DEAR EDITOR,
Mongolian gerbils (Meriones unguiculatus) are small rodents belonging to the family Muridae and widely inhabit typical steppes and desert grasslands across northern China. Yet, few viruses in wild M. unguiculatus have been reported. Using meta-transcriptomic sequencing we identified two novel RNA viruses: an astrovirus, denoted Gerbil astrovirus (GeAstV), and a paramyxovirus – Gerbil paramyxovirus (GePV) in Mongolian gerbils captured during October–November 2018 from one of the plagus zones in Shaanxi Province, China. Phylogenetic analysis based on the amino acid sequences of ORF1a, RdRp and ORF2 revealed that GeAstV clustered with rodent- and porcine-associated astroviruses, sharing the highest amino acid identity of 75.07% with a pig astrovirus in RdRp, while sharing 42.14% and 35.94% amino acid identity with a rodent astrovirus in the deduced ORF1a and ORF2 proteins, respectively, exhibiting apparent recombination and potential cross-species transmission events. For the newly discovered GePV, evolutionary analysis demonstrated that it was closely related to Tailam virus and Beilong virus, and shared 61%–77% identity in the L protein with the seven members of the genus Jeilongvirus. Notably, GeAstV was detected only in lung samples while GePV was positive only in kidney samples. In sum, we demonstrate the presence of two novel gerbil RNA viruses, expanding our understanding of viral diversity in wild Mongolian gerbils.
Rodents, representing the highest genetic diversity of mammal species, have been implicated as the natural reservoirs of zoonotic viruses including arenaviruses, hantaviruses, and Lassa viruses (Charrel & De Lamballerie, 2010; Guo et al., 2013). Rodent-associated viruses may cause severe disease in humans once crossing host barriers, despite causing little or no disease in their natural hosts. Mongolian gerbils (M. unguiculatus) are small rodents (family Muridae) widely distributed in typical steppes and desert grasslands in northern China (Liu et al., 2007). Domesticated Mongolian gerbils are extremely sensitive to several viruses including hantaviruses, and serve as excellent animal models for a wide range of infectious diseases (Bleich et al., 2012). However, little is known about the natural viral diversity in wild Mongolian gerbils, with only one hepacivirus (Gerbil hepacivirus) reported recently (An et al., 2022). Using a meta-transcriptomics approach, we report two novel RNA viruses – denoted Gerbil astrovirus and Gerbil paramyxovirus – that were discovered from Mongolian gerbils collected from a plague focus in Shaanxi Province, China. This study was approved of the Shandong First Medical University & Shandong Academy of Medical Sciences. All institutional and national guidelines for the care and use of animals were followed.
In the present study, a total of 40 apparently healthy Mongolian gerbils were collected from a known plague area of Shaanxi Province in China between October–November 2018 during the routine surveillance based on the Surveillance Standard for Plague Epizootic (No. GB16882-1997). They were divided into four groups (denoted DB01, DB02, DB03, and DB04), with each group containing ten individual gerbils. Tissue samples of kidney, lung, liver, and brain were collected immediately following humane euthanasia with ether. The same tissues of each group were pooled together and a total of 15 libraries were prepared, including four kidney, four lung, four brain, and three liver libraries (Supplementary Table S1). These samples were first homogenized with grinding beads and then subjected to total RNA extraction using TRIzol reagent (Takara Biomedical Technology, China). Ribosomal RNA (rRNA) was removed using the MGIEasy rRNA Depletion Kit (MGI Tech Co., China), with RNA sequencing libraries then constructed using the MGIEasy mRNA library Prep Kit (MGI Tech Co., China). Finally, 100 bp paired-end sequencing of the RNA libraries was performed on the BGISEQ-500RS sequencing platform (BGI, China) (Zhou et al., 2021).
