Abstract
Monitoring adverse drug events or pharmacovigilance has been promoted by the World Health Organization to assure the safety of medicines through a timely and reliable information exchange regarding drug safety issues. We aim to discuss the application of machine learning methods as well as causal inference paradigms in pharmacovigilance. We first reviewed data sources for pharmacovigilance. Then, we examined traditional causal inference paradigms, their applications in pharmacovigilance, and how machine learning methods and causal inference paradigms were integrated to enhance the performance of traditional causal inference paradigms. Finally, we summarized issues with currently mainstream correlation-based machine learning models and how the machine learning community has tried to address these issues by incorporating causal inference paradigms. Our literature search revealed that most existing data sources and tasks for pharmacovigilance were not designed for causal inference. Additionally, pharmacovigilance was lagging in adopting machine learning-causal inference integrated models. We highlight several currently trending directions or gaps to integrate causal inference with machine learning in pharmacovigilance research. Finally, our literature search revealed that the adoption of causal paradigms can mitigate known issues with machine learning models. We foresee that the pharmacovigilance domain can benefit from the progress in the machine learning field.
Key Points
| Most existing data sources and tasks for pharmacovigilance were not designed for causal inference. |
| Pharmacovigilance was lagging in adopting machine learning-causal inference integrated models. |
| Adoption of causal paradigms can mitigate known issues with machine learning models, which could further enhance the use of machine learning in pharmacovigilance tasks. |
Introduction
The World Health Organization has been promoting pharmacovigilance programs to assure the safety of medicines through a timely and reliable information exchange regarding drug safety issues, for example, adverse drug events (ADEs) [1]. An ADE is an unintended response caused by a medicine and is harmful [2]. For in-patient stays, 16.9% of the patients experienced ADEs with 6.7% categorized as serious and 0.3% as fatal [2, 3]. While medication errors (e.g., wrong/missing doses, wrong administration techniques, equipment failure) and prescription of multiple medications were considered important risk factors of ADEs [4, 5], there are still many incidences of ADEs due to undetected signals during clinical trials [3]. This may be due to limited sample sizes and stringent patient eligibility criteria in pre-approval studies [3]. Therefore, pharmacovigilance is important to the safe use of medicines. In this review, we focus on the tasks of ADE detection and monitoring (including pre-clinical prediction) in the pharmacovigilance program lifecycle because those tasks were mostly likely to be achieved with machine learning and causal inference. While we have narrowed down our scope to focus on the tasks of ADE detection and monitoring in the pharmacovigilance program lifecycle, the methodologies and examples of causal inference discussed in this paper could apply to each phase of the pharmacovigilance program.
Currently, major data sources for pharmacovigilance include spontaneous reporting systems (SRS), real-world data (RWD) such as electronic health records (EHRs), social media, biomedical literature, and knowledge bases [3]. Each data source has unique advantages and biases, which we discuss in the following sections. While data mining was applied to those data sources to enhance the efficiency of pharmacovigilance, the level of evidence from identified signals depended heavily on the chosen data source as well as the study design. Overall, we identified the following three main tasks in the field of pharmacovigilance.
Drug–event pair extraction. For this task, we usually use either structured data from EHRs [6, 7] or the natural language processing (NLP)-based machine learning/deep learning (ML/DL) method to extract drug–event co-occurrence pairs from the unstructured texts [8–10]. Note that those pairs only indicate a potential associative “relationship” between the drug and the event and cannot be considered a “confirmed” ADE yet. The symptoms experienced might be caused by a variety of medical conditions other than the ADE. Thus, we still need further proof using other statistical analyses or data sources.
Adverse drug event detection. For traditional pharmacovigilance, the most important task is to detect ADEs for these post-marketing drugs in time. The ADE detection task aims to identify and confirm ADEs from “real-world” medication usage information as early as possible. We consider ADE detection as a task providing a higher level associative relationship compared with disproportionality or NLP-based drug–event co-occurrence pair extraction. However, ADE detection is only associative without further confirmation if using SRS owing to the limitation of the data source (no control group can be matched, and no causality evaluation can be performed). Adverse drug event detection using an RWD database, however, can be evaluated for causality if a proper study design was adopted.
Adverse drug event prediction. Adverse drug event prediction, or ADE discovery, could be conducted only if the event data have accumulated to a certain amount. Thus, there was a time difference from drug launch to ADE prediction. Adverse drug event prediction focuses on discovering potential ADEs before being observed. The predictive power (forecast future events from data generated previously) of many ML/DL models made ADE prediction possible. Using literature and knowledge bases, researchers can predict ADEs at the pre-marketing stage. After launching and as more data accumulate, researchers can use RWD and social media data for post-marketing pharmacovigilance. While ADE prediction may not only depend on causal relationships, establishing causal relationships can facilitate feature selection and greatly improve model performance and generalizability.
Machine learning or a causal inference paradigm separately has been adopted for many pharmacovigilance studies [11–15]. The integration of machine learning into a causal inference paradigm was also studied, although mostly theoretically [16–20]. However, the relationship between machine learning and a causal inference paradigm in the context of pharmacovigilance has not been extensively examined. The goal of causal inference is to explain what factors lead (are influential) to the outcome. The emphasis is on investigating and explaining the role of individual factors in the outcome. On the contrary, most machine learning tasks emphasize the outcome and aim to predict whether an outcome will occur in the future. Weights in machine learning models are not equivalent to effect sizes in causal inference [21]. Pharmacovigilance involves a series of tasks: (1) predicting the outcome using drug exposure and a set of covariates and (2) understanding the causal effects between drug exposure and the outcome. The complicated nature of pharmacovigilance requires researchers to choose methods and study designs wisely in order to answer the proposed question (prediction or explanation). However, ideally, machine learning and causal inference could be combined to enhance both the predictive and explanatory power of a single study. Therefore, this article aims to discuss the application of machine learning and a causal inference paradigm in pharmacovigilance. Pharmacovigilance tasks, machine learning, and causal inference paradigms have intertwined relationships (Fig. 1). In the following sections, we discuss (1) data sources for pharmacovigilance, common methods (traditional or machine learning) used to analyze data from each data source, and the advantages and biases of each data source; the search query for this section was as follows: data source name (e.g., spontaneous reporting system, SRS, EHRs, data registry) + “machine learning” + “adverse event/adverse effect/side effect”. (2) Integration of machine learning into traditional causal inference paradigms (with examples of studies in the pharmaceutical industry); the search query for this section was: as follows: causal inference paradigm name (e.g., naranjo score, propensity score matching, instrumental variable) + “adverse event/adverse effect/side effect” + “machine learning/artificial intelligence” (optional). (3) Issues with machine learning and how a causal paradigm can address those issues; search query for this section was: “machine learning/artificial intelligence” + “generalizability/generalizable/explainability/explainable/fairness/bias” + “adverse event/adverse effect/side effect” (optional). Because of the length limit of the paper, we were not able to include all papers identified from the above queries. However, we selected the most recent papers representative of the data source/methods/combination of methods to reveal current trends of machine learning in causal inference with an application in pharmacovigilance.
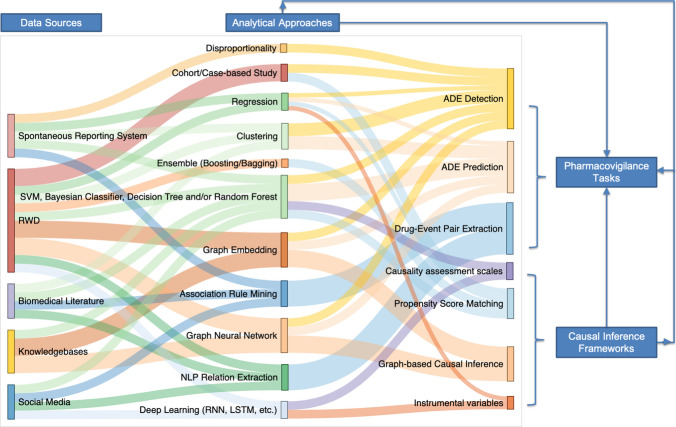
Fig. 1.
Relationships between pharmacovigilance data sources, analytical approaches, pharmacovigilance tasks, and causal inference paradigms. Each data source is commonly analyzed by specific analytical approaches depending on the characteristics of data in those data sources. Each pharmacovigilance task is also associated with specific analytical approaches. Causal inference paradigms are integrated with different analytical approaches and applied to pharmacovigilance tasks. ADE adverse drug event, LSTM long short-term memory, NLP natural language processing, RNN recurrent neural network, RWD real-world data, SVM support vector machine
Data Sources for Pharmacovigilance
Spontaneous Reporting System
The most traditional dataset for ADE detection is the SRS database, such as the FDA Adverse Event Reporting System (FAERS) [22] and WHO’s VigiBase [23]. Traditionally, statistically based methods such as disproportionality measures and multivariate analyses were used to analyze SRS data [24]. Recently, machine learning methods such as association rule mining [25, 26], clustering [11], graph mining [12], and the neural network [27] were also applied to SRS data. However, those methods were only able to detect ‘signals of suspected causality’ [27, 28]. Moreover, several studies have revealed limitations of the SRS, including reporting bias (e.g., underreporting, stimulated reporting), the lack of a population denominator, poor documentation quality [28, 29], and lower reporting rates for older products [30–32]. Important details required for a causality assessment may not be captured by the SRS, for example, comorbidities and concomitant medications. This can lead to background ‘noise’ or may generate false-positive signals [33]. Therefore, the causality of the detected signals still needs further validation from other data sources [34].
Real-World Data
Real-world data containing both structured and unstructured data, for example, insurance claims, EHRs, and registry databases offer new opportunities for pharmacovigilance as they provide a longer duration of follow-up, better ascertainment of exposure and outcomes, and a more complete collection of confounding variables such as comorbidities and co-prescribed medications [35]. We could also identify comparison groups in RWD databases using matching techniques. However, the timeliness of the RWD collection has been an issue with a claim or registry database [30]. Electronic health records were considered a better choice in terms of data timeliness. However, data quality issues such as non-random missingness and discrepancies across databases also made rapid utilization of RWD from EHRs difficult [30]. Despite the limitations, RWD databases enabled a transition from traditional “passive” surveillance toward “active” surveillance, and thus received considerable attention in the field of pharmacovigilance. Notably, RWD was superior as they offers longitudinal data for each subject. Therefore, increasing numbers of studies explored temporal relation extraction [36] using RWD to increase the confidence level of detected signals.
There has been a progression in the better utilization of RWD for observational studies in pharmacovigilance including: (1) development of common data models [37] such as the Observational Medical Outcomes Partnership [38–40] to facilitate rapid data extraction from unstructured RWD; (2) traditional epidemiologic methods (or slightly modified variants) adapted for signal detection, including a self-controlled case series study [41], a self-controlled cohort analysis [42], a tree-based scan statistic [6, 7], and a prescription symmetry analysis [43]; and (3) new ML/DL and approaches applied to a temporal analysis [36] and relational learning [44]. Patient event-level or code-level embedding was also calculated for downstream predictive modeling using RWD [45].
