Abstract
Morphological mutants of Trichoderma reesei were isolated following chemical or insertional mutagenesis. The mutant strains were shown to have reduced viscosity under industrially relevant fermentation conditions and to have maintained high specific productivity of secreted protein. This allowed higher biomass concentration to be maintained during the production phase and, consequently, increased volumetric productivity of secreted protein. The causative mutations were traced to four individual genes (designated sfb3, ssb7, seb1, and mpg1). We showed that two of the morphological mutations could be combined in a single strain to further reduce viscosity and enable a 100% increase in volumetric productivity.
Keywords: Trichoderma reesei, Viscosity, Productivity, Morphology, Mutation
Introduction
The filamentous fungus Trichoderma reesei (teleomorph, Hypocrea jecorina) is a mesophilic ascomycete. All strains of T. reesei used in industry have been derived from the original natural isolate QM6a (Mandels & Reese, 1957). Early strain improvement was performed in an academic setting and focused on increased secreted cellulase and hemicellulase production in submerged culture leading to strains such as RUT-C30 and RL-P37 (Montenecourt & Eveleigh, 1979; Peterson & Nevalainen, 2012; Sheir-Neiss & Montenecourt, 1984). Today, large-scale submerged fermentation is used to manufacture a range of commercial enzymes (Nevalainen et al., 1994). The native mix of secreted cellulases and hemicellulases can be used in the textile industry and for conversion of biomass to soluble sugars. Individual genes, encoding native secreted endoglucanases, xylanase, or glucoamylase (US FDA, 2011) may be overexpressed and genes encoding the dominant naturally secreted enzymes, such as cellobiohydrolases, may be deleted to tailor products for specific industrial purposes. Several non-native enzymes are also manufactured using these strains, chymosin and other food enzymes such as lipases (US FDA, 2019), and phytases for use in animal feed (Christensen et al., 2020). There is a continued need for strain improvement to increase the efficiency of the fermentation process and reduce costs of protein production.
Key parameters used to describe strain fermentation performance are yield on substrate (e.g., g protein/g sugar) and volumetric productivity (g product/kg broth/hr). In general, volumetric productivity can be improved by increasing specific productivity of the cells (g product/g dry cell weight/hr) or by increasing the number of cells in the broth (g dry cell weight/kg broth). Oxygen transfer is often limiting for the volumetric productivity of secreted proteins or metabolites in filamentous fungal fermentations. The high viscosity of filamentous fungal cultures requires maintaining low cell density to achieve effective mixing and thus oxygen transfer. Altered morphology and consequently broth rheology to improve oxygen transfer and allow higher cell density is clearly desirable (reviewed in Cairns et al., 2019). However, this must not come at the expense of specific productivity or there may be no net increase in volumetric productivity or yield.
The most appropriate fungal morphology in submerged culture can depend on whether secreted proteins or biochemicals are the desired product of fermentation. A pelleted macro-morphology can be encouraged by appropriate culture conditions, can reduce broth viscosity and may be suitable for biochemical production; however, dispersed mycelium is generally believed suitable for efficient secreted protein production (Cairns et al., 2019). Domingues et al. (2000) showed that a low inoculum size led to the formation of pellets but that this was associated with lower secreted enzyme production in shake flask cultures. Callow and Ju (2012) showed that addition of surfactants, particularly Triton X-100, induced pelleted growth of T. reesei in liquid and increased extracellular cellulase activity, although it was not clear that this was due to increased secretion. Bhargava et al. (2003) compared pulsed or continuous feed protocols with an engineered Aspergillus oryzae strain producing glucoamylase and fed with maltodextrin. They found that the pulsed feed strategy resulted in a lower viscosity broth with smaller fungal elements. Consequently, it was possible to feed more maltodextrin and reach higher biomass and product concentration without encountering oxygen transfer problems. Unlike A. oryzae or Aspergillus niger, for example, highest secreted protein productivity by T. reesei is observed when growth rates are slow, which can be controlled by limiting carbon source availability (Pakula et al., 2005). Consequently, a large-scale fermentation process for T. reesei will typically include a growth phase, during which biomass accumulates, followed by a production phase during which inducer is supplied and secreted protein is produced but further biomass increase is minimal. The amount of biomass generated during the growth phase depends upon the amount of glucose or other sugars supplied. However, biomass accumulation is also limited by broth viscosity and oxygen transfer. A strain with lower viscosity properties should enable higher biomass accumulation without encountering dissolved oxygen limitation. Ideally, an improved strain that exhibits reduced viscosity would not also exhibit significantly reduced growth rate that would adversely affect biomass accumulation during large-scale fermentation.
Secretion of extracellular enzymes by filamentous fungi has been considered to be hyphal tip associated (Gordon et al., 2000; Wösten et al., 1991). This led to the suggestion that a hyperbranched morphology giving more hyphal tips might lead to improved protein secretion. However, other studies suggest more distal regions of the hyphae, including septa, are involved and there may be different sites of secretion depending on the secreted protein (Nykänen et al., 1997; Valkonen et al., 2007).
Strains of filamentous fungi such as A. niger, A. oryzae, and Penicillium chrysogenum used by industry have undergone many rounds of classical mutagenesis and screening to improve performance in large-scale fermentation. The criteria for selection at each round have not been well documented, although presumably often involved observation of zones of enzyme activity around colonies on agar plates as well as fortuitous colony morphology changes. This has led to extreme alterations in morphology in some industrial lineages, with concomitant changes in submerged fermentation characteristics (reviewed in Cairns et al., 2019). As mentioned above, T. reesei has also been subjected to serial rounds of mutagenesis and selection to improve secreted protein productivity. However, there has been no dramatic alteration of colonial or hyphal morphology in these lineages and no reports of improved rheological properties during fermentation.
Many mutations affecting hyphal morphology have been identified in filamentous fungi but rarely have effects on protein secretion been investigated. However, reports describing deletion of a Rho GTPase gene (racA in A. niger, rac1 in T. reesei), which causes a hyperbranching phenotype, did include analysis of secretion. Fitz et al. (2019) concluded that rac1 deletion did not affect viscosity of T. reesei culture broth but could upregulate cellulase gene expression and increase secreted protein production dependent on carbon source. Kwon et al. (2013) concluded that racA deletion had no effect on secretion of glucoamylase by A. niger. However, in a following study, Fiedler et al. (2018) showed that that an A. niger ΔracA strain did have a greater capacity for protein secretion when glucoamylase was expressed under control of a tunable Tet-on system. It is not clear if these effects are due to the hyperbranching morphology or rac1/A deletion affecting physiology.
Bocking et al. (1999) screened for hyperbranched mutants with good protein secretion following chemical mutagenesis of A. oryzae. Highly branched mutants showed lower viscosity and produced more secreted protein during batch fermentation, although not during chemostat or fed-batch fermentations. By controlling feed rate according to dissolved oxygen during fed-batch fermentation mutants could be fed more maltodextrin, accumulated more biomass and 12% more secreted protein. The authors concluded that the mutants did not show higher specific productivity but increased volumetric productivity could be achieved. He et al. (2016) isolated a hyperbranching mutant (DES-15) derived from T. reesei RUT-C30 that exhibited an almost 70% increase in secreted cellulase concentration and 50% increased total secreted protein in batch fermentation. DES-15 biomass concentration was lower throughout fermentation relative to the parental strain suggesting that the improvement to specific productivity of secreted proteins was even higher. The causative mutation in DES-15 was not identified although the authors noted that transcript levels were reduced for several genes known to affect morphology. Recently, Gao et al. (2020) used RNAi to reduce expression of the cot1 gene in a strain of T. reesei to obtain a hyperbranched phenotype. They demonstrated that this strain exhibited slower growth but higher specific productivity, at least during the first 72 h of shake flask culture.
In this study, we sought to identify morphological mutants of T. reesei that would have improved rheological properties. We then evaluated specific productivity of the mutants to determine if protein secretion had been affected. Two possible, nonmutually exclusive, outcomes were considered. One was that we would find mutants that were not affected in specific protein productivity but had improved performance in fermenters due to lower viscosity allowing higher productive biomass accumulation. The other possibility was that altered morphology could lead to improved specific productivity. T. reesei mutant libraries were screened for altered morphology without a preconceived idea of the type of morphological change that was beneficial. Mutants were then screened for reduced viscosity in submerged culture, secreted protein productivity was checked, and the genomic mutations were identified. Many mutant strains were identified with reduced viscosity, but secreted protein productivity was often also reduced. However, some mutants that maintained good specific productivity and had significantly reduced viscosity under industry-relevant fermentation conditions were identified. Although none showed increased specific productivity, increased biomass could be maintained during fermentation leading to increased volumetric productivity.
