Abstract
Estimating the basic reproduction number of a pandemic and the changes that appear on this value over time provide a good understanding of the contagious nature of the virus and efficiency of the controlling strategies. In this paper, we focus on studying the basic reproduction number (R0) for two important variants of COVID-19 pandemic in Iran: Alpha and Delta variants. We use four different methods, three statistical models and one mathematical model, to compute R0: Exponential Growth Rate (EGR), Maximum Likelihood (ML), Sequential Bayesian (SB), and time-dependent SIR model. Alpha variant of COVID-19 was active in Iran from March 10, 2021 until June 10, 2021. Our computations indicate that total R0 of this variant according to EGR, ML, SB, and SIR model is respectively 0.9999 (95% CI: 0.9994-1), 1.046 (95% CI: 1.044-1.049), 1.06 (95% CI: 1.03-1.08), and 2.79 (95% CI: 2.77-2.81) in the whole active time interval. Moreover, during the time interval from April 3, 2021 to April 9, 2021 in which this variant was in its exponential growth in Iran, R0 of Alpha variant in Iran according to SB, EGR, ML, and SIR model is respectively 2.26 (95% CI: 2.04-2.49), 2.64 (95% CI: 2.58-2.7), 11.38 (95% CI: 11.28-11.48), and 12.13 (95% CI: 12.12-12.14). Delta variant was active in Iran during the time interval from June 22, 2021 until September 22, 2021. Our computations show that during the time interval from July 3, 2021 to July 8, 2021 in which this variant was in its exponential growth in Iran, R0 of Delta variant in Iran according to SB, EGR, ML, and SIR model is respectively 3 (95% CI: 2.34-3.66), 3.1 (95% CI: 3.02-3.17), 12 (95% CI: 11.89-12.12), and 23.3 (95% CI: 23.19-23.41). Further, total R0 of Delta variant in Iran in the whole active time interval according to EGR, ML, SB, and SIR model is respectively 1.042 (95% CI: 1.04-1.043), 1.053 (95% CI: 1.051-1.055), 0.79 (95% CI: 0.63-0.95), and 5.65 (95% CI: 5.6-5.7). As the results show Delta variant was more severe than Alpha variant in Iran. Chasing the changes in R0 during each variant shows that the controlling strategies applied were effective in controlling the virus spread.
Introduction
After the first appearance of a novel type of pneumonia in December 2019 in Wuhan, Hubei Province of China, a new type of coronavirus, initially named 2019-nCoV on January 7, 2020, was identified as the cause of the disease [1, 2]. Examining the genome sequence of this virus declared a high similarity to Severe Acute Respiratory Syndrome-related coronavirus (SARS-CoV) that swept China in 2003. Thus, on February 11, 2020, the International Committee on Taxonomy of Viruses (ICTV) renamed 2019-nCoV to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [3]. This virus got briefly referred as COVID-19 then by the World Health Organization (WHO) [4]. The most common symptoms of COVID-19 include fever, cough, fatigue, and dyspnea [5, 6]. COVID-19 started to rapidly spread not only in China, but also in all over the world. Unfortunately, in less than four months after the initiation of this virus, more than 118,000 new cases were identified in 114 different countries. Acceleration and high volume transmission of the infection of COVID-19 made this virus classified as a pandemic on March 11, 2020, by WHO [7]. Since then several different mutations of COVID-19, Alpha, Beta, Gamma, Delta, and newly appeared Omicron variants, have been emerged with difference in severity and transmission rate of this virus [8, 9]. Preventive strategies such as social distancing, mask wearing, and regular hand-washing on one side, and strengthening the immune system by following a balanced diet enriched with adequate amount of protein, vitamins, and minerals on the other side play a crucial role against COVID-19 [6, 10, 11]. Further, high rate of COVID-19 global vaccination has a vital impact on mitigating COVID-19 [12, 13]. Hopefully, several COVID-19 vaccines have been authorized so far, and some new ones are under different steps of trial [12–14]. Although efficiency of vaccination is not ignorable, sufficiency of primary vaccination is debatable. Since vaccine-induced protection from COVID-19 changes variant by variant, boosting may be ultimately needed [15]. Therefore, studying epidemiologic characteristics of variants of COVID-19 is crucial.
