Abstract
Penicillium decumbens is able to epoxidize cis-propenylphosphonic acid (cPA) to produce the antibiotic fosfomycin [FOM; also referred to as phosphonomycin and (−)-cis-1,2-epoxypropylphosphonic acid], a bioconversion of considerable commercial significance. We sought to improve the efficiency of the process by overexpression of the genes involved. A conventional approach of isolating the presumed epoxidase and its corresponding gene was not possible since cPA epoxidation could not be achieved with protein extracts. As an alternative approach, proteins induced by cPA were detected by two-dimensional gel electrophoresis. The observation that a 31-kDa protein (EpoA) was both cPA induced and overaccumulated in a strain which more efficiently converted cPA suggested that it might take part in the bioconversion. EpoA was purified, its amino acid sequence was partially determined, and the corresponding gene was isolated from cosmid and cDNA libraries with oligonucleotide probes. The DNA sequence for this gene (epoA) contained two introns and an open reading frame encoding a peptide of 277 amino acids having some similarity to oxygenases. When the gene was subcloned into P. decumbens, a fourfold increase in epoxidation activity was achieved. epoA-disruption mutants which were obtained by homologous recombination could not convert cPA to FOM. To investigate the regulation of the epoA promoter, the bialaphos resistance gene (bar, encoding phosphinothricin acetyltransferase) was used to replace the epoA-coding region. In P. decumbens, expression of the bar reporter gene was induced by cPA, FOM, and phosphorous acid but not by phosphoric acid.
Microbial conversion of precursors obtained by chemical synthesis is an attractive method for drug manufacture because it allows efficient stereoselective biosynthesis. Moreover, if genes involved in microbial conversion are identified, the converting activity can be markedly increased by gene manipulation. This study was undertaken to identify a Penicillium decumbens epoxidase that converts the chemically synthesized substrate cis-propenylphosphonic acid (cPA) into the commercially important antibiotic fosfomycin (FOM) (Fig. 1).
FIG. 1.
Conversion of cPA to fosfomycin.
FOM, first discovered in cultures of Streptomyces fradiae ATCC 21096, is structurally characterized by a phosphonate group, a carbon-phosphorus (C-P) bond, and an epoxy ring (2, 5). FOM inhibits the initial reaction in the biosynthesis of prokaryotic peptidoglycans (11) and thus has broad-spectrum antibiotic activity against gram-positive and gram-negative bacteria (24). FOM prepared by chemical epoxidation of cPA (2) yields a racemic mixture and thus requires a costly separation process to isolate the l-stereoisomer. In 1971, White et al. discovered that many Penicillium species could catalyze epoxidation of cPA (23). The microbial process had the advantage that the l-stereoisomer could be produced selectively.
We wanted to employ P. decumbens for commercial application of this bioconversion. However, the natural isolate had low conversion activities which could not be easily improved by mutation and screening. Furthermore, the classical approach of isolating the presumed rate-limiting enzyme and using reverse genetics to identify and overexpress the corresponding gene could not be used since cPA epoxidase was not detected in vitro. As an alternative strategy, we presumed that the enzyme(s) involved would accumulate in cultures most actively carrying out the conversion. Such proteins were identified by two-dimensional (2D) gel analyses. When the corresponding gene, epoA, was cloned by reverse genetics and reintroduced into P. decumbens, the goal of increasing cPA conversion yields was achieved. The similarity of the predicted EpoA protein sequence to those of other oxidases and induction of the corresponding promoter by phosphorous acid suggested that it was the cPA oxidase.
MATERIALS AND METHODS
Purification of the proteins induced by cPA and determination of their amino acid sequences.
A spore stock of P. decumbens was inoculated into 30 ml of SM-1 medium (glucose, 3%; nutrient broth, 0.8%; yeast extract, 0.2%; malt extract, 0.3%; pH 7.0) and cultured in a 250-ml flask at 28°C for 48 h with shaking. This culture (1 ml) was transferred to 30 ml of P6-1 medium (wheat germ, 3.3%; corn steep liquor, 0.8%; NH4NO3, 0.2%; FeSO4 · 7H2O, 0.025%; glucose, 10%), and cultured in a 250-ml flask at 28°C for 48 h. Mycelia were harvested by centrifugation and resuspended in 20 mM Tris-HCl buffer (pH 7.5). A cell extract was obtained by sonication (with a Sonifier Cell Disruptor 350; Branson Sonic Power Company) and subjected to 2D gel electrophoresis with a Multiphor II (Pharmacia Biotech) system. Electrophoresis was performed with Immobiline Dry Strip (pH 4 to 7) (Pharmacia Biotech) in the first dimension and with ExcelGelR sodium dodecyl sulfate gradient 8-18 (Pharmacia Biotech) in the second dimension. The protein was transferred to a polyvinylidene difluoride membrane (ProBlott; Perkin-Elmer) and applied to a model 492 protein sequencer (Perkin-Elmer), and the N-terminal amino acid sequence was determined by Edman degradation. The internal amino acid sequence of the protein was determined after digesting the protein adsorbed to the polyvinylidene difluoride membrane with lysyl endopeptidase (Wako Pure Chemical Industries, Ltd.) and fractionating peptides with a model 172 μ-Preparative high-performance liquid chromatography (HPLC) chromatograph (Perkin-Elmer).
Design of the synthetic DNA primer for PCR.
