Abstract
Various alkylbenzenes were depleted during growth of an anaerobic, sulfate-reducing enrichment culture with crude oil as the only source of organic substrates. From this culture, two new types of mesophilic, rod-shaped sulfate-reducing bacteria, strains oXyS1 and mXyS1, were isolated with o-xylene and m-xylene, respectively, as organic substrates. Sequence analyses of 16S rRNA genes revealed that the isolates affiliated with known completely oxidizing sulfate-reducing bacteria of the δ subclass of the class Proteobacteria. Strain oXyS1 showed the highest similarities to Desulfobacterium cetonicum and Desulfosarcina variabilis (similarity values, 98.4 and 98.7%, respectively). Strain mXyS1 was less closely related to known species, the closest relative being Desulfococcus multivorans (similarity value, 86.9%). Complete mineralization of o-xylene and m-xylene was demonstrated in quantitative growth experiments. Strain oXyS1 was able to utilize toluene, o-ethyltoluene, benzoate, and o-methylbenzoate in addition to o-xylene. Strain mXyS1 oxidized toluene, m-ethyltoluene, m-isoproyltoluene, benzoate, and m-methylbenzoate in addition to m-xylene. Strain oXyS1 did not utilize m-alkyltoluenes, whereas strain mXyS1 did not utilize o-alkyltoluenes. Like the enrichment culture, both isolates grew anaerobically on crude oil with concomitant reduction of sulfate to sulfide.
Sulfate-reducing bacteria are an important group of anaerobes in the global carbon and sulfur cycle. Oxidation of organic compounds coupled to the reduction of sulfate to sulfide may account for more than 50% of carbon mineralization in marine sediments (21). In oil field waters, however, the activity of sulfate-reducing bacteria may be detrimental. Their product, hydrogen sulfide, is toxic and corrosive, increases the sulfur content of oil and gas, and leads to the precipitation of ferrous sulfide, which plugs oil-bearing strata and stabilizes undesirable oil-water emulsions (8, 19, 26). Recently it was demonstrated that in a mesophilic (around 30°C) enrichment culture, sulfate-reducing bacteria can utilize various alkylbenzenes directly from crude oil (31, 34). Besides alkanes, alkylbenzenes and other aromatic hydrocarbons are major constituents of crude oil (40). A moderately thermophilic (≤65°C) sulfate-reducing isolate grew by consumption of n-alkanes from oil (34). Hence, oil hydrocarbons were believed to serve as growth substrates for sulfate-reducing bacteria under in situ conditions in oil reservoirs, at least at temperatures not higher than the optima of the respective cultures. More recent measurements with enrichment cultures under sulfate-reducing conditions at 50 to 70°C with added aromatic hydrocarbons revealed partial consumption, especially of xylenes and ethylbenzene (5). This suggested that anaerobic degradation of aromatic hydrocarbons, coupled to sulfate reduction, in oil reservoirs is in principle also possible under thermophilic conditions. Also, extremely thermophilic (85°C) sulfate-reducing members of the Archaea have been detected in oil reservoirs (22, 38); however, utilization of hydrocarbons by these organisms has not been demonstrated.
16S rRNA-targeted oligonucleotide probing of the mesophilic enrichment culture using alkylbenzenes from crude oil revealed that the major part of the bacterial population belonged to a cluster (suggested family, Desulfobacteriaceae [42]) that comprises known species of completely oxidizing sulfate-reducing bacteria within the δ subclass of the class Proteobacteria (31). Besides toluene, the enrichment culture also consumed o-xylene and m-xylene (31, 34). This paper reports on the isolation of two new types of sulfate-reducing bacteria from the enrichment culture that are able to grow on o-xylene or m-xylene. This degradative capacity has not been observed so far in pure cultures of sulfate-reducing bacteria. Among aromatic hydrocarbons, only toluene was shown before to serve as growth substrate for isolated strains of sulfate-reducing bacteria (2, 32). Anaerobic growth of pure cultures on m-xylene has been shown with denitrifying bacteria (9, 15, 20, 28, 36). However, to our knowledge, no strain of any type of anaerobic bacterium that grows on o-xylene or p-xylene has been isolated to date. In some instances, pure cultures converted these xylenes cometabolically to aromatic dead-end metabolites (2, 3, 13, 29). In contrast, enriched bacterial populations (microcosms and consortia) have been repeatedly shown to utilize the isomers of xylene (5, 10, 11, 12, 18, 45).
MATERIALS AND METHODS
Sources of bacteria.
