Abstract
Chromatin remodelers play a fundamental role in the assembly of chromatin, regulation of transcription, and DNA repair. Biochemical and functional characterizations of the CHD family of chromatin remodelers from a variety of model organisms have shown that these remodelers participate in a wide range of activities. However, because the evolutionary history of CHD homologs is unclear, it is difficult to predict which of these activities are broadly conserved and which have evolved more recently in individual eukaryotic lineages. Here, we performed a comprehensive phylogenetic analysis of 8,042 CHD homologs from 1,894 species to create a model for the evolution of this family across eukaryotes with a particular focus on the timing of duplications that gave rise to the diverse copies observed in plants, animals, and fungi. Our analysis confirms that the three major subfamilies of CHD remodelers originated in the eukaryotic last common ancestor, and subsequent losses occurred independently in different lineages. Improved taxon sampling identified several subfamilies of CHD remodelers in plants that were absent or highly divergent in the model plant Arabidopsis thaliana. Whereas the timing of CHD subfamily expansions in vertebrates corresponds to whole genome duplication events, the mechanisms underlying CHD diversification in land plants appear more complicated. Analysis of protein domains reveals that CHD remodeler diversification has been accompanied by distinct transitions in domain architecture, contributing to the functional differences observed between these remodelers. This study demonstrates the importance of proper taxon sampling when studying ancient evolutionary events to prevent misinterpretation of subsequent lineage-specific changes and provides an evolutionary framework for functional and comparative analysis of this critical chromatin remodeler family across eukaryotes.
Keywords: gene duplication, gene loss, whole genome duplication, subfunctionalization, protein domain prediction, evolutionary innovation
Significance.
Members of the CHD family of SNF2 chromatin remodelers are involved in DNA replication and in an array of transcription regulatory and epigenetic processes associated with development. Previous studies have focused on characterization in model organisms, and the conservation of homologs and their molecular functions across the tree of life remains unclear. This study reveals that the three CHD subfamilies are present in most eukaryotic lineages, but CHD evolution is highly dynamic with many lineage-specific gain and loss events, domain diversification, and structural variants that suggest that these remodelers have evolved to fulfill distinct chromatin-based roles. These findings provide the most comprehensive phylogenetic and evolutionary analysis of CHD homologs across Eukarya, expanding our understanding of the malleability of this ancient family of remodelers and reveal the existence of novel forms and thus perhaps unknown chromatin-associated activities in nonmodel organisms.
Introduction
Chromatin packaging is the complex arrangement of DNA and proteins to form nucleosomes and other higher order chromosome structure. It is one of the hallmarks of eukaryotic genomes. Complex packaging comes with a cost, as the compact structure of chromatin can prevent access of proteins involved in transcription, replication, and repair. Various chromatin remodelers are involved in the dynamic regulation of chromatin packaging and are therefore essential for organismal development (Clapier and Cairns 2009; Ho and Crabtree 2010; Ojolo et al. 2018).
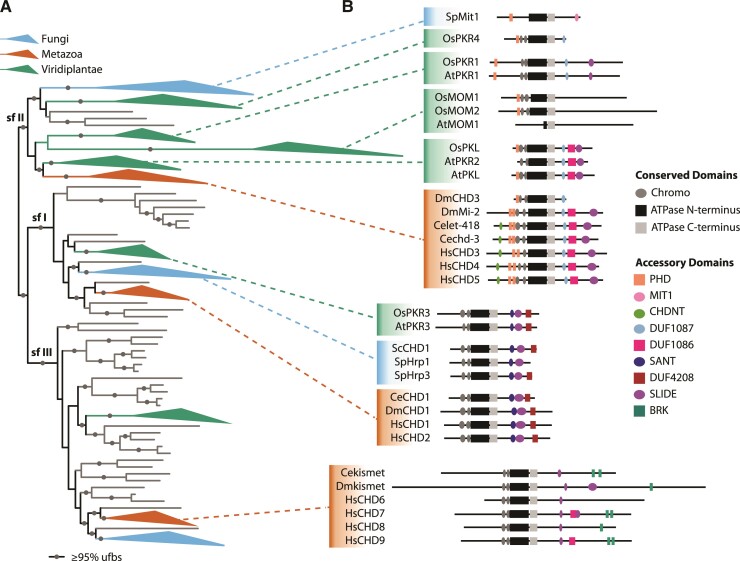
One important family of remodelers are the CHD proteins, which play an essential role in chromatin homeostasis and exhibit a diverse range of biochemical activities with nucleosomes (Marfella and Imbalzano 2007; Sims and Wade 2011). Like other ATP-dependent chromatin remodelers, CHDs contain a conserved ATPase domain, composed of SNF2_N and Helicase_C PFAM domains, that acts as a motor to power dynamic interactions with chromatin and nucleosome substrates (Clapier et al. 2017; Nodelman and Bowman 2021). The acronym of “CHD” is derived from the domains typically found in these proteins (Woodage et al. 1997): two tandemly arranged chromo domains; the ATPase domain (originally annotated as a helicase), and one or more domains associated with DNA-binding (fig. 1).
Fig. 1.
Distribution of CHD gene family across eukaryotes and model domain architecture. (A) Maximum-likelihood phylogeny of CHD homologs. Branches corresponding to subfamily (sf) I, II, and III are indicated. Grey circles indicate branches with ultrafast bootstrap support ≥ 0.95. Clades of animal (red), plant (green), or fungi (blue) are collapsed. (B) PFAM domain architecture of CHD homologs from model eukaryotes. Width of ovals and rectangles are proportional to the width of the protein domain.
CHD remodelers are typically organized into three subfamilies that possess distinct domain architectures (Flaus et al. 2006; Ho et al. 2013; Koster et al. 2015). Subfamily I is characterized by the presence of C-terminal SANT and SLIDE DNA-binding domains (Ryan et al. 2011; Sharma et al. 2011). In contrast, subfamily II CHDs typically contain 1–2 N-terminal plant homeodomains (PHDs), that have been shown to exhibit histone-binding activity and contributes to proper targeting of these remodelers (Mansfield et al. 2011; Watson et al. 2012). The accessory domain architecture of subfamily III is more variable, but often includes one or more BRK domains thought to act as a protein–protein interaction domain (Allen et al. 2007).
Most investigations into the function of different CHDs have been done in model animals and fungi. ScCHD1 is the only CHD remodeler present in the budding yeast Saccharomyces cerevisiae and belongs to subfamily I (fig. 1). ScCHD1 exhibits two distinct chromatin-associated activities: assembly of nucleosomes and nucleosome positioning (Torigoe et al. 2013). Functional characterization of ScCHD1 revealed that it contributes to chromatin assembly associated with replication and transcription (Gkikopoulos et al. 2011; Smolle et al. 2012; Zentner et al. 2013; Yadav and Whitehouse 2016). Biochemical characterization of DmCHD1 (the subfamily I remodeler from the fly Drosophila melanogaster) suggests that the nucleosome assembly and nucleosome remodeling activities of ScCHD1 and DmCHD1 are conserved (Lusser et al. 2005; Konev et al. 2007). Similarly, functional analyses of additional subfamily I remodelers from Schizosaccharomyces pombe (fission yeast) and Mus musculus (mouse) suggest that chromatin assembly associated with replication and transcription are also conserved (Hennig et al. 2012; de Dieuleveult et al. 2016).
However, in contrast to Sa. cerevisiae with its single CHD protein, mammals including Homo sapiens contain nine CHD remodelers: two in subfamily I (CHD1 and CHD2), three in subfamily II (CHD3–CHD5), and four in subfamily III (CHD6–CHD9) (Flaus et al. 2006; Sims and Wade 2011) (fig. 1). There is considerable interest in understanding the respective contributions of these remodelers to chromatin-associated processes due to the critical roles played by these factors in development and disease (Alendar and Berns 2021). For example, CHD2 mutations are associated with chronic lymphocytic leukemia in H. sapiens and M. musculus (Marfella et al. 2006; Nagarajan et al. 2009; Rodríguez et al. 2015), CHD4 and CHD5 proteins in H. sapiens and M. musculus play an important role in neurogenesis and tumor suppression (Kolla et al. 2014; Liu et al. 2021), and mutation of CHD7 and CHD8 genes in H. sapiens and M. musculus results in the congenital disease known as CHARGE syndrome and autism, respectively (Zentner et al. 2010; Liu et al. 2021). It is thus medically relevant to understand how and when data derived from studying CHD remodelers in various other organisms can be used to provide substantive insight into the function of their human homologs.
Characterization of CHDs in plants to date raises the prospect that the function of these proteins may be more malleable than previously thought. The AtPKL remodeler of Arabidopsis thaliana is in subfamily II (fig. 1) and contributes to repression of transcription much like subfamily II homologs in vertebrates (Zhang et al. 2008; Ho et al. 2013; Carter et al. 2018). However, unlike vertebrate subfamily II homologs, AtPKL primarily exists as a monomer and contributes to homeostasis of the transcriptionally-repressive histone modification H3K27me3 (Zhang et al. 2012; Jing et al. 2013; Carter et al. 2018). Moreover, recombinant AtPKL promotes prenucleosome maturation in addition to nucleosome mobilization (Ho et al. 2013; Carter et al. 2018). These in vitro activities suggest that AtPKL, a subfamily II remodeler, contributes to nucleosome assembly as well as mobility, biochemical properties previously associated only with CHD remodelers in subfamily I (Lusser et al. 2005; Fei et al. 2015). In addition, phylogenetic analyses suggest the existence of novel plant clades of CHD remodelers in subfamilies II and III that are absent in A. thaliana, raising the prospect of novel remodeling activities/roles for CHD proteins in this kingdom (Hu et al. 2013; Koster et al. 2015).