Materials and Methods
Strains
All T. reesei strains were derived from RL-P37 (Sheir-Neiss & Montenecourt, 1984). The T. reesei M1-1.1(Pyr4+) strain has genome deletions that remove or inactivate the cbh1, cbh2, egl1, and egl2 genes encoding the major secreted cellulases (cellobiohydrolases I and II, and endoglucanases I and II), which makes it useful for expressing other proteins in the absence of cellulase background activity. During construction of this strain large regions of the genome surrounding the cbh1 gene and surrounding cbh2, including all sequences between cbh2 and egl2, were deleted. Strain TrGA 29-9 is a derivative of M1-1.1(Pyr4+) that overexpresses the native glucoamylase gene due to two tandemly integrated copies of an expression cassette consisting of the cbh1 promoter region, glucoamylase coding region, cbh1 terminator and A. nidulans amdS selectable marker. The MAGI strain was generated by targeting the insertion of a reporter cassette to the orotidine 5′-monophosphate pyrophosphorylase (pyr2) locus of T. reesei M1-1.1(Pyr4+). This reporter cassette contained a codon optimized green fluorescent protein (GFP) from Ptilosarcus gurneyi and an α-amylase under the control of the T. reesei cellobiohydrolase I (cbhI) promoter and transcriptional terminator sequences. A hygromycin B phosphotransferase gene was also integrated with the reporters at the pyr2 locus. Coincident with integration of the reporter cassette, a 3′ portion of the pyr2 gene was deleted making the strain a uridine auxotroph. The T4 strain was derived from RL-P37 via strain A83 in two steps of nitrosoguanidine (NTG) mutagenesis and screening for increased cellulase production. Strains generated and used in this study are shown in Table 1.
Table 1.
T. reesei Strains Used in This Study
| Strain name | Parent strain | Modification from parent |
|---|---|---|
| M1-1.1(Pyr4+) | RL-P37 | Approximately 100 kb deletion in scaffold 29 including cbh1 and swo1 |
| Approximately 32 kb deletion in scaffold 3 including cbh2 and egl2 | ||
| Partial deletion of egl1 | ||
| TrGA 29-9 | M1-1.1(Pyr4+) | Insertion of two tandem copies of T. reesei glucoamylase expression cassette, pyr4 marker |
| TrGA 29-9 pyr2 | TrGA 29-9 | pyr2− |
| 70H2 | TrGA 29-9 | Calcofluor-sensitive mutant |
| 77B7 | TrGA 29-9 | Morphological mutant |
| Morph Δsfb3 | M1-1.1(Pyr4+) | Deletion of sfb3 |
| TrGA 29-9 Δsfb3 | TrGA 29-9 | Deletion of sfb3 |
| TrGA 29-9 ssb7 (TS1) | TrGA 29-9 | Creation of the TS1 allele of ssb7 |
| 299m4 | TrGA 29-9 pyr2 | Disruption of mpg1 |
| T4 | RL-P37 | UV and NTG mutagenesis for cellulase over-production. Mutation of nik1. |
| T4 pyr4 | RL-P37 | UV and NTG mutagenesis for cellulase overproduction. Mutation of nik1; pyr4− |
| T4 pyr4 F16 | T4 pyr4 | IM morphological mutant |
| MAGI | M1-1.1(Pyr4+) | Hygromycin resistance, GFP and α-amylase expression cassette disrupting pyr2 |
| MAGI 10-8g | MAGI | IM morphological mutant |
| Morph Δtku80 | M1-1.1(Pyr4+) | Deletion of tku80 |
| Morph Δtku80 pyr4 | Morph Δtku80 | Disruption of pyr4 |
| Morph Δtku80 Δseb1 | Morph Δtku80 pyr4 | Disruption of seb1 |
| 77B7 p28 | 77B7 | Spontaneous pyr2− |
| B7m4 | 77B7 p28 | Disruption of mpg1 |
NTG Mutagenesis and Screening for Calcofluor White Sensitivity
Conidiospores of TrGA 29-9 were mutagenized by NTG treatment (99% kill rate). A cell sorter was used to deposit individual spores onto agar plates of Vogel's medium (Vogel, 1956) with 1% glucose and 350 ppm calcofluor white (CFW). Following growth, individual colonies that showed altered radial growth were selected. Mutants from this screen were evaluated in high cell density shake flask culture using the NREL medium (described by Ouedraogo et al. 2015). The viscosity of these culture broths was compared using an AR550 rheometer (TA Instruments, New Castle, DE) using a gap setting of 450 μm and shear rate of 50 cps/s. Secreted glucoamylase activity in shake flask cultures was also measured (Nykänen et al., 1997). Mutants with low-viscosity broths and good protein productivity were evaluated further in 15 l fermenters for reduced viscosity, oxygen transfer properties, and secreted protein production. One such CFW sensitive mutant was named 70H2 and was shown to be also sensitive to Congo Red and sodium dodecyl sulfate (SDS).
NTG Mutagenesis and Screening for Morphological Mutations
NTG mutagenesis of TrGA 29-9 was performed at a dose to give 10% cell kill. The surviving cells were plated on PDA (potato dextrose agar) and allowed to sporulate. Spores were harvested, grown in YEG (0.5% yeast extract, 2% glucose) for 48–72 hr, and the resulting mycelia were sieved through a 200 µm filter that was subsequently washed with water. The filtrate was spun down, the mycelia were collected and re-inoculated into YEG. This procedure was repeated several times, except the mycelia were sieved through smaller filters each time. In the final step, after growth in YEG, the mycelia were filtered through a 40 µm filter, the mycelia were recovered from the filtrate, and plated on PDA. The resulting individual colonies were picked using a colony picker and transferred to 96 well microtiter plates. Using microscopy the libraries were screened in liquid culture, and mutants were recovered that showed alterations in morphology. Mutants from this screen were evaluated further in high cell density shake flasks. Mutants with low-viscosity broths were evaluated in 15 l fermenters for reduced viscosity, oxygen transfer properties, and protein production. Mutant strain 77B7 was isolated by these methods.
PEG-Mediated Transformation of Trichoderma
Transformation of T. reesei protoplasts by a polyethylene glycol-mediated method was performed as described by Penttilä et al. (1987) with slight modifications.
Agrobacterium-Mediated Insertional Mutagenesis
Insertional mutagenesis (IM) libraries were prepared by Agrobacterium tumefaciens-mediated transformation of T. reesei, with a DNA fragment containing either the T. reesei pyr4 or pyr2 gene as selectable marker and gene tag. The vector used for IM was based on the pPZP100 vector, which includes the left and right T-DNA border regions, a pBR322 bom site for mobilization from Escherichia coli to Agrobacterium, ColE1 and pVS1 plasmid origins for replication in E. coli and Agrobacterium, respectively, and a bacterial marker conferring chloramphenicol resistance (Hajdukiewicz et al., 1994). An insertion cassette containing the T. reesei pyr4 gene with native promoter and terminator regions was prepared and ligated into the final vector at a BamHI site between the left and right T-DNA borders. This cassette was ultimately used to mutate parental T. reesei strain T4 pyr4 using methods described in Online Resource 1. Alternatively, an insertion cassette containing the pyr2 gene of Trichoderma atroviride followed by the his1 promoter oriented to transcribe outward into the T. reesei genome insertion site was prepared and ligated into the vector left and right T-DNA borders (Online Resource 1). This insertion cassette was ultimately used to mutate T. reesei parental strain MAGI.
IM libraries of 50,000 transformants were generated.
Screening for Morphological Mutants in Submerged Culture
IM libraries were screened for mutants with more compact morphology or less elongated hyphae in liquid culture using light microscopy as follows. Following transformation, individual transformants were picked from the nitrocellulose filters using a colony picker (CP-700, Norgren Systems LLC, Fairlea, WV, USA) and transferred to 96-well microtiter plates containing PDA. Alternatively, spores from transformant colonies were combined and single spores were added to microtiter plate wells using a cell sorter. To accomplish this, spores were collected in 20 ml sterile distilled water, inoculated into a 250 ml flask containing 50 ml of minimal medium, and incubated at 28°C for 24 hr until germlings were obtained. Using high speed sorting (MoFlo sorter, Cytomation, Fort Collins, CO, USA; at an event rate of 15,000 events per second, 60 psi with a 70 μm nozzle) individual germlings were separated into microtiter plate wells containing PDA. Microtiter plates containing the transformants were incubated for 7 days at 28°C. The resulting spores were replicated into 384 well black sensoplates with glass bottoms (Greiner Bio-one, Germany) containing YEG and incubated at 20°C for 24 hr. The morphology of individual transformants was examined microscopically. Liquid cultures of transformants were sieved through 200 and 40 μm filters, as described above, to enrich for more compact growth. Differences in viscosity and settling behavior of high-density shake flask culture broths between transformants could also be observed by eye.