The new types of sulfate-reducing bacteria were isolated from a previously described mesophilic enrichment culture growing anaerobically with crude oil and sulfate in seawater medium (31, 34). The enrichment culture originated from the water phase of a North Sea oil tank in Wilhelmshaven, Germany. Desulfobacterium cetonicum (DSM 7267) was from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany.
Media, cultivation techniques, and quantitative growth experiments.
Techniques for preparation of media and for cultivation of sulfate-reducing bacteria under anoxic conditions were as described previously (42). Cultures were grown in defined bicarbonate-buffered, sulfide-reduced mineral medium, essentially having the same sodium, magnesium, potassium, calcium, chloride, and sulfate ion compositions as natural seawater (42). Butyl-rubber-sealed tubes (20 ml) containing 15 ml of medium under a headspace of N2-CO2 (90:10 [vol/vol]) were used for routine cultivation. Filter-sterilized (via solvent-resistant cellulose filters; pore size, 0.2 μm) hydrocarbons and aromatic alcohols, aldehydes, and ketones were diluted (0.5 to 5% [vol/vol]) in a carrier phase (0.5 ml per tube) of deaerated 2,2,4,4,6,8,8-heptamethylnonane to avoid the toxic effects of the pure substances (32). Anoxic, sterile crude oil (0.5 ml per cultivation tube) was added directly to the cultures without dilution in a carrier phase. The tubes with the overlaid insoluble hydrocarbon phases were incubated nearly horizontally to facilitate diffusion of substances into the aqueous medium. The orifices sealed with black rubber stoppers were kept somewhat below the surface of the medium to avoid adsorption of hydrophobic compounds from the overlying hydrocarbon phase by the rubber (1, 32). Heptamethylnonane and crude oil were deaerated, sterilized, and stored in a special flask under an atmosphere of N2 as described previously (1, 30). Water-soluble substrates were added from autoclaved or filter-sterilized aqueous stock solutions to yield the indicated concentrations.
Quantitative growth experiments with xylenes dissolved in a carrier phase (the concentrations are indicated in the descriptions of the individual experiments) were carried out by using flat glass bottles (250 and 500 ml) containing defined mineral medium (190 and 450 ml, respectively) and heptamethylnonane (5 and 15 ml, respectively) under a headspace of N2-CO2 (90:10 [vol/vol]). The bottles were incubated horizontally on a rotary shaker (65 rpm), while contact of the carrier phase with the stoppers was avoided, as described for cultivation in tubes.
All the chemicals used were analytical grade.
Isolation, purity control, and maintenance.
Strains oXyS1 and mXyS1 were isolated via repeated dilution in agar tubes (42). The agar was overlaid with 0.5 ml of heptamethylnonane containing 2% (vol/vol) o-xylene or m-xylene, respectively. The salinity (NaCl, MgCl2, MgSO4, and CaCl2) of the medium for preparation of the dilution series was increased by a factor of 1.5, so that mixing with the aqueous agar (6 ml of medium added to 3 ml of molten aqueous agar) yielded the same salt concentration as was present in the original liquid medium (42).
The purity of isolates was routinely checked by phase-contrast microscopy. In addition, cultures were supplied with yeast extract (0.5 g/liter) and glucose or fructose (both 5 mM) and examined microscopically.
For maintenance, strains oXyS1 and mXyS1 were grown on o-xylene and m-xylene, respectively, stored at 4°C, and transferred every 6 to 8 weeks.
Sequence analyses of 16S rRNA genes.
Genomic DNA of strain oXyS1 was extracted and a 16S rRNA gene sequence was amplified as described by Rainey et al. (33). The PCR products were sequenced directly. Analysis was carried out by Fred A. Rainey at the Deutsche Sammlung von Mikroorganismen und Zellkulturen. Genomic DNA of strain mXyS1 was extracted as described by Tsai and Olson (41). The 16S rRNA gene sequence was amplified with bacterial primers GM3F and GM4R (25). The amplified fragment was cloned into the pGM-T vector (Promega, Madison, Wis.). Plasmids were purified with the Wizard plasmid purification kit (Promega). Sequencing reactions were performed with the Taq Dye-Deoxy terminator cycle-sequencing kit (Applied Biosystems [ABI], Foster City, Calif.). Sequences were determined with an ABI 373S DNA sequencer as specified by the manufacturer.
New sequences were added to an alignment of about 5,300 homologous primary structures of bacterial 16S rRNA genes by using the alignment tool of the ARB program package (39). Similarity and distance matrices were calculated with the ARB-PHYL program of the same package. Phylogenetic trees were constructed by using subsets of data that included representative sequences of members of the δ subclass of the Proteobacteria (23). We used distance matrix, maximum-likelihood, and maximum-parsimony methods as implemented in the programs PHYLIP (14), ARB, and fastDNAml (23).