Understanding the contribution of a given CHD accessory domain can provide considerable insight into the contribution of a CHD remodeler to a chromatin-associated process. For example, the chromodomain of subfamily I CHDs contributes to both recognition of the correct nucleosomal substrate and gating of the remodeling activity of the enzyme (Sims et al. 2005; Hauk et al. 2010). Similarly, the PHD domains of CHD3/4/5 in vertebrates contribute to recognition/targeting of these remodelers (Mansfield et al. 2011; Musselman et al. 2012; Egan et al. 2013). These observations strongly suggest that the distinct domain architectures acquired by CHD remodelers in different lineages contribute to different functions/roles, as well as infer molecular function of uncharacterized lineage-specific remodelers.
Previous phylogenetic analyses relied on sequences from a handful of representative taxa (Flaus et al. 2006; Ho et al. 2013; Hu et al. 2013). A sequence similarity-based analysis performed by Koster et al. (2015) identified putative CHD homologs from diverse eukaryotic taxa in all three subfamilies, suggesting that these subfamilies were present in the last common ancestor (LCA) of eukaryotes. The same analysis also identified putative subfamily III homologs in plants and fungi (Koster et al. 2015), which were previously thought to lack subfamily III. However, without a full-scale phylogenetic analysis of CHDs, the taxonomic distribution of the different subfamilies as well as the timing of gene duplication and loss remains unclear.
Thanks to the proliferation of genome and transcriptome data from nonmodel eukaryotes, a phylogenetic reassessment of CHD remodeler evolution is now possible. Here, improved taxon sampling from over 1,800 species identified several clades of CHD remodelers in plants and fungi that were absent or highly derived in model species representatives A. thaliana and Sa. cerevisiae, respectively. Whole genome duplication (WGD) drove CHD gene family expansion in vertebrates as well as in the cruciferous family of plants (Brassicaceae). Our analysis also identified more recent, genus-specific gene duplication events in Schizosaccharomyces and Drosophila that were not WGD-derived. A hidden Markov model (HMM) analysis identified novel conserved sequence motifs in some CHD clades in plants and animals, suggesting that duplication of CHDs is often accompanied by diversification of domain architecture.
Results
Our analysis identified 8,042 CHD homologs in 1,894 eukaryotic taxa from 18 eukaryotic lineages (table 1; supplementary table S1, Supplementary Material online). No CHD homologs were identified outside of eukaryotes. Although the number of subfamily homologs varied across different eukaryotic species, homologs from each of the three CHD subfamilies were present in four eukaryotic supergroups: Amoebozoa; Archaeplastida (Glaucophyta, Rhodophyta, and Viridiplantae); Opisthokonta (Choanoflagellata, Filasterea, Fungi, Icthyosporea, Metazoa, and Nucleariids); and SAR (Stramenopiles, Alveolata, and Rhizaria) (table 1). If the position of the root of the eukaryotic tree of life is as hypothesized by Derelle et al. (2015), the LCA of these four supergroups corresponds to the LCA of extant eukaryotes. This result is consistent with prior work suggesting that three distinct CHD subfamilies were already present in the eukaryotic LCA (Flaus et al. 2006; Koster et al. 2015). To infer the evolutionary history of each subfamily, we constructed maximum-likelihood (ML) phylogenetic trees of the chromodomain-ATPase core of CHD homologs. Our CHD phylogeny recovered three well-supported, monophyletic clades, representing subfamilies I–III (fig. 1).
Table 1.
Summary Counts of All CHD Homologs
| Lineage | Subfamily I Counts | Subfamily II Counts | Subfamily III Counts | Combined Counts | ||||
|---|---|---|---|---|---|---|---|---|
| Species | Sequences | Species | Sequences | Species | Sequences | Species | Sequences | |
| Alveolata | 35 | 35 | — | — | 4 | 6 | 38 | 41 |
| Amoebozoa | 11 | 11 | 2 | 2 | 17 | 30 | 18 | 43 |
| Apusozoa | — | — | — | — | 1 | 1 | 1 | 1 |
| Choanoflagellata | 2 | 2 | 2 | 2 | — | — | 2 | 4 |
| Cryptophyta | — | — | 4 | 4 | 5 | 6 | 7 | 10 |
| Discoba | 1 | 2 | — | — | 4 | 8 | 4 | 10 |
| Filasterea | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 |
| Fungi | 281 | 287 | 203 | 206 | 16 | 18 | 292 | 511 |
| Microsporidia | — | — | — | — | 10 | 10 | 10 | 10 |
| Chytridiomycota | 3 | 3 | — | — | 3 | 3 | 3 | 6 |
| Mucoromycota | 4 | 4 | 3 | 3 | 3 | 5 | 4 | 12 |
| Basidiomycota | 53 | 53 | 30 | 31 | — | — | 53 | 84 |
| Ascomycota | 221 | 227 | 170 | 172 | — | — | 222 | 399 |
| Glaucocystophyceae | 2 | 2 | 1 | 2 | 1 | 1 | 2 | 5 |
| Haptophyta | — | — | 3 | 3 | 10 | 26 | 11 | 29 |
| Icthyosporea | 1 | 1 | — | — | 1 | 1 | 1 | 2 |
| Metamonada | — | — | — | — | 1 | 13 | 1 | 13 |
| Metazoa | 488 | 1,123 | 495 | 1,526 | 483 | 1,859 | 498 | 4,508 |
| Other Metazoans | 10 | 10 | 12 | 18 | 12 | 12 | 12 | 40 |
| Other Protostomes | 22 | 30 | 23 | 40 | 21 | 27 | 24 | 97 |
| Arthropods | 146 | 166 | 147 | 167 | 138 | 277 | 149 | 610 |
| Other Deuterostomes | 5 | 6 | 6 | 6 | 5 | 5 | 6 | 17 |
| Chondrichthyes | 2 | 5 | 2 | 3 | 2 | 7 | 2 | 15 |
| Other Bony Vertebrates | 78 | 231 | 79 | 376 | 79 | 425 | 79 | 1,032 |
| Amphibians | 5 | 14 | 5 | 18 | 5 | 29 | 5 | 61 |
| Reptiles | 91 | 231 | 91 | 221 | 91 | 371 | 91 | 823 |
| Mammals | 129 | 430 | 130 | 677 | 130 | 706 | 130 | 1,813 |
| nucleariids | 1 | 1 | — | — | — | — | 1 | 1 |
| Rhizaria | 5 | 9 | — | — | 9 | 13 | 9 | 22 |
| Rhodophyta | 27 | 27 | 5 | 5 | 12 | 12 | 31 | 44 |
| Stramenopiles | — | — | 8 | 8 | 85 | 167 | 86 | 175 |
| Viridiplantae | 560 | 610 | 832 | 1,910 | 72 | 100 | 891 | 2,620 |
| Chlorophyta | 71 | 76 | 45 | 54 | 30 | 52 | 94 | 182 |
| Other Streptophytes | 18 | 18 | 21 | 25 | 1 | 1 | 27 | 44 |
| Other Embryophytes | 37 | 40 | 55 | 139 | 26 | 31 | 55 | 210 |
| Lycophytes | 11 | 11 | 13 | 29 | 2 | 2 | 15 | 42 |
| Ferns | 20 | 20 | 47 | 68 | 13 | 14 | 47 | 102 |
| Gymnosperms | 37 | 37 | 59 | 112 | — | — | 59 | 149 |
| Other flowering plants | 29 | 29 | 47 | 101 | — | — | 47 | 130 |
| Monocots | 58 | 62 | 91 | 229 | — | — | 92 | 291 |
| Eudicots | 279 | 317 | 454 | 1,153 | — | — | 455 | 1,470 |
| Total | 1,415 | 2,111 | 1,556 | 3,669 | 722 | 2,262 | 1,894 | 8,042 |
note.—Main eukaryotic lineages are bolded. Sub-lineages of Fungi, Metazoa, and Viridiplantae are also listed (unbolded).
Subfamily I: The Most Conserved CHD Subfamily in Plants, Animals, and Fungi
Accessory domain architecture is tightly conserved in subfamily I and consists of three C-terminal domains: SANT, SLIDE, and a domain of unknown function, DUF4208 (fig. 2). Most lineages maintain a single subfamily I homolog, with a few notable exceptions.
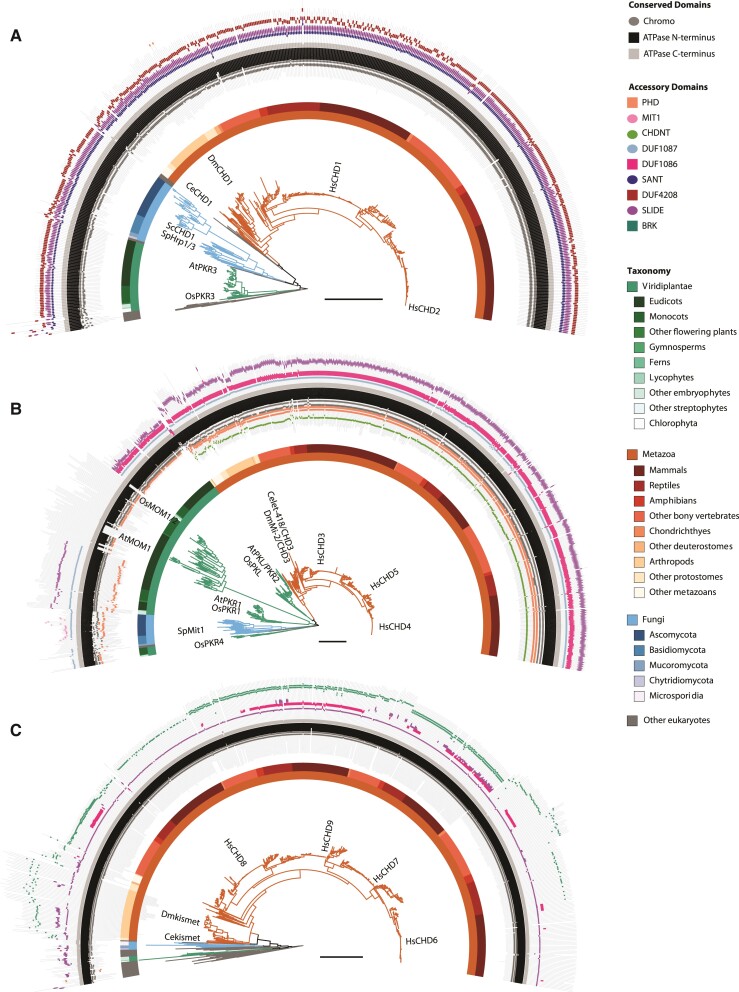
Fig. 2.