Strain Characterization in Fermenters
Parental and mutant strains were grown under identical conditions in submerged culture and their growth phenotypes were compared. Spore suspensions of T. reesei were added to 500 ml of medium in 3 l flasks with both side and bottom baffles. The medium contained 5 g/l (NH4)2SO4, 4.5 g/l KH2PO4, 1 g/l MgSO4∙7H2O, and 14.4 g/l citric acid, adjusted to pH 5.5 with 5% NaOH. After autoclaving for 30 min, sterile 60% glucose was added to a final concentration of 27.5 g/l, along with 2.5 ml/l of a trace element solution containing 175 g/l citric acid, 200 g/l FeSO4∙7H2O, 16 g/l ZnSO4∙7H2O, 3.2 g/l CuSO4∙5H2O, 1.4 g/l MnSO4∙H2O, and 0.8 g/l H3BO3. The cultures were grown for 48 hr at 34°C with shaking. The contents of each flask were added separately to 15 l fermenters containing 9.5 l of medium containing 4.7 g/l KH2PO4, 1.0 g/l MgSO4∙7H2O, 4.3 g/l (NH4)2SO4 and 2.5 ml/l of the trace element solution. These components were heat sterilized at 121°C for 30 min prior to inoculation. A solution of 60% glucose and 0.48% CaCl2∙2H2O was separately sterilized, cooled, and added to the fermenter to a final concentration of 75 g/l glucose and 0.6 g/l CaCl2∙2H2O. The medium was adjusted to pH 3.5 with 28% NH3 and the temperature was maintained at 34°C during the growth period. Media for the MAGI strain were supplemented with 2 mg/ml uridine. The amount of biomass that accumulated was dependent on the batched glucose.
A dissolved oxygen (DO2) probe was calibrated to 100% when there was no added pressure in the headspace (i.e., 0 bar gauge, 1 bar absolute). The pressure in the headspace was then set to 0.7 bar (gauge), after which the oxygen probe read 170% before the seed culture was added. The fermenter contained two, four-blade radial flow (standard Rushton) turbines that provided mixing via a variable speed motor that was initially set at 500 rpm. As the cultures grew, CO2 evolution rate (CER) increased and DO2 levels dropped, at least partly as a consequence of the increased viscosity of the broth due to the proliferation of fungal hyphae. When DO2 fell below 40%, the agitation rate was increased to maintain the dissolved oxygen at 40%. Upon reaching 750 RPM agitation DO2 was allowed to drop below 40%. If DO2 did not fall below 40% it was unnecessary to increase the agitation rate and the initial agitation rate was higher than necessary. When the glucose had been consumed, growth rate was reduced as indicated by reduced CER. The amount of biomass produced in each fermenter was measured and found to be substantially the same for both parental and mutant strains. Once glucose was exhausted, the temperature was dropped to 28°C and a slow feed of glucose and mixed disaccharides, including sophorose, was started to maintain the cells in a glucose-limited state and induce secreted protein production (England et al., 2010).
The DO2 level in each fermenter at a given level of agitation, and the amount of agitation required to maintain a given DO2 level are indirect measures of the viscosity of the different broths, due to the different strain phenotypes. Although it would be ideal to vary only one variable (i.e., DO2 or agitation) and measure the other, it is also desirable to prevent the DO2 from falling below 40% to ensure the production of sufficient biomass in each fermenter, thereby permitting a more meaningful comparison between growth of the different strains.
As the culture grows, the oxygen consumption rate of the culture increases with the cell concentration, as in Equation 1. With all other things constant, this will result in a reduction in the dissolved oxygen concentration. But if the dissolved oxygen concentration gets too low, specific growth rate (μ) of this obligate aerobic organism will be negatively impacted, following typical Monod kinetics, also as in Equation 1. If dissolved oxygen concentration is not changing rapidly, a pseudo-steady-state condition can be assumed, and oxygen uptake rate of the culture matches the oxygen transfer rate of the equipment. Per Equation 2, to keep dissolved oxygen higher, increases can be made to either the mass transfer coefficient, kLa, or the dissolved oxygen concentration at equilibrium with the exit gas, DO2*. As can be seen in Equations 3 and 4, kLa can be increased by increasing power to increase agitation speed or by increasing air flow rate. In small scale systems, the agitation power term dominates making impeller speed the more effective variable. For DO2*, increased fermenter back pressure and increased air flow to increase the oxygen concentration in the gas phase leaving the fermenter can be employed. For these fungal fermentations, the oxygen stripped from the inlet air is not that great if the air flow is relatively high, meaning pressure is the more effective term. In the fermentations described here, however, back pressure and airflow rate were held constant and only agitation rate was increased. But as the cell concentration increases, viscosity of the broth due to the filamentous nature of the culture also negatively affects kLa and thus the dissolved oxygen concentration. By rearranging Equations 2, 3, and 4, viscosity can be estimated from the remaining measured values (Equation 5). Since all the cultures achieved the same cell concentration, the relative viscosity brought about by the different strains can be estimated. This estimate can actually be better than trying to measure broth viscosity explicitly, as such measurements are notoriously difficult (Charles & Wilson, 1994). The exponents in Equation 3 can only be empirically determined. Garcia-Ochoa and Gomez (2009) reviewed the literature and found an average value for α of 0.72 and for γ of 0.64. Actual differences in these values alter the magnitude of the estimated viscosity change but do not change the conclusions on the difference between different strains and mutations.
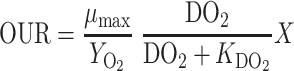 |
(1) |
where OUR is the oxygen uptake rate in mmol/l/hr, YO2 is the yield of cell mass on oxygen in gDCW/mmol, DO2 is the dissolved oxygen concentration in mmol/l, and X is the biomass concentration in gDCW/l.
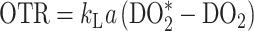 |
(2) |
where OTR is the oxygen transfer rate in mmol/l/hr, kLa is the oxygen mass transfer coefficient in h−1, and DO2* is the dissolved oxygen concentration when at equilibrium with the exit gas in mmol/l.
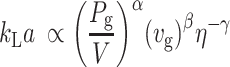 |
(3) |
where Pg is the gassed power in W, V is the broth volume in l, vg is the linear gas flow rate in cm/min, and η is the viscosity of the liquid medium in Pa s. α, β, and γ are exponent terms.
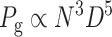 |
(4) |
where N is the agitation rate in min−1 and D is the impeller diameter in cm.
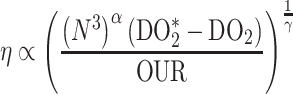 |
(5) |
The equations above do not take into account the non-Newtonian nature of filamentous fungal fermentation broth. The rheology of the broth is shear-thinning in nature, such that viscosity changes with the shear rate, as in equation (Christensen et al., 2020). The impact on the viscosity properties will be discussed further in the Results and Discussion sections.
Shear rate ∞ N2D2 (Christensen et al., 2020)
Identification of Mutations in Strain 70H2
The genomes of parental strain TrGA 29-9 and mutant 70H2 were sequenced to >40× coverage using Illumina technology (BaseClear BV, Leiden, The Netherlands). For each genome the 35 nt reads were mapped onto the assembled T. reesei QM6a genome available from the Joint Genome Institute (JGI) genome portal (T. reesei genome v2.0, http://genome.jgi-psf.org/Trire2/Trire2.home.html) and SNPs and indels unique to 70H2 were identified (GQ Life Sciences, Inc., Boston, MA). Mutations that were within ORFs, were predicted to alter amino acid sequence and had high-quality scores were prioritized. This resulted in a list of 23 mutated genes of potential interest.
Identification of the Causative Mutation (ssb7) in T. reesei 77B7
The genomes of the T. reesei TrGA 29-9 parent and the 77B7 mutant strains were sequenced (BaseClear BV, Leiden, The Netherlands). Reads were mapped to the assembled QM6a genome available at the JGI database leading to the identification of eight mutations predicted to alter a coding sequence in the 77B7 genome that were not present in TrGA 29-9 genome. For each of the eight mutations plasmids were developed containing transformation selection markers and a DNA fragment corresponding to the wild-type locus sequence with sufficient flanking sequences to ensure expression of the open reading frame contained therein. Mutant strain 77B7 protoplasts were transformed with each of these plasmids separately. Two transformants for each of the eight DNA fragments corresponding to the eight mutant loci were transferred to shake flasks with selective liquid media and grown for 3 days at 33°C to observe their morphology.
Inverse PCR to Identify Mutations in T4 pyr4 F16 (seb1) and MAGI 10-8g (mpg1)
The T4 pyr4 F16 mutant strain was obtained using IM with parent strain T4 pyr4 and MAGI 10-8g was obtained by IM of strain MAGI. Inverse PCR was used to identify the sites of T-DNA insertion in these strains (Online Resource 2).