The variant of B.1.1.7 which is also known as Alpha variant was first observed in the south of England [16]. The patients infected by B.1.1.7 had more cough, sore throat, myalgia, and fatigue. Loss of sense of smell and taste were less reported among infections of this variant [17]. Combining several behavioral and epidemiological data sources with statistical and dynamic modeling have indicated that Alpha variant of COVID-19 pandemic is approximately 43% to 100% more transmissible than the previous infections [18]. On December 18, 2020, at the same time as the designation of Alpha variant, African authorities identified a new variant of COVID-19, called B.1.351 or Beta variant, which was swiftly infecting people [19]. Study of Beta variant showed more infections in elderly people, and increase in the number of hospitalizations and deaths compared to Alpha variant [20]. Another variant of COVID-19 pandemic found in Brazil on January 21, 2021, was called P.1 or Gamma variant. Mutations in Gamma variant caused reinfection to the virus in spite of existent antibodies [21]. Following that, in April 2021, the variant of B.1.617.2 or Delta variant was first identified in India [9]. The most common symptoms in the individuals with Delta variant were headache, sore throat, and a runny nose [22]. It is reported that one dose of COVID-19 vaccine does not provide adequate resistance against Delta variant [8]. Due to faster transmission rate and less effectiveness of known public health treatments in this variant, WHO considered Delta variant as a Variant of Concern (VOC) on May 11, 2021 [9]. The number of total infections classified by variants of COVID-19 and countries are presented in Table 1.
Table 1. Number of COVID-19 infections by October 18, 2021, classified by variants and countries.
| Country | Alpha Variant | Beta Variant | Gamma Variant | Delta Variant |
|---|---|---|---|---|
| Italy | 25,979 [23] | 128 [24] | 2,585 [25] | 22,588 [26] |
| India | 4,184 [23] | 240 [24] | 5 [25] | 36,509 [26] |
| SriLanka | 430 [23] | 6 [24] | unknown | 730 [26] |
| Turkey | 1,916 [23] | 502 [24] | 169 [25] | 45,200 [26] |
| USA | 232,486 [23] | 2,998 [24] | 27,870 [25] | 588,640 [26] |
In Iran, the first case of COVID-19 infection was noticed in Qom on February 19, 2020 [27]. Since then, the number of confirmed cases has risen steeply so that by October 11, 2021, the cumulative total number of confirmed cases and deaths due to COVID-19 in Iran was respectively 5,716,394 and 122,868 [28]. Table 2 provides a detailed description about the total number of infections and deaths in two important variants of COVID-19 pandemic, Alpha and Delta variant, in Iran. The data is collected form Iranian Students News Agency (ISNA) [29]. Although acceleration of vaccination plays a vital role in deceleration of the infection, the impact of vaccination is not similar against different variants of COVID-19 pandemic. Variant by variant, COVID-19 virus empowers. The soonest this pandemic is closed, the less costs are forced on societies. Much is still unknown about the nature of this virus. Therefore, study of COVID-19 properties, such as the transmission rate of different variants can improve knowledge about nature of the virus, and can help societies with effective strategies to slow down the speed of spread or even stop this pandemic [30].
Table 2. The number of total cases and deaths in Alpha and Delta variants of COVID-19 in Iran.
| Variant | Total Cases | Total Deaths |
|---|---|---|
| Alpha variant | 1,296,553 | 20,929 |
| Delta variant | 2,596,064 | 39,787 |
One of the tools that can be utilized to evaluate the dissemination and transmission rate of a pandemic in distinctive periods is the basic reproduction number, denoted by R0. The basic reproduction number is defined as the average number of secondary cases directly generated by an individual infection [31]. It assumes that the entire population is susceptible against the infection, which is true at the beginning of the virus spread. Thus, the basic reproduction number describes the power of virus transmission under no external control. External controls such as city lockdown, isolation of confirmed cases, and traffic control can play a significant role in decreasing the transmissibility of the virus [32]. When R0 > 1, the virus is spreading. R0 becoming less and less, shows the effectiveness of the external controls, and when R0 < 1, it means the pandemic is over [33]. Estimating R0 for a long-running outbreak, such as COVID-19 pandemic, provides a better recognition of nature of the virus and is helpful for establishing and evaluating controlling strategies for the virus spread. Table 3 specifies R0 for several epidemics so far. Moreover, Table 4 estimates R0 of the ongoing COVID-19 pandemic during specific time intervals in several countries.
