Abstract
Candida auris proliferates and persists on the skin of patients, often leading to health care-associated infections with high mortality. Here, we describe 2 clinically relevant skin models and show that C. auris grows similarly on human and porcine skin. Additionally, we demonstrate that other Candida spp., including those with phylogenetic similarity to C. auris, do not display high growth in the skin microenvironment. These studies highlight the utility of 2 ex vivo models of C. auris colonization that allow reproducible differentiation among Candida spp., which should be a useful tool for comparison of C. auris clinical isolates and genetically mutated strains.
Keywords: Candida, skin, colonization, porcine, human, ex vivo, biofilm, microscopy, Candida auris
Candida aurisefficiently colonizes the skin of patients in health care settings, often leading to nosocomial outbreaks. We show that human and porcine ex vivo skin models support high-burden colonization for C. aurisbut not closely related species.
First isolated from the ear canal of a patient in 2009, Candida auris is now a major cause of nosocomial infection [1]. C. auris is an urgent public health threat due to the high mortality rate associated with invasive infection, as well as its propensity for rapid spread in health care settings [2, 3]. Major factors contributing to hospital transmission include the ability of C. auris to efficiently colonize patient skin and its capacity to persist on medical devices and hospital surfaces [3, 4]. Interestingly, persistent and long-term skin colonization appears to be a trait relatively unique to C. auris, as many other disease-causing Candida species typically colonize the gastrointestinal tract and do not cause hospital outbreaks. C. auris skin colonization models are necessary for understanding this unique characteristic and identifying mechanisms that promote colonization.
In this work, we aimed to develop a model of C. auris colonization of human skin ex vivo. We based the model on recent description of a porcine skin model of C. auris colonization [5, 6]. Porcine skin is frequently utilized as a model for human skin, as human and porcine skin exhibit many similarities, including the presence of skin layers of comparable thickness and a developed superficial vascular system [7, 8]. In addition, human and porcine skin have similar cell types and repair mechanisms [7]. Miniature swine more closely model human skin than conventional swine, which are larger with higher muscle mass. Despite these many similarities between the skins of humans and pigs, some key differences exist. For example, porcine skin is marked by the absence of eccrine sweat glands, smaller sebaceous glands, fewer pigment cells, and fewer elastic fibers when compared to human skin [7]. Therefore, here we aimed to develop a human skin colonization model and assess if the human and porcine models could distinguish the high capacity of C. auris for skin colonization from other species exhibiting poor skin colonization.
METHODS
Organisms and Inoculum
Strains (Supplementary Table 1) were maintained on yeast extract-peptone-dextrose (YPD) agar plates, grown overnight in YPD broth on an orbital shaker at 30°C, counted by hemocytometer, and adjusted to 106 or 107 cells/mL in synthetic sweat medium without fatty acids [5, 9].
Human Skin Model
Human skin samples were collected from patients who underwent reconstructive surgeries through an institutional review board-exempt protocol. Full-thickness punch biopsies were harvested from excised skin and placed in 12-well plates containing 3 mL of Dulbecco’s Modified Eagle Medium (DMEM; Lonza), supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals), penicillin (1000 U/mL), and streptomycin (1 mg/mL) [5]. After 6 hours, samples were washed with Dulbecco's phosphate buffered saline (DPBS) and moved to 12-well plates containing semisolid media (6:4 ratio of 1% agarose [BIO-RAD] in DPBS and DMEM with 10% FBS). Paraffin wax was applied as a barrier between the epidermal surface and the semisolid media. Candida spp. (10 µL at 107 cells/mL) were applied to the skin surface. Skin samples were incubated at 37°C for 24 hours and processed for viable burden determination or microscopy.
Porcine Skin Model
Porcine skin was obtained following protocols approved by the University of Wisconsin-Madison Institutional Animal Care and Use Committee, under published guidelines from the National Institutes of Health and United States Department of Agriculture. Animals were euthanized and full-thickness punch biopsies were harvested from the excised skin. Samples were processed following the protocols listed in human skin studies [5, 6].
Scanning Electron Microscopy
Skin samples were fixed overnight (4% formaldehyde, 1% glutaraldehyde, in sodium phosphate buffer), washed with sodium phosphate buffer, and dehydrated through a series of ethanol rinses [5]. After critical point drying, samples were mounted on aluminum stubs and silver paint was applied around the edge of the samples. Samples were then platinum sputter coated and imaged by scanning electron microscopy (Zeiss Gemini 450, 3 kV).