The mixed oligonucleotides N-1 (5′-AC[T/A]CCXGA[G/A]CA[G/A]AT[C/T/A]GC-3′) and N-2 (5′-AC[C/G]CCXGA[G/A]CA[G/A]AT[C/T/A]GC-3′), corresponding to the N-terminal amino acid sequence of epoA (T-P-E-Q-I-A), were used as forward primers. The mixed oligonucleotides 15-1 (5′-GC[C/T]TG[G/A]AAXCC[G/A]TT[T/A]CC-3′) and 15-2 (5′-GC[C/T]TG[G/A]AAXCC[G/A]TT[C/G]CC-3′), corresponding to the internal sequence (G-N-G-F-Q-A), were used as reverse primers for PCR amplification of the nucleotide sequence corresponding to peptide 15. X is inosine.
Cosmid cloning of genomic DNA.
P. decumbens HP147 mycelia (about 10 g) grown in SM-1 medium were washed twice with 20 mM Tris-HCl buffer (pH 7.5), frozen with liquid nitrogen, and fragmented with a homogenizer (Nihonseiki Kaisha, Ltd.). Chromosomal DNA was extracted (18), digested partially with Sau3AI, and ligated into the BamHI site of cosmid pCRB8 (constructed and provided by K. Gomi [4a]). Packaging was done with a lambda DNA in vitro packaging kit (Amersham), and libraries were screened with an ECL direct nucleic acid labelling and detection kit (Amersham).
cDNA library construction, screening, and DNA sequencing.
Cultures were grown and lysed as described above in “Purification of the proteins induced by cPA and determination of their amino acid sequences.” Poly(A)+ RNA was prepared with a QuickPrep mRNA purification kit with procedure C provided by the supplier (Pharmacia Biotech). The cDNA library was prepared in lambda gt11 with 5 μg of poly(A)+ RNA isolated from cPA-induced cultures. cDNA was made with a TimeSaver cDNA synthesis kit (Pharmacia Biotech). Screening was performed with ECL direct nucleic acid labelling and detection systems with a PCR product as probe. The target fragment from the positive clone was subcloned into the EcoRI site of pUC118 (pFOC2), and its nucleotide sequence was determined.
Transformation of P. decumbens.
P. decumbens LP3 spores were inoculated into 40 ml of S-1 medium (glycerol, 3%; nutrient broth, 0.8%; malt extract, 0.3%; yeast extract, 0.2%; sodium glutamate, 0.1%) and cultured in a 250-ml flask at 28°C for 48 h. This culture (2 ml) was diluted into a similar flask containing 40 ml of S-1 medium and grown for an additional 24 h. Mycelia in the culture were collected by centrifugation (this and subsequent centrifugations were carried out at 2,500 × g for 10 min), suspended in 35% sorbitol, and centrifuged again. Protoplasts were prepared by incubating the mycelia at 34°C for 3 h with gentle agitation in a solution containing 3 mg of achromopeptidase (Wako Pure Chemical Industries, Ltd.) per ml, 3 mg of lysing enzyme (Sigma) per ml, and 1 mg of chitinase (Sigma) per ml. After digestion, mycelia were removed by filtration through cotton, and protoplasts were recovered by centrifugation and suspended in 10 ml of 0.5 M sucrose containing 10 mM Tris-HCl buffer (pH 7.5) and 10 mM CaCl2 (0.5 M sucrose buffer). This protoplast suspension (100 μl) was mixed with 10 μl of DNA (10 μg) and 400 μl of polyethylene glycol solution (60% polyethylene glycol 4000, 10 mM Tris-HCl buffer [pH 7.5], 10 mM CaCl2). Ten milliliters of 0.5 M sucrose buffer was then added, and the mixture was stirred and centrifuged. The pellet obtained was resuspended in 1 ml of 35% sorbitol and spread on protoplast regeneration medium (potato dextrose agar [Difco], 3.9%; raffinose, 30%; hygromycin B, 25 μg/ml). A soft agar medium (2 ml; potato dextrose agar, 1.3%; raffinose, 30%) was poured over the surface, and the culture was incubated at 26°C for 5 days.
Construction and verification of epoA-disruption mutants.
P. decumbens LP3 was transformed (see “Transformation of P. decumbens,” above) with pFOM13, a plasmid containing its homologous epoA locus in which the structural gene was disrupted with the hygromycin resistance cassette (hgh). Transformants were selected with hygromycin (see “Transformation of P. decumbens,” above), purified, and grown in S-1 liquid medium (see “Purification of the proteins induced by cPA and determination of their amino acid sequences,” above). Genomic DNA was extracted and analyzed by Southern hybridization (see “Southern blot hybridization,” below).
Evaluation of the cPA-converting activity of transformed strains.
P. decumbens was grown as described above (“Purification of the proteins induced by cPA and determination of their amino acid sequences”). After cPA was added to a final concentration of 4 mg/ml, cultivation was continued at 28°C for an additional 7 days. The culture was centrifuged, and FOM was assayed in the supernatant by ion chromatography (column, TSKgel IC-Anion-PWXL; eluent, TSKeluent IC-Anion-A [Tosoh Co.]).
Southern blot hybridization.
For Southern hybridization (20), DNA probes were labelled with digoxigenin-11-dUTP (Boehringer Mannheim) by random priming. Hybridization was carried out at 42°C in DIG Easy Hyb solution buffer (Boehringer Mannheim). Membranes were washed twice at 68°C in 2× SSC (1× SSC is 0.15 M NaCl and 15 mM C6H5Na3O7) and twice in 0.1× SSC containing 0.1% sodium dodecyl sulfate. The hybridized probes were immunodetected with antidigoxigenin according to the procedure provided (Boehringer Mannheim).