Generation of Strain TrGA 29-9 Δsfb3 and M1-1.1(Pyr4+) Δsfb3
Protoplasts of strain TrGA 29-9 or M1-1.1(Pyr4+) were transformed with a deletion cassette consisting of sfb3 flanking regions and a hygromycin resistance marker (Online Resource 3) by PEG-mediated transformation and plated on Vogel's minimal medium containing glucose, hygromycin B, and sorbitol. Transformants were transferred to minimal medium containing hygromycin B and then to minimal medium or PDA containing Congo Red to assess Congo Red sensitivity. PCR analysis using primers that sit outside and within the Δsfb3 deletion cassette identified Congo Red-sensitive candidate transformants, named TrGA 29-9 Δsfb3 or M1-1.1(Pyr4+) Δsfb3, in which the deletion cassette had integrated at the sfb3 locus by homologous recombination.
Complementation of Mutant Strain 70H2 With the Wild-Type sfb3 Gene
Protoplasts of strain 70H2 were separately transformed with either of two vectors (Online Resource 4) containing the wild-type sfb3 or the mutated sfb3 gene from strain 70H2. Transformants were selected on Vogel's minimal medium with hygromycin B. Thirty candidates from each transformation were transferred to PDA with hygromycin B, and their phenotype compared to 70H2 and TrGA 29-9 transformed with the vector without sfb3 gene as controls.
Transfer of Mutated ssb7 Gene to Strain TrGA 29-9
To validate that the ssb7 mutation was causative for viscosity reduction, the ssb7 locus was specifically mutated in parental strain TrGA 29-9. The method required construction of a vector for transient expression of Streptococcus pyogenes Cas9 in T. reesei, insertion of a guide RNA (sgRNA) expression cassette into this vector, and addition of a region of ssb7 homology for HR-directed genome editing. The region of ssb7 homology introduced the ΔG mutation identified in strain 77B7, along with one synonymous codon change (relative to original frame) downstream of the frame-shift mutation (Online Resource 5). The final strain with mutated ssb7 gene was designated TrGA 29-9 ssb7(TS1).
Deletion of the seb1 Gene from T. reesei Strain Morph Δtku80
The tku80 gene was deleted from M1-1.1(Pyr4+) to decrease nonhomologous recombination and aid in the sequence-specific disruption of seb1 (gene model estEXT_GeneWisePlus.C_50617). The pyr4 gene was then disrupted so that it could be used as a transformation marker, creating Morph ∆tku80 pyr4. A seb1 disruption cassette was prepared by PCR amplification of the disrupted seb1 gene (including inserted pyr4 gene) from genomic DNA obtained from the F16 mutant, along with approximately 500 bp of 5′ seb1 flanking sequences and 500 bp of 3′ seb1 flanking sequences. Strain Morph ∆tku80 pyr4 was transformed with this deletion cassette, to produce strain Morph ∆tku80∆seb1. This strain had an osmotic stress response similar to that of the T. atroviride seb1-deleted strain described in the literature (Peterbauer et al., 2002).
Deletion of the mpg1 Gene from T. reesei Mutant 77B7
The T. reesei mpg1 (gene model estExt_fgenesh5_pg.C_130115) was deleted from T. reesei mutant strain 77B7. The mpg1 disruption cassette in plasmid pRATT249 (Online Resource 6) was amplified by PCR.
A spontaneous pyr2 mutant derivative of mutant strain 77B7 was isolated by 5-fluoro-orotic acid (FOA) selection and called 77B7 p28. This strain was transformed with the mpg1 disruption cassette and plated on Vogel's minimal medium containing sorbitol to select for candidates based on uridine prototrophy. PCR analysis was used to identify transformants in which the mpg1 disruption cassette integrated at the mpg1 locus by homologous recombination. Homologous integration of the Δmpg1 disruption cassette at the mpg1 locus was verified by amplifying DNA fragments of the expected sizes using two primer pairs. One primer pair amplified a DNA fragment starting outside the 5′ end of the disruption cassette region and ending within the 3′ region of the cassette. The other primer pair amplified a DNA fragment starting within the 5′ region of the cassette and ending outside the 3′ end of the disruption cassette region. The generated strain with confirmed homologous integration of the mpg1 disruption cassette was named B7m4.
Results
Mutation of sfb3 Gene
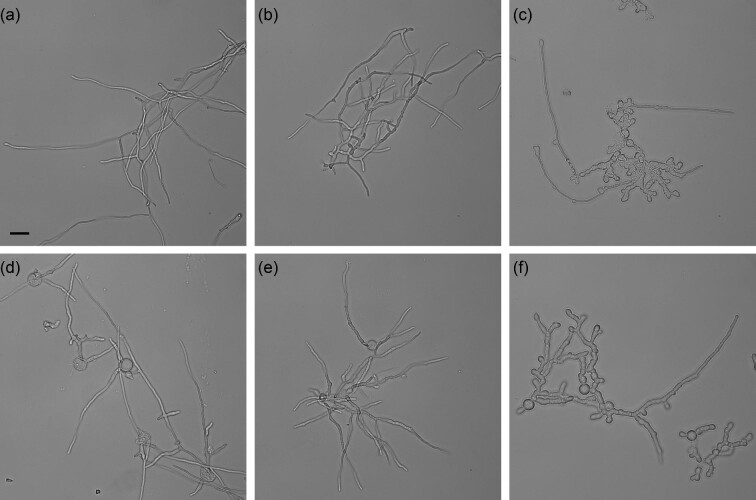
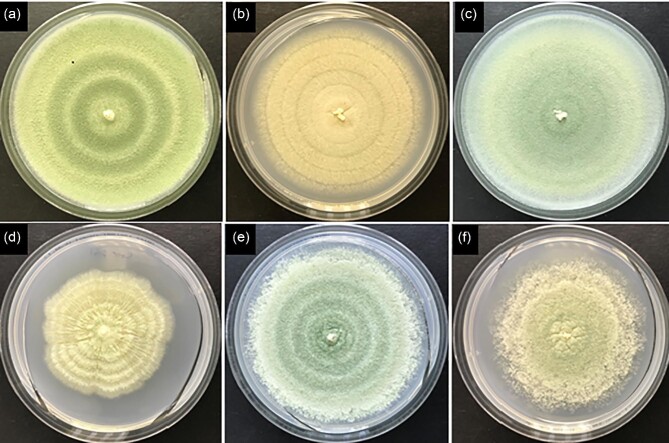
CFW sensitive mutant strain 70H2 was obtained after NTG mutagenesis of the glucoamylase overproducing strain TrGA 29-9. Mutants with altered growth on CFW were observed at a frequency of 3-5 × 10−3 surviving colonies. Secondary screening of shake flask cultures by viscometry led to identification of 70H2 as a candidate reduced viscosity mutant. Despite selection based on reduced viscosity strain 70H2 exhibited little, if any, difference in morphology compared to strain TrGA 29-9 during growth phase in fermenters or when grown for 24 hr in shake flasks with YEG medium (Fig. 1a and b). Compared to the parent strain on PDA plates 70H2 exhibited highly restricted colonial growth in the presence of CFW (Online Resource 3) and slightly restricted growth and reduced sporulation without CFW (Fig. 2a and b). Genome sequence analysis identified nonsynonymous mutations in several genes in 70H2. When attempting to determine which was the causative morphological mutation, one was of primary interest; a nonsense mutation in the sfb3 gene (Gene model: estEXT_GeneWisePlus.C_80450; PID:77570) that changed a lysine codon to a stop codon that would be expected to truncate the protein near the mid-point in the sequence. We have named this gene sfb3 due to the strong similarity of the predicted protein to Saccharomyces cerevisiae Sfb3p.
Fig. 1.
Hyphae of parental strain (a) and strains with morphological mutations sfb3 (b), ssb7 (c), seb1 (d), mpg1 (e), or mpg1 + ssb7 (f). Cultured for 20 hr in shake flasks with YEG at 25°C, 180 rpm. Black bar shown in (a) is =25 μ m.
Fig. 2.
Colonies of parental strain (a) and strains with morphological mutations sfb3 (b), ssb7 (c), seb1 (d), mpg1 (e), or mpg1 + ssb7 (f). Grown for 7 days on Vogel's minimal agar with glucose.
Sfb3p (Lst1p) has homology to the COPII coat subunit Sec24p (Shimoni et al., 2000). Like Sec24p and Sfb2p, Sfb3p is a peripheral ER membrane protein that binds to the COPII subunit Sec23p and is involved in selective export of proteins in COPII vesicles (Roberg et al., 1999). A BLAST search of the publicly available genome sequence of T. reesei using S. cerevisiae Sec24p, Sfb3p, or Sfb2p amino acid sequences as query sequences revealed that T. reesei has a single gene that is most closely homologous to yeast SEC24 (E-value = 0) and a single gene that is most closely homologous to yeast SFB3 (E-value = 1.02e−74; protein sequence alignments are provide in Online Resource 7). No other homolog was identified suggesting that T. reesei does not have a gene equivalent to yeast SFB2.
