Abstract
In an attempt to generate g.A746G substitution in the BMPR-IB gene, we unexpectedly obtained BMPR-IB homozygous knockout piglets (BMPR-IB-/-) and heterogeneous knockout piglets with one copy of the A746G mutation (BMPR-IB-/746G) via CRISPR/Cas9 editing. Polymerase chain reaction (PCR) and sequencing revealed complex genomic rearrangements in the target region. All BMPR-IB-disrupted piglets showed an inability to stand and walk normally. Both BMPR-IB-/- and BMPR-IB-/746G piglets exhibited severe skeletal dysplasia characterized by distorted and truncated forearms (ulna, radius) and disordered carpal, metacarpal, and phalangeal bones in the forelimbs. The piglets displayed more severe deformities in the hindlimbs by visual inspection, including fibular hemimelia, enlarged tarsal bone, and disordered toe joint bones. Limb deformities were more profound in BMPR-IB-/- piglets than in the BMPR-IB-/746G piglets. Proteomic analysis identified 139 differentially expressed proteins (DEPs) in the hindlimb fibula of BMPR-IB-/746G piglets compared to the wild-type (WT) controls. Most DEPs are involved in skeletal or embryonic development and/or the TGF-β pathway and tumor progression. Gene Ontology (GO) and protein domain enrichment analysis suggested alterations in these processes. Of the top 50 DEPs, a large proportion, e.g., C1QA, MYO1H, SRSF1, P3H1, GJA1, TCOF1, RBM10, SPP2, MMP13, and PHAX, were significantly associated with skeletal development. Our study provides novel findings on the role of BMPR-IB in mammalian limb development.
Keywords: BMPR-IB, A746G, Pigs, Limb deformities
INTRODUCTION
Bone morphogenetic proteins (BMPs), members of the transforming growth factor β (TGF-β) family, play important roles in the formation of bone and cartilage and the development of other organs, such as muscle, kidney, and blood vessels (Katagiri & Watabe, 2016). BMPs transduce their signals through type I and type II serine-threonine kinase receptors (BMPRI and BMPRII) (Miyazono et al., 2010). Perturbations of BMP signaling via BMPRI have been linked to multiple diseases in bone, cartilage, and muscles (Lin et al., 2016). As a type of BMPRI, activin-like kinase 6 (ALK6, also called BMPR-IB) is a critical regulator of chondrogenesis and osteogenesis (Lin et al., 2016). Patients with missense (I200K and R486W) or deletion (del359-366) mutations in BMPR-IB suffer severe limb deformations, including short stature, fibula aplasia, severe brachydactyly, and ulnar deviation of the hands, which are mainly caused by chondrodysplasia during skeletal development (Demirhan et al., 2005; Lehmann et al., 2003). In addition, null mutations of BMPR-IB in mice result in limb abnormalities, demonstrating that BMPR-IB is required for chondrocyte proliferation, differentiation, and maturation (Baur et al., 2000; Yi et al., 2000). In vitro studies have shown that continuous expression of active BMPR-IB induces mineralized bone matrix formation, while inhibition of endogenous BMPR-IB blocks BMP2-induced osteoblast differentiation and mineralized bone matrix formation, suggesting that BMPR-IB is required for osteoblast differentiation and bone formation (Chen et al., 1998). Mice expressing truncated dominant-negative BMPR-IB in target osteoblasts exhibit impaired postnatal bone formation (Zhao et al., 2002). Thus, osteoblastic BMPR-IB appears to play a necessary role during postnatal bone modeling and remodeling.
The BMPR-IB gene also affects prolificacy in sheep and plays a vital role in the control of follicular growth and development (Davis et al., 2006; Reader et al., 2012). The A746G mutation in BMPR-IB is reported to be highly associated with increased ovulation rates and litter size in sheep (Mulsant et al., 2001; Wilson et al., 2001). As the BMPR-IB A746G mutation was not detected in any pig breeds in our collection, we introduced the 746 GG mutation into the porcine genome via traditional transgenic technology in a previous study (Zhao et al., 2016), aiming to improve reproductive performance. Though the resulting transgenic boar exhibited stronger spermatogenic ability (Zhao et al., 2016), an important functional gene, NAGLU, was disrupted due to random insertion (Yang et al., 2018).
To avoid random integration of exogenous vectors, we used the CRISPR/Cas9 genome-editing platform (Cong et al., 2013) to edit AA746 into 746GG of the BMPR-IB gene in porcine fetal fibroblasts (PFFs) using a polymerase chain reaction (PCR)-amplified donor, with cloned pigs then generated by somatic cell nuclear transfer (SCNT). Unexpectedly, all nine cloned piglets from four deliveries showed severe limb deformities and were unable to walk and stand normally. Based on these ubiquitous limb deformities, we proposed that accidental gene disruption may have occurred. In this study, we first genotyped the target region across the donor sequence and found that the BMPR-IB gene was disrupted in these piglets. We then dissected the limb phenotypes and investigated molecular regulation in these pigs.
MATERIALS AND METHODS
Animals and ethics statement
The PFFs were isolated from one-month-old large white pig fetuses using 200 U/mL collagenase type IV (Sigma-Aldrich, USA). Surrogate sows were housed individually according to standard procedures. All experiments involving animals were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals formulated by the Ministry of Agriculture and Rural Affairs of the People’s Republic of China. The study was approved by the Ethics Committee of Jiangxi Agricultural University. All animal operations were performed under anesthesia to minimize suffering. Sixteen cloned piglets (nine individuals with deformed limbs and seven normal individuals) aged 3–5 days were used in this experiment.
Construction of CRISPR/Cas9 vector and double-stranded DNA donor template
The pSpCas9(BB)-2A-GFP (PX458, #48138) plasmid used in this study was purchased from Addgene (http://www.addgene.org/CRISPR/, USA). Single-guide RNA (sgRNA) targeting exon 8 of porcine BMPR-IB was designed using the Benchling online tool (https://www.benchling.com/). Here, 5'-TCATTGCTGCAGACATCAAA-3' was selected as the sgRNA, and the corresponding protospacer adjacent motif (PAM) sequence was GGG (Figure 1A). Paired synthesized oligonucleotides containing the BMPR-IB-sgRNA sequence (Supplementary Table S1) were ordered from Sangon Biotech Co., Ltd. (China); the DNA pair were annealed and ligated into the BbsI-digested PX458 vector to generate the recombinant plasmid. The recombinant plasmid DNA was transformed into Trans5ɑ competent cells (TransGen, China), and then extracted using an EndoFree® Plasmid Maxi Kit (Qiagen, Germany).
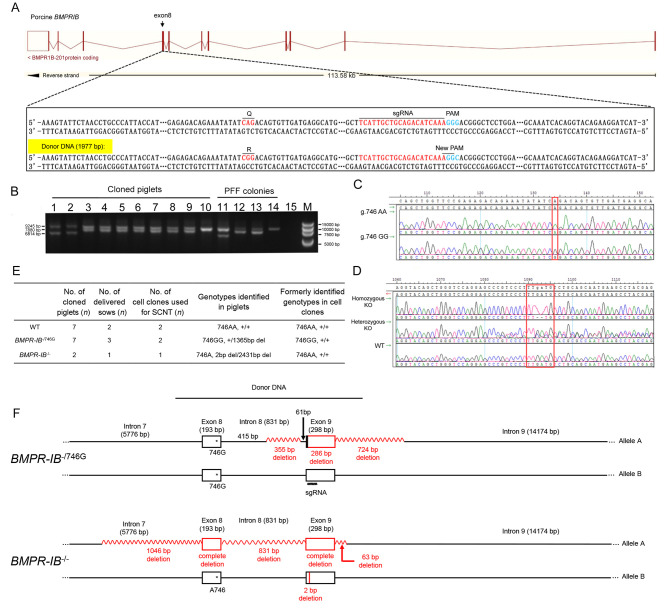
Figure 1.
Generation of BMPR-IB-modified PFFs and piglets
A: Schematic of strategy used to generate BMPR-IB-modified PFFs via linear double-stranded DNA-mediated homology-directed repair. Q, glutamine; R, arginine. B: Long-range PCR detection of BMPR-IB-deleted mutations in cloned piglets and PFF colonies. Lane 15 is a negative control (water) and M is a DNA marker. C: Sequencing chromatograms showing BMPR-IB A746G mutation in cloned piglets. Mutated nucleotide is marked by red box. D: Sequencing chromatograms showing BMPR-IB KO mutations in cloned piglets. Target site is marked by a red box. KO, knockout. E: Genotypes of newborn piglets. WT, wild-type; BMPR-IB-/746G, harboring 746G and 1 365 bp del intrans; BMPR-IB-/-, harboring 746A and 2 bp del/2 431 bp del. F: Genome rearrangements ofBMPR-IB gene in BMPR-IB-/746G and BMPR-IB-/- piglets. Deleted regions are marked in red.
