Abstract
A bacterium capable of utilizing carbaryl (1-naphthyl N-methylcarbamate) as the sole carbon source was isolated from carbaryl-treated soil. This bacterium was characterized taxonomically as Arthrobacter and was designated strain RC100. RC100 hydrolyzes the N-methylcarbamate linkage to 1-naphthol, which was further metabolized via salicylate and gentisate. Strain RC100 harbored three plasmids (designated pRC1, pRC2, and pRC3). Mutants unable to degrade carbaryl arose at a high frequency after treating the culture with mitomycin C. All carbaryl-hydrolysis-deficient mutants (Cah−) lacked pRC1, and all 1-naphthol-utilization-deficient mutants (Nat−) lacked pRC2. The plasmid-free strain RC107 grew on gentisate as a carbon source. These two plasmids could be transferred to Cah− mutants or Nat− mutants by conjugation, resulting in the restoration of the Cah and Nah phenotypes.
Each year, pesticides are manufactured and used in massive quantities, and toxicity remains a major environmental problem associated with pesticide usage. Carbamate pesticides, such as carbaryl, comprise the major proportion of agricultural pesticides used in today’s agricultural industry. Microorganisms capable of degrading carbamate pesticides are thought to play a significant role in the breaking down and detoxifying pesticides in the environment. These microorganisms have received considerable attention because of their potential use in pesticide waste detoxification as well as their effect on the fate of carbamate pesticides in the environment. Thus, there are many reports on the isolation and characterization of carbamate-pesticide-degrading microorganisms (5, 18, 25).
The involvement of naturally occurring plasmids in the degradation of synthetic organic compounds has been extensively documented (23). The dissemination of biodegradative genes by bacterial plasmids has led to the rapid evolution of certain specialized strains capable of using these xenobiotic compounds as substrates (17). The extensive use of carbamate pesticides in agriculture has induced the rapid evolution and dissemination of specific degradative pathways for the compound in soil bacteria (8). Recently, plasmids involved in pesticide degradation have also been recognized by several investigations, and the genes for the degradation of phenmedipham (22), EPTC (26), carbofuran (9, 21, 28), bromoxynil (24), and parathion (19) were identified on the plasmids. Thus, plasmids are thought to play important roles in the evolution of pesticide-degrading capabilities. However, little is known of the plasmid-mediated degradation of carbamate pesticides. Our interest has been the degradation of carbaryl (1-naphthyl N-methylcarbamate), which is one of the most commonly used carbamate pesticides for the control of a wide variety of insect pests. Carbaryl is known to be metabolized by microorganisms belonging to a variety of bacterial genera, such as Achromobacter (25), Blastobacter (12), and Pseudomonas (3, 4). However, there are no reports of plasmid-associated carbaryl degradation.
In this report, we describe the isolation and characterization of an Arthrobacter strain capable of utilizing carbaryl as the sole source of carbon. Furthermore, we demonstrate that the pathway of carbaryl degradation was encoded on two distinct plasmids.
MATERIALS AND METHODS
Isolation and culture conditions.
The bacterial strains were stored on nutrient agar plates. Minimal medium (MM) (12) containing 1.0 mM carbaryl was used (MMC), as well as MM containing 1.0 mM salicylate (MMS) and MM supplemented with 0.2% glucose, 0.4% Bacto Tryptone, and 0.2% yeast (MMGTY). Liquid cultures were shaken at 120 rpm in a reciprocal shaker at 30°C.
Strain RC100 was isolated by enrichment culture techniques using an inoculum of carbaryl-treated soil collected from agricultural fields with histories of carbaryl application. A 10-g carbaryl-treated soil sample was suspended in 100 ml of MMC. The suspension was incubated at 30°C for 2 days and then transferred to fresh MMC. Following growth in medium for two serial transfers, the culture was spread onto MMC agar plates. We selected a colony that utilized carbaryl as a sole source of carbon. This was designated isolate RC100. Spontaneous rifampin-resistant (Rifr) and streptomycin-resistant (Strr) mutants of RC100 were isolated by plating MMGTY-broth-grown cells on nutrient agar supplemented with 25 μg of rifampin or streptomycin per ml.
Bacterial strains isolated, used, and derived and their plasmids and relevant phenotypes are shown in Table 1.
TABLE 1.