Measurement of the bar-inducing activity.
A spore stock of P. decumbens LP3/pFOM8 was used to inoculate 30 ml of SM-1 medium. The culture was incubated at 28°C for 48 h with shaking, diluted 1/30 into P6-1 medium, and cultured at 28°C for 48 h. Phosphonates were added, and mycelia were collected 24 h later and used to prepare a cell extract as described above (“Purification of the proteins induced by cPA and determination of their amino acid sequences”). Phosphinothricin acetyltransferase (PAT) encoded by the bar gene was measured with phosphinothricin as the substrate at room temperature (21), and the specific activity (micromoles per minute per milligram of protein) was calculated from the rate of change in molar absorption coefficient of 5,5′-dithio-bis(2-nitrobenzoic acid) (13.6 × 106).
Bacterial strains and reagents.
cPA was chemically synthesized and used after purification (98.5% homogeneity). Other phosphonates were provided by Sigma Chemical Company. Concentrated aqueous stock solutions of these compounds were neutralized with NaOH and sterilized with a 0.45-μm-pore-size Millex filter unit.
P. decumbens LP3, a soil isolate, was subjected to repeated rounds of mutagenesis and screening to isolate a derivative able to convert cPA to FOM more efficiently (HP147).
Nucleotide sequence accession number.
The nucleotide sequence described in this paper (2,329-bp HindIII-XbaI fragment) has been submitted to the DDBJ, EMBL, and GenBank nucleotide sequence databases and assigned the accession number D73371.
RESULTS
Identification of proteins induced by cPA and determination of their amino acid sequences.
Although repeated attempts to assay cPA-epoxidizing activity in crude extracts were fruitless, two observations suggested a novel approach to identifying the enzyme(s). Bioconversion rates observed in P. decumbens cultures pregrown in cPA were apparently higher than those observed in cultures without cPA preinduction. Secondly, cPA conversion was much more efficient in the improved strain, HP147, than in the original soil isolate. Therefore, 2D gels were used to identify proteins which either were induced by cPA or accumulated at higher levels in HP147 than in its parent (wild-type P. decumbens LP3). Representative gels are shown in Fig. 2.
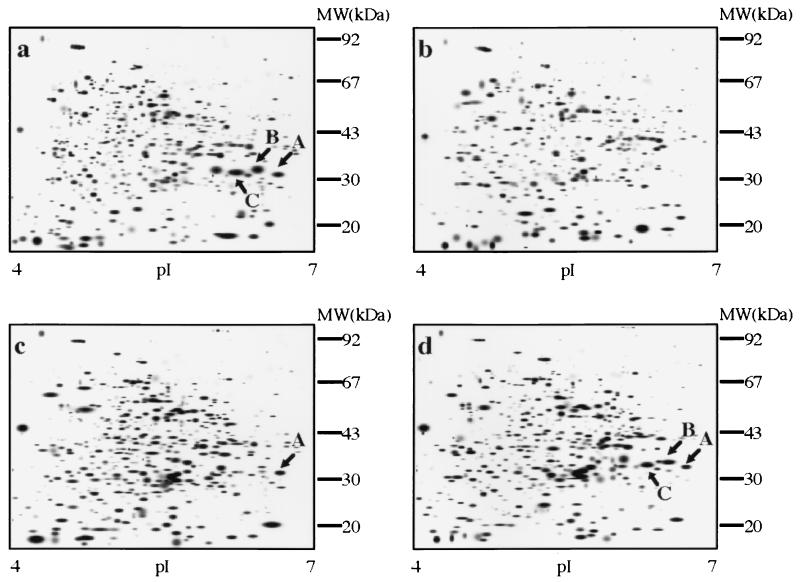
FIG. 2.
cPA induction of P. decumbens protein spots. P. decumbens wild-type LP3 (c), a higher-producing derivative (HP147) (a and b), and LP3 transformed with a plasmid containing epoA (pFOM4) (d) were induced with cPA (a, c, and d) and compared to an untreated HP147 control (b). In other words, panels a to d show HP147 plus cPA, HP147, LP3 plus cPA, and LP3/pFOM4 plus cPA, respectively. Protein spots were silver stained and then scanned to produce the smoothed gel image with an Image Master system (Pharmacia Biotech) (4). cPA-induced proteins are labelled A, B, and C. MW, molecular mass.
Comparison of uninduced HP147 (Fig. 2b) with induced HP147 (Fig. 2a) revealed three cPA-induced proteins (A, B, and C). All had the same apparent molecular mass of 31 kDa but different isoelectric points. Two of these cPA-induced spots (B and C) were also present in higher concentrations in the more efficient converter strain HP147 (Fig. 2a) than in its wild-type parent, P. decumbens LP3 (Fig. 2c).
Peptide sequence analysis was carried out as a first step toward isolation of the gene(s) corresponding to these proteins. The first 10 amino acid residues of the N termini of these three proteins were identical (MKPLTPEQIA). When spots A and C were digested with lysyl endopeptidase and fractionated by HPLC, the peptide profiles of the two proteins were identical (data not shown). Four of the eluted peptides were isolated, and their amino acid sequences were determined (peptide 3, INYK; peptide 7, DYSEGYRT; peptide 13, SEGLDLRE; and peptide 15, QPHGNGFQAHLDAPAYDHIG).