Detailed subfamily phylogenies with domains. Maximum-likelihood phylogenies for (A) subfamily I, (B) subfamily II, and (C) subfamily III. Location of CHD homologs from model eukaryotes are indicated. Branches are colored as in figure 1. Additional taxonomic resolution is provided by the color bars. The outer track indicates the PFAM domain architecture for each homolog.
Vertebrates have two subfamily I clades, CHD1 and CHD2 (fig. 2; supplementary fig. S1, Supplementary Material online). The duplication of CHD1/2 coincides with two rounds of WGD in ancestral vertebrates (Ohno et al. 1968; Abi-Rached et al. 2002; Dehal and Boore 2005). We searched the OHNOLOGS v2 database (Singh and Isambert 2020), which maintains a list of genes retained from WGD (i.e., ohnologs) in vertebrate genomes, and found that HsCHD1 and HsCHD2 are indeed WGD-derived gene pairs (weighted q-score from outgroup comparison 0.0006; weighted q-score from self-comparison 8.256E−29; lower q-scores imply more statistically significant ohnolog pairs). CHD1 and CHD2 are likely to be at least partially functionally redundant; they are recruited to common regions of the genome of mammalian cells (Siggens et al. 2015), and a dominant negative mutation of CHD1 has a more severe phenotype than a simple knockdown of CHD1 on nucleosome turnover at the promoter of transcribed genes (Skene et al. 2014).
The fission yeast Sc. pombe also has two subfamily I homologs, ScHrp1 and ScHrp3 (Jin et al. 1998; Yoo et al. 2002). Our phylogenetic analysis indicates that this duplication event occurred in an ancestor of the Schizosaccharomyces genus (fig. 2; supplementary fig. S2, Supplementary Material online). The Hrp1 clade retains all three C-terminal domains; whereas, the Hrp3 clade has either lost the region corresponding to DUF4208, or the sequence has diverged to the point that it is no longer detected by sequence similarity search (fig. 1; supplementary table S1, Supplementary Material online). In contrast to vertebrates, Schizosaccharomyces does not have a history of WGD, and a check for shared synteny between ScHrp1 and ScHrp3 was negative. This indicates that the subfamily I copies in Schizosaccharomyces arose through some other form of gene duplication, such as segmental duplication.
Subfamily II: Independent Expansions in Plants and Vertebrates
Subfamily II is the largest CHD subfamily due to multiple duplications in vertebrates and green plants (fig. 1; supplementary fig. S3, Supplementary Material online). The most common accessory domain architecture in subfamily II is the presence of one or tandem N-terminal PHD domains and three C-terminal domains: DUF1087, DUF1086, and SLIDE (figs. 1 and 2). However, the accessory domains are noticeably more variable compared to subfamily I, with one or more C-terminal domains frequently absent in different clades. Moreover, some lineages within subfamily II have acquired novel accessory domains. The animal subfamily II homologs, including HsCHD3/4/5 in humans, have a unique N-terminal CHDNT domain (fig. 1; supplementary fig. S3, Supplementary Material online). Similarly, many ascomycota subfamily II homologs, including ScMit1 from Sc. pombe, have a unique MIT1 C-terminal accessory domain (fig. 1; supplementary fig. S4A, Supplementary Material online). Investigation of ScMit1 indicates that this MIT1 domain overlaps with a region that plays a key role in formation of SHREC, the fission yeast nucleosome remodeling and deacetylation complex (Job et al. 2016). The majority of ascomycota subfamily II CHDs possess an MIT1 accessory domain (supplementary fig. S4A, supplementary table S1, Supplementary Material online), suggesting that the SHREC complex is not limited to fission yeast, but is common in the ascomycota lineage. Interestingly, ascomycota in the Saccharomycotina subdivision, including Sa. cerevisiae, have lost subfamily II consistent with the absence of the heterochromatic features associated with the SHREC complex in the Saccharomycotina.
As with CHD1/2, duplications that gave rise to ohnologs CHD3/4/5 in vertebrates can be traced back to WGD in their common ancestor (weighted q-score for HsCHD3/4/5 gene pairs was less than 1E−05 for all comparisons). In contrast, two independent single gene duplications occurred in model invertebrates Drosophila and Caenorhabditis giving rise to DmMi-2 and DmCHD3 in D. melanogaster and Celet-418 and Cechd-3 in Caenorhabditis elegans, respectively. The Celet-418 and Cechd-3 paralogs in C. elegans share the same accessory domain architecture. In contrast, sequences in the Drosophila dCHD3 clade are truncated and missing both N- and C-terminal accessory domains (fig. 1; supplementary fig. S5, Supplementary Material online). For clarity, and in agreement with prior literature (Murawska et al. 2008), we refer to these Drosophila clades as dCHD3 and dMi-2 to differentiate dCHD3 from the vertebrate clade CHD3. Further analysis of Drosophila subfamily II homologs revealed that not all Drosophila species possessed dCHD3 homologs, which was only found in a subset of species from the melanogaster group. In addition, the dCHD3 clade contains noticeably longer branches compared to the dMi-2 clade (supplementary fig. S5, Supplementary Material online), which is suggestive of elevated rates of evolution in the dCHD3 clade. We performed a PAML analysis to measure the rate of evolution within the conserved chromo and ATPase domains following the duplication that gave rise to dCHD3 and dMi-2 subclades in Drosophila. Positive selection was not detected along the branches leading to either subclade (P-value > 0.05; supplementary fig. S5; supplementary table S2, Supplementary Material online). However, both subclades have a higher proportion of sites with an elevated rate of evolution (w = 0.37 and w = 0.4 for dCHD3 and dMi-2, respectively) compared to remaining Drosophila orthologs (supplementary table S2, Supplementary Material online). These results suggest that in addition to structural changes (e.g., loss of accessory domains), relaxed selection within the core chromo and ATPase domain region may have contributed to retention and functional differences between the two copies. Although both DmCHD3 and DmMi-2 remodelers colocalize with RNA polymerase II in transcribed regions of polytene chromosomes (Murawska et al. 2008), DmCHD3 exists as a monomer rather than in a multi-subunit complex like DmMi-2 (Murawska et al. 2008; Kunert and Brehm 2009), suggesting that melanogaster group dCHD3 proteins remodel in a context that is distinct from dMi-2.
Viridiplantae (plants and green algae) comprise four distinct clades in subfamily II: PKL, PKR1, PKR4, and MOM (fig. 1). Unlike the WGD-based duplication of CHD3/4/5 in vertebrates, the origins of the four Viridiplantae clades are less clear. They do not form a single monophyletic group, as would be expected if they resulted from gene duplication in the LCA of plants. Instead, the PKL clade groups closest to animal CHDs, and PKR4 groups closest to fungi (fig. 1). To evaluate the strength of these associations, we performed alternative topology tests. The ML phylogeny presented in figure 1 was significantly better than alternative topologies that forced the plant clades to be monophyletic (P-value < 1E−5 for all comparisons; supplementary table S3, Supplementary Material online). Horizontal gene transfer, cryptic gene duplication and differential loss, convergent evolution, and methodological artifacts (e.g., long-branch attraction) are all possible explanations for the lack of plant monophyly in subfamily II. Additional sequenced genomes from the Viridiplantae sister lineages Rhodophyta and Glaucophyta could help differentiate between these alternatives.
The PKL clade is present in all lineages of green plants (table 2) and contains accessory domains similar to animal subfamily II CHDs including an N-terminal PHD domain and three C-terminal domains (DUF1087, DUF1086, and SLIDE) (fig. 2). Though functionally uncharacterized, DUF1086 contains a region of sequence and structural similarity to the SANT domain in yeast CHD1, suggesting this domain is involved in chromatin interactions, in particular nucleosomal DNA, similar to subfamily I members (Ho et al. 2013). The two A. thaliana sequences (AtPKL and AtPKR2) present in this clade have shared synteny, which, in addition to the taxonomic distribution present in both PKL and PKR2 subclades, indicates that they are ohnologs resulting from WGD at the base of the Brassicaceae family (Bowers et al. 2003). Similar to the pattern observed between the dMi-2 and dCHD3 clades in Drosophila, the Brassicaceae PKR2 sub clade was recovered in few species and is comprised of longer branches compared to the Brassicaceae PKL sub clade (supplementary fig. S4B, Supplementary Material online). PKL and PKR2 are both genetically linked to homeostasis of the transcriptionally-repressive histone modification H3K27me3 (Zhang et al. 2012; Jing et al. 2013; Huang et al. 2017; Carter et al. 2018). However, AtPKL is expressed ubiquitously in A. thaliana, whereas the expression of AtPKR2 is restricted to the seed endosperm (Carter et al. 2016).
