Abstract
Context
Primary ovarian insufficiency (POI) is a genetically heterogeneous condition associated with infertility and an increased risk of comorbidities. An increased number of genes implicated in DNA damage response pathways has been associated with POI as well as predisposition to cancers.
Objective
We sought to identify and characterize patients affected by POI caused by pathogenic variants in genes involved in DNA damage response during meiosis.
Setting
Study subjects were recruited at academic centers.
Patients or Other Participants
Individuals with a diagnosis of POI and their family members were enrolled for genetic analysis. Clinical findings, family history, and peripheral blood samples were collected.
Research design
Exome sequencing was performed on the study participants and their family members (when available). Protein conservation analysis and in silico modeling were used to obtain the structural model of the detected variants in the ZSWIM7 gene.
Main Outcome Measure(s)
Rare deleterious variants in known and candidate genes associated with POI.
Results
Homozygous deleterious variants in the ZSWIM7 gene were identified in 2 unrelated patients with amenorrhea, an absence of puberty, and prepubertal ovaries and uterus. Observed variants were shown to alter the ZSWIM7 DNA-binding region, possibly affecting its function.
Conclusions
Our study highlights the pivotal role of the ZSWIM7 gene involved in DNA damage response during meiosis on ovarian development and function. Characterization of patients with defects in DNA repair genes has important diagnostic and prognostic consequences for clinical management and reproductive decisions.
Keywords: ZSWIM7, primary ovarian insufficiency, DNA repair, DNA damage response, female infertility
Primary ovarian insufficiency (POI) encompasses heterogeneous conditions characterized by primary or secondary amenorrhea from the depletion of ovarian follicles or ovarian dysfunction before age 40 years. Several chromosomal and monogenic causes have been identified, leading to a spectrum of POI manifestations ranging from milder forms of hypogonadism to a more severe presentation such as ovarian dysgenesis (1). POI-causing genes have diverse functions, including adrenal and gonadal steroidogenesis, germ cell recruitment and folliculogenesis, DNA repair, meiosis, and neuroendocrine signaling, all of which are pivotal in the development and function of gonads and other reproductive organs.
Recent applications of exome and genome sequencing on consanguineous families with multiple affected individuals have revealed a substantial contribution of DNA repair and damage response genes to gonadogenesis and etiology of POI (2-4). Multiple genome- and transcriptome- wide studies have demonstrated a global effect of DNA damage response and repair genes on the depletion of the ovarian reserve and the age of natural menopause onset (5-7). In fetal oocytes, hundreds of DNA double-strand breaks are generated throughout the genome to promote meiotic recombination. In both somatic and germ cells, the inability to efficiently repair DNA damage compromises genomic integrity inducing cell-cycle arrest and apoptosis. Biallelic pathogenic variants in several genes involved in responses to DNA damage during meiosis such as BRCA2 (MIM*600185), HFM1 (MIM*615684), MCM8 (MIM*608187), MCM9 (MIM*610098), MEIOB (MIM*617670), MSH4 (MIM*602105), MSH5 (MIM*603382), and XRCC4 (MIM*194363) have been established to cause POI in multiple unrelated patients with primary amenorrhea and ovarian dysgenesis (1-3, 8, 9). Many of these genes are also expressed in testis. Pathogenic variants in other DNA damage response genes such as DMC1 (MIM*602721), XRCC2 (MIM*600375), and ZSWIM7 (MIM*614535) have been reported to cause DNA replication errors and aberrant meiotic recombination, resulting in spermatogenic arrest and infertility in men (8, 10, 11). However, their role in POI is not yet established.
