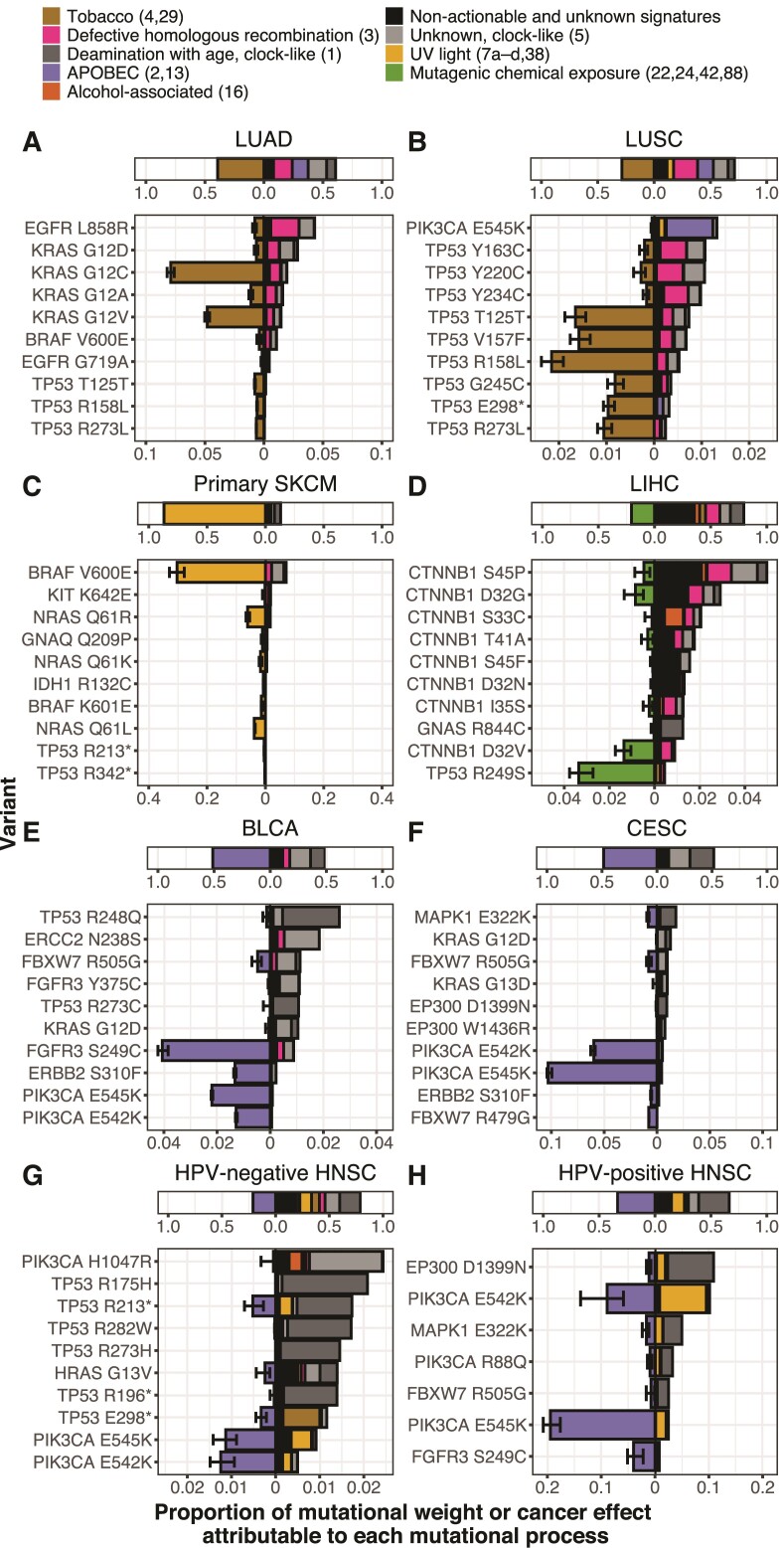
Many well-known processes have been established as major contributors to tumor mutation burden, such as tobacco in lung tissues, UV radiation in skin tissues, and APOBEC cytidine deaminases in bladder, cervical, and HNSC tissues. However, mutational processes are trinucleotide-specific, which leads to differences in underlying amino-acid mutation rates depending on the sequence context of each variant site. Moreover, the mutational process most likely to originate an oncogenic variant can not only differ from variant to variant, but can also differ from the mutational process that causes the greatest number of mutations within each tumor type (fig. 3).
Fig. 3.
Average contributions of each mutational process to the total mutation burden among 1,000 bootstrap resamples (top stacked bars), and median across bootstraps cross-tumor average cancer effects of variants classified as drivers (bottom stacked bars). Cancer effect is quantified proportionate to the total cancer effect in each tumor, and variants are filtered to include only genes classified as drivers in (Bailey et al. 2018). For each cancer type, the average contribution of the dominant preventable process (quantified by the bar width left of the x-axis origin) and the nondominant or nonpreventable processes (quantified by the bar width right of the x-axis origin) to total mutation is shown, above the top 10 variants contributing the greatest cancer effect to primary tumors of that cancer type, ordered by the average proportional effect size attributable to that variant from all of the nondominant or nonpreventable mutational processes, alongside the average contribution of the dominant preventable processes (left of the x-axis origin) and the nondominant or nonpreventable processes (right of the x-axis origin) to cancer effect (measured proportionate to the total effect in each tumor). (A) Lung adenocarcinoma (LUAD), (B) LUSC, (C) SKCM (primary tumors only), (D) Liver hepatocellular carcinoma (LIHC), (E) Bladder urothelial carcinoma (BLCA), (F) Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), (G) HPV-negative HNSC, and (H) HPV-positive HNSC. Whiskers represent 95% confidence intervals, supplemental table S1, Supplementary Material online contains all confidence intervals of average cancer effects of variants pictured.