Bacterial strains
| Strain | Plasmid content
|
Relevant phenotypea | Source | ||
|---|---|---|---|---|---|
| pRC1 | pRC2 | pRC3 | |||
| RC100 | + | + | + | Cah+ Car+ Nat+ Sal+ Gen+ | Wild-type strain |
| RC101 | + | + | − | Cah+ Car+ Nat+ Sal+ Gen+ | Spontaneous |
| RC102 | + | − | + | Cah+ Car− Nat− Sal− Gen+ | Mitomycin C treatment of RC100 |
| RC103 | − | + | + | Cah− Car− Nat+ Sal+ Gen+ | Mitomycin C treatment of RC100 |
| RC104 | + | − | − | Cah+ Car− Nat− Sal− Gen+ | Mitomycin C treatment of RC101 |
| RC105 | − | + | − | Cah− Car− Nat+ Sal+ Gen+ | Mitomycin C treatment of RC101 |
| RC1053 | − | + | − | Cah− Car− Nat+ Sal+ Gen+ Rifr Strr | Rifr, Strr derivatives of RC105 |
| RC106 | − | − | + | Cah− Car− Nat− Sal− Gen+ | Spontaneous |
| RC107 | − | − | − | Cah− Car− Nat− Sal− Gen+ | Mitomycin C treatment of RC104 |
| RC1073 | − | − | − | Cah− Car− Nat− Sal− Gen+ Rifr Strr | Rifr, Strr derivatives of RC107 |
| RC1453 | + | + | − | Cah+ Car+ Nat+ Sal+ Gen+ Rifr Strr | Conjugation (RC104 × RC1053) |
| RC1473 | + | − | − | Cah+ Car− Nat− Sal− Gen+ Rifr Strr | Conjugation (RC104 × RC1073) |
| RC1573 | − | + | − | Cah− Car− Nat+ Sal+ Gen+ Rifr Strr | Conjugation (RC105 × RC1073) |
Phenotype designations: Car+, Nat+, Sal+, and Gen+, abilities to grow on carbaryl, 1-naphthol, salicylic acid, and gentisic acid, respectively, as a sole source of carbon; Strr and Rifr, resistance to streptomycin and rifampin, respectively; Cah+, ability to hydrolyze carbaryl.
Taxonomic identification.
The strain was identified on the basis of classification schemes published in Bergey’s Manual of Systematic Bacteriology (14). The guanine plus cytosine (G+C) content of bacterial DNA was determined as described by Tamaoka and Komagata (27). The quinone type of the strain was determined by high-performance liquid chromatography (HPLC) with the reference standards. For sequencing of the 16S rRNA gene of RC100, total genomic DNA was prepared from strain RC100 by a standard phenolic extraction procedure. A portion of DNA (ca. 100 ng) was used in the PCR to amplify the 16S rRNA gene with two eubacterial primers, pA and 1492r (7). The resulting PCR products were purified with QIAquick spin columns (Qiagen, Valencia, Calif.). The nucleotide sequences of the products were determined by automated fluorescent dye primer sequencing with an SQ5500 sequencer (Hitachi Instruments Service Co., Ltd., Tokyo, Japan). The forward sequencing primers spanned positions 1094 to 1112 and the reverse primers spanned positions 536 to 518, 805 to 786, 1111 to 1093, and 1406 to 1389 (numbered as for the E. coli 16S rRNA gene) (13). Computer analysis was performed with DDBJ software packages. The sequence of the 16S rRNA of the bacteria was compared with other available 16S rRNA sequences by using the BLAST search option of the DDBJ database to determine the closest phylogenetic neighbors.
Carbaryl hydrolase activity.
Cells were grown in MMGTY for 24 h and then were harvested by centrifugation (10,000 × g, 15 min, 4°C). The cells were washed twice with 50 mM potassium phosphate buffer (pH 7.0). The cell paste was resuspended in the same buffer and then disrupted by sonication for 20 min at 0°C. Cellular debris was removed by centrifugation (10,000 × g, 15 min, 4°C). Carbaryl hydrolase in the cell extract was assayed in a reaction mixture containing 250 μM carbaryl, 50 mM potassium phosphate buffer (pH 7.0), and the cell extract in a total volume of 1.0 ml. The reaction was started by the addition of cell extract and carried out at 30°C. The reaction was stopped, and the yielded 1-naphthol was quantified by a previously described method (12). Five carbamate pesticides were tested to determine substrate specificity by using the above reaction mixture containing a pesticide instead of carbaryl. After 30 min of incubation, hydrolysis of the substrate was measured by HPLC.
Plasmid curing.
