Abstract
A peroxidase (DyP) involved in the decolorization of dyes and produced by the fungus strain Geotrichum candidum Dec 1 was purified. DyP, a glycoprotein, is glycosylated with N-acetylglucosamine and mannose (17%) and has a molecular mass of 60 kDa and an isoelectric point (pI) of 3.8. The absorption spectrum of DyP exhibited a Soret band at 406 nm corresponding to a hemoprotein, and its Na2S2O4-reduced form revealed a peak at 556 nm that indicates the presence of a protoheme as its prosthetic group. Nine of the 21 types of dyes that were decolorized by Dec 1 cells were decolorized by DyP; in particular, anthraquinone dyes were highly decolorized. DyP also oxidized 2,6-dimethoxyphenol and guaiacol but not veratryl alcohol. The optimal temperature for DyP activity was 30°C, and DyP activity was stable even after incubation at 50°C for 11 h.
The discharge to the environment of 10 to 15% of the synthetic dyes produced (42) causes environmental problems. These dyes are poorly biodegradable because of their structures, and treatment of wastewater containing dyes usually involves physical and/or chemical methods. Although these treatment methods are efficient, they may result in the production of toxic by-products and/or require high levels of energy. Microbial decolorization has been proposed as a less expensive and less environmentally intrusive alternative. Various bacteria and fungi have decolorizing abilities, and an extensive review of microbiological decolorization has been made (3); in many cases adsorption of dyes to the microbial cell surface is the primary mechanism of decolorization (22).
Azo dyes may be microbially degraded under anaerobic (28, 52) or aerobic (8, 13, 30, 32, 42, 47) conditions or in aerobic and anaerobic two-stage systems (39). Enzymes, such as lignin peroxidase (LiP), manganese peroxidase (MnP), and laccase, all of which are involved in lignin degradation, participate in the decolorization of the dyes (6–8, 13, 30, 47). Recently, another such peroxidase was purified from Pleurotus ostreatus that was found to be different from MnP, LiP, and horseradish peroxidase (HRP) (17, 41). However, few studies have been made of the enzymatic degradation of anthraquinone dyes, which are xenobiotic chemicals similar to azo dyes but different in structure (11, 22, 31, 47).
Previously, we reported that Geotrichum candidum Dec 1, a newly isolated decolorizing fungus, decolorized 21 types of reactive dyes, including azo and anthraquinone dyes (18). The broad decolorization spectrum of this strain suggested the involvement of extracellular peroxidase-type enzymes. Our objectives in this study were to purify and characterize the novel peroxidase (DyP) that is responsible for the dye-decolorizing activity of G. candidum Dec 1.
MATERIALS AND METHODS
Organism and culture media.
Geotrichum candidum Dec 1 was isolated from soil (18–20). Cells of Dec 1 from a potato dextrose agar (PDA) (Eiken Chemical Co. Ltd., Tokyo, Japan) slant stored at 4°C were transferred to fresh PDA plates and incubated at 30°C for 6 days; all of the mycelia on the PDA plates were suspended in sterile distilled water. After being filtered through gauze to remove fungal mycelia, a spore suspension of about 107 CFU/ml was prepared. Potato dextrose broth (Difco) was used for liquid cultivation.
Enzymes and chemicals.
HRP (Wako Chemical Co. Ltd., Osaka, Japan) was used for comparison with the newly purified peroxidase involved in decolorization (DyP). The main dyes used were Reactive Blue 5 (RB5), an anthraquinone dye, and model compounds of RB5 (Nippon Kayaku Co., Ltd., Tokyo, Japan), i.e., 1,4-diamino-2-sodium anthraquinone sulfonate (AQ-2′), 1-amino-4-methylamino-2-sodium anthraquinone sulfonate (AQ-2), and 1-amino-4-(3-amino-4-sodium-sulfonoanilino)-2-sodium anthraquinone sulfonate (AQ-1). The other reagents used were of the highest quality available.
Enzyme purification.
One hundred fifty milliliters of potato dextrose broth in a 500-ml flask was inoculated with 5 ml of spore suspension and shaken at 30°C at 120 strokes per min for 6 days. Unless otherwise stated, all procedures were performed at 4°C. The supernatant (4.4 × 103 ml) obtained by centrifugation of the culture broth at 7,200 × g for 20 min was passed through a glass fiber filter (GC 50; Toyo Roshi Co. Ltd., Tokyo, Japan) to remove polysaccharides produced during cultivation. The filtrate was concentrated to 60 ml by ultrafiltration with a YM 10 membrane (Amicon Grace Japan, Tokyo, Japan). The concentrate was dialyzed with 25 mM piperazine buffer with a counterion of piperazine chloride (pH 5.5) to 80 ml and then concentrated to 17 ml by ultrafiltration with Centriprep 10 (Amicon Grace Japan). The pooled fraction (17 ml) was loaded onto a Super Q 650M (Tosoh Co. Ltd., Tokyo, Japan) column (2.8 by 6.0 cm) previously equilibrated with the same buffer (pH 5.5). The column was subsequently washed with 200 ml of the same buffer. The enzyme was eluted with a linear gradient of 0 to 0.4 M NaCl in 25 mM piperazine buffer with a counterion of piperazine chloride (pH 5.5) at a flow rate of 1 ml/min, and 1-ml fractions were collected. The fractions that exhibited enzyme activity were pooled and then concentrated to 2.8 ml with Centriprep 10. The 2.8-ml of concentrate was applied to a Butyl Toyopearl (Tosoh Co. Ltd.) column (1.6 by 6.5 cm) equilibrated with 25 mM citrate buffer (pH 5) containing 0.8 M (NH4)2SO4. After the column was washed with 50 ml of the same equilibration buffer, proteins were eluted with a linear gradient of 0.8 to 0 M (NH4)2SO4 in 25 mM citrate buffer (pH 5) at an elution rate of 1 ml/min, and 1-ml fractions were collected. Fractions corresponding to the main peak that exhibited enzyme activity were collected and divided into those corresponding to the left half of the peak and those corresponding to the right half of the peak. Each of the pooled proteins was dialyzed against 25 mM citrate buffer (pH 5) and concentrated to 2.8 ml with Centriprep 10. The dialyzed proteins were preserved at 4°C before being used in enzyme characterization.