For instance, among preventable processes, mutations in lung adenocarcinoma (LUAD) and lung squamous-cell carcinoma (LUSC) were most frequently attributed to tobacco-associated mutagenesis (fig. 3A and B). The high attribution of the Kirsten rat sarcoma virus protein (KRAS) G12C mutations to this lung-specific mutagenic process explains their high frequency in LUAD compared with other RAS-driven cancer types such as pancreas or colon adenocarcinomas. Major driver variants of KRAS and TP53, in LUAD and LUSC, respectively, exhibit markedly different origination rates from tobacco-associated processes. Perhaps most notable is the minimal attribution of EGFR L858R to tobacco-associated mutagenic processes. The attribution of tobacco-associated mutagenic processes to the cancer effects of KRAS G12 variants and epidermal growth factor receptor (EGFR) L858R (fig. 3A) are consistent with—and provide an explanation for—the increased odds of KRAS mutation in tumor tissue of ever smokers compared with never smokers, as well as the increased odds of EGFR mutation in never smokers compared with ever smokers (Chapman et al. 2016). Even nucleotide variants that do not cause an amino-acid substitution have quantifiable cancer effects that can be attributed to mutagenic processes—for example, TP53 synonymous mutation T125T, which affects splicing of the TP53 transcript (Varley et al. 2001), is attributable to tobacco-associated signatures in both LUAD and LUSC (fig. 3A and B).
Distinct major driver mutations within the same cancer type can be attributed to different mutagenic processes. UV light associated Signatures 7a–d and 38 are the most likely source of the majority of the cancer effect contributed by v-raf murine sarcoma viral oncogene homolog b1 (BRAF) V600E, the most prevalent (and most strongly selected [cf. Cannataro, Gaffney, and Townsend 2018]) driver of primary SKCM (fig. 3C). In contrast, one major oncogenic variant common to SKCM (KIT K642E) is almost entirely attributable to defective homologous recombination and age-associated processes, rather than UV-associated processes (fig. 3C). In liver hepatocellular carcinoma (LIHC), the greatest proportions of cancer effect for several oncogenic somatic variants such as TP53 R249S and catenin beta 1 (CTNNB1) D32V are attributable to mutagenic chemical exposure. Nevertheless, the greatest proportions of cancer effect in several other CTNNB1 variants are attributable to processes with as-yet unknown etiology that may in the future be linked to other mutagenic chemical exposures (fig. 3D). In bladder urothelial carcinoma (BLCA), major oncogenic mutations were attributed to signatures indicative of APOBEC cytidine deaminase activity, the major contributor to tumor mutation burden in BLCA. APOBEC cytidine deaminases are thought to be activated by exposure to viruses, which may be presumed to be preventable. Although six of the top ten variants as determined by cancer effect were largely attributed to nonpreventable, age-associated processes; four known cancer-driver variants (fibroblast growth factor receptor FGFR3 S249C, erythroblastic oncogene B ERBB2 S310F, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha PIK3CA E545K, and PIK3CA E542K) were almost entirely attributed to the action of APOBEC cytidine deaminases. Cervical squamous-cell carcinoma and endocervical adenocarcinoma (CESC), HPV-negative HNSC, and HPV-positive HNSC were also bestrewn with APOBEC-associated mutations. CESC and HNSC also exhibited diversity in which process was most likely to originate each oncogenic variant (Cannataro et al. 2019); however, attributions of APOBEC-associated processes for the origination of oncogenic PIK3CA E542K and PIK3CA E545K mutations are consistent across multiple cancer types (cf., fig. 3B, E–H).