In Vitro Biofilm Burden
Organisms were suspended in synthetic sweat medium at 106 cells/mL and 200 µL was added to flat-bottom 96-well microtiter plates. Following a 24-hour incubation at 37°C, wells were rinsed with DPBS. Then, 200 µL of DBPS was added to each well, and biofilm burdens were estimated by microplate reader (Synergy H1, Bio-Tek Instruments, 600 nm) as previously described [5].
RESULTS
Establishment of an Ex Vivo Human Skin Model of C. auris Colonization
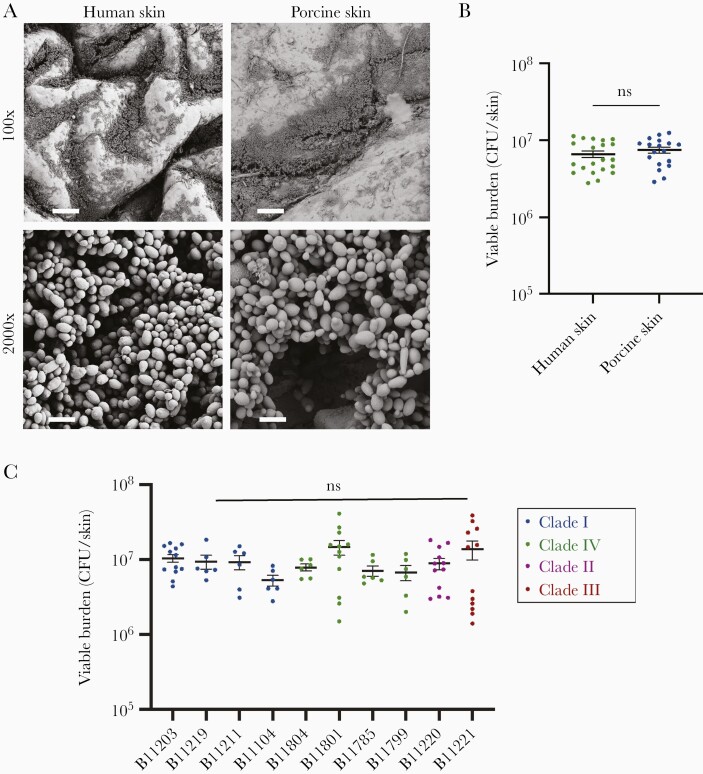
For ex vivo human skin studies, we utilized skin obtained from patients following elective reconstructive operations. We obtained full-thickness samples by punch biopsy and placed samples in semisolid media with a paraffin layer to separate the media from the epidermal surface, a design similar to an ex vivo porcine skin model [5]. We then applied C. auris suspended in synthetic sweat media to the epidermal surface, incubated the samples for 24 hours, and utilized scanning electron microscopy to visualize C. auris skin colonization. We observed diffuse growth across the entire skin surface with more abundant colonization within the skin crevices. On comparison with porcine skin, human skin contained more skin folds and irregularities across the epidermal surface (Figure 1A). Despite this difference, growth patterns for C. auris on human and porcine skin were similar, with widespread colonization and dense multilayer biofilms in the crevices. Although C. auris was adherent to the epidermal surface, the skin appeared intact, suggesting that C. auris did not invade skin tissue, consistent with previous reports [5]. Congruent with imaging, C. auris grew to nearly equivalent viable burdens in the ex vivo models (Figure 1B).
Figure 1.
Candida auris similarly colonizes pig and human skin. A, C. auris (B11203) was grown for 24 hours on porcine or human skin and imaged with scanning electron microscopy. Bars represent 100 µm and 5 µm for 100 ×, and 2000 × magnifications, respectively. B, C. auris was grown on porcine or human skin for 24 hours and colonization was assessed by viable burden via CFU counts. Data were analyzed by Student t test; mean with SEM are shown. C, C. auris isolates from 4 geographic clades were grown on porcine skin for 24 hours and colonization was assessed by viable burden via CFU counts. Data were analyzed via 1-way ANOVA with Holm-Sidak multiple comparisons, comparing all isolates to B11203; mean with are SEM shown. Abbreviations: CFU, colony-forming unit; ns, not significant; SEM, standard error of the mean.