Social Media and Biomedical Literature
Social media such as social networks, health forums, question-and-answer websites, and other types of online health information-sharing communities is another resource containing potentially useful and most timely information for pharmacovigilance. Biomedical literature, including research articles, case reports, and drug labels, was considered a more reliable source of unstructured data for pharmacovigilance compared with social media data. Association rule mining was commonly used for extraction of drug–event pair or drug–drug interaction from social media and literature [46–48]. Advancement in NLP has enabled relation extraction of drug–event pairs from the above-mentioned unstructured data sources for pharmacovigilance [49–53]. Advanced machine learning such as supervised learning was also applied to extract ADEs from social media and biomedical literature. For example, Patki et al. [54] used supervised machine learning algorithms to classify sentences into two classes: one with ADE mentions and another without, before inference of the experienced ADE. Several shared tasks based on social media and biomedical text data have significantly accelerated development for ADE detection using these two data sources, for example, Drug–Drug Interaction Extraction 2011 challenge task [55] and Social Media Mining for Health (SMM4H) shared task [56].
Knowledge Bases
With the development of ML/DL techniques, particularly on graph mining, knowledge bases have become a rising data source for pharmacovigilance study, especially for the pre-marketing phase. Drug chemical databases [57], drug target databases [58] (including a side effects database [59]), biomedical pathway databases [60], protein interaction databases [61], and drug interaction databases [62] were some of the most used knowledge bases in pharmacovigilance studies. Logistic regression, Naive Bayes, k-Nearest Neighbor, Decision Tree, Random Forest, and Support Vector Machine were commonly used algorithms for the prediction of unknown ADEs using knowledge bases. The algorithms were always compared with each other given a specific dataset before the best-performing algorithm was selected [57, 58]. Recent advancement in Graph Neural Network (GNN) has led to an increasing interest in using knowledge bases for ADE prediction as GNN has achieved superior performance compared with other machine learning algorithms. In more recent works, the graph structures of knowledge bases were integrated with RWD to enhance the causal interpretability of ADE detection [63].
Each of these data sources has its own advantage/bias and is suitable for different pharmacovigilance tasks at different phases (pre-marketing or post-marketing). We summarized this information in Table 1. Even though we discussed each of the data sources separately in Table 1, we observed that the trend in pharmacovigilance is to employ more than one type of data source [64–68]. We also observed a trend to combine multiple analytical approaches, for example, [44] combined sequence analysis with supervised learning, [69] used NLP to extract features from free text, which were later used in supervised learning, and [70] proposed a novel synthesis of unsupervised pretraining, representational composition, and supervised machine learning to extract relational information from the biomedical literature. Both data source integration and analytical approach synthesis will facilitate the design of a generalizable and causally explainable ML/DL framework.
Table 1.
Data sources for pharmacovigilance, analytical approaches, advantages, and biases
| Analytical approaches | Pharmacovigilance tasks | Advantages and biases |
|---|---|---|
| Spontaneous reporting system | ||
| Association rule mining [25, 26] |
Drug–event pair extraction [11] |
Advantages: 1. Large volume of data worldwide. Create potentials for machine learning models to be trained 2. Provide other related information such as demographic and indication data 3. More effective at detecting rare ADEs 4. Publicly accessible Biases: 1. No population denominator who takes the medications. Could not calculate incidence rates of ADEs. Limited ability to provide causal evaluation 2. Suffer from under-reporting and stimulated reporting. May cause bias in machine learning 3. Lower reporting rates for older products 4. May have duplicate reports 5. Reporters have diverse background, such as pharmaceuticals companies, physicians, patients, and lawyers, which may pose challenges in data standardization. May undermine machine learning model transportability 6. It will take a long time for data collection, thus there may be a delay in detection of ADEs |
| Disproportionality [27, 28, 31, 32] | ||
| Network analysis [12] | ||
| Clustering [11] | ||
| SVM, Bayesian classifier, decision tree and/or Random Forest [71, 72] | ||
| RWD (EHRs and registries) | ||
| Disproportionality [6, 7] |
Drug–event pair extraction [6, 7, 73] |
Advantages: 1. Provides a population denominator who has taken the same medications, which enables adoption of study designs for causal effect estimation 2. The data quality in well-curated RWD databases is better than SRS 3. Less duplicated and missing data in well-curated RWD databases 4. Less adverse event unreported rate 5. RWD databases could provide more complete clinical information such as lab test results. Provide better causal inference ability compared with SRS Biases: 1. Less sample size than SRS. May diminish predictive power of machine learning models 2. EHRs contain protected health information of the patients. Thus, it could not be opened to the public, also difficult to share between institutions 3. EHRs mainly record drug usage information in the hospital. Thus, EHRs work better in inpatient ADE detection than outpatient. May diminish generalizability of machine learning models |
| Cohort/case-based study [41, 42, 74] | ||
| Sequence/temporal analysis [36, 43, 44] | ||
| SVM, Bayesian classifier, decision tree and/or Random Forest [75, 76] | ||
| NLP relation extraction [10, 73] | ||
| Neural network [63] | ||
| Social media | ||
| Association rule mining [46] |
Drug–event pair extraction [46, 56, 69, 77, 78] ADE detection [54] |
Advantages: 1. Huge data size with rapid growth. Create potentials for machine learning models to be trained 2. Open access 3. The content is patient centric 4. Could conduct a “real-time” ADE monitor Biases: 1. The contents are not from experts, thus it may affect data quality and reliability 2. Using NLP to extract all the ADE-related data from texts is challenging. NLP techniques are essential before applying to any machine learning task or causal inference paradigm 3. Could not calculate ADE incidence rate. Limited ability to provide causal evaluation 4. Still need to be further confirmed by other evidence or analysis 5. Ethical issues may exist |
| SVM, Bayesian classifier, decision tree and/or Random Forest [54, 56, 69, 77] | ||
| NLP relation extraction [69] | ||
| Neural network [56, 78] | ||
| Biomedical literature | ||
| Clustering [70] |
Drug–event pair extraction [49, 50] ADE detection [70] |
Advantages: 1. Data quality and reliability are better 2. Literature is easily accessible. Biases: 1. Data size is smaller than social media. May diminish predictive power of machine learning models 2. Timeliness is worse because of the peer-review and publishing process 3. Detected ADEs still need to be further confirmed by other evidence or analysis |
| SVM, Bayesian classifier, decision tree and/or Random Forest [70] | ||
| NLP relation extraction [8, 65] | ||
| Neural network [49, 50] | ||
| Knowledge bases | ||
| SVM, Bayesian classifier, decision tree and/or Random Forest [57–59, 79] | ADE prediction (pre-marketing) [57–59, 66, 79] |
Advantages: 1. Most of the databases are open to the public 2. Better data structure and data standardization level. Create potentials for machine learning models to be trained Biases: 1. Need for a complicated paradigm to integrate and analyze the data 2. The graph structures in knowledge bases lack causal components, making causal interpretation difficult 3. Many false-positive results may impact the prediction accuracy 4. ADE prediction results are based on theoretical algorithms, which needs other RWD or evidence to confirm |
| Neural network [66] | ||
ADE adverse drug event, EHR electronic health record, NLP natural language processing, RWD real-world data, SRS spontaneous reporting system, SVM support vector machine
Traditional Causal Inference Paradigm and Integration with Machine Learning
Most pharmacovigilance studies are observational studies because of the nature of the data used for analysis. However, observational studies have only limited ability to prove causality, i.e., probabilities under conditions (adverse events) that are changed and induced by treatments or external interventions [80]. Conducting causal inference for observational studies required either randomization or a rigorous study design [81, 82]. In most cases of long-term pharmacovigilance, randomized trials are not feasible. Therefore, observational studies became a more favorable approach for this task. However, there are many challenges in both the design and analysis stages to draw causal conclusions from retrospective observational studies. The primary challenge is to distinguish between causal and associative relationships with observational data in the presence of confounders (i.e., factors related to both the exposure and the outcome) and colliders (i.e., factors influenced by both the exposure and the outcome). While a multivariable regression analysis was often used to adjust for potential confounders, causal effects cannot be directly estimated. Furthermore, temporal relationships are to be captured and assessed in observational studies before causal relationships can be established [83, 84]. Hill’s criteria (i.e., 1. Strength, 2. Consistency, 3. Specificity, 4. Temporality, 5. Biological gradient, 6. Plausibility, 7. Coherence, 8. Experiment, 9. Analogy between exposures and outcome) are often referenced as the standard definition for causality in epidemiology [85]. It has guided the development of many causal inference models, statistical tests, as well as machine learning tasks for the evaluation of causality.
In this section, we discuss four causal inference paradigms in the domain of pharmacovigilance: (1) causality assessment scales, (2) propensity score matching (PSM), (3) graph-based causal inference, and (4) instrumental variables (IVs). Our discussion focuses on how ML/DL was integrated into the traditional causal inference methods. We also discuss current progress in pharmacovigilance that has adopted causal inference-machine learning integration. Table 2 shows the relevant papers we reviewed.
Table 2.
Categorization of papers reviewed regarding data sources and machine learning methods used for four causal inference paradigms
| Machine learning methods | Data source | |
|---|---|---|
| Causality assessment scales | ||
| SVM, Bayesian classifier, decision tree and/or Random Forest [86–88] |
Social media [88] |
|
| Regression [89] | ||
| Neural network [90, 91] | ||
| Propensity score matching | ||
| SVM, Bayesian classifier, decision tree and/or Random Forest [92, 93] | ||
| Ensemble (boosting/bagging) [20, 92, 94, 95, 99] | ||
| Regression [20, 94, 95, 97] | ||
| Neural network [96, 98, 100] | ||
| Graph-based causal inference | ||
| SVM, Bayesian classifier, decision tree and/or Random Forest | ||
| Link prediction [101] | ||
| Recommendation systems [109] | ||
| Classification [110] | ||
| Graph embedding | ||
| Link prediction [13, 102] | ||
| Recommendation Systems [105] | ||
| Regression | ||
| Classification [103] | ||
| Neural network | ||
| Link prediction [104] | ||
| Recommendation systems [106] | ||
| Predictive modeling [107, 108] | ||
| Ensemble (boosting/bagging) | ||
| Classification [111] | ||
| Link prediction [112] | ||
| Instrumental variables | ||
| Clustering [113] |
Simulated data [16, 114, 115, 117–119] Social network data [115] |
|
| Decision tree [16] | ||
| Neural network [114–117] | ||
| New algorithms [118, 119] | ||
RWD real-world data, SRS spontaneous reporting system, SVM support vector machine
Papers for “propensity score matching” and “instrumental variables” are not applied in the field of pharmacovigilance. Papers for “graph-based causal inference” still lacks a clear causal interpretation from a graph perspective
Causality Assessment Scales
Various methods are available to assess the causal relationship between a drug and an ADE, which are based on three main approaches: (1) expert judgment-based World Health Organization-Uppsala Monitoring Centre system; (2) algorithm-based Naranjo causality assessment method; and (3) probabilistic-based Bayesian Adverse Reactions Diagnostic Instrument (BARDI) [120]. The World Health Organization-Uppsala Monitoring Centre system is relatively easy to implement, it is highly dependent on an individual expert’s judgment, thus suffering from poor reproducibility. The Naranjo algorithm is also simple and has good reproducibility. Its disadvantages include low sensitivity for the ‘uncertain’ cases and therefore a low detection rate for certain ADEs. It is also not valid for children, critically ill patients, drug toxicities, and drug–drug interaction (DDI) detection. The Bayesian approach is regarded as the most reliable approach, its complex and time-consuming nature limits its use in clinical routine practice [120].
