Abstract
Arctostaphylos (Ericaceae) species, commonly known as manzanitas, are an invaluable fire-adapted chaparral clade in the California Floristic Province (CFP), a world biodiversity hotspot on the west coast of North America. This diverse woody genus includes many rare and/or endangered taxa, and the genus plays essential ecological roles in native ecosystems. Despite their importance in conservation management, and the many ecological and evolutionary studies that have focused on manzanitas, virtually no research has been conducted on the genomics of any manzanita species. Here, we report the first genome assembly of a manzanita species, the widespread Arctostaphylos glauca. Consistent with the genomics strategy of the California Conservation Genomics project, we used Pacific Biosciences HiFi long reads and Hi-C chromatin-proximity sequencing technology to produce a de novo assembled genome. The assembly comprises a total of 271 scaffolds spanning 547Mb, close to the genome size estimated by flow cytometry. This assembly, with a scaffold N50 of 31Mb and BUSCO complete score of 98.2%, will be used as a reference genome for understanding the genetic diversity and the basis of adaptations of both common and rare and endangered manzanita species.
Keywords: California Conservation Genomics Project, California Floristic Province, CCGP, chaparral, conservation, endemic
In this genome release, we report on the first assembled genome of a member of the genus Arctostaphylos. Our genome assembly is part of the California Conservation Genomics Project (CCGP), the goal of which is to establish patterns of genomic diversity across the state of California and its many habitats. The CCGP will sequence the complete genomes of approximately 150 carefully selected species projects. Many of these taxa are threatened or endangered, and therefore in need of conservation management in the face of rapidly accelerating biodiversity decline. The combined reference genome plus landscape genomics approach of the CCGP, based on the resequencing of many individuals of each target species across the state, will allow the identification of hotspots of diversity across California and provide a framework for informed conservation decisions and management plans.
Manzanitas (Arctostaphylos Adans) are among the most conspicuous and dominant native chaparral species in the California Floristic Province (CFP), a biodiversity hotspot (Howell 1957) characterized by a Mediterranean-type climate with hot, dry summers and cool, wet winters. These plants comprise the most diverse woody genus in the CFP (Minnich and Howard 1984; Baldwin et al. 2012), and their diversity has long fascinated (and perplexed) taxonomists. Manzanitas serve essential roles in their native ecosystems, including rapidly regenerating in fired-disturbed areas, and providing food resources for pollinators and fruit-eating animals (Fulton and Carpenter 1979; Minnich and Howard 1984; Kauffmann et al. 2015). In addition, these plants are of great importance for conservation management: over half of the more than 100 morphologically defined manzanita species and subspecies are narrow endemics with highly restricted distributions and are considered rare and/or endangered (Baldwin et al. 2012; Kauffmann et al. 2015; https://www.rareplants.cnps.org/).
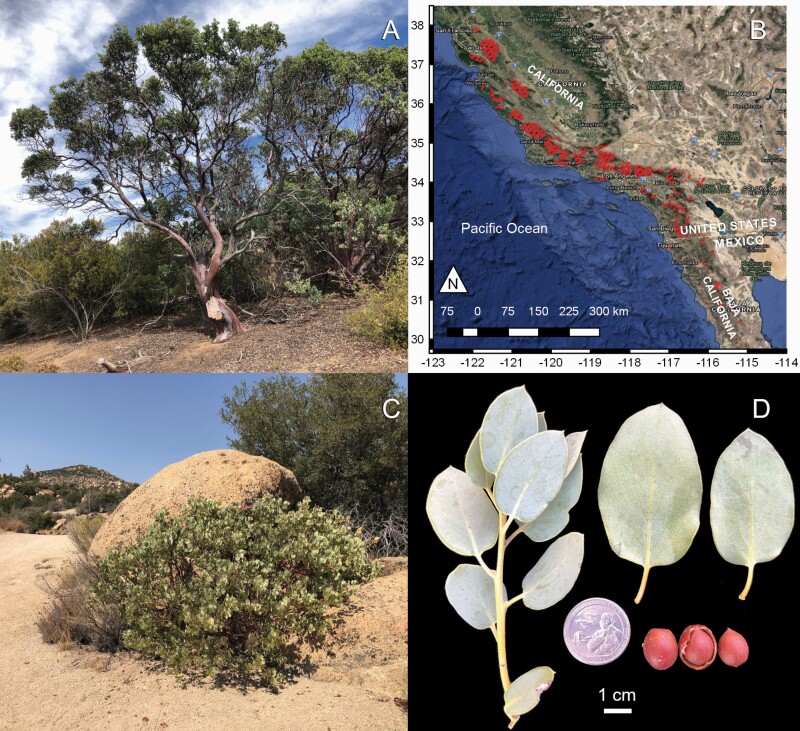
In contrast to their importance in ecology, evolution, and conservation studies, genomic resources for manzanitas are nearly nonexistent beyond investigations into karyotypes of diploid (2n = 2x = 26) and tetraploid (2n = 4x = 48) species (Wells 1968; Kruckeberg 1977; Rosatti 1981; Schierenbeck et al. 1992; Baldwin et al. 2012). In this study, we present the first genome sequence of a manzanita. Big berry manzanita, Arctostaphylos glauca (Figure 1), is a widespread diploid species common in northern Baja California and across southern and coastal central California that is hypothesized to be the progenitor of several potential hybrid manzanita species (Parker 2007). With funding and support from the CCGP, we created this scaffold-level assembly using a hybrid de novo assembly approach that combines Hi-C chromatin-proximity and PacBio HiFi long-read sequencing data. This genome assembly will provide a robust basis for studying the diversification and evolutionary history of Arctostaphylos in the CFP.
Figure 1.