Table 3. Estimated R0 for various epidemics.
Table 4. Estimated R0 for COVID-19 in different countries.
There are few works studying R0 for different variants of COVID-19 pandemic in different countries as far as we are aware are, although the general knowledge is Delta variant is more contagious than the previous variants of COVID-19 [44]. A study in Scotland reported that Delta variant doubled the risk of hospitalization compared to Alpha variant of COVID-19 pandemic especially for unvaccinated people, indicating that Delta variant caused more severe infections [45]. Hence, in this paper we focus on providing a comparative study of R0 of Alpha and Delta variants of COVID-19 pandemic in Iran.
Data source
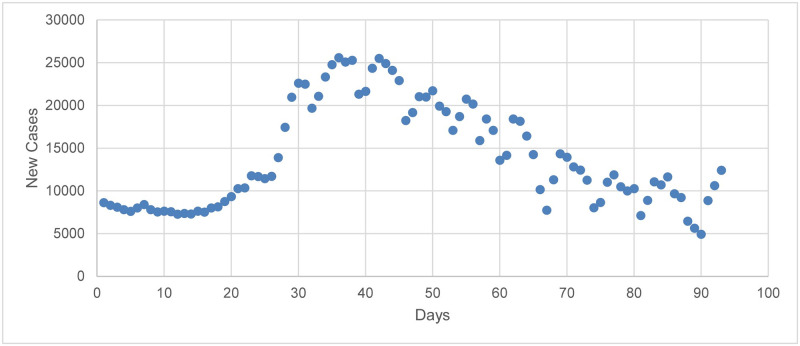
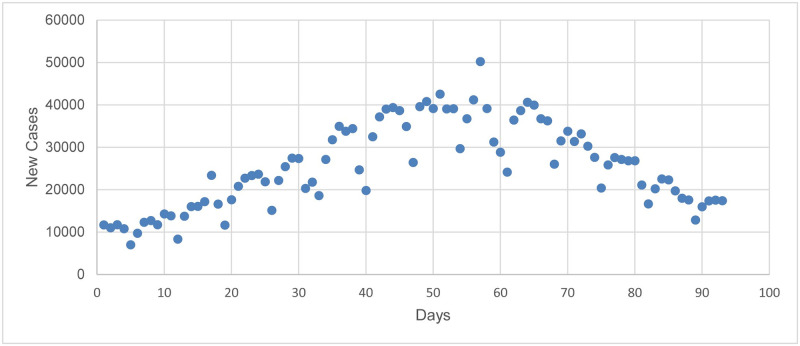
For our analysis of the basic reproduction number (R0) of variants of COVID-19 pandemic in Iran, we have collected our data set from Iranian Students News Agency (ISNA), daily outbreak news [29]. We have collected data including the number of infected cases, the number of recovered cases, and the number of deaths for two different time intervals: first from March 10, 2021 to June 10, 2021 (Alpha variant) and second from June 22, 2021 to September 22, 2021 (Delta variant). Figs 1 and 2 illustrate the diagram of daily infections in Iran classified by Alpha and Delta variants.
Fig 1. The number of daily infections for Alpha variant in Iran.
Fig 2. The number of daily infections for Delta variant in Iran.
Methods
There are various methods used for estimating R0, ranging from the epidemiological compartmental models such as SIR to a variety of statistical methods. Due to different nature of each method, the values produced by these methods can not be completely comparable with each other. However, tracking R0 value of each method can provide a good understanding of the virus behavior. In this paper, we use time-dependent SIR model and the three most famous statistical methods (maximum likelihood, exponential growth rate-based method, and sequential Baysian method) for estimating R0 of Alpha and Delta variants of COVID-19 in Iran. These methods are explained in details in the following.