Linear double-stranded donor DNA was obtained by bridge-overlap-extension PCR. First, we used primer pair two (Supplementary Table S1) to amplify the region harboring the sgRNA sequence targeting BMPR-IB in large white pigs by routine PCR, and DNA of the individual with the GGC sequence at the PAM site was selected as a template for the next amplification. Two PCR assays using primer pairs three and four (Supplementary Table S1) were then performed in a thermocycler under the following conditions: 94 °C for 2 min; 35 cycles at 98 °C for 10 s, 68 °C for 3 min; and 72 °C for 10 min. We mixed the two PCR products at a ratio of 1:1, and then conducted bridging PCR using primer pair five (Supplementary Table S1). The bridging PCR products were subjected to a final round of amplification using a touch-down PCR protocol (primer pair six for detection, Supplementary Table S1) under the following conditions: 94 °C for 5 min; 26 cycles at 94 °C for 30 s, 68 °C (−0.5 °C/cycle) for 45 s, 72 °C for 2 min; 14 cycles at 94 °C for 30 s, 55 °C for 45 s, 72 °C for 2 min; and 72 °C for 10 min. Finally, the amplified PCR product (donor DNA containing A746G mutation) was gel purified using a QIAquick PCR Purification Kit (Qiagen, Germany).
PFF transfection and selection
To obtain BMPR-IB 746G mutation cell clones, 25 μg of Cas9-sgRNA plasmid, 25 μg of purified donor DNA, and 12.5 μg of PX459 v2 plasmid were co-transfected into 3×106 PFF cells using the BTX ECM 2001 (USA) electroporation system (200 V, 1 ms, 3 pulses, 1 repeat). The electroporated PFFs were transferred into a 10 cm Petri Dish with growth medium containing Dulbecco’s Modified Eagle Medium (DMEM), 15% fetal bovine serum (FBS), 100 IU/mL penicillin and 100 µg/mL streptomycin, and SCR7 (Xcessbio, USA) to a final concentration of 1 μmol/L. After 30 h, PFFs were selected using 3.5 μg/mL puromycin (Sigma, Japan) for two days. The cells were then plated into 40 Petri Dishes (10 cm) at various cell densities for an additional 6–8 days of culture at 37 °C. The single-cell colonies in the Petri Dishes were collected and seeded in 24-well plates. After reaching 90% confluency, the cells in each colony were passaged in 6-well plates and sub-cultured at 37 °C for 48 h. About 20% of each colony was digested (56 °C, 60 min; 95 °C, 10 min) in 10 μL of lysis buffer (0.5% NP40 and 2 μg/μL of Proteinase K) to extract DNA, and the remaining cells were stored in liquid nitrogen for SCNT. Primer pair seven (Supplementary Table S1) was first used to identify the A746G locus in cell clones. The sgRNA sequence region was amplified using primer pair two (Supplementary Table S1) for A746G positive clones. The PCR products were sequenced on a 3130XL Genetic Analyzer (Applied Biosystem, USA). Colonies carrying 746GG were selected as donor cells for SCNT, and cells carrying AA746 were used as controls.
SCNT and embryo transplantation
Five cell colonies (Figure 1E) were used as nuclear donors to produce cloned pigs via SCNT, as described previously (Gong et al., 2004). Briefly, cumulus-oocyte complexes were collected and matured at 38.5 °C for 20 h in maturation medium comprised of M199 (Gibco, USA) supplemented with 10% FBS, 0.01 U/mL basal follicle stimulating hormone (bFSH), 0.01 U/mL basal luteinizing hormone (bLH), 1 μg/mL estradiol, and 1% (v:v) penicillin/streptomycin. The first polar body was then aspirated from the mature oocytes using a glass pipette, and, finally, the donor cells were fused with enucleated oocytes using BTX ECM 2001 (USA) electrofusion equipment. The reconstructed embryos were cultured in embryo-development medium at 38.5 °C for 24 h and then transferred surgically into the oviducts of estrus-synchronized pig surrogates (each recipient receiving approximate 350 embryos). Pregnancy status was determined by ultrasonography 30 days after embryo transfer. All cloned piglets were delivered by natural birth.
Genotyping cloned piglets
Genomic DNA was extracted from the ear tissues of cloned piglets using a cell/tissue genomic DNA extraction kit (Generay Biotech, Shanghai, China). Primer pair eight (Supplementary Table S1) was used for long-range PCR amplification across the donor sequence in BMPR-IB. The PCR protocol included 25 µL of 2×Gflex PCR Buffer (TaKaRa, Japan), 1 µL of each forward and reverse primer (10 µmol/L), 100 ng of genomic DNA, and ddH2O to a final volume of 50 µL. Cycling parameters were: 94 °C for 1 min; 30 cycles at 98 °C for 10 s, 60 °C for 15 s, 68 °C for 5.5 min; and 68 °C for 10 min. After sequencing the PCR products, the mutant piglets were all found to be BMPR-IB disrupted, eitherBMPR-IB-/- or BMPR-IB-/746G, as described below.
To detect whether the CRISPR plasmid vectors were integrated into the cloned piglet genomes, DNA of the cloned piglets was used as a template for PCR amplification with primer pairs nine and ten, respectively (Supplementary Table S1). The touchdown PCR conditions were 94 °C for 5 min; 26 cycles at 94 °C for 30 s, 68 °C (−0.5 °C/cycle) for 30 s, 72 °C for 45 s; 14 cycles at 94 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s; and 72 °C for 10 min. To detect whether off-target mutations existed in the BMPR-IB-disrupted piglets, the top 15 predicted off-target sites (OTS) were selected using online software (https://www.benchling.com/). Amplicons were subjected to Sanger sequencing. The primers used are listed in Supplementary Table S2.
Phenotype analysis
One BMPR-IB-/-, two BMPR-IB-/746G, and two wild-type (WT) cloned piglets were used for skeletal phenotype analyses. We used the Digital Diagnost system (Philips, Netherlands) to take X-ray pictures of the whole-body skeletons of cloned piglets. The images were taken at 50 KV with 3 mA exposure. We further analyzed the anatomical structure of limbs in the BMPR-IB-disrupted and WT cloned piglets. Briefly, we used a scalpel, scissors, and tweezers to peel and remove the skin, muscle, and related adhesion tissues of the limbs. The limb skeletons of the BMPR-IB-/- (n=1), BMPR-IB-/746G (n=2), and WT (n=2) cloned piglets were then collected and stored at −80 °C. Both forelimbs and hindlimbs of each piglet were analyzed using the Micro-CT system (Nemo NMC-100, Pingseng Healthcare Inc., China) at a resolution of 50 μm, voltage of 90 kV, and current of 60 μA. Quantification of trabecular bone was assessed by measuring a 2 mm section starting 2 mm distal to the growth plate of the forelimb radius and hindlimb tibia. For cortical bone, 3 mm sections above the midshaft of the forelimb radius and hindlimb tibia were analyzed. Based on threshold segmentation and three-dimensional (3D) measurements, quantitative analyses of trabecular and cortical bone (BV/TV, ratio of segmented bone volume to total volume; BS/TV, ratio of segmented bone surface to total volume; Tb.Sp, mean distance between trabeculae, assessed using direct 3D methods; Tb.N, average number of trabeculae per unit length, a key parameter for trabecular bone architecture; Tb.BMD, bone density measure, reflecting strength of bones as represented by calcium content; Ct.BMD, cortical bone mineral density; Ct.Th, average cortical thickness; Ct.ar/Tt.ar, cortical area fraction) in the radius and tibia were performed using Avatar v1.6.5 (Pingseng Healthcare Inc., China).
Quantitative reverse transcription PCR (qRT-PCR) and western blot analyses
Total RNA was extracted from the liver, kidney, testicle, forelimb and hindlimb cartilage, forelimb ulna, and hindlimb fibula of BMPR-IB-disrupted (BMPR-IB-/-, n=1; BMPR-IB-/746G, n=3) and WT (n=3) piglets using TRIzol Reagent (Invitrogen, USA). The mRNA was reverse transcribed into cDNA using a PrimeScriptTM RT Reagent Kit with gDNA Eraser (Takara, Japan). qRT-PCR was performed using the ABI 7900 HT Fast Real-Time PCR System (Applied Biosystems, USA), and the thermal parameters were 50 °C for 2 min; 95 °C for 10 min; 40 cycles at 95 °C for 15 s, 60 °C for 50 s; 95 °C for 15 s; 60 °C for 15 s; and 95 °C for 15 s. The 2-ΔΔCt formula was used to determine relative gene expression, which was normalized to the level of GAPDH mRNA. Each reaction was performed in technical quadruplicate. Primer pairs eleven and twelve for qRT-PCR are listed in Supplementary Table S1.