Materials and Methods
Human Subjects
The study was approved by the ethics committee of the Cukurova University Faculty of Medicine (KARAR#1; DA:KADB-13F-R.00), the institutional review boards at the University of Pittsburgh (PRO09080427), and the Center for Reproductive Medicine of Shandong University (institutional review board #77). Cohorts of POI patients and their family members were recruited by participating institutions. Informed written consent was obtained from each participating subject. Genomic DNA samples were isolated from peripheral blood samples of affected individuals (P20428 and TPOF-GII2) as well as from TPOF-G unaffected family members using a Puregene Blood Kit B (QIAGEN, Hilden, Germany).
Exome Sequencing and Data Analysis
Genomic DNA (~1.5 μg) underwent exome capture using an Agilent SureSelectXT Human All Exon V6 kit (Santa Clara, CA), and sequenced on a HiSeq X Ten platform (Illumina, San Diego, CA) with an average depth coverage of 100×. The Burrows–Wheeler Alignment tool was used to align raw data to the human reference genome sequence (UCSC Genome Browser hg19). Variant calling was performed using the Genome Analysis Toolkit (12), and ANNOVAR software was further used to annotate all variants. Common variants with minor allele frequencies (MAF) > 0.001 present in the 1000 Genomes Project (http://www.1000genomes.org/), the Genome Aggregation Database (gnomADv2, http://gnomad.broadinstitute.org/), ExAC Browser (http://exac.broadinstitute.org/, and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) were filtered out. Rare (MAF ≤ 0.001) protein-altering variants were retained for analysis. Functional effect was evaluated by SIFT (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2), and MutationTaster (http://www.mutationtaster.org) bioinformatic tools.
Genome Sequencing and Data Analysis
Raw data produced from sequencing machine were converted to fastqs using Cassava 2.20 software. The produced fastqs were uploaded to Illumina basespace using basespace API commands. After the upload was complete, DRAGEN 3.8.4 pipeline (13) was triggered through the same API to produce the vcfs, Grch38 was used to align the reads to reference genome using bwa-mem aligner. Structural variant and copy number variant callers were used to generate their respective vcfs. Single nucleotide variant (SNV), structural variant, and copy number variant VCFs were uploaded in Moon (Diploid/Invitae). Only coding variants with a read depth > 10, genotype quality > 40, and MAF < 2% in GnomAD and that segregated with the disease were further considered for interpretation. DNA samples of family members were used in segregation analysis.
In Silico Modeling
We used Alphafold (14) to obtain the structural model of ZSWIM7. No postmodeling refinement was performed. Mutations were modeled in the Pymol Molecular Graphics System (Schrödinger, Inc.) with no refinement. Images of the structures were generated in Pymol. Eighty-nine percent of the residues were modeled with very high confidence; the C-terminal 7 residues were poorly modeled.
Protein Conservation Analysis
Protein sequences of human ZSWIM7 and its homologs in Mus musculus, Danio reiro, Canis lupus, Pan troglodytes, Bos taurus, and Gallus gallus were aligned using Clustal Omega (15).
Results
The first patient, P20428 (Fig. 1A), was a 28-year-old woman from China diagnosed with POI due to elevated FSH and LH levels (45.73 and 16.65 mIU/mL, respectively) (Table 1) and amenorrhea. No parental consanguinity was reported, and family history was unremarkable. Patient had menarche at 16 years of age followed by 3 more light bleeding episodes and subsequent amenorrhea at age 18 years. Neither ovaries nor uterus was visualized on pelvic ultrasound. Prepubertal uterus was visualized by magnetic resonance imaging. On the physical examination, external genitalia had normal anatomic appearance.
Figure 1.