Enzyme assay.
Twenty-one types of dyes that were used in a previous study (18) and model compounds of RB5, AQ-1, AQ-2, and AQ-2′ were used in the assay for purified enzyme activity.
We measured DyP activity in the supernatant of the culture broth by adding 1 ml of 25 mM citrate buffer (pH 3) to 2 ml of the supernatant to adjust its pH to 3.2 and then adding 119 μM RB5.
For the enzyme assay of the samples obtained from the ultrafiltration and Super Q purification steps, crude enzyme solution (100 to 300 ng/ml) was incubated in 3 ml of 25 mM citrate buffer (pH 3.2) containing 119 μM RB5. Purified peroxidase (1.9 nM) eluted by Butyl Toyopearl chromatography was mixed with 3 ml of 25 mM citrate buffer containing each dye or model compound of RB5. DyP activity was measured with a spectrophotometer (UV-240; Shimadzu, Kyoto, Japan) at the maximum absorption wavelength of each dye and model compound at optimal pH. Measurement of DyP activity was initiated by the addition of 0.2 to 0.4 mM H2O2 at 30°C except for the assay of optimal temperature for decolorization. One unit of enzyme activity was defined as the amount of enzyme required for the decolorization of 1 μmol of RB5 or AQ-2′ per min in the reaction mixtures. The value used was an average of three experiments, and the error was ±5%.
To assay the RB5-decolorizing activity of HRP, 1.7 nM HRP (with a molecular mass of 40 kDa) was used at an optimal pH of 4.0.
The 2,6-dimethoxyphenol oxidation activity of DyP was measured by the increase in absorbance at 470 nm (A470) of the reaction mixture containing 25 mM citrate buffer (pH 4.5), 2.8 nM purified DyP, 0.2 mM 2,6-dimethoxyphenol, and 0.2 mM H2O2. The oxidation of guaiacol was measured in the same manner as that of 2,6-dimethoxyphenol except for the addition of 1 mM guaiacol instead of 2,6-dimethoxyphenol and measurement at A465.
The RB5-decolorizing activity of purified DyP was measured at different temperatures to determine the optimal temperature. To compare the thermostabilities of DyP and HRP, DyP and HRP solutions in 25 mM citrate buffer were incubated at 40, 50, and 60°C and the activities of periodically sampled DyP and HRP were measured at 30°C.
Protein concentration.
Protein concentration was measured by the Bradford method (5) with bovine gamma globulin (Bio-Rad) as the standard.
Determination of molecular mass and isoelectric point.
The apparent molecular mass of purified DyP was estimated by gel filtration chromatography on a Sephacryl S-200 column (3.1 by 95 cm) eluted in 25 mM citrate buffer (pH 5). The standard proteins (Bio-Rad) used were thyroglobulin (670 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), gamma globulin (158 kDa), and vitamin B12 (1.35 kDa).
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (type AE-6440; ATTO Co. Ltd., Tokyo, Japan) was performed in a 10% polyacrylamide gel. Reduced α-2-macroglobulin (170 kDa), phosphorylase b (97.4 kDa), glutamate dehydrogenase (55.4 kDa), lactate dehydrogenase (36.5 kDa), and trypsin inhibitor (20.1 kDa) were used as standard-molecular-mass proteins for electrophoresis (Combithek; Boehringer Mannheim Yamanouchi Co. Ltd., Tokyo, Japan).
The isoelectric point (pI) of DyP was determined by isoelectric point electrophoresis (Multiphor II 2-D; Pharmacia) with a low-pI calibration kit (pH 2.5 to 6.5) (Pharmacia) as a standard pI marker.
Assay for hemoprotein.
Purified DyP (7 μM) in 25 mM citrate buffer (pH 5) was scanned at 700 to 300 nm to identify the Soret band, and then 25 μM H2O2 was added to oxidize DyP and to allow observation of the shift of the Soret band. A small amount of Na2S2O4 was added to the oxidized DyP to obtain the reduced form (44). The pyridine hemochrome content per mole of DyP was estimated by using the molar extinction coefficient of pyridine hemochrome (33 × 103 M−1 cm−1 at 556 nm), as described for cytochrome b2 (1).
Sugar analysis.
DyP (50 μg) was dried at 100°C and then hydrolyzed in 100 μl of 2.5 M trifluoroacetic acid at 100°C for 6 h. Then it was dried to remove the trifluoroacetic acid and coupled with 2-aminopyridine, a fluorescent compound, as described by Hase et al. (14). The reactant was adjusted to pH 9 by adding NH4OH, and a two-phase separation was conducted seven times with chloroform to remove excess 2-aminopyridine. The hydrolyzed sugars were eluted with 0.25 M citrate buffer containing 1% acetonitrile (pH 4) by high-pressure liquid chromatography with ODS-120T (Tosoh Co. Ltd.) at a rate of 1 ml/min as described previously (15, 44). Glucosamine (Glc), mannose (Man), fucose (Fuc), N-acetyl-mannosamine (ManNAc), N-acetylglucosamine (GlcNAc), and N-acetylgalactosamine (GalNAc) were used as standard sugars.
Comparison of the dye-decolorizing activity of DyP and HRP.
The RB5-decolorizing activities of DyP and HRP were measured at optimal pH values of 3.2 and 4.0, respectively. To obtain pseudo-first-order kinetic constants, the necessary concentrations of enzymes were used so that the decolorization rates of the dyes were not limited by substrate concentration. The concentrations of DyP and HRP were determined to be 1.3 and 1.7 nM, respectively. The concentration of RB5 was varied from 24 to 119 μM. The concentration of H2O2 varied from 10 to 20 μM to avoid inhibiting the enzyme activity. The Km(obs) of RB5 was estimated at 0.2 mM H2O2 from a plot of enzyme activity against RB5 concentration. Km(obs) of H2O2 was estimated at a fixed RB5 concentration of 119 μM from the relationship between enzyme activity and H2O2 concentration. Kcat(obs) of DyP was estimated from Vmax obtained from the reciprocal plot between DyP activity and RB5 concentration (36). Each value was obtained in triplicate, and the average value was used. The error was ±10%.