An in-house meta-transcriptomic data analysis pipeline was employed for RNA virus discovery as previously described (Hu et al., 2021). Briefly, sequencing raw reads were trimmed for quality control using Trimmomatic (v0.36) and then de novo assembled with Trinity (version v2.5.1). Assembled sequence contigs were annotated using both Blastn search against the nucleotide (nt) database and Diamond Blastx against the non-redundant (nr) protein database available from GenBank (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST), with e-value thresholds of 1×10-10 and 1×10-4, respectively. The sequencing reads were mapped onto the predicted virus-associated contigs using Bowtie2 (v2.4.1) to evaluate the viral abundance and coverage. Virus abundance was estimated as Reads per Million (RPM, mapped viral reads/total library reads*one million). The coiled-coils and transmembrane helices were predicted using COILS (version 2.1) (https://npsa-prabi.ibcp.fr/NPSA/npsa_lupas.html) and TMHMM server (v.2.0) (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0), respectively.
Overlapping primers (Supplementary Table S2) were designed based on the assembled sequences, and RT-PCR and Sanger sequencing were performed to verify the viral sequences (Supplementary Figure S1). The 5' and 3' terminal sequences were recovered by performing random amplification of cDNA ends (RACE) using SMARTer RACE 5'/3' kit (Takara Biomedical Technology, China) according to the manufacturer's instructions with virus-specific reverse primers.
Open reading frames (ORFs) were predicted from virus genomes using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/), with gene annotation made by searching against the Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/cdd/). Amino acid sequences of the viruses identified in this study were aligned with representative reference viruses using the E-INS-i algorithm in MAFFT v7. Phylogenetic trees were then estimated using the maximum likelihood method in IQ-TREE (v1.6.1241) with the best-fit amino acid substitution model and the ultrafast bootstrap approximation (UFBoot) approach (1 000 bootstrap replicates). The nucleotide alignment was used for recombination analysis by RDP (v4.16), and six different recombination detection methods including RDP, BootScan, CHIMAERA, GENECONV, MAXCHI and SiSscan were employed.
The number of RNA-sequencing raw reads in the 15 libraries ranged from 78417434 to 175355292 (Supplementary Table S1). Analyses of these data revealed the presence of two RNA viruses, referred as gerbil astrovirus (GeAstV) and gerbil paramyxovirus (GePV) (Supplementary Table S1). GeAstV was detected in two lung libraries, while GePV was detected in two kidney libraries. We further screened previously sequenced meta-transcriptomic libraries of Mongolian gerbils for these viruses, and GePV was also detected in multiple kidney libraries (Supplementary Table S1). The highest virus abundance (RPM) of GeAstV and GePV were 281 and 263, respectively (Supplementary Table S1). In total, two complete sequences of each virus were recovered, and pairwise nucleotide comparison showed 99.72% and 99.28% similarity between the two genomes of GeAstV and GePV, respectively.
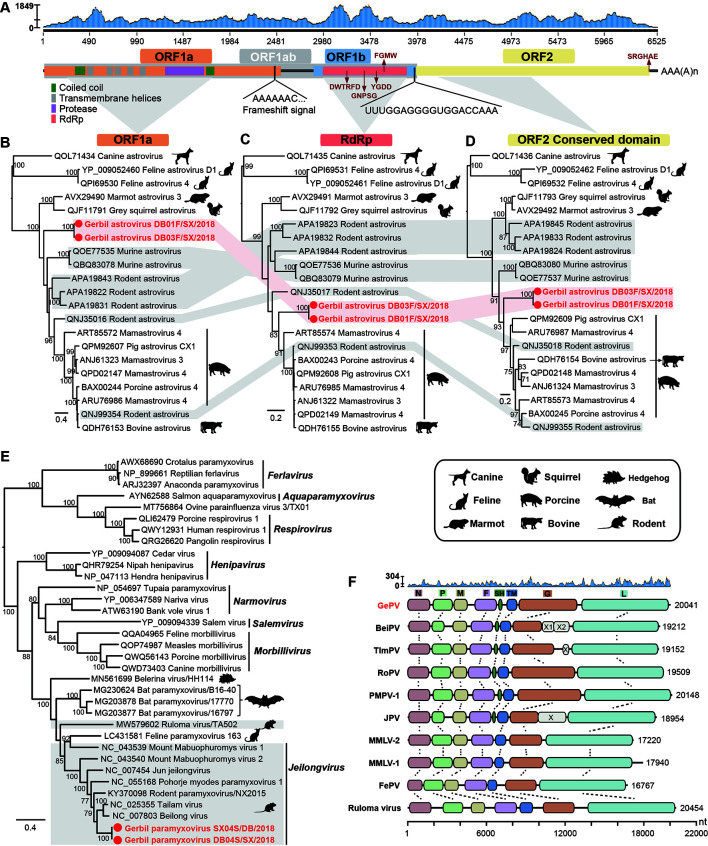