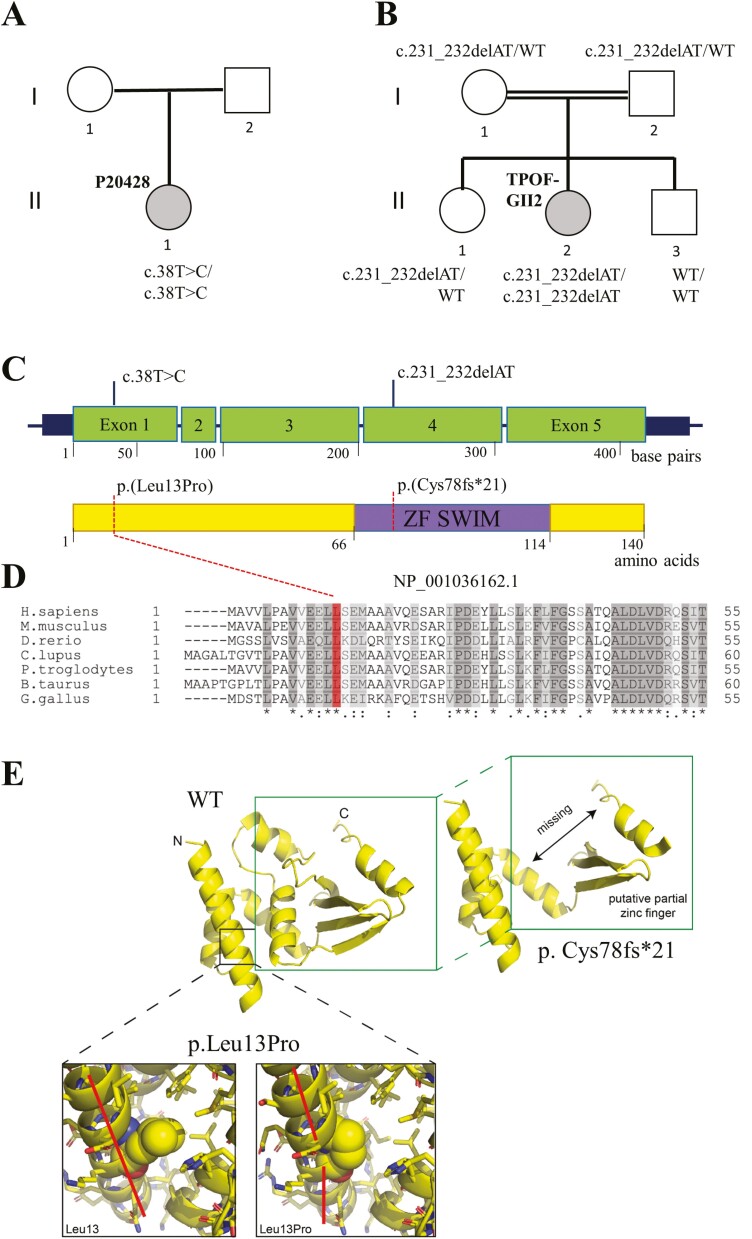
Variants in the ZSWIM7 gene. (A, B) Family pedigrees. Affected females are shown in gray-filled circles. Both patients had normal 46,XX karyotype and no history of autoimmune or iatrogenic conditions as a cause of primary ovarian insufficiency. Double horizontal lines indicate consanguinity. The ZSWIM7 genotype is provided for each individual. The wild-type allele is indicated as WT. (C) Gene structure with known protein domains of the gene product. The ZSWIM7 (zinc finger SWIM domain-containing protein 7) gene is located on chromosome 17p12 (chr17:15,879,875_ 15,903,006; hg19) and composed of 5 exons. Exons are specified as green-colored boxes. In patient P20428, the ZSWIM7 homozygous missense c.38T > C variant occurs in exon 1. In TPOF-GII2, a homozygous frameshift c.231_232delAT variant is located in exon 4, encoding the zinc finger SWIM domain (purple-shaded box). ZSWIM7, a 140-amino acid protein, contains 2 β-strands, a zinc finger SWIM (ZF SWIM) domain, and an α-helix. (D) Protein sequence alignment of human ZSWIM7 and its homologs in Mus musculus, Danio reiro, Canis lupus, Pan troglodytes, Bos taurus, and Gallus gallus. The affected Leu13 residue (marked in red) is highly conserved among all shown organisms. Residues are shaded based on similarity. Symbols denote: (*) conserved residue; (;) conservation between groups of strongly similar properties (>0.5 in the Gonnet PAM 250 matrix); (.) conservation between groups of weakly similar properties (≤0.5 in the Gonnet PAM 250 matrix). (E) In silico modeling of amino acid substitutions in ZSWIM7. The WT and 2 altered protein structures are shown. The leucine to proline change at residue 13 (p.Leu13Pro) is predicted to “break” the N-terminal helix, dividing this helix into 2, whose central axes no longer align (red lines). This could reorganize the packing of the entire molecule, including the DNA-binding region. Furthermore, if the regions surrounding position 13 still were able to form helices, the smaller proline sidechain would destabilize its interaction with neighboring amino acids. The p.Cys78fs*21 frameshift change at residue 78 that truncates the protein after 21 residues would impact the β-sheet and helix integral to supporting the zinc finger fold.