Mitomycin C was added to a final concentration of 0.6 μg/ml of MMGTY broth culture of the isolate at an optical density at 540 nm (OD540) of about 0.04. The culture was shaken at 30°C for 18 h. Samples from the culture that showed some growth were then diluted and spread on nutrient plates. Colonies able to hydrolyze carbaryl to 1-naphthol were identified by spraying the colony with 0.5 mM carbaryl and 0.01% fast blue B salt dissolved in sterilized distilled water. The carbaryl-hydrolysis-positive colony quickly turned brown. All of the mutants lacking the ability to utilize 1-naphthol also lost the ability to utilize salicylate, and the 1-naphthol-utilizing strain was not able to form a visible colony on the MM containing 1-naphthol. To test the ability of 1-naphthol utilization, the colonies on nutrient agar plate were replicated on an MMS agar plate.
Mating.
The donor and recipient strains were cultivated separately in 2 ml of MMGTY for 18 h at 25°C. The cultures were combined at a donor-to-recipient ratio of 1:1, and a 4-ml sample was filtered onto a sterile acetate cellulose filter (0.2 μm pore size). Filters were transferred to a nutrient agar plate and incubated at 25°C for 24 h. The cells on the filter were suspended in MM, and appropriate dilutions were plated on selection plates. For transfer of the carbaryl hydrolysis phenotype, selection was performed on nutrient agar or MMC agar plates containing streptomycin (25 μg/ml). For transfer of the 1-naphthol-utilization phenotype, selection was performed on MMS agar plates containing streptomycin (25 μg/ml). Carbaryl hydrolase activity was detected by the same methods as described in the curing section.
Plasmid detection.
Plasmids from strain RC100 and transconjugants were isolated by a modification of the method of alkaline-sodium dodecyl sulfate extraction (2, 11). Restriction endonuclease digestion of plasmid DNA was performed by following the instructions of the manufacturer. Electrophoresis in 0.7% agarose was carried out at 80 V for 2 h, and plasmid DNA was visualized by ethidium bromide staining.
Oxygen uptake.
Cells were harvested at the end of the logarithmic growth phase by centrifugation (15 min at 10,000 × g), washed with 20 mM potassium phosphate buffer (pH 7.0), and resuspended in the same buffer. Oxygen uptake rates were measured with an electrode (Yellow Springs Instrument Co., Yellow Springs, Ohio) mounted to a reaction vessel maintained at a constant temperature (30°C). The assay mixture contained 0.1 ml of the cell suspension (OD540 = 5) and 1.9 ml of the phosphate buffer. The reaction was started by injecting substrate as a concentrated solution (0.5 mmol per ml of acetone), and specific oxygen consumption rates were corrected for endogenous uptake. There was no effect of acetone on oxygen consumption in the concentration range examined.
Analysis.
Pesticide concentration was determined by HPLC with an ODS-C18 column (1.1 by 25 mm; Tosho Co.), and detection was based on absorption at 220 nm. The eluant for pesticide was acetonitrile-water (50:50 by volume; pH 2.0) with a flow rate of 1.0 ml/min. Metabolites of carbaryl were identified by HPLC performed with a device equipped with a diode array detector. Retention times and UV spectra of the sample peaks were compared with those of known standards prepared in MM. Optical densities of cell suspensions were measured at 540 nm.
Chemicals.
Analytical grade carbaryl, xylylcarb (3,4-xylyl-N-methylcarbamate), propoxur (2-isopropoxyphenyl-N-methylcarbamate), metolcarb (3-tolyl-N-methylcarbamate), XMC (2,4-xylyl-N-methylcarbamate), and swep (methyl-3,4-dichlorocarbanilate) were purchased from Wakojunyaku Co. (Tokyo, Japan). All other chemicals were of analytical grade and available commercially.
Nucleotide sequence accession number.
The 16S rDNA sequences of the strain RC100 have been deposited in the DDBJ under accession no. AB017354.
RESULTS
Isolation and identification.
The enrichment procedure generated a pure culture, designated RC100, that was able to grow on carbaryl as a sole source of carbon. Strain RC100 was gram positive and nonmotile. The cells were pleomorphic long rods, and some showed rather prominent branching at the log phase. V formation was observed during cell division. At the stationary phase, the cells were coccoid. The strain was catalase positive, oxidase negative, and urease positive. The strain metabolized glucose oxidatively and did not reduce nitrate. The moles percentage of G+C of bacterial DNA was 62.3%, and the quinone type was menaquinone. RC100 was able to utilize for growth the following compounds as the carbon source: glucose, mannose, melibiose, raffinose, fructose, sorbitol, xylose, glycerol, and mannitol. RC100 was not able to grow on arabinose, sorbose, erythrose, and Simmons citrate. Sequencing of the 16S rRNA gene and comparison with previously published 16S rRNA gene sequences resulted in the classification of the isolate as a member of the genus Arthrobacter (data not shown). The highest degree of similarity found was 96.9%, which was the value obtained with the 16S rRNA gene of an Arthrobacter globiformis (DDBJ accession no. X80736). Based on these observations, the isolate was identified as Arthrobacter sp. strain RC100.