Second-order kinetics.
AQ-2′, a simplified form of RB5, was used as a substrate mainly because the by-products derived from RB5 degradation inhibited DyP activity, as described previously (18), and the mechanism of DyP activity can be clarified easily by using a simplified substrate. The concentration of DyP was fixed at 2.5 nM, and that of AQ-2′ varied from 60 to 150 μM. H2O2 was used in the range of 20 to 80 μM, because the inhibitory effect of H2O2 on DyP activity was observed at low concentrations when AQ-2′ was used. DyP activity for AQ-2′ was measured at the optimal pH of 3.2. Km(AQ-2′) was obtained from the Lineweaver-Burk plot (first plot) of AQ-2′ concentration against enzyme activity. Vmax, Km(H2O2), and Kcat (turnover rate) were determined from the double-reciprocal plot (second plot) of Vmax(app) against H2O2 concentration, as described previously (17, 36). The average value of triplicate data was employed, and the error was ±10%.
RESULTS
Enzyme purification.
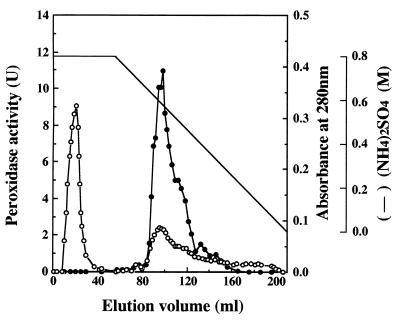
The total enzyme activity of the intact supernatant was 24 U, as shown in Table 1. The activity increased to 4.2 × 102 U after ultrafiltration with a YM 10 membrane because inhibitory substances with molecular masses of less than 10 kDa were removed (20). One peak that eluted at 0.13 M on the Super Q 650M column NaCl gradient possessed DyP activity. The elution pattern of the Butyl Toyopearl column, in which fractions that corresponded to the main peak exhibiting DyP activity were eluted at 0.57 M (NH4)2SO4, is shown in Fig. 1. Fractions 86 to 99, corresponding to the left half of the main peak, were pooled and called purified DyP. The other fractions (100 to 146), which also possessed enzyme activity, were pooled and called mixed DyPs, because they are presumed to be composed of several kinds of DyP isozymes.
TABLE 1.
Purification of peroxidase from supernatant of G. candidum Dec 1 culture
| Purification step | Protein (mg) | Total activity (U) | Sp act (U/mg) | Activity yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Supernatant | 88 | 24a | 0.28a | 1 | |
| Ultrafiltration (YM 10) | 28 | 4.2 × 102 | 15 | 100a | 55 |
| Super Q chromatography | 7.8 | 3.0 × 102 | 34 | 71 | 140 |
| Butyl Toyopearl chromatography | 1.5 | 85 | 57 | 20 | 210 |
Total and specific activities were underestimated, presumably because of the presence of inhibitors in the supernatant; thus, the activity yield was calculated based on the value after YM 10 treatment.
FIG. 1.
Butyl Toyopearl chromatography of a peroxidase produced by G. candidum Dec 1. Symbols: ●, peroxidase activity; ○, absorbance at 280 nm.
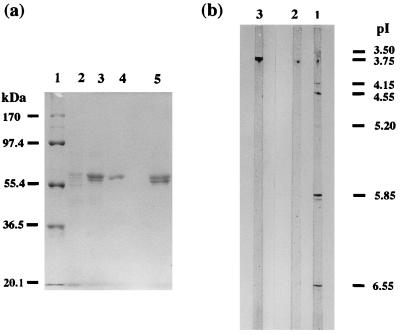
Purified DyP has a specific activity of 57 U/mg of protein and a recovery ratio of 20% (Table 1). The molecular mass of purified DyP as estimated by SDS-polyacrylamide gel electrophoresis was 60 kDa (Fig. 2a), revealing DyP to be a monomer, because the apparent molecular mass determined by gel filtration with Sephacryl S-200 was 55 kDa. The isoelectric point of DyP was 3.8 (Fig. 2b).
FIG. 2.
(a) SDS-polyacrylamide gel electrophoresis of DyP after each purification step. Lanes: 1, standard molecular mass markers; 2, sample after YM 10 ultrafiltration (amount of protein loaded, 10 μg); 3, sample after Super Q chromatography (amount of protein loaded, 10 μg); 4, purified DyP after Butyl Toyopearl chromatography (amount of protein loaded, 6 μg); 5, mixed DyPs after Butyl Toyopearl chromatography. (b) Isoelectric focusing of purified DyP and mixed DyPs obtained from Butyl Toyopearl chromatography. Lanes: 1, mixed DyPs after Butyl Toyopearl chromatography (amount of protein loaded, 5 μg); 2, purified DyP after Butyl Toyopearl chromatography (amount of protein loaded, 10 μg); 3, standard pI marker.
The mixed-DyP fraction consisted of mixed proteins whose molecular masses ranged from 55 to 60 kDa (Fig. 2a, lane 5) with a pI of 3.8 (Fig. 2b), and it had a specific activity of 57 U/mg of protein, which was about half the total activity (110 U) of DyP, corresponding to the main peak on Butyl Toyopearl chromatography.
Spectral characteristics.