Isolates cluster into distinct geographic clades, with greater levels of genetic diversity between clades than within individual clades [1, 10, 11]. C. auris isolates often display significant phenotypic diversity, including differences in virulence in animal models, capacities for biofilm formation, and patterns of drug resistance [10, 11]. While most isolates grow as individual yeast cells, some isolates clump together, propagating as large aggregates, a phenotype that has been shown to correlate with decreased virulence [11]. In light of the diversity of C. auris isolates, we considered if additional C. auris isolates would similarly colonize skin. We utilized a strain collection from the Centers for Disease Control that contains at least 1 isolate from each of 4 clades, as well as both aggregating and nonaggregating strains (Supplementary Figure 1). We examined the growth of these isolates on skin, comparing them to B11203, a clinical isolate of the South Asian clade used in Figure 1A and 1B studies and prior work [5, 6]. We found the isolates to propagate to similar viable burdens on porcine skin with no statistical differences compared to B11203 (Figure 1C). We further assessed C. auris skin colonization by histopathology, examining 1 representative isolate from each clade, and observed robust colonization of skin (Supplementary Figure 2).
Human and Porcine Skin Models Discriminate High-Burden C. auris Colonization From Poor Skin Colonization
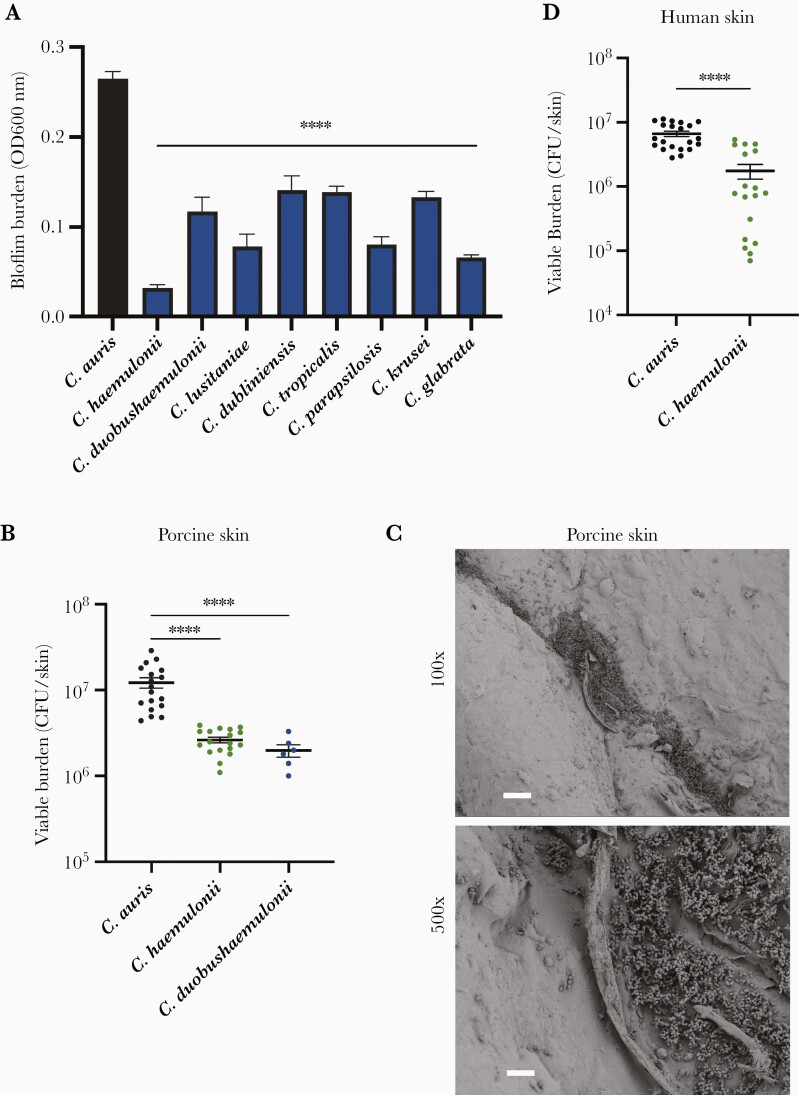
C. auris exhibits a unique capacity for growth in skin niche conditions, which promotes patient-to-patient spread in health care settings [3]. To assess if this trait could be replicated, we utilized in vitro and ex vivo skin models. We first examined clinically relevant Candida spp., including genetically similar species of the Candida haemulonii complex, C. haemulonii and Candida duobushaemulonii, in vitro using synthetic sweat media [12] (Supplementary Table 1). Strikingly, none of the Candida species were able to form biofilms close to the same burden as C. auris (Figure 2A). This is consistent with prior work showing Candida albicans to lack the ability to form biofilms in skin niche conditions [5]. We found variability in the biofilm-forming capacity among the strains, with non-auris Candida spp. forming biofilms with burdens approximately 10% to 50% of C. auris biofilm burdens (Figure 2A). The closely related C. haemulonii grew to the lowest burden of all the species, nearly 10-fold lower than C. auris. We considered the possibility that C. auris may have an overall higher replication rate. However, while C. auris propagated well in synthetic media, it was relatively slow growing in rich medium (YPD), indicating that the enhanced growth of C. auris is specific to the skin microenvironment (Supplementary Figure 3).