Arctostaphylos glauca, big berry manzanita. (A) A tall, arborescent individual in montane chaparral; (B) range map of A. glauca. The areas colored in red show the approximate range of A. glauca, based on georeferenced herbarium collections in the Consortium of California Herbaria database (CCH2.org). Satellite imagery is from Google Earth. (C) Low shrubby growth form, in a particularly dry habitat; (D) stem, leaves, and fruit. The two detached leaves show the adaxial (left) and abaxial (right) leaf surfaces. Scale bar is 1 cm long, and the coin placed for scale is a US 25 cent piece (quarter), 24.26 mm in diameter. Photograph taken on a uniform background, which was then digitally removed. Materials moved slightly in digital editing, to condense the image, while preserving scale. All photos were taken by, and are property of author GM.
Methods
Biological Materials
We selected for sequencing a naturally occurring big berry manzanita adult (34.20649°N, 117.78815°W) located in the San Gabriel Mountains, Angeles National Forest, Los Angeles County, California. We collected floral buds and inflorescences from this individual on February 19, 2018, and January 20, 2020. When possible, we extracted from floral tissue, which was easier to grind and gave higher yields of DNA than other tissues. Tissues were flash-frozen in liquid nitrogen and stored at −80 °C prior to DNA extraction. A voucher is deposited (UCR ACC. # 292491) at the herbarium of University of California, Riverside (UCR).
Nucleic Acid Library Preparation and Sequencing
Hi-C Library
A Dovetail Hi-C library was prepared in a similar manner as previously described (Lieberman-Aiden et al. 2009). For each library, chromatin was fixed in place with formaldehyde in the nucleus. Extracted, fixed chromatin was digested with DpnII, the 5′ overhangs were filled in with biotinylated nucleotides, and free blunt ends were ligated. After ligation, crosslinks were reversed, and the DNA purified from protein. Purified DNA was treated to remove biotin that was not internal to ligated fragments. The DNA was then sheared to ~350 bp mean fragment size and sequencing libraries were generated using NEBNext Ultra enzymes and Illumina-compatible adapters. Biotin-containing fragments were isolated using streptavidin beads before PCR enrichment of each library. The libraries were prepared and sequenced on an Illumina HiSeq X by Dovetail Genomics (Santa Cruz, CA).
Pacific Biosciences HiFi Library
High molecular weight (HMW) genomic DNA (gDNA) was extracted from a 750 mg sample of young floral buds following the protocol described in Workman et al. (2018) with the minor modification of using the nuclear isolation buffer (NIB) supplemented with 350 mM Sorbitol (NIB-S) to resuspend the ground tissue and during the first wash of the nuclei pellet. The integrity of the HMW DNA was evaluated using the Femto Pulse system (Agilent Technologies, Santa Clara, CA). Purity of the DNA was assessed by 260/280 and 260/230 absorbance ratios on a NanoDrop spectrophotometer.
For PacBio library preparation, 11 ug of HMW gDNA were sheared to an average size distribution of ~16 kb mode using Diagenode’s Megaruptor 3 system (Diagenode, Belgium; cat. B06010001). Sheared DNA was quantified by Quantus Fluorometer QuantiFluor ONE dsDNA Dye assay (Promega, Madison, WI; cat. E6150) and the size distribution was checked by Agilent Femto Pulse (Agilent Technologies, Santa Clara, CA; cat. P-0003-0817). The sheared gDNA was concentrated using 0.45× of AMPure PB beads (Pacific Biosciences - PacBio, Menlo Park, CA; cat. 100-265-900). Concentrated, sheared gDNA was quantified by Quantus Fluorometer QuantiFluor ONE dsDNA Dye assay (Promega, cat E6150). A HiFi library was constructed using the SMRTbell Express Template Prep Kit v2.0 (PacBio, cat. 100-938-900) according to the manufacturer’s instructions. 6 ug of sheared, concentrated DNA was used as input for the removal of single-strand overhangs at 37° for 15 min, followed by further enzymatic steps of DNA damage repair at 37° for 30 minutes, end repair and A-tailing at 20° for 10 min and 65° for 30 min, ligation of overhang adapter v3 at 20° for 1 h and 65° for 10 min to inactivate the ligase, and nuclease treatment of SMRTbell library at 37° for 1 h to remove damaged or non-intact SMRTbell templates (SMRTbell Enzyme Cleanup Kit, PacBio, cat. 107-746-400). The SMRTbell library was purified and concentrated with 1X Ampure PB beads (PacBio, cat. 100-265-900) for size selection using the BluePippin system (Sage Science, Beverly, MA; cat BLU0001). The input of 2.2 ug purified SMRTbell library was used to load into the Blue Pippin 0.75% Agarose Cassette (Sage Science, cat BLF7510) using cassette definition 0.75% DF Marer S1 3–10 kb Improved Recovery for the run protocol. Fragments >7 kb were collected from the cassette elution well. The size-selected SMRTbell library was purified and concentrated with 0.8× AMPure beads (PacBio, cat 100-265-900). The 17 kb average HiFi SMRTbell library was sequenced at UC Davis DNA Technologies Core (Davis, CA) using a single 8M SMRT Cell and Sequel II sequencing chemistry 2.0 on a PacBio Sequel II sequencer.
Genome Assembly
Nuclear Genome Assembly
We assembled the genome of the big berry manzanita following a protocol adapted from Rhie et al. (2021) as part of the CCGP assembly efforts. The CCGP assembly protocol version 1.0 uses PacBio HiFi reads and Hi-C chromatin capture data for the generation of high-quality and highly contiguous nuclear genome assemblies. The output corresponding to a diploid assembly consists of two pseudo haplotypes (primary and alternate). The primary assembly is more complete and consists of longer phased blocks. The alternate consists of haplotigs (contigs of clones with the same haplotype) in heterozygous regions and is not as complete and more fragmented. Given the characteristics of the latter, it cannot be considered on its own but as a complement of the primary assembly (https://lh3.github.io/2021/04/17/concepts-in-phased-assemblies, https://www.ncbi.nlm.nih.gov/grc/help/definitions/).