Total protein was extracted from the liver and kidney of BMPR-IB-/- piglets (n=1) and forelimb ulna and hindlimb fibula of BMPR-IB-/746G (n=3) and WT piglets (n=3) using a protein extraction kit (Applygen, China). Protein concentration was determined using the bicinchoninic acid (BCA) assay, and 30 µg of protein from each sample was resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Angle Gene, China) and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, USA). After blocking with Quick Block™ Blocking Buffer (Beyotime, China) for 1 h at room temperature, the membranes were washed in Tris Buffered Saline with Tween 20 (TBST) and incubated overnight at 4 °C with primary antibodies, including: rabbit polyclonal anti-BMPR-IB antibody (1:500; Sino Biological, China), mouse monoclonal antibodies for C1QA, SRSF1, P3H1, GJA1, TCOF1, RBM10, MMP13, and PHAX (1:100; Santa Cruz Biotechnology, USA), and β-actin (1:1 000; Abcam, UK). The membranes were washed and incubated with horseradish peroxidase (HRP)-conjugated secondary anti-rabbit or anti-mouse antibodies (1:2 000; Abcam, UK) for 1 h at room temperature. β-actin was used as the loading control. The bands were visualized with an ECL chemiluminescent kit (Beyotime, China) and scanned using the ChemiDocTM MP imaging system (Bio-Rad, USA). Band intensities were quantified using ImageJ (Rawak Software, Germany). The relative amounts of BMPR-IB, C1QA, SRSF1, P3H1, GJA1, TCOF1, RBM10, MMP13, and PHAX were calculated after normalization to β-actin.
4D label-free quantitative proteomic analysis
Proteomic analysis was conducted by PTM Bio Co., Ltd. (China). Hindlimb fibula samples from the BMPR-IB-/746G (n=3) and WT piglets (n=3) were ground into powder in liquid nitrogen and thoroughly homogenized in 8 mol/L urea containing 1% protease inhibitor cocktail. After insoluble debris was removed by centrifugation at 12 000 g for 10 min at 4 °C, the supernatant was collected and protein concentration was determined using a BCA protein assay kit (Beyotime, China). For digestion, the protein solution was reduced with 5 mmol/L dithiothreitol for 30 min at 56 °C and alkylated with 11 mmol/L iodoacetamide for 15 min at room temperature in the dark. The protein samples were then diluted with 100 mmol/L triethylammonium bicarbonate (TEAB) to a urea concentration of 2 mol/L. Finally, trypsin was added at a 1:50 trypsin-to-protein mass ratio for the first digestion overnight and a 1:100 trypsin-to-protein mass ratio for the second digestion of 4 h.
The tryptic peptides were dissolved in 0.1% formic acid and 2% acetonitrile (solvent A) and loaded onto a home-made reversed-phase analytical column (15 cm long, 75 μm i.d.). The gradient was increased from 6% to 23% solvent B (0.1% formic acid in 90% acetonitrile) over 38 min, 23% to 35% in 14 min, and to 80% in 4 min, then held at 80% for the last 4 min, all at a constant flow rate of 550 nL/min on an EASY-nLC 1000 ultra-performance liquid chromatography (UPLC) system (Thermo Fisher Scientific, USA). The peptides were subjected to nanospray ionization (NSI) source followed by tandem mass spectrometry (MS/MS) in a Q ExactiveTM Plus (Thermo Fisher Scientific, USA) coupled to the UPLC system. The electrospray voltage applied was 2.3 kV. The m/z scan range was 400 to 1 200 for full scan, and intact peptides were detected using Orbitrap at a resolution of 60 000. Peptides were then selected for MS/MS using normalized collision energy (NCE) setting as 28 and the fragments were detected using Orbitrap at a resolution of 17 500. Standard data-dependent acquisition (DDA) procedures were implemented to detect and quantify peptides, including one MS acquisition at a mass/charge ratio (m/z) of 400–1 500, with the top 20 intense precursor ions subjected to MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control (AGC) was set at 5×104 and fixed first mass was set to 100 m/z.
The resulting MS/MS data were processed using the MaxQuant search engine (v.1.5.2.8). Tandem mass spectra were searched against the Sus_scrofa_9823 database concatenated with the reverse decoy database. Trypsin/P was specified as the cleavage enzyme allowing up to four missing cleavages. Mass tolerance for the precursor ions was set to 20×10−6 in the first search and 5×10−6 in the main search, and mass tolerance for the fragment ions was set to 0.02 Da. Carbamidomethyl on Cys was specified as fixed modification and acetylation modification and oxidation on Met were specified as variable modifications. The false discovery rate (FDR) was adjusted to <1% and minimum score for modified peptides was set to >40.
Functional enrichment analysis
Proteins were classified by Gene Ontology (GO) annotation into three categories: biological process, cellular component, and molecular function. For each category, two-tailed Fisher’s exact test was employed to test differentially expressed protein (DEP) enrichment against all identified proteins. GO terms with a corrected P<0.05 were considered significant. For each DEP, the InterPro (http://www.ebi.ac.uk/interpro/) database was searched and two-tailed Fisher’s exact test was employed to test its (domain) enrichment against all identified proteins. Protein domains with a correctedP<0.05 were considered significant.
Target proteomic analysis
Parallel reaction monitoring (PRM) analysis was performed to verify and quantify the DEPs. One or two unique peptides of each target protein were selected from proteomic measurements based on thresholds of detection frequencies >50%, missed cleavage=0, and P<0.05. Finally, a spectral library of 10 selected proteins represented by 18 peptides was created. Proteins extracted from the hindlimb fibula ofBMPR-IB-/746G (n=3) and WT piglets (n=3) were alkylated and digested, as described in “4D label-free quantitative proteomic analysis”. Protein (1 μg) was injected into the liquid chromatography-tandem mass spectrometry (LC-MS/MS) system for the PRM assay (2.1 kV electrospray voltage and 35 000 resolution (AGC target 3×106, maximum injection time 50 ms)). The isolation window for MS/MS was set at 2.0 m/z. Peptide settings were: enzyme, trypsin [KR/P]; peptide length, 8–25; variable modification, carbamidomethyl on Cys and oxidation on Met; and max variable modifications, 3. Transition settings: precursor charges, 2 and 3; ion charges, 1 and 2; ion types, b, y, and p; product ions, ion 3 to last ion; and ion match tolerance, 0.02 Da. Skyline software (v3.6) was used for relative quantification for PRM study (Henderson et al., 2018).
Statistical analysis
All data are presented as mean±standard deviation (SD). Statistical analysis was performed using an independent two-tailed Student’s t-test. A null probability of P<0.05 was considered statistically significant. SPSS v17.0 was used for all analyses.
RESULTS
CRISPR/Cas9-mediated BMPR-IB A746G mutation in PFFs
To convert A746 of the BMPR-IB gene to 746G, we designed one sgRNA and 1 977 bp linear double-stranded donor (template) DNA for homology-directed repair (HDR)-mediated genome editing by CRISPR/Cas9 (Figure 1A). The linear double-stranded donor DNA, obtained from bridging PCR (Supplementary Figure S1A–C), encompassed the target point mutation (A746G) and sgRNA sequence, and was confirmed by Sanger sequencing (Supplementary Figure S1D). The donor DNA carried an altered PAM (GGG to GGC, Figure 1A; Supplementary Figure S1D). The Cas9/sgRNA expression vector and template DNA were both co-transfected into an early passage of PFFs. A total of 113 single-cell clones were obtained after 48 h of puromycin selection and subsequent subculture. These clones were genotyped via short-range PCR and Sanger sequencing. Of these 113 clones, 39 contained the A746G mutation (16 homozygotes and 23 heterozygotes), 30 contained homozygous or heterozygous deletions, and 44 were WT (Supplementary Table S3).
Generation of BMPR-IB-edited pigs
We selected two BMPR-IB/746GG and three BMPR-IB/AA746 cell colonies identified by short-range PCRs for SCNT, obtaining approximately 3 500 embryos. All embryos were surgically transferred to 10 surrogate sows, six of which carried to term and gave birth to 16 cloned piglets (Figure 1E). The genotypes of the cloned piglets were determined by Sanger sequencing of long-range PCR products across the 1 977 bp donor sequence. As shown in Figure 1B–F, seven cloned piglets carried the expected 746G mutation at the target locus, but all had a 1 365 bp heterozygous deletion of the BMPR-IB gene (referred to as “BMPR-IB-/746G”). Two piglets had a 2 bp deletion on one allele and a 2 431 bp deletion on the other allele of the BMPR-IB gene (referred to as “BMPR-IB-/-”). Piglets carrying either of the two genotypes were considered as BMPR-IB-disrupted pigs.