Figure 2.
Skin models distinguish Candida auris from poor fungal colonizers. A, Candida species were grown in synthetic sweat medium for 24 hours and biofilm burden was estimated by absorbance at 600 nm. Data were analyzed by 1-way ANOVA with Holm-Sidak multiple comparisons to C. auris, Mean with SEM are shown, n = 3. B, C. auris, C. haemulonii, and C. duobushaemulonii were grown on porcine skin for 24 hours and viable burden was determined via CFU counts. Data were analyzed by Student t test. C, C. haemulonii was grown on porcine skin for 24 hours and imaged with scanning electron microscopy. Bars represent 100 µm for 100 × and 20 µm for 500 × magnifications. D, C. auris and C. haemulonii were grown on human skin for 24 hours and viable burden was determined via CFU counts. Data were analyzed by Student t test; mean with SEM are shown, n = 3. C. auris B11203, C. haemulonii CAU-13, and C. duobushaemulonii CAU-11 were used (Supplementary Table 1). **** P < .0001. Abbreviations: CFU, colony-forming unit; OD, optical density; SEM, standard error of the mean.
We next assessed the ability of the porcine and human skin models to distinguish the high capacity of C. auris to colonize skin from other Candida spp. that do not typically propagate to high levels on skin or spread rapidly in nosocomial settings. We selected C. haemulonii and C. duobushaemulonii for analysis. After 24 hours of growth on pig skin, we found that viable burdens for these 2 species were approximately 5 to 10-fold lower than C. auris (Figure 2B). For C. haemulonii, we examined skin colonization by scanning electron microscopy. We found sparse skin colonization with fungal growth limited to only a few areas of skin folds (Figure 2C). These areas of colonization by C. haemulonii appeared less dense than the multilayer biofilms observed for C. auris on porcine skin (Figure 1A). For C. haemulonii, we also examined the capacity for human skin colonization. Similar to pig skin colonization studies, C. haemulonii grew to a significantly lower burden, with viable burdens on the human skin mirroring the results of pig skin (Figure 2D). We also considered genetic variability among isolates and tested 1 additional C. haemulonii isolate (CAU-15) and 2 additional C. duobushaemulonii isolates (CAU-12 and CAU-14) (Supplementary Table 1). We observed similar growth patterns for these isolates (Supplementary Figure 4).
DISCUSSION
It is critical to understand the propensity of C. auris for skin colonization to eradicate colonization, prevent hospital spread, and reduce invasive infections. Colonization studies require models that recapitulate high burden C. auris growth on the skin of patients. Here, we describe 2 ex vivo models of skin colonization. On comparison of porcine and human skin, we found that, despite some differences between these skin types, C. auris colonizes both similarly, including isolates from 4 clades. This finding supports porcine skin and/or human skin as relevant models for study of C. auris skin colonization of patients. On both skin types, we observed C. auris broadly colonizing the entire skin surface, with higher-burden areas in the skin folds. Both models effectively modeled the high-burden colonization of C. auris, differentiating it from the closely related C. haemulonii, which does not colonize skin as effectively as C. auris.