Table 2.
Summary Counts of Viridiplantae Sequences in Subfamily II
| Lineage | PKL Counts | PKR1 Counts | PKR4 Counts | MOM Counts | ||||
|---|---|---|---|---|---|---|---|---|
| Species | Sequences | Species | Sequences | Species | Sequences | Species | Sequences | |
| Chlorophyta | 41 | 47 | 4 | 4 | 3 | 3 | — | — |
| Other Streptophytes | 16 | 16 | 8 | 8 | 1 | 1 | — | — |
| Other Embryophytes | 54 | 70 | 26 | 30 | 37 | 39 | 1* | 1* |
| Lycophytes | 12 | 18 | 9 | 9 | 2 | 2 | 5* | 5* |
| Ferns | 47 | 47 | 21 | 21 | — | — | 6* | 7* |
| Other flowering plants | 46 | 51 | 23 | 25 | 2 | 2 | 15 | 23 |
| Gymnosperms | 59 | 62 | 18 | 19 | 25 | 25 | 6 | 6 |
| Monocots | 90 | 107 | 53 | 62 | 13 | 15 | 27 | 45 |
| Eudicots | 440 | 587 | 262 | 317 | — | — | 164 | 249 |
note.—Asterisk (*) indicates sequences that were manually added based on presence of conserved MOM motif(s), see Materials and Methods.
The PKR1 clade is also present in all lineages of green plants (table 2; supplementary table S1, Supplementary Material online) and shares the same accessory domains as PKL, except for DUF1086, which is absent. Given that DUF1086 shares sequence similarity to the SANT domain of CHD1 (Ho et al. 2013), which in conjunction with the SLIDE domain comprises the DNA-binding domain of CHD1 (Ryan et al. 2011; Sharma et al. 2011), the absence of DUF1086 may imply a substantial alteration of the DNA interaction surface in PKR1 compared to PKL. Additionally, a stretch of ∼300 amino acids separate the PHD and Chromo domains in PKR1 (figs. 1 and 2). An IUPred3 scan of PKR1 homologs suggests that these extra inter-domain regions of PKR1 homologs are composed primarily of disordered sequence rather than structural domains (supplementary fig. S6, Supplementary Material online). Although intrinsically disordered sequence lacks predicable structure, interactions with other proteins or cofactors may lead to the formation of secondary structure that influences protein function (Tompa 2002). Alternatively, the unstructured region may provide a flexible linker to extend the distance between PHD and chromodomain targets/binding or regulatory site(s) for moderating function. Previous characterization of intrinsically disordered regions is consistent with the possibility that these regions of PKR1 serve as entropic linkers between different domains of these CHD remodelers (Wright and Dyson 2015; Berlow et al. 2018; Li et al. 2018; Huang et al. 2020). The pervasive presence of these regions in PKR1 also raises the prospect that remodelers act as signal integration hubs and/or mediate scaffolding of higher order chromatin-based structures.
Previous analyses have had difficulty placing the OsPKR4 CHD homolog in Oryza sativa in the evolutionary context of other CHD sequences (synonyms OsCHR703, Os01g65850; see supplementary table S4, Supplementary Material online regarding varying nomenclature for rice CHD remodelers). One phylogenetic analysis of O. sativa and A. thaliana homologs showed OsPKR4 grouping sister to all other plant CHDs (Hu et al. 2013). A follow up analysis with additional sequences from Sa. cerevisiae, D. melanogaster, and humans had OsPKR4 grouping sister to animal subfamily III homologs, albeit with weak bootstrap support (Hu et al. 2014). In our analysis, OsPKR4 is located within a distinct Viridiplantae clade of subfamily II homologs, which we refer to as PKR4 (PICKLE related 4; fig. 1; supplementary fig. S4A, Supplementary Material online). The PKR4 clade is present in diverse Viridiplantae from green algae (e.g., Micromonas pusilla) to flowering plants including Amborella trichopoda and O. sativa (supplementary fig. S4A; supplementary table S1, Supplementary Material online). However, PKR4 is noticeably absent in eudicots (including A. thaliana) and ferns (table 2; supplementary table S1, Supplementary Material online), suggesting that the PKR4 gene was secondarily lost in those lineages. The accessory domains of PKR4 are similar to PKL and PKR1, having an N-terminal PHD domain and C-terminal DUF1087 domain (fig. 2; supplementary fig. S4A, Supplementary Material online). An analysis of transcript levels of ATP-dependent chromatin remodelers in rice (Hu et al. 2013) revealed that OsPKR4 exhibits an expression profile that is distinct from OsPKL, with tissue-specific expression highest in the endosperm (supplementary fig. S7, Supplementary Material online). In an interesting convergence of tissue-specific expression, PKR2 in A. thaliana is also expressed highest in seed unlike other CHD homologs (supplementary fig. S8, Supplementary Material online). Differing expression profiles between the CHD different remodelers in plants is consistent with the possibility that PKR4 and PKR2 each play a role that is distinct from that of PKL.
MOM1 is a Highly Divergent Subfamily II CHD Protein
The final plant clade within subfamily II is comprised of MORPHEUS’ MOLECULE (MOM) sequences, a gene family linked to DNA-methylation-independent transcriptional gene silencing based on characterization of AtMOM1 in A. thaliana (Amedeo et al. 2000; Vaillant et al. 2006). Most homologs in the MOM clade contain a N-terminal PHD domain, tandem chromodomains, and full-length ATPase domain (fig. 2; supplementary fig. S4B, Supplementary Material online), including those MOM homologs in rice (OsMOM1, Os06g01320; OsMOM2, Os02g02050) and poplar (PtMOM1, eugene3.00130053; PtMOM2, eugene3.00660276) as previously characterized (Čaikovski et al. 2008). However, the single A. thaliana sequence (AtMOM1) present in this clade bears little resemblance to other CHDs, possessing only a truncated portion of the ATPase binding domain and no canonical accessory domains (fig. 1). Loss or divergence of the N-terminal region in MOM homologs has occurred independently in different plant lineages including in Brassicales order that includes A. thaliana as well as the Phaseoleae tribe of legumes (e.g., soybean) (supplementary fig. S4B, Supplementary Material online).
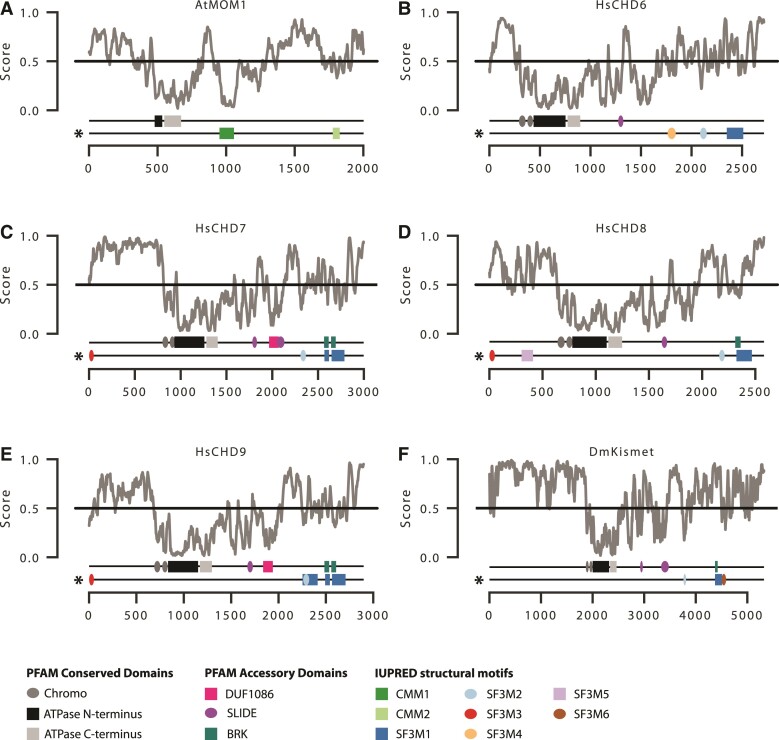
Most MOM homologs contain on average 1,037 amino acids of additional sequence downstream of the conserved ATPase domain that lacks similarity to any of the known CHD accessory domains (fig. 2; supplementary fig. S4B, Supplementary Material online). An earlier analysis, compared the MOM homologs of four species of model plants and noted the presence of conserved regions they termed conserved MOM motifs (CMMs) in this downstream region (Čaikovski et al. 2008). We performed an IUPred3 scan of all MOM homologs in our analysis to de novo identify CMMs that may correspond to uncharacterized structural domains in MOM sequences and successfully recovered CMM1 and CMM2 as described by Čaikovski et al. (2008). CMM1 spans amino acids 951–1,055 in AtMOM1 (fig. 3A). This first conserved motif has an average length of 97 amino acids and was present in 304/323 (94%) of sequences in the MOM clade (supplementary fig. S9A; supplementary table S1, Supplementary Material online) with an average amino acid pairwise identity of 47.9%. CMM2 spans 1,773–1,812 amino acids in AtMOM1 (fig. 3A). This second conserved motif has an average length of 37.2 amino acids and was identified in 225/323 (70%) of sequences in the MOM clade (supplementary fig. S9A; supplementary table S1, Supplementary Material online) with an average pairwise identity of 41.6%.
Fig. 3.