To detect whether the CRISPR plasmids were integrated into the host genome, we genotyped the Cas9 and sgRNA domains, and no integrations were found in any of the cloned piglets (Supplementary Figure S2). As off-target effects are a major concern when using the CRISPR/Cas9 system, we performed PCR amplification and sequencing for the top 15 potential OTS. No off-target effects were found in the BMPR-IB mutant piglets (Supplementary Table S4).
Phenotype characterization of BMPR-IB-disrupted pigs
The BMPR-IB-disrupted piglets were unable to stand normally, with their splayed limbs only able to move in a “paddling” motion (Video 1: https://figshare.com/s/966c57aee3cf5147604c). Close examination revealed different malformed shapes of the forelimb and hindlimb. As shown in Figure 2A, the forelimbs of BMPR-IB-disrupted piglets (BMPR-IB-/746G or BMPR-IB-/-) were severely distorted towards the abdomen, and the wrist joints could not be flexed or extended normally. In the hindlimbs of BMPR-IB-disrupted piglets, the knee joints were moderately rigid and the ankles and toes were severely bent backwards.
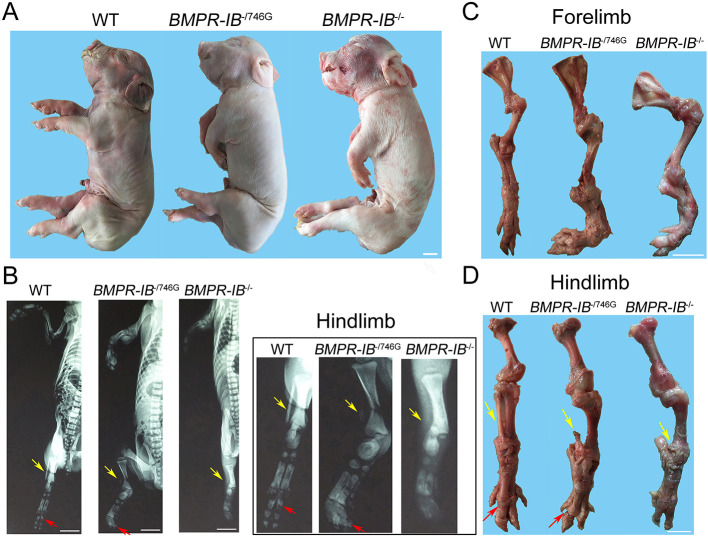
Figure 2.
Phenotypic characterization of BMPR-IB-disrupted piglets
A: Whole-body photographs of symptomatic BMPR-IB-disrupted piglets (BMPR-IB-/746G and BMPR-IB-/-) and WT piglets. B: Radiographs of forelimb and hindlimb in piglets of three different genotypes. Absence of fibula is noted in mutant piglets (yellow arrow). Red arrow indicates third proximal phalanx (os compedale). One BMPR-IB-/-, two BMPR-IB-/746G, and two WT-cloned piglets were used for radiography. C: Anatomical views of underdeveloped skeleton of forelimb in mutant piglet compared to WT piglet. D: Anatomical views of underdeveloped skeleton of hindlimb of mutant piglet compared to WT piglet. Partial or complete absence of fibula (yellow arrow) can be observed in BMPR-IB-/746G and BMPR-IB-/- piglets. Third proximal phalanx (os compedale) fell off (red arrow) in BMPR-IB-/746G piglets. White bars: 2 cm.
Radiographic and anatomical examinations showed marked skeletal hypoplasia of the limbs in BMPR-IB-disrupted piglets. In the forelimbs of the BMPR-IB-/746G and BMPR-IB-/- piglets, the ulna and radius were severely deformed and shortened. The carpal, metacarpal, and phalangeal bones of BMPR-IB-disrupted piglets were disorderedly arranged and partially absent. Moreover, bone in the wrist joint was hardened, rendering the joint unable to flex (Figure 2B, C; Supplementary Figure S3). In the hindlimbs of BMPR-IB-/746G piglets, the fibula was largely absent, tarsal bones were enlarged and partially absent, and phalangeal bones showed disordered arrangement. In addition, the ankle joint was swollen, with a large joint gap and collapsed third proximal phalanx (os compedale) (Figure 2B, D; Supplementary Figure S3). Of note, the BMPR-IB-/- piglets had nearly the same hindlimb deformity phenotypes as the BMPR-IB-/746G piglets, except for complete loss of the fibula in BMPR-IB-/- piglets (Figure 2B, D).
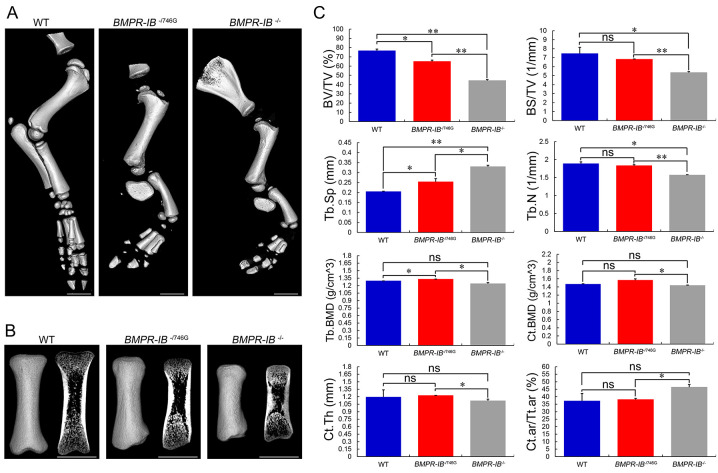
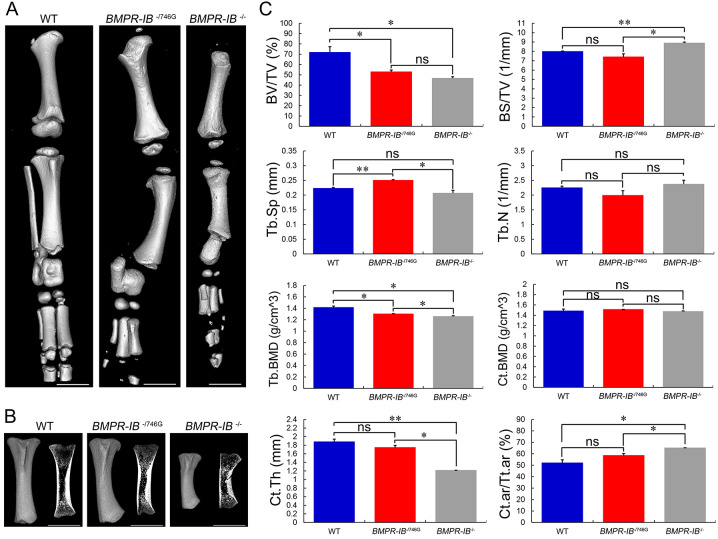
High-resolution micro-CT scans generated clearer visualization of limb bone morphology and skeletal arrangement, as well as quantitative bone morphometry (Figures 3, 4). Quantitative analysis of the forelimb radius (Figure 3C) revealed a 42% and 15% reduction in BV/TV in the BMPR-IB-/- and BMPR-IB-/746G piglets, respectively, compared with the WT piglets. Furthermore, trabecular BS/TV was 28% lower in the BMPR-IB-/- group than in the WT group, with no significant difference observed in the BMPR-IB-/746G group. In addition, compared with the WT piglets, Tb.Sp was 24% and 61% larger in the BMPR-IB-/746G and BMPR-IB-/- groups, respectively. BMPR-IB disruption did not appear to have a negative effect on Tb.BMD or Ct.BMD. In addition, no significant effects were observed in Ct.Th or Ct.ar/Tt.ar due to the large variations in WT piglets. Regarding the hindlimb tibia (Figure 4C), the BMPR-IB-/746G and BMPR-IB-/- groups had distinctly lower BV/TV (−26% and −35%, respectively) compared to the WT group. Trabecular BS/TV was 11% higher in the BMPR-IB-/- group compared to the WT, while no significant difference was found in the BMPR-IB-/746G group. Furthermore, Tb.BMD was lower in both groups (−8% and –11%, respectively), but there was no significant difference in Ct.BMD. There was a 35% reduction in Ct.Th in the BMPR-IB-/- group, while the decrease in Ct.Th was not significant in the BMPR-IB-/746G group (Figure 4C). In general, the BMPR-IB-/- group showed poorer morphometric deterioration than the BMPR-IB-/746G group.