Chemical analyses.
For routine detection of sulfide formed in enrichment cultures, a simple test with Cu2+ ions yielding CuS was applied (7). Sulfide in quantitative growth experiments was determined colorimetrically by the methylene blue formation reaction as described previously (1, 6).
Xylenes dissolved in heptamethylnonane were measured by means of an Auto System gas chromatograph (Perkin-Elmer, Norwalk, Conn.) equipped with a PVMS 54 column (length, 50 m; inner diameter, 0.32 mm) and a flame ionization detector. H2 was used as the carrier gas at a flow rate of 1.7 ml · min−1. The temperature program was run from 80°C (2-min isotherm) to 120°C at 20°C · min−1 and then from 120°C (0.1-min isotherm) to 300°C at 40°C · min−1. The temperatures at the injection port and the detector were 250 and 350°C, respectively. Defined, freshly prepared solutions of xylenes in heptamethylnonane were used for calibration.
Xylenes in the aqueous phase were determined by a high-performance liquid chromatography system (Sykam, Gilching/Munich, Germany) equipped with a Spherisorb OD S2 reverse-phase column (250 by 5 mm); the eluent was an acetonitrile-water mixture (80:20 [vol/vol]). The flow rate was 1 ml · min−1, and the temperature of the column was 25°C (28). Detection was performed by UV absorption at 265 nm. Defined, freshly prepared solutions of xylenes in acetonitrile-water (80:20 [vol/vol]) were used as standards.
The G+C content of the DNA was determined by high-performance liquid chromatography as described previously (24). The analysis was carried out by Fred A. Rainey.
Enzymatic tests.
Anoxic preparation of cell extracts and enzyme assays were carried out as described previously (1, 32). Protein was quantified by the method of Bradford (4).
Nucleotide sequence accession numbers.
The sequences determined in this study are deposited under EMBL accession no. Y17286 (strain oXyS1) and AJ006853 (strain mXyS1).
RESULTS AND DISCUSSION
Isolation.
An anaerobic enrichment culture with crude oil as the only source of organic substrates and sulfate as the electron acceptor consumed toluene, o-xylene, m-xylene, o-ethyltoluene, m-ethyltoluene, m-propyltoluene, and m-isopropyltoluene, with concomitant production of sulfide (31, 34). In counting series involving agar dilutions with benzoate or overlaid with crude oil or toluene (in carrier phases), large numbers of colonies of sulfate-reducing bacteria were formerly obtained that were also able to grow with these substrates when transferred back to liquid medium (31). Bacteria growing with defined alkylbenzenes other than toluene were not counted during the initial study. In the present study, we therefore attempted to isolate sulfate-reducing bacteria from the enrichment culture growing with oil directly by using o-xylene and m-xylene as defined hydrocarbons for dilution series. These two compounds were chosen because they were consumed the most rapidly next to toluene. Furthermore, strains of sulfate-reducing bacteria utilizing o-xylene and m-xylene would present novel metabolic types; their study would add to our knowledge of the nutritional and phylogenetic diversity of sulfate-reducing bacteria that degrade hydrocarbons. Dilution series from the enrichment culture were carried out by using agar overlaid with o-xylene and m-xylene in a carrier phase. The largest colonies developed next to the overlying hydrocarbon phase. Pure cultures were isolated by a second agar dilution series. One strain isolated with o-xylene and one strain isolated with m-xylene were chosen for further investigations. These strains were designated oXyS1 and mXyS1, respectively.
Morphological and other characteristics.
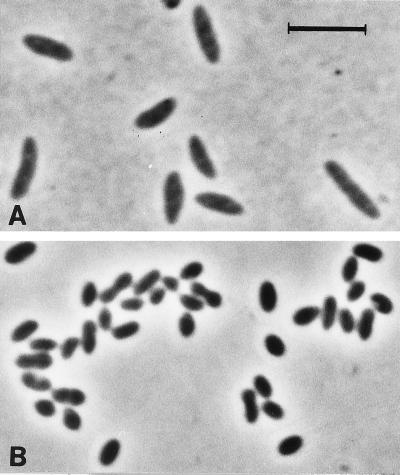
The cells of both new isolates were rod shaped. The cells of strain oXyS1 were more elongated than those of strain mXyS1 (Fig. 1). The dimensions were 0.8 to 1.0 by 2.5 to 4.0 μm for strain oXyS1 and 0.6 to 1.0 by 1 to 2 μm for strain mXyS1.
FIG. 1.