The complete genome of GeAstV was 6 525 bp in length (excluding the poly “A” tail) and contained the classical astrovirus genomic organization (Boujon et al., 2017) of three ORFs: ORF1a, ORF1b, and ORF2 (Figure 1A). A conserved ribosomal frameshift signal (AAAAAAC) was present near the 3' end of ORF1a, and a highly conserved sequence (UUUGGAGGGGUGGACCAAA) was located just before the start codon of ORF2 as observed in other AstVs (Bouzalas et al., 2016). Five potential trans-membrane domains and two coiled-coil domains were predicted based on the amino acid sequence of ORF1a (Figure 1A). The characteristic motifs DWTRFD, GNPSG, YGDD and FGMW were also identified within the RNA-dependent RNA polymerase (RdRp) domain (Figure 1A). Furthermore, the characteristic amino acid motif SRGHAE was also found at the 3' end of ORF2 (Figure 1A).
Figure 1.
Genome characterization and evolutionary relationships of GeAstV and GePV
A: Schematic representation and read abundance of the genome of GeAstV. The characteristic amino acid motifs are highlighted with red arrows. B–D: Maximum likelihood phylogenies of GeAstV and representative members of the genus Mamastrovirus were inferred based on the amino acid alignments of ORF1a (B), RdRp (C), and ORF2 (D). The two gerbil astrovirus sequences are highlighted in red with red solid circles. The grey-shaded regions show the sequences with phylogenetic incongruence and the rodent associated astrovirus. E: Maximum likelihood phylogeny of the L protein of gerbil paramyxovirus and the representative members of the Paramyxoviridae, including those from the genera Ferlavirus, Aquaparamyxovirus, Respirovirus, Henipavirus, Narmovirus, Salemvirus, Morbillivirus, and Jeilongvirus. The rodent associated viruses are highlighted with a grey background. F: The genome of GePV and nine paramyxoviruses from the genus Jeilongvirus: Beilong virus (BeiPV), Tailam virus (TlmPV), Rodent paramyxovirus (RoPV), Pohorje myodes paramyxovirus 1 (PMPV-1), Jun jeilongvirus (JPV), Mount Mabu Lophuromys virus 1 (MMLV-1) and 2 (MMLV-2), Feline paramyxovirus(FePV), and Ruloma virus. Read abundance of the genome of GePV are shown on the top, followed by open reading frames (ORFs) colored according to their putative functions. The numbers to the right of each virus indicate the length of the virus genome. The genomes are drawn to a unified length scale shown at the bottom. Homologous gene regions are connected by dotted lines. B–E: The trees were midpoint rooted for clarity. Bootstrap values of >70% are shown for key nodes.
Phylogenetic analyses of ORF1a (Figure 1B), RdRp (Figure 1C), and ORF2 (Figure 1D) of GeAstV with related astroviruses revealed that RdRp and ORF2 of GeAstV fell within a subclade containing rodent, bovine, and porcine associated astroviruses (Figure 1C, D), while ORF1a was more basal. Although the incongruous phylogenies give a signal of potential recombination events, no recombination events were identified by all the six detection methods embedded in RDP, likely due to the high level of sequence divergence between these viruses (Supplementary Figure S2A). BLASTX analysis showed that GeAstV shared the highest amino acid identity of 75.07% with a pig astrovirus – Mamastrovirus 4 (GenBank accession No. ART85572) – in RdRp (Supplementary Figure S2A), while shared 42.14% and 35.94% amino acid identity with a rodent astrovirus (GenBank accession No. QNJ35016) in the deduced ORF1a and ORF2 proteins, respectively (Supplementary Figure S2A). Taken together, these results indicated that GeAstV might represent a novel member within the genus Mamastrovirus according to the species demarcation criteria (Boujon et al., 2017).