Mineralization and metabolism of carbaryl.
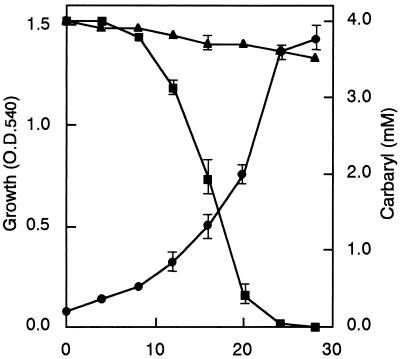
Figure 1 shows the time course of carbaryl utilization by strain RC100 in medium containing 0.4 mM carbaryl. Growth was accompanied by the degradation of carbaryl, and carbaryl completely disappeared after 48 h of cultivation. Chemical hydrolysis of carbaryl could account for disappearance of small amounts of carbaryl in the noninoculated medium. The doubling time of strain RC100 growing in MMC was estimated to be approximately 8 h. The two metabolites detected in the culture supernatants after 4 h of incubation were identified as 1-naphthol and salicylic acid on the basis of their HPLC properties and UV spectra. Utilization of aromatic compounds by the strain was examined on the basis of growth in MM containing an aromatic compound as the carbon source at 0.5 mmol/liter. Arthrobacter sp. RC100 grew on carbaryl, salicylic acid, and 1-naphthol. It additionally grew on gentisic acid, protocatechuic acid, salicylaldehyde, and 2-naphthol but did not utilize naphthalene, benzoic acid, catechol, phenol, propoxur, xylylcarb, and XMC. Strain RC100 was able to utilize methylamine as a sole carbon source or a sole nitrogen source. Methylamine was liberated by the hydrolysis of the carbamate side chain of carbaryl. The oxygen uptake responses of RC100 cells grown on carbaryl, 1-naphthol, salicylic acid, gentisic acid, and glucose are presented in Table 2. Cells of RC100 grown on carbaryl showed high oxygen consumption rates for the tested substrates except catechol, and cells grown on 1-naphthol gave similar results. Salicylic-acid-grown cells were induced for the oxidation of salicylic acid and gentisic acid, but they showed very low oxygen consumption rates for carbaryl, 1-naphthol, and salicylaldehyde. Cells grown on glucose oxidized hardly any of the tested aromatic compounds. These observations indicated that in strain RC100, the metabolism of carbaryl is initiated by its hydrolysis to 1-naphthol which is converted in several uncharacterized steps to salicylaldehyde and then through salicylate to gentisate, the substrate for oxygenase-catalyzed ring fission.
FIG. 1.
Utilization of carbaryl as the sole source of carbon for growth by Arthrobacter sp. strain RC100. Symbols: ●, OD540 of the culture; ■, carbaryl concentration in the medium; ▴, carbaryl concentration in an uninoculated control.
TABLE 2.
Oxygen consumption by whole cells of Arthrobacter sp. strain RC100
| Substratea | Rate of oxygen consumption (nmol/ml/min) after growth with:
|
||||
|---|---|---|---|---|---|
| Carbaryl | 1-Naphthol | Salicylic acid | Gentisic acid | Glucose | |
| Carbaryl | 26.1 | 12.1 | 1.9 | 0.7 | <0.1 |
| 1-Naphthol | 18.6 | 32.6 | 1.3 | 1.3 | 0.3 |
| Salicylaldehyde | 40.1 | 57.1 | 1.3 | 0.9 | 1.6 |
| Salicylic acid | 20.9 | 17.9 | 45.0 | 1.6 | 0.7 |
| Gentisic acid | 7.5 | 3.9 | 6.2 | 13.7 | <0.1 |
| Catechol | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 |
Substrate was used at a final concentration of 0.5 mM.
Hydrolysis of N-methylcarbamate.
The specific activity of carbaryl hydrolase in extract of cells grown in MMGTY was 124.3 nmol of pesticide/mg of protein/min. We tested the ability of Arthrobacter sp. RC100 to hydrolyze other carbamate pesticides. Cell extract also hydrolyzed N-methylcarbamate, propoxur (11.6 nmol of pesticide/mg of protein/min), xylylcarb (130.5), and XMC (122.2) but did not hydrolyze N-phenylcarbamate or swep, indicating that carbaryl hydrolase of RC100 is specific for molecules with a phenol-N-methyl carbamate ester linkage. However, work with purified enzyme is required to determine the correct substrate specificity and the reaction mechanisms.