Phase-contrast photomicrographs of newly isolated sulfate-reducing bacteria utilizing xylenes. (A) Strain oXyS1 isolated with o-xylene. (B) Strain mXyS1 isolated with m-xylene. Bar, 5 μm.
Growth of strain oXyS1 was observed within a temperature range of 15 to 35°C, with an optimum around 32°C, and within a pH range of 6.2 to 7.9, with an optimum around 7.5. Growth of strain mXyS1 occurred within a temperature range of 16 to 35°C, with an optimum around 30°C, and within a pH range of 5.8 to 8, with an optimum around 7.2.
Relationships based on 16S rRNA gene sequences and G+C content of DNA.
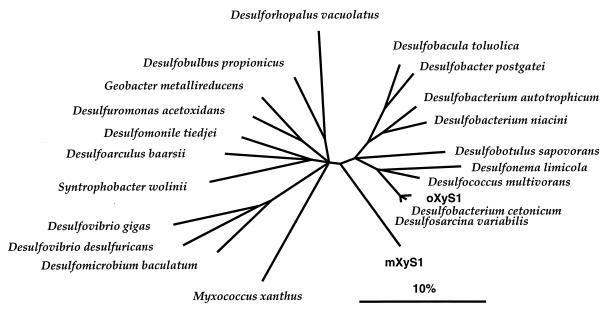
Analyses of sequences derived from the 16S rRNA genes revealed that both strains affiliated with the δ subclass of the Proteobacteria and branch within a group for which the family designation Desulfobacteriaceae has been proposed (42). This group of mesophilic sulfate-reducing bacteria contains, among a few incompletely oxidizing species, the completely oxidizing species described in the literature. Many of these are, like the presently obtained isolates, nutritionally rather versatile, and several have been reported to grow with aromatic compounds, most commonly with benzoate (42). The closest relatives of strain oXyS1 among the described species were Desulfobacterium cetonicum and Desulfosarcina variabilis, for which the similarity values were as high as 98.4 and 98.7%, respectively. As shown in Fig. 2, the branching of strain oXyS1 and its closest relatives is drawn as a multifurcation because the topology could not be resolved unambiguously by the different treeing approaches. Due to the high similarity values among the sequences from these three organisms, they may be even regarded as members of one species (17, 37). For definite classification in the future, however, additional characteristics, especially DNA-DNA hybridization data, should be taken into consideration. Strain mXyS1 did not show a specific relationship to a known species. The closest relative was Desulfococcus multivorans, for which the similarity value was 86.9% (Fig. 2). According to this relatively deep branching, strain mXyS1 may be regarded as the first representative of a so far unknown, distinct line of descent within the δ subclass of the Proteobacteria.
FIG. 2.
16S rRNA-based tree reflecting the relationships of strains oXyS1 and mXyS1 to a selected number of members of the δ subclass of the Proteobacteria. The tree is based on maximum-parsimony analysis; this includes only sequence positions that have identical residues in at least 50% of all available complete or almost complete 16S rRNA sequences from representative bacteria of this subclass (23). For most species shown in the tree, the sequences are those of the type strains. The sequence of Desulfomicrobium baculatum is from strain 9974 (DSM 1743), which is not the type strain. Desulfoarculus baarsii, Desulfobotulus sapovorans, and Desulfomicrobium baculatum are the newly suggested names of the former Desulfovibrio baarsii, Desulfovibrio sapovorans, and Desulfovibrio baculatus, respectively (for a discussion, see reference 42). The topology of the tree was evaluated and corrected on the basis of the results of distance matrix, maximum-parsimony, and maximum-likelihood analyses of various data sets. Phylogenetic positions of strains oXyS1 and mXyS1 did not differ in all treeing approaches. Multifurcations indicate topologies that could not be resolved unambiguously. The bar indicates estimated 10% sequence divergence.
The G+C contents of the DNA of strains oXyS1 and mXyS1 determined via HPLC analysis were 51 and 49.2 mol%, respectively. Reported G+C values of Desulfobacterium cetonicum and Desulfosarcina variabilis were 59 and 51 mol% (16, 42). Even though these two values were previously determined by thermal denaturation, i.e., a method different from the high-pressure liquid chromatography analysis used in the present study, the latter value is in good agreement with the close relationship between strain oXyS1 and Desulfosarcina variabilis revealed by sequence analysis of 16S rRNA genes.
Study of substrate utilization.