Figure 3.
Micro-CT analyses of forelimb bones of piglets
A: High-resolution micro-CT scans of forelimb skeletons of WT, BMPR-IB-/746G, and BMPR-IB-/- piglets. B: 3D and 2D gray-scale images of radius of three different genotype piglets. C: Quantitative results of trabecular and cortical bone at radius distal end. Data are means±SD (WT, n=4; BMPR-IB-/746G, n=4; BMPR-IB-/-, n=2). ns: P>0.05;*: P<0.05;**: P<0.01. White bars: 1 cm. BV/TV, trabecular bone volume fraction; BS/TV, trabecular bone surface density; Tb.Sp, trabecular spacing; Tb.N, trabecular number; Tb.BMD, trabecular bone mineral density; Ct.BMD, cortical bone mineral density; Ct.Th, cortical thickness; Ct.ar/Tt.ar, cortical area fraction.
Figure 4.
Micro-CT analyses of hindlimb bones of piglets
A: High-resolution micro-CT scans of hindlimb skeletons of WT, BMPR-IB-/746G, and BMPR-IB-/- piglets. B: 3D and 2D gray-scale images of tibia of three different genotype piglets. C: Quantitative results of trabecular and cortical bone at tibia distal end. Data are means±SD (WT, n=4; BMPR-IB-/746G, n=4; BMPR-IB-/-, n=2). ns:P>0.05,*: P<0.05;**: P<0.01. White bars: 1 cm.
Analysis of BMPR-IB expression in cloned piglets
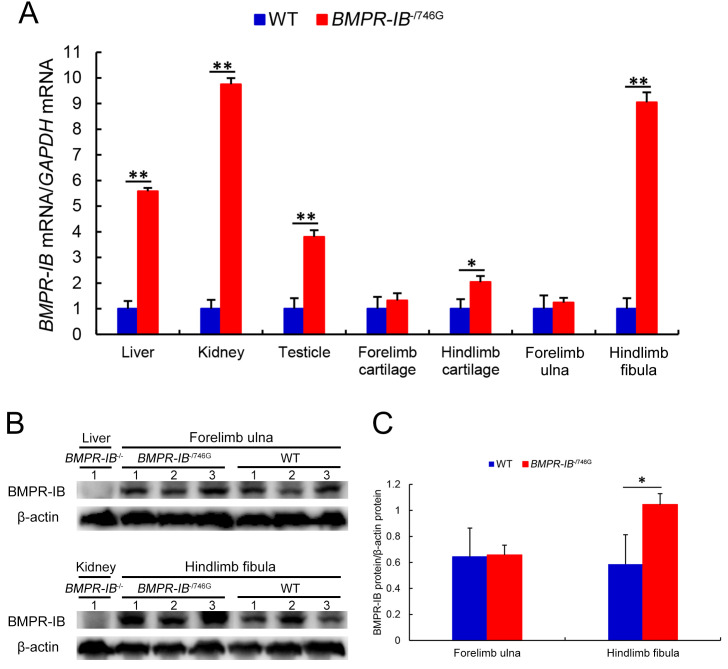
The qRT-PCR results showed that BMPR-IB gene expression was lost in all examined tissues of the BMPR-IB-/- piglets. In the BMPR-IB-/746G piglets, BMPR-IB expression was normal in the forelimb cartilage and ulna but was significantly elevated (P<0.05 orP<0.01) in the hindlimb cartilage and fibula, as well as the liver, kidney, and testes (Figure 5A).
Figure 5.
qRT-PCR and western blot analyses of BMPR-IB expression levels
A: Relative BMPR-IB mRNA expression levels in liver, kidney, testicle, forelimb cartilage, hindlimb cartilage, forelimb ulna, and hindlimb fibula of WT, BMPR-IB-/746G, and BMPR-IB-/- piglets, determined by qRT-PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a control. B: Western blots of BMPR-IB in forelimb ulna and hindlimb fibula in three BMPR-IB-/746G piglets and three WT piglets. Liver and kidney samples of one BMPR-IB-/- piglet were used as negative controls. C: Quantitative results of BMPR-IB protein using ImageJ software. β-actin was used as a control. Data are mean±SD (WT, n=3; BMPR-IB-/746G, n=3; BMPR-IB-/-, n=1). *: P<0.05;**: P<0.01. As BMPR-IB mRNA and protein expression levels were not detectable inBMPR-IB-/- piglet samples, expression level columns are not displayed in (A) and (C) in this figure.
Western blot analysis confirmed the loss of BMPR-IB in the liver and kidney of BMPR-IB-/- piglets (Figure 5B, C). The western blots showed no significant difference in BMPR-IB protein expression in the forelimb ulna between the BMPR-IB-/746G and WT piglets. However, BMPR-IB protein expression in the hindlimb fibula in the BMPR-IB-/746G piglets was significantly higher (P<0.05) than that in the WT piglets (Figure 5B, C).
Identification of DEPs
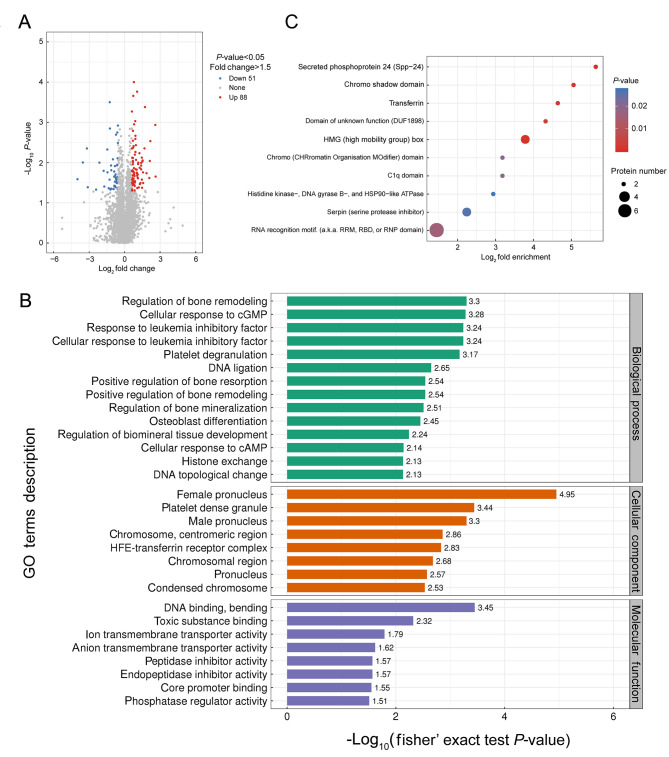
Hindlimb fibula samples from BMPR-IB-/746G (n=3) and WT piglets (n=3) were analyzed using the LC-MS/MS platform. A total of 6 412 proteins were identified and 5 957 proteins were quantified. Based on cut-off criteria (P<0.05, number of peptides>1, fold-change>1.5), a total of 139 proteins with one or more unique peptides were significantly differentially expressed in the hindlimb fibula between the two groups, including 51 down-regulated and 88 up-regulated in theBMPR-IB-/746G piglets (Figure 6A; Supplementary Table S5).
Figure 6.
DEPs in hindlimb fibula between BMPR-IB-/746G and WT groups
A: Volcano-plot distribution map. Red and blue dots denote significantly up- and down-regulated proteins, respectively. B: GO analysis of DEPs. Top 30 significantly enriched categories are shown in horizontal histogram. Horizontal axis represents –log10(P-value); vertical axis represents GO functional classification. C: Protein domain enrichment analysis of DEPs. Top 10 significantly enriched categories are shown in bubble chart. Functional classification is shown along vertical axis of bubble chart; proportion of DEPs by functional type divided by the ratio of identified proteins was log2 converted and shown along horizontal axis. Circle color indicates enriched P-value, and circle size indicates number of DEPs in functional classification.
GO and protein domain enrichment analysis
To obtain a better mechanistic understanding of the protein networks that may be related to limb deformity formation in BMPR-IB disrupted piglets, we used GO analysis to categorize DEPs. The DEPs in the hindlimb fibula were primarily involved in bone development regulation, such as regulation of bone remodeling, positive regulation of bone resorption, positive regulation of bone remodeling, regulation of bone mineralization, osteoblast differentiation, and regulation of biomineral tissue development (Figure 6B).