Table 1.
Clinical laboratory investigations in affected individuals with the ZSWIM7 variants
| Findings | Normal range | P20428 | TPOF-GII2 | Patient 1a | Patient 2a |
|---|---|---|---|---|---|
| Ethnicity | - | Han Chinese | Turkish | Turkish | Turkish |
| cDNA change: NM_001042697 | - | c.38T>C | c.231_232delAT | c.173C>G | c.173C>G |
| Protein change: NP_001036162.1 | - | p.(Leu13Pro) | p.(Cys78Phefs*21) | p.(Ser58*) | p.(Ser58*) |
| Age at menarche (y) | - | 16 | PA | PA | PA |
| Age at menopause (y) | - | 18 | - | - | - |
| FSH (mIU/mL) | 1.8-22.5 | 45.73 | 30.99 | 94.8 | 77.8 |
| LH (mIU/mL) | 1.2-100 | 16.65 | 13.88 | 17.2 | 14.3 |
| Estradiol (pg/mL) | 30 to 300 | <10 | 4 | Undetectable | Undetectable |
| Prolactin (ng/mL) | 3.3-26.7 | 11.30 | 11.93 | NR | NR |
| Uterus size | - | Prepubertal | Prepubertal | Prepubertal | Prepubertal |
| Ovarian volume (cm3) | 6.6 | Not visualized | Not visualized | streak ovaries | Not visualized |
| Tanner Stage (pubic hair) | - | 3 | 3 | NR | NR |
| Tanner Stage (breast) | - | 2 | 1 | NR | NR |
Abbreviations: NR, not reported; PA, primary amenorrhea.
aTwo affected sisters previously described in the literature (4).
Patient TPOF-GII2 (Fig. 1B) was a teenage girl of Kurdish descent from Turkey who presented with absent breast development, primary amenorrhea, and short stature. The parents were healthy first-degree cousins. Physical examination revealed cubitus valgus and a broad chest with widely spaced nipples. Both her pubic hair and axillary hair were at Tanner Stage 3. Her basal estradiol level was low with elevated LH and FSH levels (Table 1). Pelvic ultrasonography and magnetic resonance imaging showed no visible uterus or ovaries. At age 14 years, she had regular menstrual bleeding with combined estrogen-progestin replacement therapy and achieved appropriate pubic hair and breast development. Her older sister (subject TPOF-GII1) had normal breast development (Tanner Stage 4) and had regular menstrual bleeding. The patient’s 13-year-old brother (TPOF-GII3) had normal development and normal (8 mL) testicular volumes.
To identify the genetic cause of primary amenorrhea, we performed exome sequencing on patient P20428 (parental samples were unavailable) and genome sequencing on the TPOF-G family members. SNVs were filtered to retain novel and rare protein-coding changes with minor allelic frequency MAF ≤ 0.1% (GnomADv2). We prioritized variants in genes that are known to cause POI (1), followed by genes not yet reported but considered as candidates based on their involvement in gonadogenesis and oogenesis, hormonal synthesis, signaling and ovulation processes, meiotic chromosome segregation, and DNA repair in animal models (1, 8). In TPOF-GII2, we also evaluated homozygous SNVs that were absent in the patient’s sister because an autosomal-recessive inheritance was most plausible in this consanguineous family.