Studies with S. cerevisiae (Peng et al., 2000; Roberg et al., 1999; Shimoni et al., 2000; Turchini et al., 2000) supported the hypothesis that the sfb3 mutation in 70H2 was causative for the change in morphology. Chromosomal deletion of SFB3 in yeast was not lethal but inhibited transport of the plasma membrane proton-ATPase (Pma1p) to the cell surface, causing poor growth on media of low pH (Roberg et al., 1999). We have noted that growth of 70H2 is affected at low pH (results not shown). The maturation of the glycolipid-anchored plasma membrane protein Gas1p was reported to be differentially impaired in SFB3 knock-out yeast cells (Peng et al., 2000). Gas1p is a β-1,3-glucanosyltransferase involved in cross-linking cell wall glycans (Turchini et al., 2000). Deletion of GAS1 in yeast caused sensitivity to CFW and SDS, osmotic sensitivity, slower growth and morphological changes (pluribudded cells). These changes could be partially overcome by high osmolarity media (Turchini et al., 2000). Mutant strain 70H2 is also sensitive to CFW and SDS, and T. reesei and other filamentous fungi possess clear GAS1 homologs.
The 70H2 mutant phenotype could be complemented by transformation with a vector bearing the wild-type T. reesei sfb3 gene. All 30 transformants tested had a phenotype similar to parental strain TrGA 29-9 on PDA medium with hygromycin B. However, all 30 transformants with the mutated sfb3 gene derived from 70H2 retained the slower radial growth rate of 70H2 (results not shown). To confirm that the phenotype reversion was caused by the presence of the wild-type sfb3 gene and not by a change in the chromosomal DNA, transformants were transferred 4 times on PDA medium (nonselective conditions) and then back to selective medium with hygromycin B to identify unstable candidates that lost the vector. For all candidates that lost the hygromycin B resistance phenotype, the loss correlated with the reappearance of the 70H2 phenotype. These results confirm that the wild-type sfb3 was responsible for restoring the parental phenotype to 70H2.
To further confirm that mutation of sfb3 was responsible for the altered morphology of 70H2 the sfb3 gene was deleted from strain TrGA 29-9. The entire sfb3 coding sequence was replaced with a hygromycin resistance marker gene. The 70H2 and TrGA 29-9 Δsfb3 strains exhibited similar morphology on solid agar, including sensitivity to Congo Red (Online Resource 3), and in liquid culture.
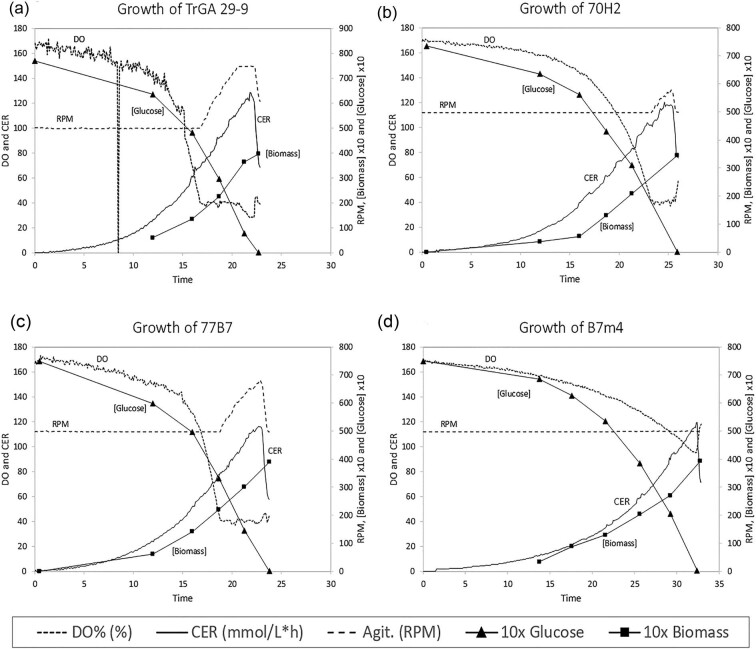
DO2, CER, agitation rate, and biomass concentration during the growth phase of representative fermentations of strains TrGA 29-9 and 70H2 are plotted in Fig. 3a and b, respectively. During growth of TrGA 29-9 agitation was increased to the maximum of 750 rpm and DO2 fell below 40%. In contrast, agitation only needed to be increased slightly above 500 rpm to prevent DO2 falling below 40% during growth of 70H2. Fermentation parameters (estimated viscosity, specific growth rate, and specific productivity) of parent strain TrGA 29-9, mutant strain 70H2 and deletion strain TrGA 29-9 Δsfb3 are reported in Table 2. It was clear that deletion of sfb3 had the same effect on viscosity (66% reduction in viscosity term) and oxygen transfer as the original truncation of the gene observed in 70H2. Specific productivity of glucoamylase was unaltered in the sfb3 mutant strains relative to the parent strain TrGA 29-9, while growth rate was 83% of the parent.
Fig. 3.
Growth characteristics in fermenters of strains (a) TrGA 29-9, (b) 70H2, (c) 77B7, and (d) B7m4. For each strain, the dissolved oxygen (DO %), carbon dioxide evolution rate (CER), agitation rate (RPM), biomass concentration, and glucose concentration were measured throughout the growth phase of fermentation.
Table 2.
Growth and Productivity Characteristics of Parental and Mutant Strains. Repeated Measurements are Shown as Mean ± Standard Deviation With the Number of Fermentations in Parentheses
| Strain | Mutation | Viscosity term (η) relative to TrGA 29-9 | Specific growth rate (μ) relative to TrGA 29-9 | Specific productivity (qp) relative to TrGA 29-9 |
|---|---|---|---|---|
| TrGA 29-9 | None | 1.00 ± 0.07 (n = 3) | 1.00 ± 0.03 (n = 3) | 1.00 |
| 70H2 | sfb3 | 0.336 ± 0.031 (n = 4) | 0.83 ± 0.03 (n = 4) | 0.97 |
| TrGA 29-9 Δsfb3 | sfb3 | 0.334 | 0.91 | 1.08 |
| 77B7 | ssb7 | 0.554 ± 0.110 (n = 2) | 0.80 ± 0.03 (n = 2) | 0.91 |
| Morph ∆tku80 | None | 1.00 | 1.05 | n/a |
| Morph ∆tku80, ∆pyr4, ∆seb1 | seb1 | 0.195 | 0.77 | n/a |
| MAGI | None | 1.07 | 0.95 | n/a |
| MAGI 10-8g | mpg1 | 0.258 | 0.95 | n/a |
| 299m4 | mpg1 | 0.775 ± 0.023 (n = 2) | 0.90 ± 0.08 (n = 2) | n/a |
| B7m4 | ssb7, mpg1 | 0.106 ± 0.011 (n = 3) | 0.76 ± 0.01 (n = 3) | 0.94 |
The estimated viscosity did not consider the shear thinning nature of non-Newtonian fungal fermentation broth. However, the shear rate in the fermentation of 70H2 was only half of that for Morph TrGA 29-9. Thus, even with reduced shear rate, the estimated viscosity was only one-third of that for the parent, further showing the improved viscosity properties brought about by the mutation.
The sfb3 gene was also deleted from strain M1-1.1(Pyr4+). The resulting Morph Δsfb3 strain was subsequently transformed with a glucoamylase expression cassette and glucoamylase productivity of transformants was measured in liquid culture in both microtiter plates and shake flasks. Integration of the expression cassette would occur at random sites in the genome and at variable copy number in these transformants, leading to variability in glucoamylase production. However, some of the transformants had productivities as high as strain TrGA 29-9, which itself was selected as a high producer among many transformants (Online Resource 3). These results confirm that deletion of sfb3 does not significantly impair the expression or secretion of a protein of interest.
Glucoamylase production during fermentation of a strain with the sfb3 mutation was compared with the TrGA 29-9 strain. In both cases, the maximum possible amount of glucose was batched initially to enable the highest possible biomass accumulation during the growth phase without DO2 limitation. Relative to TrGA 29-9, a higher level of batched glucose and a 40% increase in biomass was possible with the sfb3 mutant, leading to a 40% increase in volumetric productivity during the production phase (results not shown).