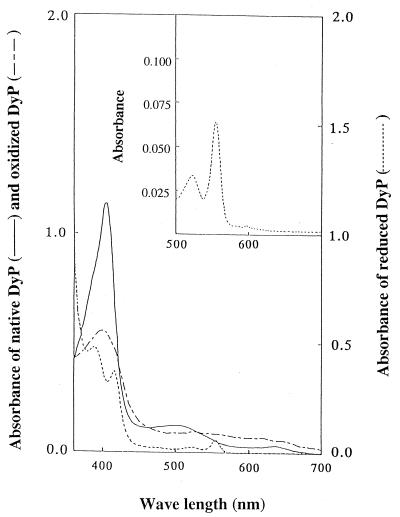
The spectral characteristics of purified DyP are shown in Fig. 3. A large Soret band was observed at 406 nm, together with two small peaks at 510 and 640 nm. The estimated molar extinction coefficient at 406 nm was 0.9 × 105 M−1 cm−1, similar to those of other peroxidases (25, 35, 40, 48). The A406/A280 (RZ) value, which reflects the purity and spectral characteristics of DyP, was 1.6 in 25 mM citrate buffer (pH 5). When H2O2 was added to the purified DyP, the peaks at 406 and 510 nm were shifted to the peak at 400 nm, with a molar extinction coefficient of 4 × 104 M−1 cm−1, and the peak at 530 nm, with a coefficient of 6.7 × 103 M−1 cm−1, respectively. When DyP that was oxidized by H2O2 was reduced by Na2S2O4, a peak at 556 nm, which corresponded to a heme-pyridine complex, appeared. From these results we concluded that DyP has a protoheme as its prosthetic group. The heme content per mole of DyP was estimated as 0.6, indicating that DyP retained a single heme.
FIG. 3.
Spectral characteristics of purified DyP, DyP oxidized by H2O2, and DyP reduced by dithionite. The inset shows the peak at 556 nm indicative of reduced DyP in the form of a heme-pyridine complex.
Substrate specificity.
Purified DyP was used to decolorize the 21 dyes (Table 2) which were decolorized by Dec 1 (18). DyP decolorized seven dyes containing azo and anthraquinone groups and three model compounds of RB5. Higher decolorizing activity was observed for anthraquinone dyes than for azo dyes. Phenolic compounds 2,6-dimethoxyphenol and guaiacol, which are substrates of MnP, were degraded by DyP, but veratryl alcohol, a well-known substrate of LiP, was not (data not shown). The oxidation of 2,6-dimethoxyphenol and guaiacol occurred without the addition of Mn2+ at optimal pH values of 4.5 and 4.0, respectively, and no enhancement of the DyP activity by the addition of Mn2+ was observed. These results confirmed that DyP had a wide degradation spectrum and that its substrate specificity differs from those of LiP and MnP.
TABLE 2.
Enzymatic activity of purified DyP on various dyes
| Color index of dyes | Chromophore | ɛ at λmaxa (liters/mM/cm) | Optimal pH | Initial concn (μM) | Decolorizing rate (μM/min) |
|---|---|---|---|---|---|
| RB5 | AQb | 7.3 | 3.2 | 119 | 24 |
| RB19 | AQ | 10 | 3.2 | 116 | 22 |
| RB114 | AQ | 8.4 | 4.0 | 151 | 12 |
| AQ-1 | AQ | 9.0 | 3.2 | 113 | 10 |
| AQ-2 | AQ | 8.3 | 3.0 | 141 | 55 |
| AQ-2′d | AQ | 7.3 | 3.2 | 148 | 20 |
| Reactive Black 5 | AZc | 37 | 3.2 | 33 | 1.1 × 10−1 |
| Reactive Red 33 | AZ | 23 | 3.2 | 60 | 4.7 × 10−1 |
| Reactive Yellow 2 | AZ | 8.1 | 3.2 | 125 | 6.2 × 10−1 |
| RB182 | AZ | 7.3 | 4.0 | 123 | 21 |
Molar extinction coefficient at maximum absorption wavelength (λmax) of each dye.
AQ, anthraquinone.
AZ, azo.
Decolorization test of AQ-2′ by Dec 1 was not conducted in the previous study (18).
Sugar analysis.
Fifty micrograms of DyP contained 8.7 μg of sugar, which was composed of GlcNAc (38 mol per mol of DyP) and Man (26 mol per mol of DyP). This result indicates that DyP is a glycoprotein containing 17% (wt/wt) sugar.
Optimal temperature for and thermostability of DyP activity.
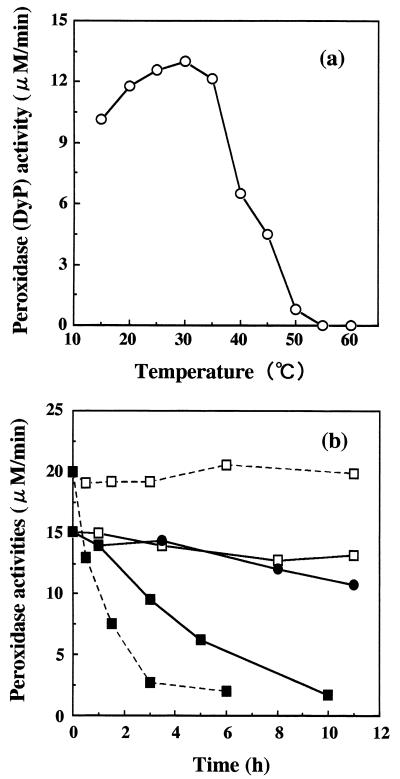
The effect of temperature on DyP activity is shown in Fig. 4. DyP activity was optimal at 30°C, and relatively high activity was maintained in the range of 15 to 35°C (Fig. 4a). The thermostabilities of DyP and HRP activities were measured at 30°C after treatment at various temperatures (Fig. 4b). DyP activity was restored at 30°C even after treatment at 40 and 50°C for 11 h. When DyP and HRP were heated at 60°C for 3 h, 35 and 90% of their initial activities respectively, were lost. These results suggest that DyP is more thermostable than HRP. Additionally, when DyP was incubated at 30 and 40°C for 14 days, the inactivation rates were 37 and 59%, respectively.
FIG. 4.
(a) Optimal decolorization temperature of DyP. (b) Thermostability of DyP and HRP; enzyme activity at 30°C after treatment with DyP at 40°C ( —— ), DyP at 50°C (●——●), DyP at 60°C (▪——▪), HRP at 40°C ( - - - ), and HRP at 60°C (▪- - -▪).
H2O2 inhibition of DyP activity.