At the time of the initial analysis, no variants in known POI-causing genes were observed in either P20428 or TPOF-GII2 patient. Thirteen homozygous variants that fulfilled our filtering and family segregation criteria were identified in the TPOF-GII2 patient (Table 2). Of those, 12 were ruled out as a cause after examination of gene expression profiles and their known function. Similarly, no variants in known POI-causing genes were initially identified in the P20428 patient.
Table 2.
Homozygous variants found in TPOF-GII2, but not in her unaffected sister
| Chr | Position, hg19 | Gene | Disorder | Effect | cDNA change | Protein effect | Ensembl transcript ID | RefSeq transcript ID | MAF gnomAD | Allele hmz | Allele htz |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 113921475 | GPAM | No associated disorder | Missense | c.1444A>C | p.Ile482Leu | 348367 | 0 | 0 | 0 | |
| 11 | 46404352 | MDK | No associated disorder | Frameshift | c.461_462insGC | p.Arg155fs | 489525 | 0.00194525 | 0 | 280 | |
| 11 | 114549328 | NXPE2 | No associated disorder | Splice_region | c.26 + 5G>A | 389586 | NM_182495.5 | 6.35E-06 | 0 | 1 | |
| 13 | 24463420 | MIPEP | Combined oxidative phosphorylation deficiency 31 | Missense | c.40G>A | p.Ala14Thr | 382172 | NM_005932.3 | 0.000398142 | 0 | 6 |
| 13 | 52971362 | THSD1 | Nonimmune hydrops fetalis, THSD1-related | Splice_region | c.1021 + 5G>T | 258613 | NM_018676.3 | 0 | 0 | 0 | |
| 14 | 81609751 | TSHR | Hyperthyroidism | Missense | c.1349G>A | p.Arg450His | 298171 | NM_000369.2 | 0.000234637 | 0 | 59 |
| 17 | 7834015 | TRAPPC1 | No associated disorder | Missense | c.347C>T | p.Pro116Leu | 303731 | NM_021210.4 | 7.96E-06 | 0 | 2 |
| 17 | 8045782 | PER1 | No associated disorder | Splice_region | c.3260-6T>A | 317276 | NM_002616.2 | 0.000302402 | 0 | 65 | |
| 17 | 15884427 | ZSWIM7 | Male infertility, ZSWIM7-related | Frameshift | c.231_232delAT | p.Cys78fs | 399277 | NM_001042698.1 | 0.000364808 | 0 | 91 |
| 17 | 17070643 | MPRIP | No associated disorder | Missense | c.3842A>G | p.Asn1281Ser | 313485 | 0 | 0 | 0 | |
| 17 | 38320381 | CASC3 | No associated disorder | Missense | c.1433C>T | p.Pro478Leu | 264645 | NM_007359.4 | 0.00110912 | 0 | 269 |
| 20 | 25457678 | NINL | No associated disorder | Missense | c.2249C>T | p.Ala750Val | 278886 | NM_025176.4 | 0.000235843 | 0 | 54 |
| 22 | 46653641 | PKDREJ | No associated disorder | Missense | c.5579C>T | p.Pro1860Leu | 253255 | NM_006071.1 | 0 | 0 | 0 |
Abbreviations: Allele hmz, number of allele counts in a homozygous state as per GnomADv2; Allele htz, number of allele counts in a heterozygous state as per GnomADv2; MAF, minor allelic frequency as per GnomADv2.