Strains oXyS1 and mXyS1 were able to grow on a variety of aromatic and aliphatic compounds (Table 1). Growth on toluene was observed for both strains, whereas utilization of other alkylbenzenes was strain specific. Strain oXyS1 used only one higher homologue of o-xylene, o-ethyltoluene, but no meta-substituted alkyltoluenes. In contrast, strain mXyS1 used two homologues of m-xylene, m-ethyltoluene and m-isopropyltoluene, but no ortho-substituted alkylbenzenes. p-Alkylbenzenes did not allow the growth of either strain. Both strains were able to grow on crude oil. One may therefore speculate that sulfate-reducing bacteria of the types represented by strains oXyS1 and mXyS1 consume toluene as well as the respective xylenes and higher homologues also from the crude oil in the enrichment culture.
TABLE 1.
Anaerobic growth tests of sulfate-reducing strains oXyS1 and mXyS1 on aromatic and nonaromatic compounds
| Compound testeda | Strainb:
|
|
|---|---|---|
| oXyS1 | mXyS1 | |
| Aromatic hydrocarbons | ||
| Toluene (2%) | + | + |
| o-Xylene (2%) | + | − |
| m-Xylene (2%) | − | + |
| o-Ethyltoluene (2%) | + | − |
| m-Ethyltoluene (2%) | − | + |
| m-Isopropyltoluene (2%) | − | + |
| Aromatic compounds with functional groups | ||
| o-Methylbenzyl alcohol (0.5, 2) | + | − |
| m-Methylbenzyl alcohol (0.5, 2) | − | − |
| Benzoate (1, 4) | + | + |
| o-Methylbenzoate (1, 2) | + | − |
| m-Methylbenzoate (1, 2) | − | + |
| Benzylsuccinate (1, 4) | + | − |
| Other compounds | ||
| Ethanol (1, 5) | + | − |
| Formate (10, 20) | + | + |
| Acetate (5, 10) | + | + |
| Propionate (5, 10) | − | + |
| n-Butyrate (5, 10) | + | + |
| Lactate (5, 10) | + | − |
| Pyruvate (1, 5) | + | + |
| Succinate (1, 5) | + | − |
| d,l-Malate (1, 5) | + | − |
Each compound was tested twice at the concentrations given in parentheses. Unless otherwise noted, concentrations are millimolar. Concentrations in percentages (vol/vol) refer to dilutions of hydrophobic compounds in heptamethylnonane as an inert carrier phase. Further compounds tested but utilized by neither strain: benzene (2%), ethylbenzene (2%), p-xylene (2%), p-ethyltoluene (2%), o-isopropyltoluene (2%), p-isopropyltoluene (2%), n-hexane (2, 5%), n-decane (5, 10%), cyclohexane (2%), methylcyclohexane (2%), phenol (0.5, 2), o-cresol (0.5, 2), m-cresol (0.5, 2), p-cresol (0.5, 2), acetophenone (1%), p-methylbenzoate (0.5, 2), methanol (5, 10), glucose (1, 5), fructose (1, 5).
Symbols: +, growth observed; −, no growth observed.
Desulfobacterium cetonicum, a very close relative of strain oXyS1, was originally isolated from an oil field by using butyrate as a substrate (16). The origin of Desulfobacterium cetonicum as well as its capacity to utilize benzoate suggested that this species can also oxidize aromatic hydrocarbons. Aromatic hydrocarbons have not been tested so far with this species. Actually, growth tests during the present study revealed utilization of toluene but not of o-xylene. Growth tests on aromatic hydrocarbons were also carried out with the other close relative of strain oXyS1, Desulfosarcina variabilis, which has been originally isolated on benzoate (27, 42). However, Desulfosarcina variabilis neither utilized o-xylene, m-xylene nor toluene. Two sulfate-reducing bacteria formerly isolated on toluene did not grow on o-xylene or m-xylene (2, 32).
Quantitative growth experiments.
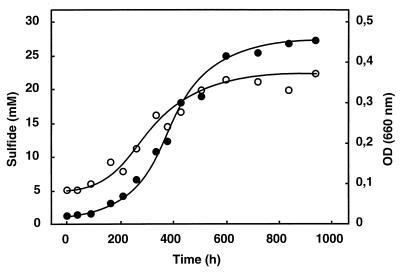
Growth of the new isolates was slower than that of the previously described toluene-degrading sulfate-reducing bacteria (2, 32). The growth curve of strain oXyS1 on o-xylene is shown in Fig. 3. The exponential growth phase was rather short, as could be seen in a semilogarithmic plot of the sulfide concentration or the optical density versus time (results not shown). Exponential growth soon turned into pronounced linear growth; i.e., the growth rate decreased steadily. The relatively short exponential phase allowed only an estimate of the doubling time, which was around 75 h for strain oXyS1 and around 55 h for strain mXyS1. The sulfate-reducing strains PROTL1 and Tol2 exhibit doubling times on toluene of 36 h (2) and 27 h (32), respectively.