Time-dependent SIR model
Time-dependent SIR model is a simple and at the same time an efficient compartmental epidemiological model for infectious diseases [33]. In this model, the population is divided into three categories: Susceptible (S), Infectious (I), and Recovered (R). At the beginning of a pandemic, all population is in the state S. Then, they move to the state I with the rate of β, and afterwards to the state R with the rate of γ. Hence, SIR model has two time-varying variables: the transmission rate β and the recovering rate γ. The basic reproduction number in this model is defined as β(t)/γ(t) since an infected person will be recovered in 1/γ days (on average), and he/she is in contact with β (on average) people during the infectious period. Thus, in time-dependent SIR model, S(t), X(t) and R(t) respectively indicates the number of susceptible, the number of infected, and the number of recovered people at time t. In order to compute R0 in this model, β(t) and γ(t) should be calculated. This model follows the following discrete difference equations [46]:
| (1) |
| (2) |
| (3) |
The parameter n is the total number of population. At the beginning of a pandemic, most of the population are in the susceptible state; as a result, we assume S(t) ≃ n, t ≥ 0. So, we can simplify Eqs (1) and (2) as follows:
| (4) |
| (5) |
From the difference equations above, β(t) and γ(t) can be easily computed as
| (6) |
| (7) |
Now, fitting the data set to these simplified equations leads to estimating R0 for the corresponding pandemic.
Maximum Likelihood method
Using Maximum Likelihood method (ML) for estimating R0 was introduced by White and Pagano [47]. This method assumes that a primary case generates the secondary cases according to a Poisson distribution with mean R0. In this method, N0, …, Nt demonstrate the incident cases, and W represents the generation time distribution. Therefore, R0 can be estimated by maximizing the log-likelihood as follows.
| (8) |
Where
| (9) |
The likelihood should be computed in a period of exponential growth. It is noteworthy to mention that maximum likelihood method has some assumptions and if any infraction happens, the result alters. These assumptions are as follows: No data should be missing and the population has to be uniformly mixed.
Exponential Growth Rate-based method
The method of Exponential Growth Rate (EGR) for calculating R0 was introduced by Wallinga and Lipsitch [48]. In this method, first a time span in which the epidemic curve has an exponential growth is selected, where r indicates the growth rate (changes in the number of new incidents), and is calculated by Poisson regression. Then, R0 is estimated using the moment generating function of the generating time distribution, M, which is:
| (10) |
Sequential Bayesian method
Bayesian time-dependant method proposed by Bettencourt and Ribeiro [49] calculates R0 using Poisson distribution in a time period with exponential growth. This method works by starting with a slightly informative prior on R0 and updates it sequentially. That is why the method is also referred as Sequential Bayesian (SB). Let Nt+1 be the number of cases in time t + 1 with an approximate Poisson distribution where N(t)e(γ(R−1)) is the mean of the distribution and specifies the average of the infection period. In this method, P(R0) is the prior, which captures information of the distribution of R0. The prior distribution of each day demonstrates the posterior distribution of the day before and will be recalculated by:
| (11) |
As a limitation of this method is the fact that this method can not be used for data sets with no new infection in some time intervals, since it leads to a Poisson mean of zero.
Results
All the the statical analysis were performed using software R version 4.1.1 and the R0 package [50]. Since we did not have access to the clinical COVID-19 data, the parameters of the serial interval distribution were not computed directly. And according to the fact that nature of COVID-19 virus is the same worldwide, we assume the fitting model is the same as the models already observed in COVID-19 pandemic worldwide, which is a Gamma distribution with a mean of 7.5 days and a standard deviation of 3.4 days [51]. Then, the basic reproduction number (R0) of Alpha and Delta variants of COVID-19 pandemic in Iran is estimated by using four different methods: EGR, ML, SB, and SIR. We have used two time intervals for each variant: time interval with exponential growth and the total active time interval. Results of computing R0 in the time spans with exponential growth are classified in Table 5.