Recently, ZSWIM7 pathogenic variants were identified in infertile males (https://preview.ncbi.nlm.nih.gov/clinvar/variation/1013608/), and mouse Zswim7 knockout model showed gonadal dysgenesis and infertility in both male and female mice (11, 16). Reanalysis of sequencing data in the TPOF-GII2 patient showed that the ZSWIM7 homozygous pathogenic c.231_232delAT (rs368517882) frameshift variant was present in TPOF-GII2, but not in her unaffected siblings (Fig. 1C). This specific variant is seen in heterozygous state in the 1000 Genomes, dbSNP, and gnomADv2.1 databases among multiple ethnicities with a global MAF = 0.0003774 (gnomADv2.1) and has been reported in the homozygous state in patients with nonobstructive azoospermia (11). In the P20428 patient, a novel homozygous ZSWIM7 c.38T > C, p.(Leu13Pro) variant (rs200271348) was identified and confirmed by Sanger sequencing. This variant has a low frequency (MAF = 0.00001464) in the human population and is present in a highly conserved residue among multiple species (Fig. 1D). The ZSWIM7 c.38T>C, p.(Leu13Pro) variant is predicted to be damaging by bioinformatic tools (CAAD score = 31). In silico modeling of the ZSWIM7 c.38T>C, p.(Leu13Pro) and c.231_232delAT (rs368517882) frameshift variants was performed to investigate the molecular impacts of these variants on the ZSWIM7 structure (Fig. 1E). Both variants were found to reorganize or truncate the DNA-binding region of ZSWIM protein, affecting the zinc finger function.
Discussion
Studies in mice show that ZSWIM7 is involved in meiotic homologous recombination repair through formation of a heterodimeric complex with SWIM-type zinc finger 7 associated protein 1 (MIM*614536) and plays a role in RAD51 and DMC1 driven repair of DNA double-strand breaks (16, 17). ZSWIM7 is a conserved protein, and the SWIM domain is required for its function. Based on our analysis, both variants are predicted to adversely affect ZSWIM7 by impacting the structural integrity of the DNA-binding domain. Inadequate repair of damaged DNA in mitotic cells results in genomic instability, growth defects, and ultimately germ cell loss. Germ cells are known to be sensitive to unrepaired double-stranded DNA breaks, as shown in knockout animal models. Male and female Zswim7-/- and Swsap1-/- mice show no obvious morphological anomalies, but have gonadal failure because of premature loss of postmeiotic germ cells (16). Mice homozygous for a knock-out allele exhibit female and male infertility with decreased testis and ovary weights, azoospermia, absent ovarian follicles, and impaired chromosomal synapsis.
Both of our patients, and recently published affected sisters from a single family with a loss-of-function ZSWIM7 variant (4), presented with early-onset amenorrhea, absence of puberty, and atrophic ovaries, a phenotype that complements one described in affected male individuals (11, 18) and mimic mouse knockout phenotype.
The presence of ZSWIM7 variants in diverse ethnic groups (11, 18) suggests that it might be a more frequent cause of gonadal dysgenesis and infertility than anticipated. Future and follow-up studies on POI and nonobstructive azoospermia individuals with pathogenic variants in ZSWIM7 and other DNA damage response genes will be necessary to define the spectrum of pathogenic variants for clinical diagnosis and human reproductive and other phenotypes attributed to mitotic and meiotic genomic instability. Reproductive problems or menstrual irregularities have not been reported in females heterozygous for the pathogenic variants in the ZSWIM7 gene; however, there are too few families identified to establish such an association. DNA repair genes have been shown to influence the reproductive lifespan regulating germ cells proliferation, the size of the initial pool of primordial follicles, and the rate of its depletion (2, 3, 5, 7). Although heterozygous variants are not associated with obvious gonadal pathology, they may confer risk for accelerated loss of ovarian reserves or infertility. The identification of genetic causes and characterization of phenotypic spectrum of patients with pathogenic variants in infertility have diagnostic and prognostic consequences for clinical management and reproductive decisions. Moreover, longitudinal studies are necessary to determine if these variants and genes contribute to additional morbidity and mortality (7) previously associated with loss of ovarian function before age 40 years.