FIG. 3.
Sulfide production (●) and increase in cell turbidity (○) in a culture of strain oXyS1 growing in defined medium on o-xylene. The growth experiment was carried out in a flat bottle (500 ml) with 450 ml of medium and 0.3 ml of o-xylene dissolved in 15 ml of heptamethylnonane. The bottle was incubated horizontally and shaken slightly. OD, optical density.
The balance of substrate consumption and sulfide formation by strains oXyS1 and mXyS1 was quantitatively determined in separate experiments with two different initial concentrations of o-xylene and m-xylene, respectively, in the presence of sulfate (28 mM). To avoid changes in the volumes of the carrier phase and the medium, samples were not taken during growth but only at the end of the experiments. The results obtained with strain mXyS1 are shown in Table 2. In the presence of cells, more than 90% of the originally added m-xylene had disappeared, whereas abiotic loss of m-xylene in cell-free controls was not observed. The amount of electrons that can be theoretically derived from the amount of m-xylene that was consumed is 13 to 17% greater than the amount of electrons required for sulfide production from sulfate. This deviation can be explained by partial utilization of the substrate for cell synthesis. Various sulfate-reducing bacteria were shown to assimilate approximately 10 to 15% of the organic substrate (32, 35, 44). Furthermore, some undetected organic metabolites may have been excreted. In conclusion, the result is in good agreement with the occurrence of complete oxidation of m-xylene to CO2. Essentially the same result was obtained with strain oXyS1 incubated on o-xylene. Again, more than 90% of the added growth substrate was consumed in inoculated bottles whereas the loss of o-xylene in sterile bottles was not measurable (further data not shown). The ability of both strains to oxidize substrates completely is also obvious from their growth on acetate, which, according to our present biochemical knowledge, necessarily leads to CO2 as the end product. The equation for complete oxidation of xylenes is as follows: (CH3)2C6H4 + 5.25SO42− + 2.5H+ + 3H2O → 8HCO3− + 5.25H2S. To our knowledge, this is the first demonstration of the mineralization of o-xylene and m-xylene in pure cultures of sulfate-reducing bacteria.
TABLE 2.
Quantification of m-xylene consumption and sulfide formation by strain mXyS1
| Expa | Amt (mmol) of:
|
||||
|---|---|---|---|---|---|
| m-Xylene added | m-Xylene consumedb | Sulfide producedc | Electrons from m-xylene consumedd | Electrons consumed by SO42− reductione | |
| Strain mXyS1 with small amount of m-xylene | 0.242 | 0.231 | 1.07 | 9.7 | 8.56 |
| Strain mXyS1 with large amount of m-xylene | 1.21 | 1.11 | 5.0 | 46.6 | 40.0 |
| Strain mXyS1 without m-xylene (control) | 0.0 | 0.0 | 0.067 | 0.0 | 0.54 |
| Sterile medium without cells (control) | 1.21 | 0.0 | 0.0 | 0.0 | 0.0 |
Experiments were carried out under anoxic conditions with flat bottles (250 ml) with a culture volume of 190 ml. The total amount of sulfate added to each bottle was 5.23 mmol (27.5 mM). The medium was overlaid with 5 ml of heptamethylnonane as the carrier phase for m-xylene. The volume of m-xylene to be added was calculated from the density (0.866 g · cm−3 at 20°C) and molecular mass (106.2 g · mol−1).
Difference between m-xylene added and m-xylene recovered in the carrier and aqueous phase at the end of the experiment.
The small amount of sulfide produced in the control with cells without substrate was subtracted from the amount of sulfide produced in experiments with cells and m-xylene.
Stoichiometrically, 42 mmol of electrons is derived from 1 mmol of m-xylene oxidized to CO2.
Stoichiometrically, 8 mmol of electrons is required for complete reduction of 1 mmol of SO42− to 1 mmol of H2S.
Key enzymes of acetyl coenzyme A oxidation.
Enzyme tests for detection of key enzymes for acetyl coenzyme A oxidation were carried out with cell extract of strain oXyS1. Carbon monoxide dehydrogenase and formate dehydrogenase were measured at specific activities (with respect to protein) of 0.12 and 0.23 μmol · min−1 · mg−1, respectively. No activity of 2-oxoglutarate dehydrogenase was detectable. The results suggest that terminal oxidation occurs via the carbon monoxide dehydrogenase pathway (reverse Wood pathway, or C1 pathway), as occurs in most species of completely oxidizing sulfate-reducing bacteria (43).