Novel conserved motifs and disordered regions in CHD proteins: (A) AtMOM1, (B) HsCHD6, (C) HsCHD7, (D) HsCHD8, (E) HsCHD9, and (F) DmKismet. IUPred score denotes the disorder tendency of each residue in the given protein, where higher values correspond to a higher probability of disorder. The top domain track for each protein indicates the location of the canonical PFAM conserved and accessory structural domains. The bottom track (*) indicates the location of predicted IUPred-derived structural domains in MOM (CMM1/2) and subfamily III (SF3M1-6). Width of ovals and rectangles are proportional to the width of the protein domain.
We queried the new custom CMM1 and CMM2 HMMs against our comprehensive protein database (see Methods) and identified 14 additional homologs from ferns, lycophytes, and a single liverwort (Pellia neesinia) (supplementary table S1, Supplementary Material online), which were previously excluded from our analysis due to low sequence similarity to known CHD domains. Therefore, we constructed a revised phylogeny for PKR1 and MOM homologs that included these additional 14 sequences (supplementary fig. S9A, Supplementary Material online). In the revised analysis, MOM sequences (i.e., those CHDs containing at least CMM1) were nested within the PKR1 clade (supplementary fig. S9B, Supplementary Material online). Moreover, 10 of the new sequences had significant hits to the canonical CHD accessory domain DUF1087 (supplementary fig. S9B, Supplementary Material online). This suggests that MOM arose via duplication early in the evolution of embryophytes from a PKR1-like progenitor, and that loss of the canonical C-terminal CHD accessory domains and gain of the MOM-specific CMM1/2 domains was a stepwise process. However, it is important to note that most CHD sequences from nonseed plants come from the oneKP transcriptome sequencing initiative (Leebens-Mack et al. 2019). These predicted proteomes from de novo transcriptome assemblies are less complete than those from genome assemblies, and discrete loci may be fragmented or collapsed. Additional whole genome sequencing of nonseed plants is required to fully resolve the evolutionary history of MOM.
Subfamily III: Evolution of Novel Accessory Domains in Animals
The majority (82%) of subfamily III sequences are from metazoans due to extensive gene family expansion in vertebrates. As in subfamilies I and II, duplications that gave rise to vertebrate CHD6/7/8/9 can be traced back to WGD in their common ancestor (supplementary fig. S10, Supplementary Material online; maximum weighted q-score for all HsCHD6/7/8/9 gene pairs = 0.0052). In addition to vertebrates, subfamily III has expanded in stramenopiles and amoebozoans; most stramenopile and amoebozoan sequences are found in three separate clades (supplementary fig. S11, Supplementary Material online).
In contrast to the extensive expansion in animals, subfamily III is noticeably absent in model plants and fungi (fig. 1). In plants, subfamily III is present in green algae, mosses, lycophytes, and ferns (table 1; supplementary fig. S11, Supplementary Material online), indicating that the subfamily was lost in the ancestor of seed plants. Similarly, subfamily III is present in some fungal lineages including Microsporidia, Chytridiomycota, and Mucoromycotina (table 1; supplementary fig. S11, Supplementary Material online), which suggests the subfamily was independently lost in the ancestor of Dikarya (the largest subkingdom of fungi).
The accessory domain architecture of subfamily III is more variable compared to the other two subfamilies. Most subfamily III homologs contain a SLIDE and one or more BRK domains (fig. 2). DUF1086 was recovered in only 20% (498/2,262) of homologs (supplementary table S1, Supplementary Material online). However, there were several vertebrate clades (e.g., CHD6/8 in fish, CHD7/9 in mammals) where DUF1086 is more common (fig. 2; supplementary fig. S10, Supplementary Material online).
Subfamily III homologs in animals are notable for long stretches of sequence outside of the canonical structural domains (fig. 1), which could correspond to inherently disordered regions (e.g., as in PKR1 in plants) or could contain novel subfamily specific structural domains (e.g., as in MOM). We performed an IUPred3 scan of subfamily III and identified six predicted globular domains, which we refer to as SF3Ms for subfamily III motifs (fig. 3). SF3M1 has an average length of 133 amino acids and is present in 1,774/1,859 (95.4%) of metazoan subfamily III homologs (supplementary table S1, Supplementary Material online). SF3M1 frequently overlaps with known BRK domains, but not always. For example, the PFAM-based BRK domain was not recovered in mammal CHD6s; yet, SF3M1 is present (fig. 3; supplementary figs. S9 and S12, Supplementary Material online). This suggests that the BRK domain, as characterized by PFAM domain PF07533, is likely too conservative to recover the full diversity of BRK-like sequences in subfamily III. Interestingly, sequence similarity to SF3M1 is also found in the related SWI/SNF transcription factor family proteins (supplementary table S5, Supplementary Material online).
The remaining SF3Ms do not overlap with canonical accessory domain predictions and represent new regions of interest for further investigation. SF3M2 has an average length of 73 amino acids and is also present in the majority of subfamily III (present in 1,789/1,859 (96.2%) of metazoan sequences; supplementary table S1, Supplementary Material online). SF3M3 is 38 amino acids on average and present at the N-terminus of 970/1,076 = 90% of vertebrate CHD7/8/9s (fig. 3; supplementary table S1, Supplementary Material online). Vertebrate CHD6 contains a shorter N-terminal region upstream of the helicase core suggesting the LCA of this clade secondarily lost SF3M3 (supplementary fig. S12, Supplementary Material online). The last three motifs SF3M4, SF3M5, and SF3M5 are unique to specific clades within subfamily III (fig. 3; supplementary fig. S12; supplementary table S1, Supplementary Material online). SF3M4 has an average length of 103 amino acids and is unique to mammal CHD6. SF3M5 has an average length of 77 amino acids and is present in the N-terminal region of vertebrate CHD8. Finally, SF3M6 is 77 amino acids on average and is unique to arthropods.
We checked if any of the newly predicted SF3Ms contained mutations associated with human diseases. Human CHD7 was the only subfamily III homolog with significant single nucleotide variants (SNVs) resulting in nonsynonymous substitutions. CHD7 SNVs were associated with CHARGE syndrome and Hypogonadotropic Hypogonadism 5 with or without anosmia (HH5). The majority of these mutations were located in two hotspots located within the two SLIDE domains (supplementary fig. S13, Supplementary Material online). Some disease associated SNVs overlapped with the newly predicted SF3M1/2/3, although the impact of these mutations on protein function is unclear.
Discussion
Several evolutionary mechanisms contribute to the retention of gene duplicates including dosage sensitivity (Edger and Chris Pires 2009), subfunctionalization (Hughes 1994; Force et al. 1999), and neofunctionalization (Lewis 1951; Ohno 1970); all three mechanisms appear to have played a role in the evolution of CHDs. Gene dosage is particularly important to the evolution of protein complexes as imbalanced levels of gene product (i.e., proteins) may be detrimental to the formation of the complex. Following whole genome duplications, proteins that function in macromolecular complexes tend to be over-retained in duplicate, because the dosage of all genes in the complex are equivalently and simultaneously increased (Edger and Pires 2009). It is thus tempting to speculate that dosage sensitivity may have been the primary driver behind the expansion of CHDs in vertebrates following WGD as these proteins are frequently components of multiprotein remodeler complexes. However, subfunctionalization has also likely played a role in the retention of multiple vertebrate CHD paralogs. For example, human subfamily II paralogs, which are known to be components of the Mi-2/NuRD complex, have also evolved different tissue specificity, with HsCHD3/4 expressed in all tissues and HsCHD5 expressed more exclusively in the brain, pituitary gland, and testis (Alendar and Berns 2021) (supplementary fig. S14, Supplementary Material online). In addition, the evolution of novel protein motifs in subfamily III (fig. 3; supplementary fig. S12; supplementary table S1, Supplementary Material online) is suggestive of neofunctionalization, although further analysis of these domains is necessary to determine their specific role.
In contrast to the biased retention of dosage-sensitive protein duplicates following WGD, proteins with less connectivity or dosage sensitivity are more often retained following smaller scale tandem or segmental duplications (Edger and Pires 2009). The duplication that gave rise to dMi-2 and dCHD3 in Drosophila, which was not WGD-derived, fits this pattern; following the duplication, DmCHD3 evolved to function as a monomer with presumably less dosage sensitivity compared to DmMi-2 (Murawska et al. 2008). In plants, AtPKL also primarily exists as a monomer (Ho et al. 2013) in distinct contrast to the animal members of subfamily II such as CHD3/4/5 from vertebrates. With regards to the other plant clades of subfamily II, gel filtration data indicates that AtMOM1 is part of a complex (Han et al. 2016), and it is unknown if the proteins in the remaining plant clades, PKR1 and PKR4, function as a monomer or as part of a complex. It is possible that plant CHD remodelers in subfamily II typically exist as monomers, in contrast to their vertebrate homologs, thereby relaxing the evolutionary constraint of dosage sensitivity and enabling the numerous duplications and expansion of plant CHD homologs in subfamily II.
The MOM1 clade is notably divergent from other subfamily II clades, possessing two unique structural domains not found in any other CHD homologs, suggesting neofunctionalization is involved in its retention. Indeed, AtMOM1 has a distinct role compared to other CHD homologs in A. thaliana (Čaikovski et al. 2008; Hu et al. 2014). However, it is important to remember that the Brassicales MOM sequences, including those in A. thaliana, have diverged substantially from other plant MOMs with the loss of additional N-terminal accessory domains as well as the majority of the ATPase domain that drives nucleosome remodeling activity (supplementary fig. S9, Supplementary Material online), and therefore are not representative of the larger MOM clade. Further investigation of the function of non-Brassicaceae MOM as well as PKR4 in monocots and PKR1 in A. thaliana and other plants is necessary to resolve the complex evolutionary history of plant subfamily II homologs.