Relative Mutagenic Input and Relative Cancer Effect are Specific to Each Tumor Type
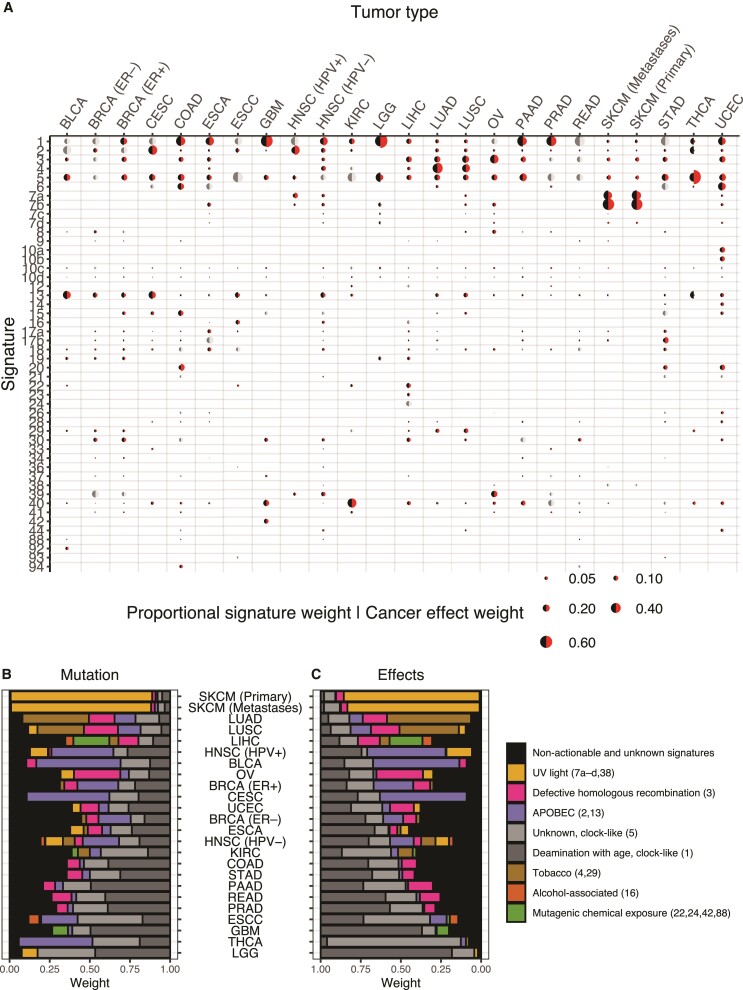
The mismatches between the proportional input to total mutations by each mutagenic source (fig. 1B) and the proportional cancer effect arising from each mutagenic source (fig. 1F) exist not only at the level of individual tumors, but also at the level of tumor types—where they indicate which mutational sources make an outsized contribution to the causation of cancer compared with their production of mutations, and vice versa. Many tumor-type mutational signature pairs exhibit statistically significant differences between the proportional input to total mutations by each mutagenic source and the proportional cancer effect arising from each mutagenic source (continuity-corrected Wilcoxon two-sided rank-sum tests, P < 0.05; fig. 4A; supplemental table S2, Supplementary Material online).
Fig. 4.
Mutational process weights and cancer effects for unknown and etiology-associated mutation signatures. (A) Relative contributions (averaged across bootstraps and tumors) of mutational signatures to total substitution burden (black half-circle) and cancer effects (red half-circle) across 24 cancer types. Cancer effect weights that are not statistically significantly different from mutation weights by a paired Wilcoxon two-sided rank-sum test (comparing only tumors with the signature present) are indicated by light and dark gray half-circles. Comparisons are only visualized and statistical associations tested if 30 or more tumors had corresponding signatures. Relative contributions to (B) total somatic SNVs and to (C) relative cancer effects (averaged among 1,000 bootstrap resamplings) of age-associated, preventable, and unknown processes across 24 cancer types.
For example, APOBEC-related Signatures 2 and 13 exhibit larger mutation weight than cancer effect across many cancer types, as do Signatures 10c–d, 17a, and 18. Some signatures contribute to higher proportional cancer effect than mutational weight, such as Signature #40, clock-like #5, defective homologous recombination associated #3, and age-associated Signature #1. These specific signatures contributed higher proportional cancer effect than mutational weight in 9, 9, 8, and 7 tumor types, respectively. In lower-grade glioma (LGG), the age-associated Signature #5 constitutes much more of the mutation weight than its cancer effect (mean 36% compared with 14%), whereas age-associated Signature 1 has the opposite relationship (47% compared with 82%). This difference between the two age-associated signatures is largely attributable to the high-effect size of isocitrate dehydrogenase 1 (IDH1) variants, which occur predominantly as a consequence of ACG → ATG mutations that are frequent in Signature 1 and rare in Signature 5. A similar contrast can be seen in thyroid adenocarcinoma, wherein the APOBEC-associated Signature 2 exhibits high mutation weight and very little cancer effect (24% compared with 2%), and wherein the aging Signature 5 exhibits much more cancer effect than mutation weight (83% compared with 29%). This contrast comes about because thyroid adenocarcinoma is often driven by BRAF V600E mutations that convey enormous cancer effects, and BRAF V600E mutations come about frequently as a consequence of GTG → GAG mutations that are found at low frequency within the aging Signature #5, but are found at extremely low frequency within APOBEC Signature #2.
Preventable Mutational Processes Contribute Substantially to Causation of Skin, Lung, Head-and-Neck, Bladder, and Cervical Cancer
Among the non-age-related etiologies are a number of mutational processes that are putatively “preventable”—in that they can be mitigated by individual behaviors or interventions (fig. 4B and C). Skin cancer, lung cancer, HPV-positive head-and-neck cancer, bladder cancer, and cervical cancer are notable for the dominant role of putatively preventable processes underlying both raw somatic SNV mutation weight (fig. 4A) and cancer effect (fig. 4B). Lower-grade glioma (LGG), glioblastoma (GBM), and prostate adenocarcinoma (PRAD) are notable for the lack of putatively preventable processes underlying both raw mutation weight and cancer effect.
In thyroid carcinoma (THCA), an average of 46% of the underlying total mutational burden is attributable to the processes associated with APOBEC activity. Thus, minimizing APOBEC mutagenesis (potentially by avoiding or suppressing viral infections) would prevent a large proportion of mutations. However, the processes attributable to the variants of highest cancer effect are nearly all associated with aging. Consequently, prevention of APOBEC-associated mutation would likely do little to prevent the majority of THCA. A contrasting case is the lung cancers: the net cancer effects of the SNVs attributable to tobacco chewing (7.6% in LUAD and 10% in LUSC) and tobacco smoking (46% in LUAD and 28% in LUSC) are larger than the mutation weights of these sources of mutagenesis (6% and 7% for tobacco chewing and 36% and 22% for smoking in LUAD and LUSC, respectively; P < 0.001 for all relationships; Wilcoxon rank-sum test).