Mutation of ssb7 Gene
Following NTG mutagenesis of strain TrGA 29-9, we screened 108,000 mutated colonies and found 38 morphological mutants by microscopy in 384 well microtiter plates. Ten of these mutants were selected by evaluation of protein production in shake flask culture and further tested in 15 l fermenters. Morphological mutant strain 77B7 was identified in this manner and exhibited hyperbranching and thickened hyphae compared to the parent strain in liquid culture (Fig. 1c). On agar plates 77B7 had similar colonial growth rate as the parent strain, but with reduced sporulation (Fig. 2c). Whole genome sequence analysis identified eight mutations within coding sequences of eight different genes in the 77B7 genome not present in TrGA 29-9. Any of these mutations alone, or in combination, might have been responsible for the altered morphology and viscosity reduction of 77B7. Complementation by transformation with vectors bearing the eight different open reading frames was used to identify the causative mutation. Transformants carrying seven of the DNA sequences retained the hyperbranching and thick hyphae phenotype observed for the 77B7 mutant strain. In contrast, transformants with one DNA sequence encoding Protein ID 108712, herein named SSB7, reversed the phenotype, at least partially, with thinner and less-branched hyphae (results not shown). Partial complementation can be attributed to transformants having the mutated form of ssb7 of strain 77B7 plus additional ectopic copies of the vector-borne wild-type ssb7. Also, the vectors would be integrated at different sites and at differing copy number in the genomes of these transformants causing variable expression levels of the wild-type ssb7.
The ssb7 gene encodes a newly predicted protein, SSB7, that is identical in part with other protein predictions on the same DNA sequence, for example, T. reesei QM6a Protein ID 108712 and T. reesei Rut-C30 Protein ID 82397, available at the Joint Genome Institute genome portal. The DNA sequences are identical at this locus between the two T. reesei strains, however, the coding regions are predicted to be slightly different. The N-terminus of the strain QM6a gene model harbors two extra exons compared to the strain RUT-C30 gene model, while the strain RUT-C30 gene model contains an additional intron. To resolve the gene model prediction at this locus, we utilized RNAseq data from T. reesei coupled with the Funannotate fungal genome annotation pipeline (Love et al., 2019). Funannotate predicted a gene model that was supported by RNAseq read mapping, while both the model predictions from JGI in QM6a and RUT-C30 had incompatible intron–exon boundaries (see Online Resource 8 for the deduced ssb7 sequence). The ssb7 gene prediction resulted in a translated protein of 1308 amino acids derived from a 4762 bp transcript containing 3 exons, a 462 bp 5′UTR, and a 373 bp 3′UTR. In the mutant 77B7, the ssb7 mutant allele had a single guanine nucleotide deletion in exon 4 (Scaffold 13, 167404) resulting in a frame-shift mutation, and a premature stop prior to the last intron of the ssb7 gene. This allele is referred to as ssb7(fs).
The T. reesei ssb7 gene encodes an uncharacterized, but highly conserved SSB7 protein, including a predicted microtubule interacting and trafficking (MIT) domain. BLAST searches showed that SSB7 protein orthologs are present in Fusarium sp., Neurospora sp., Myceliophthora sp., Talaroymyces sp., Aspergillus sp., and Penicillium sp., but not Saccharomycetes. A multiple sequence alignment of 353 Ascomycete homologs identified four regions of high amino acid identity including the MIT domain (Online Resource 9).
Strain 77B7 was evaluated in 15 l fermenters (Fig. 3). DO2, CER, agitation rate, and biomass concentration during fermentation of strain 77B7 were similar to strain 70H2 even though the hyphal morphology was very different (Fig. 1c). During growth of 77B7 agitation had to be increased from 500 rpm to prevent DO2 falling below 40% but did not reach 700 rpm. Fermentation parameters (estimated viscosity, specific growth rate, and specific productivity) of strain 77B7 are reported in Table 2. Specific growth rate and specific productivity of 77B7 were both 90% of the parent strain TrGA 29-9 and the viscosity term was reduced to less than half that of the parent. As with 70H2, the viscosity term for 77B7 was reduced significantly from TrGA 29-9, despite having a shear rate that was only two-thirds that of the parent. Once again, this only further shows the improved viscosity properties of the mutant.
To further validate that the ssb7 mutation was causative for the viscosity reduction, the ssb7 locus was specifically mutated in parental strain TrGA 29-9 using Cas9-endonuclease facilitated homologous recombination. The strain with this allele of ssb7, TrGA 29-9 ssb7 (TS1), exhibited reduced viscosity demonstrating that mutation of ssb7 alone was sufficient to confer the desired phenotype observed in strain 77B7 (Online Resource 5).
Mutation of seb1 Gene
Agrobacterium-mediated transformation of filamentous fungi has been shown to be an efficient method of IM (Sugui et al., 2005). Mutant F16 was identified following IM of parent strain T4 pyr4, screening of microtiter plate liquid cultures and sieving of liquid cultures to enrich for more compact growth. Secondary screening involved evaluation of morphological changes on solid and in liquid medium, and evaluation of protein production in shake flask cultures. On agar plates mutant F16 had clearly restricted growth compared to the T4 parent (Fig. 2d). In liquid medium, F16 was difficult to distinguish from the parent strain when grown either in shake flasks (Fig. 1d) or in fermenters.
The pyr4 marker gene insertion site in mutant strain T4 pyr4 F16 was identified by inverse PCR as being at Scaffold 5 at 1203727 in the T. reesei JGI genome database v2. The gene (gene model: estExt_GeneWisePlus.C_50617; Protein ID: 76505) found at this site has homology to the seb1 gene found in several other fungi including Aspergillus fumigatus and T. atroviride. The seb1 gene encodes an AGGGG-binding protein, which was reported to be involved in osmotic stress response (Peterbauer et al., 2002). Indeed, the T4 pyr4 F16 strain was found to have a stress response phenotype similar to that described in the literature (colonies on PDA restricted by high sorbitol or salt). Since Southern analysis showed this site to be the only T-DNA insertion in the T4 pyr4 F16 strain, it follows that disruption of the seb1 gene was responsible for the observed morphological changes. The specific productivity of total secreted protein (predominantly native cellulases and hemicellulases) during fermentation of T4 pyr4 F16 was unchanged relative to the T4 parental strain (results not shown).
The seb1 gene was disrupted in strain Morph Δtku80, re-creating the same insertion of the pyr4 marker gene in this strain as observed in strain T4 pyr4 F16. The resulting strain, Morph ∆tku80, ∆pyr4, ∆seb1, exhibited similar morphology and osmotic sensitivity as T4 pyr4 F16. DO2, CER, agitation rate, and biomass concentration during fermentation of strain Morph ∆tku80, ∆pyr4, ∆seb1 were similar to strain 70H2 and 77B7 shown in Fig. 3b and c. During growth of Morph ∆tku80, ∆pyr4, ∆seb1 DO2 did not fall below 55% even though agitation was not increased above 500 rpm. Fermentation parameters (estimated viscosity and specific growth rate) of strain Morph ∆tku80, ∆pyr4, ∆seb1 are reported in Table 2. Specific growth rate of Morph ∆tku80, ∆pyr4, ∆seb1 was 77% of the parent strain and the viscosity term was reduced to less 20% of the parent.
Mutation of mpg1 gene
Mutant strain MAGI 10-8g was obtained by IM of parent strain MAGI and was initially identified by its slow growth on transformation plates. Secondary screening involved observation of morphological changes on solid and in liquid medium, and testing protein production in shake flask cultures. On agar plates MAGI 10-8g had slightly restricted growth compared to the MAGI parent. In liquid medium, MAGI 10-8g hyphae appeared similar to the MAGI parent in shake flask culture (Fig. 1e) and in fermenters.
PCR analysis was used to show that the pyr2 marker insertion in strain mutant MAGI 10-8g resulted in deletion of the region 369089 to 370324 of Scaffold 13 in the T. reesei JGI genome database v2. The gene found at this site is the mpg1 gene (gene model: estEXT_fgenesh5_pg.C_130115, Protein ID 122551) that is found in other fungi including Aspergillus clavatus, A. fumigatus, and Neosartorya fischeri. As described by Kruszewska et al. and Zakrzewska et al. mpg1 from T. reesei encodes for a GTP:α-d-mannose-1-phoshate guanyltransferase that can play a major regulatory role in early stages of protein glycosylation (Kruszewska et al., 1998; Zakrzewska et al., 2003) Southern analysis showed that this strain contained only one copy of the pyr2 gene in addition to the native copy indicating that one disruption event had taken place.
A spontaneous pyr2 variant of strain TrGA 29-9, named TrGA 29-9 pyr2, was obtained by selection on agar plates containing FOA. The mpg1 gene was inactivated by transformation with an mpg1 disruption cassette (Online Resource 6) to create strain 299m4.
Parental strain MAGI, mutant strain MAGI 10-8g and strain 299m4 were evaluated in 15 l fermenters. DO2, CER, agitation rate, and biomass concentration during fermentation of strain MAGI 10-8g were similar to strains 70H2 and 77B7 shown in Fig. 3b and c. During growth of MAGI 10-8g agitation was only increased slightly to 513 rpm in order to maintain DO2 at or above 40%. Fermentation parameters (estimated viscosity and specific growth rate) of MAGI 10-8g are reported in Table 2. The viscosity term was approximately 25% of the parent strain while specific growth rate was reduced by 5%. The viscosity term for strain 299m4 was reduced to 77% that of the parent strain TrGA 29-9, which was not as dramatic as the reduction observed with MAGI 10-8g. It is possible that other mutations not associated with pyr2 marker insertion in MAGI 10-8g contributed to the low viscosity of this strain.