To prepare long probes for cloning the corresponding gene (designated epoA), PCR was performed with chromosomal DNA of P. decumbens HP147 and oligonucleotide probes based on the N-terminal and peptide 15 amino acid sequences. A DNA fragment of about 500 bp was amplified, and its nucleotide sequence was determined (Fig. 3). The fact that this sequence (designated “pep-3 probe”) encoded peptide 3 confirmed the amplification of the target gene.
FIG. 3.
Nucleotide and deduced amino acid sequences of epoA. A mixed PCR probe was designed based on the amino acid residue sequence enclosed by rectangles. The nucleotides with the dotted underlines indicate the DNA sequence amplified by PCR. Nucleotides with the solid underlines indicate TATAAAT and CCAAT motifs. The solid-underlined amino acids indicate lysyl endopeptidase peptides 3, 7, 13, and 15.
Cloning and sequencing of epoA and corresponding cDNA.
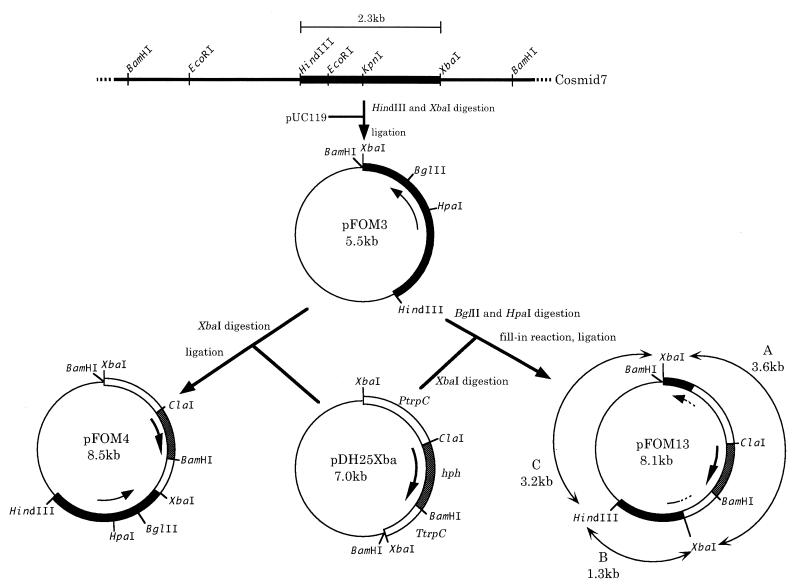
A cosmid bank was prepared from HP147 genomic DNA and screened with the pep-3 probe. Four hybridization-positive cosmid clones were obtained from about 10,000 colonies. Southern hybridization of cosmid DNA showed that the target gene was located on a HindIII-XbaI fragment of about 2.3 kb (Fig. 4). This was subcloned into pUC119 to generate plasmid pFOM3 (Fig. 4).
FIG. 4.
Construction of the plasmid for cloning and disruption of epoA. A HindIII-XbaI fragment (2.3 kb) containing epoA was isolated from cosmid 7 and inserted into pUC119 to generate pFOM3. pFOM4 was prepared by inserting the hph cassette [on an XbaI fragment isolated from pDH25Xba after EcoRI/XbaI linkers were added to pDH25 (3)] into the XbaI site of pFOM3 located downstream of epoA. To prepare pFOM13, pFOM3 was digested to eliminate an HpaI-BglII fragment encoding about 400 bp of the epoA gene. The digested plasmid was filled in and ligated to the hph cassette (isolated from pDH25Xba as a filled-in XbaI fragment). pFOM XbaI/HindIII fragments are labelled A, B, and C (observed in Southern blot of Fig. 5). Abbreviations: PtrpC, the A. nidulans trpC promoter region; TtrpC, the A. nidulans trpC terminator region; hph, the E. coli hygromycin B phosphotransferase gene.
mRNA isolated from P. decumbens HP147 was used to prepare a cDNA bank with phage vectors. Three clones which hybridized to the HindIII/XbaI fragment were isolated from about 8,000 plaques. All contained a fragment of about 1 kb which was subcloned into pUC118 (pFOC2). Analysis of the nucleotide sequences of pFOC2 and pFOM3 showed that the HindIII-XbaI fragment consisted of 2,329 bp and contained an open reading frame encoding 277 amino acids including two introns. This open reading frame encoded amino acid sequences corresponding to the N terminus and various peptides (peptides 3, 7, 13, and 15) in the cPA-induced protein (Fig. 3). A TATAAAT sequence was immediately upstream of the gene, and two CCAAT sequences were in series further upstream (Fig. 3).
Improved cPA-converting activity of strains transformed with epoA.
To examine whether multiple copies of epoA isolated from the efficiently converting strain HP147 would improve the cPA-converting activity in its parental strain, wild-type P. decumbens LP3 was transformed with pFOM4 (containing the intact epoA gene) or pFOM13 (containing an insertionally inactivated epoA allele [see below, “Insertional mutagenesis of epoA,” or Fig. 4]). Ten recombinants were cultured, and their cPA-converting activities were compared with that of the parental strain and P. decumbens containing pFOM13 (Fig. 4) as a control. The converting activities of the recombinants were 1.43- to 4.28-fold higher than that of the parental strain (P. decumbens LP3). P. decumbens LP3/pFOM13 was comparable to the parental strain (Table 1).