To generate this assembly, we removed remnant adapter sequences from the PacBio HiFi dataset using HiFiAdapterFilt [Version 1.0] (Sim 2021) and assembled the initial set of contigs with the filtered PacBio reads using HiFiasm [Version 0.13-r308] (Cheng et al. 2021) (see Table 1 for assembly pipeline and relevant software). Next, we identified sequences corresponding to haplotypic duplications and contig overlaps on the primary assembly with purge_dups [Version 1.0.1] (Guan et al. 2020) and transferred them to the alternate assembly. We aligned the Hi-C data to both primary and alternate assemblies using the Arima Genomics Mapping Pipeline (https://github.com/ArimaGenomics/mapping_pipeline) and scaffolded the genomes using SALSA [Version 2, options --e GATCGATC] (Ghurye et al. 2017; Ghurye et al. 2019). We closed the generated gaps in both assemblies using the PacBio HiFi reads and YAGCloser [commit 20e2769] (https://github.com/merlyescalona/yagcloser). The primary assembly was manually curated by iteratively generating and analyzing Hi-C contact maps. To generate the contact maps, we aligned the Hi-C data against the corresponding reference with bwa mem [Version 0.7.17-r1188, options -5SP] (Li 2013), identified ligation junctions, and generated Hi-C pairs using pairtools [Version 0.3.0] (Goloborodko et al. 2018). We generated a multi-resolution Hi-C matrix in binary form with cooler [Version 0.8.10] (Abdennur and Mirny 2020) and balanced it with hicExplorer [Version 3.6] (Ramírez et al. 2018). We used HiGlass [Version 2.1.11] (Kerpedjiev et al. 2018) and the PretextSuite (https://github.com/wtsi-hpag/PretextView; https://github.com/wtsihpag/PretextMap; https://github.com/wtsi-hpag/PretextSnapshot) to visualize the contact maps. Assemblies were then checked for contamination using the BlobToolKit Framework [Version 2.3.3] (Challis et al. 2020), and trimmed for remnants of sequence adaptors and mitochondrial contamination.
Table 1.
Assembly pipeline and software usage. Software citations are listed in the text
| Assembly | Software | Version |
|---|---|---|
| Filtering PacBio HiFi adapters | HiFiAdapterFilt https://github.com/sheinasim/HiFiAdapterFilt |
Commit 64d1c7b |
| K-mer counting | Meryl | 1 |
| Estimation of genome size and heterozygosity | GenomeScope | 2 |
| De novo assembly (contiging) | HiFiasm | 0.13-r308 |
| Long read, genome-genome alignment | minimap2 | 2.16 |
| Remove low-coverage, duplicated contigs | purge_dups | 1.0.1 |
| Scaffolding | ||
| Hi-C mapping for SALSA | Arima Genomics mapping pipeline https://github.com/ArimaGenomics/mapping_pipeline |
Commit 2e74ea4 |
| Hi-C Scaffolding | SALSA | 2 |
| Gap closing | YAGCloser https://github.com/merlyescalona/yagcloser |
Commit 20e2769 |
| Hi-C contact map generation | ||
| Short-read alignment | bwa | 0.7.17-r1188 |
| SAM/BAM processing | samtools | 1.11 |
| SAM/BAM filtering | pairtools | 0.3.0 |
| Pairs indexing | pairix | 0.3.7 |
| Matrix generation | Cooler | 0.8.10 |
| Matrix balancing | hicExplorer | 3.6 |
| Contact map visualization | HiGlass | 2.1.11 |
| PretextMap | 0.1.4 | |
| PretextView | 0.1.5 | |
| PretextSnapshot | 0.0.3 | |
| Organelle assembly | ||
| Sequence similarity search | BLAST+ | 2.10 |
| Long read alignment | Pbmm2 (https://github.com/PacificBiosciences/pbmm2) | 1.4.0 |
| Variant calling and consensus | bcftools | 1.11-5-g9c15769 |
| Extraction of sequences | seqtk | 1.3-r115-dirty |
| Circular-aware long-read alignment | racon | 1.4.19 |
| Sequence polishing | raptor | 0.20.3-171e0f1 |
| Sequence alignment | lastz | 1.04.08 |
| Gene annotation | MitoFinder | 1.4 |
| Genome quality assessment | ||
| Basic assembly metrics | QUAST | 5.0.2 |
| Assembly completeness | BUSCO | 5.0.0 |
| Merqury | 1 | |
| Contamination screening | ||
| General contamination screening | BlobToolKit | 2.3.3 |
Mitochondrial Genome Assembly
We identified a subset of mitochondrial reads from the PacBio HiFi dataset using BLAST+ [Version 2.10] (Camacho et al. 2009) by identifying regions of similarity between the reads and the mitochondrial (mito) database (National Library of Medicine (US), National Center for Biotechnology Information 2004). These mitochondrial reads were used as input in HiFiasm [Version 0.13-r308] to generate the mitochondrial assembly. Given the circularity of the mitochondrial genome, we carried out self-alignment of the sequence using lastz [Version 1.04.08] (Harris 2007) to manually identify and remove duplicated regions. We aligned the subset of mitochondrial reads to the assembly using raptor [Version 0.20.3-171e0f1] (https://github.com/isovic/raptor) and polished it with racon [Version 1.14.] (https://github.com/isovic/racon). We searched for matches of the resulting mitochondrial assembly sequence in the nuclear genome assembly using BLAST+ and filtered out scaffolds from the nuclear genome with a percentage of sequence identity >99% and size smaller than the mitochondrial assembly sequence. From the subset of mitochondrial reads used for the assembly, we analyzed the BLAST output and the species of the closest mitochondrial sequence available in the NCBI GenBank database, Vaccinium macrocarpon (Accession number: NC_023338.1). We used the mitochondrial assembly of V. macrocarpon as a guide for the mitochondrial gene annotation generated with MitoFinder [Version 1.4] (Allio et al. 2020).