We found that the relationships between machine learning and causality assessment scales are three-fold: (1) causality assessment scales serve as outcome labels in machine learning models that predict causality of extracted drug–ADE pairs. For example, in studies [86–88], researchers have utilized the World Health Organization-Uppsala Monitoring Centre to create gold-standard labels of causal drug–ADE pairs, which were later used for training supervised machine learning models to perform causal classifications on the identified drug–ADE pairs. Likewise, Rawat et al. [90] constructed a multi-task joint model using unstructured text in EHRs, using physicians’ annotation as the gold standard. These efforts demonstrated that machine learning algorithms have some ability to predict the value of a report from SRS or content from social media for causal inference. (2) Causality assessment scales serve as features in machine learning models that predict causality. A group of researchers from Roche developed a model called MONARCSi with nine features capturing important criteria from Naranjo’s scoring system, Hill’s criteria, and internal Roche safety practices [89]. Their model achieved a moderate sensitivity and high specificity with high positive and negative predictive values. However, this approach cannot be fully automated, restricting its potential for future application. Thus, automated tools for extracting features capturing important criteria from Naranjo’s scoring system or Hill’s criteria are desirable. (3) Machine learning methods were employed to extract Naranjo score features and improve the efficiency of causality assessment score calculations. As discussed above, the inability to automate the extraction of Naranjo score features restricted the adoption of the proposed decision support system by Roche. Recent work by Rawat et al. [90, 91] offered solutions to this limitation. In [90], they formulated Naranjo questions as an end-to-end question-answer task. They used Bidirectional Long short-term memory (BiLSTM) to predict the scores for a subset of Naranjo questions. Later in [91], they used Bidirectional Encoder Representations from Transformers (BERT) to extract relevant paragraphs for each Naranjo question and then used a logistic regression model to predict the Naranjo score for each drug–ADE pair. To sum up, with the availability of more data sources and the advancement of deep learning-based NLP methods for analyzing unstructured text, future researchers can better utilize unstructured data for causality assessment score calculations.
Propensity Score Matching
Matching has been widely used in observational or cohort studies for drug safety investigation [14, 121–125] through subsampling of the dataset strategically to balance the confounder distribution in the treatment and control groups so that both groups share a similar probability of receiving treatment [126]. It allows observational studies to be designed similar to randomized designs with the outcome being independent of confounders [127]. Matching methods have evolved from “exact” matching to matching on propensity scores and to algorithmic matching, where machine learning algorithms were used for the matching process [92]. Regardless of the types of matching, this approach is often used during data preprocessing or cohort construction. Matching involves two steps: (1) definition of a similarity metric (e.g., propensity score) and (2) matching controls to treatment groups based on the defined metric [128]. While some most recent algorithmic matching techniques such as Dynamic Almost-Exact Matching with Replacement (D-AEMR) [19] and DeepMatch [129] did not necessarily use a propensity score as a similarity metric, matching using a propensity score was still the most widely adopted method in observational studies. Therefore, we focus our discussion on PSM in the following paragraphs.
Propensity score matching enabled the estimation of the causal effect of treatments. However, the definition of similarity and selection of covariates before matching may sometimes hinder the causal inference power of matching [130]. In other words, it could be hard to account for all possible confounders and an inappropriate assumption of similarity is likely to undermine the matched analysis. Machine learning has inspired new methods for propensity score estimation that are hypothesis-free and thus enhance the causal inference ability of PSM. Traditional PSM mainly used logistic regression for propensity score estimation. A more recent study showed promising performance improvement by using tree-based algorithms such as Classification and Regression Trees (CART) and bagging algorithms such as Random Forest for propensity score estimation [92]. Contrary to statistical models that fit models with assumptions and estimations of parameters from the data, machine learning models tend to learn the relationship between features and outcomes without an a priori model, i.e., hypothesis-free [131]. Additionally, machine learning models were also useful in addressing the “curse of dimensionality” when the number of covariates becomes too large, which has become very common in the era of “big data” [132]. For example, Zhu et al. were able to control the number of covariates and thus balance the trade-off between bias and variance of a propensity score estimator by tuning the number of optimal trees using a tree-based boosting algorithm [20].
Integration of PSM and machine learning techniques has been found frequently in observational studies [94–96, 100], including but not limited to treatment effect estimation and outcome evaluation [93, 97–99], which all showed promising performance improvement compared with traditional PSM. Theoretical developments of PSM and a machine learning combination are also booming through the development and use of simulated datasets [133–136]. However, the application of such a combination has not yet been utilized/discussed in the domain of pharmacovigilance. Propensity score matching is important for pharmacovigilance studies [14, 137]. As more data or covariates become available for pharmacovigilance, the combination of PSM and machine learning can handle large covariate sets and reduce bias and variance compared with traditional PSM. Therefore, we foresee that machine learning-integrated PSM will empower future studies in pharmacovigilance.
Graph-Based Causal Inference
The graph is a common data structure that consists of a finite set of vertices (concepts) and a set of edges that represents relationships (semantic or associative) between the vertices. Graph-based methods are mainstream in both exploratory machine learning and causal inference paradigms. Graph-based methods also offer theoretical and systematic representations of causality that do not require an a priori model [138–140]. They can be applied to analyze integrated data from various databases, e.g., knowledge bases, molecular (multi-omics) databases, and RWD databases for causal signal detection.
In pharmacovigilance, because of the complex nature of relationships between drugs, diseases (indication, comorbidities, or adverse event), and individual characteristics (e.g., demographic, multi-omics), graph-based ML/DL methods demonstrate their strengths in modeling these complicated topologies. Graph-based methods can be applied in two separate phases of pharmacovigilance: pre-marketing and post-marketing. The rationale behind pre-marketing ADE prediction is to identify potential ADEs from a biological mechanism perspective: chemical structure, DDIs, and protein–protein interactions (PPIs). Traditionally, researchers utilized chemical structures [13, 57] or biological phenotypes [58, 103, 141] from graph knowledge bases to predict potential adverse effects of a drug candidate. More recently, Zhang et al. predicted potential adverse effects of a drug candidate using a knowledge graph embedding generated from Drugbank [142]. Dey et al. [102] developed an attention-based deep learning method to predict adverse drug effects from chemical structures using SIDER. The hidden attention scores were utilized to interpret and prioritize the associative relationships between the presence of drug substructures and ADEs. Zitnik et al. [104] applied graph convolutional neural networks to predict potential side effects induced by PPI networks [61] and DDI networks [60, 62]. Researchers have also constructed knowledge graphs through literature mining [101]. Most of the papers using graph-based methods were for pre-marketing ADE prediction because knowledge bases regarding biomarkers, drug targets, disease indications, and adverse effects are readily available.
As more clinical or observational databases become available, researchers have transited from using a single data source, for example, knowledge bases, towards combining RWD in their analysis. For example, Kwak et al. [63] predicted ADE signals via GNNs from a graph constructed combining a knowledge base and EHR data. There were several recent studies proposed to use graphs generated from both knowledge bases and EHRs for safe medication recommendations [105, 106, 109]. In [106], graph embeddings were combined with a memory network recommender system. In [105], drug–ADE pairs were identified through a link prediction task. In [109], an encoder-decoder attention-based model was proposed for sequential decision making on drug selection in a multi-morbidity polypharmacy situation. Additionally, the characteristics of RWD enabled researchers to incorporate the temporality and sparsity of the features into signal detection models [110, 111].
Machine learning/deep learning frameworks demonstrated improved performance in structure learning compared with the baseline greedy search scoring strategy [18, 143, 144] for the identification of causal graph structures with the highest score or probability. In the meantime, causal inference methods were introduced to graph-based ML/DL models to improve the explainability and generalizability of those ML/DL models. For instance, Narendra et al. adopted counterfactual reasoning for causal structural learning [145]. Lin et al. utilized a loss function based on Granger causality to provide generative causal explanations for GNN models [17]. Rebane et al. evaluated the temporal relevancy of medical events to interpret medical code-level feature importance [107]. In a more recent paper, Rebane et al. incorporated the SHAP (SHapley Additive exPlanations) framework to provide more clinically appropriate global explanations in addition to medical code-level explanations captured by attention mechanisms [108].
While the advancement of ML/DL has enabled a plethora of graph-based data mining studies in pharmacovigilance, causality interpretation was still not explicitly discussed in any of those papers. We cannot naively equate link prediction to causal inference. This is not to say that existing knowledge bases are not causal graphs, thus existing links may only be associative and have a different level of confidence in terms of causality. Among all those papers reviewed, only [17] had a clear causality evaluation. We resort to the lack of causality interpretation to the shortage of a graph-based benchmark dataset with causal components in the domain of pharmacovigilance. Currently, most studies used SIDER [102, 103] or datasets integrating multiple knowledge bases as the benchmark. In [112] for example, the author used Pauwels’s dataset [57], Mizutani’s dataset [146], and Liu’s dataset [58] as the benchmark datasets. The benchmark datasets currently prevailing lacked a causal component, for example, a level of confidence for relationships. We believe a benchmark dataset with causal components and/or with integrated information from multiple sources could significantly benefit the development of causally explainable graph mining models.
Instrumental Variables
Estimation of causal relationships through an IV can adjust for both observed and unobserved confounders. This is a big advantage over methods such as stratification, matching, and multiple regression methods, which only allow adjustment for observed confounding variables. An IV is an additional variable, Z, that is used in a regression analysis to evaluate the causal effect of an independent variable X on a dependent variable Y (Fig. 2). The assumption of Z to be a valid IV is that (1) Z is correlated with the regressor X, (2) Z is uncorrelated with the error term U, and (3) Z is not a direct cause of outcome variable Y. Therefore, Z only influences Y through its effect on X. However, IV-based methods also suffer from criticism. First, different instruments will identify different subgroups and thus obtain different numerical treatment effects. Another criticism is that one cannot rule out “mild” violations of assumptions. Finally, an IV is consistent but not unbiased.
Fig. 2.