Age-associated Mutational Processes Contribute Substantially to Causation of Glioma, Prostate, Thyroid, Pancreatic, and Colorectal Cancer
Because each mutagenic process is linked to a trinucleotide variant signature that has been identified as clock-like (ubiquitous and age-associated) or non-clock-like (not associated with age; Alexandrov et al. 2015, 2020), the proportion of total mutations attributable to clock-like processes and non-clock-like processes in each cancer type can be quantified (fig. 4B). Among tissues, the cancer types with the greatest proportion of total mutations contributed by non-clock-like processes are melanoma (primary and metastatic), lung cancers (adenocarcinoma and squamous-cell carcinoma), head-and-neck cancers, urothelial bladder carcinoma, cervical squamous-cell carcinoma, and liver hepatocellular carcinoma. Lower-grade glioma exhibits the greatest proportion of total mutations contributed by age-associated, “clock-like” processes. Moreover, with regard to the explanation of tumorigenesis and cancer incidence, the cancer effect attributable to age-associated processes and non-age-associated processes in each cancer type can be quantified (fig. 4C). Among tumor tissues, those with the greatest proportion of cancer effect contributed by age-associated processes are gliomas (LGG, GBM), thyroid cancer, and prostate adenocarcinomas, consistent with the strong association of the incidence of these cancers with age (Dubrow and Darefsky 2011; Rawla 2019), as well as pancreatic cancers. Primary and metastatic melanoma, lung adenocarcinoma, liver hepatocellular carcinoma, and HPV-positive head-and-neck squamous-cell carcinoma exhibit the greatest proportion of cancer effect contributed by non-age-associated processes, consistent with the strong association of these cancers with exogenous factors (UV exposure, smoking, mutagenic chemical exposure, and HPV infection).
Our analysis attributes an amount of cancer causation to such endogenous and non-preventable processes that varies widely among cancer types. Cancer types varied in the degree to which their causation was associated with COSMIC Signature 1, which correlates with stem-cell division in different tissues (Alexandrov et al. 2015) and represents the processes associated with the mitotic clock (fig. 3C; cf., Tomasetti and Vogelstein 2015; Tomasetti et al. 2017), ranging from small contributions (<10%) in primary and metastatic SKCM, THCA, LUAD, and LUSC, to 82% of the cancer effect in LGG. Combining the replication-associated and nonreplication-associated signatures that correlate with age, cancer effect attributable to all age-associated processes ranged from 9% in primary SKCM to 96% in LGG; in 17 of the cancer types (stomach adenocarcinoma (STAD), colon adenocarcinoma, HPV-negative and -positive HNSC, CESC, Breast invasive carcinoma (BRCA)—ER positive and negative, Kidney renal clear cell carcinoma (KIRC), Uterine Corpus Endometrial Carcinoma (UCEC), CESC, BLCA, ovarian cystadenocarcinoma (OV), LIHC, LUSC, LUAD, primary and metastatic SKCM) a minority of cancer causation was attributable to age-associated processes. Age has not been associated across cancer types with any of the signatures that have unknown etiology (Alexandrov et al. 2020). Greater than 35% of the cancer effect leading to kidney renal clear cell carcinoma (KIRC) is attributed to unknown mutational processes; breast invasive carcinoma (ER− and +), OV, esophageal carcinoma (ESCA), STAD, LIHC, and PRAD all have 20% or more of their cancer effect caused by currently unknown or unattributed mutational processes (fig. 4C).
Discussion
Here we have shown that the impact on carcinogenesis of mutagenic processes associated with SNV signatures can be quantified. This quantification is distinct from the number or proportion of mutations that can be attributed to a process, because it accounts for the extent to which each mutation contributes to the cancer phenotype—increased replicative and survival advantage in each tissue and cancer type—via SNVs. We have shown how to use the proportions of observed mutations in a tumor caused by each signature to calculate the probability that each mutational source contributed to each variant in this tumor. Each of these probabilities serves to weight the cancer effect size of each variant, yielding the probability-weighted portion of effect size for each variant attributable to each source of mutations and thus the proportion of cancer causation attributable to each source of mutations. In turn, the quantification of increased cellular replication and survival within each tumor, characterized across a population of patients, provides a reductionist molecular approach toward quantifying the degree to which a process can be held responsible for carcinogenesis in a cancer type that is wholly distinct from traditional epidemiological studies.
Our analysis of the cancer effects of single-nucleotide mutations and associated signatures has been enabled by quantitative estimates of their intrinsic mutation rates (Fousteri and Mullenders 2008; Stamatoyannopoulos et al. 2009; Lawrence et al. 2013). Deconvolution of the quantitative contributions of known mutation signatures explains the high prevalence of KRAS G12C and low prevalence of EGFR L858R in ever smokers, and the converse relationships in never smokers. It illuminates the potent role of UV light in BRAF V600E-driven melanoma. It attributes major drivers PIK3CA E542K and E545K to the potentially virally-induced action of APOBEC cytidine deaminases, and highlights unknown processes that deserve further identification such as those underlying high-cancer-effect SNVs of LIHC. Importantly, germline variants, copy-number variation, epigenetic alterations, and changes to the aging tissue microenvironment also contribute to the cancer phenotype (Mroz et al. 2015; Ramakodi et al. 2016; Liggett and DeGregori 2017; Montgomery et al. 2018; Sun et al. 2018; Laconi et al. 2020). Incorporation of signatures associated with these kinds of alterations (Macintyre et al. 2018) and of attributions of each signature to relevant sources would markedly increase the purview of inferred cancer causation, revealing a full picture of the importance of diverse mechanisms behind the spectrum of genomic alterations fueling cancer evolution.