Astroviruses (AstVs) can infect a wide range of mammals and birds, causing asymptomatic or systemic diseases (Boujon et al., 2017). Current evidence reveals that cross-species transmission of astroviruses could occur between animals living in the same ecological niche, as AstVs have high environmental stability and employ the fecal-oral transmission route (Roach & Langlois, 2021). In this study, rodent- and porcine-associated astroviruses are clustered together, suggesting potential interspecies transmission between these animals.
The full-length genome of GePV was 20 068 nucleotides in length and comprised eight genes in the order (3'-5'): nucleocapsid protein (N), phosphoprotein and proteins V and C (P/V/C), matrix protein (M), fusion protein (F), small hydrophobic protein (SH), transmembrane protein (TM), glycoprotein (G), and RNA polymerase (L) (Figure 1F). Phylogenetic analysis of the complete L protein of GePV and other representative members of Paramyxoviridae revealed that GePV fell within the genus Jeilongvirus, clustering with Tailam virus and Beilong virus (Figure 1E). Comparative analysis of the genome organizations of GePV and nine members from the same cluster showed that GePV and five Jeilongviruses contained eight predicted genes while Mount Mabu Lophuromys virus 1 (MMLV-1) and 2 (MMLV-2), Feline paramyxovirus (FePV), and Ruloma virus might not possess the SH gene (Figure 1F). With the exception of the feline paramyxovirus identified in cat, the remaining eight viruses were all detected in rodents. Regarding the N, G and L proteins, GePV shared the highest similarity (59.56%, 61.29%, and 77.04%, respectively) with Tailam virus, while it had the highest identity (82.35% and 28.57%, respectively) to Beilong virus in the M and TM proteins, and shared the highest similarity (56.35% and 73.13%, respectively) with rodent paramyxovirus in the P and F proteins (Supplementary Figure S2B). Collectively, GePV might represent a novel species within the genus Jeilongvirus, and most closely related to Tailam virus and Beilong virus.
Paramyxoviruses contain several highly pathogenic zoonotic viruses such as Measles virus, Hendra virus, and Nipah virus (Sakaguchi et al., 2020). The genus Jeilongvirus has been recently established, and members of this genus were identified in rodents, bats, and cats (Sakaguchi et al., 2020). Although zoonotic infections of jeilongviruses are rarely described, Beilongvirus has been discovered in a human kidney cell line and antibodies against Jun jeilongvirus have been found in human, pigs, and rodents.
In summary, we reported the discovery of a divergent astrovirus in lung samples and a novel paramyxovirus in kidney samples from wild Mongolian gerbils, China, suggesting that Mongolian gerbil could be an important host of rodent astrovirus and paramyxovirus. However, whether these viruses could infect humans or other animals clearly merits further investigation.
DATA AVAILABILITY
All raw reads generated here are available at the NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/) under BioProjectID PRJNA749072 with BioSample accessions SAMN22137953-SAMN22137967, as well as CNCB Genome Sequence Archive (GSA) database (https://ngdc.cncb.ac.cn/gsa/) under BioProject accession PRJCA008551 with BioSample accessions SAMC627845-SAMC627859. Four consensus genome sequences of identified viruses generated in this study have been uploaded in GenBank under accession Nos. OL345583, OL345584, OL409126, and OL409127, as well as CNCB Genome Warehouse (GWH) under accession GWHBHNY01000000 and GWHBHNX01000000.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
S.M.N., C.X.L., and W.F.S. designed the study. S.M.N., C.H.A., Y.X.S., and W.H.C. collected the samples. J.L., Y.T.W., H.Z., and L.X. performed the laboratory work. C.X.L. performed data analysis. C.X.L and W.F.S drafted and revised the paper. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We are grateful to Dr. John-Sebastian Eden (University of Sydney, Australia) for proofreading the manuscript.