Table 5. Estimated R0 of Alpha and Delta variants of COVID-19 pandemic in Iran in a time span of exponential growth.
| Variant | Time Span | EGR | ML | SB | SIR |
|---|---|---|---|---|---|
| Alpha | April 3-April 9, 2021 | 2.6422 (95% CI: 2.58–2.7) |
11.3811 (95% CI: 11.28–11.48) |
2.2680 (95% CI: 2.06–2.46) |
12.1317 (95% CI: 12.12–12.14) |
| Delta | July 3-July 8, 2021 | 3.1002 (95% CI: 3.02–3.17) |
12.0089 (95% CI: 11.89–12.12) |
3.0060 (95% CI: 2.34–3.66) |
23.3021 (95% CI: 23.19–23.41) |
Considering the estimated R0 for the total time intervals in which Alpha and Delta variants were active in Iran as shown in Table 6, proves higher contagion of Delta variant than Alpha variant in Iran. Further, in the time span that the number of infections increased exponentially, the results showed more transmissibility of each variant. Moreover, R0 for the total time interval of Alpha and Delta in Iran is respectively less than R0 of each of these variants in the time span with the exponential growth. This indicates that the controlling strategies in both variants were effective to control the virus spread. Table 6 represents the details.
Table 6. Estimated total R0 of Alpha and Delta variants in Iran.
| Variant | Total Time Interval | EGR | ML | SB | SIR |
|---|---|---|---|---|---|
| Alpha | March 10-June 10, 2021 | 0.9999 (95% CI: 0.99941–1.00043) |
1.0468 (95% CI: 1.044–1.049) |
1.0613 (95% CI: 1.034–1.089) |
2.7949 (95% CI: 2.77–2.81) |
| Delta | June 22-September 22, 2021 | 1.0426 (95% CI: 1.042–1.043) |
1.0538 (95% CI: 1.051–1.055) |
0.7970 (95% CI: 0.63–0.95) |
5.6577(95% CI: 5.6–5.7) |
Discussion
The results of estimating the basic reproduction number for each COVID-19’s variants show that Delta variant has more infectivity compared to Alpha variant, as confirmed by health organizations. However, the strategies that have been applied, have had positive impacts on controlling the virus spread of each variant.
Of the limitations of this study can be the fact that the reported number of infections is based on COVID-19 RT-PCR tests with positive results. However, this test can have a high rate of false-negative result [52]. Hence, due to failure of RT-PCR tests in precisely detecting COVID-19 and its variants, the number of infected people is not completely accurate. Therefore, using complementary tests may result in a more accurate number of infections. Moreover, the vaccination rate has had a powerful effect on controlling the spread of COVID-19, which is not considered in the models provided for estimating the basic reproduction number. Thus, further studies should take into account all contributing factors to the basic reproduction number of COVID-19 pandemic.
Conclusion
As a conclusion, R0 of Alpha variant of COVID-19 pandemic in Iran is estimated between 2.26–11.38 by the statistical methods, and 12.13 by time dependent SIR model in the time interval with exponential growth. Further, R0 of this variant in the total time interval of activation is estimated between 0.9–1.6 by the statistical methods, and 2.79 by time-dependent SIR model. For Delta variant of COVID-19 pandemic in Iran, estimated R0 is in range 3.0–12.0 by the statistical methods, and 23.3 by time-dependent SIR model in the time interval with exponential growth. And in the total time interval of activation of Delta variant in Iran, the estimated R0 is in range 0.797–1.05 by the statistical methods, and 5.65 by time-dependent SIR model. The results specify that the nature of Delta variant is more contagious than Alpha variant in Iran. Decrease in R0 in the total time interval of each variant indicates the positive effects of the controlling strategies such as traffic restrictions, distance working, school closures, and cancelation of mass gatherings in reducing the virus spread. Although the nature of Delta variant in Iran is more contagious that Alpha variant, as Table 2 confirms doubling of the number of cases in Delta variant in Iran, the proximity of R0 for the total active time of Alpha and Delta variants in Iran confirms the positive effect of accelerating the vaccination from the beginning of the Delta variant in Iran [53].
Supporting information
The dataset that was used for calculating Reproduction number for the Alpha variant.
(TXT)
The dataset that was used for calculating Reproduction number for the Delta variant.