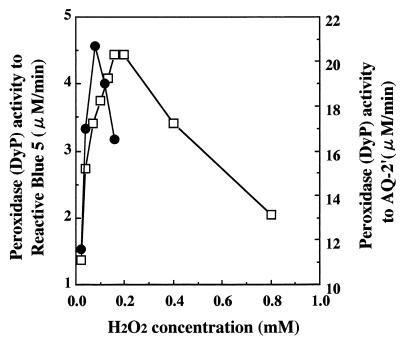
When RB5 and AQ-2′ were used as substrates, the inhibitory effect of H2O2 on DyP activity was observed (Fig. 5). DyP activity for RB5 was inhibited when the H2O2 concentration exceeded 0.2 mM at a fixed DyP concentration of 0.6 nM. At 2.8 nM of DyP, its activity for AQ-2′ decreased sharply when the H2O2 concentration exceeded 0.1 mM, and it was lower than that for RB5. The degree of inhibition by H2O2 of DyP activity differed significantly depending on the substrate used.
FIG. 5.
H2O2 inhibition of DyP activity when RB5 and its simplified model compound, AQ-2′, were used as substrates. Symbols: □, degradation activity of 0.6 nM DyP for RB5; ●, degradation activity of 2.8 nM DyP for AQ-2′.
Comparison of decolorizing activities of DyP and HRP.
Pseudo-first-order kinetics was applied to compare the rates of decolorization of RB5 by DyP and HRP (Table 3). DyP had slightly lower Km(RB5) and Km(H2O2) values than HRP, and the Kcat(obs) of 260 s−1 for DyP was 1.8 times higher than that for HRP (140 s−1). When RB5 solution was scanned after decolorization by both enzymes, the spectral patterns were considerably different at wavelengths below 450 nm, reflecting the difference in the by-products produced from RB5 by the two peroxidases.
TABLE 3.
Kinetic parameters for activities of DyP and HRPa
| Parameter | Unit(s) | Value
|
|
|---|---|---|---|
| DyP | HRP | ||
| Km(RB5) | μM | 54 | 58 |
| Km(H2O2) | μM | 26 | 36 |
| Kcat(obs) | s−1 | 260 | 140 |
| Kcat/Km(RB5) | M−1 s−1 | 4.8 × 106 | 2.4 × 106 |
| Kcat/Km(H2O2) | M−1 s−1 | 1.0 × 107 | 3.8 × 106 |
RB5 decolorization by DyP and HRP was determined by pseudo-first-order kinetics.
Second-order kinetics of DyP activity.
In the DyP turnover involving the two substrates AQ-2′ and H2O2, the apparent Km(AQ-2′) was initially obtained from the Lineweaver-Burk plot of AQ-2′ concentration and DyP activity. Then the Vmax, Km(H2O2), and apparent Kcat were determined from the reciprocal plot of Vmax(app) and H2O2 concentration. The estimated apparent kinetic constants are shown in Table 4. Kcat/Km(AQ-2′) and Kcat/Km(H2O2), which are physicochemical constants of the substrates, were 3.2 × 106 and 7.6 × 106 M−1 s−1, respectively. The apparent Kcat was similar to the Kcat(obs) when RB5 was used as the substrate, indicating a similar turnover rate of a more simply structured substrate, AQ-2′, by DyP.
TABLE 4.
Second-order kinetic constants of DyP with H2O2 and AQ-2′ as substrates
| Parameter | Unit(s) | Value |
|---|---|---|
| Km(AQ-2′) | μM | 84 |
| Km(H2O2) | μM | 36 |
| Kcat | s−1 | 270 |
| Kcat/Km(AQ-2′) | M−1 s−1 | 3.2 × 106 |
| Kcat/Km(H2O2) | M−1 s−1 | 7.6 × 106 |
DISCUSSION
A peroxidase that transformed anilines was purified from the culture broth of a strain of G. candidum (4). DyP was produced under aerobic conditions as a secondary metabolite in the stationary phase and reached its maximum level at day 6 (18). Since DyP activity is maintained as long as sugars exist in basal III mineral medium (24), a medium frequently used in LiP production, DyP of strain Dec 1 is considered to be a constitutive enzyme. LiP of Phanerochaete chrysosporium was produced under limited-nitrogen conditions (8, 9, 42). It required inducers, such as veratryl alcohol and veratryl acid, for production, and oxygen-enriched aeration (9) and low shear stress (21, 24) were essential to obtain higher activity. However, the production of peroxidase by Dec 1 is more efficient and convenient than that by P. chrysosporium, because DyP is constitutively produced during shaking cultivation (18) or in a stirred tank reactor (19) without a marked decrease in activity.
Electrophoretic analysis of DyP gave a pI of 3.8. Although molecular mass values obtained by SDS-polyacrylamide gel electrophoresis and by gel filtration differed slightly, i.e., 60 and 55 kDa, respectively, DyP is considered to be a monomer. The difference in molecular mass may be due to the gel filtration value being underestimated, presumably because DyP is not spherical or the sugar portion of DyP is interacting with the gel matrix. The mass of DyP was considerably larger than those reported previously (10, 27, 45, 48, 51), which ranged from 40 to 44 kDa. Kang et al. (17) reported a 140-kDa peroxidase with two subunits of 72 kDa. In addition to DyP, Dec 1 produced mixed DyPs that were eluted by Butyl Toyopearl chromatography and had the same pI as purified DyP. Since there are small differences in molecular mass among them, mixed DyPs are assumed to consist of proteins with amino acid sequences identical to that of purified DyP but with different sugar contents.
DyP degraded 7 of the 18 dyes that were decolorized by Dec 1, indicating that other enzymes contributed to the broad decolorizing spectrum of Dec 1. DyP showed a higher decolorizing rate for anthraquinone dyes than for azo dyes (Table 2). However, high decolorizing activity was obtained for the azo dye RB182. This is presumably because this dye contains Cu in its structure. DyP degraded phenolic compounds, such as 2,6-dimethoxyphenol and guaiacol, as well as a variety of dyes, while it did not degrade nonphenolic veratryl alcohol. Considering its substrate specificity and molecular mass, purified DyP is thought to be a novel decolorizing peroxidase, distinct from LiP, MnP, HRP, and other peroxidases reported previously. The enzymes that were involved in lignin degradation also degraded various aromatic compounds, lignin model compounds (33), and synthesized dyes (6–8, 11, 13, 22, 30, 43, 47). The wide degradation spectra may depend on the involvement of an active oxygen species and/or radicals (hydroxyl, etc.) in the degradation (2, 33). The broad degradation spectrum of DyP could also be due to the presence of an active oxygen and/or radicals initially produced by DyP.