For an individual cancer patient, calculation of the relative cancer effect of diverse sources of mutation provides an estimate of how much each mutagenic process is responsible for an individual’s cancer. From a public health perspective, these calculations constitute a bridge between molecular studies and longstanding epidemiological analyses that have associated behaviors (e.g., smoking) or professions (e.g., sun exposure) with cancer incidence. Public health intervention targeted at minimizing exposure to these preventable signatures would mitigate disease severity by preventing the accumulation of mutations that directly contribute to the cancer phenotype. Finally, our findings connect specific mutagenesis patterns and processes with cancer, providing guidance as to why an instance of cancer happened—and have promise to play a significant role in demonstrating individual as well as group-level cause for legal recourse due to carcinogenic exposure (e.g., Lee 2016).
The quantification of cancer effect attributable to specific sources of mutation has evident parallels to epidemiological results that assess the effect of risk factors on cancer causation (Shield et al. 2016). These epidemiological results often rely on correlation, and calculate an increase in the probability of cancer in relation to some behavior or exposure. Calculations of the relative cancer effect of diverse correctly identified sources of mutation relate the mutations driving tumorigenesis to mechanistic processes. However, multiple challenges impede their use at a population level in comparison to longstanding, well-crafted epidemiological studies: (1) conducting appropriate tumor sampling—most large tumor sequencing studies are sampled haphazardly, without reference to a distinct population, without stratification or even “random” sampling; (2) formulating an “apples to apples” quantitative mapping comparing proportions of effect to odds ratios; and (3) forming a discrete mapping of mutational signatures to mechanistic processes to epidemiological factors. These mechanistic annotations associated with COSMIC Signatures are critical to our interpretations of these results, and range in surety from well-established (e.g., UV #7 and smoking #4), to presumptive (e.g., indirect damage from UV light #38). Proposed etiologies reflect associations with processes—not necessarily direct causation (Koh et al. 2021).
Recent research has touched on a debate as to what extent “bad luck”—endogenous mutagenic processes that accumulate naturally with age—plays a role in the incidence of cancer arising in various tissues. Here, we addressed the question regarding the relative contributions of exogenous and endogenous sources of mutation to tumorigenesis by quantifying the strength of selection on specific variants that are driving tumorigenesis, and attributing the variants back to the mutational processes that originally fueled their creation. We found that signatures relating to aging processes (#1 and #5) were responsible for the majority of cancer effects in tumors of the brain (LGG, GBM) and tissues with large amounts of epithelial turnover (Rectum adenocarcinoma (READ), COAD, STAD, and esophageal squamous-cell carcinoma (ESCC)). Other tumors whose cancer effects could largely be attributed to aging include PRAD—a tumor type strongly associated with age (Bostwick et al. 2004), THCA (whose major single-nucleotide driver, BRAF V600E, is more likely to be caused by mutations associated with clock-like Signature 5 than by mutations associated with other signatures), and pancreatic adenocarinomas (PAAD). Several tumor types have large proportions of the cancer effect size directly attributable to mutational processes that are preventable, that is, interventions could reduce the mutations in these tissues that are responsible for the cancer-causing variants. CESC, HNSC, and BLCA are largely driven by mutations attributed to virus-induced APOBEC activity, SKCM is largely driven by UV light exposure, and mutations responsible for increased proliferation and survival of cancerous cells within lung cancers trace back to smoking.
The importance of understanding the underlying sources of mutations that are selected along the molecular evolutionary trajectory toward cancer in each and every patient is underscored by the remarkable successes of antismoking interventions against carcinogenic exposures, which have saved many lives (Holford et al. 2014). Many lives can be saved by preventing the origination of somatic mutations that lead to cancer, and quantification of the relative roles of these mutagenic processes in each cancer type provides essential guidance toward suitable strategies for prevention. In our study, some cancer types such as KIRC, ESCA, STAD, and BRCA (ER−) exhibited a large proportion of cancer effect that was attributable to signatures with unknown etiology. We are likely to gain greater insight into these mutational signatures—including whether they are endogenous or exogenous, and whether they come from sources that are preventable. As we do so, we may discover additional preventable mutational processes that can be mitigated by proactive public health interventions.
Methods
To attribute the increased cellular reproduction and survival conferred by SNVs responsible for cancer growth to their underlying mutational processes, we determined the mutational signatures within individual tumors, calculated the effect size of each single-nucleotide substitution among tumors in each tumor type, and evaluated the likelihood that each of these substitutions was the product of each mutational source within each tumor. Thus, single-nucleotide substitutions responsible for the largest influence on cellular division and survival—and hence the tumor phenotype—may be attributed to the root sources of molecular variation within each somatic tissue. We analyzed the pan-cancer whole-exome tumor sequencing dataset curated in Cannataro, Gaffney, and Townsend (2018), except all Yale-Gilead tumors that might have been treated with chemotherapies were removed (removed tumors in supplementary table S3, Supplementary Material online). Scripts used to perform these analyses are available online (Townsend-Lab-Yale).