(TXT)
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.WHO Director-General’s opening remarks at the mission briefing on COVID-19. 2020 Feb 19 [cited 2022 Feb 2]. In: WHO.int[Internet]. Available from: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-mission-briefing-on-covid-19
- 2.World Health Organization. Novel Coronavirus (2019-nCoV): situation report, 1. 2020 Jan 21 [cited 2022 Feb 2]. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf
- 3. Gorbalenya A, Baker S, Baric R, de Groot R, Drosten C, Gulyaeva A, et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. Naming the Coronavirus disease (COVID-19) and the virus that causes it. 2020 Feb 11 [cited 2022 Feb 2]. In: WHO.int[Internet]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
- 5. Alimohamadi Y, Sepandi M, Taghdir M, Hosamirudsari H. Determine the most common clinical symptoms in COVID-19 patients: a systematic review and meta-analysis. J Prev Med Hyg. 2020;61(3):E304–E312. doi: 10.15167/2421-4248/jpmh2020.61.3.1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patel SKS, Lee JK, Kalia VC. Deploying biomolecules as anti-COVID-19 agents. Indian J. Microbiol. 2020;60(3):263–268. doi: 10.1007/s12088-020-00893-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Director. WHO Director-General’s opening remarks at the media briefing on COVID-19—11 March 2020. Last update: 2020 Mar 11 [cited 2022 Feb 2]. In: WHO.int[Internet]. Available from: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020
- 8. Thakur V, Bhola S, Thakur P, Patel S, Kulshrestha S, Ratho R, et al. Waves and variants of SARS-CoV-2: understanding the causes and effect of the COVID-19 catastrophe. Infection. 2021;1–16. doi: 10.1007/s15010-021-01734-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Tracking SARS-CoV-2 variants. Last update: 2022 Feb 2 [cited 2022 Feb 2]. In: WHO.int[Internet]. Available from: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 10. Rishi P, Thakur K, Vij S, Rishi L, Singh A, Kaur IP, et al. Diet, Gut Microbiota and COVID-19. Indian J Microbiol. 2020;60(4):1–10. doi: 10.1007/s12088-020-00908-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. BourBour F, Mirzaei Dahka S, Gholamalizadeh M, Akbari ME, Shadnoush M, Haghighi M, et al. Nutrients in prevention, treatment, and management of viral infections; special focus on Coronavirus. Arch Physiol Biochem. 2020;1–10. doi: 10.1080/13813455.2020.1791188 [DOI] [PubMed] [Google Scholar]
- 12. Shah SM, Alsaab HO, Rawas-Qalaji MM, Uddin MN. A review on current COVID-19 vaccines and evaluation of particulate vaccine delivery systems. Vaccines (Basel). 2021;9(10):1086. doi: 10.3390/vaccines9101086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moghadas SM, Vilches TN, Zhang K, Wells CR, Shoukat A, Singer BH, et al. The impact of vaccination on COVID-19 outbreaks in the United States. Clin Infect Dis. 2021;73(12):2257–2264. doi: 10.1093/cid/ciab079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Q, Qin C, Liu M, Liu J. Effectiveness and safety of SARS-CoV-2 vaccine in real-world studies: A systematic review and meta-analysis. Infect Dis Poverty. 2021;10(1):132. doi: 10.1186/s40249-021-00915-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krause PR, Fleming TR, Peto R, Longini IM, Figueroa JP, Sterne JAC, et al. Considerations in boosting COVID-19 vaccine immune responses. Lancet. 2021;398(10308):1377–1380. doi: 10.1016/S0140-6736(21)02046-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burki T. Understanding variants of SARS-CoV-2. Lancet. 2021;397(10273):462. doi: 10.1016/S0140-6736(21)00298-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jewell BL. Monitoring differences between the SARS-CoV-2 B.1.1.7 variant and other lineages. Lancet Public Health. 2021;6(5):e267–e268. doi: 10.1016/S2468-2667(21)00073-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538):eabg3055. doi: 10.1126/science.abg3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization. Listings of WHO’s response to COVID-19. Last updated: 2021 Jan 29 [cited 2022 Feb 2]. In: WHO.int[Internet]. Available from: https://www.who.int/news/item/29-06-2020-covidtimeline