TABLE 1.
cPA conversion efficiency of recombinants carrying multiple copies of the cloned epoA genea
| P. decumbens strain | FOM (μg/ml) | Increase (fold) |
|---|---|---|
| LP3 | 800 | 1.00 |
| LP3/pFOM13 | 760 | 0.95 |
| LP3/pFOM16 | 780 | 0.98 |
| LP3/pFOM4 TF1 | 1,140 | 1.43 |
| LP3/pFOM4 TF2 | 1,150 | 1.44 |
| LP3/pFOM4 TF3 | 2,460 | 3.08 |
| LP3/pFOM4 TF4 | 1,960 | 2.45 |
| LP3/pFOM4 TF5 | 2,020 | 2.53 |
| LP3/pFOM4 TF6 | 2,120 | 2.65 |
| LP3/pFOM4 TF7 | 3,420 | 4.28 |
| LP3/pFOM4 TF8 | 3,090 | 3.86 |
| LP3/pFOM4 TF9 | 2,040 | 2.55 |
| LP3/pFOM4 TF10 | 2,500 | 3.13 |
The efficiency of cPA conversion in P. decumbens LP3 was compared to that in transformants containing pFOM4 (TF1 to -10, containing epoA), pFOM13 (containing disrupted epoA), or pFOM16 (vector control containing the hph cassette cloned into the XbaI site of pUC119). cPA was added (final concentration, 4 mg/ml) to P. decumbens cultures prepared as described in Materials and Methods. Incubation was continued under the same conditions for 7 days. FOM was assayed by HPLC analysis as described in Materials and Methods.
Overexpression of cPA-induced protein in transformed strains.
To examine whether cPA-induced proteins were expressed in large amounts of pFOM4-containing strains with improved converting activities, P. decumbens LP3 and the recombinant that showed the greatest increase in the converting activity (P. decumbens LP3/pFOM4 TF7) were analyzed by 2D gel electrophoresis. Compared to the untransformed host (Fig. 2c), larger amounts of 31-kDa protein spots A, B, and C accumulated in the recombinant (Fig. 2d). Northern blots showed that induction of epoA by cPA was at the transcriptional level (data not shown).
Insertional mutagenesis of epoA.
To further elucidate its function, the P. decumbens LP3 chromosomal epoA gene was inactivated by homologous recombination by the following strategy. First, the HpaI-BglII region (about 400 bp) of epoA (Fig. 3) cloned in pFOM3 was deleted and replaced with an hph cassette (pFOM13) (Fig. 4). This plasmid was used to transform P. decumbens LP3, and 50 hygromycin-resistant transformants were examined for their ability to convert cPA. Four of these could no longer epoxidize cPA to FOM.
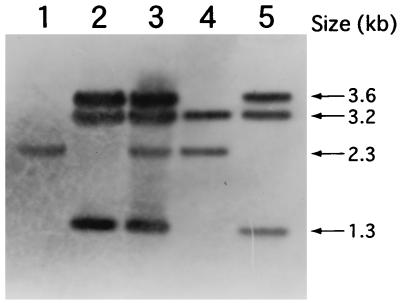
Genomic DNA from representative conversion-positive (P. decumbens LP3/pFOM13) or conversion-negative (P. decumbens LP3/pFOM13 [TF-1]) strains was digested with enzymes that do not cleave the pFOM3 epoA locus (HindIII/XbaI) and analyzed by Southern blot hybridization with pFOM13 as a probe (representative hybridization is shown in Fig. 5; refer to Fig. 4, where fragments are described and labelled A, B, and C). In the conversion-negative strain (P. decumbens LP3/pFOM13 TF1 [Fig. 5, lane 2]), the 2.3-kb fragment (Fig. 4) containing the native epoA gene (Fig. 5, lane 1) was not detected. Transformants which retained the wild-type conversion activity contained an intact 2.3-kb fragment (Fig. 5, lane 3). This provided unambiguous evidence that the epoA gene was mutagenized in conversion-negative strains. Both cPA bioconversion-active and inactive strains contained all pFOM13 fragments (Fig. 5, lane 5; 3.6, 3.2, and 1.3 kb). This indicated the presence of multimeric forms of pFOM13 inserted as tandem copies into the genome (22) at an undefined location. The precise molecular topological changes associated with this mutational event were not studied further.
FIG. 5.
Southern blot analysis of P. decumbens genomic DNA. Genomic DNA from P. decumbens transformants was double digested with HindIII/XbaI, blotted, and probed with pFOM13. Lane 1, genomic DNA from P. decumbens LP3; lane 2, genomic DNA from P. decumbens LP3/pFOM13 TF1 (cPA bioconversion-negative strain). lane 3, genomic DNA from P. decumbens LP3/pFOM13 (cPA bioconversion-positive strain); lane 4, pFOM3 double digested with HindIII/XbaI; lane 5, pFOM13 double digested with HindIII/XbaI. Four different HindIII/XbaI-sized bands were observed: 3.6 kb, identified in Fig. 4 as hph-epoA (fragment A); 3.2 kb, the pUC119 vector fragment (Fig. 4, fragment C); 2.3 kb, the nondisrupted epoA locus (Fig. 4); and 1.3 kb, hph (Fig. 4, fragment B).
Investigation of the function of EpoA.
To further investigate the function of EpoA from the molecular evolutionary point of view, proteins having similar sequences were identified by using BLASTA. These analyses drew our attention to the similarity of EpoA to l-proline 4-hydroxylase of Dactylosporangium sp. (DDBJ/EMBL/GenBank accession no. D78338). The alignment was refined by careful visual examination (Fig. 6). We then computed homology between the two aligned sequences. Of 240 positions available for comparison, 75 were occupied by the same amino acids. Taking into account evolutionary constraints (Poisson correction), 37% of the sites were conserved. This value indicates that the two sequences had evolved from a common ancestor. Moreover, as the figure shows, there were many sets of continuous residues at which the two sequences have the same amino acid. In addition, both sequences have an H-DXXXXXH motif (Fig. 6), which forms the active site of a family of oxygenases (17). These results together provide independent supporting evidence that epoA encodes an oxygenase.