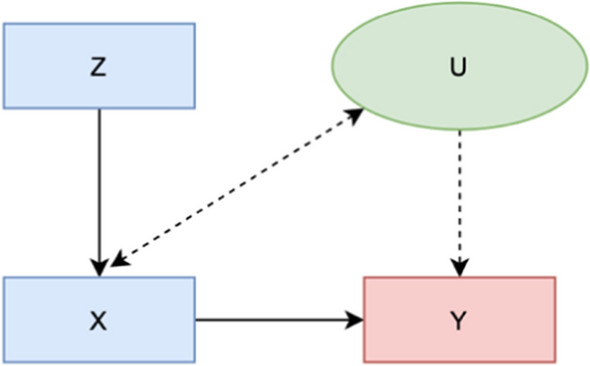
Graph representations of relationships between X, Y, Z, and U under instrumental variable assumptions
Several pharmacovigilance studies used an IV to investigate the adverse impact of certain medications. For example, Brookhart et al. [15] used physician preference of a cyclooxygenase-2 inhibitor over non-selective non-steroidal anti-inflammatory drugs as the IV to assess the adverse effect of cyclooxygenase-2 inhibitor use on gastrointestinal complications. Ramirez et al. [147] investigated the adverse effect of rosiglitazone on cardiovascular hospitalization and all-cause mortality using the facility proportion of patients taking rosiglitazone as the IV. The study found an increased risk for all-cause and cardiovascular mortality among patients taking rosiglitazone vs those who were not. Groenwold et al. [148] studied the effect of the influenza vaccine on mortality as reported in many observational studies. The study evaluated the usefulness of five IVs including a history of gout, a history of orthopedic morbidity, a history of antacid medication use, and general practitioner-specific vaccination rates in assessing the effect of influenza vaccination on mortality adverse events. They found that these IVs did not meet the necessary criteria because of their association with the outcome. In the field of causal inference for pharmacovigilance, IV-based methods have been overshadowed by PSM and graph-based methods because of the difficulty of finding a valid and unbiased IV that can serve as a randomization factor.
Recently, a few studies have explored using machine learning to improve the efficiency and fairness of IV learning from observational data. Hartford et al. [114] proposed the DeepIV framework, an approach that trained deep neural networks by leveraging IVs to minimize the counterfactual prediction error. DeepIV had two prediction tasks: first, it performed treatment prediction. In the second stage, DeepIV calculated its loss by integrating over the conditional treatment distribution. The author claimed that DeepIV estimated the causal effects by adopting the adapted loss function, which helped to minimize the counterfactual prediction errors. The proposed framework was also able to replicate the previous IV experiment with minimal feature engineering. Singh et al. [119] proposed a general framework called MLIV (machine-learned IVs) that allowed IV learning through any machine learning method and causal inference using IVs to be performed simultaneously. They showed that their method significantly improved causal inference performance through experiments from both simulation and real-world datasets. McCulloch et al. [16] proposed another framework for modeling the effects of IVs and other explanatory variables using Bayesian Additive Regression Trees (BARTs). Their results showed that when nonlinear relationships were present, the proposed method improved the performance dramatically compared with linear specifications. While these new advancements in IV learning have not yet been adopted in pharmacovigilance studies, they created new potentials when integrating with other causal inference study designs, for example, algorithmic matching [149], Mendelian randomization [113], and counterfactual prediction [118].
Issues with Machine Learning and Why Causality Matters
Machine learning/deep learning algorithms are good at identifying correlations but not causation. In many use cases, correlation suffices. However, this is not the case with pharmacovigilance, or generally speaking, the healthcare domain. Without evaluation of causality, ML/DL algorithms suffer from a myriad of issues: generalizability, explainability, and fairness. The ML/DL research society has directed increasing attention on improving generalizability, explainability, and fairness in recent years. As discussed in previous paragraphs, ML/DL has been integrated with traditional causal inference paradigms to enhance the performance of traditional paradigms. The opposite is fitting ML/DL into a causal inference paradigm can enhance the generalizability, explainability, and fairness of ML/DL models. Addressing these issues is critical to providing high-quality evidence for pharmacovigilance if machine learning were to be employed for signal detection.
Generalizability
Generalizability is the ability of a machine learning model trained on a sample dataset to perform on unseen data. Generalizability is important for the wide adoption of machine learning models. Recent work utilized cross-validation [150, 151] or eternal validation [152, 153] to examine the generalizability of their proposed machine learning model. More recently, anchor regression was proposed to deal with conditions when training data and test data distributions differed by a linear shift [154]. Anchor regression makes use of external variables to modify the least-squares loss. If anchor regression and least-squares provide the same answer (‘anchor stability’), the model can be considered invariant under certain distributional changes. Comparing different ML/DL methods using ensemble methods or robust feature selection can avoid overfitted models and thus secure model generalizability [155]. In recent work, we observed that the trend in pharmacovigilance is to employ more than one type of data source [64–68] and to compare/combine multiple analytical approaches [44, 69, 70]. We also observed that causal inference models were adopted for feature selection. For example, Rieckmann et al. presented the Causes of Outcome Learning approach, which fitted all exposures from a causal model and then used ML models to identify combinations of exposures responsible for an increased risk of a health outcome [156]. We foresee that data source integration, new analytical approaches (e.g., anchor regression to address the data shift issue), and causal feature selection will benefit the design of a generalizable ML/DL framework for pharmacovigilance.
Explainability
Explainable AI (XAI) refers to ML/DL models with the results or analytical process understandable by humans, in contrast to the “black box” design where researchers cannot explain why a model arrives at a specific output [157]. This is especially important for domains such as healthcare that require an understanding of the causal relationships between features and outcomes for decision support. Several ML/DL algorithms are inherently “explainable” using feature importance, for example, Random Forest, logistic regression, and causality explanation do not equate to feature importance or regression coefficients. As in the case of [107, 108], the authors utilized feature weight to interpret the contribution of each medical code to the predicted ADE outcome. However, a causality explanation between medical codes and ADE incidence cannot be established. Similarly, we cannot naively equate link prediction to a causality explanation although several existing graph-based XAI works were framed as a link prediction task, for example, prediction of potential PPI, DDI, or drug–ADE link given a medication [13, 101, 102, 104, 112]. Therefore, integration of causality evaluation is much needed to improve the power of XAI models. For example, the examples below integrated three different causal inference approaches to enhance the explainability of drug–event relationships for ADE detection: [17] (Granger causality), [145] (counterfactual reasoning), and [158] (combination of a transformer-based component with a do-calculus causal inference paradigm). The three causal inference approaches discussed above have not been extensively used for pharmacovigilance tasks, thus we did not discuss them in previous sections. However, future researchers might be able to integrate them with ML/DL models to enhance model explainability. Additionally, as we have discussed earlier in Sect. 3.3, a benchmark dataset (e.g., PPI, DDI, or drug–ADE network) with causal relationships between graph features, for example, level of confidence, can significantly benefit the development of XAI models for pharmacovigilance studies.
Fairness
Machine learning fairness is a recently established area that studies how certain biases (e.g., race, gender, disabilities, and sexual or political orientation) in the data and model affect model predictions of individuals. This issue has caught more attention under the current pandemic, as the health disparity issue was under public scrutiny [159]. Racial disparity is also a significant issue in ADE detection. As pointed out in a review paper, 27 out of 40 pharmacovigilance studies reviewed demonstrated the presence of a racial or ethnic disparity [160]. Therefore, Du et al. [161] proposed to adopt a kernel re-weighting mechanism to achieve the global fairness of the learned model. Several ML/DL fairness studies have leveraged feature importance to understand which feature contributes more or less to the model disparity [162, 163]. A recent study proposed to decompose the disparity into the sum of contributions from fairness-aware causal paths linking sensitive features and the predictions, on a causal graph [159]. The same group of researchers also proposed a Federated Learning framework that balanced algorithmic fairness and performance consistency across different data sources [164]. The work discussed above, however, was applied only to datasets and tasks in the general healthcare domain. We have not found any work on machine learning fairness in the pharmacovigilance domain that pointed to a new direction worthy of exploration in the future. We anticipated that the new approaches introduced in [159, 161–164] can be extended to pharmacovigilance studies as well. Furthermore, while causal inference paradigms have not been utilized to address the machine learning fairness issue, we anticipated that the integration of causal inference paradigms with machine learning algorithms may also be a potential direction.
Current Challenges, Trends, and Future Directions
To summarize the discussion from the above sections, we found that missing data and data quality posed significant issues for currently dominant pharmacovigilance data sources. Researchers have attempted to address these issues through (1) integration of multiple data sources, (2) development of analytical approaches to impute missing data and mitigate other data issues (e.g., unbalanced confounder distribution, biased samples), and (3) development of novel estimators that allow estimation through incomplete or biased data. New methodology advancements in machine learning, causal inference, and especially, the integration of the two have accelerated the progress in each of the three directions above. On the one hand, the adoption of machine learning has facilitated the efficient implementation of traditional causal inference paradigms. On the other hand, the adoption of causal inference paradigms has facilitated our understanding and thus addresses current issues with machine learning models.
High rates of underreporting and missing covariate information in SRS have undermined the power of SRS for pharmacovigilance [165]. While regulatory approaches were previously proposed to improve reporting, current approaches to address the under-reporting issue were from two directions:
Incorporating multiple data sources or data types to mine under-reported cases from additional data sources. As RWD becomes more available for pharmacovigilance, signals from RWD can complement under-estimated signals using SRS alone. Zhan et al. imputed the ADE cases using specific medicines for treating the ADE as indicators [166]. McMaster et al. developed a machine learning model to detect ADE signals using the International Classification of Diseases, 10th Revision codes [76]. However, their proposed approach only accounted for 44.5% of all ADE cases. Therefore, addressing the missing value in RWD is also unavoidable and opens new research opportunities. As quantitative clinical measurements can be indicative of ADEs, new progress in missing values imputation for quantitative clinical measurements [167–169] could potentially address ADE under-reporting issue in RWD. However, instead of imputing missing values, the author in [168] revealed that when clinical measurements have a high missing rate, the number of times they were taken by one patient is ranked as more informative than looking at their actual values.
Using machine learning to estimate under-reporting or predict and impute under-reported cases. Recent progress in machine learning has enabled the estimation of AE under-reporting rates for data quality management [170, 171]. Traditionally, missing data imputation was conducted statistically via unconditional mean imputation, k-Nearest Neighbor imputation, multiple imputation, or regression-based imputation [172, 173]. Here, we only highlighted a few more recent studies incorporating machine learning approaches. Nestsiarovich et al. [174] proposed to use supervised machine learning (classification) to impute self-harm cases that were significantly under-reported in EHRs. They demonstrated that using the combined coded and imputed cohort, the power of their analysis could be enhanced. Another work by Sechidis et al. [175] presented solutions using the m-graph, a graphical representation of missingness that incorporated a prior belief of under-reporting. They demonstrated an approach to correct mutual information for under-reporting by examining independence properties observed through the m-graph. Their work represented a recent interest in the field of machine learning towards PU learning [176], i.e., learning from positive and unlabeled data. The assumption of PU learning is that each unlabeled data point could belong to either the positive or negative class. Therefore, potential under-reported cases could be estimated from unlabeled data. Alternatively, the anchor variable framework may be adopted to reduce dependency on gold-standard labels for unlabeled cases [177–179]. These new directions in machine learning could provide potential solutions to alleviate the under-reporting issue.
In terms of machine learning for traditional causal inference paradigms, we observed that new advancements in PSM and IV learning through machine learning-causal inference integration have not yet been adopted in pharmacovigilance studies. However, theoretical advancements or successful adoptions in other domains demonstrated new potentials for future adoption of the integrated paradigm in the pharmacovigilance domain. For graph-based causal inference, while both graph databases and graph mining methods for pharmacovigilance are booming, causal interpretations from the graphs as well as the algorithm outputs are much needed, yet currently missing, for most of the studies. Even the currently prevailing benchmark datasets were mostly association-based. Relationships in knowledge bases may represent a certain level of causality but the level of confidence for a causal relationship was not represented explicitly. Therefore, we also recommend future researchers be very careful about the level of causality represented by graph edges when constructing graph databases.