- 20. Jassat W, Mudara C, Ozougwu L, Tempia S, Blumberg L, Davies MA, et al. Difference in mortality among individuals admitted to hospital with COVID-19 during the first and second waves in South Africa: a cohort study. Lancet Glob Health. 2021;9(9):e1216–e1225. doi: 10.1016/S2214-109X(21)00289-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet. 2021;22(12):757–773. doi: 10.1038/s41576-021-00408-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alexandar S, Ravisankar M, Kumar RS,Jakkan K. A Comprehensive Review on Covid-19 Delta variant. Int J of Pharmacology and Clin. Research (IJPCR). 2021;5(2):83–85. [Google Scholar]
- 23.Statista. Number of SARS-CoV-2 Alpha variant cases worldwide as of January 17, 2022, by country or territory. [cited 2022 Jan 17] In: www.statista.com[Internet]. Available from: https://www.statista.com/statistics/1246219/number-alpha-variant-worldwide-by-country/
- 24.Statista. Number of SARS-CoV-2 Beta variant cases worldwide as of January 17, 2022, by country or territory. [cited 2022 Jan 17] In: www.statista.com[Internet]. Available from: https://www.statista.com/statistics/1246243/number-beta-variant-worldwide-by-country/
- 25.Statista. Number of SARS-CoV-2 Gamma variant cases worldwide as of January 17, 2022, by country or territory. [cited 2022 Jan 17] In: www.statista.com[Internet]. Available from: https://www.statista.com/statistics/1246259/number-gamma-variant-worldwide-by-country/
- 26.Statista. Number of SARS-CoV-2 Delta variant cases worldwide as of January 17, 2022, by country or territory. [cited 2022 Jan 17] In: www.statista.com[Internet]. Available from: https://www.statista.com/statistics/1245971/number-delta-variant-worldwide-by-country/
- 27. Blandenier E, Habibi Z, Kousi T, Sestito P, Flahault A, Rozanova L. Initial COVID-19 outbreak: an epidemiological and socioeconomic case review of Iran. Int J Environ Res Public Health. 2020;17(24):9593. doi: 10.3390/ijerph17249593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Worldometers. Coronavirus Cases. Last access: [2022 Jan 18]. In: www.worldometers.info[Internet]. Available from: https://www.worldometers.info/coronavirus/country/iran/
- 29.ISNA. Last access: [2022 Jan 18]. Available from: https://www.isna.ir/service/graphic/infographic
- 30.Eyre D, Taylor D, Purver M, Chapman D, Fowler T, Pouwels KB, et al. The impact of SARS-CoV-2 vaccination on Alpha and Delta variant transmission. MedRxiv [Preprint]. 2021 MedRxiv 21264260. 10.1101/2021.09.28.21264260. Available from: https://www.medrxiv.org/content/10.1101/2021.09.28.21264260v2 [DOI]
- 31. Heesterbeek JAP, Dietz K. The concept of R0 in epidemic theory. Statistica Neerlandica. 1996;50(1):89–110. doi: 10.1111/j.1467-9574.1996.tb01482.x [DOI] [Google Scholar]
- 32. Hethcote HW. The mathematics of infectious disease. SIAM Rev. 2000;42(4):599–653. doi: 10.1137/S0036144500371907 [DOI] [Google Scholar]
- 33. Chen YC, Lu PE, Chang CS, Liu TH. A time-dependent SIR model for COVID-19 with undetectable infected persons. IEEE Trans Netw Sci Eng. 2020;7(4):3279–3294. doi: 10.1109/TNSE.2020.3024723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Oxford University Press; 1992. [Google Scholar]