Plasmid curing.
Arthrobacter sp. RC100 was studied for its extrachromosomal DNA by the procedure described in Material and Methods. The molecular sizes of the plasmids were determined by analysis of their digestion patterns after treatment with BamHI and PstI restriction enzymes. The total size of the plasmids was estimated on the basis of electrophoretic mobility of the fragments compared with those of lambda DNA digestion patterns of known sizes. The strain RC100 was observed to contain 110, 120, and 130 kbp, designated pRC1, pRC2, and pRC300, respectively.
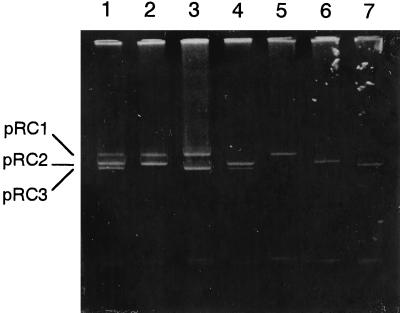
Mutants not able to utilize carbaryl as a sole source of carbon arose at a high frequency from wild-type strain RC100 by curing with mitomycin C (Fig. 2; lanes 3 to 7). The frequency of loss of the Cah+ (carbaryl-hydrolase-positive) phenotype in strain RC100 during flowing growth on MMGPY was less than 0.1%, and this increased to 4 to 6% when mitomycin C was present during growth. Agarose gel electrophoresis revealed that each of the Cah− derivatives lost a single plasmid species, pRC1. On the other hand, we also found that about 5% of colonies examined lost the Nat+ (1-naphthol-utilization) phenotype after mitomycin C treatment, and they also lost pRC2. Moreover, these strains (RC102, RC104, RC106, and RC107) lacking pRC2 also lost the ability to utilize salicylaldehyde and salicylic acid. Strain RC101, which retained both pRC1 and pRC2, could utilize carbaryl as the sole source of carbon (Table 1). There were no revertants to the Nat+ and Cah+ phenotypes from Nat− and Cah− strains, respectively. If revertants do occur, they do so at a frequency well below 10−9. RC107, a plasmid-free strain, retained the ability to utilize gentisic acid (Table 1). These results indicated that pRC1 encoded the carbaryl hydrolyzing ability, and pRC2 encoded the metabolizing pathway of 1-naphthol to gentisic acid. By the way, the plasmid-free strain RC107 and strain RC106, lacking plasmid pRC2 and pRC1, lost the ability of 1-naphthol utilization but utilized gentisic acid as the sole carbon source, indicating that the gentisic acid metabolic pathway was encoded on the chromosome.
FIG. 2.
Agarose gel electrophoresis of plasmids from strain RC100 and its derivatives. Lanes: 1, RC100 (pRC1, pRC2, pRC3); 2, RC101 (pRC1, pRC2); 3, RC102 (pRC1, pRC3); 4, RC103 (pRC2, pRC3); 5, RC104 (pRC1); 6, RC105 (pRC2); 7, RC106 (pRC3).
Conjugation.
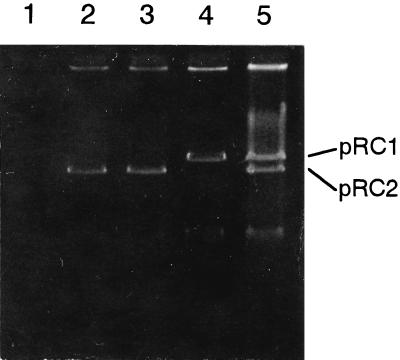
To further establish the role of pRC1 and pRC2 in the carbaryl metabolism, a mating experiment was performed by using the procedure described in Materials and Methods. Transconjugants obtained were all rifampin resistant, showing that they represented true transconjugants and not donors that had spontaneously acquired streptomycin resistance. The ability to hydrolyze carbaryl was transferred from RC104 (pRC1, Cah+) to RC1073 (plasmid free, Cah− Nat− Strr Rifr). The transconjugants which had acquired the ability to hydrolyze carbaryl contained a plasmid which was shown to be identical to pRC1 by agarose gel electrophoresis (Fig. 3). The frequency of plasmid transfer from RC104 to RC1073 ranged from 10−4 to 10−3 per donor cell. This provided additional evidence that pRC1 functionally mediated carbaryl hydrolysis. On the other hand, the Sal+ (Nat+) phenotype was introduced via filter mating into RC1073 (Cah− Nat− Strr Rifr) using RC105 donors. The frequency of plasmid transfer from RC105 to RC1073 ranged from 10−6 to 10−5 per donor cell. All of the transconjugants (Nat+ Strr Rifr) harbored pRC2, indicating that 1-naphthol degradation is mediated by pRC2. The transconjugants able to utilize carbaryl were derived from a mating experiment using RC104 and RC1053, and these bacteria carried both pRC1 and pRC2, indicating that these two plasmids were needed to completely degrade carbaryl.