Chloroplast Genome Assembly
We identified chloroplast reads from the PacBio HiFi dataset with BLAST+ using the plastids RefSeq genomes [v4.1] (O’Leary et al. 2016). From this subset, we analyzed the matches and identified the species of the closest chloroplast sequence available in the NCBI database as Camellia taliensis (NC_022264.1). Next, we found matches of the C. taliensis chloroplast genome sequence in the nuclear genome assembly with BLAST+ and filtered out scaffolds from the nuclear genome assembly with length smaller than the C. taliensis length, sequence identity >90%, and e-value <0.00001. We aligned the filtered scaffolds to the C. taliensis chloroplast genome with minimap2 (Li 2018) and generated a consensus sequence with bcftools (Li 2011). We manually curated the sequence using lastz. Finally, we polished the last assembly version using raptor and racon and annotated it using the web platform GeSeq (Tillich et al. 2017).
Genome Size Estimation, Quality Assessment and Repeat Element Identification
We generated k-mer counts (k = 21) from the PacBio HiFi reads using meryl [Version 1] (https://github.com/marbl/meryl). The generated k-mer database was then used in GenomeScope2.0 [Version 2.0] (Ranallo-Benavidez et al. 2020) to estimate genome features including genome size, heterozygosity, and repeat content. To obtain general contiguity metrics, we ran QUAST [Version 5.0.2] (Gurevich et al. 2013). To evaluate genome quality and completeness we used BUSCO [Version 5.0.0] (Simão et al. 2015; Seppey et al. 2019) with the embryophyta ortholog database (embryophyta_odb10) which contains 1614 genes. Assessment of base level accuracy (QV) and k-mer completeness was performed using the previously generated meryl database and merqury (Rhie et al. 2020). We further estimated genome assembly accuracy via BUSCO gene set frameshift analysis using the pipeline described in (Korlach et al. 2017). In addition, we used RepeatModeler (v1.0.11) and RepeatMasker (v4-0-7) to identify and classify repetitive elements in the assembled genome (Smit and Hubley 2008; Smit et al. 2015).
Results
We generated a de novo nuclear genome assembly of the big berry manzanita (ddArcGlau1) using 199 million read pairs of Hi-C data and 1.8 million PacBio HiFi reads. The latter yielded ~45-fold coverage (N50 read length 15 246 bp; minimum read length 46 bp; mean read length 14 552 bp; maximum read length of 54 291 bp). Calculation of coverage is based on a flow-cytometry estimated genome size of ~600 Mb reported in a previous study of Arctostaphylosuva-ursi (Siljak-Yakovlev et al. 2010). Assembly statistics are reported in tabular and graphical form in Table 2 and Figure 2, respectively.
Table 2.
Sequencing and assembly statistics, and accession numbers
| Bio projects & vouchers |
CCGP NCBI BioProject | PRJNA720569 | ||||
| Genera NCBI BioProject | PRJNA721387 | |||||
| Species NCBI BioProject | PRJNA734616 | |||||
| NCBI BioSample | SAMN19489519 | |||||
| Specimen identification | UCR ACC. # 292491 | |||||
| NCBI Genome accessions | Primary | Alternate | ||||
| Assembly accession | GCA_019985065.1 | GCA_019985075.1 | ||||
| Genome sequences | JAHSPW000000000 | JAHSPX000000000 | ||||
| Genome sequence | PacBio HiFi reads | Run | 1 PACBIO_SMRT (Sequel II) run: 1.8 M spots, 27.3G bases, 8.7Gb downloads | |||
| Accession | SRR14883332 | |||||
| Hi-C Illumina reads | Run | 1 Illumina HiSeq X Ten run: 199.2M spots, 59.7G bases, 37Gb download |
||||
| Accession | SRR14883331 | |||||
| Genome assembly quality metrics | Assembly identifier (Quality codea) | ddArcGlau1 (6.7.Q62) | ||||
| HiFi Read coverageb | 45X | |||||
| Primary | Alternate | |||||
| Number of contigs | 353 | 2470 | ||||
| Contig N50 (bp) | 8 041 760 | 1 739 008 | ||||
| Longest Contigs | 22 990 225 | 10 884 557 | ||||
| Number of scaffolds | 271 | 2350 | ||||
| Scaffold N50 (bp) | 31 280 158 | 3 804 428 | ||||
| Largest scaffold | 45 401 621 | 22 987 546 | ||||
| Size of final assembly (bp) | 547 548 103 | 556 397 040 | ||||
| Gaps per Gbp | 150 | 1885 | ||||
| Indel QV (Frame shift) | 48.36 | 47.39 | ||||
| Base pair QV | 62.36 | 56.24 | ||||
| Full assembly = 58.28 | ||||||
| k-mer completeness | 74.39 | 65.01 | ||||
| Full assembly = 95.59 | ||||||
| BUSCO completeness | C | S | D | F | M | |
|
(embryophyta) n = 1614 |
98.20% | 95.70% | 2.50% | 0.90% | 0.90% | |
| 85.90% | 83.30% | 2.60% | 1.30% | 12.80% | ||
| Organelles | 1 Partial mitochondrial sequence 1 Partial chloroplast sequence |
MZ779111 XXXXXX |
a Assembly quality code x.y.Q derived notation, from (Rhie et al. 2021). x = log10[contig NG50]; y = log10[scaffold NG50]; Q = Phred base accuracy QV (Quality value). BUSCO Scores. (C)omplete and (S)ingle; (C)omplete and (D)uplicated; (F)ragmented and (M)issing BUSCO genes. n, number of BUSCO genes in the set/data base. Bp: base pairs.
b Read coverage has been calculated based on a genome size of 600Mb.