- 35. Gumel AB, Ruan S, Day T, Watmough J, Brauer F, Van den Driessche P, et al. Modelling strategies for controlling SARS outbreaks. Proc Biol Sci. 2004;271(1554):2223–2232. doi: 10.1098/rspb.2004.2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. White LF, Archer B, Pagano M. Estimating the reproductive number in the presence of spatial heterogeneity of transmission patterns. Int J Health Geogr. 2013;12(35)1–10. doi: 10.1186/1476-072X-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Althaus CL. Estimating the reproduction number of Ebola virus (EBOV) during the 2014 outbreak in West Africa. PLoS Curr. 2014;6. doi: 10.1371/currents.outbreaks.91afb5e0f279e7f29e7056095255b288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao D, Lou Y, He D, Porco TC, Kuang Y, Chowell G, et al. Prevention and control of Zika as a mosquito-borne and sexually transmitted disease: a mathematical modeling analysis. Sci Rep. 2016;6:28070. doi: 10.1038/srep28070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yuan J, Li M, Lv G, Lu ZK. Monitoring transmissibility and mortality of COVID-19 in Europe. Int J Infect Dis. 2020;95:311–315. doi: 10.1016/j.ijid.2020.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dhunagna HN, Ghimire S. Modelling of reproduction number for COVID-19 in India and high incidence states. Clin Epidemiol Glob Health. 2021;9:85–86. doi: 10.1016/j.cegh.2020.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Azimi SS, Koohi F, Aghaali M, Nikbakht R, Mahdavi M, Mokhayeri Y, et al. Estimation of the basic reproduction number R0 of the COVID-19 epidemic in Iran. Med J Islam Repub Iran. 2020;34:95. doi: 10.34171/mjiri.34.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dharmaratne S, Sudaraka S, Abeyagunawardena I, Manchanayake K, Kothalawala M, Gunathunga W. Estimation of the basic reproduction number R0 for the novel coronavirus disease in Sri Lanka. Virol J. 2020;17(1):144. doi: 10.1186/s12985-020-01411-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. ULAŞ E. Prediction of COVID-19 Pandemic Before The Latest Restrictions in Turkey by Using SIR Model. Süleyman Demirel Üniversitesi Fen Edebiyat Fakültesi Fen Dergisi. 2021;(1):77–85. 10.29233/sdufeffd.852222 [DOI] [Google Scholar]
- 44. Liu Y, Rocklöv J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J Travel Med. 2021;28(7):taab124. doi: 10.1093/jtm/taab124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.FRANCE24. Covid: Delta doubles hospital risk vs Alpha variant. 2021 Aug 28 [cited 2022 Feb 2]. In: www.france24.com[Internet]. Available from: https://www.france24.com/en/live-news/20210827-covid-delta-doubles-hospital-risk-vs-alpha-variant
- 46. Newman M. Networks: An Introduction. 1st ed. UK: Oxford University Press. 2010. [Google Scholar]
- 47. White LF, Pagano M. A likelihood-based method for real-time estimation of the serial interval and reproductive number of an epidemic. Stat Med. 2008;27(16):2999–3016. doi: 10.1002/sim.3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc Biol Sci. 2007;274(1609):599–604. doi: 10.1098/rspb.2006.3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bettencourt LM, Ribeiro RM. Real time bayesian estimation of the epidemic potential of emerging infectious diseases. PLoS One. 2008;3(5):e2185. doi: 10.1371/journal.pone.0002185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Obadia T, Haneef R, Boëlle PY. The R0 package: a toolbox to estimate reproduction numbers for epidemic outbreaks. BMC Med Inform Decis Mak. 2012;12:147. doi: 10.1186/1472-6947-12-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chu J. A statistical analysis of the novel coronavirus (COVID-19) in Italy and Spain. PLoS One. 2021;16(3):e0249037. doi: 10.1371/journal.pone.0249037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pecoraro V, Negro A, Pirotti T, Trenti T. Estimate false-negative RT-PCR rates for SARS-CoV-2. A systematic review and meta-analysis. Eur J Clin Invest. 2022;52(2):e13706. doi: 10.1111/eci.13706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.ISNA. Last update: 2021 Jul 11 [cited 2022 Jan 18]. In: www.isna.ir[Internet]. Available from: https://www.isna.ir/news/1400041913551/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The dataset that was used for calculating Reproduction number for the Alpha variant.
(TXT)
The dataset that was used for calculating Reproduction number for the Delta variant.
(TXT)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.