ACKNOWLEDGMENTS
We are indebted to Fred A. Rainey, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany, for performing sequence analysis and for providing unpublished sequence data. We thank Jens Harder, Bremen, for instrumental help.
G. Harms and K. Zengler contributed equally to this study.
This work was supported by the Max-Planck-Gesellschaft and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Aeckersberg F, Bak F, Widdel F. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch Microbiol. 1991;156:5–14. [Google Scholar]
- 2.Beller H R, Spormann A M, Sharma P K, Cole J R, Reinhard M. Isolation and characterization of a novel toluene-degrading, sulfate-reducing bacterium. Appl Environ Microbiol. 1996;62:1188–1196. doi: 10.1128/aem.62.4.1188-1196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biegert T, Fuchs G. Anaerobic oxidation of toluene (analogues) to benzoate (analogues) by whole cells and by cell extracts of a denitrifying Thauera sp. Arch Microbiol. 1995;163:407–417. doi: 10.1007/BF00272130. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen C-I, Taylor R T. Thermophilic biodegradation of BTEX by two consortia of anaerobic bacteria. Appl Microbiol Biotechnol. 1997;48:121–128. doi: 10.1007/s002530051026. [DOI] [PubMed] [Google Scholar]
- 6.Cline J D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 7.Cord-Ruwisch R. A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J Microbiol Methods. 1985;4:33–36. [Google Scholar]
- 8.Cord-Ruwisch R, Kleinitz W, Widdel F. Sulfate-reducing bacteria and their activities in oil production. J Petrol Technol. 1987;1987(Jan):97–106. [Google Scholar]
- 9.Dolfing J, Zeyer J, Binder-Eicher P, Schwarzenbach R P. Isolation and characterization of a bacterium that mineralizes toluene in the absence of molecular oxygen. Arch Microbiol. 1990;154:336–341. doi: 10.1007/BF00276528. [DOI] [PubMed] [Google Scholar]
- 10.Edwards E A, Wills L E, Reinhard M, Grbić-Galić D. Anaerobic degradation of toluene and xylene by aquifer microorganisms under sulfate-reducing conditions. Appl Environ Microbiol. 1992;58:794–800. doi: 10.1128/aem.58.3.794-800.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards E A, Grbić-Galić D. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl Environ Microbiol. 1994;60:313–322. doi: 10.1128/aem.60.1.313-322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans P J, Mang D T, Young L Y. Degradation of toluene and m-xylene and transformation of o-xylene by denitrifying enrichment cultures. Appl Environ Microbiol. 1991;57:450–454. doi: 10.1128/aem.57.2.450-454.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans P J, Mang D T, Kim K S, Young L Y. Anaerobic degradation of toluene by a denitrifying bacterium. Appl Environ Microbiol. 1991;57:1139–1145. doi: 10.1128/aem.57.4.1139-1145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein J. Numerical methods for inferring phylogenetic trees. Q Rev Biol. 1992;57:379–404. [Google Scholar]
- 15.Fries M R, Zhou J, Chee-Sanford J, Tiedje J M. Isolation, characterization and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galushko A S, Rozanova E P. Desulfobacterium cetonicum sp. nov.: a sulfate-reducing bacterium which oxidizes fatty acids and ketones. Microbiology. 1995;60:742–746. [Google Scholar]
- 17.Goodfellow M, Manfio G P, Chun J. Towards a practical species concept for cultivable bacteria. In: Claridge M F, Dawah H A, Wilson M R, editors. The units of biodiversity. London, United Kingdom: Chapman & Hall; 1997. pp. 25–59. [Google Scholar]
- 18.Häner A, Höhener P, Zeyer J. Degradation of p-xylene by a denitrifying enrichment culture. Appl Environ Microbiol. 1995;61:3185–3188. doi: 10.1128/aem.61.8.3185-3188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbert B N. Reservoir souring. In: Hill E C, Shennan J L, Walkinson R J, editors. Microbial problems in the offshore oil industry. London, United Kingdom: John Wiley & Sons, Ltd.; 1987. pp. 63–71. [Google Scholar]
- 20.Hess A, Zarda B, Hahn D, Häner A, Stax D, Höhener P, Zeyer J. In situ analysis of denitrifying toluene- and m-xylene-degrading bacteria in a diesel fuel-contaminated laboratory aquifer column. Appl Environ Microbiol. 1997;63:2136–2141. doi: 10.1128/aem.63.6.2136-2141.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jørgensen B B. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature (London) 1982;296:643–645. [Google Scholar]