Figure 2.
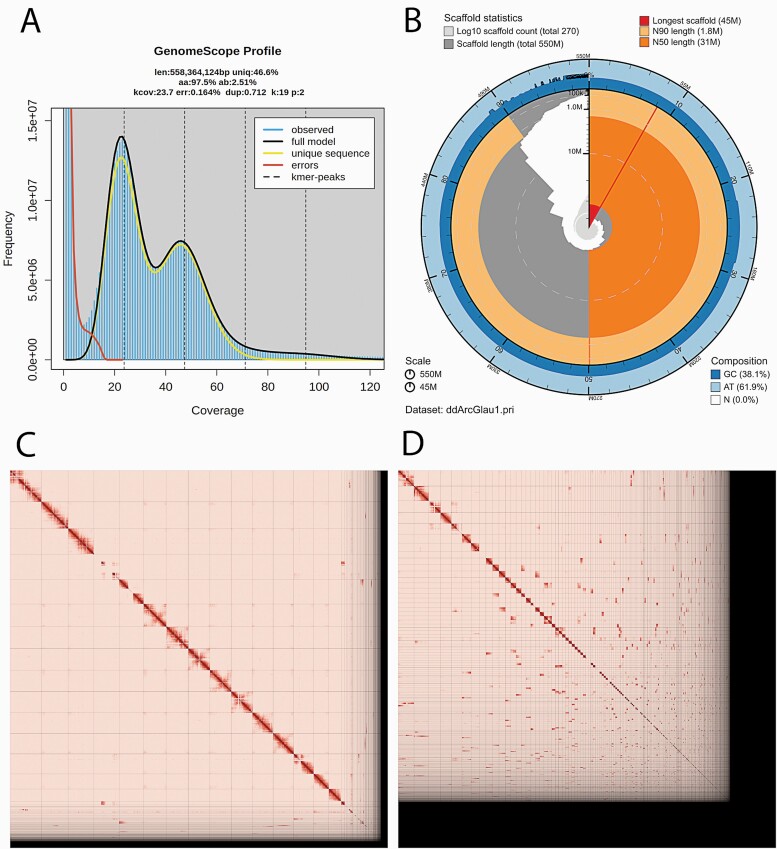
Visual overview of genome assembly metrics. (A) K-mer spectra output generated from PacBio HiFi data without adapters using GenomeScope2.0. The bimodal pattern observed corresponds to a diploid genome. K-mers covered at lower coverage but higher frequency correspond to differences between haplotypes, whereas the higher coverage but lower frequency k-mers correspond to the similarities between haplotypes; (B) BlobToolKit Snail plot showing a graphical representation of the quality metrics presented in Table 2 for the A. glauca primary assembly (ddArcGlau1). The plot circle represents the full size of the assembly. From the inside-out, the central plot covers length-related metrics. The red line represents the size of the longest scaffold; all other scaffolds are arranged in size-order moving clockwise around the plot and drawn in grey starting from the outside of the central plot. Dark and light orange arcs show the scaffold N50 and scaffold N90 values. The central light grey spiral shows the cumulative scaffold count with a white line at each order of magnitude. White regions in this area reflect the proportion of Ns in the assembly The dark vs. light blue area around it shows mean, maximum and minimum GC vs. AT content at 0.1% intervals (Challis et al. 2020); (C, D) Hi-C contact maps for the primary (C) and alternate (D) genome assembly generated with PretextSnapshot. Hi-C contact maps translate proximity of genomic regions in 3D space to contiguous linear organization. Each cell in the contact map corresponds to sequencing data supporting the linkage (or join) between two of such regions. Scaffolds are separated by black lines and higher density corresponds to higher levels of fragmentation..
The primary assembly consists of 271 scaffolds spanning 547Mb with contig N50 of 8Mb, scaffold N50 of 31Mb, largest contig of 22Mb, and largest scaffold of 44Mb. The Hi-C contact map suggests that the primary assembly is highly contiguous (Figure 2C). As expected, the alternate assembly, which consists of sequence from heterozygous regions, is less contiguous (Figure 2D). Because the primary assembly is not fully phased, we have deposited scaffolds corresponding to the alternate haplotype in addition to the primary assembly.
The final genome size (547 Mb) is close to the estimated values from the Genomescope2.0 k-mer spectra (estimated size of 558Mb, range 556.1–558.3Mb). The k-mer spectrum output shows a bimodal distribution with two major peaks, at ~24- and ~47-fold coverage, where peaks correspond to homozygous and heterozygous states respectively. This pattern corresponds to a diploid genome. Based on PacBio HiFi reads, we estimated a 0.164% sequencing error rate and 2.51% nucleotide heterozygosity rate. The assembly has a BUSCO completeness score of 98.2% using the embryophyta gene set, and a per base quality (QV) of 66. RepeatModeler indicates that the genome includes 57.71% repetitive elements. The classification of repeat elements generated by RepeatMasker is shown in Table 3.
Table 3.