In contrast to the numerous expansions of CHD subfamilies in animals and plants, some lineages appear to have lost specific subfamily homologs entirely. Independent losses of subfamily III in dikarya fungi and seed plants are the most notable, but the implications of these losses are unclear. In animals, subfamily III homologs are present at promoters and enhancers (Schnetz et al. 2010; Payne et al. 2015; Shen et al. 2015; de Dieuleveult et al. 2016) and/or interact with CTCF (Ishihara et al. 2006; Allen et al. 2007; Nguyen et al. 2008: 3) and contribute to a diverse array of processes in embryonic development (Bosman et al. 2005; Hurd et al. 2007; Nishiyama et al. 2009; Gaspar-Maia et al. 2011). These molecular phenotypes and developmental traits vary greatly or do not exist in fungi and plants, making it difficult to infer the function of subfamily III CHDs in early fungi and plants. It is possible that the molecular function(s) of these lost homologs has been compensated for through the expansion of another CHD subfamily or different chromatin remodeling family during the evolution of dikarya fungi and seed plants. Molecular characterization of additional CHD homologs from all three subfamilies in fungi and plants could help to clarify the evolution of subfamily III and changes in remodeling activities and/or machinery accompanying these loss events. Outside of plants and fungi, nine additional lineages of eukaryotes in our analysis are also missing one or more CHD subfamilies (table 1). However, we are cautious not to draw conclusions regarding gene loss in these cases, because these lineages are underrepresented in the NCBI Refseq and Taxonomy databases used in our analysis. Ongoing genome and transcriptome surveys of under sampled taxa (Richter et al. 2018; Brunet et al. 2019; Gawryluk et al. 2019; Grau-Bové et al. 2021; Van Vlierberghe et al. 2021) as well as advances in single-celled genome sequencing (Schön et al. 2021) and efforts to resolve the evolutionary relationship between eukaryotic groups (Tice et al. 2021; Irisarri et al. 2022) are enabling future investigations into the evolution and function of CHDs in these diverse eukaryotic lineages.
Analysis of predicted structural domains and disordered regions provided additional support for the role of neofunctionalization in evolution of CHD remodelers and emphasizes the potential for disordered regions in enabling this process. Our analysis identified several regions of high disorder in different clades of CHD remodelers (fig. 3; supplementary fig. S6, Supplementary Material online). These regions were particularly striking in the subfamily II PKR1 clade in plants, which maintains similar accessory domain architecture to the PKL clade interspersed with long stretches of disordered sequence (supplementary fig. S6, Supplementary Material online). Similar analysis of the plant MOM clade in subfamily II and the animal clades in subfamily III revealed disordered regions that surround small, previously unpredicted structural domains (fig. 3). The function of these novel domains remains to be determined, but the sequence conservation suggests acquisition of shared properties by the respective clades of CHD remodelers. Similarly, the conserved acquisition of disordered regions in CHD remodelers has functional implications. Such regions may act as flexible linkers, separating other domains by a specific distance for proper function of the remodeler and have the capacity to enable allosteric regulation of multidomain proteins (Berlow et al. 2018; Armache et al. 2019; Huang et al. 2020) and thereby enable recognition of the desired chromatin context by CHD proteins to enable remodeling activity or specify a particular remodeling outcome. Another possible role suggested by the presence of these domains, not necessarily exclusive, is that these remodelers play a scaffolding role in generating higher order chromatin-associated complexes (Cortese et al. 2008; Uversky 2015; Cho et al. 2021). In this light, it is intriguing to note that loss of AtMOM1 results in a chromatin-associated phenotype despite the absence of an intact ATPase domain (Čaikovski et al. 2008) (supplementary fig. S9, Supplementary Material online).
CHD proteins play a foundational role in chromatin-based processes in eukaryotes and a better understanding of their various roles is relevant to human health (Alendar and Berns 2021). Our comprehensive phylogenetic analysis has revealed new sequence features of CHD remodelers that are likely to contribute to our understanding of their function. In addition, our analysis highlights both the advantages and potential perils of using model organisms as the basis for inferring the function of proteins sharing a common ancestry. We observed that CHD evolution is highly dynamic and that the CHD repertoires of commonly used model organisms are the result of lineage-specific changes that may make it more challenging to infer the function and chromatin remodeling mechanisms of CHDs in other species. For example, due to the extensive divergence in both the accessory and core domain architecture of MOMs in the Brassicaceae, the functional characterization of AtMOM1 in A. thaliana is likely not representative of MOM function across seed plants. Similarly, PKR4 from subfamily II has been lost in eudicots, and its absence in A. thaliana precludes the characterization of this novel clade in this model system and further highlights the opportunities associated with studying chromatin-associated processes in additional model systems. Similarly, the full diversity of remodelers in subfamily III has likely been underappreciated due to its absence in model plants and fungi. In short, our study identifies new contexts for functional characterization of these architects of genome-based traits and expand our awareness of the functional potential associated with their modular structure. Broadening the organismal scope for functional characterization of these remodelers will greatly advance our knowledge of their properties and the chromatin-based processes in which they participate.
Materials and Methods
Identification of CHD Homologs
The A. thaliana CHD homolog PKL (AT2G25170) was queried against a custom protein database using phmmer, part of the HMMER v3.3.1 software package (Eddy 2009), with the following parameters: –E 0.001 –domE 1 –incE 0.01 –incdomE 0.03 –mx BLOSUM62 –pextend 0.4 –popen 0.02. The custom database primarily consisted of NCBI RefSeq (release 98) (O’Leary et al. 2016) and was supplemented with additional predicted protein sequences from the Marine Microbial Eukaryotic Transcriptome Sequencing Project (Keeling et al. 2014) and the 1,000 Plants transcriptome sequencing project (OneKP) (Matasci et al. 2014). This initial search returned 97,035 sequences (supplementary table S6, Supplementary Material online), which were queried against the two PFAM domains (SNF2_N, PF00176; Helicase_C, PF00271) corresponding to the conserved ATPase domain of chromatin remodelers using hmmsearch v3.3.1 (Eddy 2009) with default parameters. Sequences with one or more ATPase domains were retained, and the conserved sequence region was extracted. Sequences were aligned using MAFFT version v7.407 using –auto to select the best alignment strategy (Katoh and Standley 2013). FastTree v2.1.7 using default methods was used to construct an approximately ML phylogenetic tree (Price et al. 2010). The tree was midpoint rooted and the subtree containing known CHD homologs was retained.
Preliminary analysis of CHD homologs revealed that some sequences (e.g., XP_015643423 from O. sativa) had a top hit in A. thaliana to AtMOM1. However, AtMOM1 itself had been excluded earlier because it did not have a significant hit to either ATPase PFAM domains. Further investigation indicated that AtMOM1 has homologous sequence corresponding to the ATPase domains of CHDs but that the MOM1 sequence was too divergent to be detected using the PFAM ATPase domains. Therefore, full-length sequences with a significant hit to AtMOM1 (phmmer full sequence bitscore > 50) but lacking a significant hit to ATPase PFAM domains were added back into the analysis at this stage.
We performed a second round of tree building on this reduced sequence set using MAFFT and FastTree as described above. The second tree was midpoint rooted and sequences within the clade containing known CHD sequences were considered CHD homologs and retained for downstream analysis.
Protein Domain Annotation
Conserved protein domains were identified in CHD homologs using an iterative process. First, the PFAM web portal was used to annotate PFAM domains present in model CHD homologs from A. thaliana, O. sativa, H. sapiens, C. elegans, D. melanogaster, Sa. cerevisiae, and Sc. pombe (see supplementary table S1, Supplementary Material online), which identified the following domains of interest: Chromodomain (PF00385), SNF2_N (PF00176), Helicase_C (PF00271), PHD (PF00628), CHDNT (PF08073), MIT1 (PF18585), DUF1086 (PF06461), DUF1087 (PF06465), DUF4208 (PF13907), SANT (PF18375), SLIDE (PF09111), HAND (PF09110), and BRK (PF07533). Second, the representative proteome (rp15) for each PFAM domain was downloaded and queried against CHD homologs using hmmsearch v3.3.1 (Eddy 2009). Third, sequence regions in all CHD homologs corresponding to these PFAM domains (E-value cutoff 1e−5) were aligned using MAFFT (–auto) to construct custom, CHD-specific HMM protein domains using hmmbuild v3.3.1 (Eddy 2009). Finally, all CHD homologs were annotated with the custom CHD HMM domains using hmmsearch (E-value cutoff 1e−5) (supplementary table S1, Supplementary Material online).
IUPred structural domain predictions for all CHD homologs was performed with the command line version of IUPred3 using the glob analysis type and default parameters (Erdős et al. 2021). Regions corresponding to globular (i.e., structural) domains were extracted using a custom python script. Similar IUPred-predicted globular domains were identified using an all-by-all blastp search (BLAST v2.11.0+) and clustered into homologous groups with MCL v14-137 using an inflation parameter of 1.4 (Enright et al. 2002). Clustered domain sequences were aligned with MAFFT version v7.407 using the E-INS-i alignment strategy (Katoh and Standley 2013). Poorly aligned sequences were identified manually, and the alignment was repeated. The second alignment was trimmed with TrimAL v1.4.rev15 using the gappyout and terminalonly options (Capella-Gutierrez et al. 2009). Finally, custom HMMs were constructed from the trimmed alignments and HMMs were searched against the custom protein database (see above) using hmmbuild and hmmsearch v3.3.1 (Eddy 2009). All CHD homologs were annotated with the IUPred HMM domains using an E-value cutoff of 1e−5 (supplementary table S1, Supplementary Material online).