FIG. 3.
Agarose gel electrophoresis of recipient strains and transconjugants. Lanes: 1, RC1073 (recipient strain); 2, RC1053 (recipient strain); 3, RC1573 (transconjugant of RC1073 from mating with RC105); 4, RC1473 (transconjugant of RC1073 from mating with RC104); 5, RC1453 (transconjugant of RC1053 from mating with RC104).
DISCUSSION
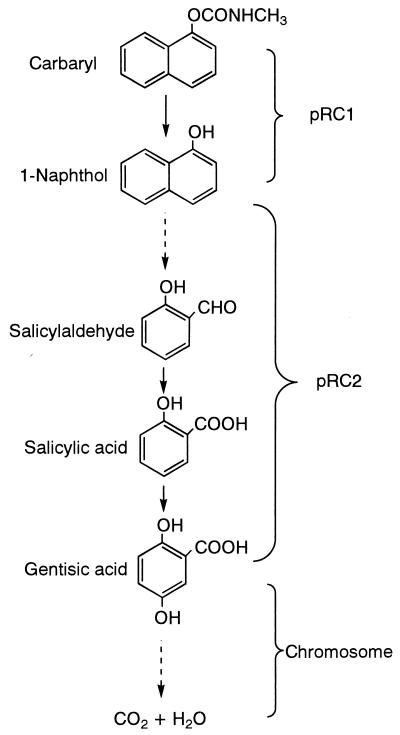
A number of carbaryl-degrading bacteria have been described, including several Pseudomonas (3, 4, 18), Blastobacter (12), and Achromobacter (15). A proposed pathway for the metabolism of carbaryl and 1-naphthol by Arthrobacter sp. strain RC100 is presented in Fig. 4. Strain RC100 grew at the expense of 1-naphthol produced by hydrolysis of carbaryl. In this respect, this microorganism is similar to other carbaryl-metabolizing bacteria, including Pseudomonas sp. (NCIB12043) (18) and Achromobacter sp. (25). Achromobacter sp. was capable of utilizing catechol, which was produced as a metabolite during carbaryl degradation. However, strain RC100 was not able to utilize catechol, and thus the 1-naphthol metabolic pathway of the strain RC100 differed from the pathway of Achromobacter sp. The pathway of carbaryl metabolism in strain RC100 closely resembled that in Pseudomonas sp. (NCIB12043) because 1-naphthol, salicylaldehyde, salicylate, and gentisate were metabolites in the metabolism of carbaryl in both organisms. Gentisate has been reported to be a metabolite in the metabolism of 1-naphthol (18) and 2-naphthol (30).
FIG. 4.
Proposed pathway of carbaryl degradation by Arthrobacter sp. strain RC100.
Members of the genus Arthrobacter that are ubiquitous in soil are capable of degrading a wide variety of naturally occurring aromatic and aliphatic compounds. Several studies have shown that a variety of synthetic organic compounds, such as pesticides, are degraded by members of the genus Arthrobacter (22, 26). The ability of Arthrobacter to degrade various organic compounds is known to be encoded often by catabolic plasmids. Plasmid-associated degradation of nicotine (1), chlorinated biphenyl (10), and hydroxypyridine (31) has been shown. For the herbicide EPTC, part or all of the degradative pathway is carried on the conjugative plasmid in the strain of Arthrobacter sp. strain TE1 (26). Single plasmid species present in these strains were responsible for the catabolic functions. However, Arthrobacter sp. strain RC100 isolated in the present study harbors three plasmids, and two of these are involved in carbaryl degradation. The functions encoded on these two distinct plasmids, pRC1 and pRC2, acted in concert to completely degrade carbaryl, although a single plasmid function was not capable of completely degrading the pesticide. Moreover, the plasmids were conjugally transmissible to Nah− and Cah− mutants. The results indicated that bacterial catabolic plasmids and their transfer in the soil microbial community play a significant role in acquiring a new metabolic pathway for the mineralization of pesticides.