Attributing Sources of Mutation within Tumors
To attribute observed sets of substitutions in tumors to the underlying sources of mutations, we used the R package MutationalPatterns v3.0.1 (Blokzijl et al. 2018) to extract version 3.2 COSMIC Signatures from each tumor’s set of nonrecurrent substitutions. We excluded recurrent variants because they are much more likely to be under selection in the cancer cell population; nonrecurrent mutations more accurately reflect mutational influx. To minimize signature bleeding because some COSMIC signatures share similar mutational profiles, we limited the number of signatures detectable in each tumor type to those signatures detected at any prevalence in tumors of that type previously by Alexandrov et al. (2020), with the addition of enabling inference of COSMIC single-base substitution signature SBS16 within esophageal squamous-cell carcinoma (Li et al. 2018). We also applied the recommended minimum threshold for the number of substitutions necessary to attribute to a signature associated with increased mutagenesis. For example, signatures attributable to defective DNA mismatch repair were only allowed in tumors with over 200 substitutions (Alexandrov et al. 2020). Some tumors analyzed exhibited fewer than 50 substitutions (supplemental fig. S2, Supplementary Material online)—a threshold below which precise deconvolution of mutational signatures has been deemed problematic with a previous signature deconstruction algorithm (Rosenthal et al. 2016). For these tumors, we mixed the MutationalPatterns estimates of the signature weights for the specific tumor with the average signature weights for the tumors with 50 or more substitutions of the same tumor type, weighting the former in proportion to the number of variants in the tumor out of 50. We constructed 1,000 resamplings of the variant sets within each tumor using the built-in bootstrapping functionality of MutationalPatterns and analyzed each to result in 1,000 estimates of the underlying signatures for each tumor. Subsequent calculations that utilize these signatures for each tumor, such as the effect size and effect size attribution calculations, were performed on each batch of the bootstrap resampling. Averages (means and medians) across bootstrap resamplings were reported as our estimate, along with 95% error bars based on the 25th and 975th sorted bootstrapped values.
As some COSMIC signatures have been attributed to artefactual processes such as sample handling and sequencing, we focused on the tumor-type-specific subset of signatures that represent biologically relevant mutational processes, B (Alexandrov et al. 2020). To determine the proportions of mutations attributable to each biological signature in each tumor, we divided the weights of each biological signature within the tumor by the sum of all biological signature weights. That is, letting represent the original fitted weights of all signatures in a tumor, we define the relative biological weights such that, for each signature i ∈ B,
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
Members of the Townsend Lab provided stimulating discussions and helpful feedback on this research. This work was supported by NIH 1P50DE030707, NIH 1R01LM012487, NIH 1R01CA215900, NIH 5R01 CA231112, the Yale Cancer Biology Training program (NIH T32 CA193200/CA/NCI HHS/United States), and the Elihu Professorship endowed research funds.
Contributor Information
Vincent L. Cannataro, Department of Biology, Emmanuel College, Boston, MA, USA.
Jeffrey D. Mandell, Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA
Jeffrey P. Townsend, Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA Department of Biostatistics, Yale School of Public Health, New Haven, CT, USA; Department of Ecology and Evolutionary Biology, Yale University, New Haven, CT, USA.
Data Availability
The datasets generated and/or analyzed during the current study are available in the Genomic Data Commons repository, https://portal.gdc.cancer.gov/, as an open-access supplement to Cannataro, Gaffney, and Townsend (2018), and all computational scripts used to analyze these data are found in our github repository here: https://github.com/Townsend-Lab-Yale/cancer_causes_and_effects. We used the cancereffectsizeR v2.3.4 package to calculate cancer effect sizes.
References
- Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, Stratton MR. 2015. Clock-like mutational processes in human somatic cells. Nat Genet. 47:1402–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, et al. 2020. The repertoire of mutational signatures in human cancer. Nature 578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Zhivagui M. 2019. Mutational signatures and the etiology of human cancers. In: Boffetta P, Hainaut P, editors. Encyclopedia of cancer: reference module in biomedical sciences. 3rd ed. London: Academic Press. p. 499–510. 10.1016/b978-0-12-801238-3.65046-8 [DOI] [Google Scholar]
- Ashford NA, Bauman P, Brown HS, Clapp RW, Finkel AM, Gee D, Hattis DB, Martuzzi M, Sasco AJ, Sass JB. 2015. Cancer risk: role of environment. Science 347:727. [DOI] [PubMed] [Google Scholar]
- Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, et al. 2018. Comprehensive characterization of cancer driver genes and mutations. Cell 174:1034–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes JL, Zubair M, John K, Poirier MC, Martin FL. 2018. Carcinogens and DNA damage. Biochem Soc Trans. 46:1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokzijl F, Janssen R, van Boxtel R, Cuppen E. 2018. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med. 10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostwick DG, Burke HB, Djakiew D, Euling S, Ho S-M, Landolph J, Morrison H, Sonawane B, Shifflett T, Waters DJ, et al. 2004. Human prostate cancer risk factors. Cancer 101:2371–2490. [DOI] [PubMed] [Google Scholar]
- Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Pontén J. 1991. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci U S A. 88:10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD. 2005. Population genetics of molecular evolution, editors. Statistical methods in molecular evolution. Statistics for biology and health. Springer New York. p. 63–99. [Google Scholar]