Incorporating causal inference paradigms to address currently prominent machine learning issues in pharmacovigilance is also considered a promising future direction. It is especially worth exploration for those less utilized (in pharmacovigilance tasks) causal study designs, for example, Granger causality, counterfactual reasoning, and do-calculus. In addition, there is a scarcity of exploration of addressing the machine learning fairness issue through the incorporation of causal paradigms, and thus may be a new direction for future pharmacovigilance studies.
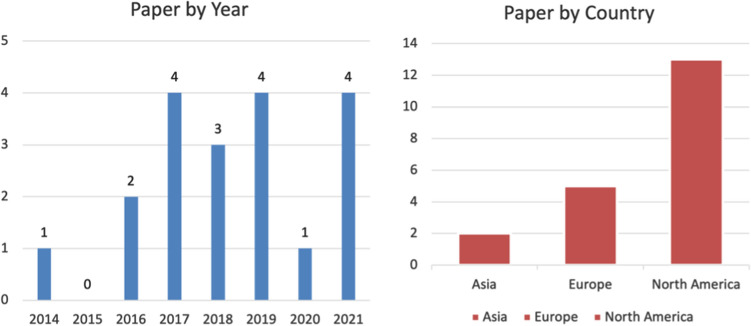
Finally, to examine the distribution and trend in this research area, we considered 19 publications to fall into the intersection of machine learning, causal inference, and pharmacovigilance [86–91, 101–112, 158]. The breakdown of the 19 papers by year and country is shown in Fig. 3. The earliest paper was published in 2014 and utilized knowledge bases to predict potential ADEs. We observed a trend that older papers mostly use databases such as knowledge bases or social media to predict or monitor, while more recent papers utilized RWD, SRS, or a combination of multiple databases. North America was dominant in this research area followed by Europe. This may be owing to the availability of datasets for analysis.
Fig. 3.
Year and continent distribution of 19 papers most relevant to the intersection of machine learning, causal inference, and pharmacovigilance
Conclusions
In this paper, we reviewed (1) data sources and tasks for pharmacovigilance, (2) traditional causal inference paradigms and integration of machine learning into traditional paradigms, and (3) issues with machine learning, and how causal designs could mitigate current issues. First, we found that most existing data sources and tasks for pharmacovigilance were not designed for causal inference. In the meantime, low data quality undermined the ability to evaluate causal relationships. As establishing a causal relationship is important in pharmacovigilance, research on enhancing data quality and data representation will be an imperative step towards high-quality study for pharmacovigilance. Second, we observed that pharmacovigilance was lagging in adopting machine learning-causal inference integrated models, which pointed to some missed opportunities. For example, machine learning-based PSM and IV learning can be further developed and refined for pharmacovigilance tasks. Finally, we recognized that attempts have been made to address currently prominent issues with correlation-based ML/DL models, especially through the incorporation of causal paradigms. Therefore, we anticipated that the pharmacovigilance domain can benefit from the progress in the ML/DL field, especially through the integration of machine learning and the causal inference paradigm.
Declarations
Funding
This article was funded by National Institutes of Health grants U01TR003528 and R01LM013337.
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Data availability
The publications reviewed in this paper are all available online.
Code availability
Not applicable.
Author contributions
YZ and YL originated and planned the scope of the study. YY drafted Sect. 1, HW drafted Sect. 2.2, YL drafted Sect. 2.3, YD drafted Sect. 2.4, and YZ drafted Sects. 2.1 and 3–6. YZ and YL revised the manuscript till its final version. All of the authors have read and approved the final manuscript.
References
- 1.World Health Organization . The importance of pharmacovigilance. Geneva: World Health Organization; 2002. [Google Scholar]
- 2.Bailey C, Peddie D, Wickham ME, Badke K, Small SS, Doyle-Waters MM, et al. Adverse drug event reporting systems: a systematic review. Br J Clin Pharmacol. 2016;82(1):17–29. doi: 10.1111/bcp.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee CY, Chen Y. Machine learning on adverse drug reactions for pharmacovigilance. Drug Discov Today. 2019;24(7):1332–1343. doi: 10.1016/j.drudis.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004;329(7456):15–19. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothschild JM, Churchill W, Erickson A, Munz K, Schuur JD, Salzberg CA, et al. Medication errors recovered by emergency department pharmacists. Ann Emerg Med. 2010;55(6):513–521. doi: 10.1016/j.annemergmed.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Schachterle SE, Hurley S, Liu Q, Petronis KR, Bate A. An implementation and visualization of the tree-based scan statistic for safety event monitoring in longitudinal electronic health data. Drug Saf. 2019;42(6):727–741. doi: 10.1007/s40264-018-00784-0. [DOI] [PubMed] [Google Scholar]
- 7.Kulldorff M, Dashevsky I, Avery TR, Chan AK, Davis RL, Graham D, et al. Drug safety data mining with a tree-based scan statistic. Pharmacoepidemiol Drug Saf. 2013;22(5):517–523. doi: 10.1002/pds.3423. [DOI] [PubMed] [Google Scholar]
- 8.Harpaz R, Callahan A, Tamang S, Low Y, Odgers D, Finlayson S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Saf. 2014;37(10):777–790. doi: 10.1007/s40264-014-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Z, Deng Y, Li X, Naumann T, Luo Y. Natural language processing for EHR-based computational phenotyping. IEEE/ACM Transact Comput Biol Bioinform. 2018;16(1):139–153. doi: 10.1109/TCBB.2018.2849968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo Y, Thompson WK, Herr TM, Zeng Z, Berendsen MA, Jonnalagadda SR, et al. Natural language processing for EHR-based pharmacovigilance: a structured review. Drug Saf. 2017;40(11):1075–1089. doi: 10.1007/s40264-017-0558-6. [DOI] [PubMed] [Google Scholar]
- 11.Harpaz R, Perez H, Chase HS, Rabadan R, Hripcsak G, Friedman C. Biclustering of adverse drug events in the FDA's spontaneous reporting system. Clin Pharmacol Ther. 2011;89(2):243–250. doi: 10.1038/clpt.2010.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ball R, Botsis T. Can network analysis improve pattern recognition among adverse events following immunization reported to VAERS? Clin Pharmacol Ther. 2011;90(2):271–278. doi: 10.1038/clpt.2011.119. [DOI] [PubMed] [Google Scholar]
- 13.Huang K, Xiao C, Hoang T, Glass L, Sun J. Caster: predicting drug interactions with chemical substructure representation. p. 702–9.
- 14.Courtois É, Pariente A, Salvo F, Volatier É, Tubert-Bitter P, Ahmed I. Propensity score-based approaches in high dimension for pharmacovigilance signal detection: an empirical comparison on the French spontaneous reporting database. Front Pharmacol. 2018;9:1010. doi: 10.3389/fphar.2018.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brookhart MA, Wang P, Solomon DH, Schneeweiss S. Evaluating short-term drug effects using a physician-specific prescribing preference as an instrumental variable. Epidemiology. 2006;17(3):268. doi: 10.1097/01.ede.0000193606.58671.c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCulloch RE, Sparapani RA, Logan BR, Laud PW. Causal inference with the instrumental variable approach and Bayesian nonparametric machine learning (2021). arXiv preprint arXiv:2102.01199.
- 17.Lin W, Lan H, Li B. Generative causal explanations for graph neural networks (2021). arXiv preprint arXiv:2104.06643.
- 18.Zhu S, Ng I, Chen Z. Causal discovery with reinforcement learning (2019). arXiv preprint arXiv:1906.04477.
- 19.Dieng A, Liu Y, Roy S, Rudin C, Volfovsky A. Almost-exact matching with replacement for causal inference. Proc Artific Intell Stat (2019). [PMC free article] [PubMed]
- 20.Zhu Y, Coffman DL, Ghosh D. A boosting algorithm for estimating generalized propensity scores with continuous treatments. J Causal Infer. 2015;3(1):25–40. doi: 10.1515/jci-2014-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shmueli G. To explain or to predict? Stat Sci. 2010;25(3):289–310. doi: 10.1214/10-STS330. [DOI] [Google Scholar]
- 22.US FDA. FDA Adverse Event Reporting System (FAERS); 2021. https://www.fda.gov/drugs/questions-and-answers-fdas-adverse-event-reporting-system-faers/fda-adverse-event-reporting-system-faers-public-dashboard. Accessed 20 Feb 2022.
- 23.Lindquist M. VigiBase, the WHO global ICSR database system: basic facts. Drug Inf J. 2008;42(5):409–419. doi: 10.1177/009286150804200501. [DOI] [Google Scholar]
- 24.Harpaz R, DuMouchel W, Shah NH, Madigan D, Ryan P, Friedman C. Novel data-mining methodologies for adverse drug event discovery and analysis. Clin Pharmacol Ther. 2012;91(6):1010–1021. doi: 10.1038/clpt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouane-Hacene M, Toussaint Y, Valtchev P. Mining safety signals in spontaneous reports database using concept analysis. p. 285–94.