Protein domain enrichment analysis was performed to identify functional domains of the DEPs. As shown in Figure 6C, secreted phosphoprotein 24 (Spp-24), high mobility group (HMG) box, chromo shadow domain, transferrin, and serpin (serine protease inhibitor) were significantly enriched in the hindlimb fibula.
Analysis of target proteins using PRM
To verify the changes in differential expression identified by 4D label-free quantitative proteomic analysis, PRM was employed using the same samples. Ten up-regulated proteins with potential involvement in bone development regulation were selected and a panel of seven proteins with one or two unique peptides were successfully detected and quantified. All seven proteins were significantly up-regulated in the hindlimb fibula of BMPR-IB-/746G piglets (P<0.05,BMPR-IB-/746G/WT ratio>1) (Table 1). Thus, the PRM results were highly consistent with those from the 4D label-free quantitative phase (Table 1).
Table 1. PRM-verified DEPs in hindlimb fibula of BMPR-IB-/746G piglets compared with WT individuals .
| Protein accession | Gene | BMPR-IB-/746G/WT ratio (PRM) |
BMPR-IB-/746G/WT
P-value |
BMPR-IB-/746G/WT ratio (LQ) |
| PRM: Parallel reaction monitoring; LQ: 4D label-free quantitative proteomic analysis. | ||||
| F1S4R5 | SRSF7 | 1.79 | 3.49E-04 | 1.69 |
| Q29101 | GJA1 | 1.52 | 1.45E-02 | 1.52 |
| P14287 | SPP1 | 1.93 | 1.52E-02 | 2.48 |
| F1SV56 | MMP13 | 1.97 | 1.72E-02 | 2.19 |
| P09571 | TF | 4.38 | 2.19E-02 | 2.78 |
| Q8HZV3 | TFRC | 1.66 | 2.31E-02 | 1.51 |
| F1SM14 | SPP2 | 3.15 | 2.52E-02 | 4.30 |
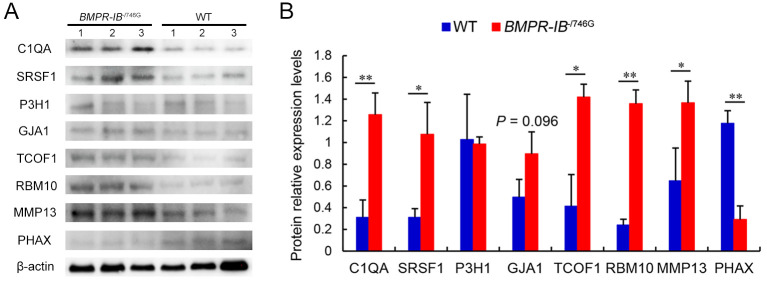
Validation of expression levels of selected DEPs
To validate the 4D label-free quantitative proteomic results, the expression levels of several DEPs in the hindlimb fibula were detected via western blotting. As shown in Figure 7, the protein expression levels of C1QA, SRSF1, TCOF1, RBM10, and MMP13 in the hindlimb fibula were significantly higher in the BMPR-IB-/746G piglets (P<0.01 orP<0.05) than in the WT piglets, and GJA1 expression was higher in theBMPR-IB-/746G piglets than in the WT piglets, but with a weakly significant effect (P=0.096). PHAX protein expression in the hindlimb fibula was significantly lower in BMPR-IB-/746G piglets (P<0.01) than in WT piglets (Figure 7). Thus, the expression levels of the selected DEPs detected by western blotting were highly consistent with the 4D label-free quantitative proteomic data (Figure 7; Supplementary Table S5).
Figure 7.
Western blot analyses of DEP expression levels in hindlimb fibula between BMPR-IB-/746G and WT groups
A: Western blots of DEPs in hindlimb fibula from three BMPR-IB-/746G piglets and three WT piglets. B: Quantitative analysis results of DEPs using ImageJ software. β-actin was used as a control. Data are mean±SD (WT, n=3; BMPR-IB-/746G, n=3). *: P<0.05;**: P<0.01.
DISCUSSION
In this study, we originally planned to produce cloned pigs with a BMPR-IB 746GG mutation. We isolated cell clones carrying this mutation via CRISPR/Cas9 techniques using bridge PCR-amplified products as the HDR template. However, our primary design had two significant defects. First, we attempted to avoid re-cutting the repair template by changing GGG (PAM) to GGC (Figure 1A). However, we overlooked the fact that an extra NGG PAM was introduced by the A746G mutation (Figure 1A), enabling Cas9 to re-cleave the repair template. Second, we implemented short-range PCR (Supplementary Figure S1) rather than long-range PCR across the whole donor region to identify isolated cell clones. This led to unknown genome rearrangements in the BMPR-IB gene in the selected colonies (Figure 1F). These two factors resulted in the accidental production of piglets carrying a disrupted BMPR-IB gene, which, to the best of our knowledge, is the first report on the generation of BMPR-IB gene disruption in pigs. Recent evidence indicates that complex rearrangements are frequently observed in CRISPR/Cas9 editing (Alanis-Lobato et al., 2021; Canaj et al., 2019; Skryabin et al., 2020). In this study, the BMPR-IB-/746G piglets carried a discontinuous 1 365 bp deletion with short fragments of intron 8 and exon 9 retained in the gene (Figure 1F). Regarding BMPR-IB-/- piglets, a 2 431 bp segment was deleted in one allele and a 2 bp segment was deleted in the other (Figure 1F). Both the 1 365 bp and 2 431 bp deletions spanned the donor region. Our study indicated that multiple PCR procedures may be a better option for detecting repair donor and flanking regions. This is because complex rearrangements can occur during genome editing, and a particular pair of primers may amplify only one allele of the target gene in cases where the other allele contains a deletion.
In our study, BMPR-IB-/746G piglets harboring compound mutations (g.746G and 1 365 bp del, Figure 1E) in theBMPR-IB gene displayed phenotypes typically observed in limbs with skeletal dysplasia. The g.746A>G mutation in theBMPR-IB gene has been previously identified in domestic sheep and is suggested to increase ovulation rates and litter size (Chu et al., 2007; Mahdavi et al., 2014; Reader et al., 2012; Roy et al., 2011; Zhou et al., 2018). In the current study, BMPR-IB mRNA and protein expression levels in five of the seven examined tissues were significantly higher in BMPR-IB-/746G piglets than in WT piglets (P<0.01 orP<0.05;Figure 5), suggesting that the expression levels of single intact BMPR-IB allele (carrying the 746G variant without deletion) in BMPR-IB-/746G individuals exceeded the sum of the transcript levels of the two alleles in the WT individuals. We speculate two possible reasons for this phenomenon. First, the A746G mutation may be located on a cis-acting element that regulates BMPR-IB expression, and it may alter the function of this element, thereby affecting gene expression. Second, genetic compensation may be induced due to deleterious mutations (El-Brolosy & Stainier, 2017). Indeed, bioinformatic analysis in this study indicated that the A746G mutation in the highly conserved intracellular kinase signaling domain of the BMP-IB receptor is likely to damage its structure and function, and intriguingly, cause distinct alterations in secondary structures in pigs and sheep (Supplementary Figure S4). As the function of the mutant BMPR-IB protein was greatly reduced but not necessarily completely lost, the BMPR-IB-/746G individuals may exhibited a genetic compensation response, in which the intact mutant allele and its translated protein were overexpressed to compensate for its functional deficits. This may also explain why BMPR-IB-/746G piglets are anatomically and pathologically similar to BMPR-IB-/- individuals but show less severe deformities in skeletal development (Figures 2D, 3, 4).
Disruption of BMPR-IB in mice and humans often results in severe limb abnormalities. For example, in BMPR-IB-/- mice, the proximal and middle phalanges are reduced and fused, the radius, ulna, and tibia lengths are normal, but the metacarpals/metatarsals are shorter, and several carpal/tarsal bones are affected (Yi et al., 2000). In the current study, however, mutant piglets not only exhibited malformation in the carpal, metacarpal, and phalangeal bones, but the ulna and radius were also severely distorted and shortened (Figures 2B, C, 3A). In humans, both I200K and R486W missense mutations are reported to cause brachydactyly type A2 (Lehmann et al., 2003). Furthermore, loss-of-function mutation (del359-366) can cause acromesomelic chondrodysplasia and genital anomalies, with hypoplasia of the femoral neck and head (Demirhan et al., 2005). The severe skeletal defects in our BMPR-IB mutant (BMPR-IB-/746G or BMPR-IB-/-) piglets, such as loss of fibula, abnormally developed tarsal bones, disorderedly arranged phalangeal bones, and severely distorted and shortened ulna and radius, are analogous to many clinical phenotypes. We also found that the forelimb wrist joint in BMPR-IB mutant piglets was hardened and could not be flexed, the hindlimb ankle joint in BMPR-IB-/746G piglets was malformed, and the third proximal phalanx (os compedale) was collapsed (Figure 2C, D). However, no defects were found in other parts of the skeleton.