FIG. 6.
Alignment of EpoA and l-proline-4-hydroxylase (D78338) sequences. The sequences are represented by the one-letter amino acid code, and a dash refers to a gap. An asterisk above a residue indicates the same amino acid in both sequences. The underlined H-D....H residues in the two sequences form a motif that resembles the active site of oxygenase.
BLASTA searches did not detect significant homology between EpoA and peroxidases; however, the two most similar proteins of known function found were hydroxylases (BLAST alignments; l-proline 4-hydroxylase, P = 0.06; phytanoyl-coenzyme A alpha hydroxylase [15], P = 0.99).
Regulation of the epoA promoter by phosphorus-containing compounds.
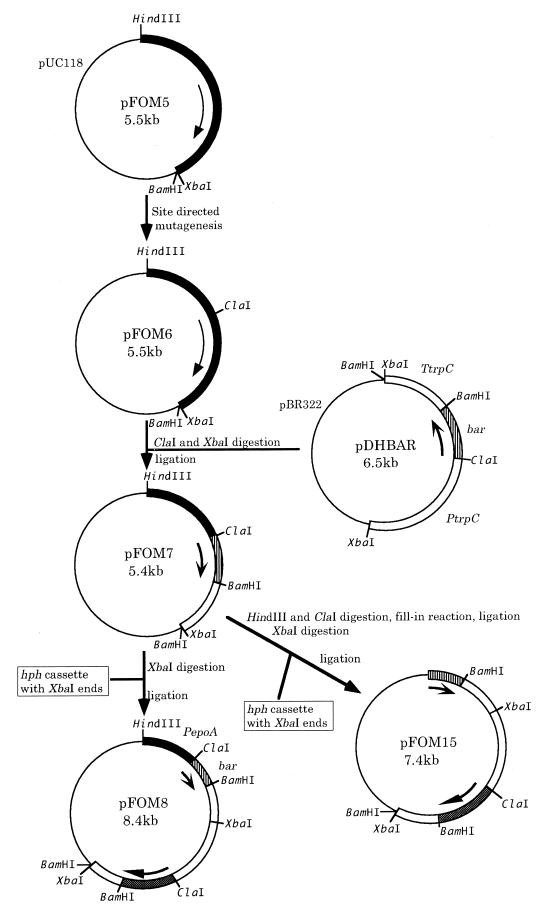
To study the regulation of the epoA promoter, the structural gene cloned in pFOM4 was replaced with a reporter gene (bar) which confers bialaphos resistance (bar) to generate pFOM8 (Fig. 7). bar gene expression was measured by the activity of its corresponding enzyme, PAT. P. decumbens LP3/pFOM8 was used to screen compounds for their ability to induce bar expression (Table 2). PAT activity was markedly induced by cPA, FOM, and phosphorous acid (also demonstrated by unpublished Northern blots). Inductions with phosphonoacetic acid, dl-2-amino-3-phosphonopropionic acid, and 2-aminoethylphosphonic acid (9) were much lower (21, 13, and 2%, respectively). No induction of PAT activity was detected with phosphoric acid or phosphocreatine (Table 2). When the putative epoA promoter region ws deleted (pFOM15 [Fig. 7]), PAT activity was not induced by cPA (P. decumbens LP3/pFOM15; Table 2).
FIG. 7.
Construction of transcriptional fusions linking the epoA promoter region to the bar reporter gene. Plasmid pFOM5 was prepared from pFOM4 (Fig. 4) by ligating a 2.3-kb HindIII-XbaI fragment including epoA into HindIII-XbaI sites of pUC118. Plasmid pFOM6 was obtained from pFOM5 by introducing a ClaI site upstream of the ATG start codon of epoA by site-specific mutation (12) with a primer (5′-AAATCCCCCATCGATGAAGCCTC-3′). The Streptomyces hygroscopicus bar gene was PCR amplified from pARK22 (19) with two primers, barU (5′-AAGGATCGATATGAGCCCAGAACGACGCCC-3′) and barR (5′-GCTTGGATCCTCAGATCTCGGTGACGGGC-3′), which included ClaI and BamHI sites. The termini were cut with ClaI and BamHI and replaced with hph from pDH25 (3) (pDHBAR). Next, a ClaI-XbaI fragment including bar and the trpC terminator (TtrpC) was removed from pDHBAR and inserted into pFOM6 cleaved with ClaI/XbaI so as to replace the epoA structural gene and put the bar reporter gene under the control of the epoA promoter (pFOM7). pFOM8 was prepared by inserting a cassette containing the hph gene into the XbaI site of pFOM7. pFOM15 was prepared by first deleting the putative epoA promoter region from pFOM7 by digestion with HindIII and ClaI and then by using the same procedure to insert the hph cassette. Abbreviations: PepoA, the P. decumbens epoA promoter region; bar, the S. hygroscopicus bialaphos resistance gene.
TABLE 2.