- 22.L’Haridon S, Reysenbach A-L, Glénat P, Prieur D, Jeanthon C. Hot subterranean biosphere in a continental oil reservoir. Nature (London) 1995;377:223–224. [PubMed] [Google Scholar]
- 23.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mesbah M, Premachandran U, Whitman W B. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol. 1989;39:159–167. [Google Scholar]
- 25.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 26.Odom J M. Industrial and environmental activities of sulfate-reducing bacteria. In: Odom J M, Singleton R Jr, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer-Verlag; 1993. pp. 189–210. [Google Scholar]
- 27.Pfennig N, Widdel F, Trüper H G. The dissimilatory sulfate-reducing bacteria. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The prokaryotes. 1st ed. I. New York, N.Y: Springer-Verlag; 1981. pp. 926–940. [Google Scholar]
- 28.Rabus R, Widdel F. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch Microbiol. 1995;163:96–103. doi: 10.1007/BF00381782. [DOI] [PubMed] [Google Scholar]
- 29.Rabus R, Widdel F. Conversion studies with substrate analogues of toluene in a sulfate-reducing bacterium, strain Tol2. Arch Microbiol. 1995;164:448–451. doi: 10.1007/BF02529744. [DOI] [PubMed] [Google Scholar]
- 30.Rabus R, Widdel F. Utilization of alkylbenzenes during anaerobic growth of pure cultures of denitrifying bacteria on crude oil. Appl Environ Microbiol. 1996;62:1238–1241. doi: 10.1128/aem.62.4.1238-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabus R, Fukui M, Wilkes H, Widdel F. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil. Appl Environ Microbiol. 1996;62:3605–3613. doi: 10.1128/aem.62.10.3605-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabus R, Nordhaus R, Ludwig W, Widdel F. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol. 1993;59:1444–1451. doi: 10.1128/aem.59.5.1444-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rainey F A, Dorsch M, Morgan H W, Stackebrandt E. 16S rDNA analysis of Spirochaeta thermophila: position and implications for the systematics of the order Spirochaetales. Syst. Appl. Microbiol. . 1992;16:224–226. [Google Scholar]
- 34.Rueter P, Rabus R, Wilkes H, Aeckersberg F, Rainey F A, Jannasch H W, Widdel F. Anaerobic oxidation of hydrocarbons in crude oil by new types of sulfate-reducing bacteria. Nature (London) 1994;372:455–458. doi: 10.1038/372455a0. [DOI] [PubMed] [Google Scholar]
- 35.Schnell S, Bak F, Pfennig N. Anaerobic degradation of aniline and dihydroxybenzenes by newly isolated sulfate-reducing bacteria and description of Desulfobacterium anilini. Arch Microbiol. 1989;152:556–563. doi: 10.1007/BF00425486. [DOI] [PubMed] [Google Scholar]
- 36.Seyfried B, Glod G, Schocher R, Tschech A, Zeyer J. Initial reactions in the anaerobic oxidation of toluene and m-xylene by denitrifying bacteria. Appl Environ Microbiol. 1994;60:4047–4052. doi: 10.1128/aem.60.11.4047-4052.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA:DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 38.Stetter K O, Huber R, Blöchl E, Kurr M, Eden R D, Fielder M, Cash H, Vance I. Hyperthermophilic archaea are thriving in deep North Sea and Alaskan oil reservoirs. Nature (London) 1993;365:743–745. [Google Scholar]
- 39.Strunk, O., and W. Ludwig. ARB: a software environment for sequence data. http://www.mikro.biologie.tu-muenchen.de. Department of Microbiology, Technische Universität München, Munich, Germany.
- 40.Tissot B P, Welte D H. Petroleum formation and occurrence. 2nd ed. Berlin, Germany: Springer-Verlag KG; 1984. [Google Scholar]
- 41.Tsai Y L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. IV. New York, N.Y: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]
- 43.Widdel F, Hansen T A. The dissimilatory sulfate- and sulfur-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. I. New York, N.Y: Springer-Verlag; 1992. pp. 583–624. [Google Scholar]
- 44.Widdel F, Pfennig N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov. sp. nov. Arch Microbiol. 1981;129:395–400. doi: 10.1007/BF00406470. [DOI] [PubMed] [Google Scholar]
- 45.Zeyer J, Kuhn E P, Schwarzenbach R P. Rapid microbial mineralization of toluene and 1,3-dimethylbenzene in the absence of molecular oxygen. Appl Environ Microbiol. 1986;52:944–947. doi: 10.1128/aem.52.4.944-947.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]