- 26.Harpaz R, Chase HS, Friedman C. Mining multi-item drug adverse effect associations in spontaneous reporting systems. p. 1–8. [DOI] [PMC free article] [PubMed]
- 27.Bate A, Lindquist M, Edwards IR, Olsson S, Orre R, Lansner A, et al. A Bayesian neural network method for adverse drug reaction signal generation. Eur J Clin Pharmacol. 1998;54(4):315–321. doi: 10.1007/s002280050466. [DOI] [PubMed] [Google Scholar]
- 28.Bate A, Evans S. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiol Drug Saf. 2009;18(6):427–436. doi: 10.1002/pds.1742. [DOI] [PubMed] [Google Scholar]
- 29.Rawlins M. Spontaneous reporting of adverse drug reactions. I: the data. Br J Clin Pharmacol. 1988;26(1):1–5. doi: 10.1111/j.1365-2125.1988.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bate A, Hornbuckle K, Juhaeri J, Motsko SP, Reynolds RF. Hypothesis-free signal detection in healthcare databases: finding its value for pharmacovigilance. London: SAGE Publications; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey S, Singh A, Azadian R, Huber P, Blum M. Prospective data mining of six products in the US FDA adverse event reporting system. Drug Saf. 2010;33(2):139–146. doi: 10.2165/11319000-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 32.Alvarez Y, Hidalgo A, Maignen F, Slattery J. Validation of statistical signal detection procedures in EudraVigilance post-authorization data. Drug Saf. 2010;33(6):475–487. doi: 10.2165/11534410-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 33.Begaud B, Moride Y, Tubert-Bitter P, Chaslerie A, Haramburu F. False-positives in spontaneous reporting: should we worry about them? Br J Clin Pharmacol. 1994;38(5):401–404. doi: 10.1111/j.1365-2125.1994.tb04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robb MA, Racoosin JA, Sherman RE, Gross TP, Ball R, Reichman ME, et al. The US Food and Drug Administration's Sentinel Initiative: expanding the horizons of medical product safety. Pharmacoepidemiol Drug Saf. 2012;21(1):9. doi: 10.1002/pds.2311. [DOI] [PubMed] [Google Scholar]
- 35.Bate A, Juniper J, Lawton AM, Thwaites RM. Designing and incorporating a real world data approach to international drug development and use: what the UK offers. Drug Discov. 2016;21(3):400–405. doi: 10.1016/j.drudis.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Whalen E, Hauben M, Bate A. Time series disturbance detection for hypothesis-free signal detection in longitudinal observational databases. Drug Saf. 2018;41(6):565–577. doi: 10.1007/s40264-018-0640-8. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y, Zhou X, Suehs BT, Hartzema AG, Kahn MG, Moride Y, et al. A comparative assessment of observational medical outcomes partnership and mini-sentinel common data models and analytics: implications for active drug safety surveillance. Drug Saf. 2015;38(8):749–765. doi: 10.1007/s40264-015-0297-5. [DOI] [PubMed] [Google Scholar]
- 38.Ryan PB, Madigan D, Stang PE, Overhage JM, Racoosin JA, Hartzema AG. Empirical assessment of methods for risk identification in healthcare data: results from the experiments of the Observational Medical Outcomes Partnership. Stat Med. 2012;31(30):4401–4415. doi: 10.1002/sim.5620. [DOI] [PubMed] [Google Scholar]
- 39.Sharma H, Mao C, Zhang Y, Vatani H, Yao L, Zhong Y, et al. Developing a portable natural language processing based phenotyping system. BMC Med Inform Decis Making. 2019;19(3):79–87. doi: 10.1186/s12911-019-0786-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen L, Brandt P, Jiang G, Kiefer R, Pacheco J, Adekkanattu P, et al. Considerations for improving the portability of electronic health record-based phenotype algorithms. [PMC free article] [PubMed]
- 41.Zhou X, Douglas IJ, Shen R, Bate A. Signal detection for recently approved products: adapting and evaluating self-controlled case series method using a US claims and UK electronic medical records database. Drug Saf. 2018;41(5):523–536. doi: 10.1007/s40264-017-0626-y. [DOI] [PubMed] [Google Scholar]
- 42.Norén GN, Bergvall T, Ryan PB, Juhlin K, Schuemie MJ, Madigan D. Empirical performance of the calibrated self-controlled cohort analysis within temporal pattern discovery: lessons for developing a risk identification and analysis system. Drug Saf. 2013;36(1):107–121. doi: 10.1007/s40264-013-0095-x. [DOI] [PubMed] [Google Scholar]
- 43.Hallas J. Evidence of depression provoked by cardiovascular medication: a prescription sequence symmetry analysis. Epidemiology. 1996;7:478–484. doi: 10.1097/00001648-199609000-00005. [DOI] [PubMed] [Google Scholar]
- 44.Page D, Costa VS, Natarajan S, Barnard A, Peissig P, Caldwell M. Identifying adverse drug events by relational learning. [PMC free article] [PubMed]
- 45.Choi E, Xiao C, Stewart WF, Sun J. Mime: multilevel medical embedding of electronic health records for predictive healthcare (2018). arXiv preprint arXiv:1810.09593.
- 46.Nikfarjam A, Gonzalez GH. Pattern mining for extraction of mentions of adverse drug reactions from user comments. p. 1019. [PMC free article] [PubMed]
- 47.Zhang R, Cairelli MJ, Fiszman M, Rosemblat G, Kilicoglu H, Rindflesch TC, et al. Using semantic predications to uncover drug–drug interactions in clinical data. J Biomed Inform. 2014;49:134–147. doi: 10.1016/j.jbi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang R, Adam TJ, Simon G, Cairelli MJ, Rindflesch T, Pakhomov S, et al. Mining biomedical literature to explore interactions between cancer drugs and dietary supplements. AMIA Jt Summits Transl Sci Proc. 2015;2015:69–73. [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Z, Yang Z, Luo L, Lin H, Wang J. Drug drug interaction extraction from biomedical literature using syntax convolutional neural network. Bioinformatics. 2016;32(22):3444–3453. doi: 10.1093/bioinformatics/btw486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahu SK, Anand A. Drug–drug interaction extraction from biomedical texts using long short-term memory network. J Biomed Inform. 2018;86:15–24. doi: 10.1016/j.jbi.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Luo Y. Recurrent neural networks for classifying relations in clinical notes. J Biomed Inform. 2017;72:85–95. doi: 10.1016/j.jbi.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo Y, Cheng Y, Uzuner O, Szolovits P, Starren J. Segment convolutional neural networks (Seg-CNNs) for classifying relations in clinical notes. J Am Med Inform Assoc. 2018;25(1):93–98. doi: 10.1093/jamia/ocx090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Jin R, Luo Y. Classifying relations in clinical narratives using segment graph convolutional and recurrent neural networks (Seg-GCRNs) J Am Med Inform Assoc. 2018;26(3):262–268. doi: 10.1093/jamia/ocy157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patki A, Sarker A, Pimpalkhute P, Nikfarjam A, Ginn R, O’Connor K, et al. Mining adverse drug reaction signals from social media: going beyond extraction. Proc BioLinkSig. 2014;2014:1–8. [Google Scholar]
- 55.Segura-Bedmar I, Martínez Fernández P, Sánchez Cisneros D. The 1st DDIExtraction-2011 challenge task: extraction of drug–drug interactions from biomedical texts (2011).
- 56.Sarker A, Belousov M, Friedrichs J, Hakala K, Kiritchenko S, Mehryary F, et al. Data and systems for medication-related text classification and concept normalization from Twitter: insights from the Social Media Mining for Health (SMM4H)-2017 shared task. J Am Med Inform Assoc. 2018;25(10):1274–1283. doi: 10.1093/jamia/ocy114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pauwels E, Stoven V, Yamanishi Y. Predicting drug side-effect profiles: a chemical fragment-based approach. BMC Bioinform. 2011;12(1):1–13. doi: 10.1186/1471-2105-12-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu M, Wu Y, Chen Y, Sun J, Zhao Z, Chen X-W, et al. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc. 2012;19(e1):e28–35. doi: 10.1136/amiajnl-2011-000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muñoz E, Nováček V, Vandenbussche P-Y. Facilitating prediction of adverse drug reactions by using knowledge graphs and multi-label learning models. Brief Bioinform. 2019;20(1):190–202. doi: 10.1093/bib/bbx099. [DOI] [PubMed] [Google Scholar]
- 60.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, et al. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347(6224):1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chatr-Aryamontri A, Breitkreutz B-J, Oughtred R, Boucher L, Heinicke S, Chen D, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43(D1):D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwak H, Lee M, Yoon S, Chang J, Park S, Jung K. Drug–disease graph: predicting adverse drug reaction signals via graph neural network with clinical data. Adv Knowl Discov Data Min. 2020;12085:633. doi: 10.1007/978-3-030-47436-2_48. [DOI] [Google Scholar]
- 64.Banda JM, Callahan A, Winnenburg R, Strasberg HR, Cami A, Reis BY, et al. Feasibility of prioritizing drug–drug–event associations found in electronic health records. Drug Saf. 2016;39(1):45–57. doi: 10.1007/s40264-015-0352-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malec SA, Wei P, Bernstam EV, Boyce RD, Cohen T. Using computable knowledge mined from the literature to elucidate confounders for EHR-based pharmacovigilance. J Biomed Inform. 2021;117:103719. doi: 10.1016/j.jbi.2021.103719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohsen A, Tripathi LP, Mizuguchi K. Deep learning prediction of adverse drug reactions using open TG-GATEs and FAERS databases (2020). arXiv preprint arXiv:2010.05411.
- 67.Sarker A, Gonzalez G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. J Biomed Inform. 2015;53:196–207. doi: 10.1016/j.jbi.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mower J, Cohen T, Subramanian D. Complementing observational signals with literature-derived distributed representations for post-marketing drug surveillance. Drug Saf. 2020;43(1):67–77. doi: 10.1007/s40264-019-00872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farooq H, Niaz JS, Fakhar S, Naveed H. Leveraging digital media data for pharmacovigilance. p. 442. [PMC free article] [PubMed]
- 70.Mower J, Subramanian D, Cohen T. Learning predictive models of drug side-effect relationships from distributed representations of literature-derived semantic predications. J Am Med Inform Assoc. 2018;25(10):1339–1350. doi: 10.1093/jamia/ocy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ietswaart R, Arat S, Chen AX, Farahmand S, Kim B, DuMouchel W, et al. Machine learning guided association of adverse drug reactions with in vitro target-based pharmacology. EBioMedicine. 2020;57:102837. doi: 10.1016/j.ebiom.2020.102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandak P, Tatonetti NP. Using machine learning to identify adverse drug effects posing increased risk to women. Patterns. 2020;1(7):100108. doi: 10.1016/j.patter.2020.100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haerian K, Varn D, Vaidya S, Ena L, Chase H, Friedman C. Detection of pharmacovigilance-related adverse events using electronic health records and automated methods. Clin Pharmacol Ther. 2012;92(2):228–234. doi: 10.1038/clpt.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai B, Murugesan S, Geier J, Bate A. Applying high dimensional propensity score (HDPS) in a exploratory data analysis with a US claims database for recent medicinal products: 24. Pharmacoepidemiol Drug Saf. 2014;23:13–14. [Google Scholar]
- 75.Zhao J, Henriksson A, Asker L, Boström H. Predictive modeling of structured electronic health records for adverse drug event detection. BMC Med Inform Decis Making. 2015;15(4):1–15. doi: 10.1186/1472-6947-15-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McMaster C, Liew D, Keith C, Aminian P, Frauman A. A machine-learning algorithm to optimise automated adverse drug reaction detection from clinical coding. Drug Saf. 2019;42(6):721–725. doi: 10.1007/s40264-018-00794-y. [DOI] [PubMed] [Google Scholar]