As the hindlimb fibula was completely missing in BMPR-IB-/- piglets (Figures 2B, D, 4A), we obtained fibula samples from BMPR-IB-/746G and WT piglets for proteomic analysis. In total, 139 DEPs (88 up-regulated, 51 down-regulated) were identified (Figure 6A; Supplementary Table S5). PRM analysis was performed to verify the quantitative data of seven randomly selected proteins (Table 1). Most DEPs were significantly involved in one or multiple functions of skeletal development, embryonic development, TGF-β pathway, and immune system, especially tumor progression (Supplementary Table S5). These results are consistent with the known functions of the BMPR-IB gene (Dituri et al., 2019; Rahman et al., 2015). A large proportion of genes in the top 50 DEPs were significantly associated with skeletal or embryonic development (Supplementary Table S5), including C1QA, MYO1H, SRSF1, P3H1, GJA1, TCOF1, RBM10, SPP2, MMP13, and PHAX. C1QA is a subunit chain of C1q, which is implicated in osteoclast development from monocytes (Teo et al., 2012). Polymorphisms in MYO1H are associated with mandibular retrognathism (Arun et al., 2016) and sagittal and vertical craniofacial skeletal patterns (Cunha et al., 2019). SRSF1 is a prototype member of the serine/arginine (SR) rich family of splicing proteins and may modulate pattern formation (including cartilage formation) by inhibiting transcription of tissue-specific genes during embryogenesis (Lee et al., 2016). P3H1, also known as LEPRE1, forms a complex with cyclophilin B in the endoplasmic reticulum (Vranka et al., 2010). P3H1-null mice display abnormalities in fibrillar collagen-rich tissues, including bones (Vranka et al., 2010), and mutations in P3H1 can cause non-lethal (Takagi et al., 2012) and lethal recessive (Cabral et al., 2012) osteogenesis imperfections. Mutations in GJA1 can lead to human oculodentodigital dysplasia, including face, eye, tooth, and limb deformities (Paznekas et al., 2009; Sargiannidou et al., 2021). TCOF1 encodes the nucleolar phosphoprotein treacle, and its mutation is responsible for Treacher Collins syndrome (Dai et al., 2016; Grzanka & Piekiełko-Witkowska, 2021). Mutations in RBM10 can lead to splicing changes that affect mouse palate development (Rodor et al., 2017). SPP2 is a highly phosphorylated and glycosylated sialoprotein and a prominent component of mineralized extracellular matrices in bone. Full-length SPP2 (24 kD) inhibits BMP-induced bone formation, while its degradation product (18.5 kD) (designated spp18.5) appears to be pro-osteogenic (Brochmann et al., 2009; Sintuu et al., 2008). We speculate that the biological function of SPP2 may be finely regulated by proteolysis, and this process may be altered in BMPR-IB-disrupted pigs. MMP13 is considered to play a crucial role in bone formation and remodeling and is expressed in both terminal hypertrophic chondrocytes in the growth plate and in osteoblasts (Li et al., 2017; Stickens et al., 2004). MMP13 is up-regulated at the onset of osteoarthritis (Li et al., 2017), and MMP13-deficient mice show abnormal development of the skeletal growth plate (Stickens et al., 2004). A research suggests that PHAX is associated with Pierre Robin syndrome, which is characterized by congenital micrognathia, glossoptosis and airway obstruction (Ansari et al., 2014). Here, GO analysis showed noticeable enrichment in biological pathways related to bone and embryonic development, including regulation of bone remodeling, positive regulation of bone resorption, positive regulation of bone remodeling, regulation of bone mineralization, osteoblast differentiation, and regulation of biomineral tissue development.
In summary, the BMPR-IB-disrupted piglets obtained via CRISPR-Cas9 and SCNT exhibited walking difficulties and severe developmental deformities in the forelimbs and hindlimbs. The BMPR-IB-/746G and BMPR-IB-/- piglets showed similar phenotypes. We identified 139 DEPs in the hindlimb fibula of BMPR-IB-/746G piglets with limb deformities, most of which are involved in skeletal and (or) embryonic development. Our study provides novel insights into BMPR-IB deficiency in a large mammal.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Conceptualization, Y.Y.X. and Q.Y.; formal analysis and experiment, Q.Y. and C.M.Q.; investigation, W.W.L. and H.Y.J.; methodology, Q.Q.J. and Y.Y.L.; resources, Y.Y.X. and J.R.; visualization, Q.Y. and C.M.Q; writing-original draft, Q.Y.; writing-review & editing, Y.Y.X. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
We would like to thank Prof. Lu-Sheng Huang at Jiangxi Agricultural University for kind suggestions during the experimental design process.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31560304) and National Key Research Programs of China (2016ZX08006-003)
References
- 1.Alanis-Lobato G, Zohren J, McCarthy A, Fogarty NME, Kubikova N, Hardman E, et al Frequent loss of heterozygosity in CRISPR-Cas9-edited early human embryos. Proceedings of the National Academy of Sciences of the United States of America. 2021;118(22):e2004832117. doi: 10.1073/pnas.2004832117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari M, Rainger JK, Murray JE, Hanson I, Firth HV, Mehendale F, et al. 2014. A syndromic form of Pierre Robin sequence is caused by 5q23 deletions encompassing FBN2 and PHAX. European Journal of Medical Genetics, 57(10): 587–595.
- 3.Arun RM, Lakkakula BVKS, Chitharanjan AB Role of myosin 1H gene polymorphisms in mandibular retrognathism. American Journal of Orthodontics and Dentofacial Orthopedics. 2016;149(5):699–704. doi: 10.1016/j.ajodo.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 4.Baur ST, Mai JJ, Dymecki SM Combinatorial signaling through BMP receptor IB and GDF5: shaping of the distal mouse limb and the genetics of distal limb diversity. Development. 2000;127(3):605–619. doi: 10.1242/dev.127.3.605. [DOI] [PubMed] [Google Scholar]
- 5.Brochmann EJ, Behnam K, Murray SS Bone morphogenetic protein-2 activity is regulated by secreted phosphoprotein-24 kd, an extracellular pseudoreceptor, the gene for which maps to a region of the human genome important for bone quality. Metabolism. 2009;58(5):644–650. doi: 10.1016/j.metabol.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Cabral WA, Barnes AM, Adeyemo A, Cushing K, Chitayat D, Porter FD, et al A founder mutation in LEPRE1 carried by 1.5% of West Africans and 0.4% of African Americans causes lethal recessive osteogenesis imperfect . Genetics in Medicine. 2012;14(5):543–551. doi: 10.1038/gim.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canaj H, Hussmann JA, Li H, Beckman KA, Goodrich L, Cho NH, et al. 2019. Deep profiling reveals substantial heterogeneity of integration outcomes in CRISPR knock-in experiments. BioRxiv,doi: 10.1101/841098.