- Cannataro VL, Gaffney SG, Sasaki T, Issaeva N, Grewal NKS, Grandis JR, Yarbrough WG, Burtness B, Anderson KS, Townsend JP. 2019. APOBEC-induced mutations and their cancer effect size in head and neck squamous cell carcinoma. Oncogene 38:3475–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannataro VL, Gaffney SG, Stender C, Zhao Z-M, Philips M, Greenstein AE, Townsend JP. 2018. Heterogeneity and mutation in KRAS and associated oncogenes: evaluating the potential for the evolution of resistance to targeting of KRAS G12C. Oncogene 37:2444–2455. [DOI] [PubMed] [Google Scholar]
- Cannataro VL, Gaffney SG, Townsend JP. 2018. Effect sizes of somatic mutations in cancer. J Natl Cancer Inst. 110:1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannataro VL, Townsend JP. 2018. Neutral theory and the somatic evolution of cancer. Mol Biol Evol. 35:1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman AM, Sun KY, Ruestow P, Cowan DM, Madl AK. 2016. Lung cancer mutation profile of EGFR, ALK, and KRAS: meta-analysis and comparison of never and ever smokers. Lung Cancer 102:122–134. [DOI] [PubMed] [Google Scholar]
- Chen C, Benjamin MS, Sun X, Otto KB, Guo P, Dong X-Y, Bao Y, Zhou Z, Cheng X, Simons JW, et al. 2006. KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU-Pr1 human bladder cancer cell line. Int J Cancer 118:1346–1355. [DOI] [PubMed] [Google Scholar]
- Doll R, Hill AB. 1950. Smoking and carcinoma of the lung; preliminary report. Br Med J. 2:739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrow R, Darefsky AS. 2011. Demographic variation in incidence of adult glioma by subtype, United States, 1992–2007. BMC Cancer 11:325. Available from: https://pubmed.ncbi.nlm.nih.gov/21801393/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LHF. 2008. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 18:73–84. [DOI] [PubMed] [Google Scholar]
- Golemis EA, Scheet P, Beck TN, Scolnick EM, Hunter DJ, Hawk E, Hopkins N. 2018. Molecular mechanisms of the preventable causes of cancer in the United States. Genes Dev. 32:868–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M. 2015. Evolutionary determinants of cancer. Cancer Discov. 5:806–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolleman JE, Díaz-Gay M, Franch-Expósito S, Castellví-Bel S, de Voer RM. 2019. Somatic mutational signatures in polyposis and colorectal cancer. Mol Aspects Med. 69:62–72. [DOI] [PubMed] [Google Scholar]
- Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. 2016. Toward a shared vision for cancer genomic data. N Engl J Med. 375:1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. 2007. Principles of population genetics. Sunderland (MA): Sinauer and Associates. [Google Scholar]
- Holford TR, Meza R, Warner KE, Meernik C, Jeon J, Moolgavkar SH, Levy DT. 2014. Tobacco control and the reduction in smoking-related premature deaths in the United States, 1964–2012. JAMA 311:164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini S-R, Diaz-Uriarte R, Markowetz F, Beerenwinkel N. 2019. Estimating the predictability of cancer evolution. Bioinformatics 35:i389–i397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kim Y. 2004. Pattern of polymorphism after strong artificial selection in a domestication event. Proc Natl Acad Sci U S A. 101:10667–10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh G, Degasperi A, Zou X, Momen S, Nik-Zainal S. 2021. Mutational signatures: emerging concepts, caveats and clinical applications. Nat Rev Cancer. 21:619–637. [DOI] [PubMed] [Google Scholar]
- Laconi E, Marongiu F, DeGregori J. 2020. Cancer as a disease of old age: changing mutational and microenvironmental landscapes. Br J Cancer 122:943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SG. 2016. Proving causation with epidemiological evidence in tobacco lawsuits. J Prev Med Public Health. 49:80–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Wang MY, Yang M, Dai HJ, Zhang BF, Wang W, Chu XL, Wang X, Zheng H, Niu RF, et al. 2018. A mutational signature associated with alcohol consumption and prognostically significantly mutated driver genes in esophageal squamous cell carcinoma. Ann Oncol. 29:938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggett LA, DeGregori J. 2017. Changing mutational and adaptive landscapes and the genesis of cancer. Biochim Biophys Acta Rev Cancer. 1867:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Harris CC. 2008. Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res. 68:6863–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre G, Goranova TE, De Silva D, Ennis D, Piskorz AM, Eldridge M, Sie D, Lewsley L-A, Hanif A, Wilson C, et al. 2018. Copy number signatures and mutational processes in ovarian carcinoma. Nat Genet. 50:1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR, Campbell PJ. 2017. Universal patterns of selection in cancer and somatic tissues. Cell 171:1029–1041.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LMF, Pepper JW, Reid BJ, Maley CC. 2006. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 6:924–935. [DOI] [PubMed] [Google Scholar]
- Montgomery ND, Selitsky SR, Patel NM, Neil Hayes D, Parker JS, Weck KE. 2018. Identification of germline variants in tumor genomic sequencing analysis. J Mol Diagn. 20:123–125. [DOI] [PubMed] [Google Scholar]
- Mroz EA, Tward AD, Hammon RJ, Ren Y, Rocco JW. 2015. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 12:e1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149:979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. 1976. The clonal evolution of tumor cell populations. Science 194:23–28. [DOI] [PubMed] [Google Scholar]
- Ostrow SL, Barshir R, DeGregori J, Yeger-Lotem E, Hershberg R. 2014. Cancer evolution is associated with pervasive positive selection on globally expressed genes. PLoS Genet. 10:e1004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons TL, Quince C. 2007. Fixation in haploid populations exhibiting density dependence I: the non-neutral case. Theor Popul Biol. 72:121–135. [DOI] [PubMed] [Google Scholar]