Combined Effect of ssb7 and mpg1 Mutations
Strain 77B7 p28 was transformed with an mpg1 disruption cassette (Online Resource 6) to create a strain with mutations in both ssb7 and mpg1. This strain exhibited a similar morphology in liquid culture to a strain having only the ssb7 mutation (Fig. 1f), whereas on agar plates it showed a more restricted colonial growth than strains having only the ssb7 or the mpg1 mutation (Fig. 2f). However, deletion of the mpg1 gene from strain 77B7 resulted in a strain (B7m4) having a further reduction in broth viscosity. DO2, CER, agitation rate, and biomass concentration during growth phase of one fermentation of strain B7m4 are shown in Fig. 3d. During growth agitation was not increased above 500 rpm while DO2 remained above 90%. Fermentation parameters (estimated viscosity, specific growth rate, and specific productivity) of strain B7m4 are reported in Table 2. Specific productivity of glucoamylase was 94% and specific growth rate was 76%, respectively, of that recorded for strain TrGA 29-9, whereas the calculated viscosity term was reduced to 11% of that for strain TrGA 29-9.
Discussion
We have developed screening methods and identified mutations that reduce the viscosity of T. reesei fermentation broth and, in one case, clearly altered hyphal morphology. The reduced viscosity improves oxygen transfer allowing strains with these mutations to accumulate greater biomass during the growth phase of the fermentation. This in turn results in higher volumetric productivity during production phase due to minimal or no reduction in specific productivity or yield. Generally, the frequency of morphological mutants was higher after NTG mutagenesis than IM (1:3,000–5,000 versus 1:10,000 mutated spores screened, respectively). Single mutations that reduced viscosity while retaining good specific productivity and good specific growth rate (>80% of the parent for both) were found. A higher proportion of IM mutants than NTG mutants retained good specific productivity. Although single mutations that conferred much lower viscosity were found, none maintained secreted protein specific productivity above 50% of the parent. It may require accumulation of several mutations to achieve a combination of very low viscosity with maintained good specific productivity. A promising step in this direction was observed in strain B7m4, which combines ssb7 and mpg1 mutations and showed the lowest viscosity and highest DO2 values combined with specific productivity >90% of strain TrGA 29-9. With strains having this degree of viscosity reduction it has been possible to increase the amount of glucose added during the growth phase such that twice as much biomass accumulated before the switch to production phase compared to that possible with the parental strain (data not shown). This has enabled a 100% increase in volumetric productivity compared to the parent strain. As a result, significant reduction in required industrial fermentation capacity can be achieved.
None of the mutants that were shown to have reduced viscosity and improved oxygen transfer in liquid medium exhibited an increase in specific productivity. This suggests that the morphological changes that we identified were not beneficial to protein production or secretion on a per biomass basis. This is similar to the result obtained by Bocking et al. (1999) with A. oryzae although the observed magnitude of improvement in volumetric productivity was much greater in the present study, possibly reflecting the different fermentation protocols employed.
The function of the ssb7 gene and possible effects of mutation are unknown. However, the other three mutations identified in this work that alter viscosity are all expected to have effects on cell wall integrity. One screen utilized in this study was for CFW sensitivity in the knowledge that this chitin-binding agent could identify mutants of filamentous fungi with altered response to osmotic stress, cell wall integrity and morphology (Hill et al., 2006). Mutation of sfb3 causes CFW and osmotic sensitivity, possibly due to altered transport or functionality of β-1,3-glucanosyltransferase involved in glucan remodeling in the cell wall as suggested by studies using yeast (Peng et al., 2000; Turchini et al., 2000). By analogy to the function of STRE-binding transcription factors MSN2/4p and their role in osmotic signaling in yeast (Martinez-Pastor et al., 1996), Seb1 was implicated in the osmotic stress response in T. atroviride. Although not essential for the osmotic stress response there is evidence that T. atroviride Seb1 has a role in glycerol accumulation (Seidl et al., 2004). MPG1 is a GTP:α-d-mannose-1-phoshate guanyltransferase that has a role in protein glycosylation and overexpression of the mpg1 gene in T. reesei was reported to increase GDP-mannose and cause hypermannosylation of secreted glycoproteins (Zakrzewska et al., 2003). Mutation of this gene may affect cell wall galactomannans or mannoproteins. Additionally, a CFW sensitive morphological mutant of A. nidulans was shown to have an altered GDP-Mannose transporter gene (Jackson-Hayes et al., 2008).
It is difficult to predict the oxygen transfer properties during fermentation based on mutant morphology in liquid culture. Strains with mutated seb1, sfb3, or mpg1, do not have dramatically altered morphology during growth in fermenters and yet show similar reductions in viscosity. The underlying basis for reduced viscosity in these strains is unknown. Conversely, the morphology of strains with mutated ssb7 are very different. Nonetheless, this mutation alone leads to a similar reduction in viscosity as the other mutations. A strain with both ssb7 and mpg1 mutations looks very similar to one with only an ssb7 mutation but this double mutant strain has a much reduced viscosity compared to any of the single mutants.
Supplementary Material
Acknowledgments
We are grateful to Jonathan M. Palmer for assistance with modeling the ssb7 gene structure. Igor Nikolaev and Paulien Kruithof are thanked for providing photographs of fungal colonies.
Contributor Information
Elizabeth Bodie, Nutrition and Biosciences, DuPont, Palo Alto, CA 94304, USA.
Aleksandra Virag, Nutrition and Biosciences, DuPont, Palo Alto, CA 94304, USA.
Robert J Pratt, Nutrition and Biosciences, DuPont, Palo Alto, CA 94304, USA.
Nicholas Leiva, Nutrition and Biosciences, DuPont, Palo Alto, CA 94304, USA.
Michael Ward, Nutrition and Biosciences, DuPont, Palo Alto, CA 94304, USA.
Tim Dodge, Nutrition and Biosciences, DuPont, Palo Alto, CA 94304, USA.
Funding
None declared.
Conflict of Interest
The authors declare no conflict of interest.
Data Availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.
References
- Bhargava S., Wenger K. S., Marten M. R. (2003). Pulsed addition of limiting-carbon during Aspergillus oryzae fermentation leads to improved productivity of recombinant enzyme. Biotechnology and Bioengineering, 82, 111–117. [DOI] [PubMed] [Google Scholar]
- Bocking S. P., Wiebe M. G., Robson G. D., Hansen K., Christiansen L. H., Trinci A. P. J. (1999). Effect of branch frequency in Aspergillus oryzae on protein secretion and culture viscosity. Biotechnology and Bioengineering, 65, 638–648. [DOI] [PubMed] [Google Scholar]
- Cairns T. C., Zheng X., Zheng P., Sun J., Meyer V. (2019). Moulding the mould: Understanding and reprogramming filamentous fungal growth and morphogenesis for the next generation cell factories. Biotechnology for Biofuels, 12, 77. 10.1186/s13068-019-1400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow N. V., Ju L.-K. (2012). Promoting pellet growth of Trichoderma reesei Rut C30 by surfactants for easy separation and enhanced cellulase production. Enzyme and Microbial Technology, 50, 311–317. [DOI] [PubMed] [Google Scholar]
- Charles M., Wilson J. (1994). Fermenter Design. In Lydersen B. K., D'Elia N. A., Nelson K. L. (Eds.), Bioprocess engineering: Systems, equipment and facilities (pp. 3–68). Wiley. [Google Scholar]
- Christensen T., Dersjant-Li Y., Sewalt V., Mejldal R., Haaning S., Pricelius S., Nikolaev I., Sorg R. A., de Kreij A. (2020). In vitro characterization of a novel consensus bacterial 6-Phytase and one of its variants. Current Biochemical Engineering, 6, 156–171. [Google Scholar]
- Domingues F. C., Queiroz J. A., Cabral J. M. S., Fonseca L. P. (2000). The influence of culture conditions on mycelial structure and cellulase production by Trichoderma reesei Rut C-30. Enzyme and Microbial Technology, 26, 394–401. [DOI] [PubMed] [Google Scholar]
- England G. R., Kelley A., Mitchinson C. (2010). Induction of gene expression using a high concentration sugar mixture. U.S. Patent No. 7,713,725.