It is ideal to have multiple options for skin colonization models, as each have advantages and limitations. One advantage of porcine skin is that pigs can be genetically manipulated and bred for particular traits. Additionally, skin can be obtained from selected areas of the pig, allowing for studies examining specific colonization sites. This is a limitation of human skin models, as the source is typically restricted to skin obtained during elective procedures that involve excision of abdominal adipose tissue. Adipose tissue has been shown to impact the skin microbiome and is linked to inflammatory skin disease, such as psoriasis [13, 14]. Therefore, skin from these sites may not fully represent the skin of patients. Despite this limitation, human skin does not have the anatomical differences of pig skin. However, as both of these ex vivo models are similar for Candida colonization, tissue availability may guide model selection. While the current studies utilize ex vivo models of skin colonization, pigs could also be used for in vivo analysis of skin colonization, including studies examining the influence of microbiome, immunity, pharmacologic modulation, and a variety of other factors. Murine models may also be selected for in vivo studies to assess colonization burden considering their smaller size and available genetic tools [15]. However, the skin of rodents differs considerably from human skin with regard to skin thickness, skin layers, cell types, and repair mechanisms [7, 8]. Therefore, the porcine skin may serve as closer model of human tissue with human skin providing an optimal patient-level validation.
Overall, these studies effectively model clinical observations of an enhanced capacity for C. auris to grow on skin beyond that observed for other Candida species. The multiple skin models represent important tools for uncovering the mechanisms by which C. auris flourishes in skin niche conditions. Additional understanding of C. auris colonization is needed to identify strategies to limit, reduce, or eradicate C. auris on skin of patients.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the National Institutes of Health (grant numbers R01AI145939 and R21AI159583); the Burroughs Wellcome Fund (grant number 1012299 to J. N.); and the National Science Foundation graduate research fellowship program (grant number DGE-1747503 to E. E.).
Potential conflicts of interest . All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 2016; 64:134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rudramurthy SM, Chakrabarti A, Paul RA, et al. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother 2017; 72:1794–801. [DOI] [PubMed] [Google Scholar]
- 3. Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 2016; 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergeron G, Bloch D, Murray K, et al. Candida auris colonization after discharge to a community setting: New York City, 2017–2019. Open Forum Infect Dis 2020; 8:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Horton MV, Johnson CJ, Kernien JF, et al. Candida auris forms high-burden biofilms in skin niche conditions and on porcine skin. mSphere 2020; 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson CJ, Eix EF, Lam BC, et al. Augmenting the activity of chlorhexidine for decolonization of Candida auris from porcine skin. J Fungi (Basel) 2021; 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y, Chen JY, Shang HT, et al. Light microscopic, electron microscopic, and immunohistochemical comparison of Bama minipig (Sus scrofa domestica) and human skin. Comp Med 2010; 60:142–8. [PMC free article] [PubMed] [Google Scholar]
- 8. Summerfield A, Meurens F, Ricklin ME.. The immunology of the porcine skin and its value as a model for human skin. Mol Immunol 2015; 66:14–21. [DOI] [PubMed] [Google Scholar]
- 9. Callewaert C, Buysschaert B, Vossen E, Fievez V, Van de Wiele T, Boon N.. Artificial sweat composition to grow and sustain a mixed human axillary microbiome. J Microbiol Methods 2014; 103:6–8. [DOI] [PubMed] [Google Scholar]
- 10. Larkin E, Hager C, Chandra J, et al. The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob Agents Chemother 2017; 61:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown JL, Delaney C, Short B, et al. Candida auris phenotypic heterogeneity determines pathogenicity in vitro. mSphere 2020; 5:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muñoz JF, Gade L, Chow NA, et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun 2018; 9:5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hegde V, Dhurandhar NV.. Microbes and obesity—interrelationship between infection, adipose tissue and the immune system. Clin Microbiol Infect 2013; 19:314–20. [DOI] [PubMed] [Google Scholar]
- 14. Wong Y, Nakamizo S, Tan KJ, Kabashima K.. An update on the role of adipose tissues in psoriasis. Front Immunol 2019; 10:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang X, Hurabielle C, Drummond RA, et al. Murine model of colonization with fungal pathogen Candida auris to explore skin tropism, host risk factors and therapeutic strategies. Cell Host Microbe 2021; 29:210–21.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.