Many carbamate-insecticide-degrading pathways have been shown to be most often initiated by hydrolysis of the methylcarbamate linkage. Recently, several N-methylcarbamate hydrolyzing enzymes were purified from Arthrobacter sp. WM111 (6, 16), Pseudomonas sp. CRL-OK (20), Blastobacter sp. M501 (12), and Pseudomonas aeruginosa (3) and were well characterized. The specificity of these enzymes (except that of P. aeruginosa) was broad with substrates of N-methylcarbamate insecticides. The cell extract of Arthrobacter sp. strain RC100 hydrolyzed N-methylcarbamate pesticides tested in the present study, indicating that the carbaryl hydrolyzing enzyme of strain RC100 is the type of the hydrolase with broad substrate specificity. The gene which encodes carbofuran hydrolase in Achromobacter sp. WM111 has been shown to be located on a 100-kb plasmid (pDL11) (28). On the basis of the observation of a methylotrophic bacterium which degrades carbofuran, the carbofuran hydrolase gene (mcd) and the plasmid pDL11 was shown to be not unique to Achromobacter sp. strain WM111 (29). Most recently, Parekh et al. (21) indicated that diverse carbofuran-degrading bacteria contained sequences homologous to that of the mcd gene. It will be interesting to determine whether the plasmid pRC1 of strain RC100 exhibits any degree of homology to the plasmid pDL11 and the mcd gene of Achromobacter sp.
ACKNOWLEDGMENT
This work was supported by Program for Promotion of Basic Research Activities for Innovative Biosciences of the Bio-oriented Technology Research Advancement Institution (Japan).
REFERENCES
- 1.Brandsch R, Hinkkanen A E, Decker K. Plasmid-mediated nicotine degradation in Arthrobacter oxidans. Arch Microbiol. 1982;132:26–30. [Google Scholar]
- 2.Casse F, Boucher C, Julliot J S, Michel M, Denarie J. Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J Gen Microbiol. 1979;113:229–242. [Google Scholar]
- 3.Chapalmadugu S, Chaudhry G R. Isolation of a constitutively expressed enzyme for hydrolysis of carbaryl in Pseudomonas aeruginosa. J Bacteriol. 1993;175:6711–6716. doi: 10.1128/jb.175.20.6711-6716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapalmadugu S, Chaudhry G R. Hydrolysis of carbaryl by a Pseudomonas sp. and construction of a microbial consortium that completely metabolizes carbaryl. Appl Environ Microbiol. 1991;57:744–750. doi: 10.1128/aem.57.3.744-750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhry G R, Ali A N. Bacterial metabolism of carbofuran. Appl Environ Microbiol. 1988;54:1414–1419. doi: 10.1128/aem.54.6.1414-1419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derbyshire M K, Karns J S, Kearney P C, Nelson J O. Purification and characterization of an N-methylcarbamate pesticide hydrolyzing enzyme. J Agric Food Chem. 1987;35:871–877. [Google Scholar]
- 7.Devereux R, Willis S G. Amplification of ribosomal RNA sequences, p3.3.1–3.3.11. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. [Google Scholar]
- 8.Felsot A S, Maddox J V, Bruce W. Enhanced microbial degradation of carbofuran in soils with histories of furadan use. Bull Environ Contam Toxicol. 1981;26:781–788. doi: 10.1007/BF01622171. [DOI] [PubMed] [Google Scholar]
- 9.Feng X, Ou L T, Ogram A. Plasmid-mediated mineralization of carbofuran by Sphingomonas sp. strain CF06. Appl Environ Microbiol. 1997;63:1332–1337. doi: 10.1128/aem.63.4.1332-1337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa K, Chakrabarty A M. Involvement of plasmids in total degradation of chlorinated biphenyls. Appl Environ Microbiol. 1982;44:619–629. doi: 10.1128/aem.44.3.619-626.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen J B, Olsen R H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978;135:227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayatsu M, Nagata T. Purification and characterization of carbaryl hydrolase from Blastobacter sp. M501. Appl Environ Microbiol. 1993;59:2121–2125. doi: 10.1128/aem.59.7.2121-2125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiraishi A, Shin Y-K, Ueda Y, Sugiyama J. Automated sequencing of PCR-amplified 16S rDNA on ‘Hydrolink’ gels. J Microbiol Methods. 1994;19:145–154. [Google Scholar]