Classification of repeat elements generated from RepeatMasker
| Number of elementsa | Length occupied (bp) | Percentage of sequence (%) | |
|---|---|---|---|
| Retroelements | 160 425 | 112 810 688 | 20.6 |
| SINEs | 17 580 | 3 242 835 | 0.59 |
| Penelope | 1155 | 97 740 | 0.02 |
| LINEs | 20 303 | 6 704 697 | 1.22 |
| CRE/SLACS | 37 | 1672 | 0 |
| L2/CR1/Rex | 4680 | 678 218 | 0.12 |
| R1/LOA/Jockey | 340 | 22 011 | 0 |
| R2/R4/NeSL | 70 | 3992 | 0 |
| RTE/Bov-B | 3670 | 1 765 787 | 0.32 |
| L1/CIN4 | 9180 | 3 982 099 | 0.73 |
| LTR elements | 122 542 | 102 863 156 | 18.79 |
| BEL/Pao | 622 | 98 414 | 0.02 |
| Ty1/Copia | 49 026 | 41 614 407 | 7.6 |
| Gypsy/DIRS1 | 62 124 | 58 039 401 | 10.6 |
| Retroviral | 3184 | 346 807 | 0.06 |
| DNA transposons | 288 141 | 58 683 503 | 10.72 |
| hobo-Activator | 43 650 | 10 654 515 | 1.95 |
| Tc1-IS630-Pogo | 4515 | 615 692 | 0.11 |
| En-Spm | 0 | 0 | 0 |
| MuDR-IS905 | 54 | 53 928 | 0.01 |
| PiggyBac | 121 | 6007 | 0 |
| Tourist/harbinger | 4780 | 1 700 494 | 0.31 |
| Other (mirage, P-element, P-element, Transib) | 695 | 43 698 | 0.01 |
| Rolling-circles | 4313 | 1 802 302 | 0.33 |
| Unclassified | 373 015 | 120 296 966 | 21.97 |
| Total interspersed repeats | 291 791 157 | 53.29 | |
| Small RNA | 19 997 | 17 205 637 | 3.14 |
| Satellites | 2056 | 628 418 | 0.11 |
| Simple repeats | 121 138 | 6 848 295 | 1.25 |
| Low complexity | 17 346 | 833 121 | 0.15 |
a Most repeats fragmented by insertions or deletions have been counted as one element.
Mitochondrial Assembly
We generated an initial mitochondrial genome assembly with HiFiasm using a subset of HiFi reads that matched to publicly available mitochondrial reference genomes. Following an initial gene annotation with MitoFinder, we manually curated the assembly and introduced 13 gaps (N sequence of 100 bp) to solve partial annotation of 12 genes and re-annotated the assembly. Final mitochondrial genome size was 592 049 bp, about average for a plant mitochondrial genome. The base composition of the final assembly version is A = 27.16%, C = 22.78%, G = 22.84%, T = 26.99%, and consists of 23 transfer RNAs and 33 protein coding genes (12 of them partial). In comparison to mitochondrial genomes from other members of the Ericaceae, this assembly is similar to that of Vaccinium macrocarpon at 459 678 bp (Fajardo et al. 2014) but considerably smaller than that of the saprophytic Hypopitys monotropa (810 116 bp; Shtratnikova et al. 2020; also known as Monotropa hypopitys) or Rhododendron simsii (802 707 bp; Yang et al. 2020).
Chloroplast Assembly
The preliminary chloroplast assembly size was 118 663 bp, which is at the beginning of the average range for a chloroplast genome (110–200 kbp; De Las Rivas et al. 2002). The base composition of the assembly is A = 31.61%, C = 18.69%, G = 18.03%, T = 31.65%, and it consists of 32 transfer RNA genes, 5 ribosomal RNA genes, and 115 protein coding genes. The chloroplast of big berry manzanita is smaller than that of R. pulchrum (136 249 bp; Shen et al. 2019), R. simsii (152 214 bp; Yang et al. 2020), V. macrocarpon, and four additional Vaccinium species (173 245 bp ~176 045 bp; Kim et al. 2020). Not surprisingly, the big berry manzanita chloroplast assembly is an order of magnitude larger than the highly reduced chloroplast genome of the saprophytic and non-photosynthesizing H. monotropa (35 336 bp; Gruzdev et al. 2016).
Discussion
Our assembly is the first sequenced genome in the genus Arctostaphylos, representing the initial step toward understanding genetics and adaptation in this highly diversified genus. In addition to enhancing ongoing phylogenetic and conservation research, this assembly will enable investigations of the genetic basis of adaptations including drought tolerance and fire resilience. These traits, which are ecological hallmarks of manzanitas, are of growing importance in the context of the increased drought and fire frequency and intensity that are occurring in California as a result of climate change. This assembly can also serve as a reference for studying the diversification and population genetics of Arctostaphylos, which may shed light on aspects of diversification in other complex groups of the CFP.
Arctostaphylos is the third genus with a genome assembly in the heath family, Ericaceae, following release of assembled genome sequences of Rhododendron (two assemblies) and Vaccinium (one assembly) (Colle et al. 2019; Soza et al. 2019; Yang et al. 2020). These two genera are of significant economic importance: many species, hybrids, and cultivars of Rhododendron, including rhododendron and azalea, are important landscape and ornamental plants, and the fruits of many Vaccinium species, which include cranberry, blueberry, and huckleberry, are consumed by humans and other animals. Species in the Ericaceae are also notable for their ability to tolerate acidic and nutrient-poor soils that often characterize boreal forests and bogs, allowing them to thrive in habitats that are inaccessible to most species. Their tolerance for these conditions is due in part to the formation of mutualistic associations between the roots of the plants and soil fungi of a type unique to the heath family known as ericoid mycorrhizae. Ericoid mycorrhizae are distinct from common mycorrhizal associations found in most angiosperms, and are far less well understood (Watkinson 2016). Complete genome sequences from three genera in this family will provide a strong foundation for investigating the basis of this unique mutualism and its ability to promote survival in inhospitable soils.