- Petljak M, Alexandrov LB. 2016. Understanding mutagenesis through delineation of mutational signatures in human cancer. Carcinogenesis 37:531–540. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP. 2015. How the environment shapes cancer genomes. Curr Opin Oncol. 27:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. 2002. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 21:7435–7451. [DOI] [PubMed] [Google Scholar]
- Poon SL, McPherson JR, Tan P, Teh BT, Rozen SG. 2014. Mutation signatures of carcinogen exposure: genome-wide detection and new opportunities for cancer prevention. Genome Med. 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos RC, Wong YT, Ryan R, Pang H, Wong JWH. 2018. Analysis of 7,815 cancer exomes reveals associations between mutational processes and somatic driver mutations. PLoS Genet. 14:e1007779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakodi MP, Kulathinal RJ, Chung Y, Serebriiskii I, Liu JC, Ragin CC. 2016. Ancestral-derived effects on the mutational landscape of laryngeal cancer. Genomics 107:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawla P. 2019. Epidemiology of prostate cancer. World J Oncol. 10:63–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. 2016. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SA, Hartl DL. 1992. Population genetics of polymorphism and divergence. Genetics 132:1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia R, Tam AS, Stirling PC. 2015. Dissecting genetic and environmental mutation signatures with model organisms. Trends Genet. 31:465–474. [DOI] [PubMed] [Google Scholar]
- Shield KD, Maxwell Parkin D, Whiteman DC, Rehm J, Viallon V, Micallef CM, Vineis P, Rushton L, Bray F, Soerjomataram I. 2016. Population attributable and preventable fractions: cancer risk factor surveillance, and cancer policy projection. Curr Epidemiol Rep. 3:201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak M, Lu J. 2016. An evolutionary genetic perspective on cancer biology. Annu Rev Ecol Evol Syst. 47:25–49. [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2020. Cancer statistics, 2020. CA Cancer J Clin. 70:7–30. [DOI] [PubMed] [Google Scholar]
- Smith MT, Guyton KZ, Gibbons CF, Fritz JM, Portier CJ, Rusyn I, DeMarini DM, Caldwell JC, Kavlock RJ, Lambert PF, et al. 2016. Key characteristics of carcinogens as a basis for organizing data on mechanisms of carcinogenesis. Environ Health Perspect. 124:713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarelli JA, Gardner H, Cannataro VL, Gunady EF, Boddy AM, Johnson NA, Fisk JN, Gaffney SG, Chuang JH, Li S, et al. 2020. Molecular biology and evolution of cancer: from discovery to action. Mol Biol Evol. 37:320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. 2009. Human mutation rate associated with DNA replication timing. Nat Genet. 41:393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Bunn P, Jin C, Little P, Zhabotynsky V, Perou CM, Hayes DN, Chen M, Lin D-Y. 2018. The association between copy number aberration, DNA methylation and gene expression in tumor samples. Nucleic Acids Res. 46:3009–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temko D, Tomlinson IPM, Severini S, Schuster-Böckler B, Graham TA. 2018. The effects of mutational processes and selection on driver mutations across cancer types. Nat Commun. 9:1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Li L, Vogelstein B. 2017. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355:1330–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B. 2015. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347:78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend-Lab-Yale . Townsend-Lab-Yale/cancer_causes_and_effects. Available from: https://github.com/Townsend-Lab-Yale/cancer_causes_and_effects
- Van den Eynden J, Larsson E. 2017. Mutational signatures are critical for proper estimation of purifying selection pressures in cancer somatic mutation data when using the dN/dS metric. Front Genet. 8:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley JM, Attwooll C, White G, McGown G, Thorncroft M, Kelsey AM, Greaves M, Boyle J, Birch JM. 2001. Characterization of germline TP53 splicing mutations and their genetic and functional analysis. Oncogene 20:2647–2654. [DOI] [PubMed] [Google Scholar]
- Venkatesan S, Birkbak NJ, Swanton C. 2017. Constraints in cancer evolution. Biochem Soc Trans. 45:1–13. [DOI] [PubMed] [Google Scholar]
- Wang X, Sun Q. 2017. TP53 mutations, expression and interaction networks in human cancers. Oncotarget 8:624–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C, Brennan P, Plummer M, Bray F, Straif K, Zavadil J. 2015. Cancer risk: role of chance overstated. Science 347:728. [DOI] [PubMed] [Google Scholar]
- Wu S, Powers S, Zhu W, Hannun YA. 2016. Substantial contribution of extrinsic risk factors to cancer development. Nature 529:43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Campbell PJ. 2012. Evolution of the cancer genome. Nat Rev Genet. 13:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z-M, Zhao B, Bai Y, Iamarino A, Gaffney SG, Schlessinger J, Lifton RP, Rimm DL, Townsend JP. 2016. Early and multiple origins of metastatic lineages within primary tumors. Proc Natl Acad Sci U S A. 113:2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H-Q, Zhou Z, Huang J, Chaudhury L, Dong J-T, Chen C. 2009. Krüppel-like factor 5 promotes breast cell proliferation partially through upregulating the transcription of fibroblast growth factor binding protein 1. Oncogene 28:3702–3713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Genomic Data Commons repository, https://portal.gdc.cancer.gov/, as an open-access supplement to Cannataro, Gaffney, and Townsend (2018), and all computational scripts used to analyze these data are found in our github repository here: https://github.com/Townsend-Lab-Yale/cancer_causes_and_effects. We used the cancereffectsizeR v2.3.4 package to calculate cancer effect sizes.