Phylogenetic Analysis
To construct robust phylogenies of CHD homologs, protein sequences corresponding to the custom chromo, ATPase N-terminus, and ATPase C-terminus domains were trimmed to ±20 residues around the conserved region. For the full CHD phylogeny, vertebrate sequences from the ALC sister family (Hu et al. 2013) were included as an outgroup. Trimmed sequences were aligned with MAFFT version v7.407 using the following parameters –bl 30 –maxiterate 0 –6merpair (Katoh and Standley 2013). FastTree v2.1.7 using default methods was used to construct an approximately ML phylogenetic tree (Price et al. 2010). Potentially spurious homologs (n = 132) on long terminal branches or those that grouped outside of the taxon’s established lineage (i.e., suspected contamination) were identified manually and removed from the analysis (see supplementary table S1, Supplementary Material online). The alignment and tree building were repeated as described above until no more long terminal branches remained.
Due to the large number of sequences in the full CHD sequence set, we also created pruned CHD phylogenies containing a reduced taxa set. To select taxa for the pruned CHD sequence set, the species phylogeny of all CHD-containing organisms was extracted from the NCBI taxonomy database using phyloT (https://phylot.biobyte.de/) (supplementary fig. S15A, Supplementary Material online). A subset of 302 species were selected to maximize taxonomic diversity while reducing polytomies (supplementary fig. S15C, Supplementary Material online). All CHD homologs within these 302 species (2,179 sequences) were extracted and aligned with MAFFT version v7.407 using the following parameters: –bl 30 –maxiterate 0 –6merpair (Katoh and Standley 2013). A ML phylogenetic tree was constructed using IQ-TREE v1.6.10 (Nguyen et al. 2015) using the built in ModelFinder (Kalyaanamoorthy et al. 2017) to determine the best-fit amino acid substitution model and performing SH-aLRT and ultrafast bootstrapping analyses with 1000 replicates each.
For both the full and pruned CHD sequence sets, clades corresponding to the three subfamilies were extracted and aligned separately with MAFFT version v7.407 using the following parameters: –bl 30 –maxiterate 1000 –retree 1 –genafpair. ML trees for each subfamily were constructed using IQ-TREE v1.6.10 (Nguyen et al. 2015) using the built in ModelFinder (Kalyaanamoorthy et al. 2017) to determine the best-fit amino acid substitution model and performing SH-aLRT and ultrafast bootstrapping analyses with 1,000 replicates each. Trees were visualized using iTOL v5.7 (Letunic and Bork 2019).
Tests of positive selection among Diptera subfamily II homologs were evaluated using codeml within the PAML v4.9 software suite (Yang 2007). Rates of evolution were defined by omega (ω), which is the rate ratio of synonymous (dS) and nonsynonymous substitutions (dN). Three models were evaluated. Model 0 determined a global ω across the whole tree (e.g., supplementary fig. S5B, Supplementary Material online). The Branch-Sites Test, Model 2 with NS_sites = 2, was performed with ω estimated or fixed at 1, representing the alternative (L1) and null (L0) hypotheses, respectively. Positive selection along the dMi-2 or dCHD3 branch was inferred by calculating the Likelihood Ratio Test [LRT = 2(lnL1–lnL0)] for each branch and using X2 distribution to determine the significance thresholds for the given degrees of freedom. Initial ω values of 0.2, 0.4, 0.6, 0.8, 1, 1.2, 1.4, 1.6, 1.8, and 5 were used to evaluate the effect on likelihood calculations, but results were identical regardless of initial value.
IQ-TREE v1.6.10 (Nguyen et al. 2015) was used to perform topology tests on subfamily II homologs, specifically the topology/relationship among clades of plant homologs. Four alternative topologies were evaluated, constraining different clades of plant homologs to be monophyletic: 1) All plant subfamily II homologs, 2) PKL, PKR1, and MOM1, 3) PKR4, PKR1, and MOM1, and 4) PKR4 and PKL. RELL approximation (Kishino et al. 1990) was used to determine whether any of the constrained trees were significantly worse than the unconstrained tree and could be rejected (supplementary table S3, Supplementary Material online).
Ohnolog Detection
To determine whether human CHD paralogs were derived from WGD, we used the OHNOLOGS v2 database (Singh and Isambert 2020). For all other species, regions of synteny were first detected using SynMap2 on the online Comparative Genomics Platform (CoGe; https://genomevolution.org/coge/) using the CoGe recommended genome for each species. SynMap2 default settings were used with the exception that the merge syntenic blocks algorithm was set to Quota Align Merge and the syntenic depth algorithm was set to Quota Align. CHD paralogs of interest were checked to see if they resided within syntenic blocks.
Supplementary Material
Acknowledgments
The authors thank members of the Wisecaver lab for helpful discussions. This work was conducted in part using the resources of the Rosen Center for Advanced Computing at Purdue University. This work was supported by the National Science Foundation (http://www.nsf.gov) under grants IOS-1401682 to J.T.T., MCB-1951698 to J.O., and DEB-1831493 to J.H.W. The authors gratefully acknowledge the Walther Cancer Foundation and support from the Purdue University Center for Cancer Research, P30CA023168.
Contributor Information
Joshua T. Trujillo, Center for Plant Biology and Department of Biochemistry, Purdue University, West Lafayette, Indiana 47907, USA
Jiaxin Long, Center for Plant Biology and Department of Biochemistry, Purdue University, West Lafayette, Indiana 47907, USA.
Erin Aboelnour, Center for Plant Biology and Department of Biochemistry, Purdue University, West Lafayette, Indiana 47907, USA; Helmholtz Pioneer Campus, Helmholtz Zentrum München, 85764 Neuherberg, Germany.
Joseph Ogas, Center for Plant Biology and Department of Biochemistry, Purdue University, West Lafayette, Indiana 47907, USA.
Jennifer H. Wisecaver, Center for Plant Biology and Department of Biochemistry, Purdue University, West Lafayette, Indiana 47907, USA
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Data Availability
All sequence alignments, tree files, and custom PFAM and IUPRED-based domain hmms are available through FigShare (https://doi.org/10.6084/m9.figshare.19350698.v1). Scripts are available through GitHub (https://github.com/JenWisecaver/CHD_evolution). iTOL phylogenies can be viewed online at: https://itol.embl.de/shared/WisecaverLab. The custom protein database used in this analysis is available from the authors as well as through the following link: https://www.datadepot.rcac.purdue.edu/jwisecav/custom-refseq/2020-02-15/.
Literature Cited
- Abi-Rached L, Gilles A, Shiina T, Pontarotti P, Inoko H. 2002. Evidence of en bloc duplication in vertebrate genomes. Nat Genet. 31:100–105. [DOI] [PubMed] [Google Scholar]
- Alendar A, Berns A. 2021. Sentinels of chromatin: chromodomain helicase DNA-binding proteins in development and disease. Genes Dev. 35:1403–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Religa TL, Freund SMV, Bycroft M. 2007. Solution structure of the BRK domains from CHD7. J Mol Biol. 371:1135–1140. [DOI] [PubMed] [Google Scholar]
- Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. 2000. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405:203–206. [DOI] [PubMed] [Google Scholar]
- Armache JP, et al. 2019. Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. eLife 8:e46057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlow RB, Dyson HJ, Wright PE. 2018. Expanding the paradigm: intrinsically disordered proteins and allosteric regulation. J Mol Biol. 430:2309–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman EA, et al. 2005. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet. 14:3463–3476. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438. [DOI] [PubMed] [Google Scholar]
- Brunet T, et al. 2019. Light-regulated collective contractility in a multicellular choanoflagellate. Science 366:326–334. [DOI] [PubMed] [Google Scholar]
- Čaikovski M, et al. 2008. Divergent evolution of CHD3 proteins resulted in MOM1 refining epigenetic control in vascular plants. PLOS Genet. 4:e1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B, et al. 2016. Cross-talk between sporophyte and gametophyte generations is promoted by CHD3 chromatin remodelers in Arabidopsis thaliana. Genetics 203:817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B, et al. 2018. The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in Arabidopsis. Plant Cell 30:1337–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B, et al. 2021. Thermodynamic models for assembly of intrinsically disordered protein hubs with multiple interaction partners. J Am Chem Soc. 143:12509–12523. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem. 78:273–304. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, Peterson CL. 2017. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 18:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese MS, Uversky VN, Dunker AK. 2008. Intrinsic disorder in scaffold proteins: getting more from less. Prog Biophys Mol Biol 98:85–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dieuleveult M, et al. 2016. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature 530:113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3:e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle R, et al. 2015. Bacterial proteins pinpoint a single eukaryotic root. Proc Natl Acad Sci U S A 112:E693–E699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform. 23:205–211. [PubMed] [Google Scholar]
- Edger PP, Chris Pires J. 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17:699–717. [DOI] [PubMed] [Google Scholar]
- Edger PP, Pires JC. 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17:699–717. [DOI] [PubMed] [Google Scholar]
- Egan CM, et al. 2013. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell 26:223–236. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdős G, Pajkos M, Dosztányi Z. 2021. IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 49:W297–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, et al. 2015. The prenucleosome, a stable conformational isomer of the nucleosome. Genes Dev. 29:2563–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DMA, Barton GJ, Owen-Hughes T. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34:2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, et al. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. 2011. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 12:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawryluk RMR, et al. 2019. Non-photosynthetic predators are sister to red algae. Nature 572:240–243. [DOI] [PubMed] [Google Scholar]
- Gkikopoulos T, et al. 2011. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333:1758–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau-Bové X, et al. 2021. Comparative proteogenomics deciphers the origin and evolution of eukaryotic chromatin. 2021.11.30.470311. Available from: https://www.biorxiv.org/content/10.1101/2021.11.30.470311v1.