- 77.Comfort S, Perera S, Hudson Z, Dorrell D, Meireis S, Nagarajan M, et al. Sorting through the safety data haystack: using machine learning to identify individual case safety reports in social-digital media. Drug Saf. 2018;41(6):579–590. doi: 10.1007/s40264-018-0641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cocos A, Fiks AG, Masino AJ. Deep learning for pharmacovigilance: recurrent neural network architectures for labeling adverse drug reactions in Twitter posts. J Am Med Inform Assoc. 2017;24(4):813–821. doi: 10.1093/jamia/ocw180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song D, Chen Y, Min Q, Sun Q, Ye K, Zhou C, et al. Similarity-based machine learning support vector machine predictor of drug–drug interactions with improved accuracies. J Clin Pharm Ther. 2019;44(2):268–275. doi: 10.1111/jcpt.12786. [DOI] [PubMed] [Google Scholar]
- 80.Spirtes P. Introduction to causal inference. J Mach Learn Res. 2010;11(5):1643–1662. [Google Scholar]
- 81.Greenland S. Randomization, statistics, and causal inference. Epidemiology. 1990;1:421–429. doi: 10.1097/00001648-199011000-00003. [DOI] [PubMed] [Google Scholar]
- 82.Rubin DB. Causal inference using potential outcomes: design, modeling, decisions. J Am Stat Assoc. 2005;100(469):322–331. doi: 10.1198/016214504000001880. [DOI] [Google Scholar]
- 83.Rubin DB. For objective causal inference, design trumps analysis. Ann Appl Stat. 2008;2(3):808–840. doi: 10.1214/08-AOAS187. [DOI] [Google Scholar]
- 84.Rubin DB. The design versus the analysis of observational studies for causal effects: parallels with the design of randomized trials. Stat Med. 2007;26(1):20–36. doi: 10.1002/sim.2739. [DOI] [PubMed] [Google Scholar]
- 85.Hill AB. The environment and disease: association or causation? London: SAGE Publications; 1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han L, Ball R, Pamer CA, Altman RB, Proestel S. Development of an automated assessment tool for MedWatch reports in the FDA Adverse Event Reporting System. J Am Med Inform Assoc. 2017;24(5):913–920. doi: 10.1093/jamia/ocx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kreimeyer K, Dang O, Spiker J, Muñoz MA, Rosner G, Ball R, et al. Feature engineering and machine learning for causality assessment in pharmacovigilance: lessons learned from application to the FDA Adverse Event Reporting System. Comput Biol Med. 2021;135:104517. doi: 10.1016/j.compbiomed.2021.104517. [DOI] [PubMed] [Google Scholar]
- 88.Pierce CE, Bouri K, Pamer C, Proestel S, Rodriguez HW, Van Le H, et al. Evaluation of Facebook and Twitter monitoring to detect safety signals for medical products: an analysis of recent FDA safety alerts. Drug Saf. 2017;40(4):317–331. doi: 10.1007/s40264-016-0491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Comfort S, Dorrell D, Meireis S, Fine J. Modified NARanjo causality scale for ICSRs (MONARCSI): a decision support tool for safety scientists. Drug Saf. 2018;41(11):1073–1085. doi: 10.1007/s40264-018-0690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rawat BPS, Li F, Yu H. Naranjo question answering using end-to-end multi-task learning model. p. 2547–55. [DOI] [PMC free article] [PubMed]
- 91.Rawat BPS, Jagannatha A, Liu F, Yu H. Inferring ADR causality by predicting the Naranjo score from clinical notes. p. 1041. [PMC free article] [PubMed]
- 92.Lee BK, Lessler J, Stuart EA. Improving propensity score weighting using machine learning. Stat Med. 2010;29(3):337–346. doi: 10.1002/sim.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Linden A, Yarnold PR. Using classification tree analysis to generate propensity score weights. J Eval Clin Pract. 2017;23(4):703–712. doi: 10.1111/jep.12744. [DOI] [PubMed] [Google Scholar]
- 94.McCaffrey DF, Ridgeway G, Morral AR. Propensity score estimation with boosted regression for evaluating causal effects in observational studies. Psychol Methods. 2004;9(4):403–425. doi: 10.1037/1082-989X.9.4.403. [DOI] [PubMed] [Google Scholar]
- 95.Wyss R, Ellis AR, Brookhart MA, Girman CJ, Jonsson Funk M, LoCasale R, et al. The role of prediction modeling in propensity score estimation: an evaluation of logistic regression, bCART, and the covariate-balancing propensity score. Am J Epidemiol. 2014;180(6):645–655. doi: 10.1093/aje/kwu181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Balanescu DV, Monlezun DJ, Donisan T, Boone D, Cervoni-Curet F, Palaskas N, et al. A cancer paradox: machine-learning backed propensity-score analysis of coronary angiography findings in cardio-oncology. J Invasive Cardiol. 2019;31(1):21–26. doi: 10.25270/jic/18.00215. [DOI] [PubMed] [Google Scholar]
- 97.Linden A, Yarnold PR. Combining machine learning and propensity score weighting to estimate causal effects in multivalued treatments. J Eval Clin Pract. 2016;22(6):875–885. doi: 10.1111/jep.12610. [DOI] [PubMed] [Google Scholar]
- 98.Monlezun DJ, Hostetter L, Balan P, Palaskas N, Lopez-Mattei J, Cilingiroglu M, et al. TAVR and cancer: machine learning-augmented propensity score mortality and cost analysis in over 30 million patients. Cardio-oncology. 2021;7(1):1–9. doi: 10.1186/s40959-021-00111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martini ML, Neifert SN, Shuman WH, Chapman EK, Schüpper AJ, Oermann EK, et al. Rescue therapy for vasospasm following aneurysmal subarachnoid hemorrhage: a propensity score-matched analysis with machine learning. J Neurosurg. 2021;136(1):134–147. doi: 10.3171/2020.12.JNS203778. [DOI] [PubMed] [Google Scholar]
- 100.Setoguchi S, Schneeweiss S, Brookhart MA, Glynn RJ, Cook EF. Evaluating uses of data mining techniques in propensity score estimation: a simulation study. Pharmacoepidemiol Drug Saf. 2008;17(6):546–555. doi: 10.1002/pds.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang M, Ma X, Si J, Tang H, Wang H, Li T, et al. Adverse drug reaction discovery using a tumor-biomarker knowledge graph. Front Genet. 2021;11:1737. doi: 10.3389/fgene.2020.625659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dey S, Luo H, Fokoue A, Hu J, Zhang P. Predicting adverse drug reactions through interpretable deep learning framework. BMC Bioinform. 2018;19(21):1–13. doi: 10.1186/s12859-018-2544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu M, Cai R, Hu Y, Matheny ME, Sun J, Hu J, et al. Determining molecular predictors of adverse drug reactions with causality analysis based on structure learning. J Am Med Inform Assoc. 2014;21(2):245–251. doi: 10.1136/amiajnl-2013-002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zitnik M, Agrawal M, Leskovec J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics. 2018;34(13):457–466. doi: 10.1093/bioinformatics/bty294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang M, Liu M, Liu J, Wang S, Long G, Qian B. Safe medicine recommendation via medical knowledge graph embedding. ArXiv e-prints (2017). arXiv:1710.05980.
- 106.Shang J, Xiao C, Ma T, Li H, Sun J. Gamenet: graph augmented memory networks for recommending medication combination. p. 1126–33.
- 107.Rebane J, Karlsson I, Papapetrou P. An investigation of interpretable deep learning for adverse drug event prediction. p. 337–42. [DOI] [PubMed]
- 108.Rebane J, Samsten I, Pantelidis P, Papapetrou P. Assessing the clinical validity of attention-based and SHAP temporal explanations for adverse drug event predictions. p. 235–40.
- 109.Zhang Y, Chen R, Tang J, Stewart WF, Sun J. LEAP: learning to prescribe effective and safe treatment combinations for multimorbidity. p. 1315–24.
- 110.Bagattini F, Karlsson I, Rebane J, Papapetrou P. A classification framework for exploiting sparse multi-variate temporal features with application to adverse drug event detection in medical records. BMC Med Inform Decis Making. 2019;19(1):1–20. doi: 10.1186/s12911-018-0717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Karlsson I, Boström H. Predicting adverse drug events using heterogeneous event sequences. p. 356–62.
- 112.Zhang W, Zou H, Luo L, Liu Q, Wu W, Xiao W. Predicting potential side effects of drugs by recommender methods and ensemble learning. Neurocomputing. 2016;173:979–987. doi: 10.1016/j.neucom.2015.08.054. [DOI] [Google Scholar]
- 113.Kuang Z, Cordova-Palomera A, Sala F, Wu S, Dunnmon J, Re C, et al. Mendelian randomization with instrumental variable synthesis (IVY). bioRxiv. 2019, 657775.
- 114.Hartford J, Lewis G, Leyton-Brown K, Taddy M. Deep IV: a flexible approach for counterfactual prediction. p. 1414–23.
- 115.Wu PA, Fukumizu K. Identifying treatment effects under unobserved confounding by causal representation learning (2020).
- 116.Jo S, Jun DB, Park S. Estimating the effect of general health checkup using uncertainty aware attention of Deep Instrument Variable 2-Stage Network. p. 883–8.
- 117.Lin A, Lu J, Xuan J, Zhu F, Zhang G. One-stage deep instrumental variable method for causal inference from observational data. p. 419–28.
- 118.Yuan J, Wu A, Kuang K, Li B, Wu R, Wu F, et al. Auto IV: counterfactual prediction via automatic instrumental variable decomposition (2021). arXiv preprint arXiv:2107.05884.
- 119.Singh A, Hosanagar K, Gandhi A. Machine learning instrument variables for causal inference. p. 835–6.
- 120.Behera SK, Das S, Xavier AS, Velupula S, Sandhiya S. Comparison of different methods for causality assessment of adverse drug reactions. Int J Clin Pharm. 2018;40(4):903–910. doi: 10.1007/s11096-018-0694-9. [DOI] [PubMed] [Google Scholar]
- 121.Inamoto T, Azuma H, Tatsugami K, Oya M, Adachi M, Okayama Y, et al. Real-world use of sorafenib for advanced renal cell carcinoma patients with cardiovascular disease: nationwide survey in Japan. Expert Rev Anticancer Ther. 2020;20(7):615–623. doi: 10.1080/14737140.2020.1773805. [DOI] [PubMed] [Google Scholar]
- 122.Tao P, Chen PE, Tao J, Yang SN, Tung TH, Chien SW. Correlation between potentially inappropriate medication and Alzheimer's disease among the elderly. Arch Gerontol Geriatr. 2020;87:103842. doi: 10.1016/j.archger.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 123.Vu M, Tortorice K, Zacher J, Dong D, Hur K, Zhang R, et al. Assessment of use and safety of edaravone for amyotrophic lateral sclerosis in the Veterans Affairs health care system. JAMA Netw Open. 2020;3(10):e2014645. doi: 10.1001/jamanetworkopen.2020.14645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Conti V, Biagi C, Melis M, Fortino I, Donati M, Vaccheri A, et al. Acute renal failure in patients treated with dronedarone or amiodarone: a large population-based cohort study in Italy. Eur J Clin Pharmacol. 2015;71(9):1147–1153. doi: 10.1007/s00228-015-1903-2. [DOI] [PubMed] [Google Scholar]
- 125.Spoendlin J, Layton JB, Mundkur M, Meier C, Jick SS, Meier CR. The risk of Achilles or biceps tendon rupture in new statin users: a propensity score-matched sequential cohort study. Drug Saf. 2016;39(12):1229–1237. doi: 10.1007/s40264-016-0462-5. [DOI] [PubMed] [Google Scholar]
- 126.Austin PC. An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivar Behav Res. 2011;46(3):399–424. doi: 10.1080/00273171.2011.568786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Austin PC, Stuart EA. Moving towards best practice when using inverse probability of treatment weighting (IPTW) using the propensity score to estimate causal treatment effects in observational studies. Stat Med. 2015;34(28):3661–3679. doi: 10.1002/sim.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Caliendo M, Kopeinig S. Some practical guidance for the implementation of propensity score matching. J Econ Surv. 2008;22(1):31–72. doi: 10.1111/j.1467-6419.2007.00527.x. [DOI] [Google Scholar]
- 129.Kallus N. Deepmatch: balancing deep covariate representations for causal inference using adversarial training. p. 5067–77.