Acknowledgments
The authors thank the patients and their family members for participating in this research study.
Glossary
Abbreviations
- MAF
minor allele frequency
- POI
primary ovarian insufficiency
- SNV
single nucleotide variant
Financial Support
This work was supported by the National Institute of Child Health and Human Development (R01HD070647 and R21HD074278 to A.R.) as well as National Science Fund for Distinguished Young Scholars (82125014) and the National Natural Science Foundation of China (82071609, 81771541 to Y.Q.).
Author Contributions
A.R., Y.Q., S.A..Y, F.G., and K.T. conceived the experiments and collected the clinical information; P.M.M., Y.Q., S.A.Y., and A.R. analyzed the next-generation sequencing data and identified the pathogenic variants; A.J.B. performed in silico modeling. The manuscript was written by S.A.Y., M.R.E., and A.R. and reviewed by P.M.M., Y.Q., A.J..B, F.G., K.T., and A.R. All authors read and approved the final manuscript.
Disclosures
All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Yatsenko SA, Rajkovic A. Genetics of human female infertility. Biol Reprod. 2019;101(3):549-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wood-Trageser MA, Gurbuz F, Yatsenko SA, et al. 9 mutations are associated with ovarian failure, short stature, and chromosomal instability. Am J Hum Genet. 2014;95(6):754-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. AlAsiri S, Basit S, Wood-Trageser MA, et al. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J Clin Invest. 2015;125(1):258-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McGlacken-Byrne SM, Le Quesne Stabej P, Del Valle Torres I, et al. ZSWIM7 is associated with human female meiosis and familial primary ovarian insufficiency. J Clin Endocrinol Metab. 2022;107(1):e254-e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruth KS, Day FR, Hussain J, et al. Genetic insights into biological mechanisms governing human ovarian ageing. Nature. 2021;596(7872):393-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi J, Wu L, Li B, et al. Transcriptome-wide association study identifies susceptibility loci and genes for age at natural menopause. Reprod Sci. 2019;26(4):496-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. .Rajkovic A, Pangas S. Ovary as a biomarker of health and longevity: insights from genetics. Semin Reprod Med. 2017;35(3):231-240. [DOI] [PubMed] [Google Scholar]
- 8. Huang C, Guo T, Qin Y. Meiotic recombination defects and premature ovarian insufficiency. Front Cell Dev Biol. 2021;9:652407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tucker EJ, Bell KM, Robevska G, et al. Meiotic genes in premature ovarian insufficiency: variants in HROB and REC8 as likely genetic causes. Eur J Hum Genet. 2021. doi: 10.1038/s41431-021-00977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He WB, Tu CF, Liu Q, et al. DMC1 mutation that causes human non-obstructive azoospermia and premature ovarian insufficiency identified by whole-exome sequencing. J Med Genet. 2018;55(3):198-204. [DOI] [PubMed] [Google Scholar]
- 11. Li Y, Wu Y, Zhou J, et al. A recurrent A ZSWIM7 mutation causes male infertility resulting from decreased meiotic recombination. Hum Reprod. 2021;36(5):1436-1445. [DOI] [PubMed] [Google Scholar]
- 12. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller NA, Farrow EG, Gibson M, et al. A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 2015;7:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McWilliam H, Li W, Uludag M, et al. . Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41(Web Server issue):W597-W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abreu CM, Prakash R, Romanienko PJ, Roig I, Keeney S, Jasin M. Shu complex SWS1- SWSAP1 promotes early steps in mouse meiotic recombination. Nat Commun. 2018;9:3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu T, Wan L, Wu Y, Chen J, Huang J. hSWS1·SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J Biol Chem. 2011;286(48):41758-41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alhathal N, Maddirevula S, Coskun S, et al. Agenomics approach to male infertility. Genet Med. 2020;22(12):1967-1975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.