The spectral characteristics of DyP are similar to those of typical peroxidases. The Soret band, which is the representative absorption peak of peroxidase (16, 34, 48), was observed at 406 nm for native DyP. When H2O2 was added to native DyP, the Soret band shifted to 400 nm, presumably due to the formation of compound I of DyP, which resembles LiP L3 from Phlebia radiata (25). Since the DyP reduced by dithionite gave a new peak at 556 nm, assigned to a pyridine hemochrome, we concluded that DyP had a protoheme as its prosthetic group, similar to HRP and LiP (40).
H2O2 inhibition of DyP activity was observed (Fig. 5). The inhibition of HRP or LiP activity by excess H2O2 is well known (26, 49). In the turnover cycles of these peroxidases, they are first oxidized by two electrons from H2O2 to compound I. Then, one electron is removed by a substrate, changing compound I to compound II. Compound II is further reduced to a resting enzyme by another electron from a substrate. However, in the presence of excess H2O2, compound II that cannot be converted to a resting enzyme is changed to compound III, an inactivated state, which decreases peroxidase activity (26, 46, 49). In the case of DyP, conversion to compound I (Fig. 3) was suggested by shifts of the peak at 406 nm to 410 nm and of the peak at 510 nm to 530 nm, which were similar to those in previous reports (25, 37). Details of H2O2 inhibition of DyP activity were not elucidated, but a mechanism similar to that described above may be involved. Furthermore, the extent of inhibition is dependent on the substrate; the inhibition was observed at a lower H2O2 concentration for the simplified substrate AQ-2′ even if the DyP concentration was higher than the RB5 concentration.
Sugar analysis revealed that DyP contained 17% (wt/wt) sugar (GlcNAc and Man) in its structure. The GlcNAc and Man contents per mole of DyP were 34 and 23 mol, respectively. HRP is composed of 16 to 18% sugar, although the sugar content differs depending on the isozyme (35, 40, 51). The sugar content of LiP varied widely, from 17% at a pI of 3.8 to 39% at a pI of 4.2 (12). The nature of the sugar linkage in DyP is not yet clear, but LiP retained both N-linked sugar chains and O-linked monosaccharides (38), while HRP containing 18% sugar had only asparagine N-linked types at eight sites per mole (51). The N-terminal amino acid of DyP was not detected by the Edman procedure, indicating that it may be blocked by sugars (17, 41). Cloning of dyp is necessary to clarify the structure of DyP.
DyP activity was maximal at 30°C, and it was maintained at a high level at temperatures ranging from 20 to 35°C (Fig. 4a). However, according to the present experimental procedure, residual DyP activity was stable even after incubation at 50°C for 11 h and DyP revealed higher thermostability than HRP, in particular, at 60°C (Fig. 4b). This is presumably due to its larger molecular mass with a sugar attached and its high thermal reversibility, because a partial recovery or a reversible thermal inactivation of HRP and peroxidase-M2 activities was also observed (29, 34, 48).
The decolorizing activities of DyP and HRP for RB5 were compared by analysis of pseudo-first-order kinetics (Table 3). Although the Km values for RB5 were almost the same for the two peroxidases, the Km of DyP for H2O2 was lower than that of HRP, indicating a higher affinity of DyP to H2O2. Kcat(obs), which represents DyP activity, was 1.8 times that of HRP. High decolorizing activity of DyP for a synthetic dye may have resulted from its higher redox potential or its affinity for a substrate. By second-order kinetic analysis of DyP activity with AQ-2′ and H2O2, the apparent kinetic parameters, Km(AQ-2′), Km(H2O2), and Kcat, were estimated (Table 4). Since the values of Kcat of LiP for veratryl alcohol and of MnP for Mn(II) were reported to range from 30 to 160 s−1 (12, 27, 30), a higher turnover rate of DyP on dyes is suggested by the results of this experiment. The apparent Kcat of AQ-2′ was similar to that of RB5. This suggests the usefulness of using AQ-2′ to elucidate the mechanism of degradation of a dye by DyP and to compare it with those of the other peroxidases (17, 23, 25, 50), because AQ-2′ is a simplified structure of RB5.
In our previous paper, we showed that G. candidum Dec 1 decolorized 18 kinds of dyes and three model compounds. Purified DyP showed high activity for anthraquinone dyes and decolorized seven dyes. Mixed DyPs containing several isozymes decolorized 15 kinds of dyes (data not shown). However, purification of each isozyme is sometimes difficult due to the structural similarities. Therefore, we plan to clone the DyP gene (dyp) and use this DNA sequence to isolate genes which encode isozymes. We expect these isozymes to be sufficient to explain the broad decolorization spectrum of this strain.
ACKNOWLEDGMENTS
We are grateful to Bayer Japan, Ltd., and Nippon Kayaku Co., Ltd., for providing the dye samples. We also thank M. Hirai, Y. Hirohashi, and A. Ferdous of the Tokyo Institute of Technology for valuable suggestions and technical assistance.