The size of the A. glauca assembly is 547Mb, which is similar to the two Rhododendron genomes and half that of V corymbosum, which is tetraploid (Supplementary Table 1). The tetraploid nature of V. corymbosum also explains the vastly greater number of duplicated genes in its assembly compared to the two diploid assemblies. The scaffold N50 of the A. glauca assembly is longer than R. williamsianum, and close to R. simsii and V. corymbosum, suggesting that the contiguity of Arctostaphylos is comparable to the other taxa (Supplementary Table 1). Analysis using RepeatModeler indicated that 57.71% of the A. glauca genome is composed of different categories of repetitive elements (Table 3). In contrast, analysis using RepeatModeler identified only 26%, 47.5%, and 44.3% of the genome comprising repeat elements in R. williamsianum, R. simsii, and V. corymbosum respectively. The BUSCO completeness assessment of the A. glauca assembly (98%) is higher than R. williamsianum (89%) and close to the V. corymbosum (97%), indicating that our final assembly is high quality (Supplementary Table 1). Overall, the A. glauca, R. simsii and V. corymbosum genomes are of comparably high contiguity and completeness. The lower contiguity and completeness of the R. williamsianum genome may be due to the lack of HiFi or other long-read data in the assembly. This explanation is consistent with other studies demonstrating improved assembly with the inclusion of longer reads (Bradnam et al. 2013; Utturkar et al. 2014).
Although the big berry manzanita is a common and widespread species, nearly half of the 60+ manzanita species are rare or threatened. Many are now represented by only one or two populations, and are thus vulnerable to complete eradication by the increasingly common and intense wildfires experienced across California each year. Our manzanita genome sequence will help fulfill the overall goal of the CCGP, serving as a key resource to assess genetic diversity in these threatened endemics and move forward with coordinated conservation programs.
Supplementary Material
Acknowledgments
The isolation of HMW DNA, the HiFi sequencing library preparation, and the PacBio long-read sequencing was carried out at the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786-01. Partial support was provided by Dovetail Genomics for Hi-C sequencing. We are grateful to the staff at UC Davis genome center.
Funding
This work was supported by the California Conservation Genomics Project, with funding provided to the University of California by the State of California, State Budget Act of 2019 (UC Award ID RSI-19-690224), and the University of California, Riverside.
Data Availability
Data generated for this study are available under NCBI BioProject PRJNA720447. Raw sequencing data for sample voucher UCR ACC. # 292491 (NCBI BioSample SAMN19489519) are deposited in the NCBI Short Read Archive (SRA) under SRR14883331 for PacBio HiFi data and SRR14883332 for Hi-C Illumina short-read data. GenBank accessions for both primary and alternate assemblies are GCA_019985065.1 and GCA_019985075.1; and for genome sequences JAHSPW000000000 and JAHSPX000000000. The GenBank organelle genome assembly accession MZ779111 is for the mitochondrial genome. GitHub repository with assembly scripts is available on https://github.com/ccgproject/ccgp_assembly.
References
- Abdennur N, Mirny LA. 2020. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 36:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F. 2020. MitoFinder: Efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 20:892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin BG, Goldman D, Keil DJ, Patterson R, Rosatti TJ, Wilken D editors. 2012. The Jepson Manual: Vascular Plants of California. Second Edition, Thoroughly Revised and Expanded edition. University of California Press [Google Scholar]
- Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, et al. 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, Cochrane G, Blaxter M. 2020. BlobToolKit--Interactive quality assessment of genome assemblies. G3: Genes Genomes Genetics. 10:1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 18:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colle M, Leisner CP, Wai CM, Ou S, Bird KA, Wang J, Wisecaver JH, Yocca AE, Alger EI, Tang H, et al. 2019. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience. 8(3):giz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Rivas J, Lozano JJ, Ortiz AR. 2002. Comparative analysis of chloroplast genomes: functional annotation, genome-based phylogeny, and deduced evolutionary patterns. Genome Res. 12:567–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo D, Schlautman B, Steffan S, Polashock J, Vorsa N, Zalapa J. 2014. The American cranberry mitochondrial genome reveals the presence of selenocysteine (tRNA-Sec and SECIS) insertion machinery in land plants. Gene. 536:336–343. [DOI] [PubMed] [Google Scholar]
- Fulton RE, Lynn Carpenter F. 1979. Pollination, reproduction, and fire in California Arctostaphylos. Oecologia. 38:147–157. [DOI] [PubMed] [Google Scholar]
- Ghurye J, Pop M, Koren S, Bickhart D, Chin CS. 2017. Scaffolding of long read assemblies using long range contact information. BMC Genomics. 18:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, Schmitt A, Selvaraj S, Pop M, Phillippy AM, Koren S. 2019. Integrating Hi-C links with assembly graphs for chromosome-scale assembly. PLoS Comput Biol. 15:e1007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Abdennur N, Venev S, hbbrandao, gfudenberg. 2018. mirnylab/pairtools: v0.2.0. Available from: https://zenodo.org/record/1490831
- Gruzdev EV, Mardanov AV, Beletsky AV, Kochieva EZ, Ravin NV, Skryabin KG. 2016. The complete chloroplast genome of parasitic flowering plant Monotropa hypopitys: extensive gene losses and size reduction. Mitochondrial DNA B Resour. 1:212–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, Howe K, Wang Y, Durbin R. 2020. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 36:2896–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 29:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved pairwise alignment of genomic DNA. Ph.D, Thesis The Pennsylvania State University.http://www.bx.psu.edu/~rsharris/rsharris_phd_thesis_2007.pdf [Google Scholar]