- 14.Jones D, Collins M D. Irregular, nonsporing gram-positive rods, genus Arthrobacter. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins Co.; 1986. pp. 1288–1301. [Google Scholar]
- 15.Karns J S, Mulbry W W, Nelson J O, Kearney P C. Metabolism of carbofuran by a pure bacterial culture. Pestic Biochem Physiol. 1986;25:211–217. [Google Scholar]
- 16.Karns J S, Tomasek P H. Carbofuran hydrolase—purification and properties. J Agric Food Chem. 1991;39:1004–1008. [Google Scholar]
- 17.Kellogg S T, Chatterjee D K, Chakrabarty A M. Plasmid-assisted molecular breeding: new technique for enhanced biodegradation of persistent toxic chemicals. Science. 1981;214:1133–1135. doi: 10.1126/science.7302584. [DOI] [PubMed] [Google Scholar]
- 18.Larkin M J, Day M J. The metabolism of carbaryl by three bacterial isolates, Pseudomonas spp. (NCIB 12042 & 12043) and Rhodococcus sp. (NCIB 12038) from garden soil. J Appl Bacteriol. 1986;60:233–242. doi: 10.1111/j.1365-2672.1986.tb01078.x. [DOI] [PubMed] [Google Scholar]
- 19.Mulbry W W, Kearney P C, Nelson J O, Karns J F. Physical comparison of parathion hydrolase plasmids from Pseudomonas diminuta and Flavobacterium sp. Plasmid. 1987;18:173–177. doi: 10.1016/0147-619x(87)90046-1. [DOI] [PubMed] [Google Scholar]
- 20.Mulbry W W, Eaton W R. Purification and characterization of the N-methylcarbamate hydrolase from Pseudomonas strain CRL-OK. Appl Environ Microbiol. 1991;57:3679–3682. doi: 10.1128/aem.57.12.3679-3682.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parekh N R, Hartmann A, Fournier J-C. PCR detection of the mcd gene and evidence of sequence homology between the degradative genes and plasmids from diverse carbofuran-degrading bacteria. Soil Biol Biochem. 1997;28:1797–1804. [Google Scholar]
- 22.Pohlenz H D, Boidol W, Schuttke I, Streber W R. Purification and properties of an Arthrobacter oxydans P52 carbamate hydrolase specific for the herbicide phenmedipham and nucleotide sequence of the corresponding gene. J Bacteriol. 1992;174:6600–6607. doi: 10.1128/jb.174.20.6600-6607.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayler G S, Hooper S W, Layton A C, Henry King J M. Catabolic plasmids of environmental and ecological significance. Microb Ecol. 1990;19:1–20. doi: 10.1007/BF02015050. [DOI] [PubMed] [Google Scholar]
- 24.Stalker D M, McBride K E. Cloning and expression in Escherichia coli of a Klebsiella ozaenae plasmid-borne gene encoding a nitrilase specific for the herbicide bromoxynil. J Bacteriol. 1987;169:955–960. doi: 10.1128/jb.169.3.955-960.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sud R K, Sud A K, Gupta K G. Degradation of sevin (1-naphthyl-N-methylcarbamate) by Achromobacter sp. Arch Microbiol. 1972;87:353–358. doi: 10.1007/BF00409134. [DOI] [PubMed] [Google Scholar]
- 26.Tam A C, Behki R M, Khan S U. Isolation and characterization of an s-ethyl-N,N-dipropylthiocarbamate-degrading Arthrobacter strain and evidence for plasmid-associated s-ethyl-N,N-dipropylthiocarbamate degradation. Appl Environ Microbiol. 1987;53:1088–1093. doi: 10.1128/aem.53.5.1088-1093.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamaoka J, Komagata K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett. 1984;25:125–128. [Google Scholar]
- 28.Tomasek P H, Karns J S. Cloning of a carbofuran hydrolase gene from Achromobacter sp. strain WM111 and its expression in gram-negative bacteria. J Bacteriol. 1989;171:4038–4044. doi: 10.1128/jb.171.7.4038-4044.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topp E, Hanson R S, Ringelberg D B, White D C, Wheatcroft R. Isolation and characterization of an N-methylcarbamate insecticide-degrading methylotrophic bacterium. Appl Environ Microbiol. 1993;59:3339–3349. doi: 10.1128/aem.59.10.3339-3349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker N, Lippert K D. Formation of gentisic acid from 2-naphthol by a pseudomonad. Biochem J. 1965;95:5C–6C. doi: 10.1042/bj0950005c. [DOI] [PubMed] [Google Scholar]
- 31.Weinberger M, Kolenbander P E. Plasmid-determined 2-hydroxypyridine utilization by Arthrobacter crystallopoietes. Can J Microbiol. 1979;25:329–334. doi: 10.1139/m79-052. [DOI] [PubMed] [Google Scholar]