- Fiedler M. R. M., Barthel L., Kubisch C., Nai C., Meyer V. (2018). Construction of an improved Aspergillus niger platform for enhanced glucoamylase secretion. Microbial Cell Factories, 17, 95. 10.1186/s12934-018-0941-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz E., Gamauf C., Seiboth B., Wanka F. (2019). Deletion of the small GTPase rac1 in Trichoderma reesei provokes hyperbranching and impacts growth and cellulase production. Fungal Biology and Biotechnology, 6, 16. 10.1186/s40694-019-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Li M., Liu W., Bai Y., Tu T., Wang Y., Zhang J., Luo H., Yao B., Huang H., Su X. (2020). RNAi-mediated gene silencing of Trcot1 induces a hyperbranching phenotype in Trichoderma reesei. Journal of Microbiology and Biotechnology, 30, 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ochoa F., Gomez E. (2009). Bioreactor scale-up and oxygen transfer rate in microbial processes: An overview. Biotechnology Advances, 27, 153–176. [DOI] [PubMed] [Google Scholar]
- Gordon C. L., Archer D. B., Jeenes D. J., Doonan J. H., Wells B., Trinci A. J. P., Robson G. D. (2000). A glucoamylase::GFP gene fusion to study protein secretion by individual hyphae of Aspergillus niger. Journal of Microbiological Methods, 42, 39–48. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Molecular Biology, 25, 989–994. [DOI] [PubMed] [Google Scholar]
- He R., Li C., Ma L., Zhang D., Chen S. (2016). Effect of highly branched hyphal morphology on the enhanced production of cellulases in Trichoderma reesei DES-15. 3 Biotech, 6, 214. 10.1007/s13205-016-0516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. W., Loprete D. M., Momany M., Harsch L. M., Livesay J. A., Mirchandani A., Murdock J. J., Vaughan M. J., Watt M. B. (2006). Isolation of cell wall mutants in Aspergillus nidulans by screening for hypersensitivity to calcofluor white. Mycologia, 98, 399–409. [DOI] [PubMed] [Google Scholar]
- Jackson-Hayes L., Hill T. W., Loprete M., Fay L. M., Gordon B. S., Nkashama S. A., Patel R. V., Sartain C. V. (2008). Two GDP-mannose transporters contribute to hyphal form and cell wall integrity in Aspergillus nidulans. Microbiology (Reading, England), 154, 2037–2047. [DOI] [PubMed] [Google Scholar]
- Kruszewska J. S., Saloheimo M., Penttilä M., Palamarczyk G. (1998). Isolation of a Trichoderma reesei cDNA encoding GTP: α-d-mannose-1-phosphate guanyltransferase involved in early steps of protein glycosylation. Current Genetics, 33, 445–450. [DOI] [PubMed] [Google Scholar]
- Kwon M. J., Nitsche B. M., Arentshorst M., Jorgensen T. R., Ram A. F. J., Meyer V. (2013). The transcriptomic signature of RacA activation provides new insights into the morphogenetic network of Aspergillus niger. PLoS One, 8, e68946. 10.1371/journal.pone.0068946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love J., Palmer J. M., Stajich J., Esser T., Kastman E., Bogema D., Winter D. (2019). Funannotate genome annotation pipeline. Available via Zenodo.org. 10.5281/zenodo.1134477. [DOI]
- Mandels M., Reese E. T. (1957). Induction of cellulase in Trichoderma viride as influenced by carbon sources and metals. Journal of Bacteriology, 73, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor M. T., Marchler G., Schuller C., Marchler-Bauer A., Ruis H., Estruch F. (1996). The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE). The EMBO Journal, 15, 2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Montenecourt B. S., Eveleigh D. E. (1979). Selective screening methods for the isolation of high yielding cellulase mutants of Trichoderma reesei. Advances in Chemistry Series, 181, 289–301. [Google Scholar]
- Nevalainen H., Suominen P., Taimisto K. (1994). On the safety of Trichoderma reesei. Journal of Biotechnology, 37, 193–200. [DOI] [PubMed] [Google Scholar]
- Nykänen M., Saarelainen R., Raudaskoski M., Nevalainen H., Mikkonen A. (1997). Expression and secretion of barley cysteine endopeptidase B and cellobiohydrolase I in Trichoderma reesei. Applied and Environmental Microbiology, 63, 4929–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouedraogo J. P., Arentshorst M., Nikolaev I., Barends S., Ram A. F. J. (2015). I-SceI-mediated double-strand DNA breaks stimulate efficient gene targeting in the industrial fungus Trichoderma reesei. Applied Microbiology and Biotechnology, 99, 10083–10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakula T. M., Salonen K., Uusitalo J., Penttilä M. (2005). The effect of specific growth rate on protein synthesis and secretion in the filamentous fungus Trichoderma reesei. Microbiology (Reading, England), 151, 135–143. [DOI] [PubMed] [Google Scholar]
- Peng R., De Antoni A., Gallwitz D. (2000). Evidence for overlapping and distinct functions in protein transport of coat protein SEC24p family members. Journal of Biological Chemistry, 275, 11521–11528. [DOI] [PubMed] [Google Scholar]
- Penttilä M., Nevalainen H., Ratto M., Salminen E., Knowles J. (1987). A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene, 61, 155–164. [DOI] [PubMed] [Google Scholar]
- Peterbauer C. K., Lischer D., Kubicek C. P. (2002). The Trichoderma atroviride seb1 (stress response element binding) gene encodes an AGGGG-binding protein which is involved in the response to high osmolarity stress. Molecular Genetics and Genomics, 268, 223–231. [DOI] [PubMed] [Google Scholar]
- Peterson R., Nevalainen H. (2012). Trichoderma reesei RUT-C30—thirty years of strain improvement. Microbiology (Reading, England), 158, 58–68. [DOI] [PubMed] [Google Scholar]
- Roberg K. J., Crotwell M., Espenshade P., Gimeno R., Kaiser C. A. (1999). LST1 is a Sec24 homologue used for the selective export of the plasma membrane ATPase from the endoplasmic reticulum. Journal of Cell Biology, 145, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl V., Seiboth B., Karaffa L., Kubicek C. P. (2004). The fungal STRE-element-binding protein Seb1 is involved but not essential for glycerol dehydrogenase (gld1) gene expression and glycerol accumulation in Trichoderma atroviride during osmotic stress. Fungal Genetics and Biology, 41, 1132–1140. [DOI] [PubMed] [Google Scholar]
- Sheir-Neiss G., Montenecourt B. S. (1984). Characterization of the secreted cellulases of Trichoderma reesei wild type and mutants during controlled fermentations. Applied Microbiology and Biotechnology, 20, 46–53. [Google Scholar]
- Shimoni Y., Kurihara T., Ravazzola M., Amherdt M., Orci L., Schekman R. (2000). Lst1p and Sec24p cooperate in sorting of the plasma membrane ATPase into COPII vesicles in Saccharomyces cerevisiae. Journal of Cell Biology, 151, 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui J. A., Chang Y. C., Kwon-Chung K. J. (2005). Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: An efficient tool for insertional mutagenesis and targeted gene disruption. Applied and Environmental Microbiology, 71, 1798–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchini A., Ferrario L., Popolo L. (2000). Increase of external osmolarity reduces morphogenetic defects and accumulation of chitin in a gas1 mutant of Saccharomyces cerevisiae. Journal of Bacteriology, 182, 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US FDA . (2011). Glucoamylase enzyme preparation from Trichoderma reesei expressing the glucoamylase gene from T. reesei (glucoamylase enzyme preparation). GRN No. 372. Available via www.cfsanappsexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=372.
- US FDA . (2019). Lipase from Aspergillus tubingensis produced in Trichoderma reesei. GRN No. 808. Available via www.cfsanappsexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=808.
- Valkonen M., Kalkman E. R., Saloheimo M., Penttilä M., Read N. D., Duncan R. (2007). Spatially segregated SNARE protein interactions in living fungal cells. Journal of Biological Chemistry, 282, 22775–22785. [DOI] [PubMed] [Google Scholar]
- Vogel H. J. (1956). A convenient growth medium for Neurospora (Medium N). Microbial Genetics Bulletin, 13, 42–43. [Google Scholar]
- Wösten H. A. B., Moukha S. M., Sietsma J. H., Wessels J. G. H. (1991). Localisation of growth and secretion of proteins in Aspergillus niger. Journal of General Microbiology, 137, 2017–2023. [DOI] [PubMed] [Google Scholar]
- Zakrzewska A., Palamarczyk G., Krotkiewski H., Zdebska E., Saloheimo M., Penttilä M., Kruszewska J. S. (2003). Overexpression of the gene encoding GTP:mannose-1-phosphate guanyltransferase, mpg1, increases cellular GDP-mannose levels and protein mannosylation in Trichoderma reesei. Applied and Environmental Microbiology, 69, 4383–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Love J., Palmer J. M., Stajich J., Esser T., Kastman E., Bogema D., Winter D. (2019). Funannotate genome annotation pipeline. Available via Zenodo.org. 10.5281/zenodo.1134477. [DOI]
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.