Induction of the epoA promoter with phosphonatesa
| P. decumbens strain | Inducer | bar activity |
|---|---|---|
| LP3 | NA | 0 |
| LP3 | cPA | 0 |
| LP3/pFOM15 | NA | 0 |
| LP3/pFOM15 | cPA | 0 |
| LP3/pFOM8 | NA | 0 |
| LP3/pFOM8 | cPA | 470 |
| LP3/pFOM8 | FOM | 192 |
| LP3/pFOM8 | Phosphorous acid | 365 |
| LP3/pFOM8 | Phosphonoacetic acid | 98 |
| LP3/pFOM8 | dl-2-Amino-3-phosphonopropionic acid | 61 |
| LP3/pFOM8 | 2-Aminoethylphosphonic acid | 9 |
| LP3/pFOM8 | N,N-Bis(phosphonomethyl)-glycine | 9 |
| LP3/pFOM8 | Phosphonocreatine | 0 |
| LP3/pFOM8 | Phosphoric acid | 0 |
P. decumbens LP3 strains containing pFOM8 (described in Fig. 7; the epoA promoter region upstream of the bar reporter gene) or pFOM15 (a vector control similar to pFOM8 but without the epoA promoter) were cultured and induced (48 h) with phosphonates (10 mM) as described in Materials and Methods. Protein extracts were prepared and assayed for reporter gene (bar) expression. bar activity, PAT specific activity, expressed as nanomoles of phosphinothricin converted/min/mg of protein. NA, not added.
DISCUSSION
In the fermentation industry, empirical approaches involving repeated rounds of mutation and screening are often required to create modified strains for efficient manufacturing of microbial products. Although the activities of some rate-limiting enzymes or transport proteins are undoubtedly increased in these modified strains (1), corresponding biochemical assays can be difficult to establish. In such cases, 2D gel electrophoresis can be used to identify these proteins (8).
We were not able to assay the enzyme system that converts cPA to FOM in P. decumbens. To overcome this problem, our approach was based on the principle that enzymes for bioconversions are often induced by corresponding substrates. We focused on three protein spots, apparently products of the same gene, which were induced by cPA under conditions where it was being converted to FOM. The fact that these spots also overaccumulated in strains that carry out the bioconversion more efficiently further indicated the important role of the corresponding gene, epoA. When epoA was reintroduced to P. decumbens LP3, the converting activity increased as much as 4.3-fold. P. decumbens lost the ability to convert cPA to FOM when the epoA gene was destroyed by homologous gene replacement (16). Thus, EpoA plays an important role in the epoxidation of this phosphonic acid derivative. cPA can also be converted to FOM in Pseudomonas and Flavobacterium spp.; however, two enzymes (bromoperoxidase and bromohydrin epoxidase) are required (10). Epoxidation of cPA is not the biosynthetic pathway used in the biosynthesis of FOM in Streptomyces wedmorensis (6). Transcriptional regulation of epoA gave some clues as to its role in phosphonate catabolism or anabolism.
In order to study cPA-induced transcriptional control, a construction was made in which the epoA promoter directed expression of the bar gene. This reporter gene was chosen because its product, PAT, is both easy to assay and selectable. The bar gene confers resistance to phosphinothricin and is widely employed as a selectable marker, primarily in plants but also in fungi. Thus, selection for increased phosphinothricin resistance in P. decumbens containing pFOM8 could be used to select for mutants which overexpress the epoA promoter. A subset of these mutants may be second-site, trans-acting alleles which also increase expression of the native epoA promoter directing transcription of this putative epoxidizing activity.
Here, we used the bar reporter construct to screen various substances for their ability to induce expression of the epoA promoter. This is the first report of a promoter whose expression can be regulated by phosphorous acid, phosphonates, and diverse derivatives. cPA-like phosphonates which induced the promoter included those that have no double bonds (FOM), one having a shorter carbon chain (phosphonoacetic acid), and several having amino radicals such as 2-aminoethylphosphoric acid.
Interestingly, while phosphoric acid (H3PO4) did not induce expression, phosphorous acid (H3PO3) was active. It is reasonable that the reduced, rather than the more oxidized, form of phosphorus induced expression of this promoter, apparently regulating an oxidative conversion. However, we cannot easily rationalize the fact that both the substrate (cPA) and the putative product of epoA (FOM) induced the promoter. Future studies of these curious observations may provide clues to the regulation of phosphonate metabolism, a field where little information is currently available (7, 14).
The TATAAAT sequence characteristic of eukaryotic promoters was found immediately upstream of epoA; the regulatory motif CCAAT appeared repeatedly further upstream. These sequences are likely to be involved in expression and induction by cPA. They play a similar role in regulation of amdS in Aspergillus nidulans (13).
The cPA-converting activity was markedly improved simply by amplifying one gene (epoA). Derivatives containing the cloned epoA gene (Fig. 2d) accumulated increased levels of three related proteins. Although these three proteins had slightly different molecular weights and isoelectric points, comparison of their N-terminal amino acid sequences and peptide maps (data not shown) suggested that they represented posttranslationally modified derivatives of EpoA.
The increase in the cPA-converting activity varied among the recombinants transformed with the same plasmid (Table 1, 1.43- to 4.28-fold). Southern hybridization of genomic DNAs (data not shown) indicated that this phenomenon was probably related to the site of plasmid insertion (a phenomenon commonly observed in fungi) rather than to differences in the number of copies of the plasmid. Further understanding of the complete enzyme system and its coordinated synthesis is expected to allow even greater increases in bioconversion efficiency.
ACKNOWLEDGMENTS
We thank K. Aoyagi for technical assistance, K. Yanai for instruction in site-directed mutation, and H. Shoun and S. Harayama for valuable discussions. We are grateful to K. Gomi for providing cosmid pCRB8 and to J. Gaskell for pDH25. A. L. Demain and D. Taylor encouraged us to submit the manuscript.