REFERENCES
- 1.Appleby C A, Morton R K. Lactate dehydrogenase and cytochrome b2of baker’s yeast. Biochem J. 1959;73:539–550. doi: 10.1042/bj0730539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backa S, Gierer J, Reitberger T, Nilsson T. Hydroxyl radical activity associated with the growth of white-rot fungi. Holzforschung. 1993;47:181–187. [Google Scholar]
- 3.Banat I M, Nigam P, Singh D, Marchant R. Microbial decolorization of textile-dye-containing effluents. A review. Biores Technol. 1996;58:217–227. [Google Scholar]
- 4.Bordeleau L M, Bartha R. Biochemical transformations of herbicide-derived anilines: purification and characterization of causative enzymes. Can J Microbiol. 1972;18:1865–1871. doi: 10.1139/m72-291. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Chivukula M, Spadaro J T, Renganathan V. Lignin peroxidase-catalyzed oxidation of sulfonated azo dyes generates novel sulfophenyl hydroperoxides. Biochemistry. 1995;34:7765–7772. doi: 10.1021/bi00023a024. [DOI] [PubMed] [Google Scholar]
- 7.Chivukula M, Renganathan V. Phenolic azo dye oxidation by laccase from Pyricularia oryzae. Appl Environ Microbiol. 1995;61:4374–4377. doi: 10.1128/aem.61.12.4374-4377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cripps C, Bumpus J A, Aust S D. Biodegradation of azo and heterocyclic dyes by Phanerochaete chrysosporium. Appl Environ Microbiol. 1990;56:1114–1118. doi: 10.1128/aem.56.4.1114-1118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faison B D, Kirk T K. Factors involved in the regulation of a ligninase activity in Phanerochaete chrysosporium. Appl Environ Microbiol. 1985;49:299–304. doi: 10.1128/aem.49.2.299-304.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiechter A. Function and synthesis of enzymes involved in lignin degradation. J Biotechnol. 1993;30:49–55. [Google Scholar]
- 11.Glenn J K, Gold M H. Decolorization of several polymeric dyes by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1983;45:1741–1747. doi: 10.1128/aem.45.6.1741-1747.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glumoff T, Harvey P J, Molinari S, Goble M, Frank G, Palmer J M, Smit J D G, Leisola M S A. Lignin peroxidase from Phanerochaete chrysosporium. Eur J Biochem. 1990;187:515–520. doi: 10.1111/j.1432-1033.1990.tb15333.x. [DOI] [PubMed] [Google Scholar]
- 13.Goszczynski S, Paszczynski A, Pasti-Grigsby M B, Crawford R L, Crawford D L. New pathway for degradation of sulfonated azo dyes by microbial peroxidases of Phanerochaete chrysosporium and Streptomyces chromofuscus. J Bacteriol. 1994;176:1339–1347. doi: 10.1128/jb.176.5.1339-1347.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hase S, Hara S, Matsushima Y. Tagging of sugars with a fluorescent compound, 2-aminopyridine. J Biochem. 1979;85:217–220. doi: 10.1093/oxfordjournals.jbchem.a132314. [DOI] [PubMed] [Google Scholar]
- 15.Iwase H, Ishii-Karakasa K I, Urata T, Saito T, Hotta K. Extraction method for preparing pyridylamino sugar derivatives and application to porcine gastric mucus glycoprotein analysis. Anal Biochem. 1990;188:200–202. doi: 10.1016/0003-2697(90)90552-k. [DOI] [PubMed] [Google Scholar]
- 16.Johansson T, Nyman P O. Isozymes of lignin peroxidase and manganese (II) peroxidase from the white-rot basidiomycete Trametes versicolor. Arch Biochem Biophys. 1993;300:49–56. doi: 10.1006/abbi.1993.1007. [DOI] [PubMed] [Google Scholar]
- 17.Kang S-O, Shin K-S, Han Y-H, Youn H-D, Hah Y C. Purification and characterization of an extracellular peroxidase from white-rot fungus Pleurotus ostreatus. Biochim Biophys Acta. 1993;1163:158–164. doi: 10.1016/0167-4838(93)90177-s. [DOI] [PubMed] [Google Scholar]
- 18.Kim S J, Ishikawa K, Hirai M, Shoda M. Characteristics of a newly isolated fungus, Geotrichum candidumDec 1, which decolorizes various dyes. J Ferment Bioeng. 1995;79:601–607. [Google Scholar]
- 19.Kim S J, Shoda M. Decolorization of molasses by a new isolate of Geotrichum candidumin a jar fermentor. Biotechnol Tech. 1998;12:497–499. [Google Scholar]
- 20.Kim S J, Shoda M. Decolorization of molasses and a dye by a newly isolated strain of the fungus Geotrichum candidumDec 1. Biotechnol Bioeng. 1999;62:114–119. doi: 10.1002/(sici)1097-0290(19990105)62:1<114::aid-bit13>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 21.Kirk T K, Schultz E, Connors W J, Lorenz L F, Zeikus J G. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch Microbiol. 1978;117:277–285. [Google Scholar]
- 22.Knapp J S, Newby P S, Reece L P. Decolorization of dyes by wood-rotting basidiomycete fungi. Enzyme Microb Technol. 1995;17:664–668. [Google Scholar]
- 23.Kuan I-C, Johnson K A, Tien M. Kinetic analysis of manganese peroxidase. J Biol Chem. 1993;268:20064–20070. [PubMed] [Google Scholar]
- 24.Linko S. Production of Phanerochaete chrysosporiumlignin peroxidase. Biotechnol Adv. 1992;10:191–236. doi: 10.1016/0734-9750(92)90003-r. [DOI] [PubMed] [Google Scholar]
- 25.Lundell T, Wever R, Floris R, Harvey P, Hatakka A, Brunow G, Schoemaker H. Lignin peroxidase L3 from Phlebia radiata; pre-steady-state and steady-state studies with veratryl alcohol and a non-phenolic lignin model compound 1-(3,4-dimethoxyphenyl)-2-(2-methoxyphenoxy)propane-1,3-diol. Eur J Biochem. 1993;211:391–402. doi: 10.1111/j.1432-1033.1993.tb17562.x. [DOI] [PubMed] [Google Scholar]
- 26.Maehly A C. Plant peroxidase. In: Colowick S P, Kaplan N O, editors. Methods in enzymology. II. New York, N.Y: Academic Press, Inc.; 1955. pp. 801–803. [Google Scholar]