- Han Y-F, et al. 2016. The SUMO E3 ligase-like proteins PIAL1 and PIAL2 interact with MOM1 and form a novel complex required for transcriptional silencing. Plant Cell 28:1215–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk G, McKnight JN, Nodelman IM, Bowman GD. 2010. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell 39:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig BP, Bendrin K, Zhou Y, Fischer T. 2012. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep. 13:997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463:474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KK, Zhang H, Golden BL, Ogas J. 2013. PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim Biophys Acta 1829:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, et al. 2013. Analysis of rice Snf2 family proteins and their potential roles in epigenetic regulation. Plant Physiol Biochem. 70:33–42. [DOI] [PubMed] [Google Scholar]
- Hu Y, Lai Y, Zhu D. 2014. Transcription regulation by CHD proteins to control plant development. Front Plant Sci. 5:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, et al. 2017. Mutants in the imprinted PICKLE RELATED 2 gene suppress seed abortion of fertilization independent seed class mutants and paternal excess interploidy crosses in Arabidopsis. Plant J. 90:383–395. [DOI] [PubMed] [Google Scholar]
- Huang Q, Li M, Lai L, Liu Z. 2020. Allostery of multidomain proteins with disordered linkers. Curr Opin Struct Biol. 62:175–182. [DOI] [PubMed] [Google Scholar]
- Hughes AL. 1994. The evolution of functionally novel proteins after gene duplication. Proc R Soc Lond. Series B: Biol Sci. 256:119–124. [DOI] [PubMed] [Google Scholar]
- Hurd EA, et al. 2007. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome 18:94–104. [DOI] [PubMed] [Google Scholar]
- Irisarri I, Strassert JFH, Burki F. 2022. Phylogenomic insights into the origin of primary plastids. Syst Biol. 71:105–120. [DOI] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M. 2006. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell 23:733–742. [DOI] [PubMed] [Google Scholar]
- Jin YH, et al. 1998. Isolation and characterization of hrp1+, a new member of the SNF2/SWI2 gene family from the fission yeast Schizosaccharomyces pombe. Mol Genet Genomics 257:319–329. [DOI] [PubMed] [Google Scholar]
- Jing Y, et al. 2013. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25:242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job G, et al. 2016. SHREC silences heterochromatin via distinct remodeling and deacetylation modules. Mol Cell 62:207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolla V, Zhuang T, Higashi M, Naraparaju K, Brodeur GM. 2014. Role of CHD5 in human cancers: 10 years later. Cancer Res. 74:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konev AY, et al. 2007. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 317:1087–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MJE, Snel B, Timmers HTM. 2015. Genesis of chromatin and transcription dynamics in the origin of species. Cell 161:724–736. [DOI] [PubMed] [Google Scholar]
- Kunert N, Brehm A. 2009. Novel Mi-2 related ATP-dependent chromatin remodelers. Epigenetics 4:209–211. [DOI] [PubMed] [Google Scholar]
- Leebens-Mack JH, et al. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574:679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47:W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. 1951. Pseudoallelism and gene evolution. Cold Spring Harbor Symp Quant Biol. 16:159–174. [DOI] [PubMed] [Google Scholar]
- Li M, Cao H, Lai L, Liu Z. 2018. Disordered linkers in multidomain allosteric proteins: Entropic effect to favor the open state or enhanced local concentration to favor the closed state? Protein Sci. 27:1600–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kang N, Guo Y, Gong P. 2021. Advances in chromodomain helicase DNA-binding (CHD) proteins regulating stem cell differentiation and human diseases. Front Cell Dev Biol. 9:710203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A, Urwin DL, Kadonaga JT. 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol. 12:160–166. [DOI] [PubMed] [Google Scholar]
- Mansfield RE, et al. 2011. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem. 286:11779–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella CGA, et al. 2006. Mutation of the SNF2 family member Chd2 affects mouse development and survival. J Cell Physiol. 209:162–171. [DOI] [PubMed] [Google Scholar]
- Marfella CGA, Imbalzano AN. 2007. The Chd family of chromatin remodelers. Mutat Res. 618:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N, et al. 2014. Data access for the 1,000 Plants (1KP) project. GigaScience 3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawska M, et al. 2008. dCHD3, a novel ATP-dependent chromatin remodeler associated with sites of active transcription. Mol Cell Biol. 28:2745–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Lalonde M-E, Côté J, Kutateladze TG. 2012. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 19:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan P, et al. 2009. Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene 28:1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P, et al. 2008. BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol Cell Biol. 28:6720–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, et al. 2009. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol. 11:172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodelman IM, Bowman GD. 2021. Biophysics of chromatin remodeling. Annu Rev Biophys. 50:73–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer Berlin Heidelberg. [Google Scholar]
- Ohno S, Wolf U, Atkin NB. 1968. Evolution from fish to mammals by gene duplication. Hereditas 59:169–187. [DOI] [PubMed] [Google Scholar]
- Ojolo SP, et al. 2018. Regulation of plant growth and development: a review from a chromatin remodeling perspective. Front Plant Sci. 9:1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary NA, et al. 2016. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucl Acids Res. 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S, et al. 2015. A critical role for the chromatin remodeller CHD7 in anterior mesoderm during cardiovascular development. Dev Biol. 405:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. Fasttree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter DJ, Fozouni P, Eisen MB, King N. 2018. Gene family innovation, conservation and loss on the animal stem lineage. eLife 7:e34226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez D, et al. 2015. Mutations in CHD2 cause defective association with active chromatin in chronic lymphocytic leukemia. Blood 126:195–202. [DOI] [PubMed] [Google Scholar]
- Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T. 2011. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains: Identification of SANT and SLIDE domains in Chd1. EMBO J. 30:2596–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz MP, et al. 2010. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLOS Genet. 6:e1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön ME, et al. 2021. Single cell genomics reveals plastid-lacking Picozoa are close relatives of red algae. Nat Commun. 12:6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Jenkins KR, Héroux A, Bowman GD. 2011. Crystal structure of the chromodomain helicase DNA-binding protein 1 (Chd1) DNA-binding domain in complex with DNA. J Biol Chem. 286:42099–42104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, et al. 2015. NSD3-Short Is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell 60:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggens L, Cordeddu L, Rönnerblad M, Lennartsson A, Ekwall K. 2015. Transcription-coupled recruitment of human CHD1 and CHD2 influences chromatin accessibility and histone H3 and H3.3 occupancy at active chromatin regions. Epigenet Chromatin 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ 3rd, et al. 2005. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 280:41789–41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JK, Wade PA. 2011. SnapShot: chromatin remodeling: CHD. Cell 144:626–626.e1. [DOI] [PubMed] [Google Scholar]
- Singh PP, Isambert H. 2020. OHNOLOGS v2: a comprehensive resource for the genes retained from whole genome duplication in vertebrates. Nucl Acids Res. 48:D724–D730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Hernandez AE, Groudine M, Henikoff S. 2014. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. Elife 3:e02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle M, et al. 2012. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol. 19:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice AK, et al. 2021. PhyloFisher: a phylogenomic package for resolving eukaryotic relationships. PLOS Biol. 19:e3001365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. 2002. Intrinsically unstructured proteins. Trends Biochem Sci. 27:527–533. [DOI] [PubMed] [Google Scholar]
- Torigoe SE, Patel A, Khuong MT, Bowman GD, Kadonaga JT. 2013. ATP-dependent chromatin assembly is functionally distinct from chromatin remodeling. Elife 2:e00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. 2015. The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett. 589:2498–2506. [DOI] [PubMed] [Google Scholar]
- Vaillant I, Schubert I, Tourmente S, Mathieu O. 2006. MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep. 7:1273–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe M, Di Franco A, Philippe H, Baurain D. 2021. Decontamination, pooling and dereplication of the 678 samples of the Marine Microbial Eukaryote Transcriptome Sequencing Project. BMC Res Notes 14:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AA, et al. 2012. The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4. J Mol Biol. 422:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. 1997. Characterization of the CHD family of proteins. Proc Natl Acad Sci U S A 94:11472–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. 2015. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 16:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav T, Whitehouse I. 2016. Replication-coupled nucleosome assembly and positioning by ATP-dependent chromatin-remodeling enzymes. Cell Rep. 15:715–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yoo EJ, et al. 2002. Hrp3, a chromodomain helicase/ATPase DNA binding protein, is required for heterochromatin silencing in fission yeast. Biochem Biophys Res Commun. 295:970–974. [DOI] [PubMed] [Google Scholar]
- Zentner GE, et al. 2010. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum Mol Genet. 19:3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tsukiyama T, Henikoff S. 2013. ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet. 9:e1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. 2008. The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J Biol Chem. 283:22637–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bishop B, Ringenberg W, Muir WM, Ogas J. 2012. The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159:418–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence alignments, tree files, and custom PFAM and IUPRED-based domain hmms are available through FigShare (https://doi.org/10.6084/m9.figshare.19350698.v1). Scripts are available through GitHub (https://github.com/JenWisecaver/CHD_evolution). iTOL phylogenies can be viewed online at: https://itol.embl.de/shared/WisecaverLab. The custom protein database used in this analysis is available from the authors as well as through the following link: https://www.datadepot.rcac.purdue.edu/jwisecav/custom-refseq/2020-02-15/.