Funding Statement
This study was supported by the Academic Promotion Programme of Shandong First Medical University (2019QL006), and the Open Research Fund Program of CAS Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology (2021SPCAS002)
Contributor Information
Ci-Xiu Li, Email: licixiu@sdfmu.edu.cn.
Wei-Feng Shi, Email: shiwf@ioz.ac.cn.
References
- 1.An CH, Li J, Wang YT, Nie SM, Chang WH, Zhou H, et al. 2022. Identification of a novel hepacivirus in Mongolian gerbil (Meriones unguiculatus) from Shaanxi, China. Virologica Sinica, 19: S1995-820X(22)00016-5.
- 2.Bleich EM, Keubler LM, Smoczek A, Mähler M, Bleich A Hygienic Monitoring of mongolian gerbils: which mouse viruses should be included? Laboratory Animals. 2012;46(2):173–175. doi: 10.1258/la.2011.011144. [DOI] [PubMed] [Google Scholar]
- 3.Boujon CL, Koch MC, Seuberlich T The expanding field of mammalian astroviruses: opportunities and challenges in clinical virology. Advances in Virus Research. 2017;99:109–137. doi: 10.1016/bs.aivir.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Bouzalas IG, Wüthrich D, Selimovic-Hamza S, Drögemuller C, Bruggmann R, Seuberlich T Full-genome based molecular characterization of encephalitis-associated bovine astroviruses. Infection, Genetics and Evolution. 2016;44:162–168. doi: 10.1016/j.meegid.2016.06.052. [DOI] [PubMed] [Google Scholar]
- 5.Charrel RN, De Lamballerie X Zoonotic aspects of arenavirus infections. Veterinary Microbiology. 2010;140(3-4):213–220. doi: 10.1016/j.vetmic.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Guo WP, Lin XD, Wang W, Tian JH, Cong ML, Zhang HL, et al Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathogens. 2013;9(2):e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu T, Li J, Zhou H, Li CX, Holmes EC, Shi WF Bioinformatics resources for SARS-CoV-2 discovery and surveillance. Briefings in Bioinformatics. 2021;22(2):631–641. doi: 10.1093/bib/bbaa386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, Wan X, Zhong W Population dynamics of the mongolian gerbils: seasonal patterns and interactions among density, reproduction and climate. Journal of Arid Environments. 2007;68(3):383–397. doi: 10.1016/j.jaridenv.2006.07.002. [DOI] [Google Scholar]
- 9.Roach SN, Langlois RA Intra- and cross-species transmission of astroviruses. Viruses. 2021;13(6):1127. doi: 10.3390/v13061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakaguchi S, Nakagawa S, Mitsuhashi S, Ogawa M, Sugiyama K, Tamukai K, et al Molecular characterization of feline paramyxovirus in japanese cat populations. Archives of Virology. 2020;165(2):413–418. doi: 10.1007/s00705-019-04480-x. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H, Ji JK, Chen X, Bi YH, Li J, Wang QH, et al Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021;184(17):4380–4391.e14. doi: 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
All raw reads generated here are available at the NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/) under BioProjectID PRJNA749072 with BioSample accessions SAMN22137953-SAMN22137967, as well as CNCB Genome Sequence Archive (GSA) database (https://ngdc.cncb.ac.cn/gsa/) under BioProject accession PRJCA008551 with BioSample accessions SAMC627845-SAMC627859. Four consensus genome sequences of identified viruses generated in this study have been uploaded in GenBank under accession Nos. OL345583, OL345584, OL409126, and OL409127, as well as CNCB Genome Warehouse (GWH) under accession GWHBHNY01000000 and GWHBHNX01000000.