- Howell JT. 1957. The California flora and its province. Leaf. West. Bot. 8:133–38 [Google Scholar]
- Kauffmann ME, Parker T, Vasey M, Bisbee J. 2015. Field Guide to Manzanitas. 1st ed. Kneeland, CA: Backcountry Press. [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, McCallum C, Dinkla K, Strobelt H, Luber JM, Ouellette SB, Azhir A, Kumar N, et al. 2018. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 19:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shin J, Oh D-R, Kim A-Y, Choi C. 2020. Comparative analysis of complete chloroplast genome sequences and insertion-deletion (Indel) polymorphisms to distinguish five vaccinium species. For Trees Livelihoods. 11:927. [Google Scholar]
- Korlach J, Gedman G, Kingan SB, Chin CS, Howard JT, Audet JN, Cantin L, Jarvis ED. 2017. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. Gigascience. 6:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruckeberg AR. 1977. Manzanita (Arctostaphylos) hybrids in the Pacific Northwest: effects of human and natural disturbance. Syst. Bot. 2:233–250. [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-bio.GN]http://arxiv.org/abs/1303.3997
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnich R, Howard L. 1984. Biogeography and prehistory of Shrublands. In: Shrublands in California: Literature Review and Research Needed for Management. Devries Johannes J., editor. Oakland, CA: California Water Resources Center, University of California Contribution No. 191. [Google Scholar]
- National Library of Medicine (US), National Center for Biotechnology Information. 2004. Mito. In: BLAST Databases [Internet]. Bethesda (MD): The National Center for Biotechnology Information (NCBI). https://ftp.ncbi.nlm.nih.gov/blast/db/mito.tar.gz. [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker VT. 2007. Diversity and evolution of arctostaphylos and ceanothus. Fremontia. 35(4):8–11. [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 9(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC. 2020. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 11:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 592:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM. 2020. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosatti TJ. 1981. A new chromosome number in Arctostaphylos uva-ursi. Can. J. Bot. 59:272–273. [Google Scholar]
- Schierenbeck KA, Stebbins GL, Patterson RW. 1992. Morphological and cytological evidence for polyphyletic allopolyploidy inArctostaphylos mewukka (Ericaceae). Osterr. bot. Z. 179:187–205. [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: Assessing genome assembly and annotation completeness. Methods Mol Biol. 1962:227–245. [DOI] [PubMed] [Google Scholar]
- Shen JS, Li XQ, Zhu XT, Huang XL, Jin SH. 2019. Complete chloroplast genome of Rhododendron pulchrum, an ornamental medicinal and food tree. Mitochondrial DNA B Resour. 4:3527–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtratnikova VY, Schelkunov MI, Penin AA, Logacheva MD. 2020. Mitochondrial genome of the nonphotosynthetic mycoheterotrophic plant Hypopitys monotropa, its structure, gene expression and RNA editing. PeerJ. 8:e9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siljak-Yakovlev S, Pustahija F, Oli EM, Boguni F, Muratovi E, Bai N, Catrice O, Brown SC. 2010. Towards a genome size and chromosome number database of Balkan Flora: C-values in 343 taxa with novel values for 242. Adv Sci Lett. 3:190–213. [Google Scholar]
- Sim S. 2021. sheinasim/HiFiAdapterFilt: First release. https://zenodo.org/record/4716418
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R. 2008-2015. RepeatModeler Open-1.0. http://www.repeatmasker.org.
- Smit AFA, Hubley R, Green P. 2013--2015. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Soza VL, Lindsley D, Waalkes A, Ramage E, Patwardhan RP, Burton JN, Adey A, Kumar A, Qiu R, Shendure J, et al. 2019. The rhododendron genome and chromosomal organization provide insight into shared whole-genome duplications across the heath family (Ericaceae). Genome Biol Evol. 11:3353–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utturkar SM, Klingeman DM, Land ML, Schadt CW, Doktycz MJ, Pelletier DA, Brown SD. 2014. Evaluation and validation of de novo and hybrid assembly techniques to derive high-quality genome sequences. Bioinformatics. 30:2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson SC. 2016. Chapter 7 - Mutualistic symbiosis between fungi and autotrophs. In: Watkinson SC, Boddy L, Money NP, editors. The Fungi (Third Edition). Boston: Academic Press. p. 205–243. [Google Scholar]
- Wells PV. 1968. New taxa, combinations, and chromosome numbers in Arctostaphylos (Ericaceae). Madroño 19:193–210. [Google Scholar]
- Workman R, Timp W, Fedak R, Duncan K, Hao S, Liu K. 2018. High molecular weight DNA extraction from recalcitrant plant species for third generation sequencing. protocolexchangehttps://protocolexchange.researchsquare.com/article/nprot-6785/v1
- Yang FS, Nie S, Liu H, Shi TL, Tian XC, Zhou SS, Bao YT, Jia KH, Guo JF, Zhao W, et al. 2020. Chromosome-level genome assembly of a parent species of widely cultivated azaleas. Nat Commun. 11:5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated for this study are available under NCBI BioProject PRJNA720447. Raw sequencing data for sample voucher UCR ACC. # 292491 (NCBI BioSample SAMN19489519) are deposited in the NCBI Short Read Archive (SRA) under SRR14883331 for PacBio HiFi data and SRR14883332 for Hi-C Illumina short-read data. GenBank accessions for both primary and alternate assemblies are GCA_019985065.1 and GCA_019985075.1; and for genome sequences JAHSPW000000000 and JAHSPX000000000. The GenBank organelle genome assembly accession MZ779111 is for the mitochondrial genome. GitHub repository with assembly scripts is available on https://github.com/ccgproject/ccgp_assembly.