REFERENCES
- 1.Anzai H, Murakami T, Imai S, Satoh A, Nagaoka K, Thompson C J. Transcriptional regulation of bialaphos biosynthesis in Streptomyces hygroscopicus. J Bacteriol. 1987;169:3482–3488. doi: 10.1128/jb.169.8.3482-3488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen B G, Leanza W J, Beattie T R, Patchett A A, Arison B H, Ormond R E, Kuehl F A, Albers-Schonberg G, Jardetzky O. Phosphonomycin: structure and synthesis. Science. 1969;166:123–125. doi: 10.1126/science.166.3901.123. [DOI] [PubMed] [Google Scholar]
- 3.Cullen D, Leong S A, Wilson L J, Henner D J. Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene. 1987;57:21–26. doi: 10.1016/0378-1119(87)90172-7. [DOI] [PubMed] [Google Scholar]
- 4.Davidsen N B. Two-dimensional electrophoresis of acidic proteins isolated from ozone-stressed Norway spruce needles (Picea abies L. Karst): separation method and image processing. Electrophoresis. 1995;16:1305–1311. doi: 10.1002/elps.11501601214. [DOI] [PubMed] [Google Scholar]
- 4a.Gomi, K., et al. Unpublished data.
- 5.Hendlin D, Stapley E O, Jackson M, Wallick H, Miller A K, Wolf F J, Miller T W, Chaiet L, Kahan F M, Foltz E L, Woodruff H B, Mata J M, Hernandez S, Mochales S. Phosphonomycin, a new antibiotic produced by strains of Streptomyces. Science. 1969;166:122–123. doi: 10.1126/science.166.3901.122. [DOI] [PubMed] [Google Scholar]
- 6.Hidaka T, Kuzuyama T, Seto H. Studies on the biosynthesis of fosfomycin. Actinomycetology. 1994;8:41–46. [Google Scholar]
- 7.Hilderbrand R L, Henderson T O. Phosphonic acids in nature. In: Hilderbrand R L, editor. The role of phosphonate in living systems. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 6–29. [Google Scholar]
- 8.Holt T G, Chang C, Laurent-Winter C, Murakami T, Garrels J I, Davies J E, Thompson C J. Global changes in gene expression related to antibiotic synthesis in Streptomyces hygroscopicus. Mol Microbiol. 1992;6:969–980. doi: 10.1111/j.1365-2958.1992.tb02163.x. [DOI] [PubMed] [Google Scholar]
- 9.Horiguchi M, Kandatu M. Isolation of 2-amino-ethane phosphonic acid from rumen protozoa. Nature. 1959;184:901–902. doi: 10.1038/184901b0. [DOI] [PubMed] [Google Scholar]
- 10.Itoh N, Kusaka M, Hirota T, Nomura A. Microbial production of antibiotic fosfomycin by a stereoselective epoxidation and its formation mechanism. Appl Microbiol Biotechnol. 1995;43:394–401. [Google Scholar]
- 11.Kahan F M, Kahan J S, Cassidy P J, Kropp H. The mechanism of action of fosfomycin (Phosphonomycin) Ann N Y Acad Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 13.Littlejohn T G, Hynes M J. Analysis of the site of the amdR product for regulation of the amdS gene of Aspergillus nidulans. Mol Gen Genet. 1992;235:81–88. doi: 10.1007/BF00286184. [DOI] [PubMed] [Google Scholar]
- 14.Metcalf W W, Wanner B L. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J Bacteriol. 1991;173:587–600. doi: 10.1128/jb.173.2.587-600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mihalik S J, Morrell J C, Kim D, Sacksteder K A, Watkins P A. Identification of PAHX, a Refsum disease gene. Nat Genet. 1997;17:185–189. doi: 10.1038/ng1097-185. [DOI] [PubMed] [Google Scholar]
- 16.Miller B L, Miller K Y, Timberlake W E. Direct and indirect gene replacements in Aspergillus nidulans. Mol Cell Biol. 1985;5:1714–1721. doi: 10.1128/mcb.5.7.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roach P L, Clifton I J, Fulop V, Harlos K, Barton G J, Hajdu J, Andersson I, Schofield C J, Baldwin J E. Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature. 1995;375:700–704. doi: 10.1038/375700a0. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 19.Sawasaki T, Seki M, Anzai H, Irifune K, Morikawa H. Stable transformation of Arabidopsis with the bar gene using particle bombardment. Transgenic Res. 1995;3:279–286. [Google Scholar]
- 20.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 21.Thompson C J, Movva N R, Tizard R, Crameri R, Davies J E, Lauwereys M, Botterman J. Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 1987;6:2519–2523. doi: 10.1002/j.1460-2075.1987.tb02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walz M, Kück U. Targeted integration into the Acremonium chrysogenum genome: disruption of the pcbC gene. Curr Genet. 1993;24:421–427. doi: 10.1007/BF00351851. [DOI] [PubMed] [Google Scholar]
- 23.White R F, Birnbaum J, Meyer R T, ten Broeke J, Chemerda J M, Demain A L. Microbial epoxidation of cis-propenylphosphonic to (−)-cis-1,2-epoxypropylphosphonic acid. Appl Microbiol. 1971;22:55–60. doi: 10.1128/am.22.1.55-60.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmerman S B, Stapley E O, Wallick H, Baldwin R. Antimicrob. Agents Chemother. 1969. 1970. Phosphonomycin. IV. Susceptibility testing method and survey; pp. 303–309. [PubMed] [Google Scholar]