- 130.Okoli G, Sanders R, Myles P. Demystifying propensity scores. Oxford: Oxford University Press; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Breiman L. Statistical modeling: the two cultures (with comments and a rejoinder by the author) Stat Sci. 2001;16(3):199–231. doi: 10.1214/ss/1009213726. [DOI] [Google Scholar]
- 132.Chao G, Luo Y, Ding W. Recent advances in supervised dimension reduction: a survey. Mach Learn Knowl Extract. 2019;1(1):341–358. doi: 10.3390/make1010020. [DOI] [Google Scholar]
- 133.King G, Lucas C, Nielsen RA. The balance-sample size frontier in matching methods for causal inference. Am J Polit Sci. 2017;61(2):473–489. doi: 10.1111/ajps.12272. [DOI] [Google Scholar]
- 134.Rassen JA, Glynn RJ, Rothman KJ, Setoguchi S, Schneeweiss S. Applying propensity scores estimated in a full cohort to adjust for confounding in subgroup analyses. Pharmacoepidemiol Drug Saf. 2012;21(7):697–709. doi: 10.1002/pds.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brooks JM, Ohsfeldt RL. Squeezing the balloon: propensity scores and unmeasured covariate balance. Health Serv Res. 2013;48(4):1487–1507. doi: 10.1111/1475-6773.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Thoemmes FJ, West SG. The use of propensity scores for nonrandomized designs with clustered data. Multivar Behav Res. 2011;46(3):514–543. doi: 10.1080/00273171.2011.569395. [DOI] [PubMed] [Google Scholar]
- 137.Leslie WD, Schousboe JT. Pharmacovigilance in the real world. Ann Intern Med. 2019;170(3):201–202. doi: 10.7326/M18-3550. [DOI] [PubMed] [Google Scholar]
- 138.Etminan M, Collins GS, Mansournia MA. Using causal diagrams to improve the design and interpretation of medical research. Chest. 2020;158(1):S21–S28. doi: 10.1016/j.chest.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 139.Tian J, Pearl J. A general identification condition for causal effects. p. 567–73.
- 140.Shpitser I, Pearl J. Complete identification methods for the causal hierarchy. J Mach Learn Res. 2008;9:1941–1949. [Google Scholar]
- 141.Luo Y, Uzuner Ö, Szolovits P. Bridging semantics and syntax with graph algorithms: state-of-the-art of extracting biomedical relations. Brief Bioinform. 2016;18(1):160–178. doi: 10.1093/bib/bbw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wishart DS, Feunang YD, Guo AC, Ej Lo, Marcu A, Grant JR. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(1):1074–1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yu Y, Chen J, Gao T, Yu M. Dag-gnn: Dag structure learning with graph neural networks. p. 7154–63.
- 144.Lachapelle S, Brouillard P, Deleu T, Lacoste-Julien S. Gradient-based neural dag learning (2019). arXiv preprint arXiv:1906.02226.
- 145.Narendra T, Agarwal P, Gupta M, Dechu S. Counterfactual reasoning for process optimization using structural causal models. p. 91–106.
- 146.Mizutani S, Pauwels E, Stoven V, Goto S, Yamanishi Y. Relating drug–protein interaction network with drug side effects. Bioinformatics. 2012;28(18):i522–i528. doi: 10.1093/bioinformatics/bts383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ramirez SP, Albert JM, Blayney MJ, Tentori F, Goodkin DA, Wolfe RA, et al. Rosiglitazone is associated with mortality in chronic hemodialysis patients. J Am Soc Nephrol. 2009;20(5):1094–1101. doi: 10.1681/ASN.2008060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Groenwold RH, Hak E, Klungel OH, Hoes AW. Instrumental variables in influenza vaccination studies: mission impossible?! Value Health. 2010;13(1):132–137. doi: 10.1111/j.1524-4733.2009.00584.x. [DOI] [PubMed] [Google Scholar]
- 149.Awan MU, Liu Y, Morucci M, Roy S, Rudin C, Volfovsky A. Interpretable almost matching exactly with instrumental variables. p. 1116–26.
- 150.Linden A, Yarnold PR, Nallamothu BK. Using machine learning to model dose–response relationships. J Eval Clin Pract. 2016;22(6):860–867. doi: 10.1111/jep.12573. [DOI] [PubMed] [Google Scholar]
- 151.Linden A, Yarnold PR. Using machine learning to evaluate treatment effects in multiple-group interrupted time series analysis. J Eval Clin Pract. 2018;24(4):740–744. doi: 10.1111/jep.12966. [DOI] [PubMed] [Google Scholar]
- 152.Wardi G, Carlile M, Holder A, Shashikumar S, Hayden SR, Nemati S. Predicting progression to septic shock in the emergency department using an externally generalizable machine-learning algorithm. Ann Emerg Med. 2021;77(4):395–406. doi: 10.1016/j.annemergmed.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Van Bronswijk SC, Bruijniks SJ, Lorenzo-Luaces L, Derubeis RJ, Lemmens LH, Peeters FP, et al. Cross-trial prediction in psychotherapy: external validation of the Personalized Advantage Index using machine learning in two Dutch randomized trials comparing CBT versus IPT for depression. Psychother Res. 2021;31(1):78–91. doi: 10.1080/10503307.2020.1823029. [DOI] [PubMed] [Google Scholar]
- 154.Rothenhäusler D, Meinshausen N, Bühlmann P, Peters J. Anchor regression: heterogeneous data meet causality. J R Stat Soc Ser B (Stat Methodol). 2021;83(2):215–246. doi: 10.1111/rssb.12398. [DOI] [Google Scholar]
- 155.Feng X, Liang Y, Shi X, Xu D, Wang X, Guan R. Overfitting reduction of text classification based on AdaBELM. Entropy. 2017;19(7):330. doi: 10.3390/e19070330. [DOI] [Google Scholar]
- 156.Rieckmann A, Dworzynski P, Arras L, Lapuschkin S, Samek W, Arah OA, et al. Causes of outcome learning: a causal inference-inspired machine learning approach to disentangling common combinations of potential causes of a health outcome. medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 157.Sample I. Computer says no: why making AIs fair, accountable and transparent is crucial. Guardian. 2017;5:1–15. [Google Scholar]
- 158.Wang X, Xu X, Tong W, Roberts R, Liu Z. InferBERT: a transformer-based causal inference framework for enhancing pharmacovigilance. Front Artific Intell. 2021;4:659622. doi: 10.3389/frai.2021.659622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pan W, Cui S, Bian J, Zhang C, Wang F. Explaining algorithmic fairness through fairness-aware causal path decomposition (2021). arXiv preprint arXiv:2108.05335.
- 160.Baehr A, Peña JC, Hu DJ. Racial and ethnic disparities in adverse drug events: a systematic review of the literature. J Racial Ethn Health Disparities. 2015;2(4):527–536. doi: 10.1007/s40615-015-0101-3. [DOI] [PubMed] [Google Scholar]
- 161.Du W, Xu D, Wu X, Tong H. Fairness-aware agnostic federated learning. p. 181–9.
- 162.Begley T, Schwedes T, Frye C, Feige I. Explainability for fair machine learning (2020). arXiv preprint arXiv:2010.07389.
- 163.Lundberg SM. Explaining quantitative measures of fairness.
- 164.Cui S, Pan W, Liang J, Zhang C, Wang F. Fair and consistent federated learning (2021). arXiv preprint arXiv:2108.08435.
- 165.Hazell L, Shakir SA. Under-reporting of adverse drug reactions. Drug Saf. 2006;29(5):385–396. doi: 10.2165/00002018-200629050-00003. [DOI] [PubMed] [Google Scholar]
- 166.Zhan C, Roughead E, Liu L, Pratt N, Li J. Detecting potential signals of adverse drug events from prescription data. Artif Intell Med. 2020;104:101839. doi: 10.1016/j.artmed.2020.101839. [DOI] [PubMed] [Google Scholar]
- 167.Van Buuren S, Groothuis-Oudshoorn K. Mice: multivariate imputation by chained equations in R. J Stat Softw. 2011;45(1):1–67. [Google Scholar]
- 168.Zhao J, Henriksson A, Asker L, Boström H. Detecting adverse drug events with multiple representations of clinical measurements. p. 536–43.
- 169.Luo Y, Szolovits P, Dighe AS, Baron JM. 3D-MICE: integration of cross-sectional and longitudinal imputation for multi-analyte longitudinal clinical data. J Am Med Inform Assoc. 2017;25(6):645–653. doi: 10.1093/jamia/ocx133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ménard T, Barmaz Y, Koneswarakantha B, Bowling R, Popko L. Enabling data-driven clinical quality assurance: predicting adverse event reporting in clinical trials using machine learning. Drug Saf. 2019;42(9):1045–1053. doi: 10.1007/s40264-019-00831-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ménard T, Koneswarakantha B, Rolo D, Barmaz Y, Popko L, Bowling R. Follow-up on the use of machine learning in clinical quality assurance: can we detect adverse event under-reporting in oncology trials? Drug Saf. 2020;43(3):295–296. doi: 10.1007/s40264-019-00894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lo AW, Siah KW, Wong CH. Machine learning with statistical imputation for predicting drug approvals. Available at SSRN 2973611 (2018).
- 173.Luo Y, Szolovits P, Dighe AS, Baron JM. Using machine learning to predict laboratory test results. Am J Clin Pathol. 2016;145(6):778–788. doi: 10.1093/ajcp/aqw064. [DOI] [PubMed] [Google Scholar]
- 174.Nestsiarovich A, Kumar P, Lauve NR, Hurwitz NG, Mazurie AJ, Cannon DC, et al. Using machine learning imputed outcomes to assess drug-dependent risk of self-harm in patients with bipolar disorder: a comparative effectiveness study. JMIR Mental Health. 2021;8(4):e24522. doi: 10.2196/24522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sechidis K, Sperrin M, Petherick ES, Luján M, Brown G. Dealing with under-reported variables: an information theoretic solution. Int J Approx Reason. 2017;85:159–177. doi: 10.1016/j.ijar.2017.04.002. [DOI] [Google Scholar]
- 176.Bekker J, Davis J. Learning from positive and unlabeled data: a survey. Mach Learn. 2020;109(4):719–760. doi: 10.1007/s10994-020-05877-5. [DOI] [Google Scholar]
- 177.Halpern Y, Choi Y, Horng S, Sontag D. Using anchors to estimate clinical state without labeled data. p. 606. [PMC free article] [PubMed]
- 178.Zhang L, Ding X, Ma Y, Muthu N, Ajmal I, Moore JH, et al. A maximum likelihood approach to electronic health record phenotyping using positive and unlabeled patients. J Am Med Inform Assoc. 2020;27(1):119–126. doi: 10.1093/jamia/ocz170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang L, Ma Y, Herman D, Chen J. Testing calibration of phenotyping models using positive-only electronic health record data. Biostatistics (2021). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The publications reviewed in this paper are all available online.