- 8.Chen D, Ji X, Harris MA, Feng JQ, Karsenty G, Celeste AJ, et al Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. Journal of Cell Biology. 1998;142(1):295–305. doi: 10.1083/jcb.142.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu MX, Liu ZH, Jiao CL, He YQ, Fang L, Ye SC, et al Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries) . Journal of Animal Science. 2007;85(3):598–603. doi: 10.2527/jas.2006-324. [DOI] [PubMed] [Google Scholar]
- 10.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, et al Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunha A, Nelson-Filho P, Marañón-Vásquez GA, de Carvalho Ramos AG, Dantas B, Sebastiani AM, et al Genetic variants in ACTN3 and MYO1H are associated with sagittal and vertical craniofacial skeletal patterns . Archives of Oral Biology. 2019;97:85–90. doi: 10.1016/j.archoralbio.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 12.Dai JW, Si JW, Wang MJ, Huang L, Fang B, Shi J, et al Tcof1-related molecular networks in treacher collins syndrome. Journal of Craniofacial Surgery. 2016;27(6):1420–1426. doi: 10.1097/SCS.0000000000002719. [DOI] [PubMed] [Google Scholar]
- 13.Davis GH, Balakrishnan L, Ross IK, Wilson T, Galloway SM, Lumsden BM, et al Investigation of the Booroola (FecB) and Inverdale (FecXI) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries . Animal Reproduction Science. 2006;92(1-2):87–96. doi: 10.1016/j.anireprosci.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Demirhan O, Türkmen S, Schwabe GC, Soyupak S, Akgül E, Taştemir D, et al A homozygous BMPR1B mutation causes a new subtype of acromesomelic chondrodysplasia with genital anomalies . Journal of Medical Genetics. 2005;42(4):314–317. doi: 10.1136/jmg.2004.023564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dituri F, Cossu C, Mancarella S, Giannelli G The interactivity between TGFβ and BMP signaling in organogenesis, fibrosis, and cancer. Cells. 2019;8(10):1130. doi: 10.3390/cells8101130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Brolosy MA, Stainier DYR Genetic compensation: a phenomenon in search of mechanisms. PLoS Genetics. 2017;13(7):e1006780. doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong GC, Dai YP, Fan BL, Zhu HB, Wang HP, Wang LL, et al Production of transgenic blastocyst by nuclear transfer from different types of somatic cells in cattle. Science in China Series C:Life Sciences. 2004;47(2):183–189. doi: 10.1360/03yc0015. [DOI] [PubMed] [Google Scholar]
- 18.Grzanka M, Piekiełko-Witkowska A The role of TCOF1 gene in health and disease: beyond treacher Collins syndrome . International Journal of Molecular Sciences. 2021;22(5):2482. doi: 10.3390/ijms22052482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson CM, Shulman NJ, MacLean B, MacCoss MJ, Hoofnagle AN Skyline performs as well as vendor software in the quantitative analysis of serum 25-hydroxy vitamin D and vitamin D binding globulin. Clinical Chemistry. 2018;64(2):408–410. doi: 10.1373/clinchem.2017.282293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katagiri T, Watabe T Bone morphogenetic proteins. Cold Spring Harbor Perspectives in Biology. 2016;8(6):a021899. doi: 10.1101/cshperspect.a021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SH, Lee HK, Kim C, Kim YK, Ismail T, Jeong Y, et al The splicing factor SRSF1 modulates pattern formation by inhibiting transcription of tissue specific genes during embryogenesis. Biochemical and Biophysical Research Communications. 2016;477(4):1011–1016. doi: 10.1016/j.bbrc.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann K, Seemann P, Stricker S, Sammar M, Meyer B, Süring K, et al Mutations in bone morphogenetic protein receptor 1B cause brachydactyly type A2 . Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):12277–12282. doi: 10.1073/pnas.2133476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Wang D, Yuan YJ, Min J New insights on the MMP-13 regulatory network in the pathogenesis of early osteoarthritis. Arthritis Research & Therapy. 2017;19(1):248. doi: 10.1186/s13075-017-1454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin SX, Svoboda KKH, Feng JQ, Jiang XQ The biological function of type I receptors of bone morphogenetic protein in bone. Bone Research. 2016;4(1):16005. doi: 10.1038/boneres.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdavi M, Nanekarani S, Hosseini SD Mutation in BMPR-IB gene is associated with litter size in Iranian Kalehkoohi sheep . Animal Reproduction Science. 2014;147(3-4):93–98. doi: 10.1016/j.anireprosci.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Miyazono K, Kamiya Y, Morikawa M Bone morphogenetic protein receptors and signal transduction. Journal of Biochemistry. 2010;147(1):35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 27.Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, et al Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Mérino ewes. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5104–5109. doi: 10.1073/pnas.091577598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paznekas WA, Karczeski B, Vermeer S, Lowry RB, Delatycki M, Laurence F, et al GJA1 mutations, variants, and connexin 43 dysfunction as it relates to the oculodentodigital dysplasia phenotype . Human Mutation. 2009;30(5):724–733. doi: 10.1002/humu.20958. [DOI] [PubMed] [Google Scholar]
- 29.Rahman MS, Akhtar N, Jamil HM, Banik RS, Asaduzzaman SM TGF-β/BMP signaling and other molecular events: regulation of osteoblastogenesis and bone formation. Bone Research. 2015;3(1):15005. doi: 10.1038/boneres.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reader KL, Haydon LJ, Littlejohn RP, Juengel JL, McNatty KP Booroola BMPR1B mutation alters early follicular development and oocyte ultrastructure in sheep . Reproduction, Fertility and Development. 2012;24(2):353–361. doi: 10.1071/RD11095. [DOI] [PubMed] [Google Scholar]
- 31.Rodor J, FitzPatrick DR, Eyras E, Cáceres JF The RNA-binding landscape of RBM10 and its role in alternative splicing regulation in models of mouse early development. RNA Biology. 2017;14(1):45–57. doi: 10.1080/15476286.2016.1247148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy J, Polley S, De S, Mukherjee A, Batabyal S, Pan S, et al Polymorphism of fecundity genes (FecB, FecX, and FecG) in the Indian Bonpala sheep . Animal Biotechnology. 2011;22(3):151–162. doi: 10.1080/10495398.2011.589239. [DOI] [PubMed] [Google Scholar]
- 33.Sargiannidou I, Christophidou-Anastasiadou V, Hadjisavvas A, Tanteles GA, Kleopa KA Novel GJA1/Cx43 variant associated with oculo-dento-digital dysplasia syndrome: clinical phenotype and cellular mechanisms . Frontiers in Genetics. 2021;11:604806. doi: 10.3389/fgene.2020.604806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sintuu C, Murray SS, Behnam K, Simon R, Jawien J, Silva JDP, et al Full-length bovine spp24 [spp24 (24–203)] inhibits BMP-2 induced bone formation. Journal of Orthopaedic Research. 2008;26(6):753–758. doi: 10.1002/jor.20580. [DOI] [PubMed] [Google Scholar]
- 35.Skryabin BV, Kummerfeld DM, Gubar L, Seeger B, Kaiser H, Stegemann A, et al Pervasive head-to-tail insertions of DNA templates mask desired CRISPR-Cas9-mediated genome editing events. Science Advances. 2020;6(7):eaax2941. doi: 10.1126/sciadv.aax2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stickens D, Behonick DJ, Ortega N, Heyer B, Hartenstein B, Yu Y, et al Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131(11):5883–5895. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takagi M, Ishii T, Barnes AM, Weis MA, Amano N, Tanaka M, et al A novel mutation in LEPRE1 that eliminates only the KDEL ER-retrieval sequence causes non-lethal osteogenesis imperfect . PLoS One. 2012;7(5):e36809. doi: 10.1371/journal.pone.0036809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teo BHD, Bobryshev YV, The BK, Wong SH, Lu JH Complement C1q production by osteoclasts and its regulation of osteoclast development. Biochemical Journal. 2012;447(2):229–237. doi: 10.1042/BJ20120888. [DOI] [PubMed] [Google Scholar]
- 39.Vranka JA, Pokidysheva E, Hayashi L, Zientek K, Mizuno K, Ishikawa Y, et al Prolyl 3-hydroxylase 1 null mice display abnormalities in fibrillar collagen-rich tissues such as tendons, skin, and bones. Journal of Biological Chemistry. 2010;285(22):17253–17262. doi: 10.1074/jbc.M110.102228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson T, Wu XY, Juengel JL, Ross IK, Lumsden JM, Lord EA, et al Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biology of Reproduction. 2001;64(4):1225–1235. doi: 10.1095/biolreprod64.4.1225. [DOI] [PubMed] [Google Scholar]
- 41.Yang Q, Zhao XY, Xing YY, Jiang C, Jiang K, Xu P, et al A model of mucopolysaccharidosis type IIIB in pigs. Biology Open. 2018;7(10):bio035386. doi: 10.1242/bio.035386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi SE, Daluiski A, Pederson R, Rosen V, Lyons KM The type I BMP receptor BMPR-IB is required for chondrogenesis in the mouse limb. Development. 2000;127(3):621–630. doi: 10.1242/dev.127.3.621. [DOI] [PubMed] [Google Scholar]
- 43.Zhao M, Harris SE, Horn D, Geng ZP, Nishimura R, Mundy GR, et al Bone morphogenetic protein receptor signaling is necessary for normal murine postnatal bone formation. Journal of Cell Biology. 2002;157(6):1049–1060. doi: 10.1083/jcb.200109012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao XY, Yang Q, Zhao KW, Jiang C, Ren DR, Xu P, et al Production of transgenic pigs with an introduced missense mutation of the bone morphogenetic protein receptor type IB gene related to prolificacy. Asian-Australasian Journal of Animal Sciences. 2016;29(7):925–937. doi: 10.5713/ajas.15.0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou SW, Yu HH, Zhao XE, Cai B, Ding Q, Huang Y, et al Generation of gene-edited sheep with a defined Booroola fecundity gene (FecBB) mutation in bone morphogenetic protein receptor type 1B (BMPR1B) via clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) 9 . Reproduction, Fertility and Development. 2018;30(12):1616–1621. doi: 10.1071/RD18086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.