- 27.Matsubara M, Suzuki J, Deguchi T, Miura M, Kitaoka Y. Characterization of manganese peroxidases from the hyperlignolytic fungus IZU-154. Appl Environ Microbiol. 1996;62:4066–4072. doi: 10.1128/aem.62.11.4066-4072.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer U. Biodegradation of synthetic organic colorants. FEMS Symp. 1981;12:371–387. [Google Scholar]
- 29.Nie G, Aust S D. Effect of calcium on the reversible thermal inactivation of lignin peroxidase. Arch Biochem Biophys. 1997;337:225–231. doi: 10.1006/abbi.1996.9770. [DOI] [PubMed] [Google Scholar]
- 30.Ollikka P, Alhonmaki K, Leppanen V-M, Glumoff T, Raijola T, Suominen I. Decolorization of azo, triphenyl methane, heterocyclic, and polymeric dyes by lignin peroxidase isozymes from Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:4010–4016. doi: 10.1128/aem.59.12.4010-4016.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasti M B, Crawford D L. Relationships between the abilities of streptomycetes to decolorize three anthron-type dyes and to degrade lignocellulose. Can J Microbiol. 1991;37:902–907. [Google Scholar]
- 32.Paszczynski A, Pasti M B, Goszczynski S, Crawford D L, Crawford R L. New approach to improve degradation of recalcitrant azo dyes by Streptomyces spp. and Phanerochaete chrysosporium. Enzyme Microb Technol. 1991;13:378–384. [Google Scholar]
- 33.Paszczynski A, Crawford R L. Potential for bioremediation of xenobiotic compounds by the white-rot fungus Phanerochaete chrysosporium. Biotechnol Prog. 1995;11:368–379. [Google Scholar]
- 34.Paszczynski A, Huynh V-B, Crawford R. Comparison of ligninase-1 and peroxidase-M2 from the white-rot fungus Phanerochaete chrysosporium. Arch Biochem Biophys. 1986;244:750–765. doi: 10.1016/0003-9861(86)90644-2. [DOI] [PubMed] [Google Scholar]
- 35.Paul K-G, Stigbrand T. Four isoperoxidases from horse radish root. Acta Chem Scand. 1970;24:3607–3617. [Google Scholar]
- 36.Philips A T. Enzymatic activity. In: Gerhardt P, editor. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 568–569. [Google Scholar]
- 37.Renganathan V, Gold M H. Spectral characterization of the oxidized states of lignin peroxidase, an extracellular heme enzyme from the white rot basidiomycete Phanerochaete chrysosporium. Biochemistry. 1986;25:1626–1631. [Google Scholar]
- 38.Schmidt B, Heimgartner U, Kozulic B, Leisola M S A. Lignin peroxidases are oligomannose type glycoproteins. J Biotechnol. 1990;13:223–228. [Google Scholar]
- 39.Seshadri S, Bishop P L, Agha A M. Anaerobic/aerobic treatment of selected azo dyes in wastewater. Waste Manag. 1994;14:127–137. [Google Scholar]
- 40.Shannon L M, Kay E, Lew J Y. Peroxidase isozymes from horseradish roots. J Biol Chem. 1966;241:2166–2172. [PubMed] [Google Scholar]
- 41.Shin K-S, Oh I-K, Kim C-J. Production and purification of Remazol brilliant blue R decolorizing peroxidase from the culture filtrate of Pleurotus ostreatus. Appl Environ Microbiol. 1997;63:1744–1748. doi: 10.1128/aem.63.5.1744-1748.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spadaro J T, Gold M H, Renganathan V. Degradation of azo dyes by the lignin-degrading fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58:2397–2401. doi: 10.1128/aem.58.8.2397-2401.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spadaro J T, Renganathan V. Peroxidase-catalyzed oxidation of azo dyes: mechanism of Disperse Yellow 3 degradation. Arch Biochem Biophys. 1994;312:301–307. doi: 10.1006/abbi.1994.1313. [DOI] [PubMed] [Google Scholar]
- 44.Takemoto H, Hase S, Ikenaka T. Microquantitative analysis of neutral and amino sugars as fluorescent pyridylamino derivatives by high-performance liquid chromatography. Anal Biochem. 1985;145:245–250. doi: 10.1016/0003-2697(85)90357-4. [DOI] [PubMed] [Google Scholar]
- 45.Tien M, Kirk T K. Lignin-degrading enzyme from Phanerochaete chrysosporium: purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc Natl Acad Sci USA. 1984;81:2280–2284. doi: 10.1073/pnas.81.8.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valli K, Wariishi H, Gold M H. Oxidation of monomethoxylated aromatic compounds by lignin peroxidase: role of veratryl alcohol in lignin biodegradation. Biochemistry. 1990;29:8535–8539. doi: 10.1021/bi00489a005. [DOI] [PubMed] [Google Scholar]
- 47.Vyas B R M, Molitoris H P. Involvement of an extracellular H2O2-dependent ligninolytic activity of the white rot fungus Pleurotus ostreatusin the decolorization of Remazol Brilliant Blue R. Appl Environ Microbiol. 1995;61:3919–3927. doi: 10.1128/aem.61.11.3919-3927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S S. Isolation and characterization of the native, thermally inactivated and regenerated horseradish peroxidase isozymes. J Food Sci. 1972;37:574–578. [Google Scholar]
- 49.Wariishi H, Gold M H. Lignin peroxidase compound III: formation, inactivation, and conversion to the native enzyme. FEBS Lett. 1989;243:165–168. [Google Scholar]
- 50.Wariishi H, Valli K, Gold M H. Manganese (II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. J Biol Chem. 1992;267:23688–23695. [PubMed] [Google Scholar]
- 51.Welinder K G. Covalent structure of the glycoprotein horseradish peroxidase. FEBS Lett. 1976;72:19–23. doi: 10.1016/0014-5793(76)80804-6. [DOI] [PubMed] [Google Scholar]
- 52.Zimmermann T, Kulla H G, Leisinger T. Properties of purified Orange II azoreductase, the enzyme initiating azo dye degradation by PseudomonasKF46. Eur J Biochem. 1982;129:197–203. doi: 10.1111/j.1432-1033.1982.tb07040.x. [DOI] [PubMed] [Google Scholar]